Abstract
The Escherichia coli dnaQ gene encodes the 3′→5′ exonucleolytic proofreading (ɛ) subunit of DNA polymerase III (Pol III). Genetic analysis of dnaQ mutants has suggested that ɛ might consist of two domains, an N-terminal domain containing the exonuclease and a C-terminal domain essential for binding the polymerase (α) subunit. We have created truncated forms of dnaQ resulting in ɛ subunits that contain either the N-terminal or the C-terminal domain. Using the yeast two-hybrid system, we analyzed the interactions of the single-domain ɛ subunits with the α and θ subunits of the Pol III core. The DnaQ991 protein, consisting of the N-terminal 186 amino acids, was defective in binding to the α subunit while retaining normal binding to the θ subunit. In contrast, the NΔ186 protein, consisting of the C-terminal 57 amino acids, exhibited normal binding to the α subunit but was defective in binding to the θ subunit. A strain carrying the dnaQ991 allele exhibited a strong, recessive mutator phenotype, as expected from a defective α binding mutant. The data are consistent with the existence of two functional domains in ɛ, with the C-terminal domain responsible for polymerase binding.
The Escherichia coli DNA polymerase III (Pol III) holoenzyme (HE) is responsible for the efficient and accurate replication of the bacterial chromosome (for a review, see references 9, 10, and 16). This multisubunit enzyme contains a core polymerase with both polymerizing and exonucleolytic activities. Unlike those of most other proofreading polymerases, these two activities of Pol III are contained in two separate polypeptides. The polymerase activity is contained in the α subunit (dnaE gene product), while the 3′→5′ exonuclease (Exo) activity, which serves as a proofreader for replication errors, is contained in the ɛ subunit (dnaQ gene product). The Pol III core also contains the θ subunit (holE gene product), but the function of this subunit is unclear (4, 20, 21). The subunits are arranged in a linear fashion, α-ɛ-θ, with ɛ binding both α and θ (21). The precise interactions, both functionally and structurally, between the core subunits and of the core subunits with the additional accessory factors of the HE are the subject of active current interest (2, 7, 14).
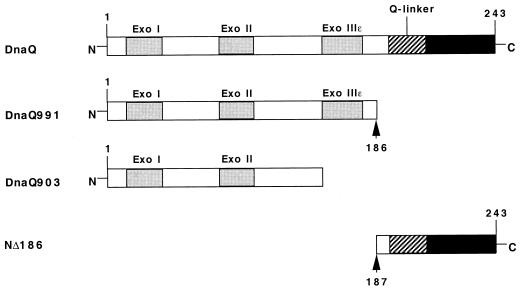
Previously, in an effort to better understand the roles of the ɛ subunit, we undertook a genetic analysis of a large collection of dnaQ mutator mutants (23). These studies confirmed and extended the importance of three conserved exonuclease motifs in the catalytic activity of the DnaQ enzyme (see Fig. 1) and also suggested that the C terminus of ɛ might be required for interaction with the α subunit. dnaQ932, a mutant containing a stop codon 3 amino acids from the C-terminal end of ɛ, was recessive in the presence of a single copy of dnaQ+, suggesting that the DnaQ932 protein does not compete efficiently with the wild-type protein for binding to α. Furthermore, the region between residues 190 and 212 has been identified as a so-called Q-linker (24). In certain proteins, Q-linkers act as a hinge to link domains that have different functions (22, 24). In ɛ, this linker may tether the N-terminal exonuclease domain to the C-terminal polymerase-binding domain (Fig. 1).
FIG. 1.
Schematic drawing of E. coli DnaQ proteins. Highlighted are the conserved Exo I, II, and IIIɛ motifs (1, 3), the proposed Q-linker (24), and the C-terminal domain. DnaQ991 is truncated at position 186, as indicated by the arrowhead. The NΔ186 protein starts at position 187, as indicated by the arrowhead. Note that DnaQ903 lacks the Exo IIIɛ motif in addition to the putative C-terminal α binding site.
The modular structure of ɛ has been probed by limited proteolysis in vitro (17). This study revealed a stable 186-amino-acid N-terminal fragment, created by proteolytic cleavage on the N-terminal side of the proposed Q-linker (Fig. 1). The 186-amino-acid fragment, still comprising the three Exo motifs, has been investigated biochemically and shown to be fully proficient in exonuclease activity but to lack interaction with the α subunit (17). Here, we present a genetic analysis of the function of the C terminus of ɛ. We used the yeast two-hybrid assay to study the interactions of the proposed N- and C-terminal domains with the α and θ subunits. In addition, we analyzed the properties of an E. coli mutant carrying the truncated N-terminal 186-amino-acid ɛ as the sole source of DnaQ protein.
The yeast two-hybrid system was used previously to probe the interactions of the α, ɛ, and θ proteins (8). For example, the ɛ subunit specified by the strong mutator allele mutD5, carrying a Thr15Ile missense mutation in the Exo I motif, was shown to be fully proficient in binding to both the α and θ subunits, consistent with its specific catalytic defect and dominant mutator phenotype (5, 6, 18), while the ɛ subunit specified by the recessive dnaQ49 mutant (15) was defective in binding to α (and, to various extents, also to θ).
Our first experiments used dnaQ991, a dnaQ allele generated by site-directed mutagenesis (Stratagene Quik Change kit) in the two-hybrid vector pGBT9-2-dnaQ (8), by converting codon 187 (Phe) into a TAA stop codon (TTT→TAA), resulting in a truncated ɛ containing only the N-terminal 186 amino acids (Fig. 1). pGBT9-2-dnaQ991 was combined in the same yeast cell with pGAD424 containing either the dnaE+ gene or the holE+ gene as described previously (8), and the relative strengths of the α-ɛ and ɛ-θ interactions were assayed by β-galactosidase activity. Table 1 shows that the ɛ subunits specified by dnaQ991 and dnaQ49 have strongly reduced α-ɛ interactions compared to those encoded by dnaQ+ or mutD5, their interaction being essentially indistinguishable from the background level. Each protein was also assessed for its ability to interact with θ. The ɛ-θ interaction occurs more efficiently in the yeast two-hybrid system, presumably due to the smaller size of θ (8 kDa) compared to the α subunit (130 kDa) (see also reference 8). The data show (Table 1) that the DnaQ991 protein is normal in the ɛ-θ interaction, in contrast to the DnaQ49 protein, which is very defective in θ binding under these conditions. The specific loss of binding of the DnaQ991 protein to the α subunit is consistent with the proposed role of the C-terminal domain of ɛ in binding to the polymerase subunit. The normal binding of DnaQ991 to the θ subunit is consistent with the 186-amino-acid fragment being properly folded, as also suggested by the biochemical demonstration of exonucleolytic proficiency (17).
TABLE 1.
Interaction of mutant ɛ subunits with Pol III core subunits α and θ as determined in the yeast two-hybrid assaya
| dnaQ allele or deletion | β-Gal activityb for the following interaction:
|
|
|---|---|---|
| α-ɛ | θ-ɛ | |
| Vectors only | 0.001 ± 0.001 | 0.001 ± 0.001 |
| dnaQ+ | 0.052 ± 0.032 | 37 ± 9 |
| mutD5 | 0.030 ± 0.009 | 46 ± 18 |
| dnaQ49 | 0.001 ± 0.001 | 0.015 ± 0.005 |
| dnaQ991 | 0.001 ± 0.001 | 48 ± 5 |
| NΔ186 | 0.098 ± 0.047 | 0.001 ± 0.001 |
Yeast transformants carrying pGBT9-2-dnaQ and either pGAD424-holE or pGAD424-dnaE were grown as described by Jonczyk et al. (8). β-Galactosidase (β-Gal) assays were performed with extracts at 30°C according to the method of Rose et al. (19), including correction for spontaneous o-nitrophenyl-β-d-galactopyranoside (ONPG) hydrolysis under the given experimental conditions. Each value represents the average (± standard deviation) for three independent transformants, each analyzed in triplicate. Similar results were obtained in several repeated experiments.
Expressed in units (amount of ONPG hydrolyzed per minute), calculated as (1,000 × OD420)/(OD600 × volume assayed × time), where OD420 and OD600 are the optical densities at 420 and 600 nm, respectively, volume is measured in milliliters, and time is measured in minutes.
To more directly substantiate the role of the C terminus in the α-ɛ interaction, we amplified the region of dnaQ encoding residues 187 to 243, the last 57 C-terminal amino acids of ɛ, ligated the PCR product into pGBT9-2 (as described in reference 8) to form plasmid pGBT9-2-NΔ186, and tested the resulting protein in the yeast two-hybrid system. Table 1 shows proficient binding of the NΔ186 fusion protein to the α subunit. This observation indicates that the C-terminal domain of DnaQ is not only required, but is likely sufficient, for interaction with the polymerase.
The in vivo properties of the dnaQ991 allele were also explored. As loss of the polymerase-binding domain would prevent ɛ from being incorporated into the Pol III core, strains carrying dnaQ991 are expected to mimic, in many respects, strains that lack ɛ entirely. Deletion dnaQ strains have been reported previously in both Salmonella typhimurium and E. coli (11, 12, 20). The strains contain (extensive) deletions from the 3′ end of dnaQ, resulting in truncated ɛ proteins lacking not only the C terminus but also part or all of the exonuclease motifs (see DnaQ903 in Fig. 1). The strains exhibit poor growth as well as highly elevated spontaneous mutation rates. Cell health is improved considerably by spontaneously occurring suppressor mutations, termed spq, that arise in the dnaE gene. We inserted the 1.6-kb SalI-SmaI dnaQ-rnh fragment from pFF588 (20) into the SalI-SmaI linearized allele exchange vector pKO3 (13) and performed site-specific mutagenesis to create dnaQ991. The pKO3-dnaQ991 plasmid was introduced into strain MG1655 (20), and a dnaQ+/dnaQ991 heterodiploid was created by plasmid integration at the dnaQ locus at the nonpermissive temperature (13). Subsequent plating of the heterodiploids on Luria-Bertani (LB)–sucrose plates at 30°C yielded dnaQ991 and dnaQ+ haploid segregants. The dnaQ991 segregants were readily distinguished by their small-colony morphology, similar to that reported previously for dnaQ deletion derivatives (11, 20), and their strong mutator phenotype. Table 2 shows the mutant frequency for rifampin-resistant mutants of the dnaQ991 haploids; dnaQ deletion (dnaQ903::tet) strains (20) are shown for comparison. The dnaQ991 mutation resulted in a mutant frequency that was dramatically (>10,000-fold) increased over the background level, similar to that seen for dnaQ903::tet. A modest (∼2-fold) but consistent increase in cell counts in stationary cultures of the dnaQ991 haploid versus the dnaQ903::tet haploid was also observed (Table 2), suggesting that the presence of active free 3′ exonuclease may be beneficial under the stressful conditions where the Pol III core consists of only the single α subunit. Like that of the dnaQ903::tet strain, the health of dnaQ991 haploids was significantly improved by the presence of the spq-2 allele, a suppressor of the small-colony phenotype of dnaQ903::tet (20), as evidenced by a much healthier colony morphology and increased cell counts in the overnight cultures (Table 2). The spq-2 allele did not affect the mutant frequency. Mutant frequencies of dnaQ+/dnaQ991 and dnaQ+/dnaQ903::tet heterodiploids were also determined (Table 3). None of the heterodiploids exhibited mutant frequencies significantly different from those of the parental wild-type strain, indicating that dnaQ991 is fully recessive. These mutational properties of dnaQ991 are consistent with lack of binding of the DnaQ991 protein to the α subunit and with the resulting absence of the ɛ subunit from the HE.
TABLE 2.
Mutability of the dnaQ991 mutant and related strainsa
| Genotype | Total no. of cells (109/ml) | No. of Rifr mutants/ml | No. of mutants/106 cells |
|---|---|---|---|
| dnaQ+ | 8.4 | 50 | 0.006 |
| dnaQ991 | 0.49 | 55,200 | 111 |
| dnaQ991 spq-2 | 3.0 | 244,000 | 82 |
| dnaQ903::tet | 0.15 | 14,640 | 99 |
| dnaQ903::tet spq-2 | 1.8 | 199,600 | 109 |
All strains are derivatives of MG1655. The dnaQ991 strains were prepared as described in the text by using either MG1655 or its spq-2 derivative. The dnaQ903::tet strains were prepared by transduction of the dnaQ903::tet/dnaQ+ heterodiploid configuration (donor provided by R. Maurer) into MG1655 and its spq-2 derivative, followed by segregation of dnaQ903 haploids as described previously (20). Strains were grown at 37°C.
TABLE 3.
Mutant frequencies in dnaQ+/dnaQ diploid strainsa
| Genotype | No. of Rifr mutants/106 cells |
|---|---|
| Wild type | 0.16 |
| dnaQ991/dnaQ+ | 0.15 |
| dnaQ991/dnaQ+ spq-2 | 0.41 |
| dnaQ903::tet/dnaQ+ | 0.10 |
| dnaQ903::tet/dnaQ+ spq-2 | 0.37 |
Strains were constructed as described in the text and the legend to Table 2. Cells were grown overnight at 43°C in LB broth, plated on LB and LB Rif plates, and incubated at 43°C overnight. Plating at 37°C yielded similar results.
In summary, our data support the contention that the ɛ subunit is composed of two distinct domains, the N-terminal domain, which contains the exonuclease site (and also the θ-binding site), and the C-terminal domain, which is necessary, and likely sufficient, for interaction with the polymerase. Our results do not rule out the existence of additional, secondary interactions between the polymerase and the N terminus of ɛ that may be important for the proper coordination of the two proteins. The discovery of a separate domain within ɛ for binding to the α subunit will aid in further understanding the precise nature of the α-ɛ interaction, which is likely important for efficient proofreading by the polymerase as well as for the structural integrity of the replication complex.
Acknowledgments
We thank Fred Perrino for sharing unpublished data, Piotr Jonczyk and Iwona Fijalkowska for the yeast two-hybrid vectors and for help with the assays, Russ Maurer for the dnaQ+/dnaQ903 heterodiploid strain, and George Church for plasmid pKO3. We also thank Dmitry Gordenin and Karin Drotschman of NIEHS for careful review of the manuscript.
REFERENCES
- 1.Barnes M H, Spacciapoli P, Li D H, Brown N C. The 3′-5′ exonuclease site of DNA polymerase III from gram-positive bacteria: definition of a novel motif structure. Gene. 1995;165:45–50. doi: 10.1016/0378-1119(95)00530-j. [DOI] [PubMed] [Google Scholar]
- 2.Bertram J G, Bloom L B, Turner J, O’Donnell M, Beechem J M, Goodman M F. Pre-steady state analysis of the assembly of wild type and mutant circular clamps of Escherichia coli DNA polymerase III onto DNA. J Biol Chem. 1998;273:24564–24574. doi: 10.1074/jbc.273.38.24564. [DOI] [PubMed] [Google Scholar]
- 3.Blanco L, Bernad A, Salas M. Evidence favouring the hypothesis of a conserved 3′-5′ exonuclease active site in DNA-dependent DNA polymerase. Gene. 1992;112:139–144. doi: 10.1016/0378-1119(92)90316-h. [DOI] [PubMed] [Google Scholar]
- 4.Carter J R, Franden M A, Aebersold R, Kim D R, McHenry C S. Isolation, sequencing and overexpression of the gene encoding the θ subunit of DNA polymerase III holoenzyme. Nucleic Acids Res. 1993;14:3281–3286. doi: 10.1093/nar/21.14.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Echols H, Liu C, Burgers P M J. Mutator strains of Escherichia coli, mutD and dnaQ, with defective exonucleolytic editing by DNA polymerase III holoenzyme. Proc Natl Acad Sci USA. 1983;80:2189–2192. doi: 10.1073/pnas.80.8.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fijalkowska I, Schaaper R M. Mutants in the Exo I motif of Escherichia coli dnaQ: defective proofreading and inviability due to error catastrophe. Proc Natl Acad Sci USA. 1996;93:2856–2861. doi: 10.1073/pnas.93.7.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hingorani M M, O’Donnell M. ATP binding to the Escherichia coli clamp loader powers opening of the ring-shaped clamp of DNA polymerase III holoenzyme. J Biol Chem. 1998;273:24550–24563. doi: 10.1074/jbc.273.38.24550. [DOI] [PubMed] [Google Scholar]
- 8.Jonczyk P, Nowicka A, Fijalkowska I J, Schaaper R M, Ciesla Z. In vivo protein interactions within the Escherichia coli DNA polymerase III core. J Bacteriol. 1998;180:1563–1566. doi: 10.1128/jb.180.6.1563-1566.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelman Z, O’Donnell M. DNA polymerase III holoenzyme: structure and function of a chromosomal replicating machine. Annu Rev Biochem. 1995;64:171–200. doi: 10.1146/annurev.bi.64.070195.001131. [DOI] [PubMed] [Google Scholar]
- 10.Kornberg A, Baker T A. DNA replication. W. H. New York, N.Y: Freeman and Company; 1992. [Google Scholar]
- 11.Lancy E D, Lifsics M R, Kehres D G, Maurer R. Isolation and characterization of mutants with deletions in dnaQ, the gene for the editing subunit of DNA polymerase III of Salmonella typhimurium. J Bacteriol. 1989;171:5572–5580. doi: 10.1128/jb.171.10.5572-5580.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lifsics M R, Lancy E D, Jr, Maurer R. DNA replication defect in Salmonella typhimurium mutants lacking the editing (ɛ) subunit of DNA polymerase III. J Bacteriol. 1992;174:6965–6973. doi: 10.1128/jb.174.21.6965-6973.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Link A J, Phillips D, Church G M. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J Bacteriol. 1997;179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marians K J, Hiasa H, Kim D R, McHenry C S. Role of the core DNA polymerase III subunits at the replication fork. α is the only subunit required for processive replication. J Biol Chem. 1998;273:2452–2457. doi: 10.1074/jbc.273.4.2452. [DOI] [PubMed] [Google Scholar]
- 15.Maruyama M, Horiuchi T, Maki H, Sekiguchi M. A dominant (mutD5) and a recessive (dnaQ49) mutator of Escherichia coli. J Mol Biol. 1983;167:757–771. doi: 10.1016/s0022-2836(83)80109-0. [DOI] [PubMed] [Google Scholar]
- 16.McHenry C S. DNA polymerase III holoenzyme. Components, structure and mechanism of a true replicative complex. J Biol Chem. 1991;266:19127–19130. [PubMed] [Google Scholar]
- 17.Perrino, F. W., S. Harvey, and S. M. McNeill. Personal communication.
- 18.Pham P T, Olson M W, McHenry C S, Schaaper R M. The base substitution and frameshift fidelity of Escherichia coli DNA polymerase III holoenzyme in vitro. J Biol Chem. 1998;273:23575–23584. doi: 10.1074/jbc.273.36.23575. [DOI] [PubMed] [Google Scholar]
- 19.Rose M D, Winston F, Hieter P. Methods in yeast genetics. A laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 20.Slater S C, Lifsics M R, O’Donnell M, Maurer R. holE, the gene coding for the θ subunit of DNA polymerase III of Escherichia coli: characterization of a holE mutant and comparison with a dnaQ (ɛ-subunit) mutant. J Bacteriol. 1994;176:815–821. doi: 10.1128/jb.176.3.815-821.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Studwell-Vaughan P S, O’Donnell M. DNA polymerase III accessory proteins. V. θ encoded by holE. J Biol Chem. 1993;268:11785–11791. [PubMed] [Google Scholar]
- 22.Sutrina S L, Reddy P, Saier M H, Jr, Reizer J. The glucose permease of Bacillus subtilis is a single polypeptide chain that functions to energize the sucrose permease. J Biol Chem. 1990;265:18581–18589. [PubMed] [Google Scholar]
- 23.Taft-Benz S A, Schaaper R M. Mutational analysis of the 3′→5′ proofreading exonuclease of Escherichia coli DNA polymerase III. Nucleic Acids Res. 1998;26:4005–4011. doi: 10.1093/nar/26.17.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wootton J C, Drummond M H. The Q-linker: a class of interdomain sequences found in bacterial multidomain regulatory proteins. Protein Eng. 1989;2:535–543. doi: 10.1093/protein/2.7.535. [DOI] [PubMed] [Google Scholar]