We reveal that the lipid droplets can emerge from ER subdomains in contact with mitochondria. The lipid transfer proteins ORP5 and ORP8 localize at these sites where they regulate the recruitment of seipin and ensure proper LD biogenesis.
Abstract
Lipid droplets (LDs) are the primary organelles of lipid storage, buffering energy fluctuations of the cell. They store neutral lipids in their core that is surrounded by a protein-decorated phospholipid monolayer. LDs arise from the endoplasmic reticulum (ER). The ER protein seipin, localizing at ER-LD junctions, controls LD nucleation and growth. However, how LD biogenesis is spatially and temporally coordinated remains elusive. Here, we show that the lipid transfer proteins ORP5 and ORP8 control LD biogenesis at mitochondria-associated ER membrane (MAM) subdomains, enriched in phosphatidic acid. We found that ORP5/8 regulates seipin recruitment to these MAM–LD contacts, and their loss impairs LD biogenesis. Importantly, the integrity of ER–mitochondria contact sites is crucial for ORP5/8 function in regulating seipin-mediated LD biogenesis. Our study uncovers an unprecedented ORP5/8 role in orchestrating LD biogenesis and maturation at MAMs and brings novel insights into the metabolic crosstalk between mitochondria, ER, and LDs at the membrane contact sites.
Introduction
Lipid droplets (LDs) are evolutionarily conserved organelles that play a primary role in regulating lipid metabolism by storing lipids in excess and releasing them upon cellular needs (Olzmann and Carvalho, 2019). They store neutral lipids in their core, mainly triacylglycerol (TG) and sterol esters, surrounded by a protein-decorated phospholipid monolayer (Thiam et al., 2013). The storage of these lipids is essential for the cells to respond to energy fluctuations. Yet, LDs are also involved in other cellular functions such as protein degradation, gene expression regulation, lipid sequestration, and membrane biosynthesis (Welte and Gould, 2017).
LDs arise from the endoplasmic reticulum (ER), following the biosynthesis and deposition of neutral lipids in the bilayer hydrophobic region (Thiam and Ikonen, 2021). The neutral lipids condense to nucleate a nascent LD, which grows and buds into a mature LD. Defects in LD biogenesis and regulation are the hallmarks of multiple metabolic and non-metabolic disorders such as type II diabetes, heart diseases, or viral infections (Gluchowski et al., 2017; Herker et al., 2021).
Seipin is a conserved ER protein forming a large oligomeric ring complex (Klug et al., 2021; Sui et al., 2018; Yan et al., 2018). It is a master regulator of LD biogenesis and plays a critical role in adipogenesis (Rao and Goodman, 2021). Mutations in seipin result in impaired lipid and calcium metabolism (Bi et al., 2014; Pagac et al., 2016) and cause lipodystrophies and neuronal disorders (Combot et al., 2022; Magre et al., 2001; Rao and Goodman, 2021; Windpassinger et al., 2004). Seipin physically marks the site of LD nucleation and mediates LD growth (Chung et al., 2019; Fei et al., 2008; Salo et al., 2019; Szymanski et al., 2007). It also regulates the physical connection of newly formed LDs to the ER at the ER–LD contact sites (Schuldiner and Bohnert, 2017) and facilitates LD growth (Choudhary et al., 2020; Grippa et al., 2015; Salo and Ikonen, 2019; Salo et al., 2019; Wang et al., 2016). In human cells, the oligomeric seipin ring consists of 11 subunits that, in vitro, bind negatively charged phospholipids, including phosphatidic acid (PA; Yan et al., 2018). In the absence of seipin, PA levels likely increase in the ER membrane (Fei et al., 2011; Han et al., 2015; Wolinski et al., 2015). Interestingly, the deletion of seipin also leads to PA accumulation in the inner nuclear envelope and the formation of nuclear LDs (Sołtysik et al., 2021). These observations support the hypothesis that seipin is recruited to ER subdomains, probably enriched in negatively charged lipids (especially PA), where it could form a scaffold that assists LD assembly.
The existence of ER subdomains promoting LD biogenesis is supported by several observations (Choudhary et al., 2020; Hariri et al., 2018; Joshi et al., 2018; Santinho et al., 2020; Wang et al., 2018). LD biogenesis can occur at the ER contact sites with vacuoles in yeast (Hariri et al., 2018) or peroxisomes, both in yeast and mammals (Joshi et al., 2018, 2021). These observations support that contact sites established between ER and other organelles could be important for LD formation. For example, such contact sites may pool key enzymes and lipid intermediates necessary for LD assembly (Choudhary et al., 2020) or locally preset optimal physical properties for the neutral lipids condensation into LDs. However, whether LD can originate at other inter-organelle contact sites is unknown.
Perturbations of the lipid composition in ER are detrimental to proper LD formation (Adeyo et al., 2011, Ben M’barek et al., 2017; Fei et al., 2011; Zoni et al., 2021), suggesting that locally editing the ER phospholipid composition, possibly by lipid transfer at specific ER contact sites, could be essential for proper LD biogenesis (Zouiouich et al., 2022). However, the existence of such a mechanism is currently unknown. Also, it is still elusive whether and how lipids are directly transferred from the ER to the LDs during and after LD formation. Therefore, mechanisms that spatially and temporally regulate LD biogenesis in the cell remain poorly understood.
The oxysterol binding protein (OSBP)-related proteins constitute a large family of lipid transfer proteins (LTPs) conserved from yeast (Osh) to humans (ORP) and localized to different subcellular sites, shown in several cases to be membrane contact sites. A common feature of all ORPs is the presence of an OSBP-related lipid-binding/transfer (ORD) domain. Most ORP proteins contain two phenylalanines (FF) in an acidic tract (FFAT)-motif that binds ER-localized VAP proteins and a pleckstrin homology (PH) domain that interacts with lipids or proteins in distinct non-ER organelle membranes. Two members of this family, ORP5 and ORP8, do not contain an FFAT motif but are directly anchored to the ER through a C-terminal transmembrane (TM) segment (Du et al., 2011).
ORP5 and ORP8 have been originally shown to localize at ER–PM contact sites where they transfer phosphatidylserine (PS) from the cortical ER to the PM, in counterexchange with the phosphoinositides phosphatidylinositol-4-phosphate (PtdIns[4]P) and phosphatidylinositol 4,5-bisphosphate (PtdIns[4,5]P; Chung et al., 2015; Ghai et al., 2017). Lately, ORP5 has been shown to also localize at ER-LD contact sites upon oleic acid (OA) treatment (Du et al., 2020) and, although an experimental demonstration is still missing, it has been proposed that, at these sites, ORP5 could also act as PS/PtdIns(4)P lipid exchanger.
We recently showed that ORP5 and ORP8 are also localized, and even enriched, at specific ER subdomains in contact with mitochondria, the so-called mitochondria-associated membranes (MAMs), where they mediate the transfer of PS and maintain mitochondrial morphology and functions(Galmes et al., 2016; Monteiro-Cardoso et al., 2022). We hypothesized that MAMs containing ORP5 and ORP8 are involved in LD biogenesis.
Here, we show that ORP5 and ORP8 localize at MAM subdomains where LDs originate and that are enriched in PA phospholipid. Oleic acid treatment leads to massive recruitment of ORP5-labeled MAMs to nascent and pre-existent LDs, suggesting a dual role in LD biogenesis and in their maintenance/turnover. We also reveal that ORP5 and ORP8 are novel players involved in LD biogenesis by regulating seipin recruitment to MAM–LD contacts.
Results
ORP5 localizes at MAM subdomains closely associated with LDs
ORP5 and ORP8 have been shown to localize and primarily interact at MAMs (Galmes et al., 2016; Monteiro-Cardoso et al., 2022). Lately, a novel localization at ER–LD contact sites upon OA treatment has been described for the two ORP5 isoforms (ORP5A and ORP5B) but not for ORP8 (Du et al., 2020). However, ORP5 distribution at the multiple ER-mediated contact sites, especially upon OA, remains controversial. Also, whether ORP8 could also localize at ER–LD contact sites is unknown.
Thus, we first investigated the localization of EGFP-tagged ORP5A, ORP5B (a natural variant of ORP5, lacking a large part of the PH domain), and ORP5ΔPH (a variant completely lacking its PH domain) by confocal microscopy in HeLa cells treated for 2 h with OA or untreated (Fig. 1, A and B).
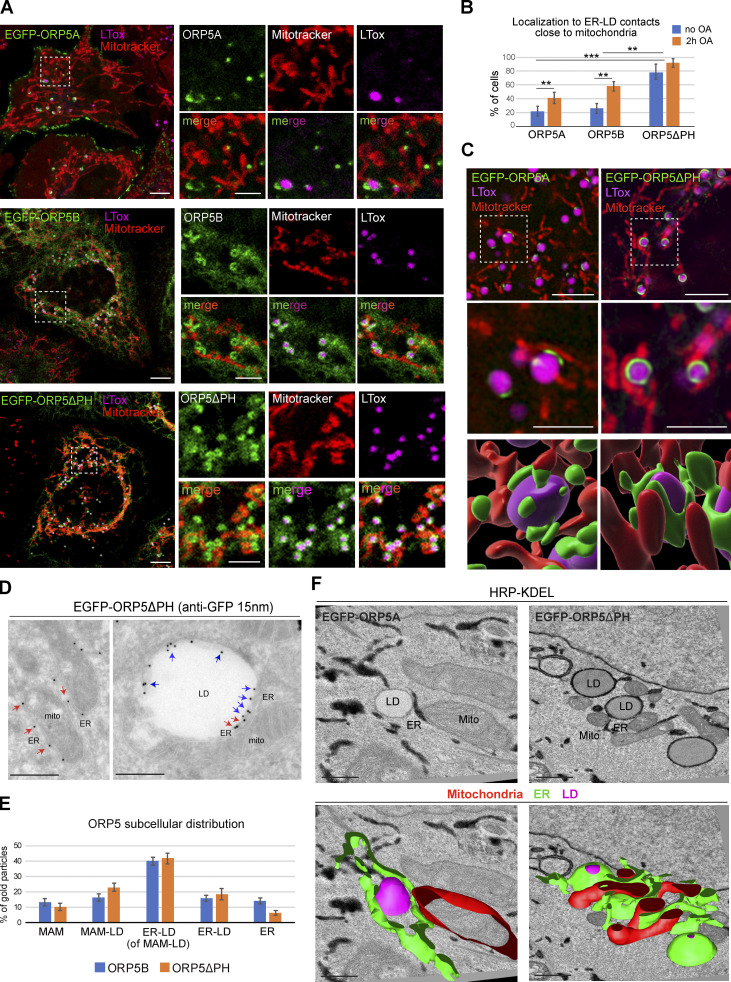
Figure 1.
ORP5 localizes to MAM subdomains in contact with LD. (A) Representative confocal images showing single focal planes of HeLa cells expressing EGFP-ORP5A, EGFP-ORP5B, or EGFP-ORP5ΔPH (green) and treated with Mitotracker (red) and LTox Deep Red (LTox, purple) to label mitochondria and lipid droplets (LDs), respectively. Scale bar, 10 μm (entire cell), or 5 μm (zoom). (B) Quantification of the % of HeLa cells showing localization of EGFP-tagged ORP5A, ORP5B, and ORP5ΔPH to ER-LD contacts close to mitochondria in the absence of oleic acid (OA) or after 2 h of 300 μM OA loading. Data represent the mean ± standard error of the mean (SEM) of n = 25 cells. **P < 0.01, ***P < 0.001, unpaired two-tailed t test. (C) SIM micrographs of HeLa cells expressing EGFP-ORP5B or EGFP-ORP5ΔPH (green), treated with OA for 2 h, and stained with Mitotracker (red) and LTox Deep Red (purple). 3D-SIM images were obtained by segmentation using Software Imaris (v 9.3, Bitplaine). Scale bar, 5 μm (entire cell), or 2.5 μm (zoom). (D) Electron micrograph of ultrathin cryosections of HeLa cells transfected with EGFP-ORP5ΔPH, treated with OA for 2 h, and immunogold stained with anti-EGFP (15 nm gold). Left: red arrows indicate EGFP-ORP5ΔPH localized to MAM. Right: blue arrows indicate ORP5ΔPH localized to ER–LD contacts and red arrows indicate ORP5ΔPH localized to MAM–LD contacts. Scale bar, 500 nm. (E) Quantification of the distribution of EGFP-ORP5B and EGFP-ORP5ΔPH immunogold particles (15 nm) to the different ER compartments: MAM, MAM–LDs, ER–LDs (associated to MAM–LDs), ER–LDs (isolated) and reticular ER, after 2 h of OA. Data are shown as % of mean ± SEM of cell profiles with n = 20 (360 and 470 gold particles analyzed in EGFP-ORP5B and EGFP-ORP5ΔPH overexpression, respectively). (F) Representative electron micrographs of HeLa cells co-overexpressing EGFP-ORP5A or EGFP-ORP5ΔPH and HRP-KDEL after 2 h OA treatment, and 3D reconstruction of 27 serial sections by 3Dmod. LD, lipid droplet; Mito, mitochondria; ER, endoplasmic reticulum. Scale bar 500 nm.
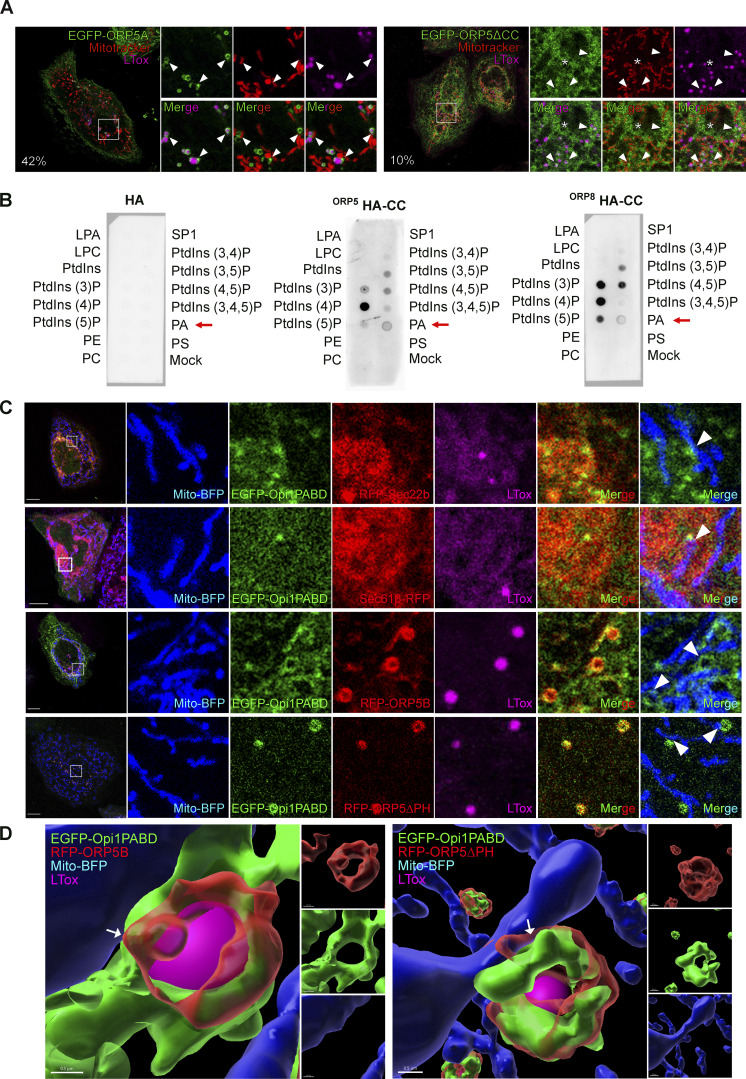
EGFP–ORP5A was detected at cortical ER in all cells analyzed (OA treated and untreated), while only a subset of cells displayed ORP5A localization at ER–LD contacts (20% for untreated and 40% for OA-treated cells). Interestingly, the ORP5A-labeled ER in contact with LDs appeared to be highly expanded as it almost completely surrounded the LD surface. EGFP-ORP5B was instead detected at reticular ER in all cells analyzed (due to the loss of PM binding), at expanded ER-LD contact sites in 22% of untreated cells, and in about 60% of OA-treated cells. Similarly, ORP5ΔPH was found at ER-LDs contact sites. However, the number of cells showing ORP5-labeled ER in contact with LD was considerably higher in the pool of cells transfected with ORP5ΔPH (80% for untreated and 96% for OA-treated cells) than in the pool of cells transfected with EGFP-ORP5B (Fig. 1 B). Interestingly, almost all the ER subdomains in contact with LDs where ORP5A, ORP5B, or ORP5ΔPH localized were found closely associated with mitochondria (Fig. 1, A and B). Super-resolution Structured Illuminated Microscopy (SIM) 3D analysis of EGFP-ORP5A and EGFP-ORP5ΔPH localization in HeLa cells treated for 2 h with OA confirmed the existence of this novel tripartite contact site association between ER, LDs, and mitochondria, which was induced by ORP5 overexpression (Fig. 1 C), and that from hereon we will define as “MAM–LD” contact sites.
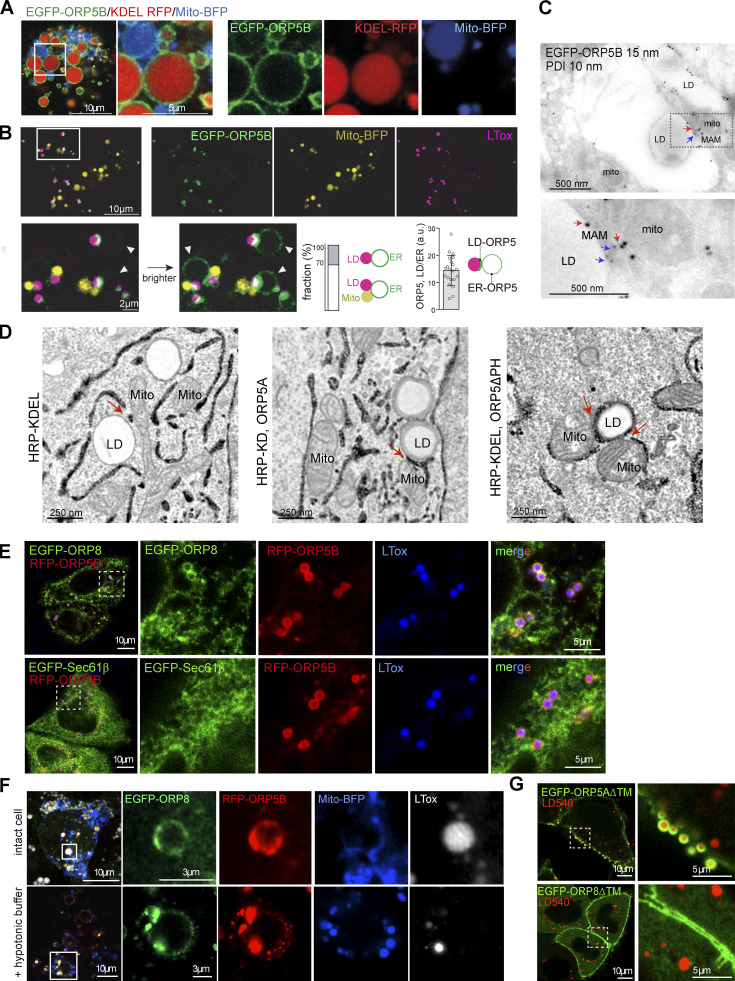
To better visualize and quantify the three organelles’ association, we proceeded with cell swelling. Cells were subjected to a hypotonic medium, which swells the bilayer-surrounded organelles only and preserves their contacts (King et al., 2020; Santinho et al., 2020). As expected, ORP5 (ORP5B) localized to KDEL-positive ER vesicles including a large pool of MAM (Fig. S1, A and B). When the cells were treated with OA for 2 h before swelling, we found that 70% of LDs formed were in close association with mitochondria and ER vesicles; the remaining 30% were in contact with ER only (Fig. S1 B). This observation confirmed the development of MAM–LD contact sites upon OA addition. Interestingly, the pool of ORP5B at the MAM–LD contacts was 15 times higher than in the ER regions, indicating that OA addition massively focalized ORP5B from the reticular ER to MAM subdomains in contact with LDs (Fig. S1 B).
Figure S1.
ORP5 localizes to MAM subdomains in contact with LD. (A) Airyscan live imaging of swollen Huh7 cells co-expressing EGFP-ORP5B (green), Mito-BFP (blue) and KDEL-RFP (red). Scale bar, 10 μm (entire cell), or 5 μm (zoom). (B) Live imaging of EGFP-ORP5B (green), Mito-BFP (yellow) and LDs (purple) 10 min after swelling Huh7 cells. Zoom area is shown below, normal and enhanced EGFP-ORP5 signal to visualize its ER membrane signal (indicated by arrowheads). The bar graphs, from left to right, show (left) the percentage of ER–LD vs. ER–LD contacts events (n = 100) and (right) the signal ratio of EGFP-ORP5 intensity at the ER–LD contacts to the ER membrane (n = 20), as depicted by the illustrating image. Scale bar, 10 μm (entire cell), or 2 μm (zoom). (C) Electron micrograph of ultrathin cryosections of HeLa cells transfected with EGFP-ORP5B treated with OA for 2 h, and immunogold stained with anti-EGFP (15 nm gold) and anti PDI (ER marker). Red arrows indicate ORP5 gold particles localized to MAM-LD contacts. Blue arrows indicate PDI-labeled ER. Scale bar, 500 nm. (D) Electron micrographs of HeLa cells overexpressing HRP-KDEL alone or together with either EGFP-ORP5A (wt) or EGFP-ORP5ΔPH after OA treatment (300 μM for 2 h). Scale bar, 250 nm. (E) Representative confocal images (single focal plane) of HeLa cells co-expressing either EGFP-ORP8 or RFP-Sec61β (green) and RFP-ORP5B (red) and stained with LTox (blue). Scale bar 10 μm (entire cell), or 5 μm (zoom). (F) Representative confocal image (single plane) of control (intact cells) and swollen (+ hypotonic buffer) Huh7 cells co-expressing EGF-ORP8, RFP-ORP5 and Mito-BFP, and treated with LTox (white) to label LDs. (G) Confocal images (single focal plane) of HeLa cells expressing EGFP-ORP5A∆TM or EGFP-ORP8∆TM (green) and treated with LD450. Scale bar, 10 μm (entire cell), or 5 μm (zoom).
Moreover, the localization of ORP5 at the expanded MAM–LD contact sites was confirmed by immunogold labeling on ultrathin cryosections of cells expressing ORP5B or ORP5ΔPH and treated with OA (Fig. 1, D and E; and Fig. S1 C). The advantage of using ORP5B and ORP5ΔPH over ORP5A relies on the higher number of cells showing their localization at MAM–LD contact sites (see Fig. 1 B), facilitating the immuno-EM analysis (Fig. 1, D and E). The bulk of ORP5B and ORP5ΔPH localized to MAM–LD contact sites and the associated ER–LD contacts (56 and 65%, respectively), while a minor pool was detected at MAMs not associated with LDs (14 and 10%, respectively) and in the reticular ER (14 and 6%, respectively; Fig. 1, D and E).
To study the morphology of these three-way MAM–LD associations and address whether they could exist also at physiological ORP5 expression levels, we performed an ultrastructural analysis by HRP-KDEL EM (carrying a horseradish peroxidase [HRP] tagged with an ER retention motif to stain the ER) in cells expressing HRP-KDEL alone or together with EGFP-ORP5A or EGFP-ORP5ΔPH (Fig. 1 F and Fig. S1 D). EM analysis confirmed that the MAM–LD contacts observed by light microscopy upon ORP5 overexpression were indeed membrane contact sites (in the range of 10–30 nm). Interestingly, MAM–LD contacts were also detected in cells expressing HRP-KDEL alone, revealing their existence even when ORP5 is not overexpressed. However, the ER involved in these contact sites was not highly-expanded around the LDs (Fig. S1 D), possibly explaining why these associations have been overlooked so far. Agreeing with our confocal data, ORP5A, and ORP5ΔPH to a greater extent, induced an expansion of the ER on the LD surface. 3D reconstruction of EM serial sections revealed the tridimensional organization of these MAM–LD contacts (Fig. 1 F).
Together, these data reveal the existence of a novel tripartite mitochondria–ER–LD contact sites junction that is regulated by ORP5 and where ORP5 also localizes. However, it is not known whether ORP8 similarly localizes at MAM–LD contact sites.
ORP8 is enriched at MAM–LD via interactions with ORP5 by its coiled-coil domain
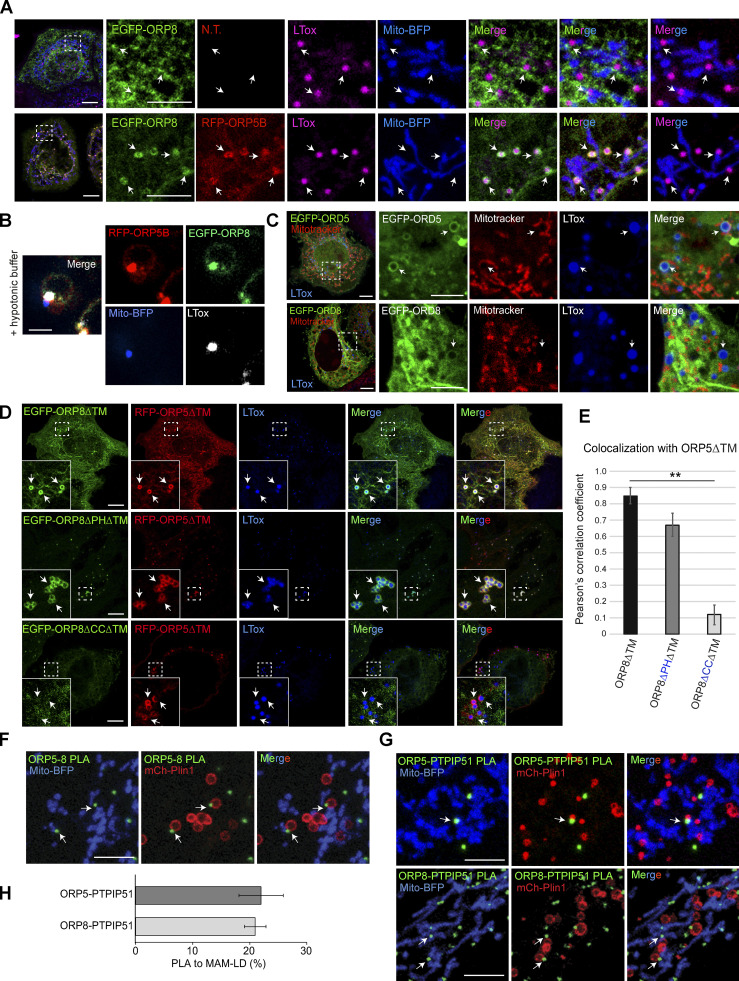
We overexpressed EGFP-ORP8 and analyzed its localization by confocal microscopy in HeLa cells treated with OA (Fig. 2 A). In contrast to ORP5, ORP8 is enriched to the reticular ER, with a minor pool additionally present at cortical ER and MAMs, as previously shown (Galmes et al., 2016). A small pool of ORP8 was detected at MAM–LD associations, but the ER elements labeled with ORP8 were not expanded around the LD surface as seen for ORP5.
Figure 2.
ORP8 localizes and interacts with ORP5 at MAM-LD contacts. (A) Representative confocal images showing localization of EGFP-ORP8 alone (green) or together with RFP-ORP5B (red) in HeLa cells co-expressing Mito-BFP (blue) and treated with OA for 2 h and stained with LTox Deep Red (purple). Each image represents a single focal plane of confocal 3D stacks. Arrows point to ORP5-labeled MAM associated with mitochondria and LD (MAM-LD contacts). Scale bar, 10 μm (entire cell) or 5 μm (zoom). (B) Zoomed confocal images showing the co-localization of ORP5, ORP8, LD, and mitochondria in swollen Huh7 cells expressing RFP-ORP5B, EGFP-ORP8, and Mito-BFP. Scale bar, 3 μm. (C) Confocal images showing the localization of EGFP-ORD5 and EGFP-ORD8 (green) in HeLa cells treated with OA for 2 h and stained with Mitotracker (red) and LTox Deep Red (blue). Each image represents a single focal plane. Scale bar, 10 μm (entire cell), or 5 μm (zoom). (D) HeLa cells co-expressing EGFP-ORP8ΔTM (green) and RFP-ORP5ΔTM (red), EGFP-ORP8ΔPHΔTM (green) and RFP-ORP5ΔTM (red), or EGFP-ORP8ΔCCΔTM (green) and RFP-ORP5ΔTM (red). Each image represents a single focal plane of confocal 3D stacks. Arrows point to ORP5 localization on the LD surface. Scale bar, 10 μm. (E) Quantitative analysis of the co-localization of either EGFP-ORP8ΔTM, EGFP-ORP8ΔPHΔTM or EGFP-ORP8ΔCCΔTM with RFP-ORP5ΔTM by Pearson’s correlation coefficient. Data represent mean ± SEM of n = 10 cells. **P < 0.001, unpaired student’s t test. (F and G) Confocal micrographs showing endogenous ORP5-ORP8, ORP5-PTPIP51, and ORP8-PTPIP51 PLA interactions (green dots) in regions of HeLa cells co-expressing Mito-BFP (blue) and mCherry-Plin1 (mCh-Plin1) and treated with 300 μM OA for 2 h. Arrows point to PLA dots associated to MAM–LD contacts. Images represents a single focal plane. Scale bar, 10 μm. (H) Quantification of endogenous ORP5-PTPIP51 and ORP8-PTPIP51 PLA interaction at MAM–LD contacts. Data is shown as % mean ± SEM of n = 36 cells (ORP5-PTPIP51) and n = 15 cells (ORP8-PTPIP51).
ORP5 and ORP8 physically interact (Chung et al., 2015; Galmes et al., 2016; Ghai et al., 2017; Monteiro-Cardoso et al., 2022). We thus reasoned that the faint signal of ORP8 detected at the MAM–LD associations when the protein is overexpressed alone could be due to the limiting pool of endogenous ORP5. We then co-overexpressed EGFP-ORP8 with RFP-ORP5B. Under this condition, ORP8 dramatically redistributes from the reticular and cortical ER to the ORP5-positive MAM–LD contact sites (Fig. 2 A). This redistribution and enrichment at MAM–LD contacts were not observed in the case of a general ER protein such as Sec61β, which, even when co-expressed with ORP5, maintained its prevalent localization to the reticular ER (Fig. S1 E). These findings confirmed the specificity of ORP8 localization at MAM–LD contacts. ORP8 and ORP5 co-localization at MAM–LD contacts was further confirmed in intact and swollen Huh7 cells co-expressing EGFP–ORP8 with RFP–ORP5B by high-resolution Airyscan confocal microscopy (Fig. 2 B and Fig. S1 F).
Altogether, these data indicate that ORP5 has a higher affinity for MAM–LD contacts than ORP8 and that ORP8 enrichment at such sites depends on its interaction with and on the expression level of ORP5.
ORP5 localization to ER–LD contacts has been shown to depend on its ORD domain (Du et al., 2020). However, whether also the ORD domain of ORP8 is able to bind LDs is still unknown. Also, the abilities of ORP5 and ORP8 ORD domains to bind LDs have not been compared yet. We thus expressed EGFP-tagged ORP5 and ORP8 ORDs in HeLa cells and found that the ORD domain of ORP5 (EGFP-ORD5) strongly binds LDs, whereas the ORD domain of ORP8 (EGFP-ORD8) only weakly binds LDs (arrows, Fig. 2 C). These experiments revealed that both ORD5 and ORD8 can bind LDs, but the binding ability of ORD8 is lower than that of ORD5. These data might explain why ORP8 overexpression does not induce an expansion of ER around the LDs and also suggest that ORP8 requires ORP5 to be enriched at the expanded MAM–LD contact sites.
To more directly test whether ORP8 binding to LDs requires ORP5 and identify the domains involved, we co-expressed an ORP5 deletion mutant lacking the ER-anchoring TM domain (RFP-ORP5ΔTM) with an ORP8 construct carrying a similar mutation (EGFP-ORP8ΔTM) or with ORP8 lacking both the TM and the PH or the coiled-coil (CC) domains (EGFP-ORP8-ΔPHΔTM or EGFP-ORP8-ΔCCΔTM) and analyzed their recruitment to LDs in HeLa cells by confocal microscopy (Fig. 2 D). The EGFP-ORP5ΔTM, expressed alone, localized to both the PM and LDs, whereas the EGFP-ORP8ΔTM mostly localized to the cytosol, to the PM, and weakly to LDs (Fig. S1 G). However, when co-expressed with RFP-ORP5ΔTM, EGFP-ORP8ΔTM strongly co-localizes with ORP5, as assessed by the Pearson’s correlation coefficient analysis, and enriches at the ORP5-labeled LDs (Fig. 2, D and E). Of note, ORP5ΔTM and ORP8ΔTM, when co-expressed, lose their PM localization and strongly redistribute to LDs, and also to cytosolic filamentous structures that resemble cytoskeleton.
The concomitant deletion of the PH domain in ORP8ΔTM does not induce significant changes in ORP8 colocalization with ORP5 at LDs. However, the deletion of the CC domain in ORP8ΔTM completely abolished ORP8 targeting the ORP5-labeled LDs (Fig. 2, D and E). These results confirm that interaction with ORP5 via the CC domain is required for the ORP8 binding to LDs.
Finally, we determined whether ORP5 and ORP8 interaction and localization at MAM–LD contacts also occur at endogenous levels. We performed proximity ligation assay (PLA) by Duolink in HeLa cells transfected with Mito-BFP to stain the mitochondria and with mCherry-Perilipin 1 (Plin1) to stain the LDs, and then analyzed by confocal microscopy. ORP5–ORP8 PLA spots were detected in the proximity of a subset of mitochondria-associated LDs (arrows, Fig. 2 F). To confirm that these sites corresponded to MAMs, we performed PLA using antibodies against either ORP5 or ORP8 and their mitochondrial binding partner PTPIP51 (Galmes et al., 2016). A similar pool of ORP8-PTPIP51 or ORP5-PTPIP51 PLA spots was observed at MAM–LD contacts (arrows, Fig. 2, G and H), confirming that ORP5-8 localize and interact at the tripartite MAM–LD contact sites in endogenous conditions.
ORP5 and ORP8 organize LD biogenesis at MAM
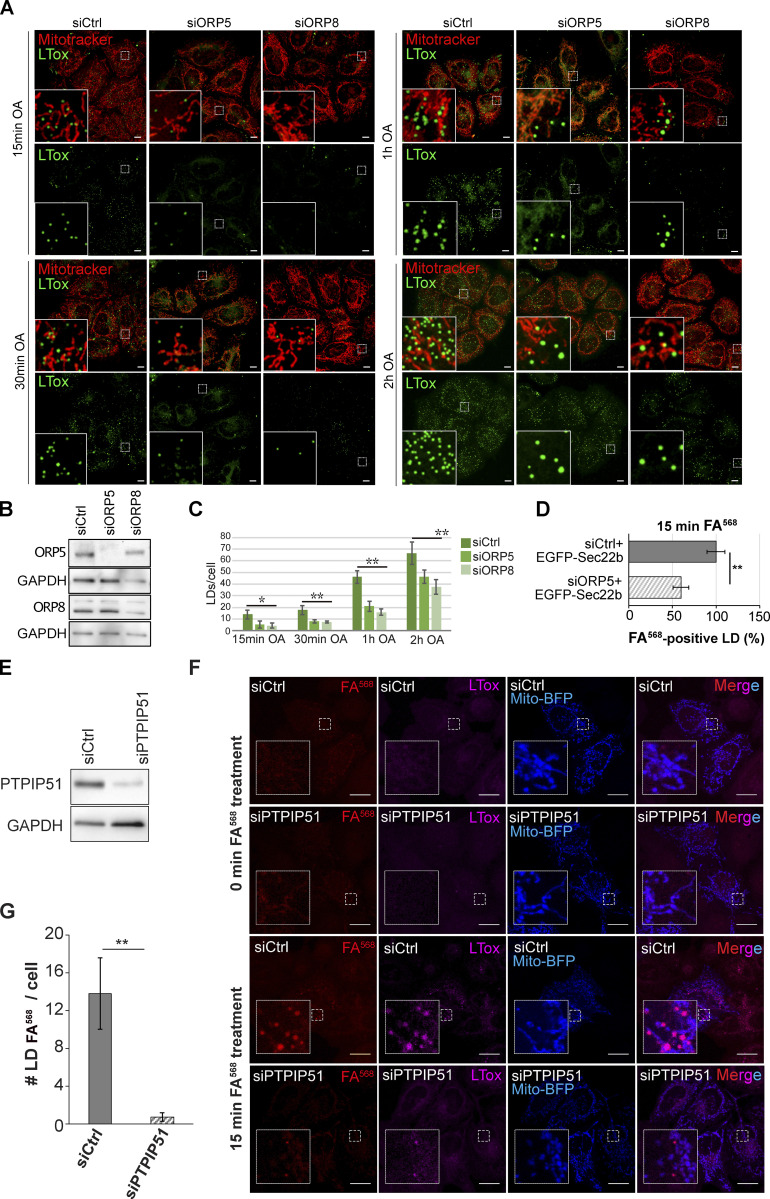
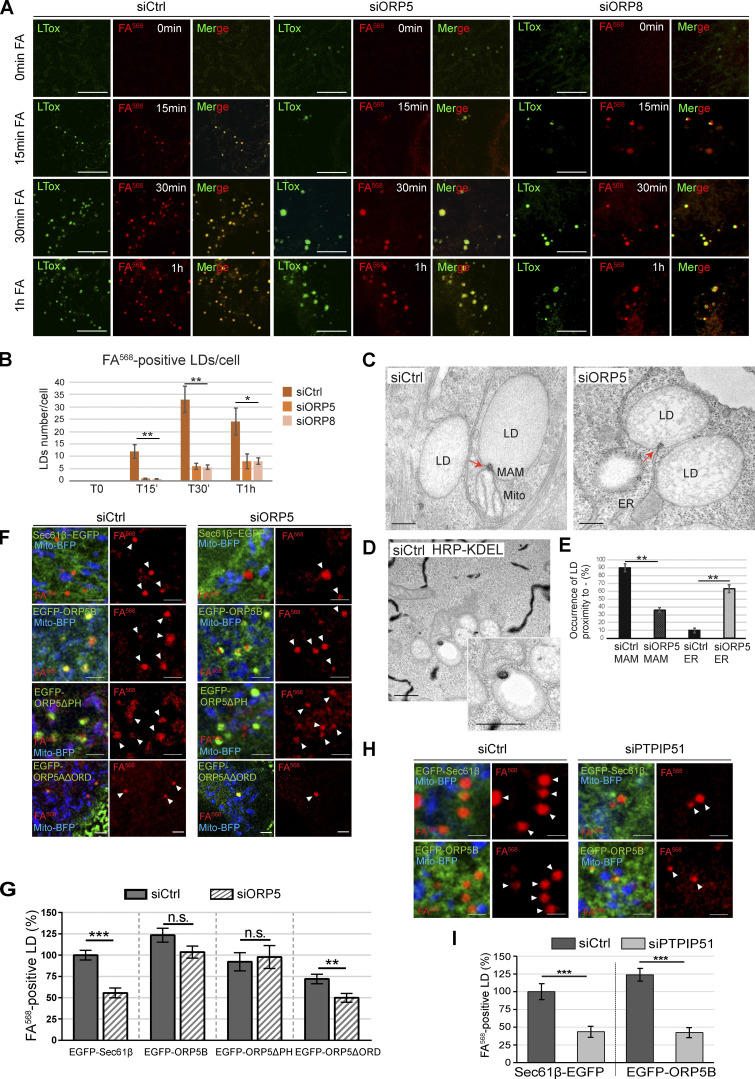
To assess the role of ORP5 and ORP8 in LD biogenesis, we depleted ORP5 and ORP8 by RNAi in HeLa cells that have been delipidated for 3 d to remove pre-existent LDs. The efficiency of the knockdown (KD) was validated by Western blot (WB; Fig. S2, A and B). LD biogenesis was induced by OA treatment, and cells were imaged by confocal microscopy at different times (15 min, 30 min, 1 h, 2 h; Fig. S2 A). The abundance of LDs, stained by LTox, in both ORP5 and ORP8 KD cells was significantly reduced at all time points, as compared with control (Ctrl) cells. The decrease in LD number was greater at earlier times (70% at 15 min, 60–65% at 30 min and 1 h, 30–40% at 2 h), suggesting a delay in LD biogenesis (Fig. S2, A and C). Additionally, after 30 min of OA loading, some bigger LD started to be observed in both ORP5 and ORP8 KD cells, and their occurrence increased with time and became more noticeable at 2 h. This finding is consistent with the presence of larger LDs upon ORP5 depletion following prolonged OA loading (Du et al., 2020).
Figure S2.
ORP5, ORP8 and PTPIP51 depletion impairs LD formation in HeLa cells. (A) LD biogenesis time-course. Confocal (single focal plane) images of control (siCtrl), and ORP5 (siORP5) or ORP8 (siORP8) siRNA-treated HeLa cells, delipidated for 72 h, and incubated with OA (300 μM) for 15 min, 30 min, 1 and 2 h. Cells were also stained with Mitotracker (red) and LTox (green). Scale bar, 10 μm. (B) WB analysis showing ORP5, ORP8 and GAPDH levels in protein lysates from Ctrl, ORP5 and ORP8 knockdown HeLa cells. (C) Quantification of the number of LTox-positive LDs in siCtrl, siORP5, or siORP8 cells in the indicated times after OA delivery. Data are shown as mean ± SEM of n = 30 cells. *P < 0.01, **P < 0.0001, unpaired two-tailed t test. (D) Analysis of FA568-positive LD in siCtrl and siORP5 HeLa cells, priorly delipidated for 72 h, and then co-transfected with Mito-BFP and EGFP-Sec22b. Data are show as % of siCtrl treated HeLa cells. n = 27 siCtrl and n = 24 siORP5. Bar indicated SEM. **P < 0.001, unpaired two-tailed t test. (E) Western blot analysis of the expression of PTPIP51 in siCtrl and siPTPIP51 HeLa cells, showing the efficiency of PTPIP51 knockdown. (F) Confocal (single focal plane) images of control (siCtrl) and PTPIP51 (siPTPIP51) siRNA-treated HeLa cells, delipidated for 72 h, and transfected with Mito-BFP (blue). Cells were treated with FA568 (1 μM) for 15 min, and stained with LTox (purple). Scale bar, 10 μm. (G) Analysis of the number of FA568-positive LDs in control and PTPIP51 knockdown cells. Data are show as mean ± SEM of n = 15 in siCtrl and n = 16 in siPTPIP51 cells. ***P < 0.001, unpaired two-tailed t test. Source data are available for this figure: SourceData FS2.
Since LTox may have marked pre-existing LDs that could have resisted delipidation, we performed parallel experiments by feeding cells with a fluorescent C12-fatty acid (referred to as FA568, associated with red fluorescence), as in Khaldoun et al. (2014). This option enabled us to track efficiently the biogenesis and the maturation of newly synthesized LDs. The efficiency of delipidation was confirmed by the almost complete disappearance of LTox-positive LDs in Ctrl, ORP5, and ORP8 KD cells at time = 0 min (Fig. 3, A and B). The FA568 treatment induced the formation of new LDs that were also labeled by LTox. The number of the FA568–containing LDs was dramatically reduced at all time points (86% at 15 min, 92% at 30 min, and 71% at 1 h) in ORP5 and ORP8 KD as compared with Ctrl cells. The decrease in newly-formed FA568–positive LDs was higher than the decrease in LTox-positive LDs, which likely included the pre-existent ones (Fig. 3 B and Fig. S2 C). Altogether, these results indicate the role of ORP5 and ORP8 in mediating proper LD biogenesis.
Figure 3.
Depletion of ORP5 and ORP8 affect LD biogenesis. (A) LD biogenesis time-course. HeLa cells delipidated for 72 h were treated with siCtrl, siORP5, or siORP8, incubated with 1 μM FA568 (red) and stained with LTox Deep Red (green). Representative confocal images of regions of HeLa cells submitted to these experimental conditions at time 0 min, 15 min, 30 min and 1 h of FA568 incubation are displayed as a single focal plane. Scale bar, 5 μm. (B) Quantification of the number of FA568-positive LD in control, ORP5 and ORP8 knockdown HeLa cells at the indicated times. Data represent mean ± SEM of n = 30 cells. *P < 0.001, **P < 0.0001, unpaired two-tailed t test. (C) Representative electron micrographs of control and ORP5 knockdown HeLa cells, evidencing (red arrows) the electrondense structure that connects the nascent LD to the ER from which it originated and sometimes also to the mitochondria (Mito) at MAM–LD contacts. Scale bar, 250 nm. (D) Representative electron micrograph of HeLa cells expressing HRP-KDEL (black), showing that the electrondense structure shown on Fig. 3 C is MAM. Scale bar 500 nm. (E) Quantification of the number of LD associated with these ER or MAM electrondense structures. Data represent mean ± SEM of n = 20 cells. **P < 0.0001, unpaired two-tailed t test. (F) Confocal (single focal plane) micrographs of regions of control and ORP5 knockdown delipidated HeLa cells co-overexpressing Mito-BFP (blue) with Sec61β-EGFP (green), EGFP-ORP5B (green), EGFP-ORP5ΔPH (green), or EGFP-ORP5AΔORD (green). Arrowheads indicate the newly formed LD. Scale bar, 2 μm. (G) Quantitative analysis of the number of FA568-positive LD in control and ORP5 knockdown delipidated HeLa cells co-overexpressing Mito-BFP and Sec61β-EGFP, siRNA-resistant EGFP-ORP5B or EGFP-ORP5ΔPH, or EGFP-ORP5AΔORD, and treated for 15 or 30 min with FA568. Data are shown as % of mean ± SEM of n = 20–85 cells. ***P < 0.001, unpaired two-tailed t test. (H) Confocal (single focal plane) micrographs of regions of control and PTPIP51 knockdown delipidated HeLa cells, co-overexpressing Mito-BFP (blue) with Sec61β-EGFP (green) or EGFP-ORP5B (green) and treated for 1 h with FA568. Arrowheads indicate the newly formed LD. Scale bar, 1 μm. (I) Quantification of the number of FA568-positive LD in control and PTPIP51 knockdown delipidated HeLa cells co-overexpressing Mito-BFP and Sec61β-EGFP, or EGFP-ORP5B and treated for 1 h with FA568. Data are shown as % of mean ± SEM of n = 20–22 cells. ***P < 0.001, unpaired two-tailed ttest.
To analyze the morphology of nascent LDs at MAMs, we performed EM analysis on control and ORP5 KD cells treated with OA. Importantly, we detected specific ER subdomains that appeared as peculiar electrondense structures associated with some LDs connected to tubular ER elements, and that likely correspond to the sites where these LD emerged from the ER (Fig. 3, C and D). These structures were found very often in contact with mitochondria in control cells and HRP-KDEL EM revealed that they indeed correspond to MAM (Fig. 3, C and D). Quantifications of these EM observations revealed that ORP5 KD induces a strong decrease in the occurrence of these MAM-emerged LDs, while the ER-emerged LD connections, not in close contact with mitochondria, were instead increased (Fig. 3 E). These data strongly suggest that ORP5 regulates LD formation from the MAM subdomains and also reveal for the first time the morphology of the MAM subdomains from which LD originates.
We then tested if re-expression of ORP5 could rescue the LD phenotype of the ORP5 KD cells. For this purpose, we re-expressed siRNA-resistant EGFP-ORP5B or EGFP-ORP5ΔPH or, as a control, the ER marker protein EGFP-Sec61β (Fig. 3, F and G) or EGFP-Sec22b (Fig. S2 D) in the delipidated ORP5 KD cells. To monitor the rescue of the LD phenotype, we performed FA568-mediated induction of LD biogenesis, as described above, and analyzed by confocal microscopy the number of FA568–positive LDs 15 min after its delivery. Remarkably, both EGFP-ORP5B and EGFP-ORP5ΔPH constructs significantly rescued the LD decrease in siORP5 cells, while EGFP-Sec61β or EGFP-Sec22b were not able to rescue the LD phenotype (Fig. 3, F and G; and Fig. S2 D). To test whether ORP5 function on LD biogenesis could depend on its lipid transfer ORD domain, we conducted the rescue experiment by overexpressing an EGFP-ORP5ΔORD construct in the ORP5 depleted cells. Overexpression of ORP5ΔORD did not rescue the LD phenotype, and even induced a decrease in LD biogenesis in control cells (Fig. 3, F and G). These results provide a causal relationship between the lack of ORP5 and the perturbation of LD formation and implicate ORP5B in the phenotype. Importantly, they also suggest that ORP5 lipid transfer activity is involved in LD biogenesis at MAMs.
To address whether the ER–Mito contact sites could play a role in LD formation, we disrupted these contacts. PTPIP51 overexpression increases ER–Mito contact sites, while its knockdown significantly reduces these contacts (Stoica et al., 2014). We performed PTPIP51 KD in cells delipidated for 3 d (Fig. S2, E and F) and loaded the cells with FA568 for 15 min to trigger the formation of LDs. We found a dramatic decrease in the number of FA568–positive LDs in the PTPIP51 depleted cells (more than 90%) as compared to control cells (Fig. S2 G). Such a decrease was not rescued by the overexpression of EGFP-ORP5B or EGFP-Sec61β as control, (Fig. 3, H and I; and Fig. S2, F and G), even though FA568 was loaded for a longer time. These data indicate that the ER–Mito contact sites integrity is required to ensure proper ORP5/8-dependent LD formation and uncover a novel role of MAMs as a key hotspot for LD formation.
ORP5 is recruited to LDs emerging from MAM subdomains and to pre-existing LDs
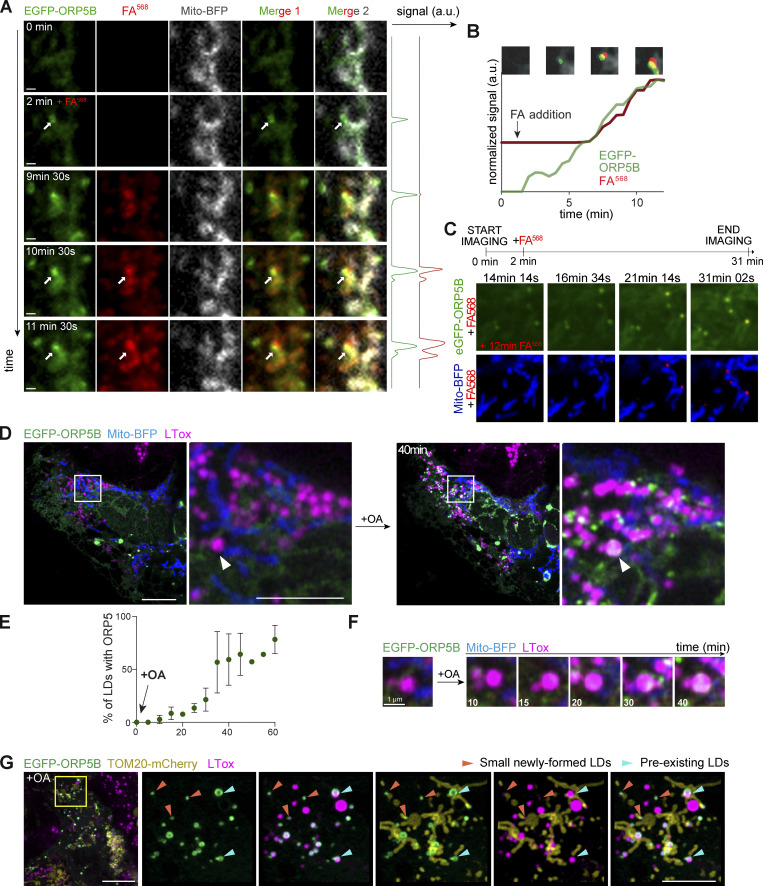
To characterize the dynamics of ORP5 localization at LD biogenesis sites, we performed confocal live-cell imaging by spinning disk microscopy in HeLa cells. Cells transfected with EGFP-ORP5B and Mito-BFP were treated with FA568 (at 2 min) and imaged for 12–31 min. Immediately after the addition of FA568, ORP5B was recruited to ER subdomains close to mitochondria where de novo LDs were formed and became visible within 8–14 min following the fatty acid addition (Fig. 4, A–C; and Videos 1, 2, and 3).
Figure 4.
ORP5 specifically localizes to ER subdomains where LDs originate and also to the preexisting lipid droplets. (A) Zoom of spinning video snapshots of HeLa cells expressing EGFP-ORP5B (green) and Mito-BFP (grey). After 2 min of acquisition, the cells were treated with FA568 (red) at 1 µM. Arrows indicate ORP5-labeled MAM-LD contacts associated to mitochondria. Scale bar, 1 µm. (B) Full time course analysis of the intensity changes for ORP5B (green) and FA568 (red) over time. (C) Additional spinning video snapshots of a region of HeLa cells expressing EGFP-ORP5B (green) and Mito-BFP (blue). After 2 min of acquisition, the cells were treated with FA568 at 1 µM. Arrows indicate ORP5-labeled MAM-LD contacts associated with mitochondria. Full cell view in Fig. S6 B. Scale bar, 1 µm. (D) Example of an Airyscan video snapshots of Huh7 cells expressing EGFP-ORP5B (green), RFP-Sec22b (red, shown in Fig. S3 A), and Mito-BFP (blue) before and after 40 min of 200 μM OA treatment. The lipid droplets were stained using LTox Deep Red (purple). Arrowheads indicate absence or presence of ORP5B at MAM-LD contacts before and after OA treatment, respectively. Full sequence in Fig. S3 A. Scale bar, 10 μm (entire cell), or 5 μm (zoom). (E) Quantification of the % of LDs with EGFP-ORP5 over the indicated time points. (F) Time course of ORP5 recruitment to a large pre-existing LDs depicted by the white arrowhead in the Huh7 cell in C. Scale bar, 1 µm. (G) Representative Airyscan snapshot of Huh7 cells expressing EGFP-ORP5B (green) and TOM20-mCherry (yellow), staining mitochondria, after 1 h 30 min of 200 μM OA treatment. The lipid droplets were stained using LTox Deep Red (purple). Scale bar, 10 μm (entire cell), or 5 μm (zoom). Orange arrowheads indicate small emerging LDs, light green arrowheads indicate pre-existing LDs.
Video 1.
LDs originate from MAM where ORP5 localizes (entire cell, cell 1). Time-lapse of confocal spinning disk microscopy (one plan taken with an objective 40×) of HeLa cells co-expressing EGFP-ORP5B (green) and Mito-BFP (gray). Images were acquired every 30 s for 12 min. After 2 min of acquisition, the cells were treated with FA568 (red) at 1 µM (related to Fig. 4 A).
Video 2.
LDs originate from MAM where ORP5 localizes (zoomed region of cell 1). Zoom of the time-lapse of confocal spinning disk microscopy (one plan taken with an objective 40×) of HeLa cells co-expressing EGFP-ORP5B (green) and Mito-BFP (gray). Images were acquired every 30 s for 12 min. After 2 min of acquisition, the cells were treated with FA568 (red) at 1 µM (related to Fig. 4 A, entire cell shown in Video 1).
Video 3.
LDs originate from MAM where ORP5 localizes (zoomed region of cell 2). Zoom of the time-lapse of confocal spinning disk microscopy (one plan taken with an objective 60×) of HeLa cells co-expressing EGFP-ORP5B (green) and Mito-BFP (blue). Images were acquired every 14 s for 31 min. After 2 min of acquisition, the cells were treated with FA568 (red) at 1 µM (related to Fig. 4 C).
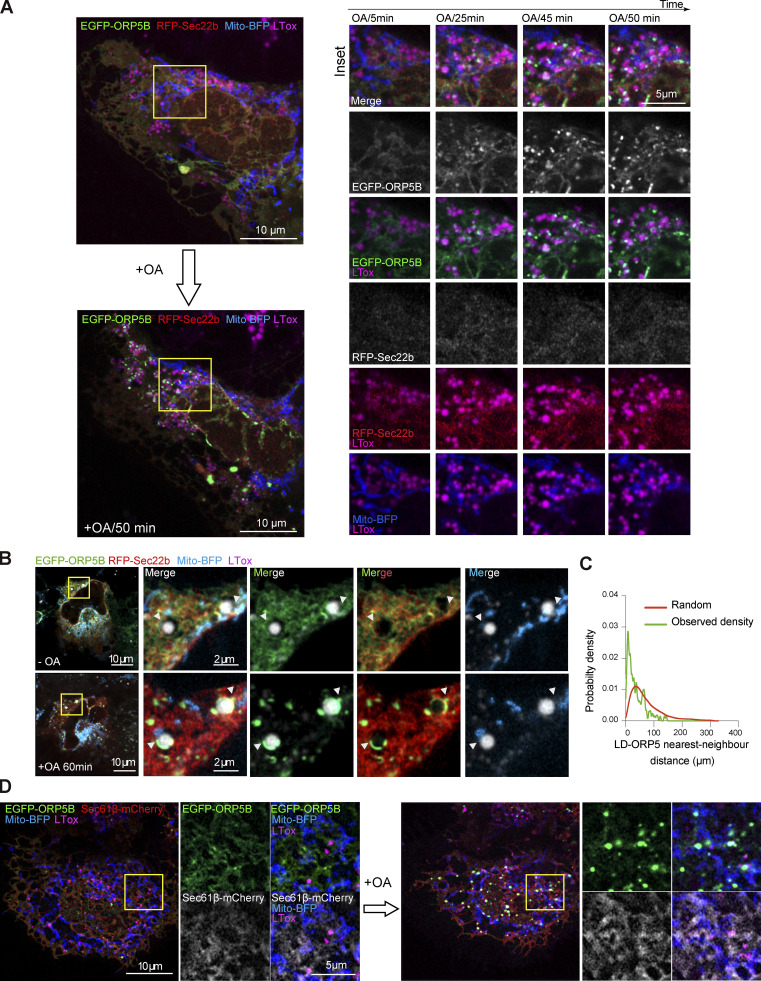
To corroborate these data in another cell model relevant to LD physiology and at a higher resolution level, we examined the dynamics of ORP5 recruitment at MAM-LDs contact sites in human hepatocytes Huh7 by Airyscan microscopy in live cells. Huh7 cells were transfected with EGFP-ORP5B, RFP-Sec22b or mCherry-Sec61β, and Mito-BFP (Fig. 4, D–F and Fig. S3, A–D) or with EGFP-ORP5B and TOM20-mCherry (Fig. 4 G). We then imaged a cell and added OA for 1 h to induce TG synthesis and de novo LD formation.
Figure S3.
ORP5 localizes to nascent LDs and the preexisting ones in Huh7. (A) Airyscan video snapshots of Huh7 cells expressing EGFP-ORP5B (green), RFP-Sec22b (red) and Mito-BFP (blue). Cells were treated with OA to induce the formation of LDs, and stained by LTox (purple). Images were taken every 5 min for 50 min. Scale bar, 10 μm (entire cell), or 5 μm (zoom). (B) Another example depicting the recruitment of ORP5 to pre-existing LDs, with mitochondria. Arrowheads indicate EGFP-tagged ORP5 at MAM-LD contacts. Scale bar, 10 μm (entire cell), or 2 μm (zoom). (C) Line graph shows the distribution of the nearest-neighbor EGFP-ORP5B puncta to LDs in the images of Huh7 cells treated with OA. The observed probability density largely deviates from randomly distributed LD and puncta EGFP-ORP5B. (D) Airyscan video snapshots of Huh7 cells expressing EGFP-ORP5B (green), Sec61β-mCherry (red) and Mito-BFP (blue). Cells were treated with OA to induce the formation of LDs, and stained by LTox (purple). Scale bar, 10 μm (entire cell), or 5 μm (zoom).
At 20–25 min from OA addition, ORP5 began to be enriched in ER subdomains often corresponding to MAMs, in contact with both LDs and mitochondria (Fig. S3, A and B). The ER-protein Sec22b or Sec61β also localized at the MAM where ORP5 was recruited (Fig. S3, B and D), confirming that these structures were indeed ER. Even after 1 h from the induction of LD formation, Sec22b or Sec61β maintained its reticular localization but was not enriched at MAM (Fig. S3 B), in contrast to ORP5, which was still strong at MAM–LD contact sites (Fig. S3, B–D).
OA triggered the strong redistribution of ORP5 from the reticular ER to MAM–LD contact regions (Fig. 4, D–G and Fig. S3), and the fraction of LDs that are positive for ORP5 increased during feeding (Fig. 4 E). Small LDs, newly emerging, had a strong ORP5 signal (arrowhead, Fig. 4 G) closely associated with mitochondria. This observation is consistent with the early accumulation of ORP5 at the MAM sites where LDs assembled in the HeLa cells (Fig. 4, A–C). Thus, data from both HeLa and Huh7 hepatocyte cells indicate that ORP5 is involved in orchestrating the early stage of LD assembly. However, OA treatment induced targeting of ORP5 not only to mitochondria-associated newly-formed small LD, but also to the preexistent larger ones (Fig. 4, D–G and Fig. S3, A–D), suggesting the role of ORP5 not only in LD biogenesis but also in their maintenance at MAMs, possibly by regulating lipid fluxes toward them to fit local cellular needs.
ORP5 is recruited to PA-enriched MAM subdomains
The ORD domain of ORP5 is involved in LD binding, while its PH domain is not required (Figs. 1, 2 C, and S1 F), as previously shown (Du et al., 2020). The CC domain, which is poorly characterized and sits just before the PH domain, has been proposed to play a role in the association of ORP5 with the plasma membrane (Ghai et al., 2017). We asked whether the ORP5 CC domain could also be relevant for the targeting of MAM–LDs and LD binding. To answer this question, we analyzed by confocal microscopy the localization of the EGFP-tagged ORP5 deletion mutant lacking the CC domain (aa 96–116; EGFP-ORP5ΔCC) in HeLa cells treated with OA (Fig. 5 A). As expected, the deletion of CC in ORP5A led to the loss of ER–PM contacts localization, indicating that the CC might be involved in the binding of PM proteins or lipids. EGFP-ORP5ΔCC localized to the reticular ER in all transfected cells and, only in very few cells (about 10%), it was detected at MAM–LD contact sites. Also, in these cells, even after OA treatment, ORP5 was not enriched at MAM–LD contacts (Fig. 5 A). This data strongly supports that the CC domain plays a key role in the association of ORPs with MAM–LDs.
Figure 5.
ORP5 localizes to LDs and ER subdomains enriched in phosphatidic acid (PA). (A) Confocal images (single focal plane of HeLa cells expressing EGFP-tagged ORP5A or ORP5∆CC (green), treated with OA (300 μM) for 2 h. The mitochondria and the LDs were stained with Mitotracker (red) and LTox (purple), respectively. Arrowhead points ORP5-labeled MAM-LD associated with mitochondria and asterisks marks ORP5 localized to reticular ER. Scale bar, 5 µm. (B) PIP strip overlay assay: PIP strips were incubated with either ORP5-HA CC or ORP8-HA CC or the HA peptide as a negative control and analyzed using the anti-HA antibody. LPA, lysophosphatidic acid; LPC, lysophosphocholine; PtdIns, phosphatidylinositol; PtdIns(3)P; PtdIns(4)P; PtdIns(5)P; PtdIns(3,4)P2; PtdIns(3,5)P2; PtdIns(4,5)P2; PtdIns(3,4,5)P3; PA, phosphatidic acid; PS, phosphatidylserine; PE, phosphatidylethanolamine; PC, phosphatidylcholine; S1P, sphingosine 1-phosphate. (C) Confocal images (single focal plane) of HeLa cells co-expressing EGFP-Opi1PABD (green) with Mito-BFP (blue) and either RFP-Sec22b (red), or Sec61β-RFP, or EGFP-ORP5B (red), or EGFP-ORP5∆PH. The LDs were stained with LTox (purple). Arrowheads points enrichment of Opi1PABD at Mito–MAM–LD contact sites. Scale bar, 10 μm (entire cell), or 3 μm (zoom). (D) 3D reconstruction of cells shown in D using IMARIS. Arrows point to the MAMs where ORP5B and ORP5∆PH co-localize with Opi1PABD at Mito–MAM–LD contact sites. Scale bar, 0.5 µm.
Since the CC domain bears multiple charges, we hypothesized it might bind to charged lipids at MAM–LDs. To assess the lipid-binding specificities of the ORPs CC domains, we performed PIP-strip binding assays. Both ORP5 and ORP8 strongly bound PtdIns(4)P, but ORP8 showed a higher preference for PtdIns(3)P and PtdIns bisphosphates (except for PtdIns[3,4]P, not recognized by the ORPs). Phosphatidic acid (PA) was the only other phospholipid bound by the CC domain, although this binding was weaker than for PtdIns (Fig. 5 B). PtdIns are not detectable in the ER or LDs under normal conditions (Dickson and Hille, 2019) as they are dephosphorylated by Sac1, for instance in the ER. In contrast, PA can be found in both the ER and LDs, especially during LD biogenesis (Gao et al., 2019). Indeed, PA, a negatively charged non-bilayer lipid, is constantly made in the ER and is essential for the synthesis of other phospholipids and TG (Gao et al., 2019). Therefore, it was interesting that ORP5 and ORP8 CCs bound PtdIns and PA very specifically as compared with other ER phospholipids (Fig. 5 B), including negatively charged ones such as PS and phosphatidylinositol (PI).
We next sought to investigate the presence of PA at MAM–LD contact sites by using a PA-specific probe, the EGFP-tagged Opi1pQ2S-PABD (hereon called EGFP-Opi1PABD), an improved version of Opi1p for sensing PA (Kassas et al., 2017). HeLa cells were co-transfected with EGFP-Opi1PABD and either RFP-Sec22b or Sec61β-RFP, RFP-ORP5B, or RFP-ORP5ΔPH and analyzed by confocal microscopy. EGFP-Opi1PABD was detected at RFP-Sec22b-positive ER subdomains, and these were often enriched in proximity to mitochondria, revealing the existence of a specific PA pool at MAMs (Fig. 5 C). When co-expressed with RFP-ORP5B, Opi1PABD was strongly enriched at the ORP5-marked MAM and MAM–LD contact sites. The enrichment at the latter contact sites was even clearer when Opi1PABD was co-expressed with RFP-ORP5ΔPH (Fig. 5, C and D), which had the highest recruitment to MAM–LD contact (Fig. 1). These data suggest a functional link between PA and ORP5 at MAMs.
Seipin localizes to MAM–LD contacts in an ORP5-dependent manner
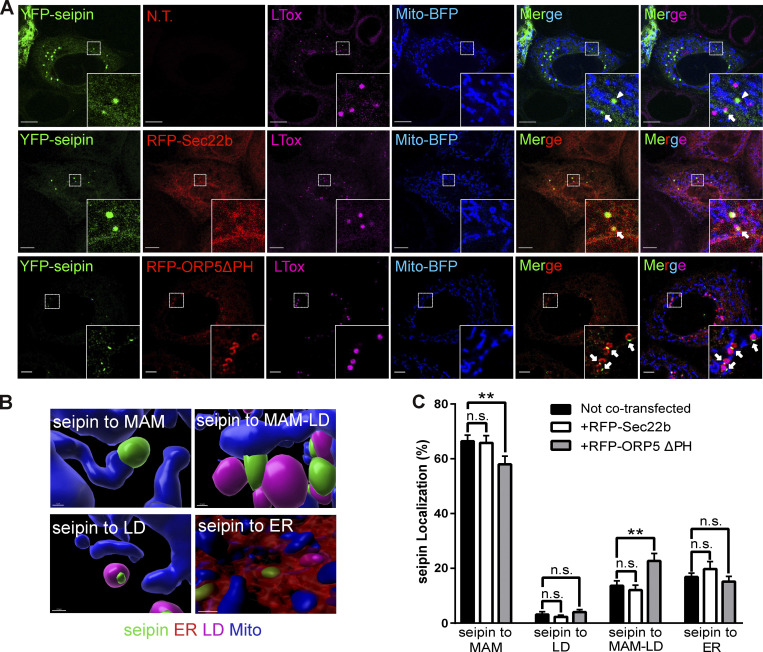
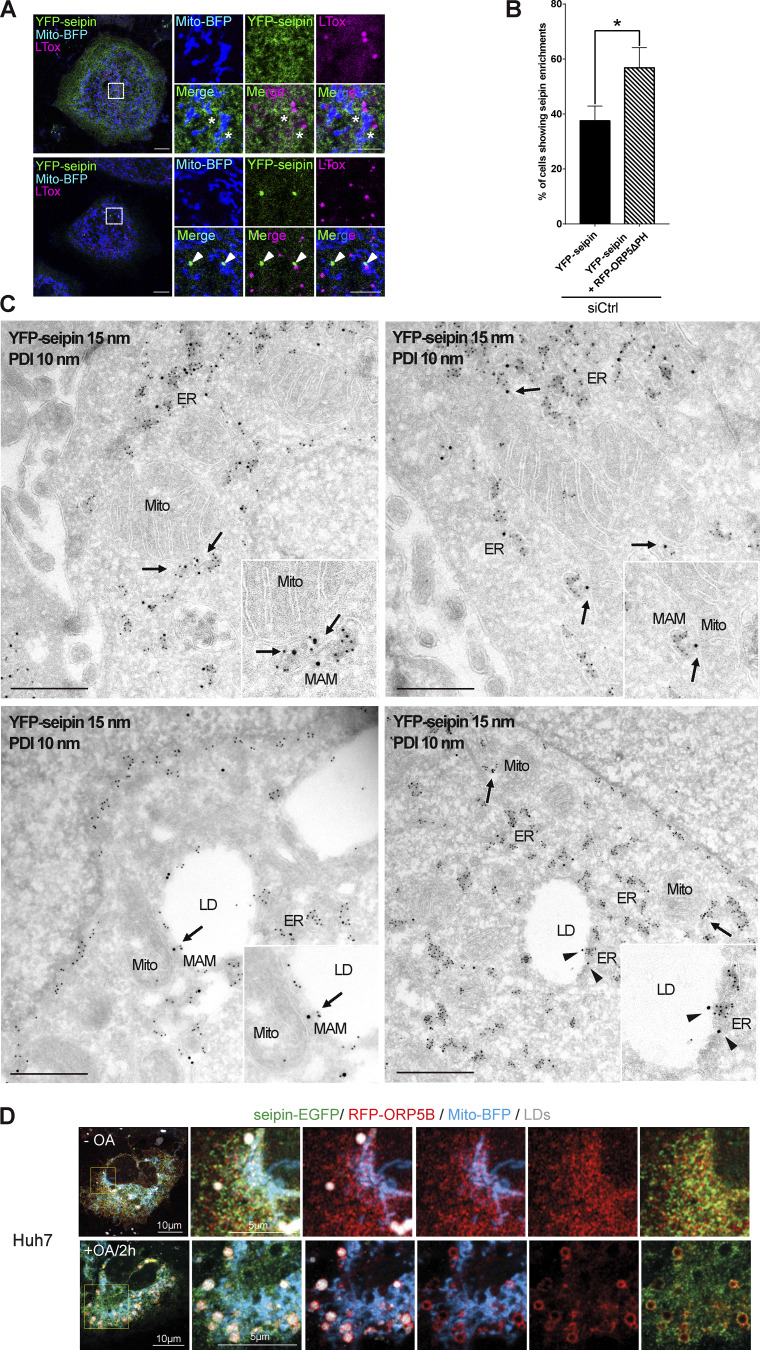
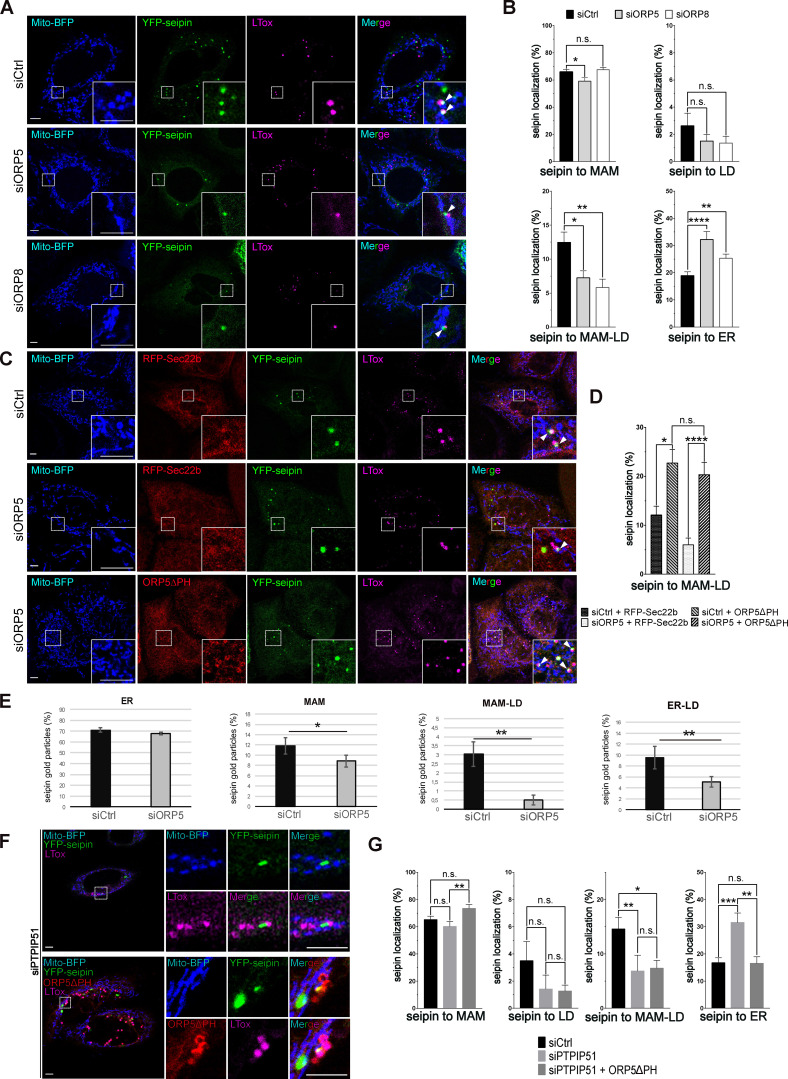
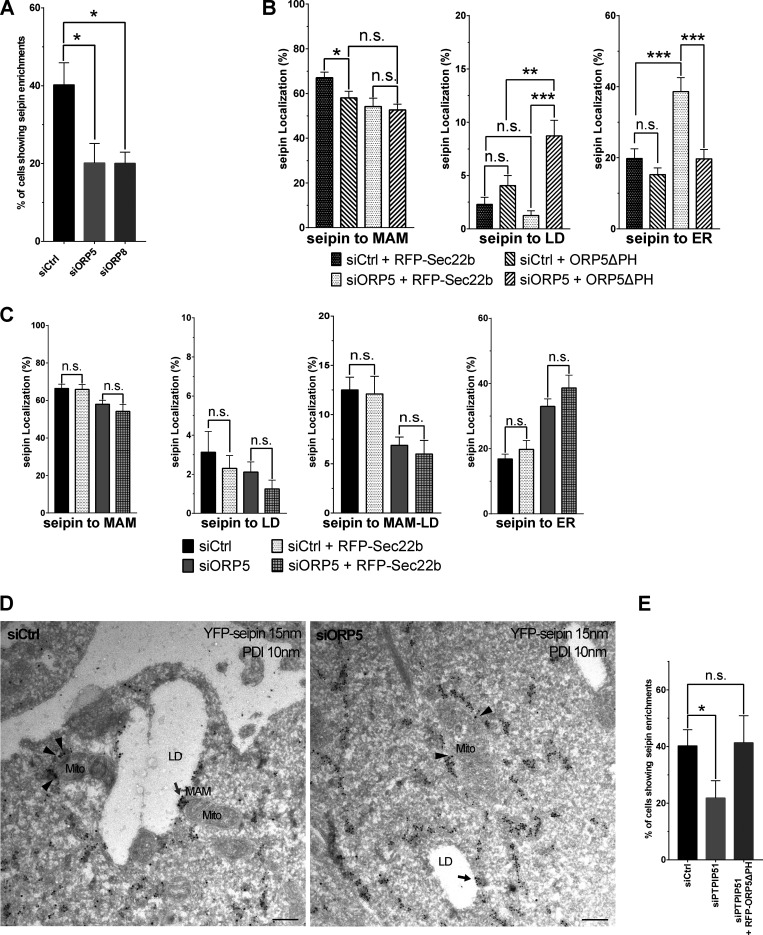
Seipin is an integral ER membrane protein playing a central role in determining where triacylglycerol LDs form. Interestingly, seipin binds anionic phospholipids, including PA (Yan et al., 2018). We asked whether seipin could also localize at MAM to facilitate LD formation in proximity to mitochondria. To address this, we co-expressed in HeLa cells a YFP-tagged mouse seipin with Mito-BFP, alone or together with RFP-Sec22b. We then analyzed seipin localization by confocal microscopy (Fig. 6 A). As expected, seipin colocalized with the ER protein Sec22b. However, a subset of cells where seipin was expressed at lower levels displayed additional enrichments of seipin to ER structures (MAM) in close proximity to mitochondria, which were often also associated with LDs (MAM–LD). We then asked whether ORP5 could regulate the localization of seipin at MAM. To address this, in parallel experiments, we co-expressed YFP–seipin with RFP-ORP5ΔPH (Fig. 6 A). Remarkably, we observed an increase in the population of cells showing the enrichment of seipin at MAM–LD contacts (Fig. S4, A and B).
Figure 6.
ORP5 over-expression induces an increase of the localization of seipin to MAM–LD contact sites. (A) Representative confocal images showing a single focal plane of HeLa cells expressing YFP-seipin (green), Mito-BFP (blue) and Sec22b (red) or ORP5∆PH (red). The LDs were stained LTox (purple). Arrowhead points seipin enrichment at MAM–mitochondria contact sites and arrows mark seipin enrichment at Mito–MAM–LD contact sites. Scale bar, 10 µm. (B) Representative 3D reconstruction images of the different categories for the classification of the localization. (C) Analysis of the localization of seipin enrichments in HeLa cells expressing seipin alone or in co-expression with Sec22b or ORP5∆PH. Data are shown as % mean ± SEM of cell of n = 56 cells in YFP-seipin (expressed alone), n = 14 cells in YFP-seipin + RFP-Sec22b, and n = 40 cells in YFP-seipin + RFP- ORP5∆PH, (** = P < 0.01; Wilcoxon-Mann-Whitney test).
Figure S4.
ORP5 over-expression induces an increase of the localization of seipin to MAM-LD contact sites. (A) Confocal images (single focal plane) of HeLa cells expressing YFP-seipin (green) and Mito-BFP (blue). The LDs were stained LTox (purple). Upper panels show a cell with a reticular staining of seipin. Lower panels show a cell with enrichment of YFP-seipin in small “clusters.” Arrowheads show the enrichment of seipin in “clusters” closely opposed to MAM-LD contact sites. Asterisks show presence of seipin at MAM-LD contacts also in cells where it has a reticular distribution. Scale bar, 10 μm (entire cell) or 5 μm (zoom). (B) Quantification of the % of cells showing seipin enrichment at MAM-LD in cells expressing seipin alone or together with ORP5∆PH. Data are shown as % mean ± SEM of cells. n = 118 cells in siCtrl, n = 121 cells in siCtrl + RFP-ORP5∆PH. (C) Representative images of electron micrographs of ultrathin cryosections of HeLa cells transfected with YFP-seipin and immunogold stained with anti-GFP (15 nm gold) to detect seipin and anti-PDI (10 nm gold) to label the ER lumen. Arrows point the localization of seipin at MAM or MAM-LDs. Arrowheads point the localization of seipin at ER–LD contacts. Mito, mitochondria; ER, endoplasmic reticulum; MAM, mitochondria-associated membranes; LD, lipid droplets. Scale bar, 250 nm. (D) Confocal images (single focal plane) of Huh7 cells expressing seipin-EGFP (green), RFP-ORP5B (red) and Mito-BFP (blue) before and after 2 h of OA treatment. The LDs were stained with LTox (white). Scale bar, 10 μm (entire cell), or 5 μm (zoom).
Quantification of the distribution of seipin-positive clusters upon segmentation of the ER, the LD, and the mitochondria by Imaris (Fig. 6, B and C) revealed that when seipin was expressed alone or with Sec22b, most of these clusters corresponded to MAM (about 78%) and only a few (20%) to the reticular ER. A fraction of the seipin-positive MAM (18%) was also closely associated with LDs (corresponding to MAM–LD contacts), and only a minimal amount of seipin-positive ER clusters (2%) was exclusively associated with LDs. When seipin was co-expressed with ORP5ΔPH, we observed a significant increase of seipin-positive MAM in contact with LD and a significant decrease of seipin-positive MAMs not associated with LD (Fig. 6, B and C). However, by confocal imaging, we could not analyze the localization of the entire ER pool of seipin, but only its local enrichment at these “clusters.”
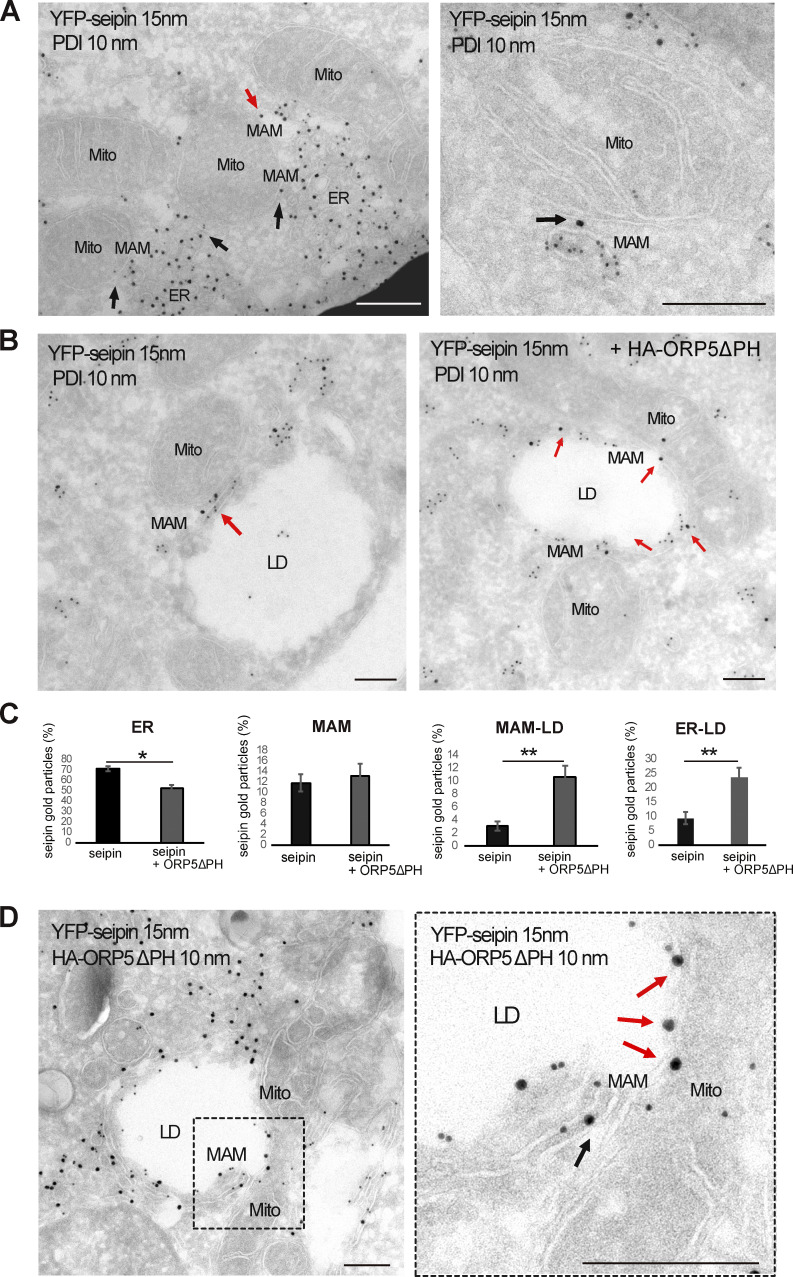
To characterize the distribution of the entire seipin pool at a high-resolution level and to also analyze the ultrastructure of these seipin “clusters,” we performed immuno-EM analysis on ultrathin cryosection (by Tokuyasu method) in HeLa cells expressing YFP-seipin alone or together with HA-ORP5ΔPH, and performed co-immunolabeling of YFP-seipin (15 nm gold) and protein disulfide isomerase (PDI; 10 nm gold) to stain the ER (Fig. 7, A and B; and Fig. S4 C). When expressed alone, seipin was mostly observed in the widely distributed reticular ER (70%), but a fraction of seipin (12%) was also found at MAMs, and a little pool (3%) was detected at MAM–LD contacts (Fig. 7, A and B, quantifications in Fig. 7 C and Fig. S4 B). Few cells displayed an accumulation of seipin-positive ER elements in contact with mitochondria that presumably correspond to the “clusters” observed and quantified by confocal. Consistent with our confocal data, these clusters were always found in close connection with mitochondria (corresponding to MAMs) and sometimes additionally with LD (corresponding to MAM–LD contacts; Fig. 7 A). Moreover, further corroborating our confocal data, co-expression with ORP5ΔPH increased the pool of seipin at MAM–LD contact sites (3–24%) accompanied by an increase of seipin at ER–LD contacts because of the ORP5-induced expansion of ER on the LD surface (from 10 to 24%). This increase was also concomitant with a significant decrease in seipin abundance at the reticular ER not associated with contact sites (from 70 to 53%; Fig. 7, B and C). Co-localization of seipin and ORP5 at the expanded MAM–LD contact sites was further confirmed by co-immunolabeling of YFP-seipin (15 nm gold) and ORP5 (10 nm gold; Fig. 7 D). Seipin recruitment to ORP5-positive MAM–LD contact sites upon OA was confirmed in Hu7h cells co-expressing seipin–EGFP, RFP-ORP5B, and Mito-BFP (Fig. S4 D). Overall, these results reveal that seipin localizes at MAM–LD contacts in an ORP5-dependent manner.
Figure 7.
ORP5∆PH increases the localization of seipin to MAM–LD and ER–LD contacts. (A) Representative images of electron micrographs of ultrathin cryosections of HeLa cells transfected with YFP-seipin and immunogold stained with anti-GFP (15 nm gold) to detect seipin and anti-PDI (10 nm gold) to label the ER lumen. Seipin localizes at MAM–LD contacts (arrows). Mito, mitochondria; ER, endoplasmic reticulum; MAM, mitochondria-associated membranes; LD, lipid droplets. Scale bar, 250 nm. (B) Representative images of electron micrographs of ultrathin cryosections of HeLa cells co-transfected with YFP-seipin alone or together with HA-ORP5. Cells were immunogold stained with anti-GFP (15 nm gold) to detect seipin and anti-PDI (10 nm gold) to label the ER. (C) Quantification of the distribution of seipin immunogold particles (15 nm). Data are shown as % mean ± SEM of cell profiles with n = 32 (750 gold particles analyzed) in seipin individual expression, and n = 50 (940 gold particle) in seipin + ORP5∆PH co-overexpression. *P < 0.001, **P < 0.0001, unpaired two-tailed t test. (D) Electron micrographs of ultrathin cryosections of HeLa cells co-transfected with YFP-seipin and HA-ORP5∆PH and immunogold stained with anti-GFP (15 nm gold) to detect seipin and anti-HA (10 nm gold) to detect ORP5. The localization of seipin at MAM-LD contacts is increased when co-expressed with ORP5∆PH (arrows). Scale bar, 250 nm.
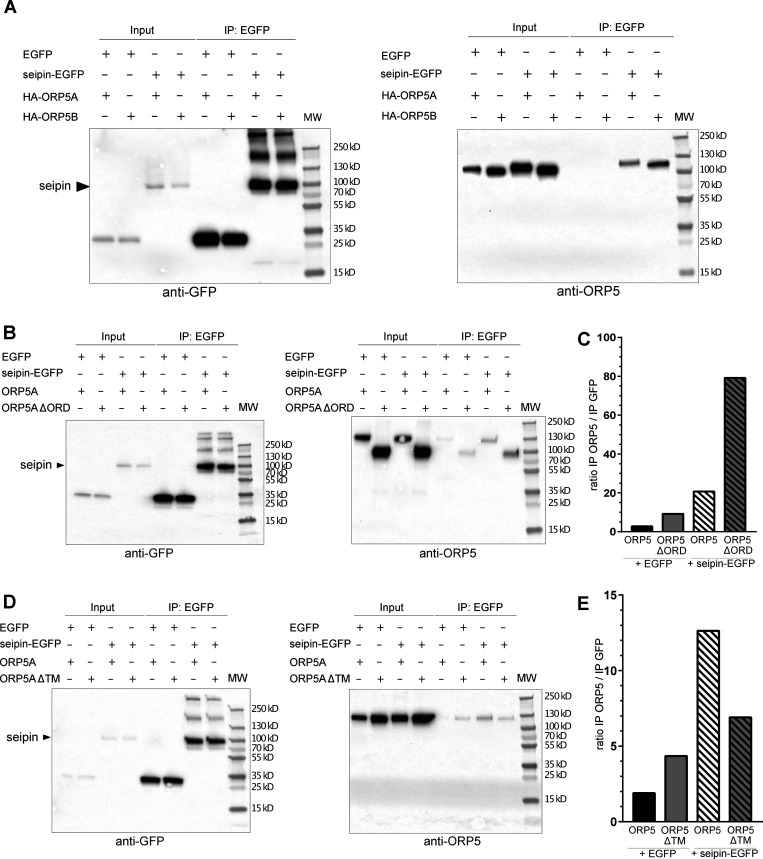
Since seipin localization to MAM–LD was ORP5-dependent, we tested whether these proteins could interact with each other. We performed GFP-pull down assays from cells co-overexpressing seipin-EGFP and HA-ORP5A or HA-ORP5B and loaded with OA for 2 h (Fig. 8 A). ORP5A and ORPB proteins were detected in seipin pull-downs revealing that these proteins biochemically interact. Interestingly, ORP5B was more enriched in the seipin immunoprecipitates as compared to ORP5A, in accord with the major localization of ORP5B than ORP5A at MAM–LD contact sites. To address the possible involvement of the ORD domain in ORP5-seipin binding, we performed similar GFP-pull down experiments in cells co-expressing seipin-EGFP and ORP5AΔORD (Fig. 8 B). ORP5AΔORD still interacted with seipin-EGFP indicating that the ORD domain is not involved in ORP5-seipin binding. On the contrary, deletion of the ORP5 TM domain decreased its interaction with seipin (Fig. 8 C), suggesting that the ER localization of ORP5 could be important for ORP5 interaction with seipin.
Figure 8.
ORP5 and seipin biochemically interact in lysates of HeLa cell. (A) Western blot analysis of ORP5A or ORP5B immunoprecipitated (IP) from lysates of HeLa cells co-expressing EGFP or seipin-EGFP with HA-ORP5A or HA-ORP5B, using antibodies against GFP (to detect seipin) or OA1 (to detect ORP5A or ORP5B). (B) Western blot analysis of co-IP products from HeLa cells co-expressing EGFP or seipin-EGFP with ORP5A or ORP5∆ORD using antibodies against GFP (to detect seipin) or OA1 (to detect ORP5A or ORP5∆ORD). (C) Relative quantification of ORP5A or ORP5∆ORD co-immunoprecipitated with seipin-EGFP or EGFP alone. (D) Western blot analysis of co-IP products from HeLa cells co-expressing EGFP or seipin-EGFP with HA-ORP5A or HA-ORP5∆TM, using antibodies against GFP (to detect seipin) or OA1 (to detect ORP5A or ORP5∆TM). (E) Relative quantification of ORP5A or HA-ORP5∆TM co-immunoprecipitated with seipin-EGFP or EGFP alone.
ORP5 role in seipin recruitment to MAM–LD contacts depends on MAM integrity
To further study the role of ORP5, and also of ORP8, in the targeting of seipin to MAM–LD contact sites, we analyzed the localization of seipin in ORP5 and ORP8 KD cells, transfected with YFP-seipin and Mito-BFP and stained by LTox (Fig. 9 A). Knockdown of ORP5 and ORP8 led to a significant decrease in the cells showing seipin clusters at MAM–LD (Fig. S5 A) accompanied by an increase in seipin association with the reticular ER (Fig. 9, A and B). Interestingly, in ORP5-depleted cells, we also detected a general decrease in the seipin pool associated with MAM, while seipin abundance at ER–LD contacts was unchanged in both ORP5- and ORP8-silenced cells (Fig. 9, A and B). These data reveal a key role of ORP5 and ORP8 in regulating the targeting of seipin to MAM–LD contacts, and a more important role for ORP5 in regulating the general recruitment of seipin to MAMs.
Figure 9.
ORP5 affects the localization of seipin in a Mito-MAM contact sites integrity dependent way. (A) Representative confocal images (single focal plane) of HeLa cells treated with siCtrl or siORP5 or siORP8 RNA oligos. The cells were then transfected with YFP-seipin (green) and Mito-BFP (blue). The LDs were stained LTox (purple). Arrowheads point to seipin enrichment at MAM–LD contact sites. Scale bar, 5 µm. (B) Analysis of the distribution of seipin enrichments in HeLa cells expressing seipin and treated with either siCtrl, siORP5, or siORP8 interfering RNAs. Data are shown as % mean ± SEM of cells with n = 64 cells in siCtrl, n = 44 cells in siORP5, and n = 32 cells in siORP8 (* = P < 0.05; ** = P < 0.01; **** = P < 0.0001; Wilcoxon-Mann-Whitney test). (C) Confocal images (single focal plane) of HeLa cells treated with siCtrl or siORP5 or siORP8 RNA oligos. The cells were then transfected with YFP-seipin (green), Mito-BFP (blue) and either RFP-Sec22b (red) or ORP5∆PH (red). The LDs were stained LTox (purple). Arrowhead points seipin enrichment at MAM–LD contact sites. (D) Analysis of the distribution of seipin enrichments to MAM-LD contact sites in HeLa cells treated with siCtrl or siORP5 RNA oligos and then co-transfected with seipin and RFP-Sec22b or siRNA-resistant ORP5∆PH. Cells were treated with OA (300 μM) for 2 h before analysis. Data are shown as % mean ± SEM of cells. n = 14 cells in siCtrl + Sec22b, n = 17 cells in siORP5 + Sec22b, n = 41 cells in siCtrl + ORP5∆PH rescue and n = 19 cells in siORP5 + ORP5∆PH rescue (* = P < 0.05; **** = P < 0.0001; Wilcoxon-Mann-Whitney test). (E) Quantification of the distribution of the immunogold particles (15 nm) staining seipin in HeLa cells treated with siCtrl or siORP5. Data are shown as % mean ± SEM of cell profiles with n = 79 (1,500 gold particles analyzed) in siCtrl, and n = 64 (1,800 gold particle) in siORP5. * = P < 0.001, ** = P < 0.0001, unpaired two-tailed t test. (F) Representative confocal images (single focal plane) of HeLa cells treated with siPTPIP51 or siCtrl RNAs. The cells were then transfected with YFP-Seipin (green) alone or in co-expression with ORP5∆PH rescue (red). The LDs were stained with LTox (purple). Scale bar, 5 µm. (G) Quantification of the distribution of seipin immunogold particles in HeLa cells treated with siCtrl or siPTPIP51 and expressing seipin alone or in co-expression with siRNA-resistant ORP5∆PH. Data are shown as % mean ± SEM of cells with n = 41 cells in siCtrl, n = 25 cells in siPTPIP51, and n = 23 cells in siPTPIP51 + ORP5∆PH (* = P < 0.05; ** = P < 0.01; *** = P < 0.001; Wilcoxon-Mann-Whitney test).
Figure S5.
ORP5 affects the localization of seipin in a ER-mitochondria contact sites integrity dependent way. (A) Quantification analysis of confocal data showing the percentage of YFP-seipin–expressing siCtrl or siORP5 or siORP8 cells displaying seipin enrichment at MAM-LD. Data are shown as % mean ± SEM of cell of n = 118 cells in siCtrl, n = 80 cells in siORP5 and n = 134 cells in siORP8. (B) Analysis of seipin localization to the indicated compartments (MAM = ER–Mito contacts, ER–LD = ER–LD contacts, ER = reticular ER) in siCtrl or siORP5 HeLa cells transfected with YFP-seipin and either RFP-Sec22b or siRNA-resistant ORP5∆PH. Data are shown as % mean ± SEM of cells. n = 14 cells in siCtrl + Sec22b, n = 17 cells in siORP5 + Sec22b, n = 41 cells in siCtrl + ORP5∆PH rescue and n = 19 cells in siORP5+ ORP5∆PH rescue (* = P < 0.05; ** = P < 0.01; *** = P < 0.001; Wilcoxon-Mann-Whitney test). (C) Quantitative analysis of the distribution of seipin enrichments in the indicated compartments (MAM = ER–Mito contacts, ER–LD = ER–LD contacts, MAM–LD = Mito–ER–LD contacts, ER = reticular ER) in HeLa cells treated with siCtrl or siORP5 and expressing YFP-seipin alone or together with RFP-Sec22b. Data are shown as % mean ± SEM of cell of n = 56 cells in siCtrl, n = 14 cells in siCtrl + Sec22b, n = 58 cells in siORP5 and n = 17 cells in siORP5 + Sec22b (n.s. = not significant; Wilcoxon-Mann-Whitney test). (D) Representative electron micrographs of ultrathin cryosections of HeLa cells transfected with YFP-seipin and immunogold stained with anti-GFP (15 nm gold) to detect seipin and anti-PDI (10 nm gold) to label the ER lumen. Arrows point to seipin localization at MAM–LD (red arrow) or ER–LD (black arrow). Arrowheads point to seipin localization at MAM. Mito, mitochondria; ER, endoplasmic reticulum; MAM, mitochondria-associated membranes; LD, lipid droplets. Scale bar, 250 nm. (E) Quantification of the % of siCtrl or siPTPIP51 cells transfected with YFP-seipin or of siPTPIP51 cells co-transfected with YFP-seipin and RFP-ORP5∆PH, showing seipin enrichment at MAM–LD. Data are shown as % mean ± SEM of cell of n = 118 cells in siCtrl, n = 91 cells in siPTPIP51, and n = 58 cells in siPTPIP51 + RFP-ORP5∆PH.
Next, we wanted to rescue the decrease of seipin at MAM–LDs induced by the loss of ORP5. For this purpose, we chose to overexpress ORP5ΔPH in the ORP KD background. We transfected siRNA-resistant ORP5ΔPH, or RFP-Sec22b as a control, in the ORP5 depleted cells and analyzed seipin localization by confocal microscopy (Fig. 9 C). Re-expression of ORP5ΔPH (detected by immunofluorescence using an antibody against ORP5), but not of RFP-Sec22b, completely rescued the levels of seipin at MAM and even increased the seipin pool at MAM–LD, confirming the specificity of ORP5 activity in controlling seipin localization to MAMs, including those associated with LDs (Fig. 9, C and D; and Fig. S5, B and C).
To further confirm these observations, we analyzed the entire seipin pool localization at a high-resolution level by performing immuno-EM analysis on ultrathin cryosection in HeLa cells expressing YFP-seipin. We co-immunolabeled YFP-seipin (15 nm gold) and the luminal ER protein PDI (10 nm gold; Fig. 9 E and Fig. S5 D). A dramatic decrease of seipin localized at MAM–LDs (of about 84%) was observed in ORP5-silenced cells. This decrease was accompanied by a decrease in seipin at ER–LDs and also a slight but statistically significant decrease in seipin at MAMs (Fig. 9 E). These results further validated the key role of ORP5 in regulating the levels of seipin at MAM and MAM–LD contacts.
Finally, to assess whether ORP5 requires intact ER–mitochondria contacts to regulate seipin targeting at MAMs, we depleted PTPIP51, which we had found involved in LD biogenesis (see Fig. 3, H and I; and Fig. S2, E–G). We then analyzed seipin localization by confocal microscopy. PTPIP51 knockdown induced a dramatic decrease (50%) in the number of cells showing seipin localization at MAM–LD contacts (Fig. S5 E). Also, seipin localization at MAM–LD was greatly reduced (50%), and, in a similar proportion, seipin localization within the reticular ER was increased (Fig. 9, F and G). Moreover, the overexpression of ORP5ΔPH in PTPIP51-depleted cells did not rescue seipin decrease at MAM–LDs, indicating that intact ER–mitochondria contacts are required for ORP5 function in seipin recruitment at MAM–LD contacts.
Discussion
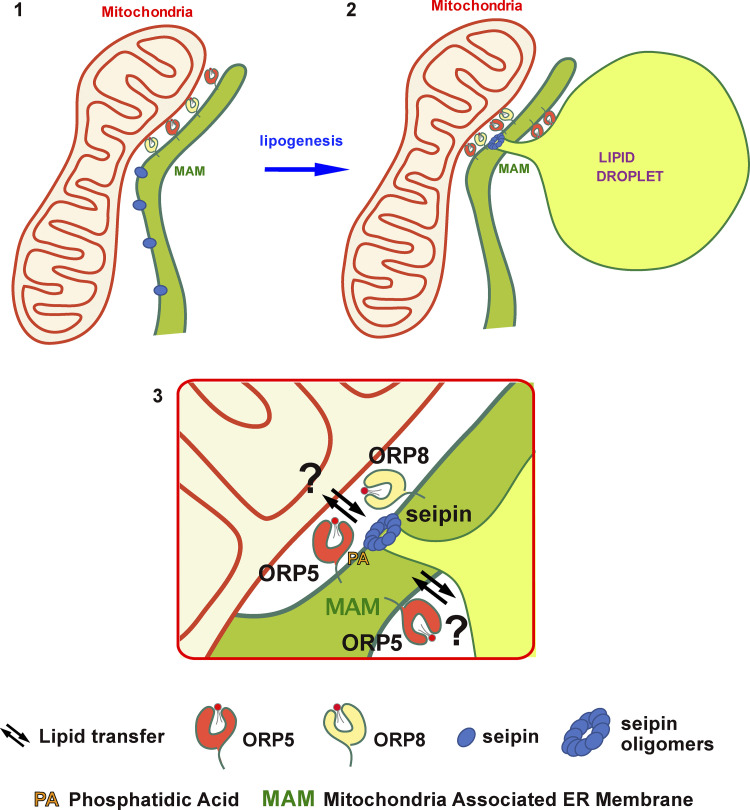
In this study, we found a novel function of the LTPs ORP5 and ORP8 in regulating LD biogenesis and growth at MAMs. We showed that the ORP5/8 complex localizes at MAM subdomains enriched in PA lipid where LDs originate and that loss of ORP5/8 impairs LD biogenesis (Fig. 10). We then revealed that ORP5 and ORP8 interact with seipin and regulate its recruitment to the newly identified MAM–LD contacts. Importantly, ER–mitochondria contact site integrity was required to ensure ORP5/8 function in proper seipin-mediated LD biogenesis.
Figure 10.
ORP5 and ORP8 orchestrate LD biogenesis at MAMs. (1 and 2) The ORP5/8 complex localizes at MAM subdomains (1) where LDs originate (2). (3) During lipogenesis, ORP5 and ORP8 interact with seipin and regulate its recruitment to MAM subdomains enriched in PA phospholipid (3) to sustain proper LD biogenesis. Our data suggest that ORP5/8 could regulate lipid transport pathways across the mitochondria—MAM—LD junctions.
Amongst all ORP proteins, ORP2 was first identified to localize to LDs to regulate cellular sterol homeostasis (Hynynen et al., 2009; Jansen et al., 2011). Recently, ORP5, but not ORP8, was described at ER–LD contacts (Du et al., 2020). These two proteins have been previously shown to localize at ER–PM and ER–mitochondria contact sites (Chung et al., 2015; Galmes et al., 2016) and to exist as a protein complex, mainly at MAMs (Chung et al., 2015; Galmes et al., 2016; Ghai et al., 2017; Monteiro-Cardoso et al., 2022). Yet, ORP5 and ORP8 distribution across all these contact sites remains controversial. Here, we have shown that ORP5 and ORP8 localize and interact at MAM subdomains in contact with LDs, highlighting the existence of a novel tripartite junctional interface between mitochondria, MAM, and LDs where these two LTPs localize. However, the ability of ORP8 to enrich at MAM–LD contact sites depends on ORP5 levels and it is increased by interaction with ORP5 through its CC domain. These findings also provide novel insights on the role of the CC in ORP5/8, whose function was so far poorly understood in regulating ORP5/8 localization and interaction at MAM–LD contact sites. Interestingly, we have revealed that LD biogenesis and growth occur at ER–mitochondria contact sites and depend on both ORP5/8-activity and in particular on their ORD domain. Also, the alterations in LD biogenesis induced by the disruption of ER–mitochondria contact sites (PTPIP51 KD) were not rescued by the overexpression of ORP5, indicating that ORP5/8 functions in this process require intact ER–mitochondria contacts.
The metabolic crosstalk between LDs and mitochondria is well-established for lipid oxidation or storage purposes (Olzmann and Carvalho, 2019; Veliova et al., 2020). Contact sites between LDs and mitochondria form in response to starvation (Herms et al., 2015; Rambold et al., 2015; Wang et al., 2011). During starvation-induced autophagy, DGAT1 (diacylglycerol acyltransferase 1)-dependent LD biogenesis protects mitochondria function by converting fatty acids into TG stored in LDs, to prevent the accumulation of toxic lipids in mitochondria (Nguyen et al., 2017). Recent studies indicate that LD biogenesis can occur at ER subdomains in contact with catabolic organelles, such as the yeast vacuole or the peroxisomes (Hariri et al., 2018; Joshi et al., 2018). Interestingly, DGAT2, one of the two diacylglycerol acyltransferase enzymes converting diacylglycerols into TG, also localizes to MAMs (Stone et al., 2009) and might induce the formation of specific LDs originating from MAMs.
Recently, the mitochondrial protein Mitoguardin 2 has been shown to link mitochondria, LD, and ER to promote de novo lipogenesis in adipocytes from non-lipid precursors (Freyre et al., 2019). Also, interesting, in brown adipocytes, a specific mitochondria subset with restricted dynamics is bound to LDs (Benador et al., 2018). These peridroplet mitochondria support the growth of LDs by providing ATP molecules necessary for TG synthesis (Benador et al., 2018). Based on this knowledge, it may not be surprising that mitochondria transiently or permanently interact locally with the ER membrane to support LD biogenesis and maintenance, e.g., by providing ATP or molecules used to synthesize TG. Currently, molecular mechanisms regulating the functional crosstalk between LD–ER–mitochondria organelles remain largely unknown.
The ER phospholipid composition is important for LD formation (Ben M’barek et al., 2017; Thiam and Forêt, 2016; Zoni et al., 2021). ORP5/8 may regulate LD biogenesis by regulating the phospholipid composition at MAM. ORP5/8 have been shown to counterexchange PS and PtdIns(4)P or PtdIns(4,5)P at ER–PM contacts (Chung et al., 2015; Ghai et al., 2017). Recently, ORP5 has been proposed to play a similar role at ER–LD contact sites (Du et al., 2020). However, that ORP5/8 systematically counter exchange PS with PtdIns(4)P is not established. For instance, we recently found that ORP5/8 can transfer PS from the ER to mitochondria at MAMs despite the lack of PtdIns(4)P on mitochondria. This result suggests that ORP5/8 might carry out multiple lipid transfer activities and that the underlying mechanisms might be different depending on the local lipid composition. In particular, several pieces of evidence link PA to LD biogenesis (Fei et al., 2011; Gao et al., 2019; Pagac et al., 2016) and, interestingly, seipin can bind to PA in vitro (Yan et al., 2018). Our confocal and immuno-EM analysis uncovers that seipin localizes at MAMs and LD-associated MAMs, in addition to its previously reported localization at ER–LD contact sites (Salo and Ikonen, 2019; Salo et al., 2019; Szymanski et al., 2007; Wang et al., 2016). Here we have shown that, in overexpression conditions, seipin biochemically interacts with ORP5 and that its localization to MAMs is dependent on ORP5: knockdown of ORP5 decreased seipin at MAM–LD junctions, while the overexpression of an ORP5 variant enriched at MAM–LD contacts induced seipin accumulation at these sites. Moreover, this impact of ORP5 function in seipin recruitment to MAM–LD contacts depended on the integrity of the ER–mitochondria contacts. These data identify ORP5 and ORP8 as novel critical players in LD biogenesis by regulating seipin targeting at MAMs.
Based on the above observation, one hypothesis is that ORP5/8 regulate seipin recruitment at MAM–LD contacts by forming a protein complex at these contact sites (Fig. 10). ORP5 interaction with seipin does not depend on the ORD (lipid transfer and LD binding) domain, although it requires its anchoring to the ER membranes via the TM domain. Another intriguing hypothesis is that ORP5/8 could be involved in PA dynamics at MAMs. ORP5/8 could enrich and cooperate with PA at MAM sites via their lipid transfer activity (e.g., from Mito-to-ER) and/or regulate the localization of seipin or PA biosynthetic enzymes. Indeed, we found that the MAM subdomains where ORP5 localize are enriched in PA lipid, providing the first evidence of the enrichment of PA at MAM. Such acute regulation of local PA levels at MAMs could be important for better control LD nucleation and growth within the MAM subdomains where ORP5 localizes, by fueling TG from the ER to the newly formed or pre-existing LDs (Fig. 10).
Finally, our EM analysis unveils the morphological features of the ER subdomains from which LDs emerge. In cells treated with OA, we observed electron-dense membrane structures, partially invaginated into the LDs, that link the LDs to the tubular ER elements and that are often found close to mitochondria. Although their origin was unknown, similar electron-dense structures were recently observed at LD–mitochondria contact sites (Ma et al., 2021). We reveal that these structures correspond to HRP-KDEL stained MAMs and are strongly decreased in ORP5-depleted cells. The decrease in the LD population originating from MAM is accompanied by an increase in the LD population still linked to the ER but not in direct contact with mitochondria.
To conclude, our study uncovers an unprecedented role of ORP5 and ORP8 in orchestrating LD biogenesis at MAMs. Our findings offer exciting perspectives in a more profound understanding of LDs formation in cells, lipodystrophies, and neuronal disorders. Indeed, ORP5/8, seipin, and MAMs, which we now establish to localize in the same subdomains, are critical players of cellular lipid and calcium homeostasis, dysregulated in the onset of the above disorders.
Material and methods
Cell culture and transfection
HeLa cells were cultured in DMEM (Life Technologies) containing GlutaMax (Life Technologies) and supplemented with 10% FBS (Life Technologies), 1% penicillin/streptomycin (Life Technologies), and 1% non-essential amino acids (Life Technologies) at 37°C and 5% CO2. For LD biogenesis experiments, HeLa cells were cultured in DMEM (Life Technologies) containing GlutaMax (Life Technologies) and supplemented with 5% lipoprotein-deficient serum FCS (Life Technologies) and 1% non-essential amino acids (Life Technologies) at 37°C and 5% CO2 for 72 h, and then treated with BODIPY 558/568 (FA568) or oleic acid in serum-depleted DMEM. For imaging, HeLa cells were seeded in 13-mm glass coverslips. HeLa cells were transfected with the indicated plasmids for 3 h in serum depleted medium (Opti-MEM; Thermo Fisher Scientific) using lipofectamine 2000 (Life Technologies) according to the manufacturer’s protocol. Cells were imaged 16–24 h post-transfection.
Human hepatocarcinoma cells, Huh7, were maintained in high glucose with stabilized glutamine and with sodium pyruvate Dulbecco’s Modified Eagle’s Medium (DMEM; Dutscher) supplemented with 10% heat-inactivated fetal bovine serum and 1% penicillin/streptomycin (GibcoBRL). The Huh7 cells were transfected with indicated plasmid using Polyethyleneimine HCl MAX (Polysciences) following the manufacturer’s instructions. For the swelling experiments, the Huh7 cells were first transfected with the plasmids and loaded with oleic acid for 24 h. Then, the culture media was next replaced by a hypotonic DMEM culture media and diluted 20 times by water. Cells were then imaged 5 min after the hypotonic medium addition.
siRNAs oligonucleotides
Transient ORP5, ORP8, and PTPIP51 knockdowns in HeLa cells were performed by transfection of siRNA oligos using oligofectamine (Life Technologies) for 5 h in serum depleted medium (Opti-MEM; Thermo Fisher Scientific), according to manufacturer’s protocol. Cells were imaged 48 h post-transfection.
Double-stranded siRNAs were derived from the following references described in Table 1.
Table 1.
siRNAs and corresponding references
| siRNA | Company, reference |
|---|---|
| OSBPL8 | Dharmacon, J-009508-06 (Galmes et al., 2016) |
| OSBPL8 | Dharmacon, J-009508-05 (Galmes et al., 2016) |
| OSBPL5 | Dharmacon, J-009274-10 (Galmes et al., 2016) |
| OSBPL5 | Dharmacon, J-009274-11 (Galmes et al., 2016) |
| PTPIP51 | Dharmacon, J-020973-10-0020 (Stoica et al., 2014) |
| Non-targeting | Dharmacon, D-001810-10 |
Plasmids and cDNA clones
EGFP-ORP5A, EGFP-ORP8, EGFP-ORP5ΔPH, EGFP-ORP5ΔTM, EGFP-ORD5, and EGFP-ORD8 were described in Galmes et al. (2016). mCh-Plin1 was described in Ajjaji et al. (2019) and YFP-Seipin was described in Santinho et al. (2020). GFP-Sec22b and RFP-Sec22b were described in Gallo et al. (2020). GFP-Sec61β and ssHRP-KDEL were kindly gifted by T. Rapoport (Harvard University, Cambridge, MA) and D. Cutler (Schikorski et al., 2007), respectively. Mito-BFP (plasmid #49151; Addgene; http://n2t.net/addgene:49151), Seipin-EGFP (plasmid #129719; Addgene; http://n2t.net/addgene:129719), mCherry-Sec61β (plasmid # 90994; Addgene; https://www.addgene.org/90994) and TOM20-mCherry (plasmid #55146; Addgene; http://n2t.net/addgene:55146) were purchased from Addgene.
Generation of the ORP5B variant by mutagenesis
The ORP5B natural variant (Du et al., 2020), partially depleted of its PH domain, was generated by site-directed mutagenesis (Quickchange II-XL; Stratagene). The oligos used were: EGFP-ORP5BΔ134-201_Fw: 5′-CCTTTGGGGCCCTTCAGGCTGTCAGCCA-3′; EGFP-ORP5BΔ134-201_Rv: 5′-TGGCTGACAGCCTGAAGGGCCCCAAAGG-3′.
Generation of the ORP5/8 deletion mutants by mutagenesis
The CC and ORD domains of ORP5 or the TM domain of ORP8 were deleted using site-directed mutagenesis (Quickchange II-XL; Stratagene) to generate EGFP-ORP5ΔCC and EGFP-ORP5ΔORD or EGFP-ORP8ΔTM.
The following primers were used:
EGFP-ORP5ΔORD_Fw: 5′-CAGAGGAGAACAAGAGTCTGGAGGACCACAGCCCCTGGGAC-3′; EGFP-ORP5ΔORD_Rv: 5′-GTCCCAGGGGCTGTGGTCCTCCAGACTCTTGTTCTCCTCTG-3′; EGFP-ORP5ΔCC 93-123_Fw: 5′-CCCACCGCCAGGCCCAGCGTGGTC-3′; EGFP-ORP5ΔCC 93-123_Rv: 5′-GACCACGCTGGGCCTGGCGGTGGG-3′; EGFP-ORP8ΔTM_Fw: 5′-TATTTTCTGCAACAAAAAGACTAGGGGCCCGGGATC-3′; EGFP-ORP8ΔTM_Rv: 5′-GATCCCGGGCCCCTAGTCTTTTTGTTGCAGAAAATA-3′. The EGFP-ORP5ΔCCΔTM and the EGFP-ORP5ΔPHΔTM were generated by deletion of the CC and the PH in the EGFP-ORP5ΔTM using site-directed mutagenesis (Quickchange II-XL; Stratagene). The oligos used to delete the CC are listed above and the oligos used to delete the PH domain were described in Galmes et al. (2016).
Cloning of the RNAi-resistant ORP5 variants
RNAi-resistant EGFP-ORP5A and EGFP-ORP5B were generated by introducing four silent point mutations in the region targeted by the two siRNA oligos (#10 and #11) by site-directed mutagenesis (Quickchange II-XL; Stratagene) and the following primers: RESCUE ORP5_siRNA10_Fw: 5′-GGGAAGGTCACCATCGAATGCGCGAAGAACAACTTCCAGGCC-3′; RESCUE ORP5_siRNA10_Rv: 5′-GGCCTGGAAGTTGTTCTTCGCGCATTCGATGGTGACCTTCCC-3′; RESCUE ORP5_siRNA11_Fw: 5′-GAAGCCCAAGGGAATCAAGAAACCCTACAACCCCATCCTGGGGG-3′; RESCUE ORP5_siRNA11_Rv: 5′-CCCCCAGGATGGGGTTGTAGGGTTTCTTGATTCCCTTGGGCTTC-3′.
The untagged ORP5ΔPH was generated from the RNAi-resistant EGFP-ORP5ΔPH by excision of the EGFP using the enzymes NheI and HindIII followed by Klenow polymerase treatment and ligation using a T4 DNA ligase.
Cloning of HA-ORP5A and HA-ORP5ΔPH
To generate HA-ORP5A, the PCR product, carrying the HA tag at the N-terminus of ORP5, was ligated between AgeI and XhoI in the pEGFP-C1 vector (Clontech) to replace the GFP- with the HA-tag. The oligos used are:
ORP5 HA Age1_Fw: 5′-GGCGGCACCGGTCGCCACCATGTACCCATACGATGTTCCAGATTACGCTATGAAGGAGGAGGCCTTCCTC-3′; ORP5 Xho1_Rv: 5′-GGCCTCGAGCTATTTGAGGATGTGGTTAATG-3′.
To generate the HA-ORP5ΔPH, the PH domain in HA-ORP5A was deleted by site-directed mutagenesis (Quickchange II-XL; Stratagene) as in Galmes et al. (2016).
Cloning of Opi1 pQ2S-PABD
Opi1 pQ2S-PABD probe was generated by inserting the fragment 113–168 from Opi1p (GeneBank M57383.1) using yeast DNA (gift from Dr. S. Friant, Université de Strasbourg, France) in BglII and EcoRI digested pEGFP-C1. Specificity for biding PA was established using a liposome biding assay as described in Kassas et al. (2017).
Cloning of RFP-ORP5∆TM and RFP-ORP5B
The insert RFP was recovered from a plasmid mTAG-RFP digested with the enzymes NheI (Fast Digest; Thermo Fisher Scientific)/BsGrI(Fast Digest; Thermo Fisher Scientific) or NheI/XhoI (Fast Digest; Thermo Fisher Scientific). Meanwhile, the plasmids EGFP-ORP∆TM and EGFP-ORP5B were respectively digested with the enzymes NheI/BsrGI and NheI/XhoI to remove the tag EGFP. The insert RFP was then ligated on the plasmid without the tag.
Antibodies, probes, and reagents
Primary antibodies used in this study were as follows: rabbit anti-ORP5 (HPA038712; Sigma-Aldrich), mouse anti-ORP8 (sc-134409; Santa Cruz), rabbit anti-PTPIP51 (RDM3, HPA009975; Sigma-Aldrich), mouse anti-PTPIP51 (FAM82C, SAB1407626; Sigma-Aldrich), mouse anti-GAPDH (GTX627408; Genetex), rabbit anti-GFP (A11122; Invitrogen), mouse anti-PDI (GTX30716; GeneTex), and mouse anti-HA (H3663; Sigma-Aldrich). Dilutions are detailed in Table 2.
Table 2.
Dilutions
| Antibodies | Application/dilution |
|---|---|
| Rabbit anti-ORP5 | WB 1:1,000 PLA 1:200 |
| Rabbit anti-ORP8 | WB 1:1,000 |
| Mouse anti-ORP8 | PLA 1:200 |
| Rabbit anti-PTPIP51 | PLA 1:200 WB 1:800 |
| Mouse anti-PTPIP51 | PLA 1:200 |
| Mouse anti-GAPDH | WB 1:10,000 |
| Rabbit anti-GFP | IEM 1:100 |
| Mouse anti-GFP | WB 1:1,000 |
| Mouse anti-HA | IEM 1:500 |
| Mouse anti-PDI | IEM 1:500 |
| Mitotracker | 1 μM |
| LipidTOXTM (LTox) | 10 μM |
| BODIPYTM 558/568 (FA568) | 1 μM |
| Oleic acid (OA) | 300 μM |
| LD450 | 10 μM |
Mitotracker red (Thermo Fisher Scientific) and LipidTox (LTox; Thermo Fisher Scientific) were used as probes for the mitochondrial network and the LDs, respectively, following the manufacturer’s instructions. Additionally, in LD biogenesis experiments, LDs were labeled by BODIPY 558/568 (FA568, fluorescent-tagged oleic acid; Thermo Fisher Scientific) or LD540 (Spandl et al., 2009). Oleic acid–albumin from bovine serum (OA, 03008; Sigma-Aldrich) was used to induce LD production.
Mitochondria and LD labeling
Labeling of mitochondrial network with Mitotracker was performed by incubating HeLa cells seeded in glass coverslips with a Mitotracker red 1 μM in Opti-MEM (Thermo Fisher Scientific) for 30 min at 37°C and 5% CO2. Cells fixation was carried out by incubation with 4% PFA/PBS for 30 min at room temperature. Coverslips were then washed in PBS and incubated with 50 mM NH4Cl for 15 min at room temperature. To label LDs, fixed cells were incubated with 10 μM LTox in 1xPBS for 30 min at room temperature. After washing with PBS, coverslips were mounted with Vectashield (Vectro Laboratories) on microscopy slides. Images were acquired on a confocal inverted microscope SP8-X (DMI 6000; Leica).
LD biogenesis analysis and oleic acid treatment
LD biogenesis was induced in delipidated HeLa cells grown on coverslips by treatment with FA568 or OA in serum depleted DMEM. For LD biogenesis time-course experiments and LD biogenesis rescue experiments, delipidated HeLa cells (control and knockdown for ORP5, ORP8 or PTPIP51) expressing Mito-BFP alone or Mito-BFP together with EGFP tagged-ORP5 or Sec61β were treated with 1 μM FA568 and then fixed in 4% PFA at different times. After fixation, LDs were stained with LTox for 30 min in PBS and mounted for observation. Additional LD biogenesis time-course experiments were performed in delipidated HeLa cells (control and knockdown for ORP5 or ORP8) treated with 300 μM OA and in which the mitochondrial network was labeled with Mitotracker. For all other experiments, non-delipidated HeLa cells were treated with 300 μM OA for 2 h before fixation.
Confocal microscopy
Images of immunostained cells or cells expressing fluorescent-tagged proteins were acquired on Confocal inverted microscope SP8-X (DMI 6000; Leica). Optical sections were acquired with a Plan Apo 63× oil immersion objective (N.A. 1.4; Leica) using the LAS-X software. Fluorescence was excited using either a 405-nm laser diode or a white light laser and later collected after adjusting the spectral windows with GaAsP PMTs or Hybrid detectors. Images from a mid-focal plane are shown. For Huh7 experiments, images were acquired by the ZEISS LSM800 Airyscan.
Live-cell imaging
HeLa cells were seeded on glass bottom ibidi chambers (µ-slide 2 wells) 2 d before imaging. The day after seeding, EGFP-ORP5B and Mito-BFP plasmids were transfected with lipofectamine 2000(Invitrogen), according to the manufacturer’s instructions. Cell imaging was performed on an inverted Nikon Ti Eclipse E microscope coupled with a Spinning Disk (CSU-X1-A1; Yokogawa) and cage incubator to control both temperature and CO2 (37°C, 5% CO2). After excitation with a 405 nm (Vortran, 100 mW), 491 nm (Vortran, 150 mW), and 561 nm laser (Coherent, 100 mW), fluorescence from the different fluorescent compounds was detected with a 40× oil immersion objective (PLAN FLUOR; NA: 1.30; Nikon) or a 60× oil immersion objective (APOTIRF; NA:1.49; Nikon), an emission filter Quad bandpass 440/40 nm, 521/20 nm, 607/34 nm, 700/45 nm (Semrock), and a Prime 95B sCMOS camera (Photometrics). Images were captured every 30 s for 15 min. Approximately 2 min after the start of captures, FA568 was added to a final concentration of 1 µM to induce the formation of lipid droplets. For live imaging, Huh7 cells were grown in MatTek 3.5 mm coverslip bottom dishes and imaged on the ZEISS LSM800 Airyscan microscope.
3D structured illumination microscopy (SIM)
Super-resolution light microscopy was performed on a Zeiss ELYRA PS.1 SIM microscope equipped with a Plan-Apochromat 63×/1.40 NA oil-immersion objective (Carl Zeiss). The illumination patterns of the 488, 561, and 642 nm lasers were projected into the sample. The emitted fluorescence light was detected with an EMCCD camera (iXon 885; Andor Technology). Five phase translations and three rotations of the illumination pattern were recorded at each z-plan, and image stacks (120-nm increment along z axis) were acquired. The 3D stacks were then computationally reconstructed with the ZEN imaging software package (algorithm of Heintzmann and Cremer) to generate super-resolution 3D SIM images with twofold extended resolution in the three axes (reconstructed image format = 1,904 × 1,900 pixels, representing voxels of 0.04 × 0.04 × 0.12 µm).
In situ proximity ligation assay (PLA)
The protein–protein interactions in fixed HeLa cells were assessed using in situ PLA (Duolink SIGMA) according to the manufacturer’s instructions. After fixation, HeLa cells co-expressing Mito-BFP and mCh-Plin1 were incubated with primary antibodies, mouse anti-ORP8 (1:200) plus rabbit anti-ORP5 (1:200), mouse anti-ORP8 (1:200) plus rabbit anti-PTPIP51 (1:200), or rabbit anti-ORP5 (1:200) plus mouse anti-PTPIP51 (1:200), in blocking solution (1% BSA, w/v 0.1% saponin, w/v, in PBS) for 1 h at room temperature. PLUS and MINUS PLA probes (anti-murine and anti-rabbit IgG antibodies conjugated with oligonucleotides, 1:5 in blocking solution) were then incubated with the samples for 1 h at 37°C. Coverslips were thereafter washed in 1× wash buffer A and incubated with ligation solution (5× Duolink Ligation buffer 1:5, ligase 1:40 in high purity water) for 30 min at 37°C. After the ligation step, the cell samples were washed in 1× wash buffer A and incubated with the polymerase solution (5× amplification buffer 1:5, polymerase 1:80 in high purity water) for 1 h 40 min at 37°C. Polymerase solution was washed out from the coverslips with 1× wash buffer B and 0.01× wash buffer B. Vectashield Mounting Medium (Vector Laboratories) was used for mounting.
Imaging quantifications
Colocalization analysis
For co-localization analysis of fluorescent signals, the acquired images were processed using the JACoP plugin in ImageJ to assess the Pearson’s correlation coefficient. The obtained values, ranging from 0 to 1 (1 = max correlation), indicated the association between the signals analyzed.
Organelles/structures count
LTox- and FA-positive LDs and ORP5 labeled-ER were counted in maximal projection confocal images using ImageJ software.
Nearest-neighbor distance analysis
The probability densities of distance distribution in Fig. S5 D were determined by the Mosaic Interaction analysis plug-in for Fiji (Shivanandan et al., 2013).
Imaris analysis
Quantification of PLA and seipin spots: To quantify PLA spots and categorize seipin spots in HeLa cells, confocal images were treated with the surface and spots function of the software IMARIS (Bitplane, v9.3.1). For PLA quantification, 3D images (PLA foci identified as “spots,” mitochondria identified as “surfaces”) were generated from confocal Z-stack images and the shortest distance between each spot center and the nearest point of the surface or cell object was calculated based on a 3D distance map. Spots objects (PLA dots) with a distance smaller than 380 nm from surfaces (mitochondria) objects were considered at a close proximity of these objects. The threshold of 380 nm was used as an estimation of the PLA reaction precision including both primary and secondary antibodies (30 nm) plus half the FWHM of the PLA amplification signals (350 nm). Similary, 3D segmented images of HeLa cells co-expressing seipin and Mito-BFP and stained with LTox were generated from z-stack confocal images (seipin, mitochondria, and LD identified as “surfaces”). Seipin 3D surfaces were then classified into four different categories (“seipin to MAM,” “seipin to LD,” “seipin to MAM-LD,” and “seipin to ER”) according to their proximity to the labeled compartments using a 3D distance map. The results as shown as the percentage of the category to the total number of seipin green surfaces.
Electron microscopy analysis
Conventional EM
For conventional EM, cells grown on 13 mm glass bottom coverslips (Agar Scientific) were fixed with 2.5% glutaraldehyde and 2% PFA in 0.1 M cacodylate and 0.05% CaCl2 buffer for 24 h. After several washes with 0.1 M cacodylate buffer, the cells were post-fixed with 1% OsO4 and 1.5% potassium ferricyanide in 0.1 M Cacodylate for 1 h. After several washes with 0.1 M cacodylate buffer and H2O, the cells were stained with 0.5% uranyl acetate for 24 h. After several washes with H2O, the cells were dehydrated in ethanol and embedded in Epon while on the coverslips. Ultrathin sections were prepared, counterstained with uranyl acetate, and observed under a 80 kV JEOL 1400 microscope equipped with a Orius High speed (Gatan) camera.
HRP detection
HeLa cells expressing HRP-KDEL were fixed on coverslips with 1.3% glutaraldehyde in 0.1 M cacodylate buffer, washed in 0.1 M ammonium phosphate (pH 7.4) buffer for 1 h, and the HRP was visualized with 0.5 mg/ml DAB and 0.005% H2O2 in 0.1 M ammonium phosphate (pH 7.4) buffer. Development of HRP (DAB dark reaction product) took between 5 min and 20 min and was stopped by extensive washes with cold water. Cells were post-fixed in 2% OsO4+1% K3Fe(CN)6 in 0.1 M cacodylate buffer at 4°C for 1 h, washed in cold water, and then contrasted in 0.5% uranyl acetate for 2 h at 4°C, dehydrated in an ethanol series and embedded in Epon as for conventional EM. Ultrathin sections were contrasted with 4% uranyl acetate and observed under a FEI Tecnai 12 microscope equipped with a OneView 4 k Gatan camera.
Serial sectioning and 3D reconstruction
Ultrathin serial sections (50 nm) of HRP–KDEL-transfected HeLa cells were cut and deposited on slot grids (EMS) with formvar (1%) and then contrasted with 4% uranyl acetate. 13 and 14 serial sections for ORP5wt and ORP5ΔPH, respectively, were collected and observed under a 80 kV JEOL 1400 microscope equipped with a Orius High speed (Gatan) camera. Image alignment was done with imod, and segmentation was done manually by using 3Dmod.
Immunogold labeling and quantifications
HeLa cells were fixed with a mixture of 2% PFA and 0.125% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) for 2 h and processed for ultracryomicrotomy, as described previously (Slot and Geuze, 2007). Ultrathin cryosections were single- or double-immunogold-labeled with antibodies and protein A coupled to 10 or 15 nm gold (CMC, UMC Utrecht, The Netherlands), as indicated in the legends to the figures. Immunogold-labeled cryosections were observed under a FEI Tecnai 12 microscope equipped with a OneView 4 k Gatan camera. For the quantification of the distribution of ORP5 or seipin immunogold labeling on ultrathin cryosections, gold particles (15 nm) were counted on acquired micrographs of randomly selected cell profiles (the number of cell profiles and the gold particle is indicated in the figure legends). All data are presented as mean (%) ± SEM of three technical replicates.
Co-immunoprecipitation of seipin-ORP5
HeLa cells co-transfected with either EGFP or EGFP-tagged seipin together with HA-ORP5A, HA-ORP5B, ORP5A, ORP5 ΔORD, or ORP5 ΔTM, were washed in cold PBS, and lysed on ice in lysis buffer (50 mM Tris, 120 mM NaCl, 40 mM Hepes, 0.5% digitonin, 0.5% CHAPS, pH 7.36, and protease inhibitor cocktail [Roche]). Cell lysates were then centrifuged at 21,000 g for 20 min at 4°C. Supernatants were then incubated with Chromotek GFP-trap agarose beads (Allele Biotech) for 1 h at 4°C under rotation. Subsequently, the beads were extensively washed in cold lysis buffer, and the immunoprecipitated proteins bound to the beads were incubated in sample buffer (containing 2% SDS), boiled for 1 min at 97°C, and separated in 10% SDS–PAGE gel for immunoblotting analysis.
Western blotting
For immunoblotting, cells were resuspended in lysis buffer (50 mM Tris, 150 mM NaCl, 1% Triton X-100, 10 mM EDTA, pH 7.2, and protease inhibitor cocktail [Roche]). Cell lysates were then centrifuged at 21,000 g for 20 min at 4°C. The supernatants were boiled in reducing SDS sample buffer. Proteins isolated from HeLa cells or obtained by immunoprecipitation were subjected to SDS-PAGE gels for electrophoresis separation. The separated proteins were transferred to 0.45 μm nitrocellulose membrane (GE Healthcare). The membrane was blocked by 5% non-fat milk in TBST buffer (TBS buffer with 0.1% Tween 20) for 1 h at room temperature and washed three times, for 5 min each, with TBST. Then the membrane was incubated with the primary antibodies (antibodies and dilutions listed in the table displayed in section “Antibodies, probes, and reagents”) at 4°C overnight. The membrane was washed three times with TBST, incubated with the peroxidase-conjugated anti-rabbit IgG secondary antibody (NA934V, 1:10,000; GE Healthcare, in 5% milk in TBST) or anti-mouse IgG secondary (NA931V, 1:10,000; GE Healthcare, in 5%milk in TBST) at room temperature for 1 h, followed with washing and detection using the enhanced chemiluminescence (ECL) detection kit (Cytiva). For Western blot quantification, bands of protein of interest were detected using ChemiDoc Imaging Systems (Life Science Research; Bio-Rad) and analyzed using Image Lab Software.
Interfacial tension measurements
Interfacial tension measurements were performed using a drop tensiometer device designed by Teclis Instruments (Tracker, Teclis-IT Concept, France) to measure the interfacial tension of oil—water interfaces. In our experiments, the pendant drop is the triolein lipid phase formed in the aqueous HKM buffer. The triolein—water interface stabilizes at ∼34 ± 1 mN/m. Adsorption of ORD5/8 translated into a decrease in tension as the protein got recruited to the oil–water interface. Throughout the adsorption kinetics to either a triolein—water interface, the drop area was constant.
PIP-strip
PIP-strip membranes (Echelon Biosciences) were blocked with 3% BSA FFA dissolved in phosphate-buffered saline (PBS) containing 0.1% Tween 20 (3% BSA FFA PBS-T) at room temperature for 60 min.
Blocked PIP Strips were incubated with the same buffer containing the ORP5HA-CC, ORP8HA-CC, or HA peptide (negative control) at the final concentration 0.35 µM for 1 h. The PIP strips were then washed, and the bound proteins were detected with rabbit anti HA antibody.
Statistical analysis
The Kolmogorov–Smirnov test was used for the tensiometer studies. The Wilcoxon–Mann–Whitney test was used for the quantification analysis of seipin by confocal imaging. The unpaired two-tailed t test was used for all the other experiments.
Online supplemental material
Fig. S1 contains additional information in support of Figs. 1 and 2, regarding the localization of ORP5 and ORP8 to MAM-LD contacts in both HeLa and Huh7 cells. Fig. S2 contains data in support of Fig. 2 regarding the effects of siORP5, siORP8 and siPTPIP51 in LD biogenesis. Fig. S3 (in support of Fig. 4) shows the recruitment of ORP5 to nascent and pre-existent LDs in Huh7 cells. Fig. S4 contains additional information in support of Figs. 6 and 7 regarding the localization of seipin to MAM-LD contacts where ORP5 localizes. Fig. S5 has additional data that support Fig. 9 showing the effects of ORP5 knockdown or overexpression on seipin localization. Video 1: LDs originate from MAM where ORP5 localizes (entire cell, cell 1). Video 2: LDs originate from MAM where ORP5 localizes (zoomed region of cell 1). Video 3: LDs originate from MAM where ORP5 localizes (zoomed region of cell 2).
Supplementary Material
is the source file for Fig. S2.
Acknowledgments
We thank Dr. Renaud Legouis, Dr. Emmanuel Culetto, and Dr. Etienne Morel for stimulating discussion. We are grateful to Romain Le Bars for his help on live-cell imaging and consultation on Duolink analysis. Our apologies to colleagues whose important work around this subject could not be cited due to space limitations.
The present work has benefited from Imagerie-Gif core facility supported by the Agence Nationale de la Recherche (ANR-11-EQPX-0029/Morphoscope, ANR-10-INBS-04/FranceBioImaging; ANR-11-IDEX-0003-02/Saclay Plant Sciences). For the immuno-EM, we acknowledge the ImagoSeine facility, a member of the France BioImaging infrastructure supported by grant ANR-10-INBS-04 from the French National Research Agency. This work was supported by the ANR Jeune Chercheur (ANR0015TD), the ATIP-Avenir Program (n°R16071LS-RSE17006LSA), the FSER (FSER 201903008955, FRM n°206548) and the Fondation Vaincre Alzheimer (CNRS LS 212527) to F. Giordano. A.R. Thiam was supported by ANR-21-CE13-0014-LIPDROPER, ANR-21-CE14-0024-MAMA, and ANR-17-CE11-0003-NANODROP.
The authors declare no competing financial interests.
Author contributions: F. Giordano and A.R. Thiam designed the work. V. Guyard, VC and M. Omrane performed and analyzed most of the confocal imaging experiments. V. Guyard and M. Omrane performed the live cell imaging experiments. VC performed and analyzed the LD biogenesis experiments and the PLA. V. Guyard performed the seipin localization analysis by Imaris. M. Omrane performed the imaging on Huh7 and the PIP strip experiments. C. Sauvanet performed and analyzed some of the LD biogenesis experiments. A. Houcine, V. Guyard and VC generated ORP5 mutants and performed confocal analysis of their localization. A. Houcine and F. Giordano performed and analyzed the electron microscopy experiments. C. Boulogne performed the serial sectioning-3D EM. O. Faklaris performed the structured illuminated microscopy. N. Vitale generated and provided tools for PA imaging analysis. N.E. Khallouki assisted in Western blot analysis, electron microscopy and generated some of the constructs for mammalian cell expression. F. Giordano and A.R. Thiam wrote the manuscript and all authors contributed to improve the manuscript.
References
- Adeyo, O., Horn P.J., Lee S., Binns D.D., Chandrahas A., Chapman K.D., and Goodman J.M.. 2011. The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J. Cell Biol. 192:1043–1055. 10.1083/jcb.201010111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajjaji, D., Ben M’barek K., Mimmack M.L., England C., Herscovitz H., Dong L., Kay R.G., Patel S., Saudek V., Small D.M., et al. 2019. Dual binding motifs underpin the hierarchical association of perilipins1-3 with lipid droplets. Mol. Biol. Cell. 30:703–716. 10.1091/mbc.E18-08-0534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben M’barek, K., Ajjaji D., Chorlay A., Vanni S., Forêt L., and Thiam A.R.. 2017. ER membrane phospholipids and surface tension control cellular lipid droplet formation. Dev. Cell. 41:591–604.e7. 10.1016/j.devcel.2017.05.012 [DOI] [PubMed] [Google Scholar]
- Benador, I.Y., Veliova M., Mahdaviani K., Petcherski A., Wikstrom J.D., Assali E.A., Acín-Pérez R., Shum M., Oliveira M.F., Cinti S., et al. 2018. Mitochondria bound to lipid droplets have unique bioenergetics, composition, and dynamics that support lipid droplet expansion. Cell Metab. 27:869–885.e6. 10.1016/j.cmet.2018.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi, J., Wang W., Liu Z., Huang X., Jiang Q., Liu G., Wang Y., and Huang X.. 2014. Seipin promotes adipose tissue fat storage through the ER Ca²⁺-ATPase SERCA. Cell Metab. 19:861–871. 10.1016/j.cmet.2014.03.028 [DOI] [PubMed] [Google Scholar]
- Choudhary, V., El Atab O., Mizzon G., Prinz W.A., and Schneiter R.. 2020. Seipin and Nem1 establish discrete ER subdomains to initiate yeast lipid droplet biogenesis. J. Cell Biol. 219:e201910177. 10.1083/jcb.201910177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, J., Torta F., Masai K., Lucast L., Czapla H., Tanner L.B., Narayanaswamy P., Wenk M.R., Nakatsu F., and De Camilli P.. 2015. PI4P/phosphatidylserine countertransport at ORP5-and ORP8-mediated ER–plasma membrane contacts. Science. 349:428–432. 10.1126/science.aab1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, J., Wu X., Lambert T.J., Lai Z.W., Walther T.C., and Farese R.V. Jr. 2019. LDAF1 and Seipin Form a Lipid Droplet Assembly Complex. Dev. Cell. 51:551–563.e7. 10.1016/j.devcel.2019.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combot, Y., Salo V.T., Chadeuf G., Holtta M., Ven K., Pulli I., Ducheix S., Pecqueur C., Renoult O., Lak B., et al. 2022. Seipin localizes at endoplasmic-reticulum-mitochondria contact sites to control mitochondrial calcium import and metabolism in adipocytes. Cell Rep. 38:110213. 10.1016/j.celrep.2021.110213 [DOI] [PubMed] [Google Scholar]
- Dickson, E.J., and Hille B.. 2019. Understanding phosphoinositides: Rare, dynamic, and essential membrane phospholipids. Biochem. J. 476:1–23. 10.1042/BCJ20180022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, X., Kumar J., Ferguson C., Schulz T.A., Ong Y.S., Hong W., Prinz W.A., Parton R.G., Brown A.J., and Yang H.. 2011. A role for oxysterol-binding protein-related protein 5 in endosomal cholesterol trafficking. J. Cell Biol. 192:121–135. 10.1083/jcb.201004142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, X., Zhou L., Aw Y.C., Mak H.Y., Xu Y., Rae J., Wang W., Zadoorian A., Hancock S.E., Osborne B., et al. 2020. ORP5 localizes to ER-lipid droplet contacts and regulates the level of PI(4)P on lipid droplets. J. Cell Biol. 219:e201905162. 10.1083/jcb.201905162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei, W., Shui G., Gaeta B., Du X., Kuerschner L., Li P., Brown A.J., Wenk M.R., Parton R.G., and Yang H.. 2008. Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J. Cell Biol. 180:473–482. 10.1083/jcb.200711136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei, W., Shui G., Zhang Y., Krahmer N., Ferguson C., Kapterian T.S., Lin R.C., Dawes I.W., Brown A.J., Li P., et al. 2011. A role for phosphatidic acid in the formation of “supersized” lipid droplets. PLoS Genet. 7:e1002201. 10.1371/journal.pgen.1002201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyre, C.A.C., Rauher P.C., Ejsing C.S., and Klemm R.W.. 2019. MIGA2 links mitochondria, the ER, and lipid droplets and promotes de novo lipogenesis in adipocytes. Mol. Cell. 76:811–825.e14. 10.1016/j.molcel.2019.09.011 [DOI] [PubMed] [Google Scholar]
- Gallo, A., Danglot L., Giordano F., Hewlett B., Binz T., Vannier C., and Galli T.. 2020. Role of the Sec22b-E-Syt complex in neurite growth and ramification. J. Cell Sci. 133:jcs247148. 10.1242/jcs.247148 [DOI] [PubMed] [Google Scholar]
- Galmes, R., Houcine A., van Vliet A.R., Agostinis P., Jackson C.L., and Giordano F.. 2016. ORP5/ORP8 localize to endoplasmic reticulum–mitochondria contacts and are involved in mitochondrial function. EMBO Rep. 17:800–810. 10.15252/embr.201541108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, M., Huang X., Song B.-L., and Yang H.. 2019. The biogenesis of lipid droplets: Lipids take center stage. Prog. Lipid Res. 75:100989. 10.1016/j.plipres.2019.100989 [DOI] [PubMed] [Google Scholar]
- Ghai, R., Du X., Wang H., Dong J., Ferguson C., Brown A.J., Parton R.G., Wu J.-W., and Yang H.. 2017. ORP5 and ORP8 bind phosphatidylinositol-4, 5-biphosphate (PtdIns [4, 5] P 2) and regulate its level at the plasma membrane. Nat. Commun. 8:757. 10.1038/s41467-017-00861-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluchowski, N.L., Becuwe M., Walther T.C., and Farese R.V. Jr. 2017. Lipid droplets and liver disease: From basic biology to clinical implications. Nat. Rev. Gastroenterol. Hepatol. 14:343–355. 10.1038/nrgastro.2017.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grippa, A., Buxó L., Mora G., Funaya C., Idrissi F.-Z., Mancuso F., Gomez R., Muntanyà J., Sabidó E., and Carvalho P.. 2015. The seipin complex Fld1/Ldb16 stabilizes ER–lipid droplet contact sites. J. Cell Biol. 211:829–844. 10.1083/jcb.201502070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, S., Binns D.D., Chang Y.-F., and Goodman J.M.. 2015. Dissecting seipin function: The localized accumulation of phosphatidic acid at ER/LD junctions in the absence of seipin is suppressed by Sei1p ΔNterm only in combination with Ldb16p. BMC Cell Biol. 16:29. 10.1186/s12860-015-0075-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariri, H., Rogers S., Ugrankar R., Liu Y.L., Feathers J.R., and Henne W.M.. 2018. Lipid droplet biogenesis is spatially coordinated at ER–vacuole contacts under nutritional stress. EMBO Rep. 19:57–72. 10.15252/embr.201744815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herker, E., Vieyres G., Beller M., Krahmer N., and Bohnert M.. 2021. Lipid droplet contact sites in health and disease. Trends Cell Biol. 31:345–358. 10.1016/j.tcb.2021.01.004 [DOI] [PubMed] [Google Scholar]
- Herms, A., Bosch M., Reddy B.J., Schieber N.L., Fajardo A., Rupérez C., Fernández-Vidal A., Ferguson C., Rentero C., Tebar F., et al. 2015. AMPK activation promotes lipid droplet dispersion on detyrosinated microtubules to increase mitochondrial fatty acid oxidation. Nat. Commun. 6:7176. 10.1038/ncomms8176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynynen, R., Suchanek M., Spandl J., Back N., Thiele C., and Olkkonen V.M.. 2009. OSBP-related protein 2 is a sterol receptor on lipid droplets that regulates the metabolism of neutral lipids. J. Lipid Res. 50:1305–1315. 10.1194/jlr.M800661-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, M., Ohsaki Y., Rega L.R., Bittman R., Olkkonen V.M., and Ikonen E.. 2011. Role of ORPs in sterol transport from plasma membrane to ER and lipid droplets in mammalian cells. Traffic. 12:218–231. 10.1111/j.1600-0854.2010.01142.x [DOI] [PubMed] [Google Scholar]
- Joshi, A.S., Nebenfuehr B., Choudhary V., Satpute-Krishnan P., Levine T.P., Golden A., and Prinz W.A.. 2018. Lipid droplet and peroxisome biogenesis occur at the same ER subdomains. Nat. Commun. 9:2940. 10.1038/s41467-018-05277-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi, A.S., Ragusa J.V., Prinz W.A., and Cohen S.. 2021. Multiple C2 domain–containing transmembrane proteins promote lipid droplet biogenesis and growth at specialized endoplasmic reticulum subdomains. Mol. Biol. Cell. 32:1147–1157. 10.1091/mbc.E20-09-0590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassas, N., Tanguy E., Thahouly T., Fouillen L., Heintz D., Chasserot-Golaz S., Bader M.F., Grant N.J., and Vitale N.. 2017. Comparative characterization of phosphatidic acid sensors and their localization during frustrated phagocytosis. J. Biol. Chem. 292:4266–4279. 10.1074/jbc.M116.742346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaldoun, S.A., Emond-Boisjoly M.A., Chateau D., Carriere V., Lacasa M., Rousset M., Demignot S., and Morel E.. 2014. Autophagosomes contribute to intracellular lipid distribution in enterocytes. Mol. Biol. Cell. 25:118–132. 10.1091/mbc.E13-06-0324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, C., Sengupta P., Seo A.Y., and Lippincott-Schwartz J.. 2020. ER membranes exhibit phase behavior at sites of organelle contact. Proc. Natl. Acad. Sci. USA. 117:7225–7235. 10.1073/pnas.1910854117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug, Y.A., Deme J.C., Corey R.A., Renne M.F., Stansfeld P.J., Lea S.M., and Carvalho P.. 2021. Mechanism of lipid droplet formation by the yeast Sei1/Ldb16 Seipin complex. Nat. Commun. 12:5892. 10.1038/s41467-021-26162-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X., Qian H., Chen A., Ni H.-M., and Ding W.-X.. 2021. Perspectives on mitochondria–ER and mitochondria–lipid droplet contact in hepatocytes and hepatic lipid metabolism. Cells. 10:2273. 10.3390/cells10092273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magré, J., Delépine M., Khallouf E., Gedde-Dahl T. Jr, Van Maldergem L., Sobel E., Papp J., Meier M., Mégarbané A., Bachy A., et al. 2001. Identification of the gene altered in Berardinelli–Seip congenital lipodystrophy on chromosome 11q13. Nat. Genet. 28:365–370. 10.1038/ng585 [DOI] [PubMed] [Google Scholar]
- Monteiro-Cardoso, VF, Rochin L., Arora A, Houcine A, Jääskeläinen E, Kivelä AM, Sauvanet C, Le Bars R, Marien E, Dehairs J, et al. 2022. ORP5/8 and MIB/MICOS Link ER-mitochondria and intermitochondrial contacts for non-vesicular transport of phosphatidylserine. BioRxiv. 10.1101/695577 [DOI] [PubMed] [Google Scholar]
- Nguyen, T.B., Louie S.M., Daniele J.R., Tran Q., Dillin A., Zoncu R., Nomura D.K., and Olzmann J.A.. 2017. DGAT1-dependent lipid droplet biogenesis protects mitochondrial function during starvation-induced autophagy. Dev. Cell. 42:9–21.e5. 10.1016/j.devcel.2017.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann, J.A., and Carvalho P.. 2019. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 20:137–155. 10.1038/s41580-018-0085-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagac, M., Cooper D.E., Qi Y., Lukmantara I.E., Mak H.Y., Wu Z., Tian Y., Liu Z., Lei M., Du X., et al. 2016. SEIPIN regulates lipid droplet expansion and adipocyte development by modulating the activity of glycerol-3-phosphate acyltransferase. Cell Rep. 17:1546–1559. 10.1016/j.celrep.2016.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambold, A.S., Cohen S., and Lippincott-Schwartz J.. 2015. Fatty acid trafficking in starved cells: Regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev. Cell. 32:678–692. 10.1016/j.devcel.2015.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, M.J., and Goodman J.M.. 2021. Seipin: Harvesting fat and keeping adipocytes healthy. Trends Cell Biol. 31:912–923. 10.1016/j.tcb.2021.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salo, V.T., and Ikonen E.. 2019. Moving out but keeping in touch: Contacts between endoplasmic reticulum and lipid droplets. Curr. Opin. Cell Biol. 57:64–70. 10.1016/j.ceb.2018.11.002 [DOI] [PubMed] [Google Scholar]
- Salo, V.T., Li S., Vihinen H., Hölttä-Vuori M., Szkalisity A., Horvath P., Belevich I., Peränen J., Thiele C., Somerharju P., et al. 2019. Seipin facilitates triglyceride flow to lipid droplet and counteracts droplet ripening via endoplasmic reticulum contact. Dev. Cell. 50:478–493.e9. 10.1016/j.devcel.2019.05.016 [DOI] [PubMed] [Google Scholar]
- Santinho, A., Salo V.T., Chorlay A., Li S., Zhou X., Omrane M., Ikonen E., and Thiam A.R.. 2020. Membrane curvature catalyzes lipid droplet assembly. Curr. Biol. 30:2481–2494.e6. 10.1016/j.cub.2020.04.066 [DOI] [PubMed] [Google Scholar]
- Schikorski, T., Young S.M. Jr., and Hu Y.. 2007. Horseradish peroxidase cDNA as a marker for electron microscopy in neurons. J. Neurosci. Methods. 165:210–215. 10.1016/j.jneumeth.2007.06.004 [DOI] [PubMed] [Google Scholar]
- Schuldiner, M., and Bohnert M.. 2017. A different kind of love: Lipid droplet contact sites. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 1862:1188–1196. 10.1016/j.bbalip.2017.06.005 [DOI] [PubMed] [Google Scholar]
- Shivanandan, A., Radenovic A., and Sbalzarini I.F.. 2013. MosaicIA: An ImageJ/Fiji plugin for spatial pattern and interaction analysis. BMC Bioinformatics. 14:349. 10.1186/1471-2105-14-349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slot, J.W., and Geuze H.J.. 2007. Cryosectioning and immunolabeling. Nat. Protoc. 2:2480–2491. 10.1038/nprot.2007.365 [DOI] [PubMed] [Google Scholar]
- Sołtysik, K., Ohsaki Y., Tatematsu T., Cheng J., Maeda A., Morita S.Y., and Fujimoto T.. 2021. Nuclear lipid droplets form in the inner nuclear membrane in a seipin-independent manner. J. Cell Biol. 220:e202005026. 10.1083/jcb.202005026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spandl, J., White D.J., Peychl J., and Thiele C.. 2009. Live cell multicolor imaging of lipid droplets with a new dye, LD540. Traffic. 10:1579–1584. 10.1111/j.1600-0854.2009.00980.x [DOI] [PubMed] [Google Scholar]
- Stoica, R., De Vos K.J., Paillusson S., Mueller S., Sancho R.M., Lau K.F., Vizcay-Barrena G., Lin W.L., Xu Y.F., Lewis J., et al. 2014. ER-mitochondria associations are regulated by the VAPB-PTPIP51 interaction and are disrupted by ALS/FTD-associated TDP-43. Nat. Commun. 5:3996. 10.1038/ncomms4996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, S.J., Levin M.C., Zhou P., Han J., Walther T.C., and Farese R.V.. 2009. The endoplasmic reticulum enzyme DGAT2 is found in mitochondria-associated membranes and has a mitochondrial targeting signal that promotes its association with mitochondria. J. Biol. Chem. 284:5352–5361. 10.1074/jbc.M805768200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui, X., Arlt H., Brock K.P., Lai Z.W., DiMaio F., Marks D.S., Liao M., Farese R.V., and Walther T.C.. 2018. Cryo–electron microscopy structure of the lipid droplet–formation protein seipin. J. Cell Biol. 217:4080–4091. 10.1083/jcb.201809067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski, K.M., Binns D., Bartz R., Grishin N.V., Li W.-P., Agarwal A.K., Garg A., Anderson R.G., and Goodman J.M.. 2007. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl. Acad. Sci. USA. 104:20890–20895. 10.1073/pnas.0704154104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiam, A.R., Farese R.V. Jr., and Walther T.C.. 2013. The biophysics and cell biology of lipid droplets. Nat. Rev. Mol. Cell Biol. 14:775–786. 10.1038/nrm3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiam, A.R., and Forêt L.. 2016. The physics of lipid droplet nucleation, growth and budding. Biochim. Biophys. Acta. 1861:715–722. 10.1016/j.bbalip.2016.04.018 [DOI] [PubMed] [Google Scholar]
- Thiam, A.R., and Ikonen E.. 2021. Lipid droplet nucleation. Trends Cell Biol. 31:108–118. 10.1016/j.tcb.2020.11.006 [DOI] [PubMed] [Google Scholar]
- Veliova, M., Petcherski A., Liesa M., and Shirihai O.S.. 2020. The biology of lipid droplet-bound mitochondria. Semin. Cell Dev. Biol. 108. 55–64. 10.1016/j.semcdb.2020.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Sreenivasan U., Hu H., Saladino A., Polster B.M., Lund L.M., Gong D.W., Stanley W.C., and Sztalryd C.. 2011. Perilipin 5, a lipid droplet-associated protein, provides physical and metabolic linkage to mitochondria. J. Lipid Res. 52:2159–2168. 10.1194/jlr.M017939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Becuwe M., Housden B.E., Chitraju C., Porras A.J., Graham M.M., Liu X.N., Thiam A.R., Savage D.B., Agarwal A.K., et al. 2016. Seipin is required for converting nascent to mature lipid droplets. Elife. 5:e16582. 10.7554/eLife.16582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S., Idrissi F.-Z., Hermansson M., Grippa A., Ejsing C.S., and Carvalho P.. 2018. Seipin and the membrane-shaping protein Pex30 cooperate in organelle budding from the endoplasmic reticulum. Nat. Commun. 9:2939. 10.1038/s41467-018-05278-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte, M.A., and Gould A.P.. 2017. Lipid droplet functions beyond energy storage. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 1862:1260–1272. 10.1016/j.bbalip.2017.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windpassinger, C., Auer-Grumbach M., Irobi J., Patel H., Petek E., Hörl G., Malli R., Reed J.A., Dierick I., Verpoorten N., et al. 2004. Heterozygous missense mutations in BSCL2 are associated with distal hereditary motor neuropathy and Silver syndrome. Nat. Genet. 36:271–276. 10.1038/ng1313 [DOI] [PubMed] [Google Scholar]
- Wolinski, H., Hofbauer H.F., Hellauer K., Cristobal-Sarramian A., Kolb D., Radulovic M., Knittelfelder O.L., Rechberger G.N., and Kohlwein S.D.. 2015. Seipin is involved in the regulation of phosphatidic acid metabolism at a subdomain of the nuclear envelope in yeast. Biochim. Biophys. Acta. 1851:1450–1464. 10.1016/j.bbalip.2015.08.003 [DOI] [PubMed] [Google Scholar]
- Yan, R., Qian H., Lukmantara I., Gao M., Du X., Yan N., and Yang H.. 2018. Human SEIPIN binds anionic phospholipids. Dev. Cell. 47:248–256.e4. 10.1016/j.devcel.2018.09.010 [DOI] [PubMed] [Google Scholar]
- Zoni, V., Khaddaj R., Campomanes P., Thiam A.R., Schneiter R., and Vanni S.. 2021. Pre-existing bilayer stresses modulate triglyceride accumulation in the ER versus lipid droplets. Elife. 10:e62886. 10.7554/eLife.62886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouiouich, M., Di Mattia T., Martinet A., Eichler J., Wendling C., Tomishige N., Grandgirard E., Fuggetta N., Fromental-Ramain C., Mizzon G., et al. 2022. MOSPD2 is an endoplasmic reticulum-lipid droplet tether functioning in LD homeostasis. J. Cell Biol. 221:e202110044. 10.1083/jcb.202110044 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
is the source file for Fig. S2.