Abstract
Strain PS of Methanococcus voltae (a methanogenic, anaerobic archaebacterium) was shown to generate spontaneously 4.4-kbp chromosomal DNA fragments that are fully protected from DNase and that, upon contact with a cell, transform it genetically. This activity, here called VTA (voltae transfer agent), affects all markers tested: three different auxotrophies (histidine, purine, and cobalamin) and resistance to BES (2-bromoethanesulfonate, an inhibitor of methanogenesis). VTA was most effectively prepared by culture filtration. This process disrupted a fraction of the M. voltae cells (which have only an S-layer covering their cytoplasmic membrane). VTA was rapidly inactivated upon storage. VTA particles were present in cultures at concentrations of approximately two per cell. Gene transfer activity varied from a minimum of 2 × 10−5 (BES resistance) to a maximum of 10−3 (histidine independence) per donor cell. Very little VTA was found free in culture supernatants. The phenomenon is functionally similar to generalized transduction, but there is no evidence, for the time being, of intrinsically viral (i.e., containing a complete viral genome) particles. Consideration of VTA DNA size makes the existence of such viral particles unlikely. If they exist, they must be relatively few in number;perhaps they differ from VTA particles in size and other properties and thus escaped detection. Digestion of VTA DNA with the AluI restriction enzyme suggests that it is a random sample of the bacterial DNA, except for a 0.9-kbp sequence which is amplified relative to the rest of the bacterial chromosome. A VTA-sized DNA fraction was demonstrated in a few other isolates of M. voltae.
Much progress has been made in the last 20 years in the understanding of the biochemistry of bacterial methanogenesis. The recognition of methanogens as archaebacteria (16) made them very interesting from a broader biological perspective as well. A recent comprehensive review (44) covers the molecular biology of methanogens. Unfortunately, their slow growth and strict anaerobiosis have tended to discourage experimental approaches that require much culture manipulation. Among methanogens, Methanobacterium and Methanococcus are the two best-studied genera. They differ in several respects, the most striking being the absence of a true cell wall in Methanococcus which, like numerous other archaebacteria, has nothing but a thin S-layer covering the plasma membrane. Concerning Methanococcus voltae, the object of the present work, a comprehensive review up to 1989 is available (29). Later developments may be traced from recent publications (2, 27, 32) (see also references 8 and 58 for the related M. maripaludis).
Strain PS of M. voltae has been known to undergo low-frequency, natural DNA transformation (6). Another, more efficient natural gene transfer mechanism, discovered several years ago (4, 5), is examined in this study. The agent responsible for this activity is referred to here as VTA (voltae transfer agent). The phenomenon may be described as generalized transduction in which, however, the bacteriophage component has not been detected and is probably absent, defective, or different in size from the transducing particles or, in any case, is present in very small amounts.
A variety of defective lysogenic systems among eubacteria have been described. Of particular interest in connection with VTA are systems that accomplish transduction in the absence of “true,” i.e., lytically self-reproducing, viral particles, as first found in Rhodopseudomonas capsulata (now Rhodobacter capsulatus) (37, 66) and later found in Desulfovibrio desulfuricans (43), Serpulina hyodysenteriae (24), and possibly Myxococcus xanthus (53). The Rhodobacter example, for which the term “capsduction” has been introduced, is particularly interesting because of the small size of the bacterial DNA fragments transferred, almost equal to that observed here for VTA. Also, capsduction has recently found application as a tool in the analysis of the Rhodobacter genome sequence (33). A more thorough comparison of VTA with the above-mentioned systems can be found in the Discussion.
The only other example of generalized transduction among archaebacteria has been reported for Methanobacterium thermoautotrophicum (39) and involves a typical, medium-sized bacteriophage.
MATERIALS AND METHODS
Anaerobic techniques, storage of bacterial strains, medium composition, bacterial DNA extraction, and optical density (OD) measurements are described elsewhere (6). Whenever a method evolved over time in the course of the work, only the most satisfactory version is described.
Bacterial strains.
Most bacterial strains were derivatives of strain PS of M. voltae and are listed in Table 1. M. voltae C1, C2, C4, and A3 (62, 64) were obtained from W. B. Whitman, University of Georgia, Athens; M. vannielii (52) was obtained from J. N. Reeve, Ohio State University, Columbus.
TABLE 1.
Derivatives of M. voltae PS
| Strain | DSMa | Genotypeb | Source or reference |
|---|---|---|---|
| PS-1 | 1537 | Wild type | 61 |
| PS-2 | bes | 6 | |
| PS-3 | 4254 | his | 6 |
| PS-5 | his mtp | 6 | |
| PS-6 | 4310 | bes pur cbl | 6 |
| PS-9 | bes cbl | 6 | |
| PS-12 | Subclone of PS-1 (6) | ||
| PS-13 | bes pur cbl x? | From PS-6 (see Materials and Methods, “Resistance to BES”) | |
| PS-15 | bes pur | From experiment J2 in Table 2 |
DSM, Deutsche Sammlung von Mikroorganismen, Braunschweig, Germany.
cbl, his, and pur, requirement for vitamin B12, histidine, and purine, respectively; bes and mtp, resistance to BES and 5-methyl-tryptophan, respectively.
Bacteriophages.
φX174 (50) and P2 lg del1 del2 (7) were used as markers in sedimentation. For mixtures of the two phages, Escherichia coli C-1172, a P2-resistant mutant of C-1055 (63), was used to detect φX174, and E. coli K-221 (23) was used to detect P2.
Cultures.
M. voltae cultures (5 ml in Bellco anaerobic culture tubes or 25 ml in 125-ml serum bottles) were grown in WM medium plus needed supplements at 30 to 33°C as described previously (6). They were used for experiments at an OD at 600 nm (OD600) of approximately 0.3 to 0.7. In calculations, a quantity of 3.5 × 107 total bacteria per ml for an OD600 of 0.1 was assumed. For the strains most frequently used, fully grown cultures were stored at room temperature in an anaerobic hood for several weeks, repressurized occasionally with H2–CO2 to delay lysis (6), and used as inocula (diluted 1:30 to 1:300) for new cultures. Stock cultures of each strain were stored frozen and retrieved when needed as described previously (6). Viable, easily accessible inocula have been successfully maintained by this method for more than 12 years.
“Tumbler” cultures (265 ml in 500-ml serum bottles) were grown at 18 lb/in2 from a large (1/20 volume) inoculum of exponentially growing bacteria and were used at an OD600 of 0.1 to 0.4. To avoid foaming with minimum stress to the cells, gas exchange was facilitated by a procedure in which bottles that were lying down were gently rolled back and forth, hitting the walls of a container about four times per minute (“tumbling”).
Media.
WM medium (6, 61) was generally used, but without resazurin. l-Histidine (final concentration, 0.5 mM), hypoxanthine (final concentration, 0.2 mM), and cyanocobalamin (final concentration, 100 nM) were added at inoculation as needed for the growth of strains carrying the mutation his, pur, or cbl (6). 2-Bromoethanesulfonate (Na salt) (BES) was added for some selections, but BES-resistant strains were generally grown without it. For frozen storage and recovery of cultures and, on occasion, to facilitate growth from old inocula or in the large tumbler cultures, the medium was supplemented with 1/20 to 1/200 volume of a concentrated solution of Difco Casamino Acids, yeast extract, and l-tryptophan (10, 5, and 0.1% [wt/vol], respectively).
Viable counts.
Viable counts were obtained by the thin-layer, soft-agar, pour plate technique (6), which does not require preparing plates ahead of time. The agar concentration was critical; satisfactory results were obtained with 0.5% Difco Noble agar as well as with 0.34% Fisher Scientific laboratory-grade agar (6). Before inoculation, each tube of melted soft agar (already containing added sulfide solution and any other needed supplements) was removed from the 45 to 48°C heating block so it could cool down for 3 to 4 min. The plates were incubated for 10 to 12 days at 30 to 33°C. At variance from the method used earlier (6), no CaCl2 was added to the canister. Also, instead of H2S gas, an open container with a few milliliters of a 20% Na2S solution was placed at the bottom of the canister (9). Colony size was strongly affected by crowding; when present in large numbers, colonies were quite small and, except for a few spreaders, did not merge with each other. For some crowded plates, colony counts were obtained under a microscope at an appropriate magnification for a number of randomly chosen fields on the plate, averaged, and multiplied by the known plate/field area ratio.
Resistance to BES.
M. voltae PS is sensitive to BES; a stationary-phase inoculum grew with a 1-day lag in a liquid culture with 1 μM BES. At 10 and 100 μM, the growth rate was reduced four- and eightfold, respectively, and there was no growth at 1 mM BES. When samples were plated in agar with 1 mM BES, the efficiency of colony formation was usually well below 10−5.
The BES-resistant strain PS-6 grew normally in a liquid culture with 1 mM BES and with a small delay with 2 mM BES. It did not grow at 10 mM BES. In agar, the efficiency of colony formation by stationary-phase inocula was only slightly affected at 1 mM BES but was greatly reduced (<10−3) at concentrations higher than 5 mM BES.
A highly resistant strain (PS-13) was isolated as follows. Several 0.5-ml aliquots of a PS-6 culture, fully grown in the presence of 3 mM BES, were UV irradiated (6) (avoiding photoreactivation) with a dose expected to inactivate 96 to 99% of the cells, added to 5 ml of WM medium with the required supplements and 2 mM BES, and incubated to turbidity. From each culture, 1 ml was plated with 7.5 or 9 mM BES. Very variable numbers of colonies were obtained. In some cases the colonies were obviously heterogeneous in morphology, as though mutants were present. Single colonies were picked and tested for growth in liquid cultures with 9 mM BES. PS-13 is one of such isolates that, unlike its parent strain, grew satisfactorily with 9 mM BES and formed colonies with a thin halo of less compact growth. It was reisolated in the absence of BES and retested. It grew well in a liquid culture with 10 mM BES, although with a 1- to 2-day lag; it also grew, although more slowly, with 15 mM BES. However, when the resistance to BES of PS-13 was compared to that of the parent strain by plating for colonies in BES-containing agar (with stationary-phase inocula), no convincing difference in resistance levels was noted. This finding has not been investigated further.
VTA assays.
VTA assays were done in essentially the same way as viable-count assays. In early experiments, recipient bacteria and filtrate samples were introduced directly into the melted-soft-agar tube by use of disposable syringes and Becton-Dickinson 22-gauge needles and by measurement of volumes in drops (holding the syringe vertically gives approximately 14 μl per drop). Later (this is now the preferred method), a measured volume of donor filtrate (between 50 and 200 μl, diluted if necessary) was placed in a 1.5-ml Eppendorf disposable vial, which was kept on ice in air and transferred to an anaerobic hood minutes before plating. A drop of a fresh culture of recipient bacteria (OD600, 0.4 to 0.7) was delivered by needle and syringe to a (10-ml) melted-soft-agar tube through the stopper. Immediately afterward, by use of a new needle and syringe, about 0.5 ml was removed from the tube and gently blown into the sample in the vial. The whole sample was sucked up again into the syringe and delivered through the stopper to the tube. To completely deliver the sample, the syringe was rinsed out into the tube by filling it with melted agar and emptying it once more. The stopper was removed with forceps, and the agar was poured into a petri dish. Recipient cultures, kept at room temperature and repressurized with H2–CO2, were sometimes used again in new platings; after a few days, however, the efficiency of gene transfer decreased significantly.
Sterile filtrates.
Fresh cultures were filtered by hand pressure with either Gelman Sciences Acrodisc (0.2-μm-pore size, 26-mm-diameter) polysulfone filter cartridges or Schleicher & Schuell UNIFLO (0.2-μm-pore size) cellulose acetate filter cartridges; they were equally satisfactory. Filters became clogged after passage of 5 to 15 ml of culture, depending on the bacterial concentration. Some other types of commercial membrane filters, also of 0.2-μm porosity, did not yield as much VTA as the two mentioned above. Filters of 0.45-μm-pore size clarified the cultures more rapidly but occasionally allowed passage of a small fraction (below any visible turbidity) of Methanococcus cells and were therefore avoided. The filters were not usually washed before use. The error in filtrate absorbance readings due to material leached from the membrane filters was negligible.
VTA stability measurements.
VTA preparations were usually titrated the day they were made. Later assays supplied estimates of VTA stability upon storage and/or dilution. VTA inactivation was expressed as the number of lethal hits, H = −lnS, where S is the VTA survival ratio (titer at time t2 to titer at time t1). More specifically, H0 is the estimate based on the first titration after the beginning of storage (or after dilution into a different suspension medium) compared with the initial titer, whereas Hmo = −(365/12)(lnS/d) (where d = t2 − t1 days of storage) is the rate (lethal hits per month) based on any two successive titrations past the initial one. When more than one Hmo estimate was available for a given preparation, an average of the n estimates, Hmo(n), was calculated. Alternatively, Hmo(n) was calculated as the linear regression coefficient of the logarithm of the VTA titer versus days of storage.
VTA preparation and concentration.
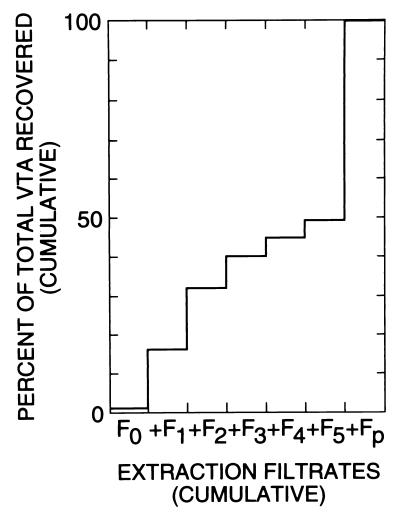
The simplest method for preparing VTA was direct filtration of cultures. Since most of the VTA in a culture was found to be associated with the cells (see Results), another simple method consisted of sedimenting the bacteria, resuspending them in a buffer, and filtering them. At earlier stages of the work, however, numerous attempts were made to separate VTA from the bacteria by what was presumed would be desorption, as in the following example. Donor bacteria were grown (e.g., two bottles, 53-ml total volume) to an OD600 of 0.3 to 0.5 (about 3 days of incubation of a 1:50 inoculum). They were transferred in a hood to polypropylene centrifuge tubes and spun anaerobically for 5 min at 10,000 rpm (Sorvall SS-34 rotor) in the cold. The tubes were opened in the hood, and the supernatants were discarded; the tubes were drained well. The pellets were gently resuspended with a Squeezette (soft plastic transfer pipette) and pooled in a total volume of 10 ml of cold 0.3 M NaCl with 5 mM sodium citrate. The suspension was spun again as described above. The new supernatant was collected and filtered (F1). The new pellet was resuspended and centrifuged again as before. The next supernatant was collected and filtered (F2). This process could be repeated. In the end, the pellet was resuspended as before and filtered (Fp). For this last filtration, because of the high bacterial concentration, more than one filter often had to be used. Bovine serum albumin (BSA) (final concentration, 1 or 2 mg/ml) was added to the filtrates, and they were stored on ice. Of the total VTA recovered, about half was in the last filtrate (see Fig. 3). The total active VTA yield, measured as his+ transfer, varied between 2 × 104 and 3 × 105 per ml of culture.
FIG. 3.
VTA cumulatively recovered in successive extractions (washings) of a culture of M. voltae PS-2. The recipient strain was PS-3. Selection was for histidine independence. F0, filtered culture supernatant; F1 through F5, filtrates of supernatants obtained in five washing cycles; Fp, filtrate of the last resuspended pellet. The culture (52 ml, 2 × 108 cells per ml), was resuspended each time in 10 ml of 0.3 M NaCl–0.015 M sodium citrate. The total his+ VTA recovered was 4.3 × 106.
Concentration and purification were problematic because of the inherent instability of VTA and because of the laboriousness of filtration, especially at high bacterial concentrations. Direct sedimentation of VTA by centrifugation resulted in large activity losses (possibly due to pressure effects [18], sticking to the tube walls, or irreversible aggregation), except when a sucrose cushion was used. Centrifuge-aided concentration in Centricon 500 filters (Amicon, Inc.) was slow (about 5 ml per h for filtrates of culture supernatants) and resulted in a low recovery (e.g., 20%) of activity. In preliminary experiments, VTA was precipitated by polyethylene glycol (PEG) (5 to 30% [wt/wt]), but only 5% of the activity was recovered from the sediment, even though stability upon storage at such PEG concentrations was about average for VTA suspensions in saline (see Table 3). Of other procedures tested, two are described below. The first aimed at concentrating VTA that was (as much as possible; see Results) set free from cells prior to filtration. The second procedure is the more satisfactory one for rapid concentration of VTA without much activity loss and could be used as the starting point for purification of the material.
TABLE 3.
Decay rates Hmo (n) and H0 (see Materials and Methods) for VTA (as his+ transfer) under various storage conditions
| Prepn | Gas phasea | Temp (°C) | Mob | Mediumc | Hmo(n) [range] | H0 |
|---|---|---|---|---|---|---|
| 1 | Anox | −70 | 31.5 | WM + 10% PEG | 0.13 (4) | |
| 2a | Air | 0 | 0.8 | WM | 6.9 (2) | 1.0 |
| 2b | Air | 0 | 0.8 | WM + 0.05% BSA | 5.0 (2) | 0.4 |
| 3a | Anox | +5 | 4.1 | 0.75 M NaCl | 0.64 (10) ± 0.15d | 2.1 |
| 3b | Air | +5 | 0.4 | As above | 0.73 (1) | 1.4 |
| 4 | Anox | +5 | 4.4 | 0.3 M NaCl | 1.1 (8) ± 0.15d | |
| 5a | Air | 0 | 3.0 | 0.3 M NaCl + 0.1% BSA | 0.82 (3) [0.5–1.4] | 1.4 |
| 5b | Air | 0 | 3.0 | ∼0.3 M NaCl + MgCl2e + 0.1% BSA | 0.65 (8) [0.3–1.3] | 1.5 |
| 6a | Air | 0 | 3.0 | 0.3 M NaCl + 0.1% BSA + 10% glycerol | 1.9 (5) [1.6–2.2] | −0.2 |
| 6b | Air | −70 | 4.3 | As above | 0.62 (4) [0.1–1.8] | 0.6 |
| 7af | Air | 0 | 4.3 | 0.3 M NaCl + ≥0.1% BSA | 0.52 (4) [0.1–1.8] | |
| 7bf | Air | 0 | 5.8 | As above | 0.33 (5) [0.0–0.6] | |
| 8 | Air | 0 | 1.1 | 0.3 M NaCl + 20% glycerol | 1.9 (4) [0.9–2.9] | |
| 9a | Air | 0 | 1.2 | 0.3 M NaCl | 2.4 (2) | 2.9 |
| 9b | Air | 0 | 2.7 | 0.3 M NaCl + 0.1% BSA | 1.6 (3) | 0.7 |
| 9c | Air | 0 | 2.7 | 0.3 M NaCl + 1% BSA | 0.75 (3) | 1.1 |
| 10ag | Air | −80 | 6.9 | 0.3–0.8 M NaCl + 10% glycerol; BSA present | 0.56 (3) | 0.6 |
| 10bg | Air | −80 | 6.9 | NaCl and BSA at concn lower than above + 10% glycerol | 0.59 (3) | 1.4 |
| 10cg | Air | −80 | 6.9 | As above | 0.22 (3) | 1.2 |
| 11h | Air | −18 | 2.3 | ∼0.2 M NaCl + 10% PEG | 1.1 (1) | 1.2 |
Anox, gas from the anaerobic hood: N2 with small amounts of CO2, H2, and probably traces of H2S.
Total duration of storage over which titrations were made.
Filterable debris from bacteria lysed in the course of filtration might affect VTA stability. WM (the standard culture medium) refers here to whole culture filtrates. Bacteria were also present at filtration when preparations 10 and 11 were made by the PEG-bag method. For all other preparations, bacteria were removed by centrifugation before filtration (see Materials and Methods). When NaCl is indicated (with the exception of preparation 5), either citrate (5 to 15 mM [pH 6.6]) or Tris (20 mM [pH 7.5 to 7.9], with up to 2 mM EGTA) was also present. There was no evidence of significant differences in VTA stability with these additions.
Calculated standard error.
Various concentrations of MgCl2 (between 14 and 41 mM) at a constant ionic strength. Pooled data.
A 1:6 dilution of a VTA preparation concentrated 200-fold in a Centricon 500 centrifugal concentrator with BSA present.
Undiluted VTA preparations (PEG-bag method). The concentrations of the components of the suspension medium are thus not precisely known. Preparation 10a is the first sample collected from the dialysis bag after concentration; preparations 10b and 10c are samples collected from the second distilled water rinse from the same bag. Titers (as his+ transfer) at collection were 6 × 106/ml in 10a and 5 × 106/ml in 10b and 10c.
A 1:21 dilution of the first distilled water rinse in the PEG-bag preparation method.
In the first procedure (procedure 1), pooled cultures (210 ml; OD600, 0.3) were centrifuged (Sorvall GSA rotor, 7,000 rpm, 10 min) anaerobically. The pellet was resuspended in 2 ml of 0.3 M NaCl with MgCl2 (5 mM) and DNase (5 μg/ml). Shortly afterward, buffer (0.3 M NaCl, 20 mM Tris, 1 mM EGTA [pH 7.6]) was added to 20 ml, and the suspension was centrifuged again (Sorvall SS-34 rotor, 10,000 rpm, 5 min). The supernatant was collected; the pellet was resuspended in 20 ml of buffer and centrifuged again, and the supernatant was pooled with the first one. The new pellet was resuspended in 2 ml of 0.3 M NaCl–5 mM MgCl2 and again treated briefly with DNase, buffer was added to 20 ml, and the suspension was centrifuged. The three pooled supernatants were filtered and then spun (Spinco SW28 rotor, 18,000 rpm, 5 h, 8°C) on a cushion of 1 ml of 66% (wt/wt) sucrose. The bottom 1.5 ml from each tube was collected after the supernatant was removed from the top; these samples were pooled and dialyzed against buffer (without EGTA) to remove the sucrose. A dialysis bag was then placed in a bed of dry PEG (molecular weight, 6,000) for 30 min to reduce the volume. The suspension was again treated with DNase (added to obtain 6 μg per ml, with MgCl2 to 10 mM) for 1 h and then with RNase (added to obtain 150 μg per ml) for 30 min and was spun (Spinco SW50.1 rotor, 32,000 rpm, 4 h, 8°C) on a 1-ml cushion of sucrose (as described above). Fractions were collected dropwise, and their refraction index was measured. Based on prior calibration, those expected to contain most of the band of VTA activity were pooled and dialyzed against 100 mM NaCl–10 mM Tris–1 mM EDTA (pH 7.8). This procedure reduced the original culture volume 420 times. The activity recovered was only a small percentage of the original, however.
In the second procedure (PEG-bag method), fresh cultures at about 108/ml total counts were spun (Sorvall GSA rotor, 7,000 rpm, 10 min, 5°C) anaerobically. The well-drained pellets were resuspended in buffer (0.3 M NaCl, 20 mM Tris, 1 mM EGTA [pH 7.5]) at about two thirds the original volume and filtered. The filtrate was transferred to dialysis tubing (flat width, 25 mm; prepared by heating at 80°C for 30 min in 10 mM EDTA made at neutral pH, rinsing with buffer, standing for a few hours before use, and filling with a 10-mg/ml BSA solution in buffer), which was then encased in PEG 6000 crystals, wrapped in plastic film, and stored in a refrigerator (9 to 10 h) until most of the fluid in the tubing had been absorbed by the PEG. The tubing was then rapidly rinsed with running water on the outside to remove PEG stuck to it, opened at one end, and laid flat on a clean sheet of Parafilm or other water-repellent surface. The contents were squeezed out onto the surface by rolling a thick glass rod from the sealed end of the tubing to the other end and collected. A very small volume of water was added to the tubing and mixed as well as possible with whatever was adsorbed to the inside surface of the tubing; the contents were squeezed out as described above and collected. This operation was repeated once more. The VTA concentrate obtained was rich in BSA and salt but showed 40 to 100% recovery of VTA activity with a 100-fold volume reduction. It was stored frozen (see Table 3).
Short-cut preparation of VTA DNA.
Bacteria from two tumbler cultures were pelleted anaerobically at 9,000 × g (Sorvall GSA rotor, 7,000 rpm, 10 min). All supernatant fluid was removed. The bacteria were resuspended in 1 ml of 0.3 M NaCl–20 mM Tris (pH 7.7) with BSA (1 mg/ml), pooled, and lysed by the addition of 200 ml of ice-cold distilled water, with rapid mixing. Small volumes of NaCl, MgCl2, and DNase (Sigma DN-25) solutions were added to obtain final concentrations of 50 mM, 1 mM, and 2 μg/ml, respectively, and the mixture was warmed to room temperature. After 20 min, RNase was added to a final concentration of 25 μg/ml. Forty minutes later, EDTA was added to a final concentration of 2 mM. VTA and bacterial debris were immediately pelleted at 44,000 × g (Spinco SW28 rotor, 18,000 rpm, 4.5 h, 8°C), resuspended in 3 to 4 ml of STE2 buffer (100 mM NaCl, 10 mM Tris, 2 mM EDTA [pH 8.0]), extracted with phenol (with hydroxyquinoline and β-mercaptoethanol) through chloroform (45), and concentrated by precipitation with cold ethanol (45) in a thick-walled glass centrifuge tube. Mussel glycogen was added at 20 μg/ml before the ethanol as a carrier (19, 56). After 1 day at −20°C, the ethanol mixture was centrifuged (Sorvall SS-34 rotor, 10,000 rpm, 1 h, near 0°C), and the pellet was dried and suspended in the smallest possible volume of STE buffer (45). No residual DNase activity was detectable in the final preparation. With strains PS-1 and PS-13, total yields of about 1 μg of VTA DNA could be reproducibly obtained. VTA DNA was further purified and concentrated by cutting bands out of agarose gels after electrophoresis and extracting the DNA by the QIAGEN method.
Other methods.
Other nucleic acid methods were standard (11, 45). Gel electrophoresis was usually performed with 0.8% agarose gel slabs (25 ml, 95 by 65 mm) horizontally submerged in buffer containing 40 mM Tris, 5 mM sodium acetate, and 1 mM EDTA (pH 7.4) at 100 V (8-V/cm gradient) for 80 min. Some gels (see Fig. 6G and Fig. 7) contained 1% agarose and were run at 50 V for 200 min. The gels were stained in a solution of ethidium bromide (1 μg/ml) for 20 min and destained with 1 mM MgSO4 for 40 min before being photographed. Restriction enzymes were used as recommended by the commercial enzyme suppliers. DNase represents DNase I and RNase represents RNase A throughout.
FIG. 6.
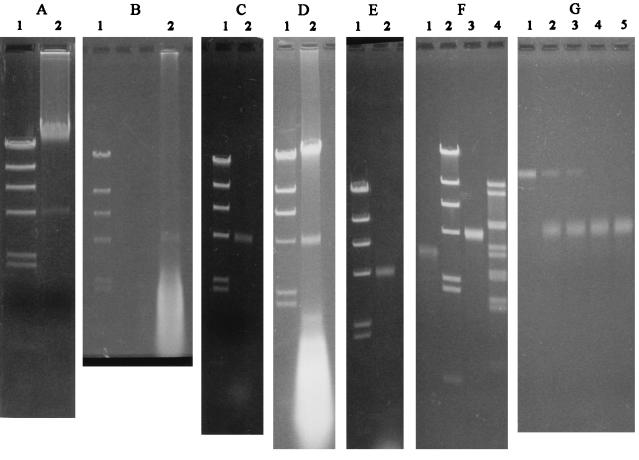
Agarose gel electrophoresis of phenol extracts showing the VTA DNA band. Size markers (lanes 1 in A, B, C, D, and E and lane 2 in F) are fragments of a HindIII digest of phage λ DNA (23.1, 9.4, 6.6, 4.4, 2.3, 2.0, and 0.6 kbp). (A) Lane 2 contains DNA from whole bacteria (strain PS-6). (B) Lane 2 contains an extract from partially purified (see Results) VTA (strain PS-6). (C) Lane 2 contains an extract from partially purified (procedure 1 in Materials and Methods) VTA (strain PS-13). (D) Lane 2 contains an extract from a concentrated filtrate (PEG-bag method) (strain PS-2). The sample applied was the equivalent of about 8 ml of culture. (E) Lane 2 contains DNA from a short-cut preparation (see Materials and Methods) (strain PS-13). (F) Lane 1, VTA DNA (strain PS-1) exposed to 85°C for 3 min; lane 3, unexposed control VTA DNA; lane 4, HindIII digest of phage λ DNA exposed to 85°C for 3 min. (G) Lane 1, unexposed control VTA DNA (strain PS-1); lanes 2 through 5, VTA DNA exposed for 1 min to 78, 80, 82, and 84°C, respectively. See Fig. 7C, lane 2, for size markers for panel G (part of the same gel slab). Note that in panel G, the agarose concentration was higher and the voltage applied was lower than those in panels A through F.
FIG. 7.
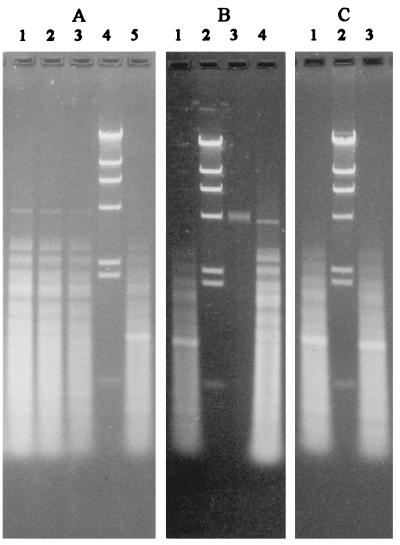
Fragment size distribution determined by agarose gel electrophoresis of M. voltae DNA and concentrated VTA DNA after exhaustive digestion with restriction endonuclease AluI. Size markers are as in Fig. 6. Electrophoresis conditions are as in Fig. 6G. (A) Lanes 1, 2, and 3, decreasing amounts (4:2:1 ratio) of M. voltae DNA; lane 4, size markers; lane 5, concentrated VTA DNA. (B) Lane 1, VTA DNA; lane 2, size markers; lane 3, undigested VTA DNA (the amount applied is approximately 1/100 that in lane 1); lane 4, M. voltae DNA. (C) Lane 1, concentrated VTA DNA; lane 2, size markers; lane 3, same as lane 1 but exposed to RNase before application to the gel.
RESULTS
VTA activity.
When the histidine-requiring PS-3 mutant strain of M. voltae was grown in mixed culture with the purine-requiring PS-6 mutant strain in fully supplemented medium, large numbers (e.g., 104/ml) of His+ Pur+ colonies were obtained upon plating in the appropriate selective medium. The frequency of prototrophs in pure cultures of either strain was extremely small (<50/ml). Large numbers of prototrophs were also found in mixed cultures to which DNase had been added at inoculation; such prototrophs could hardly have resulted from the low level of DNA-mediated, natural transformation that was demonstrated earlier for M. voltae PS (6). These observations suggested the presence of some other kind of genetic recombination process and were extended as follows.
Large numbers of prototrophs were routinely obtained by plating bacteria of the one strain with a bacteriologically sterile filtrate of the other strain (Table 2). Gene transfer was thus mediated by a filterable, subcellular agent, referred to here as VTA.
TABLE 2.
Transfer of prototrophy to M. voltae auxotrophic mutants by culture filtrates
| Expta | Recipient strain (OD)b | Donor strain (OD)b | Preparation and treatment of filtratec | Vol of filtrate per plated | Selectione | No. of colonies per platef |
|---|---|---|---|---|---|---|
| A1 | PS-5 (0.55) | None | 0 | His+ | 1 | |
| A2 | PS-6 (0.68) | A, ST | 1; 10 | His+ | ∼3,140; ∼30,000 | |
| A3 | SUP, A | 1 | His+ | 105 | ||
| A4 | PS-6 (0.68) | None | 0 | Pur+ | 0 | |
| A5 | PS-5 (0.55) | A, ST | 1; 10 | Pur+ | 188; ∼1,980 | |
| B1 | PS-6 | None | 0 | Pur+ | 1 | |
| B2 | PS-12 (0.80) | A | 2 | Pur+ | ∼1,000 | |
| B3 | As above, heated for 2 min at ∼89°C | 2 | Pur+ | 0 | ||
| C1 | PS-3 (0.56) | None | 0 | His+ | 0 | |
| C2 | PS-6 (0.39) | A | 1; 10g | His+ | 66, 98; 488 | |
| C3 | As above, diluted 1:20 in WM medium | 1g | His+ | 6, 4 | ||
| C4 | Same as in C2, treated with DNaseh at 25 μg/ml for 10 min | 1g | His+ | 61 | ||
| C5 | As above, but with DNase at 5 μg/ml | 10 | His+ | 354 | ||
| D1 | PS-6 (0.63) | None | 0 | Pur+ | 0 | |
| D2 | PS-1 (0.60) | U | 3 | Pur+ | 157 | |
| D3 | PS-12 (0.83) | U | 3 | Pur+ | 141 | |
| E1 | PS-6 (0.52) | None | 0 | Pur+ | 0 | |
| E2 | PS-13 (0.58) | None | 0 | Pur+ | 0 | |
| E3 | PS-6 | PS-12 (0.72) | A, ST | 1 | Pur+ | 1,030 |
| E4 | PS-13 | As above | 1 | Pur+ | 760 | |
| E5 | PS-6 | SUP, A, ST | 1 | Pur+ | 13 | |
| E6 | PS-13 | As above | 1 | Pur+ | 6 | |
| E7 | PS-6 | PELLET (suspension in WM medium), A, ST | 1 | Pur+ | 549 | |
| E8 | PS-13 | As above | 1 | Pur+ | 400 | |
| E9 | PS-6 | Same as in E3 | 1 | Pur+ (Cbl+) | 522i | |
| E10 | As above | 1 | (Cbl+) | 883ij | ||
| E11 | As above, exposed to 90°C for 2 min | 1 | Pur+ (Cbl+) | 0i | ||
| E12 | As above | 1 | (Cbl+) | 57ij | ||
| F1 | PS-3 (0.66)k | PS-6 | A, first milliliter of filtrate | 20 μl | His+ | 19 |
| F2 | A, next 14 ml of filtrate | 20 μl | His+ | 805 | ||
| F3 | A, last milliliter of filtrate | 20 μl | His+ | 137 | ||
| G1 | PS-3 | None | 0 | His+ | 0 | |
| G2 | PS-15 | A, ST | 10 μl | His+ | 549, 468 | |
| G3 | As above, diluted 1:100 in WM medium | 1 ml | His+ | 472, 439 | ||
| G4 | As above, diluted 1:100 in distilled water | 1 ml | His+ | 398, 524 | ||
| H1 | PS-6 (0.48) | None | 0 | Pur+ | 0 | |
| H2 | PS-3 | A | 20 μl | Pur+ | 100 | |
| H3 | SUP, A | 20 μl | Pur+ | 4 | ||
| I1 | PS-3 | None | 0 | His+ | 0 | |
| I2 | PS-6 (∼0.3) | A | 10 μl; 200 μl | His+ | 103; ∼1,340 | |
| I3 | SUP, A | 10 μl; 200 μl | His+ | 2; 17 | ||
| I4 | PELLET (in 0.6 M NaCl–0.06 M sodium citrate) kept for 5 h in ice, then SUP, A | 10 μl; 200 μl | His+ | 57; 1,089 | ||
| J1 | PS-6kl | None | 0 | Cbl+ | 13; 4; 10l | |
| J2 | PS-12 (0.34) | A | 1 | Cbl+ | 75; 40; 42l | |
| J3 | As above, heated for 5 min at nearly 100°C | 1 | Cbl+ | 13; 7; 14l | ||
| J4 | PS-3 (0.36)k | As above | 1 | His+ | 0 | |
| J5 | Same as in J2 | 1 | His+ | 20 | ||
| K1 | PS-3 | PS-13 | U | 10 μl | His+ | 1,201 |
| K2 | As above, sucked in and out 30 times with syringe through 22-gauge, 1-in. needle | 10 μl | His+ | 623 |
Procedures for VTA assays were described in Materials and Methods. Unless otherwise noted, the amount of recipient bacteria per plate was one syringe drop of a fresh, visibly turbid, undiluted culture.
In some cases, the OD was not recorded. Within each experiment a blank in this column means that the same strain indicated above the blank was used.
Filters used: A, Acrodisc; U, UNIFLO (see Materials and Methods). Filtrates were prepared anaerobically and used shortly thereafter. ST, the filtrate was checked for sterility by inoculating appropriately supplemented WM medium. At times, the culture was first spun at low speed to remove the bacteria, and the supernatant only was filtered and used (SUP); the pellet was resuspended in the same volume of fresh medium and filtered (PELLET); or the new suspension was centrifuged as described above and the second supernatant was filtered and used (PELLET then SUP).
The volume is given as the number of syringe drops, unless otherwise specified. One drop may be assumed to be, on average, 14 μl. The composition of the medium and several other factors obviously affect drop volume.
The selection medium was WM agar. Supplements: cyanocobalamin in A, B, D, E1 through E8, and H; hypoxanthine in E10, E12, J1, J2, and J3.
Values for duplicate plates are separated by a comma and a space.
Drops of recipient culture and filtrate were mixed 20 to 40 min before the addition of melted-soft-agar medium.
Sigma crude DN-25. Under exactly the same conditions, the enzyme was shown to be fully active on an M. voltae DNA preparation.
See the text.
In addition to the colonies recorded here, very large numbers (uncountable) of very tiny colonies were present on this plate. These were presumably derived from untransformed recipient cells trying to grow in the absence of cyanocobalamin. They were absent in E9 and E11, where the much tighter requirement for purine did not allow any residual growth.
Two drops of recipient culture were used.
Three cultures of the recipient, grown with 1/2, 1/20, and <1/200 of the standard (100 nM) cyanocobalamin concentration, were used in parallel. Their ODs were, respectively, 0.33, 0.31, and 0.20. Colony numbers obtained from each are given in the same sequence (last column). From the first plate in J2, one of the larger colonies (thus presumably not a revertant) was isolated as strain PS-15.
For any given filtrate and recipient culture, the number of prototrophs obtained was linearly related to the volume of filtrate plated over a wide range. Lower efficiencies of transfer (possibly due to saturation of receptors or cell killing) were observed (data not shown) when the recipient and an excess of VTA were mixed and incubated before selective plating.
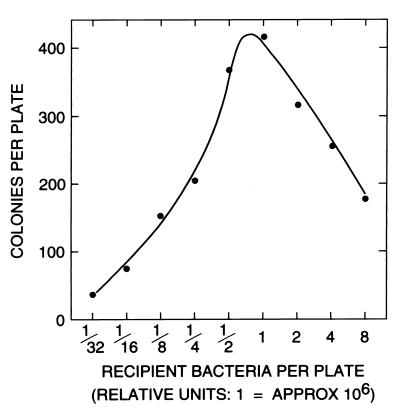
The number of prototrophs varied with the concentration of the recipient in the agar (Fig. 1). At very high bacterial concentrations, no transformant colonies were obtained. The residual growth of the auxotrophs present in excess apparently interfered with the growth of the few transformed cells; in reconstruction experiments, cells of prototrophic strain PS-12 did not form colonies when mixed with an excess of PS-3 auxotrophs in the absence of histidine, but they did when the PS-3 bacteria were first heat inactivated. As the bacterial concentration decreased (Fig. 1), the number of transformants also decreased, as one might expect for a time cutoff (irreversible incapacitation of the auxotrophic, recipient bacteria and/or spontaneous inactivation of VTA versus diffusion and adsorption rates). It was at first surprising that transformed colonies could be obtained with a recipient concentration in the agar as low as 104/ml. Calculations based on Schlesinger’s adsorption equation (46, 46a) showed, however, that even at such a low bacterial concentration, most VTA particles would be adsorbed in less than 48 h if the adsorption constant were in the range of values established for bacteriophages. For the same marker (his+) selection, the optimal input of recipient bacteria varied among cultures between 106 and 107 per plate.
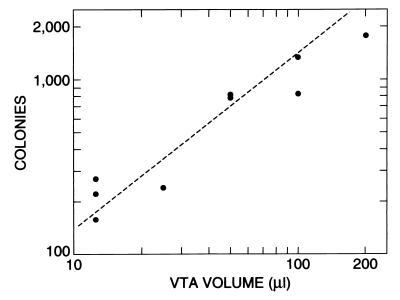
FIG. 1.
Histidine-independent colonies resulting from a constant amount of filtrate and variable recipient bacterial input. The donor strain was PS-13, and the recipient strain was PS-3. The standard pour plate technique was used (10 ml of agar per plate).
All three auxotrophic mutations available, his, pur, and cbl, were transformed to the wild type by appropriate filtrates (Table 2). For cbl, results were at first ambiguous (Table 2, compare E10 and E12 with E9 and E11), presumably because the amount of cyanocobalamin carried over with the inoculum was sufficient (see Fig. 6 in reference 6) to produce substantial amounts of residual growth, accompanied by some back-mutation, in the selection plates. (For the same reason, the colonies from Table 2, line E9, ought to be interpreted as transformed only to pur+.) Transfer of cbl+ was confirmed with a more critical experiment (Table 2, J1, J2, and J3) in which the capacity of the clb+ cells for residual growth was reduced by use of recipient cultures grown at lower concentrations of cyanocobalamin.
The levels of VTA activity for the individual filtrates shown in Table 2 were very variable, from 1.4 × 103 to 2.2 × 105 per ml of filtrate, with an overall average of 3.9 × 104. The averages for the transfer of cbl+, his+, and pur+ were, respectively, 3.1 × 104 (n = 2), 6.4 × 104 (n = 7), and 2.0 × 104 (n = 8) per ml of culture. In first approximation, assuming that 3.5 × 107 bacteria per ml corresponds to an OD of 0.1 (6) and that the average OD of the donor cultures used in Table 2 was 0.6, the average VTA activity for any given nutritional marker in a donor culture would be about 0.02% per bacterium. For his+ transfer, this value could be increased 5- to 10-fold in later, better-controlled experiments, where the bacterial concentration of the donor culture at filtration was kept at 1 × 108 to 2 × 108 per ml.
The same strain could be used as a donor or as a recipient, provided an appropriate marker could be selected. All combinations of derivatives of M. voltae PS tested gave similar results (Table 2). Thus, VTA is present in all derivatives of strain PS. On the other hand, filtrates of M. voltae A3 (64) and C1, C2, and C4 (62) did not seem to be able to transfer his+ to PS-3 or pur+ to PS-13. Likewise, no transfer of pur+ to PS-6 could be obtained with a filtrate of M. vannielii.
To show that VTA also acts on properties other than nutritional requirements, the transfer of resistance to BES was studied. Results were negative when selection for BES resistance was applied directly to sensitive cells mixed in agar with VTA from a bes strain. This result is what one might expect if bes were a recessive mutation. When BES-sensitive bacteria were plated with a filtrate from a BES-resistant strain, allowed to grow for awhile, and then challenged with BES, the transfer of bes could indeed be measured (Fig. 2). The frequency of transfer of bes was much lower than that of nutritional markers, however; for four VTA preparations from PS-2, it ranged from 0.3% to 1.9% that of his+.
FIG. 2.
VTA-mediated transfer of bes to BES-sensitive bacteria. The recipient strain was PS-1 (about 1.6 × 106 bacteria per plate). VTA was from strain PS-2. Each point represents the colonies from one plate. A 5-ml agar overlay with BES was added after 2 days of incubation, establishing upon diffusion a 1.1 mM BES concentration. Two control plates without VTA yielded 12 and 18 resistant colonies; the data as plotted are corrected for this background. The broken line represents the best fit for direct proportionality.
VTA was inactivated by heating (Table 2, B3 and E11). It was quite resistant to the syringe and needle manipulations required for diluting and plating (Table 2, K1 and K2).
Whereas M. voltae cells were promptly lysed when diluted in distilled water (reviewed in reference 62), VTA activity survived well such dilution (Table 2, G4).
VTA was not affected by DNase (Table 2, C4 and C5). Likewise, DNase added to donor cultures before filtration did not reduce VTA activity (and improved somewhat the rate of filtration for more dense cultures). RNase, at as high a concentration as 1 mg/ml, did not affect filtrate transfer activity.
VTA could also be demonstrated following the removal of bacteria by centrifugation and filtration of the supernatant. The activity was always much lower than when the whole culture was filtered (Table 2, A3, E5, E6, H3, and I3). Apparently, most of VTA was not free. Resuspension of the bacteria in saline appeared to set free some VTA activity prior to filtration (Table 2, I4) without visible cell lysis. On the other hand, interactions at the filter, presumably shearing effects, were clearly important; the VTA titer of cultures was always very low in approximately the first milliliter of filtrate, rapidly increased to a maximum, and then decreased when the filter became overloaded with bacteria and the flow rate was strongly reduced (Table 2, F1 through F3 and other experiments not shown). Calculations suggest that the setting free of VTA at the filter started approximately when the equivalent of a monolayer of bacteria was trapped on the filter surface. A difference in VTA activity between the first fraction and the rest of the filtrate was also noted when the filter was extensively washed prior to use with the same medium in which the bacteria were suspended, in order to obviate possible ionic effects at the filter.
Attempts to induce increased VTA production by transiently exposing donor cultures to heat, UV light, or mitomycin C were unsuccessful. Autolysis following starvation for H2 (6) did not seem to affect the VTA titer.
Mode of VTA liberation.
The bulk of VTA is associated with bacterial cells, and very little is free in culture supernatants (see above). A larger fraction of VTA could be recovered in supernatants through “washing” (pelleting of cells in a centrifuge, resuspension in buffer, pelleting again, and filtration of the supernatant). Although salt solutions of diverse compositions and concentrations were tried (0.15 to 1.5 M NaCl, with or without 15 to 60 mM sodium citrate, 8 to 60 mM Tris, 0.5 to 5 mM EGTA, and pH 6 to 8), there were no striking differences in the results; even with repeated washings, longer waiting times or higher temperatures after resuspension, or lower cell concentrations (to reduce possible readsorption of VTA), only rarely (e.g., Table 2, I4) could more than half of the total VTA be recovered by this procedure, the rest being recoverable only upon filtration of the bacteria themselves (Fig. 3). For 26.5-ml cultures of strain PS-2 or PS-13 (1 × 108 to 3 × 108 bacteria per ml), this procedure resulted in a total his+ VTA yield of 5 × 105 to 8 × 106 (average, 3.4 × 106 [n = 17], or approximately 6 × 10−4 his+ VTA per cell).
Filtration caused substantial bacterial lysis, as indicated by the amount of UV-absorbing material detectable in the filtrate. For example, a culture of strain PS-1 grown to an OD600 of 0.32, washed and resuspended in 0.3 M NaCl, and filtered had an A260 of 0.16 and an A280 of 0.11 in the filtrate. The same washed culture, totally lysed by resuspension in distilled water, had an A260 of 0.38 and an A280 of 0.24. One would conclude that nearly half of the bacterial proteins and nucleic acids were set free and not retained by the filter. For comparison, the A260 of a similarly prepared filtrate of an E. coli C culture (grown aerobically in mineral medium plus glucose) was only a small percentage of the original A600, and no peak at A260 was evident. Phenol extracts confirmed the presence of RNA and high-molecular-weight DNA in M. voltae filtrates (see Fig. 6D).
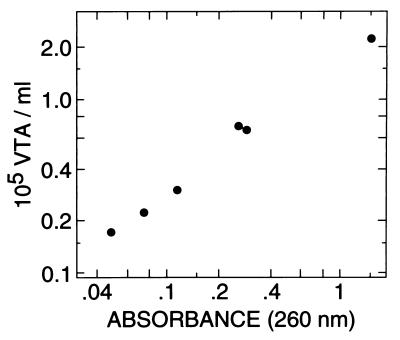
Similarly, a high absorbance in the UV range was noted in VTA preparations obtained by washing (as defined above). This result suggests that some cell lysis may also occur as a result of the centrifugation and resuspension process. Indeed, the amounts of cell lysis (measured as solubilized, UV-absorbing material) and of VTA activity recovered were correlated within a given experiment (Fig. 4).
FIG. 4.
Relationship between VTA titer and concentration of UV-absorbing material in the filtrates shown in Fig. 3 (F0 excluded). The mean A280/A260 ratio was 0.55 (range, 0.51 to 0.59).
Other properties of VTA.
Most of the experiments shown in Table 2 were done with freshly prepared culture filtrates. The VTA activity in such preparations, even under refrigeration, decayed quite rapidly. At first, VTA was routinely stored under anaerobic conditions. It was soon realized, however, that the gain in stability over that obtained with storage in air, if any, was small. All preparative operations with VTA following filtration were thereafter done without anaerobic precautions.
Longer time series for VTA titrations suffered from variability in the efficiency of plating, which is difficult to control, since a different recipient culture has to be used each time. The data available for a number of VTA preparations were tentatively interpreted as showing a bimodal decay over time: a sharp drop in titer over the first 1 or 2 days of storage, presumably due to equilibration to the storage conditions, followed by a slower decay, which was assumed to follow a simple exponential function and was expressed as lethal hits per month, Hmo (see Materials and Methods). Thus, for Hmo = 1, roughly two thirds of VTA activity would be lost during a 1-month storage period. Some examples are given in Table 3. The initial inactivation, H0 (see Materials and Methods), was not correlated with Hmo.
Conditions affecting VTA stability were not studied systematically. The still exploratory data available suggest that VTA (as his+ transfer) stability is little affected by variations in pH (between 6.7 and 7.9) or NaCl concentration (between 0.15 and 1.0 M) or by the addition of MgCl2, glycerol (5 to 20%), or chelating agents (5 to 75 mM sodium citrate, 1 mM EDTA, or 1 or 2 mM EGTA). VTA was rapidly inactivated in the presence of high concentrations of CsCl or RbCl. It tolerated well rapid dilution but not storage in H2O. It tolerated high concentrations of sucrose. BSA at ≥1 mg/ml and possibly PEG at 100 mg/ml improved VTA stability somewhat. In the presence of glycerol and/or high protein concentrations, VTA tolerated fairly well freezing and thawing; nevertheless, some decay continued to occur in the frozen state at the temperatures used (Table 3).
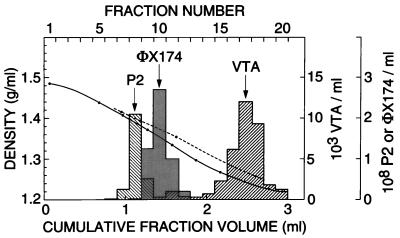
Attempts were made to establish the buoyant density of his+ VTA in a CsCl density gradient. Because of the rapid inactivation of VTA, however, the density estimate obtained, 1.28 g/ml (Fig. 5), with the recovered VTA activity being only 1% of input, may not be fully representative. In an earlier CsCl equilibrium density experiment (data not shown), in which the recovered activity was even lower (<10−4), the few his+ colonies obtained in the assays were distributed over a density range of 1.34 to 1.37 g/ml. In the gradient of Fig. 5, there seems to be a small, possibly significant VTA peak at a density of 1.38 g/ml (at a 1.6-ml cumulative fraction volume). This finding raises the possibility of two forms of VTA with different buoyant densities and different sensitivities to CsCl, one of them being rather tightly complexed to lighter material (see Discussion).
FIG. 5.
Isopycnic banding of his+ VTA and two bacteriophage markers in a CsCl density gradient (Spinco SW50.1 rotor, 41,500 rpm, 27 h, 8°C). The expected buoyant densities for P2 lg del1 del2 and for φX174 are, respectively, 1.42 (3, 7) and 1.40 (50) g/ml. The VTA sample was prepared from strain PS-2 by the PEG-bag method; hence, high concentrations of NaCl (final concentration in the centrifuge tube, 0.5 M), BSA (tentatively 8 mg/ml), and added glycerol (0.6 M) were present in addition to cell components set free by filtration. The solid line for density is from refractometer readings for a control centrifuge tube containing only CsCl in pure water. The broken line shows the gravimetrically determined densities for reconstructed solutions of corresponding concentrations of CsCl with added NaCl and glycerol. A comparison of refractometer readings for such reconstructed solutions with those taken over the experiment gradient (data not shown) suggests that BSA and/or cell proteins were beginning to band in the upper part of the gradient. The recoveries of activities were 1% of input for VTA, 65% for φX174, and >28% for P2.
VTA DNA.
Gel electrophoresis of total DNA preparations of M. voltae PS revealed, in addition to high-molecular-weight, chromosomal DNA, an extremely faint, rather broad secondary band at about 4.4 kbp (as calculated for linear, double-stranded DNA) (Fig. 6A). DNase eliminated both the chromosomal DNA and the 4.4-kbp band. Both were unaffected by RNase. The band could be demonstrated in six DNA preparations (from strains PS-1, PS-6, PS-9, and PS-12 and two subisolates) of eight made by slightly different methods over a number of years. The presence of such a band had not been noted in work from other laboratories (28, 49, 51, 62) that used M. voltae PS DNA for a variety of purposes. Presumably, the amount of DNA in the band is affected by the preparation method and by the history of the culture. For two DNA preparations of known concentrations, made from unwashed bacterial pellets, the amount of DNA in the 4.4-kbp band, roughly estimated by comparison with the DNA size standards, amounted to less than 1% of the total DNA (0.1 and 0.4%, respectively, or approximately one copy of 4.4-kbp DNA, on average, per M. voltae chromosome of 1,880 kbp [51]).
The possibility that the 4.4-kbp DNA might be involved in VTA activity was confirmed by the presence of DNA of the same size in partially purified VTA preparations. In an early experiment, the VTA particles in a culture filtrate were separated from soluble cell material by centrifugation over a sucrose cushion, further concentrated by membrane filtration (always without the addition of DNase or RNase), and extracted with phenol. The extract yielded a 4.4-kbp DNA band plus some material of lower molecular weight, presumably contaminant RNA (Fig. 6B). A similar result (Fig. 6C) was obtained with a different VTA concentration method (procedure 1 in Materials and Methods), aimed at preventing cell lysis. In this case, any free DNA was eliminated by DNase. Both procedures were cumbersome, and the recoveries of VTA activity just before DNA extraction were very low. For the isolation of VTA DNA, a more efficient method was developed (short-cut preparation of VTA DNA); this method was based on the resistance of VTA to DNase and on the assumption that VTA particles, if sedimented together with cell debris, might physically better survive pelleting (Fig. 6E). Finally, enrichment for the material in the 4.4-kbp band was also evident in extracts of concentrated (but far from pure) VTA preparations made by the PEG-bag method (Fig. 6D).
At temperatures above 78°C (in 10 mM Tris–0.6 mM EDTA, pH 8 at 20°C), VTA DNA was promptly denatured to a faster-moving, broader band in gel electrophoresis (Fig. 6F and G), indicating separation of the strands of linear (or nicked-circular), double-stranded DNA molecules. (M. voltae DNA is known [62] to have a very low GC content.)
With the short-cut preparation method, DNA bands of 4.4 kbp could also be demonstrated in preparations from M. voltae C1, C2, and A3. For A3, several slower-moving, very faint bands were also present; these could be interpreted as forms of plasmid pURB600, known to be present (64) in this strain, that were somehow protected from the DNase treatment.
No discrete restriction fragments were obtained from VTA DNA with several restriction endonucleases (AvaI, BamHI, BglII, EcoRV, HpaI, NdeI, PstI, and SalI); the VTA DNA band was still present, in some cases less intense and with a faint smear on its low-molecular-weight side. (BamHI and BglII reduced the electrophoretic mobility of the DNA band, presumably due to stable complex formation with the DNA.) The VTA DNA band disappeared after digestion with AluI, whose recognition site is only 4, rather than 6, nucleotides long. Digestion of total M. voltae DNA with AluI resulted in a complex pattern of bands (Fig. 7A, lanes 1, 2, and 3), all of which, with the exception of the slowest, AluI band A, at 4.2 kbp, ought to be interpreted as collections of individual bands. That the largest bacterial DNA fragment, AluI band A, is so near in size to VTA DNA may at first be surprising but is not beyond rational expectations for a random distribution of AluI restriction sites. Digestion of highly concentrated preparations of VTA DNA also resulted in a complex band pattern (Fig. 7A and B) which could be considered identical to that for total M. voltae DNA, except in two respects: AluI band A was absent, and a new band (or collection of bands of nearly equal sizes), AluI band ϕ, was present at 920 bp (range, 800 to 1,020 bp). The disappearance of AluI band A is not unexpected since, given its size, the probability of its inclusion, uncut, in the 4.4-kbp VTA DNA fragment would be quite low. Band ϕ was not affected by RNase (Fig. 7C). It could not be seen following double digestion of highly concentrated VTA DNA with AluI and DraI.
DISCUSSION
Based on the evidence presented here, M. voltae PS spontaneously generates DNase-resistant particles, referred to here as VTA, that contain 4.4 kbp of DNA and are able to transfer to other cells any of four genetic markers tested. It is fair to assume that VTA will transfer almost any genetic marker on the M. voltae chromosome. It was shown elsewhere (15) that partially purified preparations of VTA contain bacteriophage-like (in the morphological sense) particles with an isometric head and a tail, the head being of an appropriate size for harboring a 4.4-kbp molecule of DNA.
Is this a case of generalized transduction, where the appropriate host strain for bacteriophage plaque formation is missing? Circumstantial evidence made this idea unlikely and discouraged launching the serious effort required to try to isolate a host strain that may be attacked and lysed by a presumptive bacteriophage accompanying VTA.
First, in generalized transduction (36, 38, 55), as exemplified by bacteriophages P1 and P22, there is typically a mixed population of morphologically identical particles, some of which, the plaque formers, contain a viral genome, while others, the potentially transducing particles, contain similarly sized fragments of the donor host chromosome. The proportion R of potentially transducing particles is generally small. This fact also applies to the mechanistically different transduction by bacteriophage Mu. [Based on physical measurements, R values are 0.3% for P1 vir1 (25), 6% for P1 kc (17), and at most 2% for wild-type P22 (13). For phage SP10 in Bacillus subtilis (41), an R value of 14% may be calculated. With specially selected phage mutants, higher R values may be obtained (47). More commonly, the information available for the numerous examples of generalized transduction across the bacterial spectrum is at best the ratio of transductants to phage-producing, singly infected cells. Technically, this measurement may be affected by a low efficiency of selection for the markers used and by killing of potential transductants by secondary phage infection. As an estimate of R, such a measurement would have to be corrected for abortive transduction, uneven sampling in DNA packaging along the host chromosome, and the size of the transducing fragments relative to the host chromosome, the latter being a ratio that may vary from 1/27 for φCR30 in Caulobacter crescentus to 1/170 for Mx4 and Mx8 in Myxococcus xanthus (as calculated from sources quoted in reference 40). Raw transduction frequencies across the spectrum of materials are very variable but are rarely (39) higher than 10−4. Higher values may be obtained (10, 47, 59) if special mutations are introduced.] For the purpose of discussion, the R value for VTA in M. voltae may be estimated as follows. VTA DNA corresponds in size to 1/423 of the 1,880-kbp (51) M. voltae chromosome. A culture of 2 × 108 bacteria per ml yields (by the short-cut preparation method) about 2 ng of VTA DNA per ml, equivalent to 4.2 × 108 VTA copies. Such a culture would produce an activity (as his+ transfer) of about 1.3 × 105 per ml (an average for several similar experiments of the type exemplified in Fig. 3 for strains PS-2 and PS-13). The efficiency of transfer by VTA particles for his+ may then be calculated as 1.3 × 105/4.3 × 108, or 0.031% and, by correction for a sampling factor of 423 (see above) (tentatively assumed to be constant over the bacterial chromosome) to have an R value of 13%. The corresponding calculation for the transfer of another auxotrophic marker, pur+, would result in an R value of 4.1%. These figures do not take into account the possibility of inefficient integration (still undemonstrated in this system) of the transferred DNA fragments. If the efficiency of integration is assumed to be on the order of 1/10, as in P22 (42), the above estimates of R would encompass 100%, a result clearly unlike that for typical generalized transduction.
Second, the DNA molecules carried by VTA particles are quite small (4.4 kbp) compared to those of any double-stranded DNA bacteriophage (known range, 12 to 750 kbp). The genes necessary for the specification of particle structure, the control of replication, the sizing of host DNA fragments, and their incorporation into the capsid could hardly fit into 4.4 kbp of DNA. (However, note the arguable counterexample of polyomavirus and simian virus 40, which have 5 kbp of circular DNA and where “pseudovirions” may contain host cell DNA fragments.)
Third, routine examination of VTA DNA after digestion with a variety of restriction enzymes suggested at first that it was nothing but a random sample of the bacterial DNA of M. voltae, and the possibility that VTA might represent a new case of capsduction, described (37, 60, 66) for strains of R. capsulata, was considered. These strains spontaneously produce particles (called GTA, for gene transfer agent) with 30-nm isometric heads and short tails, containing 4.5 kbp of DNA, and able to transduce any bacterial marker. Only bacterial DNA was found in the GTA particles (66). Their production would seem to be controlled and specified by bacterial genes, without any differential DNA replication. A similar, but rather poorly characterized, example was later described (53) for Myxococcus. Additional examples were described for strains of D. desulfuricans (43) and for the spirochete S. hyodysenteriae (24), although in these the particles are larger, containing 13.5 and 7.5 kbp, respectively, of DNA. The interesting question of whether capsduction evolved in the absence or independently of “true” (i.e., infectious, virally reproducing) bacteriophage was raised (66). Which came first: capsduction and then bacteriophage or bacteriophage and then transduction, followed, through loss of function, by capsduction?
Defective phage systems have been known for many years, although they usually have been detected (e.g., in many Pseudomonas and Bacillus strains) by evidence of viral or subviral particles (35), recognized either morphologically by electron microscopy or through their bactericidal activity, rather than by evidence of gene transfer experiments. For the much-studied PBSX defective prophage (approximately 33 kbp in size) present in B. subtilis and in other species of the genus Bacillus (1, 48, 54, 65), heterologous cell killing seems to have supplanted the gene transfer function, even though the particles contain a random sample of bacterial DNA (in fragments of 13 kbp) which, after extraction, is capable of transfection. Similar examples are those of PBLA (22) in B. licheniformis (producing particles containing 51-kbp host DNA fragments) and of PBND8 (57) in B. natto (particles with 8-kbp DNA fragments). In the absence of comparative studies of particle formation and its control, the difference between such defective lysogeny and capsduction may be one of degree only, depending on the extent to which viral functions are present.
An effort was made to test stringently to what extent VTA DNA was representative of M. voltae DNA. When VTA DNA was concentrated as much as possible and then digested with AluI (which statistically may be expected to cut M. voltae DNA approximately once every 360 bp), a new band of 0.9 kbp was observed (band ϕ; Fig. 7); this band was not visible in similarly digested bacterial DNA preparations. This band presumably represents a DNA sequence that is either amplified through replication relative to the rest of the bacterial genome or preferentially packaged in the course of the production of VTA particles. It remains an open question whether it is part of a presumptive prophage sequence.
Both PBSX of B. subtilis and GTA of R. capsulata are present in a majority of the strains classified within the respective species. The fact that VTA-like DNA could be demonstrated in three other isolates of M. voltae, C1, C2, and A3 (note, however, that C1 and C2 were isolated [62] from the same site), suggests that VTA may likewise be fairly typical of M. voltae strains. The negative results for attempted gene transfer from these three strains to PS would suggest the presence of exclusion mechanisms, probably DNA restriction and modification. Since the chromosomal DNA of strain PS includes (64) some sequences that hybridize with the DNA of a plasmid present in strain A3, further work ought to consider the possibility that this plasmid (or parts of it) is the repository for the genes necessary for VTA production.
VTA has been rather unstable under all conditions tested to date. From Table 3, the average Hmo (see Materials and Methods) at ordinary refrigerator temperatures is 1.8. This value seems to be on the same order of magnitude as that for the least stable E. coli bacteriophages, e.g., Mu. The comparable Hmo for a moderately stable E. coli bacteriophage, wild-type P2, would be 0.04 (unpublished data).
The mechanism of VTA production is very puzzling. Most of the VTA in a culture is found somehow adsorbed to, or complexed with, the cells (or with large, easily sedimentable cell debris), and much of it is efficiently set free only upon filtration, presumably as a result of shear and/or cell disruption. This situation might reflect either the process by which VTA is produced or what happens to VTA after it is produced. In bacteria with an external S-layer, bacteriophages specifically adsorb to it (14, 21, 26, 34), but the reversibility of this step has not been studied. Perhaps injection of the DNA (an irreversible step) occurs only when the S-layer is locally in proper contact with the plasma membrane. Such a situation would be compatible with the recovery of VTA activity upon desorption or filtration. Observations reported elsewhere (15) make it very probable that VTA consists of tailed, bacteriophage-like particles. The buoyant density of VTA in CsCl gradients is exceptionally low compared with those reported in the literature for bacteriophage particles with similar structures. This fact might be explained if the particles are easily but still reversibly entrapped by cell surface structures. This hypothesis would also explain the stubborn association of the bulk of VTA with the cells. More speculatively, M. voltae might be capable of generating membrane vesicles, for example, like the ones that have been experimentally obtained (20), with VTA inclusions. These might then remain, at least for a time, associated with the cells that produced them. Such particles would obviously have a significantly lower buoyant density than compact, phage-like particles. Vesicular structures derived from cell membranes and other surface material are known for several eubacteria: blebs in Neisseria (12), transformosomes in Haemophilus (31), predatory membrane vesicles in Pseudomonas (30), and others. Some of these structures are known to protect DNA from DNase and to participate in the transfer of DNA to other cells.
Cases of apparent reversion to the wild type, noted in the course of the isolation and purification of auxotrophic mutants from M. voltae (see Discussion in reference 6), may well have been due to the then-unsuspected presence of VTA.
From the point of view of microbiological techniques, the setting free by filtration of a gene transfer agent largely bound to the cells is relevant to the interpretation of cases of gene transfer as being due to conjugation; the control experiments in such cases ought to use a culture filtrate and not simply (the filtrate of) a culture supernatant.
ACKNOWLEDGMENTS
This work was supported in part by the Biocatalysis Program of the Energy Conversion and Utilization Technology Division of the U.S. Department of Energy and by NASA Code E contract 961524.
The gel electrophoresis work was initiated in collaboration with L. Elizabeth Bertani and profited from the technical assistance of Gayane A. Kazarians.
REFERENCES
- 1.Anderson L M, Bott K F. DNA packaging by the Bacillus subtilis defective bacteriophage PBSX. J Virol. 1985;54:773–780. doi: 10.1128/jvi.54.3.773-780.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berghöfer Y, Klein A. Insertional mutations in the hydrogenase vhc and frc operons encoding selenium-free hydrogenases in Methanococcus voltae. Appl Environ Microbiol. 1995;61:1770–1775. doi: 10.1128/aem.61.5.1770-1775.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertani G. Deletions in bacteriophage P2. Circularity of the genetic map and its orientation relative to the DNA denaturation map. Mol Gen Genet. 1975;136:107–137. doi: 10.1007/BF00272034. [DOI] [PubMed] [Google Scholar]
- 4.Bertani G. Abstracts of the 8th European Meeting on Genetic Transformation. Uppsala, Sweden: Swedish Society for Microbiology; 1986. Gene transfer in a methanogen; p. 44. [Google Scholar]
- 5.Bertani G. Abstracts of the 89th Annual Meeting of the American Society for Microbiology 1989. 1989. Transduction-like gene transfer in a methanogen, abstr. I-30; p. 222. [Google Scholar]
- 6.Bertani G, Baresi L. Genetic transformation in the methanogen Methanococcus voltae PS. J Bacteriol. 1987;169:2730–2738. doi: 10.1128/jb.169.6.2730-2738.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertani G, Chattoraj D K. Tandem pentuplication of a DNA segment in a derivative of bacteriophage P2: its use in the study of the mechanism of DNA annealing. Nucleic Acids Res. 1980;8:1339–1356. doi: 10.1093/nar/8.6.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blank C E, Kessler P S, Leigh J A. Genetics in methanogens: transposon insertion mutagenesis in a Methanococcus maripaludis nifH gene. J Bacteriol. 1995;177:5773–5777. doi: 10.1128/jb.177.20.5773-5777.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowen T L, Whitman W B. Incorporation of exogenous purines and pyrimidines by Methanococcus voltae and isolation of analog-resistant mutants. Appl Environ Microbiol. 1987;53:1822–1826. doi: 10.1128/aem.53.8.1822-1826.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campos J M, Geisselsoder J, Zusman D R. Isolation of bacteriophage MX4, a generalized transducing phage for Myxococcus xanthus. J Mol Biol. 1978;119:167–178. doi: 10.1016/0022-2836(78)90431-x. [DOI] [PubMed] [Google Scholar]
- 11.Dillon J-A, Nasim A, Nestmann E R, editors. Recombinant DNA methodology. New York, N.Y: John Wiley & Sons, Inc.; 1985. [Google Scholar]
- 12.Dorward D W, Garon C F, Judd R C. Export and intercellular transfer of DNA via membrane blebs of Neisseria gonorrhoeae. J Bacteriol. 1989;171:2499–2505. doi: 10.1128/jb.171.5.2499-2505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ebel-Tsipis J, Botstein D, Fox M S. Generalized transduction by phage P22 in Salmonella typhimurium. I. Molecular origin of transducing DNA. J Mol Biol. 1972;71:433–448. doi: 10.1016/0022-2836(72)90361-0. [DOI] [PubMed] [Google Scholar]
- 14.Edwards P, Smit J. A transducing bacteriophage for Caulobacter crescentus uses the paracrystalline surface layer protein as a receptor. J Bacteriol. 1991;173:5568–5572. doi: 10.1128/jb.173.17.5568-5572.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eiserling, F., A. Pushkin, M. Gingery, and G. Bertani. Bacteriophage-like particles associated with the gene transfer agent of Methanococcus voltae PS. Submitted for publication. [DOI] [PubMed]
- 16.Fox G E, Magrum L J, Balch W E, Wolfe R S, Woese C R. Classification of methanogenic bacteria by 16S ribosomal RNA characterization. Proc Natl Acad Sci USA. 1977;74:4537–4541. doi: 10.1073/pnas.74.10.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanks M C, Newman B, Oliver I R, Masters M. Packaging of transducing DNA by bacteriophage P1. Mol Gen Genet. 1988;214:523–532. doi: 10.1007/BF00330490. [DOI] [PubMed] [Google Scholar]
- 18.Heden C-G. Effects of hydrostatic pressure on microbial systems. Bacteriol Rev. 1964;28:14–29. doi: 10.1128/br.28.1.14-29.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helms C, Graham M Y, Dutchik J E, Olson M V. A new method for purifying lambda DNA from phage lysates. DNA. 1985;4:39–49. doi: 10.1089/dna.1985.4.39. [DOI] [PubMed] [Google Scholar]
- 20.Hoppert M, Mayer F. Electron microscopy of native and artificial methylreductase high-molecular weight complexes in strain Gö 1 and Methanococcus voltae. FEBS Lett. 1990;267:33–37. doi: 10.1016/0014-5793(90)80281-m. [DOI] [PubMed] [Google Scholar]
- 21.Howard L, Tipper D J. A polypeptide bacteriophage receptor: modified cell wall protein subunits in bacteriophage-resistant mutants of Bacillus sphaericus P-1. J Bacteriol. 1973;113:1491–1504. doi: 10.1128/jb.113.3.1491-1504.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang W M, Marmur J. Characterization of inducible bacteriophages in Bacillus licheniformis. J Virol. 1970;5:237–246. doi: 10.1128/jvi.5.2.237-246.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hudnik-Plevnik T, Bertani G. Recombination in bacteriophage P2: recA dependent enhancement by ultraviolet irradiation and by transfection with mixed DNA dimers. Mol Gen Genet. 1980;178:131–141. doi: 10.1007/BF00267221. [DOI] [PubMed] [Google Scholar]
- 24.Humphrey S B, Stanton T B, Jensen N S, Zuerner R L. Purification and characterization of VSH-1, a generalized transducing bacteriophage of Serpulina hyodysenteriae. J Bacteriol. 1997;179:323–329. doi: 10.1128/jb.179.2.323-329.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ikeda H, Tomizawa J. Transducing fragments in generalized transduction by phage P1. I. Molecular origin of the fragments. J Mol Biol. 1965;14:85–109. doi: 10.1016/s0022-2836(65)80232-7. [DOI] [PubMed] [Google Scholar]
- 26.Ishiguro E E, Ainsworth T, Harkness R E, Kay W W, Trust T J. A temperate bacteriophage specific for strains of Aeromonas salmonicida possessing A-layer, a cell surface virulence factor. Curr Microbiol. 1984;10:199–202. [Google Scholar]
- 27.Jarrell K F, Bayley D P, Florian V, Klein A. Isolation and characterization of insertional mutations in flagellin genes in the archaeon Methanococcus voltae. Mol Microbiol. 1996;20:657–666. doi: 10.1046/j.1365-2958.1996.5371058.x. [DOI] [PubMed] [Google Scholar]
- 28.Jarrell K F, Julseth C, Pearson B, Kuzio J. Paucity of the Sau3AI recognition sequence (GATC) in the genome of Methanococcus voltae. Mol Gen Genet. 1987;208:191–194. [Google Scholar]
- 29.Jarrell K F, Koval S F. Ultrastructure and biochemistry of Methanococcus voltae. Crit Rev Microbiol. 1989;17:53–87. doi: 10.3109/10408418909105722. [DOI] [PubMed] [Google Scholar]
- 30.Kadurugamuwa J L, Beveridge T J. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J Bacteriol. 1995;177:3998–4008. doi: 10.1128/jb.177.14.3998-4008.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kahn M E, Barany F, Smith H O. Transformasomes: specialized membranous structures that protect DNA during Haemophilus transformation. Proc Natl Acad Sci USA. 1983;80:6927–6931. doi: 10.1073/pnas.80.22.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konisky J, Lynn D, Hoppert M, Mayer F, Haney P. Identification of the Methanococcus voltae S-layer structural gene. J Bacteriol. 1994;176:1790–1792. doi: 10.1128/jb.176.6.1790-1792.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar V, Fonstein M, Haselkorn R. Bacterium genome sequence. Nature. 1996;381:653–654. doi: 10.1038/381653a0. [DOI] [PubMed] [Google Scholar]
- 34.Lewis L O, Yousten A A. Bacteriophage attachment to the S-layer proteins of the mosquito-pathogenic strains of Bacillus sphaericus. Curr Microbiol. 1988;17:55–60. [Google Scholar]
- 35.Lotz W. Defective bacteriophages: the phage tail-like particles. Prog Mol Subcell Biol. 1976;4:54–102. [Google Scholar]
- 36.Margolin P. Generalized transduction. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 1154–1168. [Google Scholar]
- 37.Marrs B. Genetic recombination in Rhodopseudomonas capsulata. Proc Natl Acad Sci USA. 1974;71:971–973. doi: 10.1073/pnas.71.3.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masters M. Generalized transduction. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W D, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 2421–2441. [Google Scholar]
- 39.Meile L, Abenschein P, Leisinger T. Transduction in the archaebacterium Methanobacterium thermoautotrophicum Marburg. J Bacteriol. 1990;172:3507–3508. doi: 10.1128/jb.172.6.3507-3508.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller J F, editor. Methods in enzymology. 204. Bacterial genetic systems. New York, N.Y: Academic Press, Inc.; 1991. [Google Scholar]
- 41.Okubo S, Stodolsky M, Bott K, Strauss B. Separation of the transforming and viral deoxyribonucleic acids of a transducing bacteriophage of Bacillus subtilis. Proc Natl Acad Sci USA. 1963;50:679–686. doi: 10.1073/pnas.50.4.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ozeki H. Abortive transduction in purine-requiring mutants of Salmonella typhimurium. Carnegie Inst Washington Publ. 1956;612:97–106. [Google Scholar]
- 43.Rapp B J, Wall J D. Genetic transfer in Desulfobacterium desulfuricans. Proc Natl Acad Sci USA. 1987;84:9128–9130. doi: 10.1073/pnas.84.24.9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reeve J N. Molecular biology of methanogens. Annu Rev Microbiol. 1992;46:165–191. doi: 10.1146/annurev.mi.46.100192.001121. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 46.Schlesinger M. Ueber die Bindung des Bakteriophagen an homologe Bakterien. I Z Hyg. 1932;114:136–148. [Google Scholar]
- 46a.Schlesinger M. Ueber die Bindung des Bakteriophagen an homologe Bakterien. II Z Hyg. 1932;114:149–160. [Google Scholar]
- 47.Schmieger H. Phage P22 mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119:75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- 48.Seaman E, Tarmy E, Marmur J. Inducible phages of Bacillus subtilis. Biochemistry. 1964;3:607–613. doi: 10.1021/bi00893a001. [DOI] [PubMed] [Google Scholar]
- 49.Sibold L, Henriquet M. Cloning of the trp genes from the archaebacterium Methanococcus voltae: nucleotide sequence of the trpBA genes. Mol Gen Genet. 1988;214:439–450. doi: 10.1007/BF00330478. [DOI] [PubMed] [Google Scholar]
- 50.Sinsheimer R L. Purification and properties of bacteriophage φX174. J Mol Biol. 1959;1:37–42. doi: 10.1016/s0022-2836(63)80017-0. [DOI] [PubMed] [Google Scholar]
- 51.Sitzmann J, Klein A. Physical and genetic map of the Methanococcus voltae chromosome. Mol Microbiol. 1991;5:505–513. doi: 10.1111/j.1365-2958.1991.tb02134.x. [DOI] [PubMed] [Google Scholar]
- 52.Stadtman T C, Barker H A. Studies on the methane fermentation. X. A new formate-decomposing bacterium, Methanococcus vannielii. J Bacteriol. 1951;62:269–280. doi: 10.1128/jb.62.3.269-280.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Starich T, Cordes P, Zissler J. Transposon tagging to detect a latent virus in Myxococcus xanthus. Science. 1985;230:541–543. doi: 10.1126/science.2996138. [DOI] [PubMed] [Google Scholar]
- 54.Steensma H Y, Robertson L A, van Elsas J D. The occurrence and taxonomic value of PBSX-like defective phages in the genus Bacillus. Antonie Leeuwenhoek. 1978;44:353–366. doi: 10.1007/BF00394312. [DOI] [PubMed] [Google Scholar]
- 55.Sternberg N L, Maurer R. Bacteriophage-mediated generalized transduction in Escherichia coli and Salmonella typhimurium. Methods Enzymol. 1991;204:18–43. doi: 10.1016/0076-6879(91)04004-8. [DOI] [PubMed] [Google Scholar]
- 56.Tracy S. Improved rapid methodology for the isolation of nucleic acids from agarose gels. Prep Biochem. 1981;11:251–268. doi: 10.1080/00327488108061767. [DOI] [PubMed] [Google Scholar]
- 57.Tsutsumi Y, Hirokawa H, Shishido K. A new defective phage containing a randomly selected 8 kilobase-pair fragment of host chromosomal DNA inducible in a strain of Bacillus natto. FEMS Microbiol Lett. 1990;72:41–46. doi: 10.1016/0378-1097(90)90341-m. [DOI] [PubMed] [Google Scholar]
- 58.Tumbula D L, Bowen T I, Whitman W B. Characterization of pURB500 from the archaeon Methanococcus maripaludis and construction of a shuttle vector. J Bacteriol. 1997;179:2976–2986. doi: 10.1128/jb.179.9.2976-2986.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wall J D, Harriman P D. Phage P1 mutants with altered transducing abilities for Escherichia coli. Virology. 1974;59:532–544. doi: 10.1016/0042-6822(74)90463-2. [DOI] [PubMed] [Google Scholar]
- 60.Wall J D, Weaver P F, Gest H. Gene transfer agents, bacteriophages and bacteriocins of Rhodopseudomonas capsulata. Arch Microbiol. 1975;105:217–224. doi: 10.1007/BF00447140. [DOI] [PubMed] [Google Scholar]
- 61.Whitman W B, Ankwanda E, Wolfe R S. Nutrition and carbon metabolism of Methanococcus voltae. J Bacteriol. 1982;149:852–863. doi: 10.1128/jb.149.3.852-863.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Whitman W B, Shieh J, Sohn S, Caras D S, Premachandran U. Isolation and characterization of 22 mesophilic Methanococci. Syst Appl Microbiol. 1986;7:235–240. [Google Scholar]
- 63.Wiman M, Bertani G, Kelly B, Sasaki I. Genetic map of Escherichia coli strain C. Mol Gen Genet. 1970;107:1–31. doi: 10.1007/BF00433220. [DOI] [PubMed] [Google Scholar]
- 64.Wood A J, Whitman W B, Konisky J. Isolation and characterization of an archaebacterial virus-like particle from Methanococcus voltae A3. J Bacteriol. 1989;171:93–98. doi: 10.1128/jb.171.1.93-98.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wood H E, Dawson M T, Devine K M, McConnell D J. Characterization of PBSX, a defective prophage of Bacillus subtilis. J Bacteriol. 1990;172:2667–2674. doi: 10.1128/jb.172.5.2667-2674.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yen H C, Hu N T, Marrs B L. Characterization of the gene transfer agent made by an overproducer mutant of Rhodopseudomonas capsulata. J Mol Biol. 1979;131:157–168. doi: 10.1016/0022-2836(79)90071-8. [DOI] [PubMed] [Google Scholar]