Abstract
This study aims to investigate the function and mechanism of microRNA-106b-5p (miR-106b-5p) in cervical cancer (CC). Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was performed to determine miR-106b-5p expression in CC tissues and normal gastric tissues. Cell counting kit-8 (CCK-8) and colony formation assays were used to analyze the regulatory effects of miR-106b-5p on CC cells’ proliferative ability. Wound healing and Transwell assays were conducted to detect the effects of miR-106b-5p on cell migration and invasion. Besides, TargetScan was used to predict the potential target genes of miR-106b-5p. The interaction between miR-106b-5p and fibroblast growth factor 4 (FGF4) was proved by qRT-PCR, Western blot, and dual-luciferase reporter gene assay. MiR-106b-5p expression was down-regulated in CC tissues compared to non-tumorous tissues. The expression of miR-106b-5p was associated with the lymphatic node metastasis, FIGO stage and differentiation of CC. Functional assays revealed that miR-106b-5p overexpression suppressed CC cell proliferation, migration and invasion while miR-106b-5p inhibitor had the opposite effects. In addition, FGF4 was identified as a target gene of miR-106b-5p, and FGF could be negatively regulated by miR-106b-5p. MiR-106b-5p may serve as a tumor suppressor in CC, which can inhibit CC growth and metastasis by down-regulating FGF4 expression.
Keywords: miR-106b-5p, FGF4, Cervical cancer, Proliferation, Metastasis
Introduction
Cervical cancer (CC) is one of the most common malignancies. According to statistics, there are approximately 570,000 new cases and more than 310,000 deaths around the world each year (Bray et al. 2018). Even though early screening and human papillomavirus vaccination have greatly reduced the burden of CC in developed countries, CC remains to be a health care problem worldwide. Therefore, it is extremely crucial to clarify the underlying molecular mechanisms of CC and develop novel therapies.
MicroRNAs (miRs) are a group of non-coding, single-stranded RNAs consisting of approximately 20 nucleotides, which can suppress the expression of target gene by binding to the 3′ untranslated regions (3′UTRs) of messenger RNA (Aurora and Slack 2006). Accumulating studies suggest that miRNAs are abnormally expressed in various tumors and associated with carcinogenesis, cancer cell proliferation and metastasis (Galasso et al. 2012; Acunzo and Croce 2015; Nahand et al. 2020). For instance, miR-34a/b/c can inhibit intestinal carcinogenesis (Jiang and Hermeking 2017); miR-17-3p regulates the proliferative ability and survival ability of colonic cancer cells (Lu et al. 2018); and miR-532 promotes the migration and invasion of gastric cancer cells (Hu et al. 2017). Furthermore, there are a lot of miRs playing a key role in CC (Nahand et al. 2019). For example, miR-216b inhibits the proliferative ability of CC cells, and its high expression is related to a better prognosis of CC patients (He et al. 2017); suppressed expression of miR-138-5p by long non-coding RNA H19 in CC facilitates tumor growth (Ou et al. 2018). MiR-106b-5p regulates the proliferation and metastasis of a variety of tumors. It can inhibit the migration and invasion of colorectal cancer cells by targeting cathepsin A (Ni et al. 2018) and promote stem cell-like properties of hepatocellular cancer cells by targeting phosphatase and tensin homolog deleted on chromosome ten via phosphatidylinositol-3 kinase/Akt pathway (Shi et al. 2018). In renal cell carcinoma, miR-106b-5p promotes its aggressiveness and stem-cell-like phenotype by activating Wnt/β-catenin signaling (Lu et al. 2017). However, there are few reports focusing on the expression and function of miR-106b-5p in CC.
Fibroblast growth factors (FGFs) are a group of paracrine/autocrine proteins that bind to tyrosine kinase fibroblast growth factor receptors (FGFRs), triggering an endocellular signaling cascade that regulates cellular biological activities. FGFs are involved in a variety of biological processes including embryonic development, cell growth and metastasis (Presta et al. 2017). Fibroblast growth factor 4 (FGF4) is a member of FGFs, and its expression is up-regulated in various kinds of human tumors. For example, FGF4 can promote the metastasis of breast cancer cells and hepatocellular carcinoma cancer cells (Arao et al. 2013; Shi et al. 2016). Moreover, FGF4 evokes epithelial-mesenchymal transition by inducing store-operated calcium entry in lung adenocarcinoma cells (Qi et al. 2016).
This study was performed to clarify the role of miR-106b-5p in regulating the malignant phenotypes of CC cells and substantiate its regulatory function on FGF4, which was predicted as a potential target gene of miR-106b-5p by bioinformatics analysis.
Methods and materials
Tissue specimens and cell lines
This research was approved by the Ethical Committee of the Second Hospital of Dalian Medical University. CC tissues and adjacent tissues were obtained during surgery from 80 patients in the Second Hospital of Dalian Medical University. The inclusion criteria were: (1) The patients were diagnosed as CC by two experienced pathologists; (2) none of them had received any neoadjuvant anti-cancer treatment of CC. The exclusion criteria were: (1) patients with other chronic diseases; (2) patients with other malignancies. Of the collected cases, 71 were squamous cell carcinomas (SCC), 5 were adenocarcinomas and 4 were adenosquamous carcinomas. Written informed consent was obtained from all patients. All patients with CC were histopathologically confirmed, and the clinical information of the patients was summarized in Table 1.
Table 1.
Correlation between miR-106b-5p expression and pathological parameters of cervical cancer patients
| Pathological parameters | Number | miR-106b-5p expression | Chi-Square | p-value | |
|---|---|---|---|---|---|
| Low | High | ||||
| Age | 0.823 | 0.364 | |||
| < 55 | 32 | 21 | 11 | ||
| ≥ 55 | 48 | 36 | 12 | ||
| Histology | 0.211 | 0.646 | |||
| SCC | 71 | 50 | 21 | ||
| Others | 9 | 7 | 2 | ||
| FIGO stage | 8.14 | 0.004 | |||
| III | 67 | 52 | 15 | ||
| I + II | 13 | 5 | 8 | ||
| Differentiation | 5.73 | 0.016 | |||
| Poorly | 57 | 45 | 12 | ||
| Moderately-well | 23 | 12 | 11 | ||
| Lymphatic metastasis | 4.216 | 0.040 | |||
| Yes | 61 | 47 | 14 | ||
| No | 19 | 10 | 9 | ||
Human CC cell lines (SiHa, C-33A, ME-180, MS-751, HCC-94 and HeLa) and HEK-293 T cell line were purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). Human normal cervical epithelial cell line H8 was obtained from American Type Culture Collection (ATCC, MD, USA). All cell lines were cultured in Dulbecco’s modified Eagle medium (DMEM; Invitrogen, Shanghai, China) supplemented with 10% fetal bovine serum (FBS, Gibco, Rockville, MD), 100U/ml penicillin and 100 μg/ml streptomycin (Invitrogen, Shanghai, China) in a humidified atmosphere at 37 °C in 5% CO2. MiR-106b-5p mimics, miR-106b-5p inhibitors (miR-106b-5p-in), FGF4 small interfering RNA (siRNA) (Invitrogen, Shanghai, China) and their controls were transfected into the cells using Lipofectamine™ 2000 (Invitrogen, Shanghai, China) according to the manufacturer’s instruction.
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
TRIzol reagent (Invitrogen, Shanghai, China) was used to extract total RNA from tissues and cells according to the supplier’s instructions. RNA was reversely transcribed into cDNA using a PrimeScript RT reagent kit (Takara Bio Inc, Dalian, China). The quantitative amplification reactions for miR-106b-5p and for FGF4 were conducted by a TapMan MicroRNA Assays Real-time PCR kit (Invitrogen, Shanghai, China) and a SYBR Green Master Mixture kit (Roche, Basel, Switzerland), respectively, on ABI Prism 7300 System (Thermo Fisher Scientific, Waltham, MA, USA). The expressions of miR-106b-5p and FGF4 mRNA were calculated using the 2(−ΔΔCt) method. U6 served as the internal control for miR-106b-5p, and GAPDH acted as the internal reference for FGF4. The primer sequences are as follows: miR-106b-5p: 5′‐GCGTAAAGTGCTGACAGTGCAGAT‐3′; U6: 5′-CAAATTCGTGAAGCGTTCCATAT-3′; FGF4: 5′- CTCGCCCTTCTTCACCGATG-3′ (forward); 5′- GTAGGACTCGTAGGCGTTGTA-3′ (reverse); GAPDH: 5′- GGAGCGAGATCCCTCCAAAAT-3′ (forward), and 5′- GGCTGTTGTCATACTTCTCATGG-3′ (reverse).
Cell proliferation assay
After 24 h of transfection, the proliferation of transfected cells was detected using the cell counting kit-8 (CCK-8; Dojindo, Kumamoto, Japan). In brief, the cells were seeded in the 96-well plates (1 × 103 cells/well) and cultured in 100 μl of culture medium. After 24 h, the cells were incubated with 10 μl of CCK-8 solution for 1 h. After the termination of the culture, the 96-well plate was placed in a microplate reader, and the absorbance of each well at a wavelength of 450 nm was measured.
Colony formation assay
After transfection, HeLa cells were seeded in 6-well culture plates at a density of 1 × 103 cells per well. The cells were cultured for 2 weeks before the colonies were fixed with 4% paraformaldehyde solution and stained with 0.1% crystal violet. Then, the number of colonies was counted and recorded.
Wound healing assay
In brief, the transfected cells were seeded in 6-well plates and cultured until the cells covered the bottom of the wells, and subsequently a scratch was created with a pipette tip, and then the cells were cultured with serum-free medium. Then, the images of the scratches were captured at 0 h, 24 h and 48 h using a light microscope. After that, the migration capability of the cells was measured.
Transwell assay
Transwell assay was performed to measure the migration ability and invasion ability of cells with Transwell chambers (8 µm pore size; Corning, Beijing, China). For migration assay, cells were resuspended in serum-free medium, and the density was adjusted to 1 × 105 cells/ml. Then, 200 μl of cell suspension was added into the upper chamber, and 500 μl of medium containing 10% FBS was added to the lower chamber. Then the cells were cultured in an incubator at 37 °C for 24 h, and the cells on the underside of the membrane were fixed with 4% paraformaldehyde for 10 min and then stained with 0.1% crystal violet for 10 min, and their number was counted under a microscope. For invasion assay, Matrigel was pre-coated on the Transwell membrane, and the other experimental procedures were the same as in the migration assay.
Luciferase reporter assay
The sequence of FGF4 3′UTR carrying the binding site for miR-106b-5p was amplified by PCR, and subcloned into pGL3 vector (Promega, Madison, WI, USA) to construct the luciferase reporter vector. Then, the recombinant vectors and miRNA mimics were transfected into HEK-293 T cells. 48 h after the transfection, the luciferase activity of the cells was measured using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA).
Western blot
RIPA lysis buffer (Beyotime, Shanghai, China) was used to extract the protein from cells, and polyacrylamide gel electrophoresis was used to separate protein samples. Then, the proteins were transferred onto polyvinylidene difluoride membranes (Millipore, Boston, MA, USA). After that, the membranes were incubated with anti-FGF4 polyclonal antibody (ab65974, 1:2000; Abcam, Cambridge, UK) and anti-GAPDH antibody (ab9485, 1:1000; Abcam, Cambridge, UK) at 4 °C for 12 h, followed by the incubation with HRP labeled goat anti-rabbit secondary antibody (ab150077, 1:6000; Abcam, Cambridge, UK) at room temperature for 1 h. Next, the membranes were rinsed with tris buffered saline Tween for 3 times. Subsequently, the signals were detected by a chemiluminescence phototope-HRP kit (Pierce Biotechnology, Rockford, USA).
Statistical analysis
Each experiment was repeated 3 times, and all data were expressed as “mean ± standard deviation” and statistically analyzed using SPSS version 20.0 (SPSS, Chicago, IL, USA). Data in different groups were compared using Student’s t-test or one-way ANOVA. The relationship between miR-106b-5p expression and clinicopathologic features was analyzed by Chi-square test. P < 0.05 was considered statistically significant.
Results
MiR-106b-5p expression was down-regulated in CC tissues and cell lines
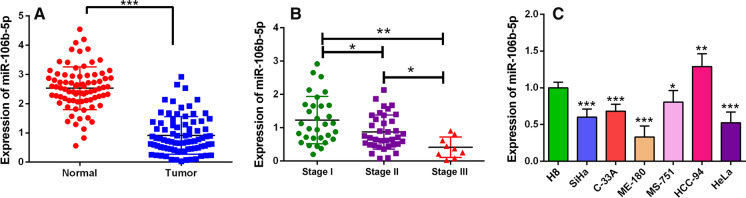
To determine whether the expression of miR-106b-5p was dysregulated in CC, we performed qRT-PCR to detect miR-106b-5p expression in 80 cases of CC tissues and 80 cases of normal tissues. As shown, miR-106b-5p expression was down-regulated in CC tissues compared with that in normal tissues (P < 0.05, Fig. 1A). When 80 CC patients were classified based on TNM stage, miR-106b-5p expression was found to be negatively correlated with the TNM stage (P < 0.05, Fig. 1B). We then measured the miR-106b-5p expression in 6 CC cell lines and a normal cervical cell line by qRT-PCR. It was found that in HeLa, C-33A, SiHa, ME-180 and MS-751 CC cell lines, miR-106b-5p expression was significantly lower than that of H8 cells (P < 0.05, Fig. 1C). However, there was an exception that the expression of miR-106b-5p in HCC-94 cells was higher than that of H8 cells (P < 0.05, Fig. 1C).
Fig. 1.
MiR-106b-5p expression was down-regulated in CC tissues and cells. A MiR-106b-5p expression in 80 paired CC samples and normal cervical tissues was determined by qRT-PCR. B qRT-PCR was performed to detect the expression of miR-106b-5p in CC tissues with different clinical stage. C MiR-106b-5p expression in CC cell lines (SiHa, C33A, ME-180, MS-751, HCC-94 and HeLa) and H8 cells were detected by qRT-PCR. All of the experiments were performed in triplicate. *P < 0.05, **P < 0.01 and ***P < 0.001
MiR-106b-5p suppressed cell proliferation of CC
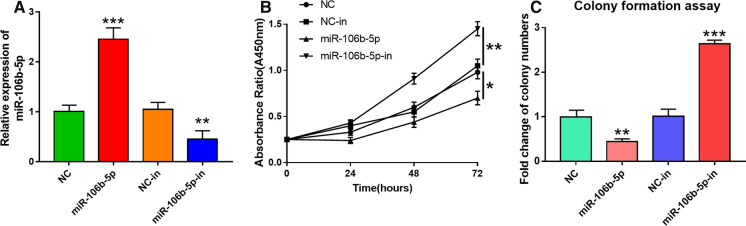
We chose HeLa cell line for the subsequent experiments because HeLa cells had moderate miR-106b-5p expression. MiR-106b-5p mimic or miR-106b-5p inhibitor was transfected into HeLa cells, and then the transfection efficiency was confirmed by qRT-PCR (P < 0.05, Fig. 2A). To explore whether miR-106b-5p regulated the growth of CC cells, CCK-8 and colony formation assays were performed. CCK-8 assay showed that compared with NC group, the proliferative ability of HeLa cells transfected with miR-106b-5p mimics was markedly suppressed (P < 0.05 on 48 h and 72 h, Fig. 2B). Colony formation assay revealed a significant decrease of HeLa cell colonies on d 14 in miR-106b-5p group compared with NC group (P < 0.05, Fig. 2C). Additionally, miR-106b-5p inhibition induced a dramatic increase in cell proliferative ability and colony number compared with the NC-in group (P < 0.05, Fig. 2B–C). These results suggested that miR-106b-5p could modulate the proliferative ability of CC cells.
Fig. 2.
MiR-106b-5p suppressed the proliferative ability of HeLa cells. A qRT-PCR was performed to detect the impacts of miR-106b-5p mimics and inhibitors on miR-106b-5p expression in HeLa cells. B CCK-8 assay showed that the transfection of miR-106b-5p mimics inhibited the proliferation of HeLa cells whereas miR-106b-5p inhibitors facilitated the proliferation. C Colony formation assay showed that, miR-106b-5p overexpression markedly repressed the colony formation ability of HeLa cells whereas miR-106b-5p inhibitors enhanced the colony formation ability. The number of colonies containing > 50 cells was counted. All of the experiments were performed in triplicate. **P < 0.01 and ***P < 0.001
MiR-106b-5p inhibited the migration and invasion of CC cells
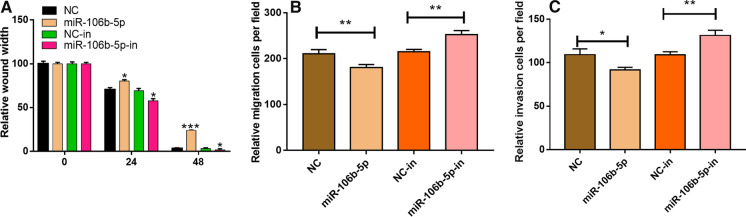
To investigate whether miR-106b-5p was associated with tumor metastasis in CC, we detected the migration and invasion ability of CC cells by wound healing assay and Transwell assay. The result of wound healing assay showed that the overexpression of miR-106b-5p could inhibit the migration of HeLa cells; on the contrary, the inhibition of miR-106b-5p significantly promoted the migration of HeLa cells (P < 0.05, Fig. 3A), and Transwell assay showed consistent results (P < 0.05, Fig. 3B–C). Collectively, these data indicated that miR-106b-5p suppressed the migration and invasion abilities of CC cells.
Fig. 3.
MiR-106b-5p inhibited the migration and invasion of HeLa cells. A Migration ability of HeLa cells transfected with NC, miR-106b-5p, miR-106b-5p-in or NC-in was detected by wound healing assay. B Transwell migration assay was used to detect the migration ability of HeLa cells transfected with NC, NC-in, miR-106b-5p mimics or miR-106b-5p-in. C Transwell invasion assay was used to detect the invasion ability of HeLa cells after the transfection with NC, NC-in, miR-106b-5p mimics or miR-106b-5p-in. All of the experiments were performed in triplicate. *P < 0.05 and **P < 0.01
FGF4 expression was negatively regulated by miR-106b-5p in CC
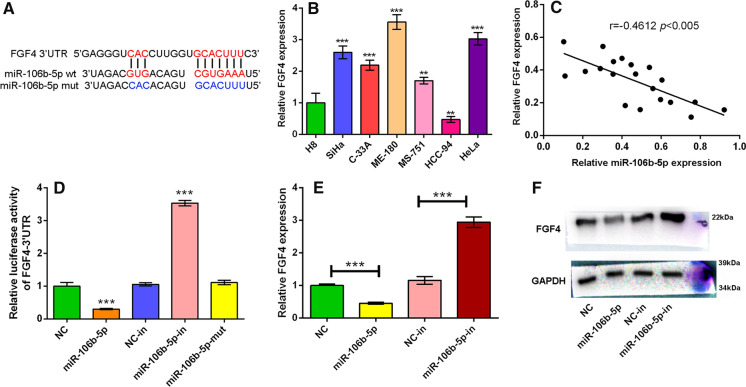
We searched TargetScan database and found that miR-106b-5p could potentially directly regulate FGF4 expression by binding to its 3′UTR (Fig. 4A). We then detected FGF4 expression in H8 cells and different CC cell lines including SiHa, C-33A, ME-180, MS-751, HCC-94 and HeLa cells by qRT-PCR. It was found that in SiHa, C-33A, ME-180, MS-751 and HeLa cells, FGF4 expression was markedly higher than that in H8 cells (P < 0.05, Fig. 4B). Then, we randomly selected 20 tumor samples from 80 CC patients to verify the relationship between miR-106b-5p expression and FGF4 expression, and the results showed that the expressions of miR-106b-5p and FGF4 mRNA were negatively correlated (Fig. 4C). Accordingly, to determine whether miR-106b-5p could regulate FGF4’s expression via binding to its 3′-UTR, we constructed pGL3 luciferase reporter vector containing FGF4 3′-UTR. It was confirmed that miR-106b-5p markedly suppressed the luciferase activity of the FGF4 3′UTR-luciferase reporter compared to the NC group (P < 0.05); inhibiting miR-106b-5p expression remarkably increased the luciferase activity of FGF4 3′UTR-luciferase reporter (P < 0.05); in contrast, miR-106b-5p mimics with mutated sequence had no significant effect on the luciferase activity of the FGF4 3′UTR-luciferase reporter (Fig. 4D). Western blot showed that the transfection of miR-106b-5p mimics into HeLa cells decreased the expression of FGF4 while miR-106b-5p inhibitors increased FGF4 expression (Fig. 4E–F). Collectively, it was concluded that FGF4 was a direct target of miR-106b-5p.
Fig. 4.
FGF4 expression was negatively regulated by miR-106b-5p in CC. A Bioinformatics was performed to predict the complementary binding sites between miR-106b-5p and FGF4 3′UTR. B qRT-PCR was performed to detect the FGF4 expression in H8 cell line and CC cell lines (SiHa, C33A, ME-180, MS-751, HCC-94 and HeLa). C The expression levels of FGF4 mRNA and miR-106b-5p in tissues of 20 CC patients were determined by qRT-PCR, and correlation analysis confirmed the negative correlation between miR-106b-5p and FGF4. D Dual-luciferase reporter assay was conducted to verify the targeting relationship between miR-106b-5p and FGF4. E–F Western blot was used to detect FGF4 expression in HeLa cells transfected with miR-106b-5p mimics or miR-106b-5p inhibitors, compared with corresponding control cells (NC and NC-in). GAPDH served as an internal control. All of the experiments were performed in triplicate. **P < 0.01 and ***P < 0.001
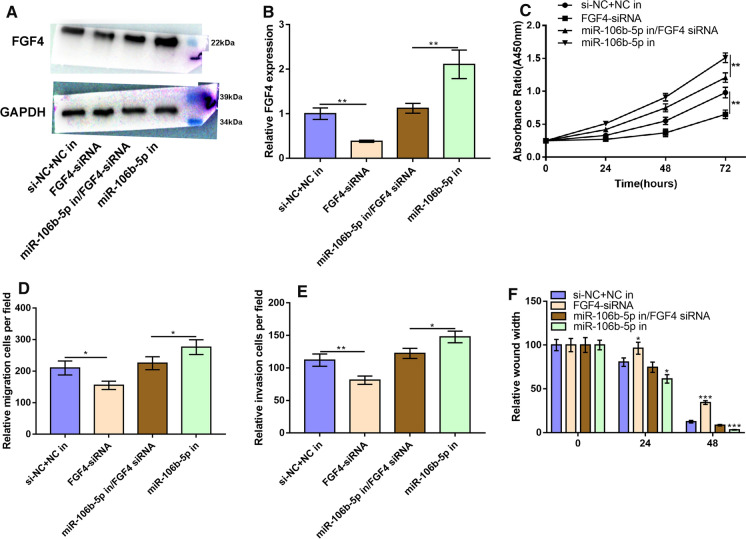
MiR-106b-5p inhibited the cell proliferation and metastatic potential of CC cells by repressing FGF4 expression
To verify that miR-106b-5p inhibited cell proliferation, migration and invasion of CC cells through suppressing FGF4, we transfected FGF4 siRNA or co-transfected FGF4 siRNA and miR-106b-5p inhibitor into HeLa cells. The result of Western blot implied that FGF4 expression was down-regulated in FGF4-siRNA group, and FGF4-siRNA effectively blocked the enhancement of FGF4 expression caused by miR-106b-5p inhibitor (Fig. 5A–B). The results of CCK-8 assay showed that FGF4 expression suppressed the proliferation of HeLa cells; meanwhile, in miR-106b-5p-in group, cell proliferation was markedly enhanced whereas FGF4-siRNA could reverse the effect of miR-106b-5p-in on the proliferation of HeLa cells (P < 0.05, Fig. 5C). Consistently, Transwell and wound healing assays also supported that inhibiting miR-106b-5p expression could promote the migration and invasion of HeLa cells, and knocking down endogenous FGF4 expression could reverse this effect (P < 0.05, Fig. 5D–F). To sum up, miR-106b-5p suppressed CC cell proliferation and metastatic potential by repressing FGF4 expression.
Fig. 5.
MiR-106b-5p suppressed the malignant phenotypes of HeLa cells by inhibiting FGF4 expression. A–B HeLa cells were transfected with FGF4-siRNA, miR-106b-5p-in or both, and then FGF4 expression was measured by Western blot. C CCK-8 assay was used to detect the proliferation ability of HeLa cells after the transfection with FGF4-siRNA, miR-106b-5p-in or both. D Transwell migration assay was used to detect the migration ability of HeLa cells after the transfection with FGF4-siRNA, miR-106b-5p-in or both. E Transwell invasion assay was used to detect the invasion ability of HeLa cells after the transfection with FGF4-siRNA, miR-106b-5p-in or both. F Migration ability of HeLa cells transfected with FGF4-siRNA, miR-106b-5p-in or both was detected by wound-healing assay. All of the experiments were performed in triplicate. *P < 0.05, **P < 0.01 and ***P < 0.001
Higher expression of miR-106b-5p was related to better prognosis in CC patients
To expound whether miR-106b-5p expression was correlated with clinical-pathological features of CC patients, 80 CC patients were classified into two groups according to the mean of miR-106b-5p expression in tumor tissues: the high miR-106b-5p group (n = 57, > mean value) and the low miR-106b-5p group (n = 23, < mean value). Correlation analysis confirmed that low miR-106b-5p expression was significantly related to advanced clinical stage, poor differentiation of tumor tissues and lymphatic metastasis (Table 1). However, the expression of miR-106b-5p was not associated with other clinical parameters, such as age and histological subtypes. These results implied that the low expression of miR-106b-5p was associated with the worse prognosis of CC patients.
Discussion
The oncogenic or tumor-suppressive role of miR-106b-5p has been reported in several studies. In colorectal cancer, miR-106b-5p can enhance the metastatic potential of tumor cells by targeting metastasis-associated lung adenocarcinoma transcript 1 (Zhuang et al. 2019). MiR-106b-5p down-regulates YT521-B homology domain family 2 expression and participates in breast cancer development (Liu et al. 2018). In colonic cancer, circ_005625 could facilitate cancer cells growth via repressing miR-106b-5p (Zhang et al. 2019). Decreased exosomal miR-106b-5p expression promotes angiogenesis in endothelial cells by targeting angiopoietin 2 (Li et al. 2018). Herein, our results suggested that miR-106b-5p played a pivotal role in the progression of CC. It was found that miR-106b-5p expression was markedly down-regulated in CC and was associated with lymphatic node metastasis, clinical stage and tumor tissue differentiation status of the patients. We confirmed that miR-106b-5p could regulate the proliferation, migration and invasion of CC cells. To our best knowledge, this is the first work to investigate the function and clinical significance of miR-106-5p in CC.
The expression of FGFs is closely associated with the occurrence and development of multiple tumors, and their roles vary depending on the type of tumor. It is reported that tumor cell-derived FGFs promote osteoclast activity and contribute to the development of metastatic lesions in breast cancer (Aukes et al. 2017). FGF1 and IGF1-conditioned 3D culture system enhance the growth and cancer stemness of lung cancer cells (Liu et al. 2017). FGF2 disrupts mitotic stability in prostate cancer cells through modulating the intracellular trafficking protein centrosomal protein 57 (Cuevas et al. 2013). FGF-1/-3/FGFR4 signaling in cancer-associated fibroblasts facilitates colonic cancer progression through activating Erk signaling and increasing the expression of matrix metalloproteinase 7 in cancer cells (Bai et al. 2015). Moreover, FGF4 expression is up-regulated in several cancers, and it participates in regulating the malignant biological behaviors of cancer cells. For example, oncoprotein hepatitis B X-interacting protein up-regulates FGF4 expression via activating transcriptional factor Sp1 to enhance the migration of breast cancer cells (Shi et al. 2016). Our research confirmed that FGF4 was a target gene of miR-106b-5p in CC cells; additionally, miR-106b-5p inhibited the malignant phenotypes of CC cells by regulating the expression of FGF4. Our findings suggest that miR-106b-5p/FGF4 axis may be an important mechanism in the progression of CC.
This study has several limitations. First of all, in this work, only in vitro models were constructed, and in vivo experiments are essential to further verify our findings in the following work. Secondly, the other downstream target genes of miR-106b-5p remain to be verified in the future, which will be helpful to further clarify the mechanism of miR-106b-5p in CC. Additionally, to further validate the prognostic value of miR-106b-5p for CC patients, more patients should be enrolled, and the relationship between the expression of miR-106b-5p and the survival time of the patients should be analyzed.
In conclusion, our data show that miR-106b-5p expression is reduced in CC, and its low expression was associated with the unfavorable pathological characteristics of CC patients. MiR-106b-5p regulates CC cell proliferation and metastatic potential by inhibiting FGF4 expression. Thus, miR-106b-5p may serve as a potential biomarker for CC, and the restoration of miR-106b-5p may be a therapeutic strategy for the treatment of CC.
Acknowledgements
We thank Hubei Yican Health Industry Co., Ltd. (Wuhan, China) for its linguistic assistance during the preparation of this manuscript.
Abbreviations
- 3′UTRs
3′ Untranslated regions
- CC
Cervical cancer
- CCK-8
Cell counting kit-8
- FGF4
Fibroblast growth factor 4
- FGFs
Fibroblast growth factors
- MiR-106b-5p
MicroRNA-106b-5p
- miRs
MicroRNAs
- qRT-PCR
Quantitative reverse transcription-polymerase chain reaction
Author contributions
Conceived and designed the experiments: XXY and FZJ; Performed the experiments: LHW, LJ and XXY; Analyzed the data: LHW and LJ; Wrote the paper: LHW and LJ.
Funding
This study is supported by the Natural Science Foundation of Dalian Medical University.
Data availability
The data used to support the findings of this study are available from the corresponding author upon request.
Declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
Our study was approved by the ethics review board of the Second Hospital of Dalian Medical University.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lei Hongwei and Li Juan both have contributed equally to this work.
Contributor Information
Xu Xiaoying, Email: Xuxiaoying1972@sohu.com.
Fan Zhijun, Email: fanzhijundl@sina.com.
References
- Acunzo M, Croce CM. MicroRNA in cancer and Cachexia–a mini-review. J Infect Dis. 2015;212:S74–S77. doi: 10.1093/infdis/jiv197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arao T, Ueshima K, Matsumoto K, Nagai T, Kimura H, Hagiwara S, Sakurai T, Haji S, Kanazawa A, Hidaka H, Iso Y, Kubota K, Shimada M, Utsunomiya T, Hirooka M, Hiasa Y, Toyoki Y, Hakamada K, Yasui K, Kumada T, Toyoda H, Sato S, Hisai H, Kuzuya T, Tsuchiya K, Izumi N, Arii S, Nishio K, Kudo M. FGF3/FGF4 amplification and multiple lung metastases in responders to sorafenib in hepatocellular carcinoma. Hepatology. 2013;57:1407–1415. doi: 10.1002/hep.25956. [DOI] [PubMed] [Google Scholar]
- Aukes K, Forsman C, Brady NJ, Astleford K, Blixt N, Sachdev D, Jensen ED, Mansky KC, Schwertfeger KL. Breast cancer cell-derived fibroblast growth factors enhance osteoclast activity and contribute to the formation of metastatic lesions. PLoS ONE. 2017;12:e185736. doi: 10.1371/journal.pone.0185736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurora EK, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Bai YP, Shang K, Chen H, Ding F, Wang Z, Liang C, Xu Y, Sun MH, Li YY. FGF-1/-3/FGFR4 signaling in cancer-associated fibroblasts promotes tumor progression in colon cancer through Erk and MMP-7. Cancer Sci. 2015;106:1278–1287. doi: 10.1111/cas.12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- Cuevas R, Korzeniewski N, Tolstov Y, Hohenfellner M, Duensing S. FGF-2 disrupts mitotic stability in prostate cancer through the intracellular trafficking protein CEP57. Cancer Res. 2013;73:1400–1410. doi: 10.1158/0008-5472.CAN-12-1857. [DOI] [PubMed] [Google Scholar]
- Galasso M, Sandhu SK, Volinia S. MicroRNA expression signatures in solid malignancies. Cancer J. 2012;18:238–243. doi: 10.1097/PPO.0b013e318258b5f4. [DOI] [PubMed] [Google Scholar]
- He S, Bing L, Deng Y, Chang S, Tuo J, Liu J, Yao S, Lin X. MiR-216b inhibits cell proliferation by targeting FOXM1 in cervical cancer cells and is associated with better prognosis. BMC Cancer. 2017;17:673. doi: 10.1186/s12885-017-3650-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Zheng Q, Wu H, Wang C, Liu T, Zhou W. MiR-532 promoted gastric cancer migration and invasion by targeting NKD1. Life Sci. 2017;177:15–19. doi: 10.1016/j.lfs.2017.03.019. [DOI] [PubMed] [Google Scholar]
- Jiang L, Hermeking H. MiR-34a and miR-34b/c suppress intestinal tumorigenesis. Cancer Res. 2017;77:2746–2758. doi: 10.1158/0008-5472.CAN-16-2183. [DOI] [PubMed] [Google Scholar]
- Li Y, Liang J, Hu J, Ren X, Sheng Y. Down-regulation of exosomal miR-106b-5p derived from cholesteatoma perimatrix fibroblasts promotes angiogenesis in endothelial cells by overexpression of Angiopoietin 2. Cell Biol Int. 2018;42:1300–1310. doi: 10.1002/cbin.11002. [DOI] [PubMed] [Google Scholar]
- Liu P, Zhang R, Yu W, Ye Y, Cheng Y, Han L, Dong L, Chen Y, Wei X, Yu J. FGF1 and IGF1-conditioned 3D culture system promoted the amplification and cancer stemness of lung cancer cells. Biomaterials. 2017;149:63–76. doi: 10.1016/j.biomaterials.2017.09.030. [DOI] [PubMed] [Google Scholar]
- Liu M, Zhou S, Wang J, Zhang Q, Yang S, Feng J, Xu B, Zhong S (2018) Identification of genes associated with survival of breast cancer patients. Breast Cancer-Tokyo 1–9.
- Lu J, Wei JH, Feng ZH, Chen ZH, Wang YQ, Huang Y, Fang Y, Liang YP, Cen JJ, Pan YH, Liao B, Chen WF, Chen W, Luo JH. MiR-106b-5p promotes renal cell carcinoma aggressiveness and stem-cell-like phenotype by activating Wnt/beta-catenin signalling. Oncotarget. 2017;8:21461–21471. doi: 10.18632/oncotarget.15591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Tang L, Zhuang Y, Zhao P. MiR-17-3P regulates the proliferation and survival of colon cancer cells by targeting Par4. Mol Med Rep. 2018;17:618–623. doi: 10.3892/mmr.2017.7863. [DOI] [PubMed] [Google Scholar]
- Nahand JS, Taghizadeh-Boroujeni S, Karimzadeh M, Borran S, Pourhanifeh MH, Moghoofei M, Bokharaei-Salim F, Karampoor S, Jafari A, Asemi Z, Tbibzadeh A, Namdar A, Mirzaei H. microRNAs: new prognostic, diagnostic, and therapeutic biomarkers in cervical cancer. J Cell Physiol. 2019;234:17064–17099. doi: 10.1002/jcp.28457. [DOI] [PubMed] [Google Scholar]
- Nahand JS, Vandchali NR, Darabi H, Doroudian M, Banafshe HR, Moghoofei M, Babaei F, Salmaninejad A, Mirzaei H. Exosomal microRNAs: novel players in cervical cancer. Epigenomics. 2020;12:1651–1660. doi: 10.2217/epi-2020-0026. [DOI] [PubMed] [Google Scholar]
- Ni S, Weng W, Xu M, Wang Q, Tan C, Sun H, Wang L, Huang D, Du X, Sheng W. MiR-106b-5p inhibits the invasion and metastasis of colorectal cancer by targeting CTSA. Onco Targets Ther. 2018;11:3835–3845. doi: 10.2147/OTT.S172887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou L, Wang D, Zhang H, Yu Q, Hua F. Decreased expression of miR-138-5p by lncRNA h19 in cervical cancer promotes tumor proliferation. Oncol Res. 2018;26:401–410. doi: 10.3727/096504017X15017209042610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presta M, Chiodelli P, Giacomini A, Rusnati M, Ronca R. Fibroblast growth factors (FGFs) in cancer: FGF traps as a new therapeutic approach. Pharmacol Ther. 2017;179:171–187. doi: 10.1016/j.pharmthera.2017.05.013. [DOI] [PubMed] [Google Scholar]
- Qi L, Song W, Li L, Cao L, Yu Y, Song C, Wang Y, Zhang F, Li Y, Zhang B, Cao W. FGF4 induces epithelial-mesenchymal transition by inducing store-operated calcium entry in lung adenocarcinoma. Oncotarget. 2016;7:74015–74030. doi: 10.18632/oncotarget.12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Li Y, Feng G, Li L, Fang R, Wang Z, Qu J, Ding P, Zhang X, Ye L. The oncoprotein HBXIP up-regulates FGF4 through activating transcriptional factor Sp1 to promote the migration of breast cancer cells. Biochem Biophys Res Commun. 2016;471:89–94. doi: 10.1016/j.bbrc.2016.01.174. [DOI] [PubMed] [Google Scholar]
- Shi DM, Bian XY, Qin CD, Wu WZ. MiR-106b-5p promotes stem cell-like properties of hepatocellular carcinoma cells by targeting PTEN via PI3K/Akt pathway. Onco Targets Ther. 2018;11:571–585. doi: 10.2147/OTT.S152611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Liu H, Zhao P, Zhou H, Mao T. Has_circ_0055625 from circRNA profile increases colon cancer cell growth by sponging miR-106b-5p. J Cell Biochem. 2019;120:3027–3037. doi: 10.1002/jcb.27355. [DOI] [PubMed] [Google Scholar]
- Zhuang M, Zhao S, Jiang Z, Wang S, Sun P, Quan J, Yan D, Wang X. MALAT1 sponges miR-106b-5p to promote the invasion and metastasis of colorectal cancer via SLAIN2 enhanced microtubules mobility. EBioMedicine. 2019;41:286–298. doi: 10.1016/j.ebiom.2018.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.