Abstract
PBP1b can be found as a dimer in Escherichia coli. Previous results suggested that dimerization involved the cysteine(s) in an intermolecular disulfide bond. We show that either deletion mutants or a mutant without cysteines is fully active and still binds penicillin and that the latter can also form dimers.
Penicillin-binding proteins (PBPs) are a set of membrane-bound enzymes involved in the late stages of peptidoglycan biosynthesis (for reviews, see references 4 and 13). PBPs in connection with several other enzymes, including hydrolases and synthetases, are essential in the control of bacterial morphology, elongation, and cell division (2, 3, 10) and must have activities coordinated in both time and space. Cell division is directed by a multimeric structure, the divisome (12). Recent findings established that some PBPs belong to this enzymatic complex and take part in the septum formation (3, 11). Moreover, it has been shown that some PBPs interact with each other and with lytic transglycosylases, suggesting the existence of murein-synthesizing enzyme complexes (14, 19). However, the composition and the functions of these complexes remain speculative and require further investigation.
PBP1b is the major enzyme for peptidoglycan synthesis in Escherichia coli (18) and should thus play a prominent part in these complexes. This transmembrane protein is organized into two catalytic domains harboring transpeptidase and transglycosylase activities (8, 16). It is usually present in the cell membrane in three molecular forms, termed α, β, and γ, with slightly different electrophoretic mobilities (15). The β component derives from PBP1bα by proteolytic release of the 24 N-terminal residues, whereas the γ component arises from a translation start site downstream from the PBP1bα translation start site of the ponB gene (17). Earlier studies have established that PBP1b can occur as dimers which can be detected by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) at a position corresponding to a molecular mass of about 140 kDa (23, 24). Two-dimensional gel analysis confirmed that this high-molecular-weight band consists of two PBP1b monomers (23). Two classes of dimers have been characterized as having different biochemical properties, but both need β-mercaptoethanol for dissociation, suggesting a stabilization by intermolecular disulfide bonds involving one or both cysteines of PBP1b (C776 or C794), located at the carboxy-terminal end of the protein. In this study, we confirmed that PBP1b can exist in a stable dimeric form. However, we also show that disulfide bridges are dispensable for PBP1b activities in vitro and in vivo and that PBP1b mutants without cysteine residues are still able to dimerize.
PBP1b dimers can dissociate under mild denaturation or solubilization conditions.
PBP1b was produced in strain QCB1, which was transformed with vector pPONB (8). QCB1 is an E. coli strain with a deletion in gene ponB, which codes for PBP1b (1). Plasmid pPONB expresses PBP1b with a tag peptide (YPYDVPDYA) from the hemagglutinin HA1 epitope of influenza virus (21) at its C-terminal end. The expressed PBP1b can be visualized by Western blotting, performed on E. coli membrane preparations with monoclonal antibody 12CA5 raised against the tag peptide, as described by Lefèvre et al. (8).
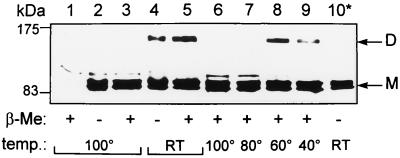
Membrane fractions (8) from QCB1(pPONB) cells grown at 37°C were incubated for 10 min in sample buffer (7) with or without β-mercaptoethanol, at either room temperature or 100°C, and applied to SDS-PAGE gels for Western blot analysis. Three major bands were detected at positions corresponding to the three molecular forms of PBP1b commonly found by SDS-PAGE (PBP1bα, -β, and -γ) (Fig. 1, lanes 2 through 5). With samples incubated at room temperature, an additional band reacted with the antitag antibody at about 150 kDa (Fig. 1, lanes 4 and 5), corresponding to the previously described dimeric form of PBP1b (23, 24). This band was never detected with boiled samples (Fig. 1, lanes 2 and 3). Furthermore, incubation of samples containing β-mercaptoethanol at several temperatures for 10 min showed that the dimeric form is stable at 60°C but almost totally dissociated at 80°C (Fig. 1, lanes 6 through 9). We observed the same pattern of dissociation in samples without β-mercaptoethanol (data not shown). These results indicate that dissociation of the dimer occurs when temperature increases, with or without β-mercaptoethanol, thus suggesting that disulfide bonds do not take part in the stabilization of the dimer.
FIG. 1.
PBP1b dimer stability. Membrane fractions prepared from strain QCB1 (lane 1) or QCB1(pPONB) (lanes 2 through 10) were incubated for 10 min in sample buffer with 5% or no β-mercaptoethanol (β-Me) at temperatures ranging from room temperature (RT) to 100°C. Lane 10* shows a soluble membrane fraction after treatment with Nonidet P-40–NaCl (see text). Samples were submitted to SDS-PAGE (7). Western blotting with the antitag 12CA5 antibody was performed with the Enhanced Chemiluminescence detection system (Amersham). M, monomeric forms of PBP1b; D, dimeric forms of PBP1b; temp., temperature, in degrees Celsius.
In an attempt to solubilize the dimeric form of PBP1b in a native conformation, membranes were incubated with 0.5% Nonidet P-40–0.5 M NaCl for 30 min at 25°C and centrifuged for 30 min (14,000 × g, 4°C). Most of the PBP1b was recovered in the soluble fraction. This solubilized PBP1b was still able to bind fluorescein 6-amino-penicilloic acid (6-APA-Flu) (8), indicating a correct fold (data not shown). The solubilized fraction was incubated for 10 min at room temperature in sample buffer without β-mercaptoethanol prior to SDS-PAGE and Western blot analysis with the antitag antibody. Under these conditions, we failed to detect the dimeric form of the protein (Fig. 1, lane 10*). Since the mild solubilization conditions used in this experiment are unlikely to break a covalent bond, these observations give further support to the idea that PBP1b dimers are not associated by a disulfide bond.
PBP1b mutants without cysteine are active in vitro and in vivo.
To further investigate the role of a possible disulfide bridge in PBP1b activities, we constructed several mutants by the inverse-PCR technique (20). These constructs were checked by DNA sequencing. pV775ΔCt and pL793ΔCt mutants carry deletions of the sequence corresponding to the C-terminal region of PBP1b. They were made by fusing the V775 codon of pV775ΔCt or the L793 codon of pL793ΔCt directly to the tag sequence. The truncated protein produced by plasmid pL793ΔCt still contains one cysteine residue (C776), whereas the one which is produced by pV775ΔCt does not. A third mutant (the pCys− mutant) was made by replacement of the C776 and C794 codons by alanine codons. These constructs were transformed in the QCB1 strain, and activities of the resulting PBP1b mutants were examined.
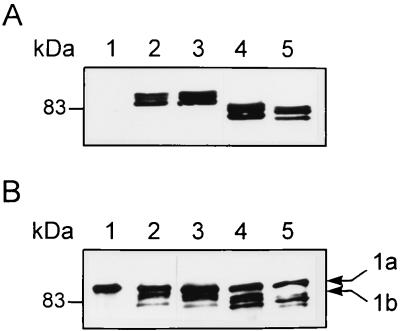
Membrane fractions were prepared from QCB1 harboring the mutated plasmids. Western blot analysis with antitag antibody 12CA5 revealed that the three PBP1b mutants are produced at the same level as the wild-type protein in bacterial cells (Fig. 2A, lanes 2 through 5). The three PBP1b mutants also bind fluorescent penicillin as efficiently as the wild type, indicating a proper folding and a functional transpeptidase activity (Fig. 2B, lanes 2 through 5).
FIG. 2.
PBP1b mutants. (A) Production of the PBP1b mutants. Membrane fractions from QCB1 (lane 1) or QCB1 transformed with pPONB (lane 2), pCys− (lane 3), pL793ΔCt (lane 4), and pV775ΔCt (lane 5) were analyzed by Western blotting with the antitag 12CA5 antibody. (B) Binding of 6-APA-Flu by the PBP1b mutants. Membrane fractions from QCB1 (lane 1) or QCB1 transformed with pPONB (lane 2), pCys− (lane 3), pL793ΔCt (lane 4), and pV775ΔCt (lane 5) were incubated with 6-APA-Flu and analyzed by Western blotting with an antifluorescein antibody. 1a, PBP1a; 1b, PBP1b.
The ability of these mutants to restore the PBP1b activities in the synthesis of the peptidoglycan was tested by the disc agar diffusion method (8, 9). Growth inhibition diameters were also measured on agar plates with 0.3 μg of cephaloridine per ml. Under these conditions, PBP1a is specifically inactivated and cell growth thus relies solely on PBP1b (22). QCB1 harboring the mutated plasmids exhibits the same antibiogram patterns as the wild-type strain MC6-RP1 or QCB1 harboring pPONB (Table 1). The PBP1b mutant resulting from the expression of pCys− is fully active in vivo, indicating that replacement of the two cysteine residues by alanine does not affect peptidoglycan synthesis. This result demonstrates that dimeric forms of PBP1b stabilized by a disulfide bridge are not required for peptidoglycan synthesis. The mutant resulting from the expression of pL793ΔCt in strain QCB1 is as active as the wild-type PBP1b, confirming that the last 51 amino acids of PBP1b are not required for in vivo activity. This result is not surprising, since it has been shown that the C-terminal residues down to M780 are dispensable for the in vitro or in vivo activity of PBP1b (5, 6). Interestingly, the pV775ΔCt mutant in strain QCB1 is slightly less active than the wild type. It has been shown that the deletion of the C-terminal domain of PBP1b down to D762 yields a mutant that is inactive in vitro and in vivo (8). Combined with our previous alanine stretch scanning results (8), our present results strongly suggest a crucial role in PBP1b structure for residues 766 through 772.
TABLE 1.
Growth inhibition diameters of antibiotic disc assays of strainsa
| Strain | Growth inhibition diam
|
|||||
|---|---|---|---|---|---|---|
| TIC | AMX | CTX | ATM | CF | MEC | |
| MC6-RP1 | ||||||
| − CD | 32 | 25 | 35 | 37 | 18 | 25 |
| + CD | 35 | 25 | 35 | 40 | 20 | 25 |
| QCB1 | ||||||
| − CD | 40 | 31 | 47 | 48 | 28 | 32 |
| + CD | D | D | D | D | D | D |
| QCB1(pPONB) | ||||||
| − CD | 29 | 21 | 34 | 35 | 23 | 26 |
| + CD | 31 | 24 | 33 | 37 | 24 | 25 |
| QCB1(pL793ΔCt) | ||||||
| − CD | 28 | 22 | 33 | 35 | 22 | 25 |
| + CD | 30 | 24 | 32 | 35 | 24 | 25 |
| QCB1(pV775ΔCt) | ||||||
| − CD | 34 | 26 | 37 | 37 | 20 | 26 |
| + CD | 35 | 26 | 34 | 38 | 20 | 26 |
| QCB1(pCys−) | ||||||
| − CD | 29 | 21 | 34 | 35 | 23 | 25 |
| + CD | 31 | 23 | 33 | 37 | 24 | 26 |
Results are expressed in millimeters. Abbreviations: TIC, ticarcillin; AMX, amoxicillin; CTX, cefotaxime; ATM, aztreonam; CF, cefalotin; MEC, mecillinam; −CD, grown without cephaloridine; +CD, grown in medium containing cephaloridine (0.3 μg/ml); D, death (no bacteria grew on the plates).
pCys− PBP1b is able to form dimers.
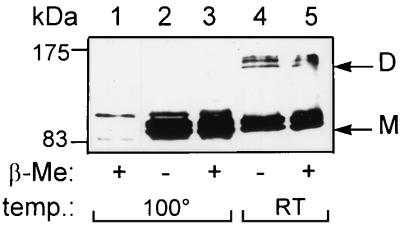
Membrane fractions of strain QCB1 harboring pCys− were incubated in sample buffer, with or without β-mercaptoethanol, either at room temperature or 100°C for 10 min, and analyzed by Western blotting with the antitag antibody 12CA5. Results clearly show that this mutant can dimerize and dissociate under the same experimental conditions as the wild-type protein (Fig. 3, lanes 2 through 5). Furthermore, the stability of the dimeric form of the PBP1b mutant expressed from pCys− was tested at several temperatures. The dimer remains stable after incubation for 10 min at 60°C with sample buffer containing β-mercaptoethanol but is almost totally dissociated at 80°C. The pattern of dissociation was similar when the experiment was performed with sample buffer without β-mercaptoethanol (data not shown). Both deletion mutants are also able to form dimers (not shown).
FIG. 3.
Dimers of the pCys− PBP1b mutant. Membrane fractions prepared from strain QCB1 (lane 1) or QCB1(pCys−) (lanes 2 through 5) were incubated with sample buffer with 5% or no β-mercaptoethanol (β-Me) at room temperature (RT) or 100°C for Western blotting analysis with antitag 12CA5 antibody. Abbreviations are the same as in Fig. 1.
Zijderveld et al. (23, 24) have shown that PBP1b exists as a dimer in E. coli. These authors characterized two classes of dimers, one bound to the inner membrane and a second associated with the peptidoglycan. Our results show that although the PBP1b dimers are fairly stable, being detected in SDS-PAGE and requiring a 10-min incubation at 80°C to be dissociated, the dimerization does not rely on the formation of disulfide bonds as previously hypothesized. Moreover, we demonstrated that the two cysteine residues of PBP1b are dispensable for both its activity in vivo and its dimerization. Mild solubilization with detergents suffices to dissociate the dimers, but we failed to detect any effect of zinc on the dimer stability as was previously shown (24). This discrepancy may result from differences in the preparation of membranes. PBP1b consists of three components (α, β, and γ) with different mobilities on SDS-PAGE gels. Two-dimensional gel analysis suggests that only homodimers, α-α, β-β, and γ-γ, occur (23). We have not observed these different forms, perhaps due to their close migration in SDS-PAGE. We noticed, however, that the 150-kDa band sometimes appeared as a doublet. Whether this doublet corresponds to two classes of dimers is under investigation.
The biological role of these dimers still remains to be experimentally demonstrated, although the recent models of peptidoglycan synthesis imply that PBP1b acts as a dimer within a multienzymatic complex (4, 13). The above-described experiments exclude models in which PBP1b is linked by disulfide bridges. Nevertheless, these dimers involve strong noncovalent interactions. Further investigations are necessary to elucidate the precise nature of these interactions and their roles in PBP1b activities.
REFERENCES
- 1.García del Portillo F, de Pedro M A. Differential effect of mutational impairment of penicillin-binding proteins 1A and 1B on Escherichia coli strains harboring thermosensitive mutations in the cell division genes ftsA, ftsQ, ftsZ, and pbpB. J Bacteriol. 1990;172:5863–5870. doi: 10.1128/jb.172.10.5863-5870.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghuysen J M. Penicillin-binding proteins. Wall peptidoglycan assembly and resistance to penicillin: facts, doubts and hopes. Int J Antimicrob Agents. 1997;8:45–60. doi: 10.1016/s0924-8579(96)00358-5. [DOI] [PubMed] [Google Scholar]
- 3.Höltje J-V. Molecular interplay of murein synthetases and murein hydrolases in Escherichia coli. Microb Drug Resist. 1996;2:99–103. doi: 10.1089/mdr.1996.2.99. [DOI] [PubMed] [Google Scholar]
- 4.Höltje J-V. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kato J I, Suzuki H, Hirota Y. Overlapping of the coding regions for α and γ components of penicillin-binding protein 1b in Escherichia coli. Mol Gen Genet. 1984;196:449–457. doi: 10.1007/BF00436192. [DOI] [PubMed] [Google Scholar]
- 6.Kato J I, Suzuki H, Hirota Y. Dispensability of either penicillin-binding protein 1A or 1B involved in the essential process for cell elongation in Escherichia coli. Mol Gen Genet. 1985;200:272–277. doi: 10.1007/BF00425435. [DOI] [PubMed] [Google Scholar]
- 7.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 8.Lefèvre F, Rémy M-H, Masson J-M. Topographical and functional investigation of Escherichia coli penicillin-binding protein 1b by alanine stretch scanning mutagenesis. J Bacteriol. 1997;179:4761–4767. doi: 10.1128/jb.179.15.4761-4767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lenfant F, Labia R, Masson J-M. Probing the active site of β-lactamase R-TEM-1 by informational suppression. Biochimie. 1990;72:495–503. doi: 10.1016/0300-9084(90)90073-p. [DOI] [PubMed] [Google Scholar]
- 10.Massova I, Mobashery S. Kinship and diversification of bacterial penicillin-binding proteins and β-lactamases. Antimicrob Agents Chemother. 1998;42:1–17. doi: 10.1128/aac.42.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuhashi M. Utilization of lipid-linked precursors and the formation of peptidoglycan in the process of cell growth and division: membrane enzymes involved in the final steps of peptidoglycan synthesis and the mechanism of their integration. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Biomedical Press; 1994. pp. 55–71. [Google Scholar]
- 12.Nanninga N. Cell division and peptidoglycan assembly in Escherichia coli. Mol Microbiol. 1991;5:791–795. doi: 10.1111/j.1365-2958.1991.tb00751.x. [DOI] [PubMed] [Google Scholar]
- 13.Nanninga N. Morphogenesis of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:110–129. doi: 10.1128/mmbr.62.1.110-129.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Romeis T, Höltje J-V. Specific interaction of penicillin-binding proteins 3 and 7/8 with the soluble lytic transglycosylase in Escherichia coli. J Biol Chem. 1994;269:21603–21607. [PubMed] [Google Scholar]
- 15.Suzuki H, Nishimura Y, Hirota Y. On the process of cellular division in Escherichia coli: a series of mutants of E. coli altered in the penicillin-binding proteins. Proc Natl Acad Sci USA. 1978;75:664–668. doi: 10.1073/pnas.75.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki H, Heijenoort Y, Tamura T, Mizoguchi J, Hirota Y, Van Heijenoort J. In vitro peptidoglycan polymerization catalysed by penicillin binding protein 1B of Escherichia coli K12. FEBS Lett. 1980;110:245–249. doi: 10.1016/0014-5793(80)80083-4. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki H, Kato J-I, Sakagami Y, Mori M, Suzuki A, Hirota Y. Conversion of the α component of penicillin-binding protein 1b to the β component in Escherichia coli. J Bacteriol. 1987;169:891–893. doi: 10.1128/jb.169.2.891-893.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamaki S, Nakajima S, Matsuhashi M. Thermosensitive mutation in Escherichia coli simultaneously causing defects in penicillin-binding protein-1Bs and in enzyme activity for peptidoglycan synthesis in vitro. Proc Natl Acad Sci USA. 1977;74:5472–5476. doi: 10.1073/pnas.74.12.5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Rechenberg M, Ursinus A, Höltje J-V. Affinity chromatography as a means to study multi-enzyme-complexes involved in murein synthesis. Microb Drug Resist. 1996;2:155–157. doi: 10.1089/mdr.1996.2.155. [DOI] [PubMed] [Google Scholar]
- 20.Weiner M P, Costa G L, Schoettlin W, Cline J, Mathur E, Bauer J C. Site-directed mutagenesis of double-stranded DNA by the polymerase chain reaction. Gene. 1994;151:119–123. doi: 10.1016/0378-1119(94)90641-6. [DOI] [PubMed] [Google Scholar]
- 21.Wilson I A, Niman H L, Houghten R A, Cherenson A R, Connoly M L, Lerner R A. The structure of an antigenic determinant in a protein. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 22.Yousif S Y, Broome-Smith J K, Spratt B G. Lysis of Escherichia coli by beta-lactam antibiotics: deletion analysis of the role of penicillin-binding proteins 1A and 1B. J Gen Microbiol. 1985;131:2839–2845. doi: 10.1099/00221287-131-10-2839. [DOI] [PubMed] [Google Scholar]
- 23.Zijderveld C A L, Aarsman M E G, den Blaauwen T, Nanninga N. Penicillin-binding protein 1B of Escherichia coli exists in dimeric forms. J Bacteriol. 1991;173:5740–5746. doi: 10.1128/jb.173.18.5740-5746.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zijderveld C A L, Aarsman M E G, Nanninga N. Differences between inner membrane and peptidoglycan-associated PBP1B dimers of Escherichia coli. J Bacteriol. 1995;177:1860–1863. doi: 10.1128/jb.177.7.1860-1863.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]