Abstract
A new Saccharomyces cerevisiae gene, XPT1, was isolated as a multicopy suppressor of a hypoxanthine phosphoribosyl transferase (HPRT) defect. Disruption of XPT1 affects xanthine utilization in vivo and results in a severe reduction of xanthine phosphoribosyl transferase (XPRT) activity while HPRT is unaffected. We conclude that XPT1 encodes XPRT in yeast.
Saccharomyces cerevisiae can use preformed purine bases from the growth medium to synthesize nucleotides. Purine bases are transported into the cell, and the corresponding monophosphate nucleotides are synthesized by phosphoribosyl transferases (Fig. 1). Mutations affecting adenine phosphoribosyl transferase (APRT) and hypoxanthine-guanine phosphoribosyl transferase (HGPRT) in S. cerevisiae have been described previously (10, 12, 13). APRT is encoded by the APT1 gene (2), while APT2, a gene encoding a putative PRT highly similar to Apt1p, has been reported (14), although no enzymatic activity could be associated with this protein (1). HGPRT is encoded by the HPT1 gene (6). An ade2 hpt1 double mutant is unable to grow on hypoxanthine as a purine source (doubling time, >20 h) but grows on adenine. We took advantage of this phenotype to seek multicopy suppressors of the ade2 hpt1 double mutant. A multicopy plasmid library (3) was introduced into the Y816 (MATα ura3-52 leu2-3,112 lys2Δ201 his3Δ200 ade2 hpt1::URA3) strain. About 104 transformants selected on synthetic complete medium (SC) without leucine and with adenine (11) were replica plated to the same medium where adenine was replaced by hypoxanthine. Twenty-four colonies growing on hypoxanthine were selected, and the corresponding multicopy plasmids were placed into three groups according to similarities in their restriction patterns. The first two groups corresponded to plasmids carrying the HPT1 or the ADE2 genes (11 and 8 plasmids, respectively). Three members of the third group were sequenced to determine the boundary of the yeast insert. The three inserts defined an overlapping region of chromosome X containing two open reading frames (ORFs). One of them, YJR133w, encodes a polypeptide 56% identical to Hpt1p. We cloned a BbuI-KpnI restriction fragment carrying the YJR133w ORF in the YEplac181 vector (5). Overexpression of this ORF alone was sufficient to restore some hypoxanthine utilization to the ade2 hpt1 double mutant (doubling time, 280 min) compared to the same strain transformed with an HPT1-containing plasmid or with the control vector (doubling times, 210 and >840 min, respectively). This new gene was called XPT1. The most conserved region (amino acids 107 to 132 in the Xpt1p sequence) corresponds to the potential PRPP binding site proposed by Hershey and Taylor (8).
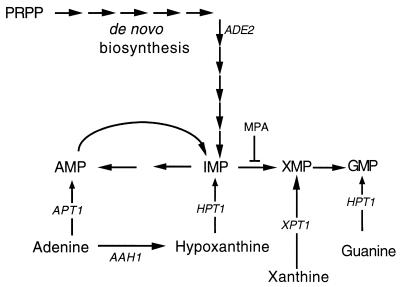
FIG. 1.
Schematic representation of purine interconversion in yeast. Abbreviations: PRPP, 5-phosphoribosyl-1-pyrophosphate; IMP, inosine 5′-monophosphate; XMP, xanthosine 5′-monophosphate; MPA, mycophenolic acid. Genes are indicated in italics and encode the following enzymatic activities: AAH1, adenine deaminase; ADE2, 5′-phosphoribosyl-5-amino-4-imidazolecarboxylate carboxy-lyase; APT1, adenine phosphoribosyl transferase; HPT1, hypoxanthine-guanine phosphoribosyl transferase; XPT1, xanthine phosphoribosyl transferase. For purposes of simplification, purine nucleosides are not represented.
To gain insight into XPT1 function, the XPT1 gene was disrupted by the method of Eberhardt and Hohmann (4). The entire XPT1 coding sequence was replaced by the HIS3 gene. An EcoRV-BbuI DNA fragment carrying the xpt1::HIS3 construct was used to transform the yeast strain Y642 (MATα ura3-52 leu2-3,112 lys2Δ201 his3Δ200 trp1::hisG). Transformants were selected for histidine prototrophy. Correct integration was verified by PCR using oligonucleotides from outside the recombination site. No obvious phenotype could be detected as a result of XPT1 disruption.
Since it has been previously shown that yeast carries a xanthine phosphoribosyl transferase (XPRT) activity that is not affected by hpt1 mutations (12), we tested whether XPT1 could encode XPRT. Both hypoxanthine phosphoribosyl transferase (HPRT) and XPRT activities from the wild-type and isogenic XPT1 disrupted strains were assayed in crude extracts using 8-3H hypoxanthine (Amersham) and 14C xanthine (Sigma) as substrates as described previously (7). HPRT activity was found to be the same in the wild type (5.28 U) and in the xpt1 strain (6.00 U), while XPRT activity in the xpt1 disrupted strain was decreased to 6% of wild-type activity (0.043 and 0.760 U, respectively). These values are the means of three independent assays, each assay being done in triplicate. For each value, the standard deviation was <12%.
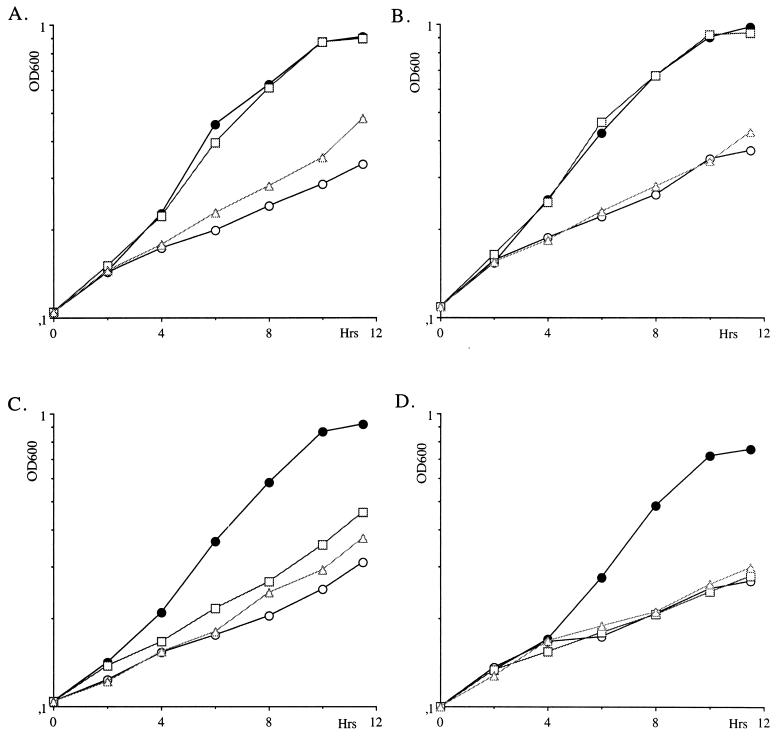
This result prompted us to evaluate whether disruption of the XPT1 gene would affect xanthine utilization in vivo. This was done by blocking XMP synthesis with mycophenolic acid (MPA) (100 mg/liter). Under these conditions, yeast cells cannot synthesize GMP so they stop growing. This phenotype can be rescued by extracellular guanine, which can be transformed to GMP by HGPRT, the product of the HPT1 gene (12). The effect of MPA can also be bypassed by extracellular xanthine if this base is transformed to XMP by XPRT. We have tested the growth capacity of hpt1 and xpt1 isogenic mutant strains in the presence of MPA supplemented with guanine or xanthine. Growth of the wild-type strain (Fig. 2A) was inhibited by MPA, and this inhibition was rescued totally by guanine and partially by xanthine, suggesting that the latter was not very efficiently transported or transformed to XMP. In the xpt1 mutant strain, inhibition by MPA was totally rescued by guanine but not at all by xanthine (Fig. 2B). Therefore, XPT1 appears necessary for xanthine utilization but does not significantly affect guanine utilization. In the hpt1 mutant (Fig. 2C), guanine could only partially rescue MPA inhibition, confirming that HPT1 encodes most of the HGPRT activity. In this mutant, xanthine was able to rescue MPA inhibition just as in the wild-type strain. This result is in good agreement with the fact that XPRT activity is unaffected in the hpt1 mutant (12). Finally, in the hpt1 xpt1 double mutant (Fig. 2D), neither guanine nor xanthine affected growth in the presence of MPA. We conclude from these experiments that XPT1 is required for xanthine utilization in yeast and for optimal utilization of guanine, at least in an hpt1 mutant strain (compare Fig. 2C and D).
FIG. 2.
Effect of mutations in the HPT1 and XPT1 genes on guanine and xanthine utilization. Growth was monitored by recording the optical density at 600 nm (OD600) as a function of time on SC (black dots), SC plus MPA (open circles), SC plus MPA plus guanine (squares), and SC plus MPA plus xanthine (triangles). Growth of the following strains was assayed: Y642 (HPT1 XPT1) (A), Y652 (HPT1 xpt1) (B), Y670 (hpt1 XPT1) (C), and Y700 (hpt1 xpt1) (D).
This study completes the identification of purine phosphoribosyl transferases in S. cerevisiae. In this organism, it therefore appears that three genes are responsible for synthesizing monophosphate nucleotides from the corresponding purine bases. HPT1 and APT1 encode HGPRT and APRT, respectively (2, 6). In this study, we identify the XPT1 gene encoding XPRT. It is noteworthy that an aspartic acid residue that has been shown to be critical for xanthine recognition by Tritricomonas foetus HGXPRT (9) is found in Xpt1p at position 192 but not in Hpt1p, where a glutamic acid is found at the equivalent position (position 194 in HGPRT). Finally, our data suggest that Xpt1p can also utilize hypoxanthine as a substrate, since overexpression of XPT1 can partially alleviate the growth defect of a mutant lacking HGPRT. Nevertheless, utilization of hypoxanthine by Xpt1p does not allow full complementation of the ade2 hpt1 growth defect.
Acknowledgments
We thank V. Denis for his critical reading of the manuscript.
This work was supported by grants from Association pour la Recherche sur le Cancer (4032), Fondation pour la Recherche Médicale, Conseil Régional d’Aquitaine and CNRS (UPR9026). M.L.G. was supported by a Ministère des Affaires Etrangères fellowship.
REFERENCES
- 1.Alfonzo J D, Crother T R, Guetsova M L, Daignan-Fornier B, Taylor M W. APT1, but not APT2, codes for a functional adenine phosphoribosyltransferase in Saccharomyces cerevisiae. J Bacteriol. 1999;181:347–352. doi: 10.1128/jb.181.1.347-352.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfonzo J D, Sahota A, Deeley M C, Ranjekar P, Taylor M W. Cloning and characterization of the adenine phosphoribosyltransferase-encoding gene (APT1) from Saccharomyces cerevisiae. Gene. 1995;161:81–85. doi: 10.1016/0378-1119(95)00239-3. [DOI] [PubMed] [Google Scholar]
- 3.Daignan-Fornier B, Reisdorf P, Nguyen C C, Lemeignan B, Bolotin-Fukuhara M. MBR1 and MBR3, two related yeast genes that can suppress the growth defect of hap2, hap3 and hap4 mutants. Mol Gen Genet. 1994;243:575–583. doi: 10.1007/BF00284206. [DOI] [PubMed] [Google Scholar]
- 4.Eberhardt I, Hohmann S. Strategy for deletion of complete open reading frames in Saccharomyces cerevisiae. Curr Genet. 1995;27:306–308. doi: 10.1007/BF00352097. [DOI] [PubMed] [Google Scholar]
- 5.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 6.Guetsova M L, Lecoq K, Daignan-Fornier B. The isolation and characterization of Saccharomyces cerevisiae mutants that constitutively express purine biosynthetic genes. Genetics. 1997;147:383–397. doi: 10.1093/genetics/147.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hershey H V, Taylor M W. Purification of adenine phosphoribosyltransferase by affinity chromatography. Prep Biochem. 1978;8:453–462. doi: 10.1080/00327487808061662. [DOI] [PubMed] [Google Scholar]
- 8.Hershey H V, Taylor M W. Nucleotide sequence and deduced amino acid sequence of Escherichia coli adenine phosphoribosyl-transferase and comparison with other analogous enzymes. Gene. 1986;43:287–293. doi: 10.1016/0378-1119(86)90218-0. [DOI] [PubMed] [Google Scholar]
- 9.Munagala N R, Wang C C. Altering the purine specificity of hypoxanthine-guanine-xanthine phosphoribosyltransferase from Tritrichomonas foetus by structure-based point mutations in the enzyme protein. Biochemistry. 1998;37:16612–16619. doi: 10.1021/bi9818974. [DOI] [PubMed] [Google Scholar]
- 10.Sahota A, Ranjekar P K, Alfonzo J, Lewin A S, Taylor M W. Mutants of Saccharomyces cerevisiae deficient in adenine phosphoribosyltransferase. Mutat Res. 1987;180:81–87. doi: 10.1016/0027-5107(87)90069-8. [DOI] [PubMed] [Google Scholar]
- 11.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 12.Woods R A, Roberts D G, Friedman T, Jolly D, Filpula D. Hypoxanthine:guanine phosphoribosyltransferase mutants in Saccharomyces cerevisiae. Mol Gen Genet. 1983;191:407–412. doi: 10.1007/BF00425755. [DOI] [PubMed] [Google Scholar]
- 13.Woods R A, Roberts D G, Sein D S, Filpula D. Adenine phosphoribosyltransferase mutants in Saccharomyces cerevisiae. J Gen Microbiol. 1984;130:2629–2637. doi: 10.1099/00221287-130-10-2629. [DOI] [PubMed] [Google Scholar]
- 14.Yuriev A, Corden J L. A Saccharomyces cerevisiae gene encoding a potential adenine phosphoribosyltransferase. Yeast. 1994;10:659–662. doi: 10.1002/yea.320100510. [DOI] [PubMed] [Google Scholar]