Pldo/ZSWIM8 is a conserved protein that promotes linear actin polymerization at the expense of branched actin in several contexts, ranging from Drosophila morphogenesis to human cell migration.
Abstract
Actin filament polymerization can be branched or linear, which depends on the associated regulatory proteins. Competition for actin monomers occurs between proteins that induce branched or linear actin polymerization. Cell specialization requires the regulation of actin filaments to allow the formation of cell type–specific structures, like cuticular hairs in Drosophila, formed by linear actin filaments. Here, we report the functional analysis of CG34401/pelado, a gene encoding a SWIM domain–containing protein, conserved throughout the animal kingdom, called ZSWIM8 in mammals. Mutant pelado epithelial cells display actin hair elongation defects. This phenotype is reversed by increasing actin monomer levels or by either pushing linear actin polymerization or reducing branched actin polymerization. Similarly, in hemocytes, Pelado is essential to induce filopodia, a linear actin-based structure. We further show that this function of Pelado/ZSWIM8 is conserved in human cells, where Pelado inhibits branched actin polymerization in a cell migration context. In summary, our data indicate that the function of Pelado/ZSWIM8 in regulating actin cytoskeletal dynamics is conserved, favoring linear actin polymerization at the expense of branched filaments.
Introduction
The regulation of actin dynamics is essential in each cell to establish cell shape and drive cell division, migration, or morphological changes of whole tissues. Actin polymerization is regulated by several proteins that bind to either monomeric or filamentous actin (Vitriol et al, 2015; and rev. in Revenu et al [2004], Campellone and Welch [2010], Bogdan et al [2013], Davidson and Wood [2016], Carlier and Shekhar [2017], Rottner et al [2017], and Hohmann and Dehghani [2019]). Over 150 proteins have been described to date, to participate as actin-binding proteins, promoting or inhibiting specific features of its dynamic polymerization (Luo et al, 1997; Mullins et al, 1998; Machesky et al, 1999; Zallen et al, 2002; Fricke et al, 2009; Ito et al, 2011; Chen et al, 2014; Gurung et al, 2016 and rev. in Frankel and Moosekert [1996], Goley and Welch [2006], Kovar [2006], Takenawa and Suetsugu [2007], Pollitt and Insall [2009], Fletcher and Mullins [2010], Rotty et al [2012], and Bogdan et al [2013]). Actin polymerization can be classified as either branched or linear, depending on the associated regulatory proteins (rev. in Campellone and Welch [2010] and Suarez and Kovar [2016]). Linear actin polymerization generally requires the activity of the formin protein family, like Diaphanous/Dia (rev. in Campellone and Welch [2010] and Bogdan et al [2013]), whereas branched polymerization requires the activation of the Arp2/3 complex, which is regulated among others, by nucleator promoting factors (NPFs) from the WASP family, including Scar/WAVE or Wasp (Mullins et al, 1998; and rev. in Frankel and Mooseker [1996], Le Clainche and Carlier [2008], Campellone and Welch [2010], Jockusch [2017], and Gautreau et al [2022]). Although cells have a high concentration of actin monomers (Koestler et al, 2009; Burke et al, 2014; and rev. in Pollitt and Insall [2009]), most of them are sequestered by different regulatory proteins, establishing a competition for actin monomers between the two distinct forms of polymerization (Burke et al, 2014; Henson et al, 2015; Suarez et al, 2015; and rev. in Suarez and Kovar [2016]). In most cells, there are a larger number of proteins that induce branched actin polymerization, and thus, competition for actin monomers is often tilted toward branched polymerization arrangements (Suarez et al, 2015). Profilin (chickadee/chic in Drosophila) is pivotal in the polymerization selection process with Chic/Profilin both favoring linear actin polymerization, mediated by the formin protein family, and also inhibiting branched polymerization by preventing the binding of NPFs to actin monomers (Suarez et al, 2015).
Cell specialization requires actin cytoskeleton regulation to allow the formation of specific structures, like for example cuticular hairs in Drosophila. Cuticular epithelial cells in Drosophila form a single cuticular actin hair at the apico-distal vertex of each cell. Hair formation depends on the Frizzled planar cell polarity (Fz/PCP) pathway (rev. in Seifert and Mlodzik [2007], Vladar et al [2009], Adler [2012], Peng and Axelrod [2012], Carvajal-Gonzalez and Mlodzik [2014], Devenport [2014], Hale and Strutt [2015], Yang and Mlodzik [2015, Aw and Devenport [2017], Humphries and Mlodzik [2018], and Harrison et al [2020]), which controls the positioning and polarized actin accumulation by regulating the hierarchical activity of groups of genes that constitute it: core PCP genes (frizzled/fz, disheveled/dsh, diego/dgo, Van gogh/Vang, prickle/pk, and flamingo/fmi) and planar polarity effector (PPE) genes, which either inhibit actin polymerization at the proximal side of wing epithelial cells (e.g., inturned, fritz, fuzzy, and multiple wing hairs/mwh) or promote actin polymerization as positive effectors of the Fz/Dsh/Dgo complex (Axelrod et al, 1998; Shimada et al, 2001; Jenny et al, 2003, 2005; Das et al, 2004; Collier et al, 2005; Park et al, 2006; Wu et al, 2008; Wu & Mlodzik, 2008; Yan et al, 2008, 2009). One of these downstream effectors of the Fz/Dsh complex is RhoA (also known as Rho1 in Drosophila), which locally activates its effector proteins, including in this context the Drosophila Rho-associated kinase, Drok (Strutt et al, 1997; Winter et al, 2001). Drok activity focuses actin polymerization but does not affect the location or orientation of wing hairs (Winter et al, 2001; Yan et al, 2009; and rev. in Adler [2012], Gao [2012], Strutt and Strutt [2009], and Carvajal-Gonzalez and Mlodzik [2014]). Actin hair localization and formation is widely used as a proxy to evaluate the Fz/PCP pathway functionality (Mitchell et al, 1983; Lee & Adler, 2004; Lu et al, 2010; Gault et al, 2012; Sagner et al, 2012; Carvajal-Gonzalez et al, 2016; and rev. in Strutt and Strutt [2009], Adler [2012], Gao [2012], and Carvajal-Gonzalez and Mlodzik [2014]).
Actin hair formation also requires the activity of the Shavenbaby/Shavenoid pathway that regulates the actin cytoskeleton (Delon et al, 2003; Ren et al, 2006). Because the hair structure, trichome, is formed by linear actin polymerization, it is a useful and well-characterized model system to identify and analyze proteins that regulate actin dynamics (Eaton et al, 1996; Turner & Adler, 1998; Winter et al, 2001; Guild et al, 2005; He et al, 2005; Ren et al, 2007; Yan et al, 2009; Adler et al, 2013; Matis et al, 2014; Adler, 2018). In particular, the wing epithelial cells in Drosophila are uniquely suited for such analyses (Mitchell et al, 1983; Klein et al, 2006; Sagner et al, 2012; and rev. in Strutt and Strutt [2009], Adler [2012], and Singh and Mlodzik [2012]). As the trichome is present in every cell and requires a specific and exquisite interplay of regulatory factors of actin dynamics, it serves as an excellent model to identify and investigate novel genes required in this process (Eaton et al, 1996; Turner & Adler, 1998; Guild et al, 2005; He et al, 2005; Ren et al, 2007; Fang & Adler, 2010; Adler et al, 2013; Fagan et al, 2014; Adler, 2018). Importantly, trichome formation is critically linked to linear actin polymerization and the associated formin, Diaphanous (Dia) (Mitchell et al, 1983; Lu & Adler, 2015). Despite accumulating knowledge of the mechanisms and proteins regulating polarized actin accumulation during trichome formation, many open questions remain, and particularly, the mechanisms regulating linear versus branched actin polymerization in the cuticular epithelial cells, and in other contexts, remain largely unknown (Turner & Adler, 1998; Delon et al, 2003; Guild et al, 2005; He et al, 2005; Ren et al, 2005, 2007; Fang & Adler, 2010; Fang et al, 2010; Gault et al, 2012).
Here, we describe the identification and functional characterization of CG34401/pelado (pldo), a gene encoding a multidomain protein that is conserved throughout the animal kingdom, called ZSWIM8 in mammals (Shi et al, 2020; the gene named dorado in this case). We show that it is required for the formation of the actin-based trichome formation in Drosophila cuticle cells, most notably on wings and the notum. This gene has also been characterized in Caenorhabditis elegans, as an E3 ubiquitin ligase, named EBAX-1, and is part of a ubiquitin ligase complex that includes elongins B and C and Cullin2 (Wang et al, 2013). Although this E3 ligase function was also described in specific cell lines (Shi et al, 2020), our data suggest that in the context of actin dynamics, Pldo/ZSWIM8 may act independently of its E3 ligase activity.
Our functional genetics and epistasis analyses demonstrate that Pldo favors linear actin polymerization–at the expense of branched polymerization–in Drosophila and human A549 cell line and thus that this function of Pldo in regulating the actin dynamics is conserved. Immunoprecipitation assays suggest that Pldo might be part of a multiprotein complex that includes Scar/WAVE, Diaphanous, and Profilin. Structure function studies reveal that the N-terminal region of Pldo, containing the highly conserved SWIM, FH1, and WH2 domains, is sufficient to promote the formation/induction of filopodia in general and that the C-terminal part might be required for cell type–specific actin-related functions, for example, in epithelial cells.
Noteworthily, the pelado gene was also identified in a screen related to human diseases (microcephaly) (Yamamoto et al, 2014), without a described mechanistic function in that context, which makes it a very interesting protein to analyze. Consistent with our data on actin regulation established below, pldo function in the central nervous system might be related to axon/dendrites formation or radial glial migration that give rise to the neurons that populate the different brain layers, being all processes that requires specific actin polymerization regulation (Kullmann et al, 2020; and rev. in Rust et al [2012], Cooper [2013], and Chou and Wang [2016]). Taken together, our studies suggest that pldo is an essential regulator of actin dynamics in Drosophila and human cells, with a likely link in the disease context and even potential roles related to neurodegenerative diseases.
Results
Pldo, a novel gene required for cellular hair formation in Drosophila
In an ENU mutant suppressor genetic screen, designed to identify the gene responsible for a wing growth phenotype produced by the overexpression driven by an EP line (EP361, [Cruz et al, 2009]), we generated a recessive embryonic lethal mutation on the X chromosome. To study the cellular phenotype in homozygous mutant cells, we used the Mosaic Analysis with a Repressible Cell Marker (MARCM) technique (Wu & Luo, 2006). Strikingly, homozygous mutant epithelial cells from different adult tissues, including the wing and notum, displayed a lack of the trichome (cellular actin–based hairs) phenotype (Fig 1A–C; also Fig S1A, B, G, and H). To identify the affected locus, we employed first a complementation strategy using several genomic duplications spanning the X chromosome, followed by small deficiencies to further restrict the chromosomal region associated with the phenotype. Subsequently, we knocked down the expression of candidate genes present within this chromosomal region by using dsRNA constructs and screened for the absence of trichomes, as seen in the phenotype obtained in homozygous mutant wing cells (Fig S1C, D, G, and H). This approach identified the previously uncharacterized gene CG34401 (specific target region of the RNAi construct is shown in Fig 1D) as responsible for the “loss-of-hairs” phenotype. As the defect associated displayed a hairless, or “bald,” phenotype, we named this gene pelado (pldo, which is the Spanish word for “bald”).
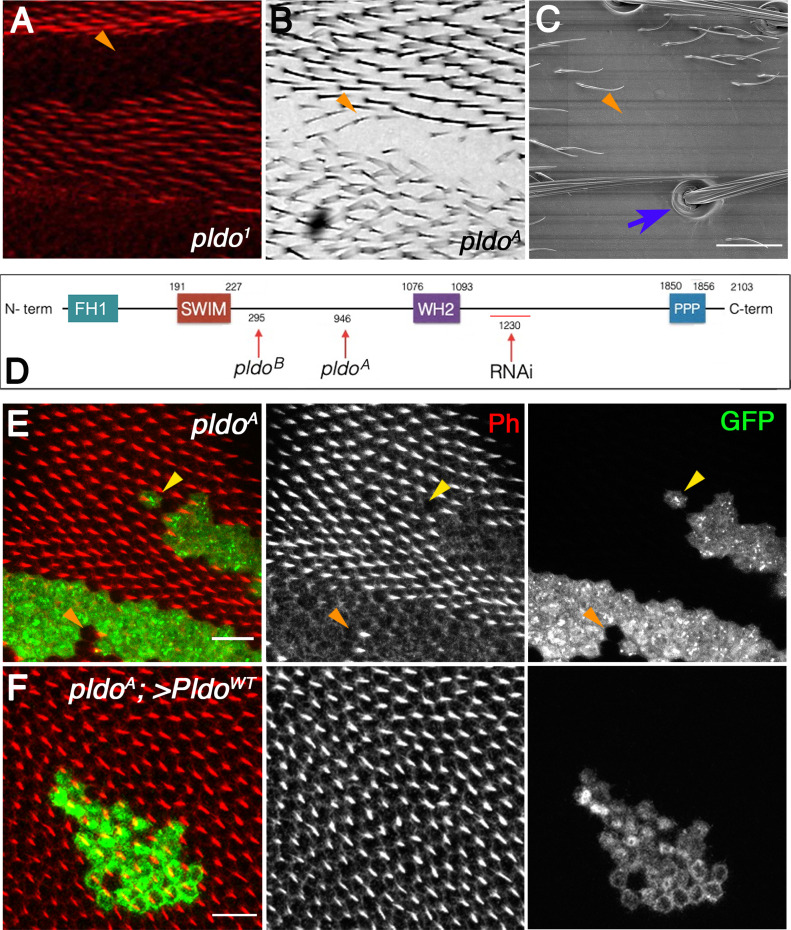
Figure 1. Pldo is required for epithelial cuticle hair formation.
(A, B, C, D) General description of CG34401/pldo-mutant phenotypes and protein sequence. Unmarked pldo1 clones showing the absence of trichomes (actin hairs) in a pupal wing (A), adult wing (B, pldoA clone), and adult thorax (C, SEM micrograph of thorax with pldo1 clone). (A, B, C) Scale bars correspond to 100 μm in (A) and 50 μm in (B) and (C). Note that cuticular cellular hairs are missing (orange arrowheads) but that sensory bristles are not affected (blue arrow). (D) Schematic presentation of the Pldo protein sequence (D) with the main conserved domains indicated (residue numbers are above the protein sequence line). Below the line are the characterized mutant alleles indicated, pldoA and pldoB (available in the Bloomington Stock Center), all showing the same phenotypes. These two alleles have point mutations: pldoA: G18874432A generates a nonsense mutation (W946Stop codon); and pldoB: C18872412T generating the change T295M. pldo1 is not indicated because its mutation has not been identified. The RNAi construct sequence (BL 18553) is also shown. (E) Pupal wings with MARCM pldo-mutant clones (mutant cells marked in green with GFP). Cellular hairs were visualized by rhodamine phalloidin staining (red). Note that the pldo LOF phenotype is fully cell autonomous, as observed in either a single-cell clone (yellow arrowhead) or a two-cell wild-type patch (orange arrowhead) displaying the absence or presence of hairs, respectively. Scale bars correspond to 50 μm. (F) pldo loss-of-function hair phenotype is fully rescued by the expression of PldoWT (via UAS in the MARCM clones), with hairs showing normal polarity and length. Scale bar represents 50 μm.
Figure S1. Pldo is required for hair and laterals formation.
(A, B) Characterization of CG34401/pldo LOF phenotypes. Adult wings with unmarked pldo clones showing the absence of hairs phenotype. In these cases, different alleles for pldo were used (pldo1 and pldoB). Scale bars correspond to 100 μm. (C, D) Adult wings showing the pldo LOF phenotype generated by the expression of RNAi (and RNAi against GFP as a control) in the ptcGal4 expression domain in the wing (note that ptc is expressed between veins 2 and 3 in the wing along the A/P compartment boundary, see schematic of ptc expression in Fig 3A). Scale bars correspond to 50 μm. (E, F) pldo LOF phenotype in adult antenna, generated by ey-Gal4 driven RNAi expression (and control RNAi for comparison). Scale bars correspond to 50 μm. (G, H) Adult thorax showing the absence of cuticular hairs generated by the expression of RNAi (and RNAi against GFP as a control) in the pannierGal4 expression domain in the notum (note that bristles are not affected). The scale bar corresponds to 100 μm. (I, J) Quantification of embryo and larvae mutants for pldo (GFP negative) or control (GFP positive). In each experiment, at least 20 embryo/larvae were considered per genotype, n = 3. Two-way ANOVA plus Sidak’s multiple comparison test was used for statistical analysis, **** indicates P < 0.0001; ** indicates P = 0.0039; and * indicates P = 0.0259. (K) pldo is located on the X chromosome at position X: 18,863,950… X: 18,884,075, and BLAST analysis indicates that pldo is conserved from Caenorhabditis elegans to humans. Protein sequence alignment of main predicted domains in Pldo from different species: Cel: C. elegans, Dros: Drosophila, Drer: Danio rerio, and Hsap: Homo sapiens. Alignment was obtained through the software package Clustal Omega (RRID:001591).
The CG34401/pelado gene (pldo) is located on the X chromosome at position 17F2 and encodes a large protein of 2103 amino acids, approx. 250 kD in size (Fig 1D). In silico analysis of the Pldo sequence (see the Materials and Methods section) revealed the presence of several conserved domains (Figs 1D and S1). The SWIM domain (Swi2/SNF2 and MuDR zinc-binding), which is defined as a domain that can interact with both DNA and other proteins (Ko et al, 2014; Godin et al, 2015), gives the name to the human ortholog of Pldo, ZSWIM8 (Zinc finger SWIM-type containing 8; www.flybase.org). Interestingly, other conserved domains present in Pldo include the FH1 (formin homology 1 domain), a Wiskott–Aldrich homology 2 domain (WH2), and a polyproline region (Fig 1D, also Fig S1K), which are also present in several proteins that regulate actin cytoskeleton dynamics, including Scar/WAVE and the formin Diaphanous/Dia (Watanabe et al, 1997; Miki et al, 1998; and rev. in Paunola et al [2002] and Carlier et al [2013]). In particular, the WH2 domain binds to monomeric actin (rev. in Paunola et al [2002]), and FH1 and polyproline domains can bind to Profilin (Chicadee/Chic in Drosophila) (rev. in Faix and Grosse [2006]).
After the identification of pldo as the gene of interest (the specific location of the mutation in the pldo1 allele is unknown), we identified two additional mutant alleles via complementation analyses. These alleles were generated in a genetic screen in the lab of Hugo Bellen intended to look for mutants related to human diseases (Haelterman et al, 2014; Yamamoto et al, 2014; both available in the Bloomington Drosophila Stock Center/BDSC with stock numbers 52333 and 52334). We subsequently refer to these molecularly characterized alleles as pldoA and pldoB, respectively (Fig 1D), with one being a nonsense mutation that produces a stop codon at residue 946, W946STOP (pldoA), and the other a point mutation, T295M (pldoB; see Fig 1D for details). In accordance with the analysis of our original allele (pldo1), homozygous mutant clones of these characterized alleles also displayed loss of cellular hairs/trichomes (Figs 1E and S1A and B). This phenotype is cell-autonomous (see examples in Fig 1E, with very small clones displaying the respective features). To unequivocally confirm the identity of the gene responsible for the loss of trichomes, we generated a transgenic line to express the pldoWT coding sequence in the pldoA mutant background using the MARCM technique (Wu & Luo, 2006). Indeed, the expression of a pldoWT transgene completely rescued the hair loss defects, with no other evident phenotype in the mutant clones (Fig 1F). Because these BDSC alleles are molecularly characterized and showed the same phenotype as our original allele, we used the pldoA allele for further analyses.
As pldo is a recessive lethal mutation, we next wished to establish the stage of lethality. To determine this, we generated pldo-mutant stocks balanced with FM7 (marked with GFP expressed under twist control), and thus, embryos homozygous mutant for pldo can be identified as GFP-negative embryos or larvae. The number of GFP-negative embryos (pldo mutants) was considerably lower than that of controls. We were also able to observe a small number of mutant larvae (at 24 h AEL [after egg-laying]) with more than half of these being already dead, and those still alive showed severe motor defects (see summary of these results in Fig S1I and J). Taken together with maternal expression of pldo (www.flybase.org), these results suggest an important function of pldo during early embryonic developmental stages, which is obscured by maternal contribution.
Pldo is required for actin hair formation and elongation
Actin hair formation in cuticular epithelial cells takes place in three stereotyped stages: apical actin accumulation, prehair formation, and hair elongation (Guild et al, 2005). Each stage is characterized by specific actin distributions and hair features. At around 28 h after puparium formation (APF), actin starts accumulating apically in the distal vertex of every epithelial wing cell (Mitchell et al, 1983; Turner & Adler, 1998). At 32 h APF, the structure called “prehair” appears, and finally at 36 h APF, the hair is almost completely formed or elongated (Classen et al, 2008; all time points relate to a growing temperature of 25°C). To address the potential role of Pldo in these stages, we generated pldo-mutant clones at third instar larvae and followed the cellular maturation and hair growth in the wing epithelia at the time points of pupal development mentioned above. Although at 28 h APF there were no obvious differences in actin distribution between pldo-mutant clones (GFP-positive cells) and control cells (Fig 2A), clear alterations in actin distribution and defects in (pre)hair formation were observed at 32 and 36 h APF (Fig 2B and C). In pldo-mutant cells, the prehair formation process was impaired, and defects were most evident from early stages of hair elongation. Thus, the core PCP-regulated hair formation process, which requires localized linear actin polymerization (Mitchell et al, 1983; Lu & Adler, 2015), was severely impaired in the pldo-mutant cells.
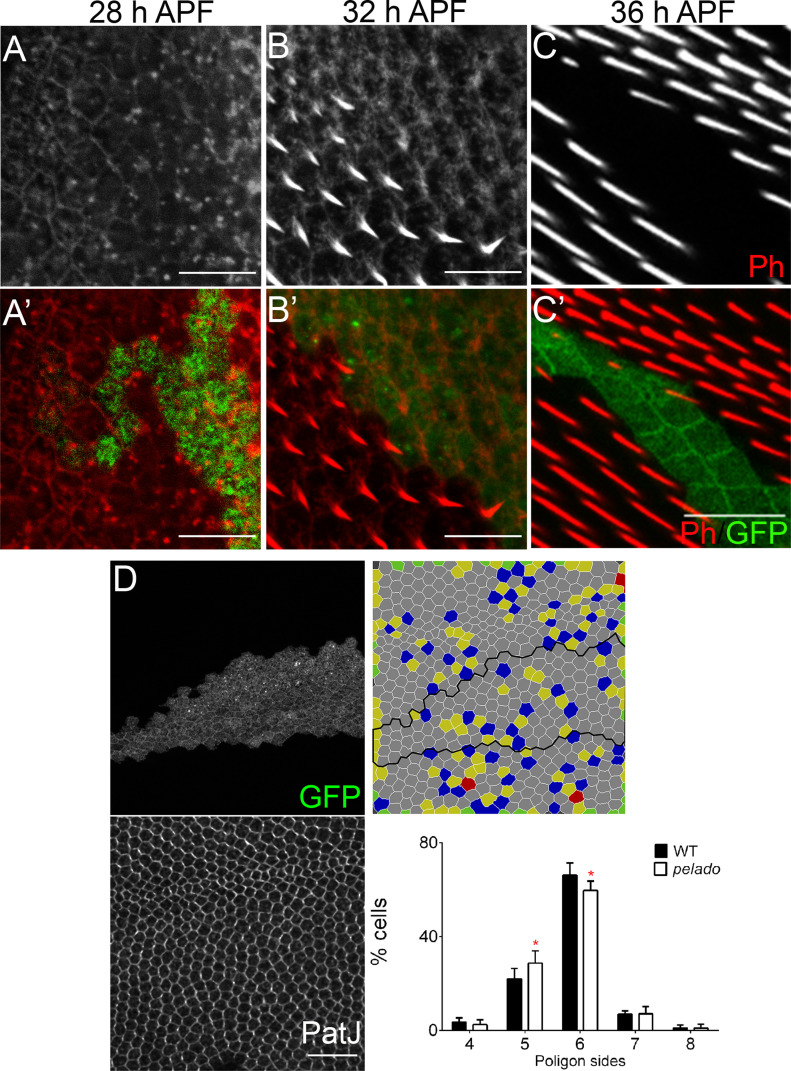
Figure 2. Pldo is required for actin hair formation and cell shape maturation.
(A, B, C) Temporal characterization of the mutant pldo phenotype during cellular hair formation. Confocal micrographs showing pldo MARCM clones (marked by GFP, green) during actin hair formation (labeled via rhodamine phalloidin, Ph). (A, B, C) Monochrome images showing actin alone (Ph) and (A′, B′, C′) displaying clonal marker (green) and actin (red). Note the absence of hair elongation from 30 to 32 h APF onward, in mutant GFP marked cells. (A, A′) Note accumulation of actin appears normal in control and mutant cells at 28 h APF. All the images are oriented with distal in the lower right corner. Scale bars correspond to 50 μm. (D) Evaluation of cell shape in pldo-mutant cells, at 28–32 h APF, as compared with neighboring wild-type cells. Note the significant reduction in the number of hexagonal cells in pldo-mutant clones (marked by GFP; cell outline displayed via PatJ staining, a junctional marker). Six different individuals were analyzed, with around 100 cells in each case. P = 0.0366 via t test. Scales bar correspond to 100 μm.
To further address the role of pldo in the hair formation process, we tested its relationship to the PCP pathway as Fz/PCP signaling is critical to focus actin polymerization to the distal vertex. Earlier stages of PCP establishment, preceding hair formation, are characterized by the asymmetric localization of its core components (e.g., Adler et al, 2013; Matis et al, 2014; and rev. in Strutt and Strutt [2009] and Carvajal-Gonzalez and Mlodzik [2014]). We thus evaluated if the asymmetric core PCP complex localization is affected. Fmi serves as the anchor of both distal and proximal PCP complexes, and its localization reflects this feature as a regular zig-zag pattern enriched in the proximal–distal axis, which can be visualized and quantified (Aigouy et al, 2010). We did not detect any changes in Fmi localization in wing cells mutant for pldo, as compared with neighboring control cells (Fig S2), indicating that Pldo is neither upstream nor part of the regulatory circuit of the core PCP pathway. This observation and interpretation are consistent with the phenotypes seen by altering the expression of core PCP factors or direct regulators of this pathway, with the actin hairs often being formed, but in abnormal number and/or locations (Yan et al, 2008; Lu et al, 2015).
Figure S2. Pldo does not affect the core Fz/PCP pathway.

Asymmetric Flamingo (Fmi) localization, as a proxy for Fz/PCP signaling, is shown. Note that it is not affected in pldo-mutant cells, as compared with surrounding control wt cells, in pupal wings (as shown in the graphs). Clones of mutant cells were generated by the MARCM technique, with mutant cells marked with GFP (green). Polarity strength lines of Fmi staining are shown as orange lines. Six different individuals were used, and around 100 cells were analyzed in each case. Scale bar represents 100 μm.
A second feature regulated by the core PCP pathway, during pupal wing development and in general, is the organization of the epithelial cells themselves with their arrangement into a hexagonal pattern (Wong & Adler, 1993; Sánchez-Gutiérrez et al, 2013; Etournay et al, 2015). This terminal shape establishment follows a cell shape elongation along the proximo–distal axis, when PCP is reorganized from a radial polarity arrangement to a proximal–distal axis alignment (Aigouy et al, 2010). We thus asked whether pldo might be required in this context and compared the polygon distribution of pldo-mutant cells to surrounding wild-type control cells in pupal wing cells at 28–32 h APF. Interestingly, cells mutant for pldo display a reduction in the proportion of hexagonal shape, compared with surrounding control cells (Fig 2D), suggesting a role for Pldo in the cortical actin bundle function.
Taken together with the actin hair phenotype, these data suggest that Pldo likely acts as an effector of the core PCP pathway to regulate actin cytoskeleton–associated functions like cell shape definition and actin accumulation during hair formation and elongation. Consistent with this notion, pldo encodes protein domains associated with actin binding and regulation of its polymerization, such as WH2, FH1, and polyproline domains.
Pldo promotes linear actin polymerization during hair outgrowth
To gain insight into the underlying regulatory mechanism(s) of how Pldo might function in actin filament dynamics, we asked whether the pldo-mutant phenotype can be genetically modified by genes encoding proteins regulating linear and/or branched actin polymerization. Such genes fall into the category of formins, including Dia or the anti-formin Mwh, or the WASP family, including Scar/WAVE and Wasp, among others (a list of actin regulators tested is shown in Fig S3C). Specifically, for example, Dia and Enabled/Ena promote linear actin polymerization, whereas Scar and Wasp induce branched actin polymerization by means of activating the Arp2/3 complex (Machesky et al, 1999; Kang et al, 2010; and rev. in Paunola et al [2002], Goley and Welch [2006], Le Clainche and Carlier [2008], Campellone and Welch [2010], Rotty et al [2012], and Carlier et al [2013]). Mwh, a functional anti-formin inhibits Dia function, preventing linear actin polymerization (Lu & Adler, 2015). To assay for potential interactions, we expressed pldo-IR under the control of ptcGal4 (Fig 3A), causing a reduction of trichomes within the ptc expression domain and thus mimicking the pldo loss-of-function (LOF) phenotype. In this context, a phenotypic rescue of pldo was detected by increasing levels of proteins that induce linear actin polymerization, for example, Dia, and Ena in the ptcGal4>pldo-IR background (Figs 3B and C and S3C). Strikingly, the pldo LOF knockdown phenotype was also “rescued” by reducing Mwh levels (Figs 3C and S3A and B). Noteworthily, a mwh knockdown reversed the pldo LOF phenotype and produced the well-described “multiple wing hair” defects, characterized by the appearance of more than one short hair per cell (Fig S3A and B) (Yan et al, 2008; Lu et al, 2015). As mentioned, Shavenoid/Sha is another component required for trichome formation. sha LOF alleles show either the absence of hairs or the appearance of more than one tiny hair and thus has been described as an inducer of linear actin polymerization (Delon et al, 2003; Ren et al, 2006; Adler, 2018). We therefore tested whether increasing Sha protein levels allowed hairs to form in a ptcGal4>pldo-IR background. Indeed, Sha co-(over)expression in this background restored hair formation (Fig 3B and C). Interestingly, pldo LOF phenotypes in the antenna laterals (Fig S1E and F) are similar to the phenotype observed in shavenoid mutant flies (He & Adler, 2002). Taken together, our data indicate that promoting linear actin polymerization by increasing the levels of Dia, Ena, Sha, and Profilin/Chic in the ptcGal4>pldo-IR background restored hair formation and rescued the pldo LOF phenotype. A similar “rescue” was observed by reducing Mwh levels, an inhibitor of Dia activity.
Figure S3. Pldo mediates competition for monomeric actin during trichome formation, extended.
(A) pldo LOF phenotype reversion evaluated in pupal and adult wings. Confocal images of pupal wings expressing ptcGal4, UAS-Dcr, and UAS-pldoIR in combination with the indicated transgenes. Rhodamine conjugated to phalloidin was used to stain actin filaments, and GFP was used to mark the ptcGAL4 domain in the wing. The scale bar corresponds to 50 μm. (B) Additional adult wings showing the interaction between ptcGal4, UAS-Dcr, and UAS-pldoIR and the indicated transgenes (see Fig 3C for quantification). Quantification was performed as described in the main text Fig 3C. The scale bar corresponds to 50 μm. (C) Table displaying all interactions assayed with ptcGAL4, UAS-Dcr, and UAS-pldoIR. Data are based on scoring at least 10 individuals in each case.
Figure 3. Pldo mediates competition for monomeric actin during trichome formation.
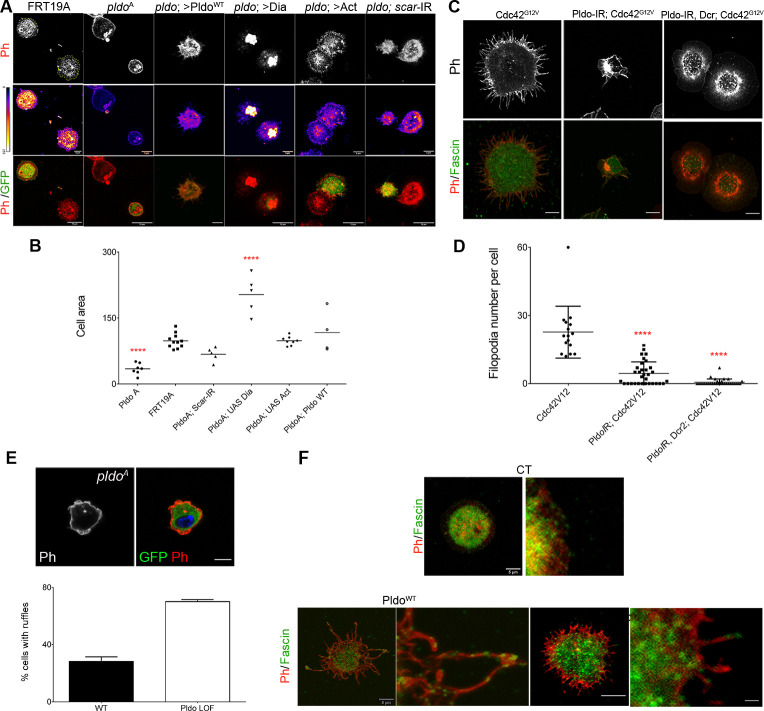
(A) Schematic displaying the ptcGal4 expression pattern (green area) used in pupal and adult wings. (B) Square in drawing corresponds to the region shown in the lower panel (wt control) and (B) panels. (B) Scale bar corresponds to 50 μm (also for panel B). (B, C) Genetic interactions between pldo-IR (knockdown) and factors that regulate actin cytoskeleton polymerization with phenotypic appearance as evaluated in adult wings. (A) Squares display high magnification views of wing area (boxed in schematic in A) expressing ptcGal4, UAS-Dcr, and UAS-pldo-IR in combination with the indicated transgenes, either knocking down additional factors or overexpressing a gene of interest. (A, B) Comparing the control phenotype of ptcGal4, UAS-Dcr, UAS-pldo-IR, and UAS-GFP-IR (upper left panel) to wt in (A) and the genotypes of interest (B). Potential reversion of the pldo-IR (LOF) phenotype was evaluated according to the size of the area in adult wings, which lost the trichomes. (C) Quantification was performed by scoring each wing with either “wild-type” (wt, normal hair number), “partial loss of trichomes” (partial loss), or “complete loss of trichomes” (hair loss) as shown in graph in (C). (B, C) Percentages of the respective appearance are shown in (C), see (B) for phenotypic reference: complete reversion to wild-type as seen with UAS-scar-IR and partial reversion displaying small areas with trichome loss as seen with UAS-Capu (Fig S3B) and no reversion (large area with no trichome) as seen in control UAS-GFP-IR or UAS-wasp-IR. Quantification is based on scoring >10 individuals in each case. (D) Reversion of the pldo (LOF) phenotype as seen in pldo-mutant MARCM clones in pupal wings to confirm the extent of phenotypic rescue. Note that actin and Dia overexpression/gain of function (GOF) showed a complete reversion of pldo-mutant clones in pupal wings. Scale bar corresponds to 25 μm.
Competition for actin monomers in the regulation of linear versus branched actin polymerization has been described (Jaiswal et al, 2013; Burke et al, 2014; Suarez et al, 2015; Vitriol et al, 2015; Davidson et al, 2018; and rev. in Davidson and Wood [2016] and Suarez and Kovar [2016]), and we thus asked whether a similar competitive nature exists during trichome formation. To address this, we asked whether reducing branched actin polymerization by knocking down scar, arp2, arp3, and wasp has an effect in the ptcGal4>pldo-IR background. Strikingly, we observed a reversion of the phenotype in most cases, including scar, arp2, and arp3 knockdowns (Figs 3B and C and S3A and B). The exception was wasp knockdown (Fig 3B and C). However, as Scar and Wasp have specific roles in different cell types (Zallen et al, 2002; Berger et al, 2008; Rajan et al, 2009; Koch et al, 2012), these results suggest that Scar function is relevant in pupal wing cells, but Wasp is not required there. Taken together, these results suggest that competition for actin monomers during trichome formation in Drosophila is a critical component of its regulatory circuitry. To further corroborate this notion, we evaluated the effect of increasing actin monomer levels in the ptcGal4>pldo-IR background. Strikingly, a ptcGal4>pldo-IR, >Act genotype displayed a complete reversion of the trichome loss (Fig 3B and C), consistent with the hypothesis that Pldo influences the competition for actin monomers. Importantly, these phenotypic rescues were also observed when assayed in pldo MARCM clones and analyzed in pupal wings (see Fig 3D for examples).
In summary, these data suggest that the function of Pldo is to promote linear actin polymerization required during trichome formation, which could be either through the inhibition of branched actin polymerization or by directly inducing linear actin polymerization. The combination of either increasing actin itself and linear actin filament promoting factors or the reduction of branched filament promoting factors suggest that Pldo promotes linear actin polymerization by positively acting on its regulators and/or represses branched actin regulators. As such, Pldo functions in this context as a balance tipping factor toward linear actin polymerization, either by a direct promotion of linear actin polymerization or by inhibiting branched actin polymerization or both.
Pelado is required for filopodia formation in hemocytes
Because the actin cytoskeleton is essential in every cell, and pldo-mutant animals die early in development, we assumed that Pldo is required in many if not all cells. The actin cytoskeleton is also important to regulate and maintain cell shape and changes in cell shape in pldo-mutant cells were observed in wing epithelial cells (Fig 2D). We thus next asked whether pldo is required to regulate cell morphology in hemocytes, a migratory Drosophila blood cell similar to mammalian macrophages (Moreira et al, 2013) (Fig 4A). pldo-mutant hemocytes (generated by the MARCM technique) displayed reduced attachment cell area and a significant increase in structures known as ruffles (Fig 4A, bottom panel, and Fig S4E). This phenotype was rescued by expressing PldoWT in this experimental setting (Fig 4B, rescued cell is marked by GFP). Exogenous expression of PldoWT in the pldo-mutant cells, likely an overexpression relative to its endogenous levels, did in fact not just rescue the LOF defects but caused an increase in the formation of filopodia-like protrusions (Fig 4B). This observation was consistent with its proposed role in favoring linear actin polymerization (which could be a direct effect or by preventing branched actin polymerization). Moreover, similar to the wing hair formation assay (Fig 3), the pldo hemocyte defects were also rescued by co-(over)expression of Dia, actin, or by IR-mediated LOF of scar (Fig S4A and B).
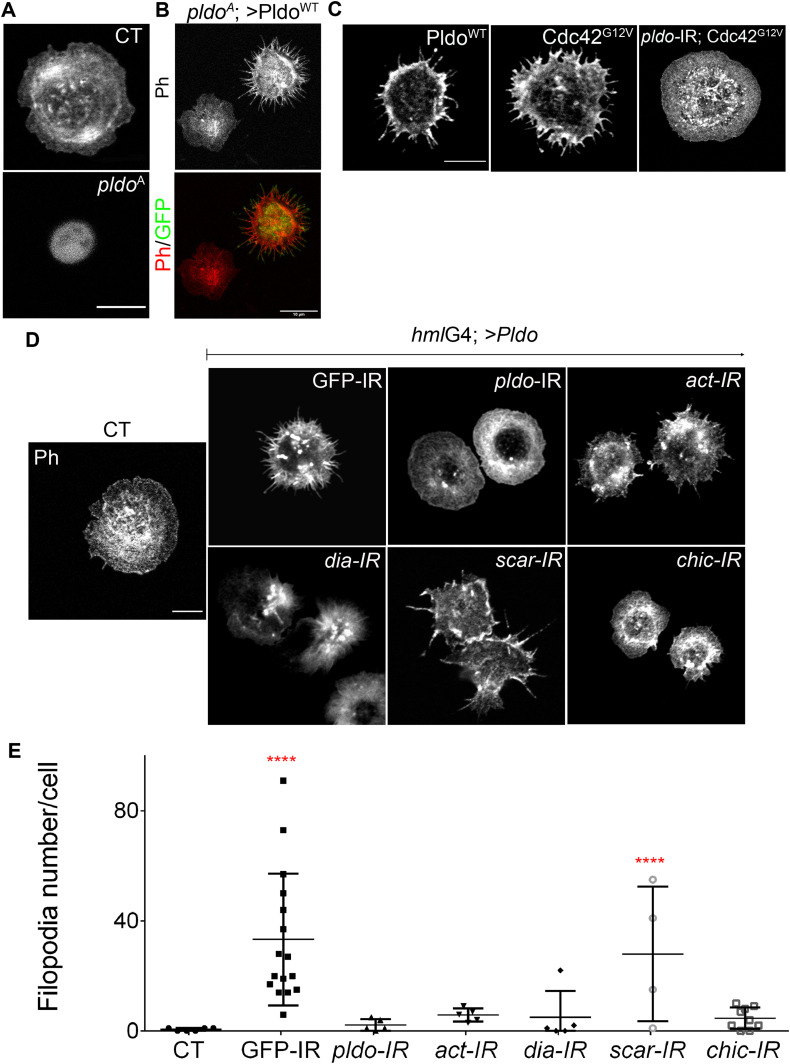
Figure 4. Pldo induces formation of filopodia in hemocytes.
(A) Confocal microscopy images of hemocytes stained with rhodamine phalloidin to visualize actin filaments. Control wild-type hemocyte (CT, top) is shown in comparison to pldo-mutant cells, which show a reduction in the cell attachment area (pldoA, bottom panel). Scale bar = 5 μm. (B) Reduction in the cell attachment area was rescued by co-expression of PldoWT. Note that such expression of PldoWT even creates a GOF phenotype, inducing the formation of filopodia-like protrusions. Rescued cell is marked by GFP (green) with the MARCM technique. Scale bar corresponds to 10 μm. (C) PldoWT expression in hemocytes induce filopodia formation, similar to Cdc42G12V expression, a known inducer of filopodia. Formation of filopodia in Cdc42G12V background depends on pldo function (also Fig S4D for quantification). Scale bar corresponds to 5 μm. (D, E) Filopodia formation induced by Pldo overexpression in hemocytes as driven by hml-Gal4 driver (second panel from left, GFP-IR control, cf. to wt in the left most panel) was suppressed by knockdown (via IR) of either pldo, act, Dia, or chic (Profilin) and enhanced by scar knockdown. Scale bar corresponds to 5 μm. (E) Quantification of the number of filopodia per cell. At least 10 cells were quantified in each genotype in three independent experiments. Statistical analysis was performed by two-way ANOVA, Tukey posttest with ***P < 0.0001.
Figure S4. Pldo is essential to induce filopodia formation and to sustain cell shape in hemocytes.
(A) pldo-mutant cells were generated by the MARCM technique (expressing GFP, green cells). An FRT19A chromosome was used as a wild-type control. Note that pldo-mutant cells show a reduction in the cell attachment area that was rescued by the expression of either Pldo, Dia, or Act or by knocking down scar. Scale bars correspond to 10 μm. (B) Cell area was quantified using the phalloidin staining (red and heatmap), using at least 10 cells per genotype in each experiment (and three independent experiments). Statistical analysis was one-way ANOVA, Tukey posttest P < 0.0001. (C, D) Filopodia formation induced by the expression of Cdc42G12V in hemocytes, was evaluated by knocking down pldo. With or without Dicer expression in the LOF of Pldo, there was a significant reduction in filopodia formation by Cdc42G12V. Scale bars represents 5 μm. Number of filopodia per cell was quantified in at least 10 cells per genotype in each experiment (and three independent experiments). Statistical analysis was one-Way ANOVA, Tukey posttest P < 0.0001. (E) Ruffle formation in pldo-mutant cells. The ruffle presence was quantified in at least 10 cells per genotype in each experiment (three independent experiments). The scale bar represents 5 μm. (F) Fascin (green, staining bundled linear actin) serves as a marker for mature filopodia. Higher magnification of filopodia showing the fascin presence in the distal filopodial region in Pldo GOF or Cdc42G12V. The scale bar represents 5 and 20 μm in high magnification views.
Pldo overexpression in hemocytes induces a significant increase in filopodia-like protrusions formation (Fig 4C, left panel). To confirm that these protrusions were filopodia, we stained the cells with anti-Fascin, a known marker of mature filopodia (Vignjevic et al, 2006). We observed Fascin staining in some of the filopodia, in all such conditions (Fig S4F). cdc42 is a well-defined inducer of filopodia formation (Leung et al, 1998). As the Pldo-induced protrusions resembled the effect of expressing constitutively active Cdc42 (Fig 4C, middle and left panel, respectively), we thus asked whether Cdc42G12V-induced filopodia in hemocytes depend on Pldo function. Strikingly, an RNAi knockdown of pldo caused a total reversion of the Cdc42G12V induced filopodia formation (Fig 4C, right panel, and Fig S4C and D). Together, these data indicate that Pldo is essential for filopodia formation in hemocytes downstream of Cdc42 and strongly supports the notion that Pldo directly favors linear actin polymerization.
Similar to the rescue experiments of trichome formation in pupal wings, Pldo-induced filopodia formation depends on regulators of linear actin polymerization, and as such, its filopodia-inducing effect was reversed by reducing the levels of Dia, Actin, or Chic/Profilin and enhanced by reduction in the scar gene, promoting branched actin (Fig 4D and E). Again, this is consistent with the notion that Pldo promotes linear actin polymerization in the cellular competition with actin monomers.
Taken together, the wing cuticle epithelial cell and hemocyte studies indicate that Pldo possesses a general function in regulating the competition for actin monomers and favoring linear actin polymerization over branched filaments.
N-terminal region of Pldo is sufficient to induce filopodia formation
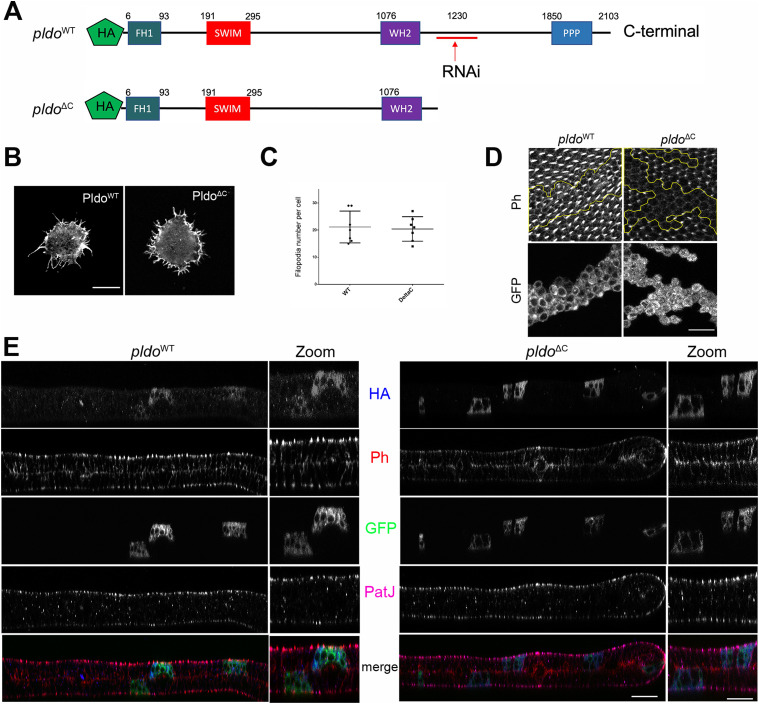
To evaluate which regions of the Pldo protein were essential for its function in actin polymerization, we generated a mutant isoform deleting its C-terminal portion (PldoΔC) (Fig 5A). We first compared the gain-of-function (GOF) phenotypes of PldoWT and PldoΔC in the hemocyte assay, noting that both PldoWT and PldoΔC induce filopodia formation equally well (Figs 5B and C and S5B).
Figure 5. N-terminal region of Pldo is sufficient to induce filopodia formation.
(A) Schematic of Pldo highlighting the deletion isoform with C-terminal deletion, PldoΔC (note that the RNAi sequences are absent in the PldoΔC construct). Both constructs have an HA tag at the N-terminus. (B, C) PldoΔC was sufficient to induce formation of filopodia in hemocytes, to the same extent as full-length PldoWT. (B, C) Examples shown in (B) and quantification in (C), showing the number of filopodia per cell. Scale bar represents 10 μm. At least 10 cells were analyzed in each condition in three independent experiments. Statistical analysis with t test, P = 0.8028, nonsignificant. (D) Expression of PldoΔC in MARCM pldo-mutant cells (right panels) in pupal wings was not able to rescue hair formation, compared to pldoWT expression (left panels). Distal is right. Scale bar represents 25 μm. (E) Subcellular localization (z-plane) analyses of HA-tagged PldoWT (left panels) and PldoΔC (right panels), displaying rhodamin phalloidin (Ph, red) staining actin filaments, clonal marker (GFP, green, via MARCM, mutant cells), PatJ (magenta) as a junctional (apical) cell marker, and HA-Pldo (blue) showing Pldo localization. Note that there is no detectable difference in the localization of PldoWT and PldoΔC (HA staining, blue) in pupal wing cells. Right panels (zoom) show higher magnification. Scale bar represents 25 μm.
Figure S5. N-terminal region of Pldo is sufficient to induce filopodia formation.
(A) PldoΔC was not capable to rescue the lack of trichome phenotype observed by pldo LOF in adult wings, when co-expressing with the UAS-pldo-IR in the ptcGAL4 expression domain. No change in the phenotype was observed, confirming the pldo LOF mutant defects, as seen in pupal wings (Fig 5D). The scale bar represents 50 μm. (B) Localization of Pldo was evaluated in confocal microscopy images, through the HA-tag staining, in hemocytes, observing no notable differences. These experiments were performed at 18°C, with near wild-type expression levels to prevent overexpression of the transgenes. Scale bar represents 5 μm. (C, D) Localization of Pldo was evaluated in confocal microscopy images, through the HA-tag staining, in pupal wings, before and after hair formation, observing no notable differences. (C, D) These experiments were performed at 18°C, at 64 (C), or 72 h (D) APF, with near wild-type expression levels to prevent overexpression of the transgenes. Note at the bottom the colocalization between Pldo-HA and Scar. The scale bar represents 25 μm.
In the context of actin hair formation in wing cells, however, PldoWT fully rescued the pldo LOF mutant phenotype, but expression of PldoΔC in mutant clones did not rescue the loss-of-hair defect (Fig 5D). Similarly, PldoΔC expression did not rescue the loss-of-hair phenotype in the ptcGal4>pldo-IR background (Fig S5A). These results indicate that the N-terminal portion of Pldo is sufficient to induce linear actin polymerization, for example, in hemocytes, but that its C-terminal region is required in more complex situations in epithelial cells, like trichome formation, likely because of a regulatory function or for subcellular localization. To test for the latter, we asked whether the C-terminal portion of Pldo is required to localize the protein to a specific region within the wing cells. Surprisingly, we did not detect a difference in the localization between PldoWT and PldoΔC in either wing cells (Figs 5E and S5C and D), or hemocytes (Fig S5B). Similar to Scar (Zallen et al, 2002; Kunda et al, 2003; Beli et al, 2008; Rodriguez-Mesa et al, 2012), Pldo localization is detected throughout the cell, which has also been noted for its human ortholog ZSWIM8 (Okumura et al, 2021).
In this context, we also tried to identify the minimal portion of Pldo required to induce linear actin polymerization by generating additional C-terminal deletions (Pldo1-685 and Pldo1-515 isoforms, as compared with 1–1,105 on PldoΔC). However, these constructs were cell lethal, in both Drosophila S2 cells and human A549 cells.
Pldo function in actin cytoskeletal regulation is conserved in mammalian cells
We next wished to determine whether the function of Pldo in actin regulation is a conserved feature of the protein and used a cell migration assay in human cells to this end. Cell migration is important in many different developmental and physiological processes and requires specific actin cytoskeletal dynamics in a highly regulated process (rev. in Le Clainche and Carlier [2008]). Different cells use distinct mechanisms for migration and can also change between different types of migration. The common component in the distinct mechanisms of cell migration is actin. Similarly, a key feature during cell migration is the up-regulation of the Arp2/3 complex activity (rev. in Swaney and Li [2016], Jockusch [2017], and Shellard and Mayor [2020]). We thus used a cell migration assay as a tool for an initial insight whether Pldo function in actin regulation is conserved in human cells.
To this end, we used the human A549 cell line (kindly provided by the Bartel laboratory [Shi et al, 2020]). We established a clonal knockout cell line for ZSWIM8−/−, with ZSWIM8 being the human pldo ortholog, and analyzed its effect in a cell migration wound healing assay. Strikingly, cells mutant for pldo/ZSWIM8 showed an increase in the migration rate, as compared with wild-type control A549 cells (Fig 6A and B). Importantly, this effect was fully reversed by the expression of Drosophila Pldo in those cells (Fig 6A and B), confirming that ZSWIM8/Pldo also regulates actin polymerization in mammalian cells and that the Drosophila Pldo protein is functionally equivalent in this context. These data are very similar to experiments demonstrating that Arp2/3 complex activity and, as consequence, branched actin filament formation are essential in the wound healing assay (Zhao et al, 2020; using the same cell line) and thus suggest that Pldo’s function is also to prevent branched actin polymerization.
Figure 6. Pldo function in actin cytoskeletal regulation is conserved in human cells.
(A, B) Pldo/ZSWIM8 function was evaluated in a scratch wound migration assay in human cells (cell line A549). pldo/ZSWIM8 mutant cells showed a significant increase in the rate of migration during assay. This effect was reversed by expressing the Drosophila pldo gene (independent experiments: n = 3). Scale bar represents 50 μm. (B) Quantification of % wound closure over time. Statistical analysis was performed via a two-way ANOVA, Tukey posttest, P < 0.0001, “a” indicates ** between CT and zswim8−/−; “b” indicates *** between CT+Pldo and zswim8−/−; and “c” indicates *** between zswim8−/− and zswim8−/− +Pldo. (C) Immunoprecipitation (IP) assay documenting an interaction of Pldo with different proteins that regulate actin cytoskeletal dynamics in Drosophila S2 cells transfected with HA-PldoWT and immunoprecipitated with anti-HA. Note that Dia, Scar, and Chic/Profilin all co-IP with Pldo, suggesting that they form a complex. Actin was not co-IPed with Pldo. The vertical red line indicates a blot crop bringing the molecular weight markers in proximity of the IP-ed proteins. (D) Schematic of proposed models for Pldo function: either forming a multiprotein complex with Scar, Dia, and Chic/Profilin during the regulation of actin dynamics (left side) or binding to these proteins independently (right side).
Source data are available for this figure.
Taken together, our data indicate that the Pldo/ZSWIM8 function in the regulation of actin cytoskeletal elements is conserved between Drosophila and human cells and further confirm the notion that these proteins function in general to favor linear actin polymerization over branched filaments.
Pldo physically associates with regulators of actin polymerization
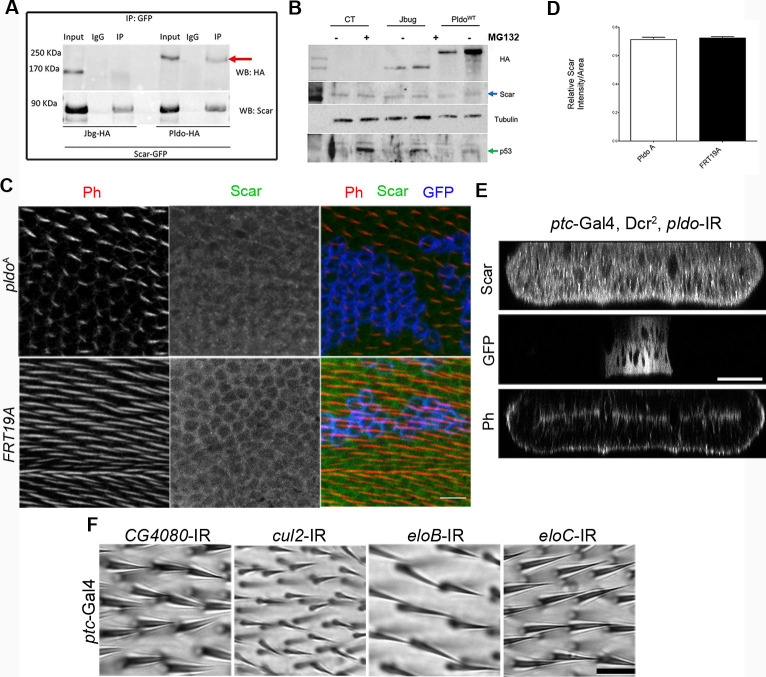
The actin polymerization regulators mDia2 and Scar/WAVE are known to be part of the same molecular complex and that the binding of Cdc42 modulates their interaction to induce filopodia formation (Krugmann et al, 2001; Beli et al, 2008; Goh et al, 2012). We thus asked whether Pldo is also associated with these and/or other proteins that regulate actin cytoskeleton dynamics. To test this hypothesis, we used the Drosophila S2 R+ cell line (Schneider, 1972; which is a macrophage-like lineage derived from late embryos), and we observed that Pldo was indeed part of a molecular complex with Dia, Scar/WAVE, and Chic/Profilin (Fig 6C). We also evaluated the potential colocalization of Pldo and Scar in pupal wing cells. These data are however not informative as both proteins are detected throughout the whole cell (Fig S5C). Together, these observations suggest that Pldo could be regulating the activity of these proteins as part of the described complex (Miki et al, 2000; Krugmann et al, 2001; Beli et al, 2008; Goh et al, 2012), including Dia and Scar/WAVE.
Two reports characterizing EBAX-1, the pldo ortholog in C. elegans, and human ZSWIM8, respectively (Wang et al, 2013; Shi et al, 2020), suggest that it can function as an E3 ubiquitin ligase, being part of a ubiquitin ligase complex including elongins B and C, and Cullin-2 (Wang et al, 2013). Because pldo LOF is reversed by reducing Scar levels and Pldo and Scar are mutually co-immunoprecipitated (Fig 6C, also Fig S6A) and colocalized in pupal wings (Fig S5C), we hypothesized that Scar could be a target of such a Pldo function. Accordingly, if Scar were the target of Pldo in the context of a ubiquitin ligase complex, we would expect a change in Scar levels in mutant cells for pldo. However, we did not observe an effect of pldo on Scar protein levels or localization, as compared with wild-type control cells (Fig S6C–E). To further test this hypothesis, we evaluated Scar levels in S2 cells with or without the proteasome inhibitor MG132. If Pldo was promoting Scar degradation, we would expect an increase in Scar levels upon MG132 treatment. However, MG132 did not cause an accumulation of Scar (Fig S6B), whereas p53 (as a control protein known to be degraded by the proteasome) (Scheffner et al, 1993; Maki et al, 1996; Haupt et al, 1997; Honda et al, 1997; Ashcroft & Vousden, 1999; Yang et al, 2017; and rev. in Pant and Lozano [2014]) was increased. To address this further in vivo during trichome formation, we tested whether the knockdown of components of the ubiquitin ligase complex (described to act with EBAX-1 in C. elegans) would have an effect on hair formation (similar to the pldo LOF phenotype). However, LOF knockdown of elongins B/C or Cullin2 showed no defects in hair formation (Fig S6F). The same was observed for the knockdown of CG4080, a described E2 ligase that might act with Pldo (Fig S6F). These results indicate that the ubiquitin ligase activity is not required for linear actin polymerization during trichome/hair formation in Drosophila.
Figure S6. Pldo function in actin cytoskeleton regulation is not related to the ubiquitin ligase complex activity.
(A) Two independent publications have shown that the Pldo homologs EBAX-1/ZSWIM8 form part of a ubiquitin ligase complex serving an E3 ligase function, recognizing the substrate targets to be degraded by the proteasome. We did not observe such a role of the gene in the actin dynamics context. IP assay evaluating the interaction of Pldo and Scar. S2 cells were co-transfected with Pldo-HA and Scar-GFP and were immunoprecipitated with anti-GFP. Jitterbug-HA was used as a control (Jbug-HA). Note Scar immunoprecipitated with PldoWT, suggesting that they form a complex. (B) Scar levels were evaluated in S2 cell protein extracts by Western blot in the presence or absence of the proteosomal inhibitor MG132. Scar levels were compared between untreated cells, Jbug expressing cells as control, and Pldo expressing cells, each of them with and without MG132. No significant difference was observed in either case. Levels of p53 were used as a positive control. (C, D, E) Scar levels were tested in vivo as a potential target of the ubiquitin ligase complex, but no detectable differences were observed. FRT19 clones of wild-type were generated as a control (equivalent to wild-type tissue). (D) Quantification is shown in (D): At least three individuals were considered in each experiment and each condition: independent experiments n = 3. Statistical analysis was done via a t test. Scale bars correspond to 25 μm. (E) Scar levels and localization were also analyzed in Z-sections in pupal wing cells, in the ptc-Gal4 > pldo-IR background, using confocal Z-line tool; the pldo knockdown region is marked with GFP. No observable differences in Scar levels/localization between the pldo LOF region and the rest of the wing were detected. Experiments were performed at 25°C and 29°C. Scale bar represents 50 μm. (F) The effect of other components of the ubiquitin ligase complex was evaluated in adult wings, using the ptc-Gal4 driver to express RNAi transgenes of the different components of the ubiquitin ligase complex (as linked to EBAX in in Caenorhabditis elegans). Two different RNAi constructs were used for EloB, EloC, and Cul2, and all of them tested at 25°C and 29°C. None showed altered hair formation. Scale bar represents 25 μm.
Source data are available for this figure.
In summary, our data are consistent with a model where Pldo regulates actin dynamics inducing linear actin polymerization, either by promoting delivery of actin monomers toward linear actin polymerization or by promoting Dia function and/or inhibiting Scar/WAVE. Pldo might be functioning as part of a multiprotein complex, where it regulates both proteins; alternatively, it may interact independently with either Dia or Scar, depending on the cellular requirements (see model in Fig 6D). Pldo could be thus acting as a molecular switch to regulate actin polymerization, either toward branched or linear filaments, in different cell contexts and/or by binding to distinct actin regulators (see the Discussion section).
Discussion
Here, we describe a lethal recessive mutation in the pelado (pldo) gene that, when analyzed in mosaic animals, results in external body regions lacking actin-based (hair) structures. Functional analyses of Drosophila Pelado (Pldo, also known as ZSWIM8 in humans and other species) in the context of the regulation of actin polymerization dynamics indicate that it is an essential component required for linear actin polymerization. Our studies support a conserved function of Pldo/ZSWIM8 in promoting linear actin polymerization—at the expense of or in competition of branched polymerization—in several cell types, ranging from Drosophila epithelial cells to a human cell line. Pldo/ZSWIM8 proteins share multiple domains that have been implicated in the regulation of actin filament dynamics, and Pldo/ZSWIM8 physically interacts with actin polymerization regulators, including Scar/WAVE, the formin Diaphanous, and Profilin. Functional dissection reveals specific domains of Pldo/ZSWIM8 being required for filopodia formation and other protein regions functioning in specific cellular contexts. Taken together, our data indicate that the N-terminal portion of Pldo/ZSWIM8 has a conserved regulatory function that is critical for linear actin polymerization. Although ZSWIM8 has been linked to ubiquitination in some contexts (Wang et al, 2013; Shi et al, 2020), our data argue against such a requirement during the regulation of actin dynamics.
Pldo/ZSWIM8 is required for linear actin polymerization
During development and morphogenesis, Drosophila cuticular epithelial cells differentiate to produce the adult phenotypic adaptations as part of the exoskeleton. Wing development, leading to largely hexagonal cells with each cell producing an actin-based hair, serves as an excellent model to study the role of cellular morphogenesis and the regulation of cytoskeletal dynamics (Lu & Adler, 2015). The actin-based cellular hair, or trichome, covers all wing cells and most parts of the external body in Drosophila with the function of the trichome presumably related to a passive role, directing airflow over the body and wing (Guild et al, 2005).
The positioning of the actin hairs, in the distal vertex of each cell, depends on the core Fz/PCP pathway and its effectors, which control the accumulation and polymerization of actin fibers at the apico-distal region of every cell (rev. in Strutt and Strutt [2009], Adler [2012], and Carvajal-Gonzalez and Mlodzik [2014]). Seminal work revealed that hair formation is highly dependent on linear actin polymerization (Mitchell et al, 1983). Hair outgrowth in the wing epithelia has been divided in three different stages: at 28 h APF, actin starts to accumulate in the apico-distal zone of wing cells, aided by tubulin-dependent transport and vesicle recycling (Gault et al, 2012), directed by the activity of the core PCP pathway; at 30–32 h APF, the prehair structure becomes apparent corresponding to the early stages of hair growth, which is focused in one distal spot because of the inhibitory activity of Mwh (rev. in Adler [2012] and Carvajal-Gonzalez and Mlodzik [2014]). Subsequently, at 32–36 h APF, the hairs extend and are completely formed (at 36 h APF), mainly by elongation of linear actin filaments, arising from the “prehair” foci. A detailed analysis of pldo/zswim8 mutants during the different stages of hair growth suggests that it is required for the formation of the prehair and linear actin extension within the hair. In addition, pldo/zswim8 appears to affect cell shape morphogenesis with a reduction in hexagonal cell shapes, consistent with a role in linear cortical actin filaments. The observation that overexpression of the formin Dia can rescue the loss-of-function (LOF) pldo/zswim8 phenotype resulting in loss of hairs, supports the notion that Pldo/ZSWIM8 regulates formin-dependent linear actin polymerization. The other described genes whose LOF generates the absence of cellular hairs in Drosophila are shavenbaby (svb) and shavenoid (sha). Svb is a transcription factor that induces the expression of Sha, which has been defined as a protein that regulates the actin cytoskeleton. However, sha is not conserved in the animal kingdom, and its presence is specific to insects and arthropods (He & Adler, 2002; www.flybase.org).
Actin hair formation, both number and location, is regulated by the core Fz/PCP pathway, but little is known about the mechanisms that regulate the actual actin polymerization during this process. Core PCP factor polarization, as exemplified by Fmi, is not affected in pldo/zswim8 mutant cells, suggesting that Pldo/ZSWIM8 is a downstream effector of the PCP pathway. Dsh accumulates at the apico-distal zone of Drosophila wing cells, where it is thought to recruit and activate RhoA, Rac, and Daam1 (disheveled-associated activator of morphogenesis 1, originally identified in Xenopus) (Habas et al, 2001). Daam1 and likely also other formins, like Dia (as Daam1 appears to be redundant in this wing hair context), interact with Profilin to induce actin polymerization (Matusek et al, 2008; Barko et al, 2010; Yaniv et al, 2020; and rev. in Goodrich and Strutt [2011], Adler [2012], and Singh and Mlodzik [2012]). The pldo/zswim8 LOF phenotype is rescued by Dia or Sha overexpression, whereas different isoforms of dDaam1, including activated ones (Matusek et al, 2006), do not rescue pldo/zswim8 LOF defects, consistent with the notion that Dia and not Daam1 is the critical formin family member in this context, in analogy to Scar and Wasp with each acting in a cell type–specific manner or process. A direct link of Dia and the Fz-Dsh complex has not yet been reported, and thus, the connection between Pldo/ZSWIM8 and core PCP complexes remains unclear. Further work will be required to establish a better understanding of the information flow from core PCP factors to the regulatory interactions at the actin polymerization level.
Besides the actin hair/trichome, pldo/zswim8–mutant epithelial cells also display cell shape defects, which—based on its link to actin—are likely caused by defects in cortical actin regulation. As the cortical actin cytoskeleton fulfills an essential role in all cells, it is consistent that a pldo/zswim8 mutant is embryonic lethal, albeit with very variable phenotypes. It should be noted that the highest levels of pldo/zswim8 message are maternal in the early embryo, but germline clones of pldo/zswim8 do not survive to allow egg development and deposition, likely because of defects during oogenesis.
Pldo/ZSWIM8 affects the balance of linear versus branched actin filament polymerization
Loss of the actin hair in pldo/zswim8– mutant cells is not only rescued by the overexpression of the formin Dia and actin itself but also by the knockdown of scar and arp2/3. These “synthetic” genetic interactions suggest that Pldo/ZSWIM8 affects the balance of linear versus branched actin polymerization, with either an excess of actin monomers or the reduction of the capability of the cell to promote branched actin, changing the balance back to linear actin. Thus, Pldo/ZSWIM8 facilitates the process by favoring linear polymerization. Notably, wasp knockdown did not reverse the pldo/zswim8 mutant phenotype, as compared with scar, which efficiently reversed the pldo/zswim8 defect. It has been described that Scar and Wasp act independently, regulating branched actin polymerization in different developmental contexts, as for example, Scar is important for morphogenetic cell processes, whereas Wasp being essential during muscle and sensory organ development (Brüser & Bogdan, 2017). Interestingly, a knockdown of mwh, an anti-formin inhibiting Dia, allowed also a partial phenotypic reversion of the pldo/zswim8 defect, suggesting that Dia function is rate-limiting in pldo/zswim8 mutants (mwh is epistatic to pldo/ZSWIM8 as the mwh LOF hair phenotype is manifested in the double knockdown, with 3+ hairs forming per cell; Yan et al, 2008).
Although cells generally have a high concentration of actin monomers (Koestler et al, 2009; Burke et al, 2014; and rev. in Pollitt and Insall [2009]), most of it is sequestered by different regulatory proteins, like Profilin and Thymosinβ4, establishing a competition for actin monomers between the two distinct forms of polymerization (Burke et al, 2014; Henson et al, 2015; Suarez et al, 2015; Suarez & Kovar, 2016). With most of actin polymerization tilted toward branched, linear actin polymerization is more regulated than branched filaments.
The notion that Pldo/ZSWIM8 favors linear actin polymerization is also supported by the phenotype observed in hemocytes. Here, pldo/zswim8–mutant and Pldo/ZSWIM8 overexpression backgrounds generate cell morphology changes involving rearrangements in actin-dependent structures, like filopodia and lamellipodia. Pldo/zswim8–mutant cells display a rounded morphology with less surface contacts and increased ruffles, structures mainly based on branched actin polymerization (Legg et al, 2007; and rev. in Campellone and Welch [2010]). Similar observations were made for Scar gain-of-function (GOF) scenarios in BG3 cells from Drosophila (Cloud et al, 2019). It is noteworthy that cell adhesion to the matrix is based in focal adhesion–related structures, with focal adhesions directly interacting with the substrate and being associated with linear actin filaments, known as stress fibers (rev. in Jockusch [2017] and Hohmann and Dehghani [2019]). [These observations in hemocytes reinforce the idea that Pldo/ZSWIM8 functions to promote linear actin polymerization. Accordingly, Pldo/ZSWIM8 overexpression in hemocytes induces the formation of filopodia, structures essentially formed by long linear actin filaments. Importantly, Pldo/ZSWIM8 LOF in hemocytes expressing the constitutive active form of Cdc42, prevented filopodia formation, strongly suggesting that Pldo/SWIM8 has a direct role in inducing linear actin polymerization. To further address this, we looked for potential binding sites of Pldo to Cdc42 or other small GTPases (through the CRIB domain), but we did not detect such domains in the Pldo sequence. This could indicate that Pldo is not a direct target of Cdc42 or other small GTPases but that it functions as a downstream effector of a direct target of these GTPases.
Taken together, Pldo/ZSWIM8 serves a “balance” function promoting linear actin polymerization, probably via Dia, as a formin, inducing linear actin polymerization, in competition with Scar, inducing branched actin polymerization. Based on our data, the balance between linear and branched actin polymerization appears to be a critical determinant in the trichome formation process. The effect of Pldo/ZSWIM8 may be comparable to the one described for Profilin (Suarez et al, 2015), which favors linear actin polymerization over branched filaments in fission yeast cells.
Pldo/ZSWIM8 is required for filopodia and related structures formation
The actin hair outgrowth process in Drosophila epithelial cells is similar to filopodia formation, with both being protrusions of the plasma membrane driven by linear actin filament extension. Similar to the epithelial actin hair context, pldo/zswim8–mutant defects in hemocytes are reversed by enhancing linear actin polymerization either by (i) increasing formin activity by overexpressing Dia, (ii) reducing branched polymerization by knocking down Scar, or (iii) by increasing actin monomer availability via overexpressing actin itself.
Importantly, this function of Pldo/ZSWIM8 is conserved from Drosophila to mammals. In a well-established wound healing cell migration assay in the human A549 cell line (Piao et al, 2014; Han et al, 2016; Wang et al, 2017; de Oliveira Rodrigues et al, 2020; Kim et al, 2020; Liao & Peng, 2020; Zhao et al, 2020), we observed that pldo/zswim8 knockout cells migrate significantly faster than control cells, suggesting that these cells have an increase in branched actin polymerization, allowing a faster migration. Strikingly, this effect is rescued by providing the Drosophila Pldo/ZSWIM8 protein to these knockout human A549 cells, which would increase the competition for actin monomers, reducing the branched polymerization. This not only suggests a conserved function but also confirms that the Drosophila factor is functionally equivalent to the human protein. Very similar results were described by knocking down WDR63, an inhibitor of the Arp2/3 complex, causing an increase in branched polymerization and cell migration in A549 cells (Zhao et al, 2020), opposite to the effect observed when arp2 is knocked down. These results are fully consistent with our data in Drosophila contexts, indicating that Pldo/ZSWIM8 changes the balance between linear and branched actin polymerization. This might be mediated possibly by either reducing Scar/WAVE activity (and in consequence Arp2/3–dependent branched actin nucleation) or by promoting Dia/formin function and hence linear actin polymerization, or both.
In summary, using multiple different cell types and experimental strategies, we have demonstrated that Pldo/ZSWIM8 serves a conserved function to promote linear actin polymerization at the expense of branched actin. Our results are all consistent with the notion that Pldo/ZSWIM8 functions in the same direction as Dia and Profilin/Chic activity and opposed to Scar and Arp2/3 activities in promoting linear actin polymerization and filopodia-like structures. Pldo/ZSWIM8 favors actin monomer competition and recruitment toward linear filament polymerization. Altogether, our results suggest that Pldo/ZSWIM8 is able to induce linear actin polymerization, as is required for filopodia formation in hemocytes, at the expense of branched actin polymerization (as shown in pldo/zswim8 LOF experiments in hemocytes and human cells).
Pldo/ZSWIM8 as a part of a molecular complex
Besides the predicted domains observed in the Pldo sequence (Fig 1D), there are also several tyrosine-binding motifs to SH2 (Src homology domain 2) and proline-rich SH3 domains detected in its sequence. In addition, a couple of binding motifs for EVH1 domain (enabled/vasodilator-stimulated phosphoprotein homology 1) are present.
mDia2, Arp2/3, and Scar/WAVE can form a multimolecular complex, and these interactions are modulated by Cdc42 (Miki et al, 2000; Krugmann et al, 2001; Beli et al, 2008). We tested for co-immunoprecipitation of Pldo/ZSWIM8 with different proteins that regulate actin filament dynamics. Pldo/ZSWIM8 appears to be part of a complex with Dia, Scar, and Profilin, suggesting that Pldo/ZSWIM8 is part of a related molecular complex to the one described (Miki et al, 2000; Krugmann et al, 2001; Beli et al, 2008). Accordingly, Pldo could be interacting with the Enabled (Ena) protein through its EVH1 domain (Ball et al, 2000; Hwang et al, 2021). IRSp53, which contains SH3 domains, has been described to interact with cytoskeletal regulatory proteins, like Scar/WAVE, Dia, and Mena (Miki et al, 2000; Jung et al, 2001; Krugmann et al, 2001; Suetsugu et al, 2006; Abou-Kheir et al, 2008; Goh et al, 2012; Stark et al, 2017; Pipathsouk et al, 2021). Thus, it is possible that the Pldo interaction with actin regulators might be indirect, mediated by other proteins that contain SH3 domains, like IRSp53. Interactions with SH2 and SH3 domains are important as these domains are frequently employed in proteins regulating several biological processes, including cytoskeletal rearrangements and cell migration, for example (Tatárová et al, 2012; and rev. in Kaneko et al [2008] and Mayer [2015). The physiological relevance of proteins with these domains is described in podocytes, where actin reorganization is mediated by interactions of NCK1 SH3 domain to N-WASP and NCK1 SH2 domain to phosphorylated Nephrin (Jones et al, 2006), and consequently, these interactions will regulate Arp2/3 complex activity. Of note, most of the SH2-binding motifs in Pldo are located at the N-terminal region of the protein, whereas the SH3-binding motifs are mainly found on the C-terminal portion. Such segregation could explain the different behavior observed in rescue experiments in hemocytes and hair formation with full-length Pldo and PldoΔC, suggesting differential regulation of distinct parts of the Pldo protein.
Along the above lines, the Pldo sequence reveals the presence of multiple serine/threonine phosphorylation sites, some of them clustered, and potential targets of different kinases, adding another level of context dependent regulation of Pldo. Several PCP proteins share this feature with Pldo. For example Vang, which requires phosphorylation for its polarized asymmetric localization, is phosphorylated on such serine/threonine clusters (Gao et al, 2011; Kelly et al, 2016). Similarly, Disheveled is heavily phosphorylated to channel its function to either canonical Wnt-signaling, PCP, or other functions, and it has been suggested that it contains a “barcode” of phosphorylation and that its activity is regulated in this manner (Bernatik et al, 2011; Yanfeng et al, 2011; Bernatík et al, 2014; Weber & Mlodzik, 2017; Hanáková et al, 2019). These examples suggest that PCP proteins share phosphorylation regulation, and thus, Pldo might participate in such regulatory input as a putative effector of the PCP pathway.
Phosphorylation could thus add a layer of regulation of Pldo, to both affect its subcellular function/requirement with, for example regulatory input missing in PldoΔC and hence its lack of activity in the hair formation rescue assay (despite the fact that PldoΔC was still very potent to induce filopodia formation in hemocytes). Similarly, phosphorylation might regulate the function of Pldo/ZSWIM8 in either actin dynamics or ubiquitin ligase activity contexts (see below).
Pldo/ZSWIM8 does not act as part of a ubiquitination complex in actin regulation
ZSWIM8 (EBAX-1 in C. elegans) has been characterized as an E3 ligase and a component of a ubiquitin ligase complex (Wang et al, 2013; Shi et al, 2020), and thus, we tested whether Pldo/ZSWIM8 shares the same function in the context of actin filament dynamics. Because a scar knockdown reversed the pldo/zswim8 LOF phenotype, Scar seemed to be a potential degradation target of Pldo/ZSWIM8 in the actin context. However, we did not detect any changes to Scar levels, although Scar and Pldo/ZSWIM8 are found in the same molecular complex (as seen via bidirectional IP experiments). Scar levels were also evaluated by Western blot in the presence or absence of the proteasome inhibitor MG132 and with or without coexpression of Pldo, revealing, however, no difference in any of these scenarios (whereas control proteins, like p53, showed protein-level differences). These studies demonstrate that Pldo is not regulating Scar levels. To further address this, we analyzed the effect of knockdown of components of the ubiquitin ligase complex associated with Pldo described in C. elegans, EloB, EloC, Cul-2, and the potential E2 ligase CG4080. None of these, assayed in LOF experiments in adult wings, showed any defects on hair formation, indicating that the ubiquitin ligase complex activity is not required in this process.
Nonetheless, although we did not detect any evidence for a ubiquitin-mediated degradation function of Pldo/ZSWIM8 in the actin cytoskeleton regulation context, we cannot rule out that Pldo/ZSWIM8 might have a function as a ubiquitin ligase in a different context. It will be interesting to determine potential regulatory conditions and mechanisms that select one function of Pldo/ZSWIM8 over the other or if there is a cell type or stage specificity to potential distinct molecular functions of this interesting protein family.
Pldo/ZSWIM8 and a potential role in human disease
As suggested by Yamamoto, Bellen, and colleagues (Yamamoto et al, 2014), Pldo/ZSWIM8 appears to be linked to a genetic disorder in humans affecting the nervous system. This observation could be explained by our data regarding its function in the regulation of actin cytoskeletal dynamics as biogenesis of neurites requires a highly regulated actin polymerization network (Ben-Yaacov et al, 2001; Korobova & Svitkina, 2008; Matusek et al, 2008; Tahirovic et al, 2010; Yaniv et al, 2020; and rev. in Da Silva and Dotti [2002]). For example, linear actin polymerization is required for radial glial migration and in the initial stages of neurite formation and branched polymerization is essential for their maturation. As such, Pldo/ZSWIM8 levels, when altered, could easily affect this process leading to neurological defects. It is also important to consider that there are five isoforms of Pldo/ZSWIM8 in humans, making it more difficult to analyze. In addition, taken together with the data presented here, Pldo/ZSWIM8 could also have a role in cancer cells because its LOF might enhance the migratory capacity of invasive metastatic cells.
Materials and Methods
Drosophila melanogaster strains and phenotypic analysis in adult wings
Drosophila flies were grown on standard food at 25°C, and phenotypes were analyzed at this temperature, unless otherwise indicated.
The following fly strains were used:
Bloomington (BL): BL5137 UAS-CD8::GFP,
BL56751 UAS-Diaphanous, BL7310 UAS-Actin,
BL18553 UAS-dsRNA pldo,
BL52333 pldoA,
BL52334 pldoB,
BL41552 UAS-dsRNA GFP,
BL1709 FRT19A,
BL34523 UAS-dsRNA chic,
BL5905 w1118, BL24651 UAS-Dicer2,
BL4854 UAS-Cdc42G12V.
VDRC strains (UAS-dsRNA constructs)
21,908 scar, 13,759 wasp, 41,514 mwh, 102,759 chic, 27,623 quail, 101,438 actin, 9,026 CG4080, 12,953 and 101,542 elonginB, 15,303 and 105,740 elonginC, 19,297 and 105,101 cullin2. UAS Scar-GFP was a gift from S. Bogdan (Stephan et al, 2011). Overexpression of cDNA transgenes or RNAi (IR) was performed using the Gal4/UAS system (Brand & Perrimon, 1993). The Gal4 expression drivers used were as follows: patched-Gal4, Hml-Gal4, pannier-Gal4, and ey-Gal4. Where indicated, UAS-Dicer2 was included with UAS-IR expression to increase RNAi efficiency. All stocks employed in experiments were generated by standard crosses.
MARCM analysis
For pupal wing clone generation, offspring were subjected to heat shock (at 37°C) for 1 h at 80 ± 12 h after egg-laying. Pupal clones were thus marked by GFP expression. Third instar larvae possessing clones (GFP positive) were selected and determined the 0 time (white prepupae) to proceed with immunofluorescence stainings at specific times during animal development (and hair formation).
In hemocytes, the offspring were subjected to heat shock at 48, 72, and 96 h after egg laying for 1 h, then incubated at 18°C for 1 h.
For analysis of adult wing trichomes, adult wings were removed, incubated in wash buffer (PBS and 0.1% Triton X-100), and mounted on a slide in 80% glycerol in PBS.
In the trichome rescue assay, classification of the phenotypic severity of the absence of hairs in adult wings was performed by visual inspection. Criteria for classification were the extent of area without hairs. Phenotypes were classified as rescued (no area without hairs), partial rescue (small area without hairs), and no rescue (large area without hairs).
To analyze trichomes in adult nota (dorsal thorax), flies were partially dissected, incubated at 95°C in 10% KOH for 10 min to clear fat tissue, washed (PBS and 0.1% Triton X-100), and then placed in 80% glycerol in PBS. Nota were fully dissected and mounted on a slide in 80% glycerol in PBS. Adult wings and nota were imaged at RT on a Axioplan (Carl Zeiss) microscope. Images were acquired with a Zeiss AxioCam (Color type 412-312) and AxioCam software.
Sequence analysis
Sequence analysis and predictions were made at online programs Psipred and ELM, the eukaryotic linear motif resource for functional sites in proteins.
Immunofluorescence
For analysis in pupal wings, white pupae were collected (0 h APF) and aged at 25°C until dissection. Dissections were performed as follows: in brief, pupae were immobilized on double-sided tape, removed from the pupal case, and placed into PBS, in which pupae were partially dissected to remove fat tissue, fixed in 4% paraformaldehyde in PBS for 45 min at RT or overnight at 4°C, and washed 3× in PBS and 0.1% Triton X-100. Wing membranes were removed, and immunostaining was performed by standard techniques. In brief, tissue was incubated in wash buffer containing 10% normal goat serum overnight for primary antibody (4°C), washed 3× with PBS, and incubated with the secondary antibody for at least 2 h (25°C) and fluorescent phalloidin for staining the actin cytoskeleton. Wings were washed 3× with PBS and mounted in Mowiol. Pupal-wing images were acquired at RT using a confocal microscope, either Zeiss LSM 710 or Leica SP8. Images were processed with ImageJ (National Institute of Health).
For hemocyte immunostaining, primary culture was performed as described (Tirouvanziam et al, 2004). Briefly, third instar larvae were washed, and a small incision on the posterior side of larvae was made to obtain hemocytes, which were incubated for 1 h 15 min, fixed, and stained. Samples were maintained in VECTASHIELD until pictures were taken. Confocal images were captured using either a Zeiss LSM 710 or a Leica SP8 confocal microscopes. Images were processed using ImageJ.
Antibodies
The following antibodies were used:
mouse anti-Fmi (1:10; Developmental Studies Hybridoma Bank),
mouse anti-Fascin (1:10; Developmental Studies Hybridoma Bank),
mouse anti-Scar (1:50; Developmental Studies Hybridoma Bank),
rabbit anti-Dia (1:1,000, donated by P Adler),
mouse anti-Chic (1:10; Developmental Studies Hybridoma Bank),
rabbit anti-PatJ (1:500), mouse anti-HA (1:50; BioLegend),
mouse anti-α-actin (1:1,000; Sigma-Aldrich).
All fluorophore-conjugated secondary antibodies were used at 1:200 and obtained from Jackson ImmunoResearch Laboratories, Inc. Rhodamine-phalloidin was from Invitrogen and used at 1:400.
Image analysis
Individual channels from color images were converted to gray scale using ImageJ. Fluorescent intensity levels were measured on maximal projections of image stacks using ImageJ.
DNA construct generation
Pelado ΔC was generated by PCR using the following primers: Fw: 5′-CAC CAT GGA CCG CTT CAG CTT CG-3′, Rv 1: 5′-CTC TCC TCG CGT CTT AAA GGT TCC-3′, RV 2: 5′-GAA AAT AAA CGT GGC CAG GTT AAT AGG CAA TAG-3′, RV 3: 5′-GCT GAG TGT AAC TAG GGC ATC GAA AAT AAA CGT GGC-3′. The PCR product was purified and cloned into a pENTR vector, sequenced, and using the Gateway kit, was cloned into the pUASt vector, with the coding sequence of HA tag at the 5′ of pldo gene. Transgenesis was performed at BestGene. The deletions Pldo1-685 and Pldo1-515 were generated by restriction enzymes KflI and BsiWI, respectively. To express Pldo in mammalian cells, the coding sequence was cloned into a VVPW/BC vector (donated by L Gusella), using XbaI and EcoRI restriction sites.
Cell culture, transfection, and scratch wound healing assay
Drosophila S2R+ cells were obtained directly from the Drosophila Genomics Resource Center (DGRC), Indiana. Cells were grown in Schneider’s medium, supplemented with 10% FBS and maintained at 25°C, and transfected with Effectene reagent, following standard protocols. Cells were lysed or immunostained 48 h after transfection in lysis buffer.
Human A549 cells were obtained from the Bartel Laboratory and were grown in DMEM medium, supplemented with 10% FBS and puromycin (10 μg/μl) and maintained in 5% CO2 at 37°C. Monoclonal cell line was generated by seeding individual cells in a 96-well plate, monitoring cells for growth, identifying individual colonies, and expanding monoclonal lines of interest. Clonal cells knocked out for pldo/zswim8 were identified by PCR with the primers described (Shi et al, 2020). Cells were transfected with Effectene reagent, following standard protocols.
Cell cultures were plated to a confluence of 50–70% and transfected the next day. After 8–12 h, the medium was changed for fresh medium. When confluence was reached, the scratch was performed with a yellow plastic tip. Pictures were taken at times 0, 18, 22, and 24 h after the scratch was made.
For MG132 treatment, the drug was used from a stock at 1 mg/ml, and the final concentration in each well was 10 μM. MG132 was added 36 h after transfection and maintained for 6 h. After 6-h incubation with the drug, media was changed and proceeded to protein extraction.
Western blotting and immunoprecipitation
S2R+ cells (grown in Schneider medium using standard procedures) were transfected with HAPldo and GFP-Scar, using Effectene reagent. After 48 h, protein extraction was performed. Immunoprecipitation (IP) and co-IP experiments were performed by incubating lysates with anti-HA or anti-GFP antibody, at 4°C overnight followed by Agarose-A beads incubation, 5× washing steps, and elution in SDS sample buffer. Samples were boiled at 95°C for 10 min and proceeded to Western blotting.
Statistical analyses
Quantifications were made in ImageJ. To determine statistical differences between data sets of continuous data, we performed nonparametric ANOVA or t test analyses on GraphPad. Specific analysis is indicated each time, and P-values lower than 0.05 were considered significant. Significance is indicated by asterisks. Sample sizes are described for each experiment in the figure legends. All experiments were performed on a minimum of three animals per condition. The statistical parameters are reported in figure legends. The error bars represent the SEM.
Supplementary Material
Acknowledgements
We thank Sven Bogdan, David Bartel, Luca Gusella, and Steven Wassarman for generously sharing Drosophila strains, cell lines, and other reagents. We are most grateful to Luca Gusella for support and sharing equipment during the cell culture experiments. We gratefully acknowledge the Bloomington Drosophila Stock Center and the Vienna Drosophila RNAi Center for supplying fly stocks. We thank all Mlodzik lab members for helpful discussions and advice. This work was supported by grant R35 GM127103 from the NIGMS/NIH to M Mlodzik; FONDECYT grant 1190119, ANID, the Center of Genome Regulation, Fondap 15200002, ANID, and ICN2021_044 Millennium Institute Center for Genome Regulation, ANID to A Glavic; and Biomedical Neuroscience Institute, Iniciativa Científica Milenio ICM P09-015F to P Olguin. C Molina-Pelayo was in part supported by a National PhD scholarship number 21140101, CONYCIT/ANID.
Author Contributions
C Molina Pelayo: conceptualization, data curation, formal analysis, validation, investigation, visualization, methodology, and writing—original draft, review, and editing.
P Olguin: conceptualization, data curation, formal analysis, investigation, and writing—original draft.
M Mlodzik: conceptualization, formal analysis, supervision, funding acquisition, project administration, and writing—original draft, review, and editing.
A Glavic: conceptualization, formal analysis, supervision, funding acquisition, investigation, methodology, writing—original draft, and project administration.
Conflict of Interest Statement
The authors declare that they have no conflict of interest.
References
- Abou-Kheir W, Isaac B, Yamaguchi H, Cox D (2008) Membrane targeting of WAVE2 is not sufficient for WAVE2 dependent actin polymerization: A role for IRSp53 in mediating the interaction between rac and WAVE2. J Cell Sci 121: 379–390. 10.1242/jcs.010272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler PN (2018) A cytoskeletal activator and inhibitor are downstream targets of the frizzled/starry night planar cell polarity pathway in the Drosophila epidermis. Prog Biophys Mol Biol 137: 69–75. 10.1016/j.pbiomolbio.2018.04.001 [DOI] [PubMed] [Google Scholar]
- Adler PN, Sobala LF, Thom D, Nagaraj R (2013) Dusky-like is required to maintain the integrity and planar cell polarity of hairs during the development of the Drosophila wing. Dev Biol 379: 76–91. 10.1016/j.ydbio.2013.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler PN (2012) The frizzled/stan pathway and planar cell polarity in the Drosophila wing. Curr Top Dev Biol 101: 1–31. 10.1016/B978-0-12-394592-1.00001-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aigouy B, Farhadifar R, Staple DB, Sagner A, Röper JC, Jülicher F, Eaton S (2010) Cell flow reorients the Axis of planar polarity in the wing epithelium of Drosophila. Cell 142: 773–786. 10.1016/j.cell.2010.07.042 [DOI] [PubMed] [Google Scholar]
- Ashcroft M, Vousden KH (1999) Regulation of p53 stability. Oncogene 18: 7637–7643. 10.1038/sj.onc.1203012 [DOI] [PubMed] [Google Scholar]
- Aw WY, Devenport D (2017) Planar cell polarity: Global inputs establishing cellular asymmetry. Curr Opin Cell Biol 44: 110–116. 10.1016/j.ceb.2016.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod JD, Miller JR, Shulman JM, Moon RT, Perrimon N (1998) Differential recruitment of dishevelled provides signaling specificity in the planar cell polarity and Wingless signaling pathways. Genes Dev 12: 2610–2622. 10.1101/gad.12.16.2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball LJ, Kühne R, Hoffmann B, Häfner A, Schmieder P, Volkmer-Engert R, Hof M, Wahl M, Schneider-Mergener J, Walter U, et al. (2000) Dual epitope recognition by the VASP EVH1 domain modulates polyproline ligand specificity and binding affinity. EMBO J 19: 4903–4914. 10.1093/emboj/19.18.4903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barko S, Bugyi B, Carlier MF, Gombos R, Matusek T, Mihály J, Nyitrai M (2010) Characterization of the biochemical properties and biological function of the formin homology domains of Drosophila DAAM. J Biol Chem 285: 13154–13169. 10.1074/jbc.m109.093914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beli P, Mascheroni D, Xu D, Innocenti M (2008) WAVE and Arp2/3 jointly inhibit filopodium formation by entering into a complex with mDia2. Nat Cell Biol 10: 849–857. 10.1038/ncb1745 [DOI] [PubMed] [Google Scholar]
- Ben-Yaacov S, Le Borgne R, Abramson I, Schweisguth F, Schejter ED (2001) Wasp, the Drosophila Wiskott-Aldrich syndrome gene homologue, is required for cell fate decisions mediated by Notch signaling. J Cell Biol 152: 1–14. 10.1083/jcb.152.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S, Schäfer G, Kesper DA, Holz A, Eriksson T, Palmer RH, Beck L, Klämbt C, Renkawitz-pohl R, Önel SF (2008) WASP and SCAR have distinct roles in activating the Arp2/3 complex during myoblast fusion. J Cell Sci 121: 1303–1313. 10.1242/jcs.022269 [DOI] [PubMed] [Google Scholar]
- Bernatik O, Ganji RS, Dijksterhuis JP, Konik P, Cervenka I, Polonio T, Krejci P, Schulte G, Bryja V (2011) Sequential activation and inactivation of dishevelled in the Wnt/β-catenin pathway by casein kinases. J Biol Chem 286: 10396–10410. 10.1074/jbc.m110.169870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernatík O, Šedová K, Schille C, Ganji RS, Ćervenka I, Trantírek L, Schambony A, Zdráhal Z, Bryja V (2014) Functional analysis of dishevelled-3 phosphorylation identifies distinct mechanisms driven by casein kinase 1ε and Frizzled5. J Biol Chem 289: 23520–23533. 10.1074/jbc.m114.590638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdan S, Schultz J, Grosshans J, Neurobiologie I, Münster U, Würzburg U, Biochemie I (2013) Physiological roles of Diaphanous (Dia) in actin dynamics Formin’ cellular structures. Commun Integr Biol 6: e27634. 10.4161/cib.27634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. 10.1242/dev.118.2.401 [DOI] [PubMed] [Google Scholar]
- Brüser L, Bogdan S (2017) Molecular control of actin dynamics in vivo: Insights from Drosophila. Handb Exp Pharmacol 235: 285–310. 10.1007/164_2016_33 [DOI] [PubMed] [Google Scholar]
- Burke TA, Christensen JR, Barone E, Suarez C, Sirotkin V, Kovar DR (2014) Homeostatic Actin cytoskeleton networks are regulated by assembly factor competition for monomers. Curr Biol 24: 579–585. 10.1016/j.cub.2014.01.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Pernier J, Avvaru BS (2013) Control of actin filament dynamics at barbed ends by WH2 domains: From capping to permissive and processive assembly. Cytoskeleton 70: 540–549. 10.1002/cm.21124 [DOI] [PubMed] [Google Scholar]
- Campellone KG, Welch MD (2010) A nucleator arms race: Cellular control of actin assembly. Nat Rev Mol Cell Biol 11: 237–251. 10.1038/nrm2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Shekhar S (2017) Global treadmilling coordinates actin turnover and controls the size of actin networks. Nat Rev Mol Cell Biol 18: 389–401. 10.1038/nrm.2016.172 [DOI] [PubMed] [Google Scholar]
- Carvajal-Gonzalez JM, Mulero-Navarro S, Smith M, Mlodzik M (2016) A novel frizzled-based screening tool identifies genetic modifiers of planar cell polarity in drosophila wings. G3 (Bethesda) 6: 3963–3973. 10.1534/g3.116.035535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal-Gonzalez JM, Mlodzik M (2014) Mechanisms of planar cell polarity establishment in Drosophila. F1000Prime Rep 6: 98. 10.12703/p6-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XJ, Squarr AJ, Stephan R, Chen B, Higgins TE, Barry DJ, Martin MC, Rosen MK, Bogdan S, Way M (2014) Ena/VASP proteins cooperate with the WAVE complex to regulate the actin cytoskeleton. Dev Cell 30: 569–584. 10.1016/j.devcel.2014.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou F-S, Wang P-S (2016) The Arp2/3 complex is essential at multiple stages of neural development. Neurogenesis 3: e1261653. 10.1080/23262133.2016.1261653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Classen A, Aigouy B, Giangrande A, Eaton S (2008) Imaging Drosophila pupal wing morphogenesis. Methods Mol Biol 420: 265–275. 10.1007/978-1-59745-583-1_16 [DOI] [PubMed] [Google Scholar]
- Cloud V, Thapa A, Morales-Sosa P, Miller TM, Miller SA, Holsapple D, Gerhart PM, Momtahan E, Jack JL, Leiva E, et al. (2019) Ataxin-7 and non-stop coordinate scar protein levels, subcellular localization, and actin cytoskeleton organization. Elife 8: e49677. 10.7554/elife.49677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier S, Lee H, Burgess R, Adler P (2005) The WD40 repeat protein fritz links cytoskeletal planar polarity to Frizzled subcellular localization in the Drosophila epidermis. Genetics 169: 2035–2045. 10.1534/genetics.104.033381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JA (2013) Mechanisms of cell migration in the nervous system. J Cell Biol 202: 725–734. 10.1083/jcb.201305021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz C, Glavic A, Casado M, De Celis JF (2009) A gain-of-function screen identifying genes required for growth and pattern formation of the Drosophila melanogaster wing. Genetics 183: 1005–1026. 10.1534/genetics.109.107748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva JS, Dotti CG (2002) Breaking the neuronal sphere: Regulation of the actin cytoskeleton in neuritogenesis. Nat Rev Neurosci 3: 694–704. 10.1038/nrn918 [DOI] [PubMed] [Google Scholar]
- Das G, Jenny A, Klein TJ, Eaton S, Mlodzik M (2004) Diego interacts with Prickle and Strabismus/Van Gogh to localize planar cell polarity complexes. Development 131: 4467–4476. 10.1242/dev.01317 [DOI] [PubMed] [Google Scholar]
- Davidson AJ, Amato C, Thomason PA, Insall RH (2018) WASP family proteins and formins compete in pseudopod- and bleb-based migration. J Cell Biol 217: 701–714. 10.1083/jcb.201705160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson AJ, Wood W (2016) Unravelling the actin cytoskeleton: A new competitive edge? Trends Cell Biol 26: 569–576. 10.1016/j.tcb.2016.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Rodrigues R, Yaochite JNU, Sasahara GL, Albuquerque AA, da Cruz Fonseca SG, de Vasconcelos Araújo TD, Santiago GMP, de Sousa LM, de Carvalho JL, Alves APNN, et al. (2020) Antioxidant, anti-inflammatory, and healing potential of ethyl acetate fraction of Bauhinia ungulata L. (Fabaceae) on in vitro and in vivo wound model. Mol Biol Rep 47: 2845–2859. 10.1007/s11033-020-05332-7 [DOI] [PubMed] [Google Scholar]
- Delon I, Chanut-Delalande H, Payre F (2003) The Ovo/Shavenbaby transcription factor specifies actin remodelling during epidermal differentiation in Drosophila. Mech Dev 120: 747–758. 10.1016/s0925-4773(03)00081-9 [DOI] [PubMed] [Google Scholar]
- Devenport D (2014) The cell biology of planar cell polarity. J Cell Biol 207: 171–179. 10.1083/jcb.201408039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton S, Wepf R, Simons K (1996) Roles for Rac1 and Cdc42 in planar polarization and hair outgrowth in the wing of Drosophila. J Cell Biol 135: 1277–1289. 10.1083/jcb.135.5.1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etournay R, Popović M, Merkel M, Nandi A, Blasse C, Aigouy B, Brandl H, Myers G, Salbreux G, Jülicher F, et al. (2015) Interplay of cell dynamics and epithelial tension during morphogenesis of the Drosophila pupal wing. Elife 4: e07090. 10.7554/elife.07090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan JK, Dollar G, Lu Q, Barnett A, Jorge JP, Schlosser A, Pfleger C, Adler P, Jenny A (2014) Combover/CG10732, a novel PCP effector for Drosophila wing hair formation. PLoS One 9: e107311. 10.1371/journal.pone.0107311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faix J, Grosse R (2006) Staying in shape with formins. Dev Cell 10: 693–706. 10.1016/j.devcel.2006.05.001 [DOI] [PubMed] [Google Scholar]
- Fang X, Adler PN (2010) Regulation of cell shape, wing hair initiation and the actin cytoskeleton by Trc/Fry and Wts/Mats complexes. Dev Biol 341: 360–374. 10.1016/j.ydbio.2010.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Lu Q, Emoto K, Adler PN (2010) The Drosophila Fry protein interacts with Trc and is highly mobile in vivo. BMC Dev Biol 10: 40. 10.1186/1471-213x-10-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher DA, Mullins RD (2010) Cell mechanics and the cytoskeleton. Nature 463: 485–492. 10.1038/nature08908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel S, Mooseker MS (1996) The actin-related proteins. Yeast 8: 30–37. 10.1016/s0955-0674(96)80045-7 [DOI] [PubMed] [Google Scholar]
- Fricke R, Gohl C, Dharmalingam E, Grevelhorster A, Zahedi B, Harden N, Kessels M, Qualmann B, Bogdan S (2009) Drosophila Cip4/Toca-1 integrates membrane trafficking and actin dynamics through WASP and SCAR/WAVE. Curr Biol 19: 1429–1437. 10.1016/j.cub.2009.07.058 [DOI] [PubMed] [Google Scholar]
- Gao B, Song H, Bishop K, Elliot G, Garrett L, English MA, Andre P, Robinson J, Sood R, Minami Y, et al. (2011) Wnt signaling gradients establish planar cell polarity by inducing Vangl2 phosphorylation through Ror2. Dev Cell 20: 163–176. 10.1016/j.devcel.2011.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B (2012) Wnt regulation of planar cell polarity (PCP). Curr Top Dev Biol 101: 263–295. 10.1016/B978-0-12-394592-1.00008-9 [DOI] [PubMed] [Google Scholar]
- Gault WJ, Olguin P, Weber U, Mlodzik M (2012) Drosophila CK1-γ, gilgamesh, controls PCP-mediated morphogenesis through regulation of vesicle trafficking. J Cell Biol 196: 605–621. 10.1083/jcb.201107137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautreau AM, Fregoso FE, Simanov G, Dominguez R (2022) Nucleation, stabilization, and disassembly of branched actin networks. Trends Cell Biol 32: 421–432. 10.1016/j.tcb.2021.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godin SK, Meslin C, Kabbinavar F, Bratton-Palmer DS, Hornack C, Mihalevic MJ, Yoshida K, Sullivan M, Clark NL, Bernstein KA (2015) Evolutionary and functional analysis of the invariant SWIM domain in the conserved Shu2/SWS1 protein family from Saccharomyces cerevisiae to Homo sapiens. Genetics 199: 1023–1033. 10.1534/genetics.114.173518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh WI, Lim KB, Sudhaharan T, Sem KP, Bu W, Chou AM, Ahmed S (2012) mDia1 and WAVE2 proteins interact directly with IRSp53 in filopodia and are involved in filopodium formation. J Biol Chem 287: 4702–4714. 10.1074/jbc.m111.305102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goley ED, Welch MD (2006) The ARP2/3 complex: An actin nucleator comes of age. Nat Rev Mol Cell Biol 7: 713–726. 10.1038/nrm2026 [DOI] [PubMed] [Google Scholar]
- Goodrich LV, Strutt D (2011) Principles of planar polarity in animal development. Development 138: 1877–1892. 10.1242/dev.054080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guild GM, Connelly PS, Ruggiero L, Vranich KA, Tilney LG (2005) Actin filament bundles in Drosophila wing hairs: Hairs and bristles use different strategies for assembly. Mol Biol Cell 16: 3620–3631. 10.1091/mbc.e05-03-0185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung R, Yadav R, Brungardt JG, Orlova A, Egelman EH, Beck MR (2016) Actin polymerization is stimulated by actin cross-linking protein palladin. Biochem J 473: 383–396. 10.1042/bj20151050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habas R, Kato Y, He X (2001) Wnt/Frizzled activation of Rho regulates vertebrate gastrulation and requires a novel formin homology protein Daam1. Cell 107: 843–854. 10.1016/s0092-8674(01)00614-6 [DOI] [PubMed] [Google Scholar]
- Haelterman NA, Jiang L, Li Y, Bayat V, Sandoval H, Ugur B, Tan KL, Zhang K, Bei D, Xiong B, et al. (2014) Large-scale identification of chemically induced mutations in Drosophila melanogaster. Genome Res 24: 1707–1718. 10.1101/gr.174615.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale R, Strutt D (2015) Conservation of planar polarity pathway function across the animal kingdom. Annu Rev Genet 49: 529–551. 10.1146/annurev-genet-112414-055224 [DOI] [PubMed] [Google Scholar]
- Han ML, Zhao YF, Tan CH, Xiong YJ, Wang WJ, Wu F, Fei Y, Wang L, Liang ZQ (2016) Cathepsin L upregulation-induced EMT phenotype is associated with the acquisition of cisplatin or paclitaxel resistance in A549 cells. Acta Pharmacol Sin 37: 1606–1622. 10.1038/aps.2016.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanáková K, Bernatík O, Kravec M, Micka M, Kumar J, Harnoš J, Ovesná P, Paclíková P, Rádsetoulal M, Potěšil D, et al. (2019) Comparative phosphorylation map of Dishevelled 3 links phospho-signatures to biological outputs. Cell Commun Signal 17: 170. 10.1186/s12964-019-0470-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison C, Shao H, Strutt H, Strutt D (2020) Molecular mechanisms mediating asymmetric subcellular localisation of the core planar polarity pathway proteins. Biochem Soc Trans 48: 1297–1308. 10.1042/bst20190404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M (1997) Mdm2 promotes the rapid degradation of p53. Nature 387: 296–299. 10.1038/387296a0 [DOI] [PubMed] [Google Scholar]
- He B, Adler PN (2002) The genetic control of arista lateral morphogenesis in Drosophila. Dev Genes Evol 212: 218–229. 10.1007/s00427-002-0229-0 [DOI] [PubMed] [Google Scholar]
- He Y, Fang X, Emoto K, Jan YN, Adler PN (2005) The Tricornered Ser/Thr protein kinase is regulated by phosphorylation and interacts with Furry during Drosophila wing hair development. Mol Biol Cell 16: 689–700. 10.1091/mbc.e04-09-0828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson JH, Yeterian M, Weeks RM, Medrano AE, Brown BL, Geist HL, Pais MD, Oldenbourg R, Shuster CB (2015) Arp2/3 complex inhibition radically alters lamellipodial actin architecture, suspended cell shape, and the cell spreading process. Mol Biol Cell 26: 887–900. 10.1091/mbc.e14-07-1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann T, Dehghani F (2019) The cytoskeleton-A complex interacting meshwork. Cells 8: 362. 10.3390/cells8040362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R, Tanaka H, Yasuda H (1997) Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett 420: 25–27. 10.1016/s0014-5793(97)01480-4 [DOI] [PubMed] [Google Scholar]
- Humphries AC, Mlodzik M (2018) From instruction to output: Wnt/PCP signaling in development and cancer. Curr Opin Cell Biol 51: 110–116. 10.1016/j.ceb.2017.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang T, Parker SS, Hill SM, Ilunga MW, Grant RA, Mouneimne G, Keating AE (2021) A distributed residue network permits conformational binding specificity in a conserved family of actin remodelers. Elife 10: e70601. 10.7554/elife.70601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Narita A, Hirayama T, Taki M, Iyoshi S, Yamamoto Y, Maéda Y, Oda T (2011) Human Spire interacts with the barbed end of the actin filament. J Mol Biol 408: 18–25. 10.1016/j.jmb.2010.12.045 [DOI] [PubMed] [Google Scholar]
- Jaiswal R, Stepanik V, Rankova A, Molinar O, Goode BL, Mccartney BM (2013) Drosophila homologues of Adenomatous Polyposis Coli (APC) and the formin Diaphanous collaborate by a conserved mechanism to stimulate actin filament assembly. J Biol Chem 288: 13897–13905. 10.1074/jbc.m113.462051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny A, Darken RS, Wilson PA, Mlodzik M (2003) Prickle and Strabismus form a functional complex to generate a correct axis during planar cell polarity signaling. EMBO J 22: 4409–4420. 10.1093/emboj/cdg424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny A, Reynolds-Kenneally J, Das G, Burnett M, Mlodzik M (2005) Diego and Prickle regulate frizzled planar cell polarity signalling by competing for Dishevelled binding. Nat Cell Biol 7: 691–697. 10.1038/ncb1271 [DOI] [PubMed] [Google Scholar]
- Jockusch Brigitte M (2017) The actin cytoskeleton. Handbook of Experimental Pharmacology 235. 10.1007/978-3-319-46371-1 [DOI] [Google Scholar]
- Jones N, Blasutig IM, Eremina V, Ruston JM, Bladt F, Li H, Huang H, Larose L, Li SSC, Takano T, et al. (2006) Nck adaptor proteins link nephrin to the actin cytoskeleton of kidney podocytes. Nature 440: 818–823. 10.1038/nature04662 [DOI] [PubMed] [Google Scholar]
- Jung G, Remmert K, Wu X, Volosky JM, Hammer JA (2001) The Dictyostelium CARMIL protein links capping protein and the Arp2/3 complex to type I myosins through their SH3 domains. J Cell Biol 153: 1479–1498. 10.1083/jcb.153.7.1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T, Li L, Li SSC (2008) The SH3 domain- A family of versatile peptide- and protein-recognition module. Front Biosci 13: 4938–4952. 10.2741/3053 [DOI] [PubMed] [Google Scholar]
- Kang H, Wang J, Longley SJ, Tang JX, Shaw SK (2010) Relative actin nucleation promotion efficiency by WASP and WAVE proteins in endothelial cells. Biochem Biophys Res Commun 400: 661–666. 10.1016/j.bbrc.2010.08.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly LK, Wu J, Yanfeng WA, Mlodzik M (2016) Frizzled-induced van gogh phosphorylation by CK1ε promotes asymmetric localization of core PCP factors in Drosophila. Cell Rep 16: 344–356. 10.1016/j.celrep.2016.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BN, Ahn DH, Kang N, Yeo CD, Kim YK, Lee KY, Kim TJ, Lee SH, Park MS, Yim HW, et al. (2020) TGF-β induced EMT and stemness characteristics are associated with epigenetic regulation in lung cancer. Sci Rep 10: 10597. 10.1038/s41598-020-67325-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein TJ, Jenny A, Djiane A, Mlodzik M (2006) CKIε/discs overgrown promotes both Wnt-fz/β-catenin and fz/PCP signaling in Drosophila. Curr Biol 16: 1337–1343. 10.1016/j.cub.2006.06.030 [DOI] [PubMed] [Google Scholar]
- Ko KK, Powell MS, Hogarth PM (2014) ZSWIM1: A novel biomarker in T helper cell differentiation. Immunol Lett 160: 133–138. 10.1016/j.imlet.2014.01.016 [DOI] [PubMed] [Google Scholar]
- Koch N, Dharmalingam E, Westermann M, Qualmann B, Thomas U, Kessels MM (2012) Abp1 utilizes the Arp2/3 complex activator Scar/WAVE in bristle development. J Cell Sci 125: 3578–3589. 10.1242/jcs.101451 [DOI] [PubMed] [Google Scholar]
- Koestler SA, Rottner K, Lai F, Block J, Vinzenz M, Small JV (2009) F- and G-actin concentrations in lamellipodia of moving cells. PLoS One 4: e4810. 10.1371/journal.pone.0004810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobova F, Svitkina T (2008) Arp2/3 complex is important for filopodia formation, growth cone motility, and neuritogenesis in neuronal cells. Mol Biol Cell 19: 1561–1574. 10.1091/mbc.e07-09-0964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR (2006) Molecular details of formin-mediated actin assembly. Curr Opin Cell Biol 18: 11–17. 10.1016/j.ceb.2005.12.011 [DOI] [PubMed] [Google Scholar]
- Krugmann S, Jordens I, Gevaert K, Driessens M, Vandekerckhove J, Hall A (2001) Cdc42 induces filopodia by promoting the formation of an IRSp53:Mena complex. Curr Biol 11: 1645–1655. 10.1016/s0960-9822(01)00506-1 [DOI] [PubMed] [Google Scholar]
- Kullmann JA, Meyer S, Pipicelli F, Kyrousi C, Schneider F, Bartels N, Cappello S, Rust MB (2020) Profilin1-dependent F-Actin assembly controls division of apical radial glia and neocortex development. Cereb Cortex 30: 3467–3482. 10.1093/cercor/bhz321 [DOI] [PubMed] [Google Scholar]
- Kunda P, Craig G, Dominguez V, Baum B (2003) Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr Biol 13: 1867–1875. 10.1016/j.cub.2003.10.005 [DOI] [PubMed] [Google Scholar]
- Le Clainche C, Carlier MF (2008) Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol Rev 88: 489–513. 10.1152/physrev.00021.2007 [DOI] [PubMed] [Google Scholar]
- Lee H, Adler PN (2004) The grainy head transcription factor is essential for the function of the Frizzled pathway in the Drosophila wing. Mech Dev 121: 37–49. 10.1016/j.mod.2003.11.002 [DOI] [PubMed] [Google Scholar]
- Legg JA, Bompard G, Dawson J, Morris HL, Andrew N, Cooper L, Johnston SA, Tramountanis G, Machesky LM (2007) N-WASP involvement in dorsal ruffle formation in mouse embryonic fibroblasts. Mol Biol Cell 18: 678–687. 10.1091/mbc.e06-06-0569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung T, Chen X-Q, Tan I, Manser E, Lim L (1998) Myotonic dystrophy KinaseRelated cdc42-binding kinase acts as a Cdc42 effector in promoting cytoskeletal reorganization. Mol Cell Biol 18: 130–140. 10.1128/mcb.18.1.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M, Peng L (2020) MiR-206 may suppress non-small lung cancer metastasis by targeting CORO1C. Cell Mol Biol Lett 25: 22. 10.1186/s11658-020-00216-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Adler PN (2015) The diaphanous gene of Drosophila interacts antagonistically with multiple wing hairs and plays a key role in wing hair morphogenesis. PLoS One 10: e0115623. 10.1371/journal.pone.0115623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Schafer DA, Adler PN (2015) The Drosophila planar polarity gene multiple wing hairs directly regulates the actin cytoskeleton. Development 142: 2478–2486. 10.1242/dev.122119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Yan J, Adler PN (2010) The Drosophila planar polarity proteins Inturned and Multiple wing hairs interact physically and function together. Genetics 185: 549–558. 10.1534/genetics.110.114066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Lee T, Tsai L, Tang G, Jan LY, Jan YN (1997) Genghis Khan (Gek) as a putative effector for Drosophila Cdc42 and regulator of actin polymerization. Proc Natl Acad Sci U S A 94: 12963–12968. 10.1073/pnas.94.24.12963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Mullins RD, Higgs HN, Kaiser DA, Blanchoin L, May RC, Hall ME, Pollard TD (1999) Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc Natl Acad Sci U S A 96: 3739–3744. 10.1073/pnas.96.7.3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki CG, Huibregtse JM, Howley PM (1996) In vivo ubiquitination and proteasome-mediated degradation of p53(1). Cancer Res 56: 2649–2654. [PubMed] [Google Scholar]
- Matis M, Russler-Germain DA, Hu Q, Tomlin CJ, Axelrod JD (2014) Microtubules provide directional information for core PCP function. Elife 3: e02893. 10.7554/elife.02893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matusek T, Djiane A, Jankovics F, Brunner D, Mlodzik M, Mihály J (2006) The Drosophila formin DAAM regulates the tracheal cuticle pattern through organizing the actin cytoskeleton. Development 133: 957–966. 10.1242/dev.02266 [DOI] [PubMed] [Google Scholar]
- Matusek T, Gombos R, Szécsényi A, Sánchez-Soriano N, Czibula Á, Pataki C, Gedai A, Prokop A, Raskó I, Mihály J (2008) Formin proteins of the DAAM subfamily play a role during axon growth. J Neurosci 28: 13310–13319. 10.1523/jneurosci.2727-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer BJ (2015) The discovery of modular binding domains: Building blocks of cell signalling. Nat Rev Mol Cell Biol 16: 691–698. 10.1038/nrm4068 [DOI] [PubMed] [Google Scholar]
- Miki H, Suetsugu S, Takenawa T (1998) WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J 17: 6932–6941. 10.1093/emboj/17.23.6932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Yamaguchi H, Suetsugu S, Takenawa T (2000) IRSp53 is an essential intermediate between Rac and WAVE in the regulation of membrane ruffling. Nature 408: 732–735. 10.1038/35047107 [DOI] [PubMed] [Google Scholar]
- Mitchell HK, Roach J, Petersen NS (1983) The morphogenesis of cell hairs on Drosophila wings. Dev Biol 95: 387–398. 10.1016/0012-1606(83)90040-4 [DOI] [PubMed] [Google Scholar]
- Moreira CGA, Jacinto A, Prag S (2013) Drosophila integrin adhesion complexes are essential for hemocyte migration in vivo. Biol Open 2: 795–801. 10.1242/bio.20134564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins RD, Heuser JA, Pollard TD (1998) The interaction of Arp2/3 complex with actin: Nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci U S A 95: 6181–6186. 10.1073/pnas.95.11.6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura F, Oki N, Fujiki Y, Ikuta R, Osaki K, Hamada S, Nakatsukasa K, Hisamoto N, Hara T, Kamura T (2021) ZSWIM8 is a myogenic protein that partly prevents C2C12 differentiation. Sci Rep 11: 20880. 10.1038/s41598-021-00306-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant V, Lozano G (2014) Limiting the power of p53 through the ubiquitin proteasome pathway. Genes Dev 28: 1739–1751. 10.1101/gad.247452.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park TJ, Haigo SL, Wallingford JB (2006) Ciliogenesis defects in embryos lacking inturned or fuzzy function are associated with failure of planar cell polarity and Hedgehog signaling. Nat Genet 38: 303–311. 10.1038/ng1753 [DOI] [PubMed] [Google Scholar]
- Paunola E, Mattila PK, Lappalainen P (2002) WH2 domain: A small, versatile adapter for actin monomers. FEBS Lett 513: 92–97. 10.1016/s0014-5793(01)03242-2 [DOI] [PubMed] [Google Scholar]
- Peng Y, Axelrod JD (2012) Asymmetric protein localization in planar cell polarity: Mechanisms, puzzles and challenges. Curr Top Dev Biol 101: 33–53. 10.1016/B978-0-12-394592-1.00002-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao YL, Wu Y, Seo SY, Lim SC, Cho H (2014) Wound healing effects of new 15hydroxyprostaglandin dehydrogenase inhibitors. Prostaglandins Leukot Essent Fat Acids 91: 325–332. 10.1016/j.plefa.2014.09.011 [DOI] [PubMed] [Google Scholar]
- Pipathsouk A, Brunetti RM, Town JP, Graziano BR, Breuer A, Pellett PA, Marchuk K, Tran NHT, Krummel MF, Stamou D, et al. (2021) The wave complex associates with sites of saddle membrane curvature. J Cell Biol 220: e202003086. 10.1083/jcb.202003086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollitt AY, Insall RH (2009) WASP and SCAR/WAVE proteins: The drivers of actin assembly. J Cell Sci 122: 2575–2578. 10.1242/jcs.023879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan A, Tien AC, Haueter CM, Schulze KL, Bellen HJ (2009) The Arp2/3 complex and WASp are required for apical trafficking of Delta into microvilli during cell fate specification of sensory organ precursors. Nat Cell Biol 11: 815–824. 10.1038/ncb1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren N, Charlton J, Adler PN (2007) The flare Gene, which encodes the AIP1 protein of Drosophila, functions to regulate F-Actin disassembly in pupal epidermal cells. Genetics 176: 2223–2234. 10.1534/genetics.107.072959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren N, He B, Stone D, Kirakodu S, Adler PN (2006) The shavenoid gene of Drosophila encodes a novel actin cytoskeleton interacting protein that promotes wing hair morphogenesis. Genetics 172: 1643–1653. 10.1534/genetics.105.051433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren N, Zhu C, Lee H, Adler PN (2005) Gene expression during Drosophila wing morphogenesis and differentiation. Genetics 171: 625–638. 10.1534/genetics.105.043687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenu C, Athman R, Robine S, Louvard D (2004) The co-workers of actin filaments: From cell structures to signals. Nat Rev Mol Cell Biol 5: 635–646. 10.1038/nrm1437 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Mesa E, Abreu-Blanco MT, Rosales-Nieves AE, Parkhurst SM (2012) Developmental expression of Drosophila Wiskott-Aldrich Syndrome family proteins. Dev Dyn 241: 608–626. 10.1002/dvdy.23742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottner K, Faix J, Bogdan S, Linder S, Kerkhoff E (2017) Actin assembly mechanisms at a glance. J Cell Sci 130: 3427–3435. 10.1242/jcs.206433 [DOI] [PubMed] [Google Scholar]
- Rotty JD, Wu C, Bear JE (2012) New insights into the regulation and cellular functions of the ARP2/3 complex. Nat Rev Mol Cell Biol 14: 7–12. 10.1038/nrm3492 [DOI] [PubMed] [Google Scholar]
- Rust MB, Kullmann JA, Witke W (2012) Role of the actin-binding protein profilin1 in radial migration and glial cell adhesion of granule neurons in the cerebellum. Cell Adhes Migr 6: 13–17. 10.4161/cam.19845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagner A, Merkel M, Aigouy B, Gaebel J, Brankatschk M, Julicher F, Eaton S (2012) Establishment of global patterns of planar polarity during growth of the Drosophila wing epithelium. Curr Biol 22: 1296–1301. 10.1016/j.cub.2012.04.066 [DOI] [PubMed] [Google Scholar]
- Sánchez-Gutiérrez D, Sáez A, Pascual A, Escudero LM (2013) Topological progression in proliferating epithelia is driven by a unique variation in polygon distribution. PLoS One 8: e79227. 10.1371/journal.pone.0079227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M, Huibregtse JM, Vierstra RD, Howley PM (1993) The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75: 495–505. 10.1016/0092-8674(93)90384-3 [DOI] [PubMed] [Google Scholar]
- Schneider I (1972) Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morphol 27: 353–365. 10.1242/dev.27.2.353 [DOI] [PubMed] [Google Scholar]
- Seifert JRK, Mlodzik M (2007) Frizzled/PCP signalling: A conserved mechanism regulating cell polarity and directed motility. Nat Rev Genet 8: 126–138. 10.1038/nrg2042 [DOI] [PubMed] [Google Scholar]
- Shellard A, Mayor R (2020) All roads lead to directional cell migration. Trends Cell Biol 30: 852–868. 10.1016/j.tcb.2020.08.002 [DOI] [PubMed] [Google Scholar]
- Shi CY, Kingston ER, Kleaveland B, Lin DH, Stubna MW, Bartel DP (2020) The ZSWIM8 ubiquitin ligase mediates target-directed microRNA degradation. Science 370: eabc9359. 10.1126/science.abc9359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y, Usui T, Yanagawa S, Takeichi M, Uemura T (2001) Asymmetric colocalization of Flamingo, a seven-pass transmembrane cadherin, and Dishevelled in planar cell polarization. Curr Biol 11: 859–863. 10.1016/s0960-9822(01)00233-0 [DOI] [PubMed] [Google Scholar]
- Singh J, Mlodzik M (2012) Planar Cell Polarity Signaling: Coordination of cellular orientation across tissues. Wiley Interdiscip Rev Dev Biol 1: 479–499. 10.1002/wdev.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark BC, Lanier MH, Cooper JA (2017) CARMIL family proteins as multidomain regulators of actin-based motility. Mol Biol Cell 28: 1713–1723. 10.1091/mbc.e17-01-0019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan R, Gohl C, Fleige A, Klämbt C, Bogdan S (2011) Membrane-targeted WAVE mediates photoreceptor axon targeting in the absence of the WAVE complex in Drosophila. Mol Biol Cell 22: 4079–4092. 10.1091/mbc.E11-02-0121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt DI, Weber U, Mlodzik M (1997) The role of RhoA in tissue polarity and Frizzled signalling. Nature 387: 292–295. 10.1038/387292a0 [DOI] [PubMed] [Google Scholar]
- Strutt H, Strutt D (2009) Asymmetric localisation of planar polarity proteins: Mechanisms and consequences. Semin Cell Dev Biol 20: 957–963. 10.1016/j.semcdb.2009.03.006 [DOI] [PubMed] [Google Scholar]
- Suarez C, Carroll RT, Burke TA, Christensen JR, Bestul AJ, Sees JA, James ML, Sirotkin V, Kovar DR (2015) Profilin regulates F-Actin network homeostasis by favoring formin over Arp2/3 complex. Dev Cell 32: 43–53. 10.1016/j.devcel.2014.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez C, Kovar DR (2016) Internetwork competition for monomers governs actin cytoskeleton organization. Nat Rev Mol Cell Biol 17: 799–810. 10.1038/nrm.2016.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu S, Kurisu S, Oikawa T, Yamazaki D, Oda A, Takenawa T (2006) Optimization of WAVE2 complex-induced actin polymerization by membrane-bound IRSp53, PIP3, and Rac. J Cell Biol 173: 571–585. 10.1083/jcb.200509067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney KF, Li R (2016) Function and regulation of the Arp2/3 complex during cell migration in diverse environments. Curr Opin Cell Biol 42: 63–72. 10.1016/j.ceb.2016.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahirovic S, Hellal F, Neukirchen D, Hindges R, Garvalov BK, Flynn KC, Stradal TE, Chrostek-grashoff A, Brakebusch C, Bradke F (2010) Rac1 regulates neuronal polarization through the WAVE complex. J Neurosci 30: 6930–6943. 10.1523/jneurosci.5395-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenawa T, Suetsugu S (2007) The WASP–WAVE protein network: Connecting the membrane to the cytoskeleton. Nat Rev Mol Cell Biol 8: 37–48. 10.1038/nrm2069 [DOI] [PubMed] [Google Scholar]
- Tatárová Z, Brábek J, Rösel D, Novotny M (2012) SH3 domain tyrosine phosphorylation - sites, role and evolution. PLoS One 7: e36310. 10.1371/journal.pone.0036310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirouvanziam R, Davidson CJ, Lipsick JS, Herzenberg LA (2004) Fluorescence-activated cell sorting (FACS) of Drosophila hemocytes reveals important functional similarities to mammalian leukocytes. Proc Natl Acad Sci U S A 101: 2912–2917. 10.1073/pnas.0308734101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner CM, Adler PN (1998) Distinct roles for the actin and microtubule cytoskeletons in the morphogenesis of epidermal hairs during wing development in Drosophila. Mech Dev 70: 181–192. 10.1016/s0925-4773(97)00194-9 [DOI] [PubMed] [Google Scholar]
- Vignjevic D, Kojima S, Aratyn Y, Danciu O, Svitkina T, Borisy GG (2006) Role of fascin in filopodial protrusion. J Cell Biol 174: 863–875. 10.1083/jcb.200603013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitriol EA, McMillen LM, Kapustina M, Gomez SM, Vavylonis D, Zheng JQ (2015) Two functionally distinct sources of actin monomers supply the leading edge of lamellipodia. Cell Rep 11: 433–445. 10.1016/j.celrep.2015.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladar EK, Antic D, Axelrod JD (2009) Planar cell polarity signaling: The developing cell’s compass. Cold Spring Harb Perspect Biol 1: a002964. 10.1101/cshperspect.a002964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang H, Pan T, Li L, Li J, Yang H (2017) STIM1 silencing inhibits the migration and invasion of A549 cells. Mol Med Rep 16: 3283–3289. 10.3892/mmr.2017.7010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Hou Y, Guo X, van derVoet M, Boxem M, Dixon JE, Chisholm A, Jin Y (2013) The EBAX-type Cullin-RING E3 ligase and Hsp90 guard the protein quality of the SAX-3/Robo receptor in developing neurons. Neuron 79: 903–916. 10.1016/j.neuron.2013.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Madaule P, Reid T, Ishizaki T, Watanabe G, Kakizuka A, Saito Y, Nakao K, Jockusch BM, Narumiya S (1997) P140mDia, a mammalian homolog of Drosophila Diaphanous, is a target protein for Rho small GTPase and is a ligand for Profilin. EMBO J 16: 3044–3056. 10.1093/emboj/16.11.3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber U, Mlodzik M (2017) APC/CFzr/Cdh1-dependent regulation of planar cell polarity establishment via Nek2 kinase acting on Dishevelled. Dev Cell 40: 53–66. 10.1016/j.devcel.2016.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter CG, Wang B, Ballew A, Royou A, Karess R, Axelrod JD, Luo L (2001) Drosophila Rho-associated kinase (Drok) links Frizzled-mediated planar cell polarity signaling to the actin cytoskeleton. Cell 105: 81–91. 10.1016/s0092-8674(01)00298-7 [DOI] [PubMed] [Google Scholar]
- Wong LL, Adler PN (1993) Tissue Polarity Genes of Drosophila regulate the subcellular location for prehair initiation in pupal wing cells. J Cell Biol 123: 209–221. 10.1083/jcb.123.1.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Jenny A, Mirkovic I, Mlodzik M (2008) Frizzled-Dishevelled signaling specificity outcome can be modulated by Diego in Drosophila. Mech Dev 125: 30–42. 10.1016/j.mod.2007.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Mlodzik M (2008) The frizzled extracellular domain is a ligand for van gogh/Stbm during nonautonomous planar cell polarity signaling. Dev Cell 15: 462–469. 10.1016/j.devcel.2008.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JS, Luo L (2006) A protocol for mosaic analysis with a repressible cell marker (MARCM) in Drosophila. Nat Protoc 1: 2583–2589. 10.1038/nprot.2006.320 [DOI] [PubMed] [Google Scholar]
- Yan J, Huen D, Morely T, Johnson G, Gubb D, Roote J, Adler PN (2008) The multiple-wing-hairs gene encodes a novel GBD-FH3 domain-containing protein that functions both prior to and after wing hair initiation. Genetics 180: 219–228. 10.1534/genetics.108.091314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Jaiswal M, Charng WL, Gambin T, Karaca E, Mirzaa G, Wiszniewski W, Sandoval H, Haelterman NA, Xiong B, et al. (2014) A Drosophila genetic resource of mutants to study mechanisms underlying human genetic diseases. Cell 159: 200–214. 10.1016/j.cell.2014.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Lu Q, Fang X, Adler PN (2009) Rho1 has multiple functions in Drosophila wing planar polarity. Dev Biol 333: 186–199. 10.1016/j.ydbio.2009.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanfeng WA, Berhane H, Mola M, Singh J, Jenny A, Mlodzik M (2011) Functional dissection of phosphorylation of Disheveled in Drosophila. Dev Biol 360: 132–142. 10.1016/j.ydbio.2011.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Gong Y, Wang Q, Wang L, Zhang X (2017) MIR-100 antagonism triggers apoptosis by inhibiting ubiquitination-mediated p53 degradation. Oncogene 36: 1023–1037. 10.1038/onc.2016.270 [DOI] [PubMed] [Google Scholar]
- Yang Y, Mlodzik M (2015) Wnt-frizzled/planar cell polarity signaling: Cellular orientation by facing the wind (Wnt). Annu Rev Cell Dev Biol 31: 623–646. 10.1146/annurev-cellbio-100814-125315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaniv SP, Meltzer H, Alyagor I, Schuldiner O (2020) Developmental axon regrowth and primary neuron sprouting utilize distinct actin elongation factors. J Cell Biol 219: e201903181. 10.1083/jcb.201903181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallen JA, Cohen Y, Hudson AM, Cooley L, Wieschaus E, Schejter ED (2002) SCAR is a primary regulator of Arp2/3-dependent morphological events in Drosophila. J Cell Biol 156: 689–701. 10.1083/jcb.200109057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Wang D, Zhao X, Wang C, Gao Y, Liu K, Wang F, Wu X, Wang X, Sun L, et al. (2020) WDR63 inhibits Arp2/3-dependent actin polymerization and mediates the function of p53 in suppressing metastasis. EMBO Rep 21: e49269. 10.15252/embr.201949269 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Source Data for Figure 6LSA-2022-01484_SdataF6_FS6.pdf (677.4KB, pdf)
Source Data for Figure S6LSA-2022-01484_SdataF6_FS6.pdf (677.4KB, pdf)