Abstract
In previous studies we demonstrated that mutations in the genes cysB, cysE, and cls (nov) affect resistance of Escherichia coli to novobiocin (J. Rakonjac, M. Milic, and D. J. Savic, Mol. Gen. Genet. 228:307–311, 1991; R. Ivanisevic, M. Milic, D. Ajdic, J. Rakonjac, and D. J. Savic, J. Bacteriol. 177:1766–1771, 1995). In this work we expand this list with mutations in rpoN (the gene for RNA polymerase subunit ς54) and the tRNA synthetase genes alaS, argS, ileS, and leuS. Similarly to resistance to the penicillin antibiotic mecillinam, resistance to novobiocin of tRNA synthetase mutants appears to depend upon the RelA-mediated stringent response. However, at this point the overlapping pathways of mecillinam and novobiocin resistance diverge. Under conditions of stringent response induction, either by the presence of tRNA synthetase mutations or by constitutive production of RelA protein, inactivation of the cls gene diminishes resistance to novobiocin but not to mecillinam.
The coumarin antibiotics novobiocin, coumermycin A1, and chlorobiocin inhibit DNA supercoiling reactions by blocking the B subunit (GyrB) of DNA gyrase (4, 5). Escherichia coli mutations that confer resistance to high concentrations of novobiocin map at the gyrB gene (5). In the past several years, we have identified several classes of gyrB-independent mutants which affect E. coli resistance to novobiocin, the mutations mapping within genes of well-established function: cls, the gene that codes for cardiolipin synthase (9, 14, 22, 23), and cysB and cysE, genes involved in biosynthesis of cysteine (10, 13).
In this study, we add to this list the genes argS, alaS, ileS, leuS, and rpoN, which, when mutated, increase the resistance of E. coli to novobiocin.
Isolation and characterization of novobiocin-resistant mutants.
Strains and plasmids used in this study are listed in Table 1. All novobiocin-resistant (Novr) mutants were isolated in E. coli C600, whose native resistance to novobiocin was established at 12.5 μg/ml (see Table 2). Diluted samples, approximately 108 cells per ml, of 10 independent overnight cultures in Luria-Bertani (LB) broth (12), were spread onto LA (LB with 1.5% agar) plates containing novobiocin at 400 μg/ml. Novobiocin-resistant colonies appeared after 2 days of incubation at 37°C. One thousand single colonies were picked and streaked on the same type of plates. Screening of these colonies on minimal medium plates supplemented with threonine, leucine, and cysteine (to eliminate Novr cysB and cysE mutants [13]) showed that 71 colonies (7.1%) were auxotrophic mutants. Further analysis by auxonography (testing for growth on minimal medium plates supplemented with different combinations of amino acids) showed that among the 71 novobiocin-resistant auxotrophs, two required isoleucine (2.8%), two required glutamine (2.8%), and one required adenosine (1.4%). The natures of the other auxotrophs could not be determined by the auxonography test. Further analyses were carried out with isoleucine and glutamine mutants only.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype | Source or reference |
|---|---|---|
| E. coli K-12 strains | ||
| C600 | thi-1 thr-1 leuB6 lacY1 tonA21 supE44 rfbD1 | 1 |
| GC2553 | ftsR1 prototroph | 25 |
| GC3743 (KL231) | leuS3(Ts) thyA6 rpsL120 | 11 |
| GC3802 | GC2553 ftsR+ argS201 | 25 |
| GC3814 | GC2553 ftsR+ alaS21 | D. Vinella |
| SY433 | C600 ileS1001 (isolated as novobiocin-resistant mutant) | This work |
| SY434 | C600 ileS1002 (isolated as novobiocin-resistant mutant) | This work |
| SY437 | SY433 car96::Tn10 (P1 transduction from CAG12093) | This work |
| SY438 | SY434 car96::Tn10 (P1 transduction from CAG12093) | This work |
| SY439 | SY435 zha-6::Tn10 (P1 transduction from CAG12153) | This work |
| SY440 | SY436 zha-6::Tn10 (P1 transduction from CAG12153) | This work |
| SY444 | C600 relA1 | Laboratory collection |
| SY456 | C600 cls::mini-Tn10 | Laboratory collection |
| SY468 | SY434 cls::mini-Tn10 (P1 transduction from SY456) | This work |
| SY500 | GC3814 fuc-3072::Tn10 (P1 transduction from CAG12079) | This work |
| SY501 | C600 alaS21 fuc-3072::Tn10 (P1 transduction from SY500) | This work |
| SY502 | GC3802 zeu-3069::Tn10 by (P1 transduction from CAG12074) | This work |
| SY503 | C600 argS201 zeu-3069::Tn10 (P1 transduction from GC3802) | This work |
| SY504 | GC3814 cls::mini-Tn10 (P1 transduction from SY456) | This work |
| SY505 | GC3802 cls::mini-Tn10 (P1 transduction from SY456) | This work |
| SY506 | C600 F′ [proAB+ lacIqlacZΔM15 Tn10] by conjugation from XL1-Blue | This work |
| SY508 | SY456 F′ [proAB+ lacIqlacZΔM15 Tn10] by conjugation from XL1-Blue | This work |
| SY510 | SY444 alaS21 (P1 transduction from SY501) | This work |
| SY511 | SY444 argS201 (P1 transduction from SY503) | This work |
| XL1-Blue | endA1 hsdR17 (rK− mK+) supE44 thi-1 recA1 gyrA96 relA1 ΔlacF′[proAB+ lacIqlacZΔM15 Tn10] | 2 |
| Plasmids | ||
| pSYC988 | Derivative of pMT521; contains an insertion mutation in the lsp gene generated by repairing the protruding tetranucleotides of XbaI-digested pMT521 DNA with E. coli DNA polymerase I and by ligating this blunt-ended DNA | 8 |
| pSM11 | Recombinant plasmid formed by inserting the truncated relA gene in expression vector pKK223-3 | 15 |
TABLE 2.
Resistance of ileS, alaS, argS, and leuS(Ts) mutants to novobiocin and mecillinam and effects of relA alleles on novobiocin resistance
| Strain | Relevant genotype | Resistance (μg/ml) to:
|
|
|---|---|---|---|
| Novobiocina | Mecillinamb | ||
| C600 | ileS+ relA+ | 12.5 | 0.05 |
| SY506 | C600 F′[proAB+ lacIqlacZΔM15 Tn10] | 12.5 | 0.05 |
| SY701 | SY506(pKK223-3) | 12.5 | 0.05 |
| SY433 | ileS1001 | 500 | >10 |
| SY434 | ileS1002 | 300 | >10 |
| SY501 | alaS21 | 300 | >10 |
| SY503 | argS201 | 250 | >10 |
| GC3743 | leuS3(Ts) | 250 | >10 |
| SY444 | relA1 | 12.5 | |
| SY510 | relA1 alaS21 | 40 | |
| SY511 | relA1 argS201 | 30 | |
| SY702 | SY506(pSM11) | 80 | |
Aliquots of diluted overnight cultures were plated on LB plates containing the corresponding amount of novobiocin. The appropriate dilution spread on plates of LB agar alone served to assess the numbers of cells on antibiotic plates. Survival of more than 50% of cells plated on plates with antibiotic (10 plates per the given concentration of antibiotic) was recorded as resistance to novobiocin. Preliminary experiments were performed to determine the approximate resistance of the strains. The amounts of novobiocin on the plates were increased from 0 to 15 μg/ml by increments of 2.5 μg/ml per plate for strains C600, SY444, SY506, and SY701; from 0 to 100 μg/ml by increments of 10 μg/ml per plate for strains SY510, SY511, and SY702; and from 0 to 600 μg/ml by increments of 50 μg/ml per plate for the rest of the strains.
Resistance to mecillinam was measured by streaking undiluted overnight cultures onto LA plates supplemented with 10 μg of mecillinam per ml. The sensitive strain C600 was tested by streaking undiluted overnight cultures on plates containing 0.01, 0.025, 0.05, and 0.1 μg of mecillinam per ml.
Subsequent analysis showed that novobiocin-resistant ile and gln auxotrophs did not demonstrate increased resistance to other hydrophobic antibiotics: coumermycin (a dimer of novobiocin), rifampin, and chloramphenicol (results not presented).
Mapping of isolated mutations was performed with a subset of CAG strains that carry Tn10 transposons inserted approximately every 0.5 min around the E. coli chromosome (16). For mapping of isoleucine mutations, a stock of phage P1vir was prepared on strain CAG12093, which carries the mutation car::Tn10 in the proximity of the ileS gene at min 0.75 of the E. coli map (16). Among Tetr transductants of both Ile− recipients, Ile+ cotransductants were obtained at a frequency of about 65%, indicating that the ile mutations very possibly map within ileS, the gene that codes for isoleucyl-tRNA synthetase. The Tetr ile+ transductants regained the native level of resistance to novobiocin of the parental strain C600 (12.5 μg/ml), while Tetr ile transductants retained high resistance to the antibiotic (400 μg/ml). Finally, when ile mutations were backcrossed from the Tetr ile transductants into the parental strain C600, the level of resistance to novobiocin changed from 12.5 to 400 μg/ml. These results were corroborated in a parallel experiment in which the presumed ileS mutations were complemented with plasmid pMT521 (20), carrying wild-type alleles of the ileS and lsp genes (Fig. 1). To prevent plasmid integration into the chromosome by homologous recombination, the recA1 mutation was introduced by P1 transduction from strain SY396 (recA1 srl::Tn10) into the ileS mutants. When transformed with plasmid pMT521, both ileS mutants became prototrophs and were again sensitive to novobiocin.
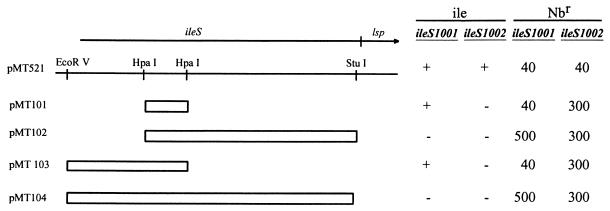
FIG. 1.
Mapping of ileS mutations by complementation with plasmid pMT521 and its deletion derivatives, pMT101 to pMT104. The ileS-lsp loci (arrow) placed in pMT521 and the adjacent 5′ plasmid region are indicated, and a partial restriction map of the region is presented. Open boxes represent regions deleted in the corresponding plasmids. Complementation of isoleucine auxotrophy (ile) in ileS1001 and ileS1002 mutants with each plasmid is denoted as positive (+) or negative (−). Resistance of ileS1001 and ileS1002 mutants to novobiocin (Nbr), measured by the efficiency-of-plating test, after complementation with the respective plasmids is indicated (in micrograms per milliliter).
The gene lsp is a part of the ileS-lsp operon and codes for prolipoprotein signal peptidase, the dysfunction of which may affect cellular resistance to antibiotics (19–21). Thus, one might imagine that ileS mutations, besides making E. coli auxotrophic for isoleucine, might affect its resistance to novobiocin by exerting a polar effect on expression of the downstream lsp gene. That this is not the case was demonstrated by complementation experiments with derivatives of plasmid pMT521 (ileS+ lsp+). Introduction of plasmid pSYC988, which carries the intact ileS allele but a partially mutated lsp gene (30% of the activity of the wild-type product [8]), into both ileS mutants resulted in (full) complementation of ile auxotrophy and resistance to novobiocin (Fig. 1). To ensure that the 30% active Lsp product could not complement novobiocin resistance, a set of plasmids (pMT101 to pMT104) harboring deletions of different ileS regions (Fig. 1), in which the product of the lsp gene has 34 to 64% enzymatic activity (21), was employed in the complementation studies. The deletion derivatives of pMT521 neither complemented ile auxotrophy nor decreased novobiocin resistance (Fig. 1). These results confirm our original presumption that the wild-type allele of the ileS gene alone is sufficient for effective complementation of the dual phenotype of isoleucine auxotrophy and increased resistance to novobiocin in both ile mutants.
The same strategy was employed to map the glutamine mutations. Transduction experiments with phage P1 prepared on strain CAG12153 (zca-6::Tn10 [16]) located both mutations at min 70.0, and complementation experiments with plasmid pTH7, carrying the wild-type allele of the rpoN gene (from B. Magasanik), narrowed their location to within the gene that codes for the alternative RNA polymerase sigma factor ς54 (strains SY435 [C600 rpoN1001] and SY436 [C600 rpoN1002]). This result was confirmed in a control experiment with strain YMC18 (rpoN::Tn10) (from B. Magasanik), which we also found to be both glutamine auxotrophic and resistant to novobiocin. When the rpoN mutation from YMC18 was transferred by P1 transduction into strain C600, the resulting tetracycline-resistant transductants became glutamine auxotrophic and resistant to novobiocin (results not presented).
Two classes of novobiocin-resistant ileS mutants.
In the course of characterizing novobiocin-resistant ileS mutants, we found that the two independently isolated mutants (ileS1001 and ileS1002 mutants) exhibited different characteristics of resistance to novobiocin and growth rate. They grew slowly in LB medium, with generation times of 173 and 84 min, respectively, and their effective resistances to novobiocin also showed a significant difference (Table 2).
The relative positions of the ileS1001 and ileS1002 mutations were deduced from the recombinational patterns (IleS+ versus IleS−) obtained upon introduction of plasmids pMT101 to pMT104 into recombination-proficient ileS1001 and ileS1002 mutants. The results suggest that the ileS1001 mutation maps on the HpaI-StuI fragment, while ileS1002 mutation may be located on the HpaI-HpaI fragment of the plasmid (pMT521) (Fig. 1).
ileS mutants are resistant to mecillinam.
It is known that some aminoacyl-tRNA synthetase mutants are resistant to the β-lactam antibiotic mecillinam (24). Mecillinam targets penicillin-binding protein 2 (PBP2), which is required for lateral cell elongation and maintenance of rod cell shape (17, 18). Several classes of mutations lead to mecillinam resistance, and one class of mutations maps within aminoacyl-tRNA synthetase genes (e.g., alaS and argS [24]). To investigate whether novobiocin-resistant ileS mutants are also resistant to mecillinam, we tested the strains carrying mutations ileS1001 and ileS1002 by plating fully grown cultures on LA plates with mecillinam. Both ileS mutants showed increased resistance to mecillinam (Table 2). Thus, it was logical to assume that other aminoacyl-tRNA synthetase mutants are resistant to novobiocin. To test this hypothesis, we analyzed the alaS21, argS201, and leuS3(Ts) tRNA synthetase mutants, previously shown to have acquired resistance to mecillinam (11, 24). The alaS and argS alleles were introduced into strain C600 by transduction with phage P1vir grown on strains SY500 (alaS21 Tn10) and SY502 (argS201 Tn10), and the obtained constructs (strains SY501 and SY503) were tested for resistance to novobiocin. The results presented in Table 2 show that the mutants tested are resistant to novobiocin.
RelA contributes to novobiocin resistance.
Previous studies on tRNA synthetase mutants showed a causative correlation between the activity of the stringent factor, the product of the relA gene, RelA-dependent synthesis of nucleotide ppGpp (3), and resistance to mecillinam (24). To examine a possible connection between the novobiocin resistance phenotype of tRNA synthetase mutants and the stringent response, we performed a comparative analysis employing alaS21 relA1 (SY510) and argS201 relA1 (SY511) double mutants and an isogenic pair of mutants with proficient stringent response capacities (alaS21 relA+ [SY501] and argS201 relA+ [SY503]). For unknown reasons, we did not manage to introduce either of the ileS mutations into a relA1 mutant of C600 (strain SY444). The results showed an eightfold decrease in resistance to novobiocin of double mutants relative to synthetase mutants with a wild-type relA allele (Table 2). In addition, a direct correlation between RelA activity and novobiocin resistance was also demonstrated. Introduction of plasmid pSM11, which encodes a truncated, hyperactive form of RelA (15), into strain SY506 (SY702) significantly increased its resistance to novobiocin (Table 2). Resistance to novobiocin remained unchanged in the control strains SY506 and SY701, showing that increased resistance to novobiocin is due neither to F-borne material nor to the pKK223-3 vector (Table 2). We conclude that RelA activity and the consequent overproduction of ppGpp induce resistance not only to mecillinam (24) but also to novobiocin.
Correlation between the stringent response and cls gene activity.
Cardiolipin synthase (cls) mutants decrease the native resistance of cells to novobiocin (9, 14, 23). To address the question of how the activity of cls correlates with increased resistance of aminoacyl-tRNA synthetase mutants to both novobiocin and mecillinam, we introduced by P1vir transduction the inactivated cls allele (cls::mini-Tn10) into ileS80, alaS21, and argS201 recipients (strains SY468, SY504, and SY505, respectively). The results of efficiency-of-plating tests showed that inactivation of cls in aminoacyl-tRNA synthetase mutants causes a decrease in resistance to novobiocin relative to synthetase mutants containing the wild-type allele of the cls gene (Tables 2 and 3). On the other hand, all of the aminoacyl-tRNA synthetase mutants tested demonstrated unchanged, high-level resistance to mecillinam regardless of the presence of either the wild-type or mutated cls gene form (Tables 2 and 3).
TABLE 3.
Effects of the cls::mini-Tn10 mutation on resistance of ileS, alaS, and argS mutants to novobiocin and mecillinam
| Strain | Relevant genotype | Resistance (μg/ml) toa:
|
|
|---|---|---|---|
| Novobiocin | Mecillinam | ||
| C600 | cls+ | 12.5 | 0.05 |
| SY456 | cls::mini-Tn10 | 2.0 | 0.05 |
| SY513 | ileS80 cls::mini-Tn10 | 40 | >10 |
| SY504 | alaS21 cls::mini-Tn10 | 60 | >10 |
| SY505 | argS201 cls::mini-Tn10 | 50 | >10 |
| SY703 | SY508(pSM11) | 7.0 | ND |
Amounts of novobiocin were increased from 0 to 10 μg/ml by increments of 0.5 μg/ml per plate for strains SY456 and SY703 and from 0 to 70 μg/ml by increments of 10 μg/ml per plate for the other strains. Other experimental details are as described in Table 2, footnotes a and b. ND, not determined.
A correlation between the cls gene and the stringent response was further confirmed in an experiment in which the pSM11-encoded hyperactive RelA′ stringent factor was shown not to increase cellular resistance to novobiocin substantially in a cls::mini-Tn10 mutant (Table 3) as it did in its cls+ counterpart (Table 2).
Conclusions.
In this study, we lengthen the list of gyrB-unrelated genes that affect E. coli resistance to novobiocin. The relationship between inactivation of rpoN, the gene for the ς54 subunit of RNA polymerase specific for the expression of genes involved in N utilization pathways, and resistance to novobiocin remains for the moment unknown. The ileS gene, analyzed in more detail, belongs to a class of aminoacyl-tRNA synthetase mutants shown previously to be responsible for mecillinam resistance (24). Similar results obtained with other tRNA synthetase mutants (alaS, argS, leuS) give credence to the belief that resistance to novobiocin (and mecillinam) is common to all tRNA synthetase mutants.
We also showed that induction of the RelA-mediated stringent response in tRNA synthetase mutants is a common denominator not only for resistance to mecillinam (24) but also for resistance to novobiocin. Introduction of the relA1 mutation significantly decreases resistance to novobiocin in aminoacyl-tRNA mutants but does not bring it down to the resistance level of either wild-type C600 or the relA1 single mutant. This difference could be a consequence of the action of the spoT gene (3), leakiness of the relA1 mutation, or other factors and mechanisms that participate in this complex phenotype.
The data obtained with a cls null mutant suggest a relationship between deficiency in tRNA synthetases, the RelA-controlled stringent response, and resistance to novobiocin mediated by the cls gene. At this point the pathways of resistance to novobiocin and mecillinam diverge, since the cls gene is clearly not implicated in cellular resistance to mecillinam. Inactivation of cls alone decreases resistance to novobiocin in the wild-type strain, in the strain producing hyperactive RelA, and in aminoacyl-tRNA synthetase mutants. This effect, however, does not eliminate the existing differences in novobiocin resistance of these strains when they carry the cls+ allele. This implies that some other gene(s), besides cls, might be found downstream in this pathway.
One plausible model that can be drawn from the present results is that the increased resistance of synthetase mutants to novobiocin is due to the accumulation of ppGpp, which in some way controls expression of the cls gene in a positive fashion. This model is in good accord with accumulated data on the regulation of the cls gene (22). These experiments indicated that expression of cls increases as cells progress from early exponential to late exponential phase (6) and that cardiolipin synthase activity increases 10-fold in stationary phase relative to exponential phase (7). In other words, induction of the stringent response in tRNA synthetase mutants could be considered a simulation of a physiological response in stationary phase elicited by a failure of the capacity for tRNA aminoacylation to keep up with the demand for protein synthesis (3). It is tempting to speculate, in this context, that different levels of resistance to novobiocin of two leaky ileS mutants (Table 2), as well as their different growth rates, reflect their abilities to facilitate the translation process. Finally, considering that these studies do not clearly distinguish between genetic and enzymatic regulation (22), further experiments are needed to demonstrate a direct relationship between the accumulation of ppGpp and cls gene activity.
One weakness of this hypothesis is the observation that host-controlled resistance to novobiocin is also lowered by about the same factor (e.g., compare strain C600 and strain SY456 [Table 3]). E. coli sensitivity to novobiocin, as related to the cls gene, is most likely due to an alteration in membrane permeability caused by a reduction in the steady-state level of cardiolipin in the mutant (22). In another model, the increased concentration of ppGpp might have an effect on cellular resistance to novobiocin by targeting some other, as-yet-unknown gene. In this scenario, the cls gene would play only a marginal role, by affecting membrane permeability to the antibiotic.
Acknowledgments
We are very grateful to R. D’Ari, B. Magasanik, and H. Wu for kindly providing bacterial strains and plasmids and to R. D’Ari for the gift of mecillinam and for help in preparing the manuscript. This work was supported by grant MSTRS 03E12.
REFERENCES
- 1.Appleyard R K. Segregation of new lysogenic types during growth of a double lysogenic strain derived from Escherichia coli K12. Genetics. 1954;39:440–452. doi: 10.1093/genetics/39.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bullock W O, Fernandez J M, Short J M. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques. 1987;5:376–378. [Google Scholar]
- 3.Cashel M, Gentry D R, Hernandez V J, Vinella D. The stringent response. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Vol. 1. Washington, D.C: American Society for Microbiology; 1996. pp. 1458–1496. [Google Scholar]
- 4.Contreras A, Maxwell A. gyrB mutations which confer coumarin resistance also affect DNA supercoiling and ATP hydrolysis by Escherichia coli DNA gyrase. Mol Microbiol. 1992;6:1617–1624. doi: 10.1111/j.1365-2958.1992.tb00886.x. [DOI] [PubMed] [Google Scholar]
- 5.Gellert M, O’Dea M H, Itoh T, Tomizawa J. Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc Natl Acad Sci USA. 1976;73:4474–4478. doi: 10.1073/pnas.73.12.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heber S, Tropp B E. Genetic regulation of cardiolipin synthase in Escherichia coli. Biochim Biophys Acta. 1991;1129:1–12. doi: 10.1016/0167-4781(91)90206-2. [DOI] [PubMed] [Google Scholar]
- 7.Hiraoka S, Matsuzaki H, Shibuya I. Active increase in cardiolipin synthesis in the stationary phase and its physiological significance in Escherichia coli. FEBS Lett. 1993;336:221–224. doi: 10.1016/0014-5793(93)80807-7. [DOI] [PubMed] [Google Scholar]
- 8.Innis M A, Tokunaga M, Williams M E, Loranger J M, Chang S Y, Chang S, Wu H C. Nucleotide sequence of the Escherichia coli prolipoprotein signal peptidase (lsp) gene. Proc Natl Acad Sci USA. 1984;81:3708–3712. doi: 10.1073/pnas.81.12.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ivanisevic R, Milic M, Ajdic D, Rakonjac J, Savic D J. Nucleotide sequence, mutational analysis, transcriptional start site, and product analysis of nov, the gene which affects Escherichia coli K-12 resistance to the gyrase inhibitor novobiocin. J Bacteriol. 1995;177:1766–1771. doi: 10.1128/jb.177.7.1766-1771.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kredich M N. Biosynthesis of cysteine. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Vol. 1. Washington, D.C: American Society for Microbiology; 1996. pp. 514–527. [Google Scholar]
- 11.Low B, Gates F, Goldstein T, Soll D. Isolation and characterization of temperature-sensitive Escherichia coli mutants with altered leucyl- and seryl-transfer ribonucleic acid synthetases. J Bacteriol. 1971;108:742–750. doi: 10.1128/jb.108.2.742-750.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 13.Rakonjac J, Milic M, Savic D J. cysB and cysE mutants of Escherichia coli K12 show increased resistance to novobiocin. Mol Gen Genet. 1991;228:307–311. doi: 10.1007/BF00282481. [DOI] [PubMed] [Google Scholar]
- 14.Rakonjac J, Milic M, Ajdic-Predic D, Santos D, Ivanisevic R, Savic D J. nov: a new genetic locus that affects the response of Escherichia coli K-12 to novobiocin. Mol Microbiol. 1992;6:1547–1553. doi: 10.1111/j.1365-2958.1992.tb00876.x. [DOI] [PubMed] [Google Scholar]
- 15.Schreiber G, Metzger S, Aizenman E, Shmuel R, Cashel M, Glaser G. Overexpression of the relA gene in Escherichia coli. J Biol Chem. 1991;266:3760–3767. [PubMed] [Google Scholar]
- 16.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spratt B G, Pardee A B. Penicillin-binding protein and cell shape in E. coli. Nature (London) 1975;254:515–517. doi: 10.1038/254516a0. [DOI] [PubMed] [Google Scholar]
- 18.Spratt B G. The mechanism of action of mecillinam. J Antimicrob Chemother. 1977;3(Suppl. B):13–19. doi: 10.1093/jac/3.suppl_b.13. [DOI] [PubMed] [Google Scholar]
- 19.Tokunaga M, Loranger J M, Wolfe P, Wu H C. Prolipoprotein signal peptidase in Escherichia coli is distinct from M13 procoat protein signal peptidase. J Biol Chem. 1982;257:9922–9925. [PubMed] [Google Scholar]
- 20.Tokunaga M, Loranger J M, Wu H C. Isolation and characterization of an Escherichia coli clone overproducing prolipoprotein signal peptidase. J Biol Chem. 1983;258:12102–12105. [PubMed] [Google Scholar]
- 21.Tokunaga M, Loranger J M, Chang S Y, Regue M, Chang S, Wu H C. Identification of prolipoprotein signal peptidase and genomic organization of the lsp gene in Escherichia coli. J Biol Chem. 1985;260:5610–5615. [PubMed] [Google Scholar]
- 22.Tropp B E. Cardiolipin synthase from Escherichia coli. Biochim Biophys Acta. 1998;1348:192–200. doi: 10.1016/s0005-2760(97)00100-8. [DOI] [PubMed] [Google Scholar]
- 23.Tropp B E, Ragolia L, Xia W, Dowhand W, Milkman R, Rudd K E, Ivanisevic R, Savic D J. Identity of the Escherichia coli cls and nov genes. J Bacteriol. 1995;177:5155–5157. doi: 10.1128/jb.177.17.5155-5157.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vinella D, D’Ari R, Bouloc P. Penicillin binding protein 2 is dispensable in Escherichia coli when ppGpp synthesis is induced. EMBO J. 1992;11:1493–1501. doi: 10.1002/j.1460-2075.1992.tb05194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vinella D, D’Ari R. Thermoinducible filamentation in Escherichia coli due to an altered RNA polymerase β subunit is suppressed by high levels of ppGpp. J Bacteriol. 1994;176:966–972. doi: 10.1128/jb.176.4.966-972.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]