Abstract
Inosine 5′‐monophosphate dehydrogenase (IMPDH) is an evolutionarily conserved enzyme that mediates the first committed step in de novo guanine nucleotide biosynthetic pathway. It is an essential enzyme in purine nucleotide biosynthesis that modulates the metabolic flux at the branch point between adenine and guanine nucleotides. IMPDH plays key roles in cell homeostasis, proliferation, and the immune response, and is the cellular target of several drugs that are widely used for antiviral and immunosuppressive chemotherapy. IMPDH enzyme is tightly regulated at multiple levels, from transcriptional control to allosteric modulation, enzyme filamentation, and posttranslational modifications. Herein, we review recent developments in our understanding of the mechanisms of IMPDH regulation, including all layers of allosteric control that fine‐tune the enzyme activity.
Keywords: allosteric regulation, enzyme filamentation, IMP dehydrogenase, protein structure and function, purine nucleotide biosynthesis
1. INTRODUCTION
Adenine and guanine nucleotides are essential molecules in biology since they constitute the building blocks of nucleic acids, play central roles in metabolism, and are involved in multitude of cellular processes, being the energy source of microtubule polymerization, mRNA translation, signal transduction, axon guidance, and so on. Therefore, both de novo and salvage biosynthetic guanine nucleotide pathways must be tightly controlled to maintain an appropriate balance between adenine and guanine nucleotide pools, as well as an optimal energy status for the cell. 1
Inosine 5′‐monophosphate dehydrogenase (IMPDH, EC 1.1.1.205) is the enzyme that catalyzes the oxidative conversion of inosine 5′‐monophosphate (IMP) into xanthosine 5′‐monophosphate (XMP), coupled to the reduction of NAD+ to NADH. XMP is subsequently transformed into GMP by the enzyme GMP synthase. The reaction catalyzed by IMPDH constitutes the first step after the separation of the ATP and GTP de novo biosynthetic routes and is a rate‐limiting step for GTP biosynthesis (Figure 1). Therefore, IMPDH controls the gateway to guanine nucleotides, playing critical roles in cell proliferation and immune response. 2 The inhibition of its catalytic activity causes a reduction in the guanine nucleotide pools and an imbalance between adenine and guanine nucleotides. 1 IMPDH inhibitors, including mycophenolic acid (CellCept®), mizoribine (Bredinin®), and ribavirin (Virazole® and Rebetol®), are widely used at present in the clinic as antivirals and immunosuppressants. 3 , 4 All these inhibitors bind to the catalytic site and compete with the substrates and, therefore, we will not review here their mechanisms of action. Instead, this review will focus on the allosteric regulation of IMPDH, its implications in disease, and its therapeutic potential.
FIGURE 1.
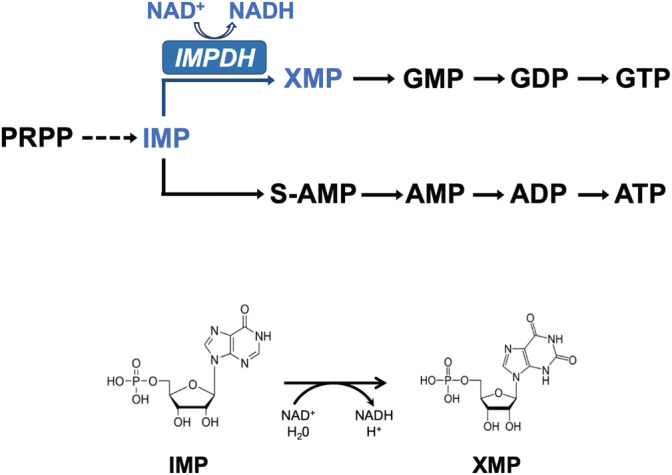
Purine nucleotide biosynthesis simplified pathway. The reaction catalyzed by IMPDH is indicated in blue color. The formulas of the substrate IMP and product XMP involved in the chemical reaction are shown below
2. STRUCTURE
An IMPDH monomer contains two well‐defined structural domains: a catalytic domain consisting of a (β/α)8 barrel, which represents the archetypal triose‐phosphate isomerase fold known as TIM barrel 5 (Figure 2; colored in light gray), and a regulatory domain inserted within a loop of the catalytic domain and composed of two cystathionine β‐synthase (CBS) motifs, that constitutes a Bateman domain 6 (Figure 2; colored in dark blue). The regulatory domain is not required for catalytic activity but is essential for allosteric regulation. 7 , 8 , 9 An additional special feature of the IMPDH family of proteins is the presence of a twisted beta sheet (Figure 2; colored in orange) that protrudes from the C‐terminal face of the TIM barrel. This structure, denoted as “finger domain,” 10 is present in all known IMPDH and is essential for allosteric modulation. 8 Within the finger domain, there exists a functionally essential loop (Figure 2; colored in green), denoted as “catalytic flap,” that acquires different conformations, 11 during the catalytic cycle, as described next. In solution, IMPDH always associates with tetramers, which are formed by the interaction of the catalytic domains (Figure 2c).
FIGURE 2.
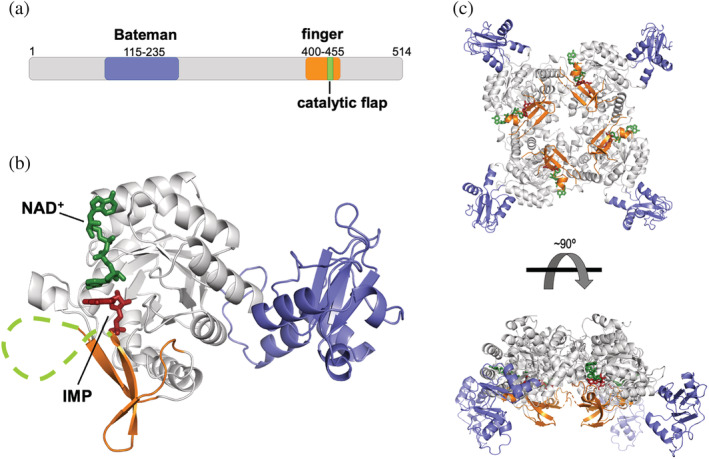
Structure of IMPDH. (a) Schematic representation of the structural and functional domains of IMPDH. Numbers correspond to the canonical variant of the human IMPDH1 enzyme. (b) Ribbon representation of a monomer of IMPDH with the two substrates (IMP in red sticks and NAD+ in green sticks) bound. The catalytic flap is shown with a discontinuous green line to indicate that it is unstructured. The structural model was generated by homology modeling, using PDBID 1MEW as template. (c) Ribbon representation of a tetramer of IMPDH with the two substrates bound. Color codes are the same for all three panels and they will be maintained throughout the rest of the figures in the manuscript (except Figure 10)
3. CATALYTIC MECHANISM
The mechanism of reaction of the IMPDH enzyme has been extensively characterized in different organisms, and proceeds through a two‐step reaction that includes fast redox transfer and rate‐limiting hydrolysis (reviewed in 9 ). Briefly, a catalytic cysteine residue attacks the C2 of IMP and the hydride is transferred to NAD+. Then, a thioimidate covalent intermediate E‐XMP* is formed and reduced NADH departs from the active site. After NADH departure, the catalytic flap occupies the vacant dinucleotide site to properly situate an Arg‐Tyr catalytic dyad that activates a water molecule through proton abstraction and hydrolyzes the E‐XMP* intermediate. Therefore, a structural reorganization of the active site of IMPDH allows the catalysis of two different chemical reactions. This reorganization implies two mutually exclusive conformations of the catalytic flap 11 : an open conformation for ligand binding, redox reaction, and product release (Figure 3a), and a closed conformation used for the hydrolysis of the covalent intermediate E‐XMP* (Figure 3b).
FIGURE 3.
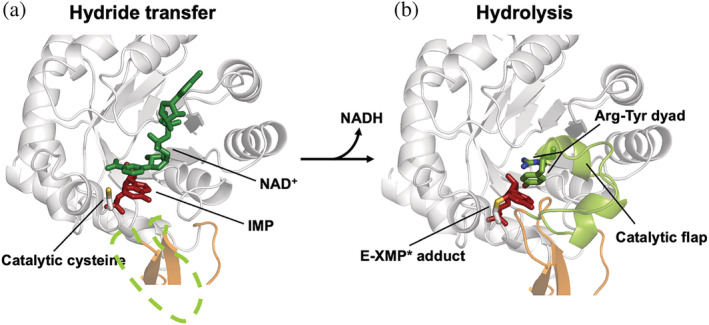
Conformations of the catalytic site of IMPDH. (a) Ribbon representation of the catalytic site of a monomer of IMPDH with the substrates bound (represented in green and red sticks) and the catalytic flap disordered (discontinuous green line). The side chain of the catalytic cysteine, shown in sticks, is positioned for the hydride transfer reaction. (b) A different conformation of the catalytic site, where the E‐XMP* covalent adduct is formed and the catalytic flap is positioned for the hydrolysis reaction. Structural models were generated by homology modeling using PDBID 1MEW and 3TSB/4XTD as templates, for (a) and (b) panels, respectively
4. THE NUCLEOTIDE‐CONTROLLED CONFORMATIONAL SWITCH
As stated before, the basic unit of IMPDH in solution is a tetramer but the binding of the adenine nucleotides (AMP, ADP, or ATP) to the Bateman domain drives the head‐to‐head interaction of two tetramers to form octamers. 7 , 12 ATP‐induced IMPDH octamers adopt an extended conformation with a hollow globular shape with approximate dimensions of 115 × 135 × 135 Å. In these octamers, the active sites, the finger domains, and the catalytic flaps remain exposed to the bulk of the solvent within the cavity (Figure 4a). All IMPDH enzymes studied so far bind adenine nucleotides, although this has sometimes been unnoticed because, in most cases, the catalytic activity of ATP‐induced octamers is similar to the activity of the tetramers in vitro. 7 , 8 , 13 , 14 , 15
FIGURE 4.
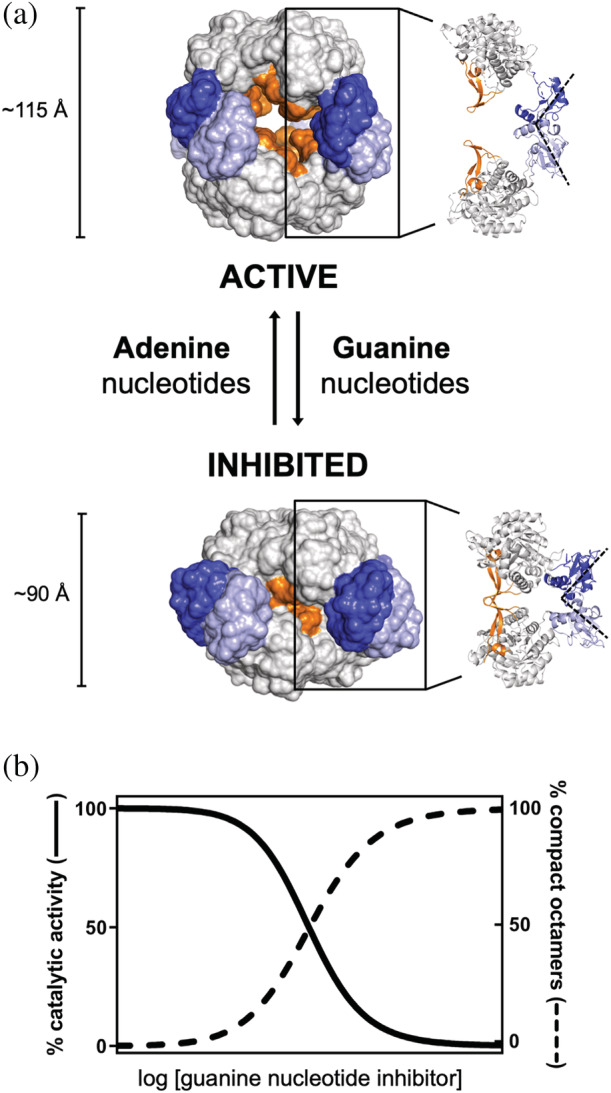
Nucleotide‐controlled conformational switch of IMPDH. (a) Surface representation of the active and inhibited octameric conformations of IMPDH. Approximate dimensions along the quaternary symmetry axis are indicated. A ribbon representation of two opposite monomers within the octamer is shown on the right to indicate the relative positions of the functional domains with these octameric conformations. The Bateman domains of the monomers in the upper and lower tetramers are shown in dark and light blue, respectively. (b) Simulated plot representing the correlation between octamer compaction and catalytic inhibition with increasing concentrations of guanine nucleotides inhibitors. This plot represents a general trend observed for the IMPDH of different organisms
The physiological relevance of AMP and ADP binding to IMPDH is unclear, given that they must compete with ATP which is much more abundant in cells. 16 , 17 Thereby, from now on, we will only refer to ATP binding to IMPDH. Given that the binding affinity of ATP for IMPDH lies in the micromolar range and the intracellular concentrations of ATP are millimolar, 16 , 17 it is reasonable to expect that ATP‐bound IMPDH holo‐octamers will be the predominant species in cells. This argument challenges the physiological relevance of previous reports showing ATP‐induced activation in vitro, 7 , 18 , 19 although in most cases, ATP has no significant effects on the catalytic activity in vitro. 12 , 13 , 20 , 21 , 22
A variety of guanine nucleotides compete with ATP for the allosteric sites in the Bateman domain and induce the compaction of the octamers along the quaternary symmetry axis by changing the relative orientation of the catalytic and regulatory domains (Figure 4a). The compaction of the octamers is correlated with the inhibition of the catalytic activity (Figure 4b). Therefore, the competition between adenine and guanine nucleotides controls a conformational switch that alternates the extended and compact IMPDH octamers to modulate their catalytic activity. Interestingly, this nucleotide‐controlled conformational switch is universally conserved from bacteria to humans. 8 , 12 , 20 , 23 , 24 , 25 , 26 , 27 , 28 , 29
The guanine nucleotide‐induced compaction of the octamers forces the finger domains and the catalytic sites of opposing tetramers to interact (Figure 4a). The interaction of the finger domains forms an interdigitated pseudo‐beta‐barrel that blocks the conformational dynamics of the active site, impeding the structural reorganization required to complete the catalytic cycle (Figure 2b). As a result, the affinity for the substrate is significantly reduced and the catalytic activity is inhibited. 8
Whether the switch between the two octameric conformations occurs in a concerted or sequential manner is an interesting and challenging issue that remains unclear. Recent cryo‐EM data suggest that the switch occurs following the Koshland–Némethy–Filmer sequential model, 30 since mixed states have been observed within filaments of human IMPDH2 (see below). This means that, at subsaturating concentrations of guanine nucleotide inhibitors, mixtures of compressed and extended protomers can be found within the same octamer. 25
5. DIVERSITY OF THE BATEMAN DOMAIN AND ALLOSTERIC MODULATORS
Bateman domains are widespread protein domains that have no defined catalytic function, but they regulate the activity of a wide variety of proteins in all kingdoms of life. 19 , 31 , 32 The amino acid sequence of the Bateman domain is the most different part of IMPDH and has divergently evolved to generate a variety of nucleotide‐binding sites to adapt allosteric modulation to the specific requirements of each organism 7 , 8 , 12 , 20 , 24 (Figure 5).
FIGURE 5.
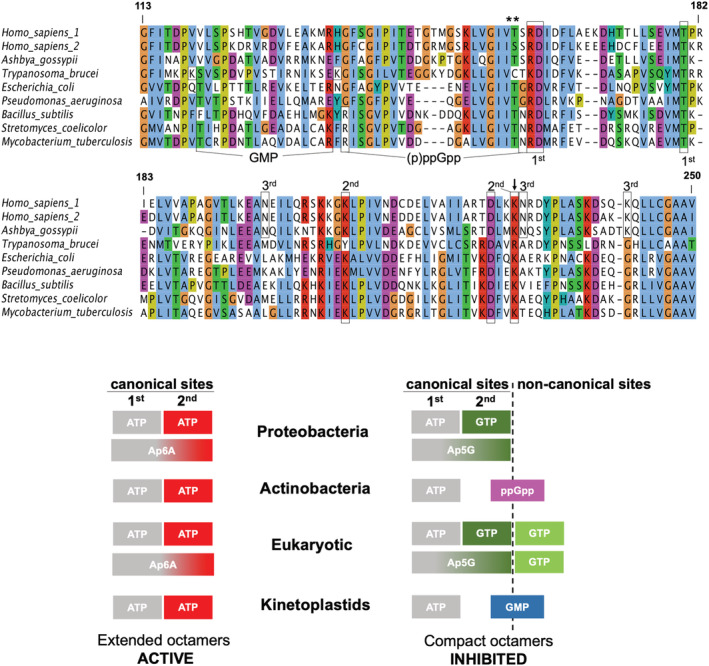
Diversity of nucleotide‐binding allosteric sites in the Bateman domain. Multiple protein sequence alignment of the Bateman domain of selected organisms. Boxes indicate key residues in the different nucleotide‐binding allosteric sites: canonical sites (first and second), third non‐canonical GTP eukaryotic site (third), (p)ppGpp bacterial site, and the GMP site in kinetoplastids. Selected residues are also marked with asterisks (phosphorylation sites) and an arrow (potential acetylation site). Numbers correspond to the canonical variant of the human IMPDH1 enzyme. The schemes below indicate which molecules bind to each of the allosteric sites in the different taxa
5.1. Canonical sites
Bateman domains feature two major cavities, related by a symmetry axis parallel to the central beta‐sheets, that constitute the canonical nucleotide‐binding sites. 31 , 32 These sites are present in all studied IMPDHs and are named according to the evolutionarily conserved Asp residues whose side chains coordinate the hydroxyls of the ribose of the nucleotides: Asp162 for the canonical site 1 and Asp226 for the canonical site 2 (these two Asp residues are framed inside boxes in Figure 5 and numbered according to the sequence of the human IMPDH1 canonical enzyme). As stated above, in all IMPDH enzymes studied so far, the two canonical sites bind ATP and induce an extended‐active conformation.
The two canonical sites are the only allosteric sites described in the Bateman domain of proteobacterial IMPDH, where site 1 binds preferentially ATP and site 2 can bind either ATP or GTP/GDP 20 (Figures 5 and 6). When ATP is bound to both canonical sites, IMPDH adopts an extended conformation that retains full catalytic activity. In contrast, when site 1 is occupied by ATP, but GTP/GDP are bound to site 2, IMPDH octamers acquire a compact conformation, and the catalytic activity is significantly inhibited (Figure 4). Thereby, in proteobacteria, ATP and GTP/GDP compete for the second canonical site to control the conformational switch that modulates the catalytic activity of IMPDH.
FIGURE 6.
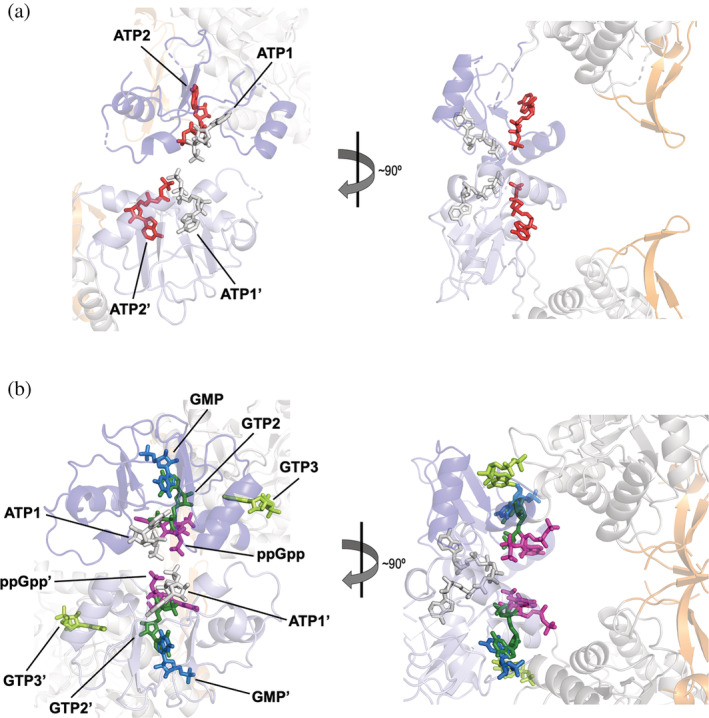
Diversity of allosteric modulators binding to the Bateman domain. (a) Conserved canonical adenine nucleotide‐binding sites in the active octameric extended conformation (PDBID 5MCP): two ATP molecules are shown in sticks, bound to the first (ATP1, light gray) and the second (ATP2, red) canonical sites. (b) Guanine nucleotide‐binding sites in the inhibited octameric compact conformation: ATP is shown in light gray sticks bound to the first canonical site (ATP1), GTP in dark green sticks bound to the second canonical site (GTP2), GTP in light green sticks bound to the third non‐canonical eukaryotic site (GTP3), ppGpp in pink sticks, and GMP in blue sticks bound to the kinetoplastidial‐specific site. PDBIDs used for creating this figure are 5TC3 (ATP1, GTP2, and GTP3), 7PMZ (ppGpp), and 6RFU (GMP). The Bateman domains of the monomers in the upper and lower tetramers are shown in dark and light blue, respectively
Similar to the case of adenine nucleotides stated above, in cells, GTP is about an order of magnitude more abundant than GDP, 16 , 17 and it is reasonable to assume that GTP, rather than GDP, is the main physiological allosteric inhibitor of IMPDH. Thereby, from now on, we will only refer to GTP binding to IMPDH.
It has been proposed that this is an essential mechanism to control the flux through the GTP de novo biosynthetic pathway and to maintain and appropriate ATP/GTP intracellular balance in Proteobacteria. 20 Accordingly, the deletion of the Bateman domain of IMPDH in Escherichia coli generates a strong imbalance in ATP and GTP intracellular pools and, thereby, it plays an essential role to maintain the physiological concentrations of adenylate and guanylate nucleotides. 13 , 33
Interestingly, both canonical sites can be simultaneously occupied by a single molecule of adenine and guanine dinucleoside polyphosphates. 24 These are ubiquitous biomolecules where two nucleosides, generally two adenosines, or an adenosine and a guanosine, are linked by a chain of two to seven phosphate moieties. Dinucleoside polyphosphates have been described to participate in a variety of cellular processes, including DNA replication and repair, cell division, neurotransmission, apoptosis, analgesia, vasoconstriction, and platelet aggregation (reviewed in references 34, 35, 36). These molecules occupy the two canonical sites simultaneously with identical binding modes than the corresponding mononucleotides (Figure 7). Nonetheless, the affinities of the former are 2 orders of magnitude higher than the latter, due to the simultaneous reduction of the entropic penalty of binding, molecularity change, and electrostatic repulsion. 24 Thereby, despite the intracellular concentration of dinucleoside polyphosphates lies in the range of low/sub‐micromolar range, they can readily compete with mononucleotides and play important roles on the physiological regulation of IMPDH. Nevertheless, this hypothesis remains to be experimentally corroborated in vivo.
FIGURE 7.
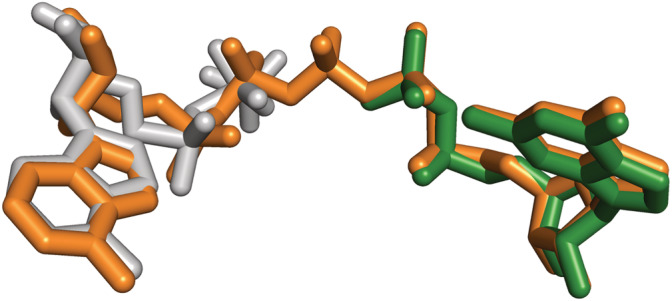
Dinucleoside polyphosphates bind to the Bateman domain of IMPDH. Structural superimposition of the mononucleotides ATP (light gray sticks) and GDP (dark green sticks) bound to the first and second canonical sites (extracted from PDBID 5TC3), respectively, and the dinucleoside polyphosphate Ap5G (orange sticks) simultaneously bound to both sites (extracted from PDBID 6RPU)
5.2. Bacterial (p)ppGpp binding site
In contrast to Proteobacteria, in most other bacterial phyla, including Firmicutes and Actinobacteria, GTP does not bind to the second canonical site and, therefore, does not affect IMPDH activity. 20 Instead, the Bateman domain of the IMPDH in these bacterial phyla contains an extra (non‐canonical) binding pocket exclusive for the (p)ppGpp alarmones. These molecules have well‐documented regulatory roles in gene expression and protein translation, and they also induce rapid changes in cellular metabolism by regulating the activity of a variety of enzymes. 37 , 38 The (p)ppGpp binding site in IMPDH partially overlaps the second canonical site (Figures 5 and 6), allowing a direct competition between (p)ppGpp and ATP to control the conformational switch and modulate the catalytic activity. An evolutionary analysis has proposed that the bacterial IMPDH ancestor contained the (p)ppGpp binding site, but this site was lost during the evolution of the proteobacterial lineage that, instead, modified the second canonical site to allow the competition between ATP and GTP. 20
5.3. Eukaryotic third guanine nucleotide non‐canonical site
Most eukaryotic IMPDH enzymes possess a unique non‐canonical site exclusive for the guanine nucleotides GTP and GDP. Nonetheless, as stated above for the second canonical site, from now on, we will only refer to GTP binding. This site is located at the interface between the catalytic and Bateman domains and is physically linked to the second canonical site 8 (Figure 6). Therefore, there exists strong cooperativity between allosteric sites 2 and 3. 24 Indeed, the incubation of IMPDH with mixtures of ATP and GTP results in complexes with ATP bound to site 1 and GTP bound to sites 2 and 3. 12 , 25 , 26 , 27
In eukaryotic IMPDH, the occupancy of the second canonical and the third non‐canonical nucleotide‐binding sites of the Bateman domain determines the conformation of eukaryotic IMPDH octamers and, therefore, their catalytic activity. Remarkably, missense mutations that map in these two allosteric sites abrogate the allosteric inhibition and are associated with retinopathies and neuropathies in humans, 39 as will be described in detail below.
5.4. Protozoan GMP biding site
The high‐resolution crystallographic structure of Trypanosoma cruzi IMPDH revealed the presence of ATP and GMP bound to the Bateman domain. Given that the crystals were obtained by in cellulo crystallization in the cytoplasm of Sf9 insect cells, these nucleotides were proposed to be the genuine allosteric cofactors of T. cruzi IMPDH. 40 In this structure, ATP occupies the first canonical site, while GMP is bound to a non‐canonical pocket that partially overlaps to the second canonical site (Figures 5 and 6). This site locates at the interface between the catalytic and Bateman domains, allowing GMP to establish direct interactions with both domains and to stabilize the compact inhibited IMPDH conformation.
In contrast to most other eukaryotic IMPDH, T. cruzi IMPDH adopts the compact conformation without any nucleotide bound to the GTP third non‐canonical site. Indeed, all the key residues in the eukaryotic GTP non‐canonical site are not conserved in either T. cruzi (Figure 5) or in any other kinetoplastid IMPDH enzyme. 41 Nonetheless, in vitro enzyme kinetics, as well as in vivo experiments, are required to validate the GMP site and its physiological significance.
6. FINE‐TUNE MODULATION OF ALLOSTERIC CONTROL
On top of allosteric modulation, several mechanisms that provide additional layers of regulatory control for fine‐tuning the activity of IMPDH have been described. These mechanisms include posttranslational modifications, enzyme filamentation, and splice variants. All of them have no significant effects on the catalytic activity but they make the enzyme more resistant to feedback allosteric inhibition, that is, these mechanisms shift the sensitivity of IMPDH to guanine nucleotide inhibitors (Figure 8). They occur under physiological conditions that require the expansion of guanine nucleotide pools, for instance, in conditions of high‐rate cellular growth 42 or in response to light during the visual cycle in retinal photoreceptors. 43 , 44
FIGURE 8.
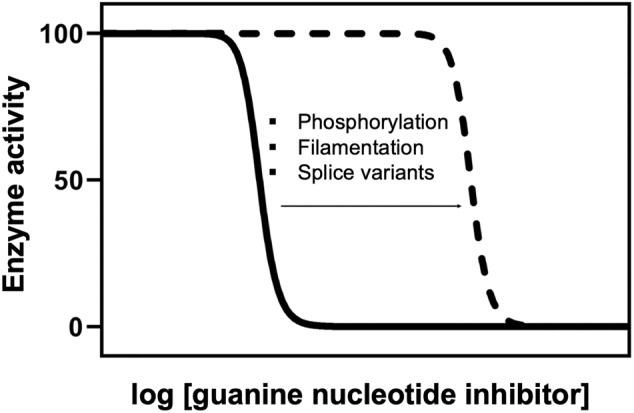
Fine‐tune of the allosteric regulation of IMPDH. Plot representing the reported effect of posttranslational modifications, enzyme filamentation, and splice variants on the allosteric inhibition of IMPDH by guanine‐nucleotides. These three mechanisms do not significantly affect enzyme activity but, instead, shift the sensitivity to the allosteric inhibition by guanine nucleotides
6.1. Posttranslational modifications
Protein posttranslational modifications are a fast and versatile mechanism used by cells to regulate the function of proteins in response to environmentally changing conditions. 45 With respect to IMPDH, this takes on special relevance in retinal cells where the regulation of the de novo guanine nucleotide biosynthetic pathway is essential for maintaining appropriate levels of cGMP, the key signaling molecule in phototransduction. 46 Moreover, the expression of the enzymes from the purine salvage biosynthetic pathways is low and, thus, most of the guanine‐nucleotide biosynthesis rely on the de novo pathway. 47
Recently, the phosphorylation of three residues in the mammalian IMPDH1 enzyme was reported in retinal cells in response to the illumination conditions. 28 Two of the phosphorylation sites are located in the Bateman domain (Thr159 and Ser160; marked with an asterisk in Figure 5) and are preferentially phosphorylated upon light exposure in vivo, while the third phosphorylation site (Ser477) locates in the catalytic domain and is phosphorylated in dark‐adapted retinal cells. 28 Phosphorylation of these amino acid residues does not affect enzyme activity but, instead, tunes the sensitivity of the enzyme to GTP‐mediated allosteric inhibition (Figure 8).
Residues Thr159 and Ser160 in the human IMPDH1 protein are directly involved in the binding of nucleotides at the canonical site 1 (Figure 5) and, thereby, their light‐dependent phosphorylation in retina abrogates GTP‐mediated allosteric inhibition. Accordingly, the overall metabolic flux toward the de novo guanine nucleotide synthesis is increased upon light exposure, facilitating the elevated GTP levels required for phototransduction. 28 In contrast, phosphorylation of residue Ser477 occurs preferentially in dark‐adapted retinas 28 and it is proposed to increase the sensitivity of IMPDH to GTP‐feedback inhibition 27 , 28 (more details below) to adapt the allosteric regulation of retinal IMPDH to the low GTP demand of photoreceptors in the dark. 43 , 44
It has been recently proposed that posttranslational modifications might also tune the allosteric regulation of IMPDH in bacteria. 20 According to biological public databases (PLMD 48 and dbPSP 49 ), lysine acetylation is a recurrent modification within the Bateman domain of bacterial IMPDH enzymes. The Lys‐to‐Gln acetylation‐mimicking mutation of an evolutionarily conserved residue significantly compromised allosteric inhibition of E. coli and B. subtilis IMPDH enzymes in vitro. 20 Remarkably, this residue (Lys229 in human IMPDH1; marked with an arrow in Figure 5) is located in the second canonical site, where it participates in the binding of GTP and (p)ppGpp. 8 , 20 Thereby, its acetylation might shift the sensitivity of IMPDH to these allosteric inhibitors. Moreover, acetylation of this residue has also been reported for a variety of organisms, including E. coli, 50 , 51 B. subtilis, 52 , 53 S. coelicolor, 54 Corynebacterium glutamicum, 55 Spiroplasma eriocheiris, 56 Bacillus amyloquefaciens, 57 and Mus musculus. 58 These data strongly suggest that allosteric regulation of bacterial IMPDH might also be tuned by posttranslational modifications, as it occurs in eukaryotic cells.
6.2. Enzyme filamentation
Enzyme filaments are defined as reversible, functional, linear self‐assemblies of a single type of enzyme. Filamentation has recently gained relevance as a new mode of enzymatic regulation. It is now clearly accepted that enzyme filamentation serves as a general mechanism for the regulation of metabolism by fine‐tuning protein functional and structural properties. 59 , 60 Perhaps the most representative examples of filament‐forming enzymes are those that catalyze the limiting steps in the de novo biosynthetic pathways of pyrimidine and purine nucleotides, CTP synthase, and IMPDH, respectively. 61 , 62
In vertebrate cells, IMPDH has evolved to acquire the capacity to form mesoscale filaments (spicules or cytoophidia) in conditions that require a strong increase in IMPDH activity to keep with high GTP demand, such as high rates of proliferation, Gln/Ser/folate‐metabolite deficiency, T‐cell activation, or retinal exposition to light. 23 , 28 , 42 , 63 IMPDH filamentation has also been observed in cultured cells upon ectopic overexpression 29 (Figure 9).
FIGURE 9.
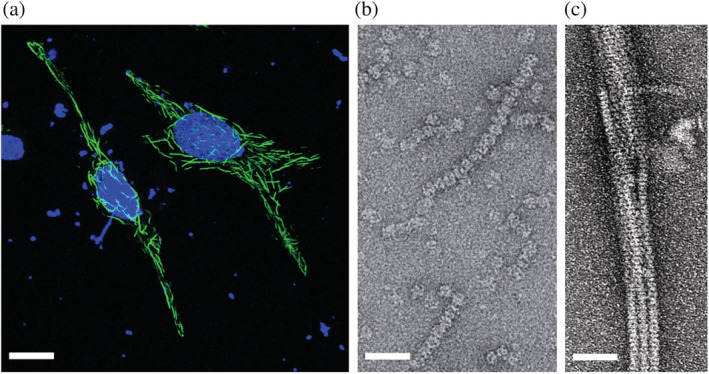
IMPDH filamentation in vertebrate cells. (a) Immunofluorescence micrograph showing human IMPDH1 filaments (green) upon overexpression in HeLa cells. Nuclei (blue) are stained with DAPI. (b, c) Negative‐stained electron microscopy micrographs showing spontaneous filamentation of human IMPDH1 in vitro in the presence of ATP (b) and filament bundling in macromolecular crowding conditions (150 mg/ml Ficoll‐70, (c)). Scale bars correspond to 5 μm (panel (a)) and 50 nm (panels (b) and (c))
In humans, there are two genes of IMPDH, that result in enzymes sharing more than 80% sequence identity: IMPDH1, which plays a housekeeping role in most tissues, and IMPDH2, which is typically upregulated in proliferating cells. 64 Thorough in vitro characterization of the filaments of both proteins by electron microscopy has revealed that nucleotide binding to the Bateman domain promotes self‐assembly of protofilaments made of stacked octamers 25 , 26 , 27 , 29 , 65 (Figure 9). Moreover, macromolecular crowding conditions induce protofilament bundling in vitro, 29 to form structures that resemble those observed in cells 66 (Figure 9).
According to recent cryo‐EM data, GTP binding to the second canonical and the third non‐canonical sites in the Bateman domain of both human IMPDH1 and IMPDH2 enzymes induces two conformational changes: (a) compression of the octamer along the quaternary symmetry axis (Figure 4), and (b) protomer tilting about 5° relative to the fourfold symmetry axis, such that the tetramers become more bowed than the flat tetramers observed in the ATP‐induced octamers (Figure 10). 25 , 27 Remarkably, a similar flat‐bowed conformational change can also be deduced by comparing the high‐resolution structures of non‐polymerizing IMPDH enzymes (4XWU vs. 4XTI, 14 shown in Figure 10, 3USB vs. 3TSB, 11 or 5AHM vs. 5AHL 67 ). Of note, the NAD+ binding pocket lies at the monomer interface within the tetramers (Figure 10), and it is plausible that the flat‐bowed conformational change might play a role in cofactor binding and release during the catalytic cycle, although its functional significance is unclear.
FIGURE 10.
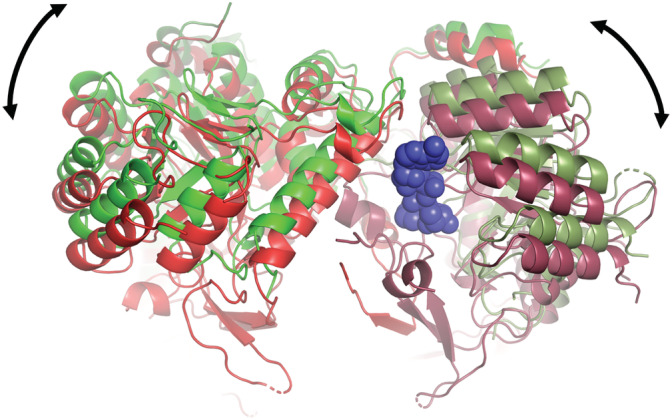
Flat‐bowed conformational change. Structural comparison of tetramers with flat (green ribbons) and bowed (red ribbons) conformations. PDBID 1WXU (flat conformation; Ashbya gossypii IMPDH with no substrate in the catalytic site) versus PDBID 1XTI (bowed conformation; A. gossypii IMPDH bound to IMP, not shown in the figure). NAD is shown in blue spheres bound at the interface between two monomers (note that red and green color tones vary between the monomers)
ATP binding to the canonical sites of the Bateman domain of the human IMPDH2 enzyme induces polymers composed of extended active octamers with flat tetramers (Figure 11). The binding of GTP to IMPDH2 polymers induces the compaction of the octamers, but the polymer lattice forces the tetramers to remain flat, which allows polymeric IMPDH2 to remain partially active. Thus, IMPDH2 polymerization reduces sensitivity to GTP inhibition. Nonetheless, at high GTP concentrations, the polymers disassemble into compressed inhibited octamers with bowed tetramers 25 , 29 (Figure 11).
FIGURE 11.
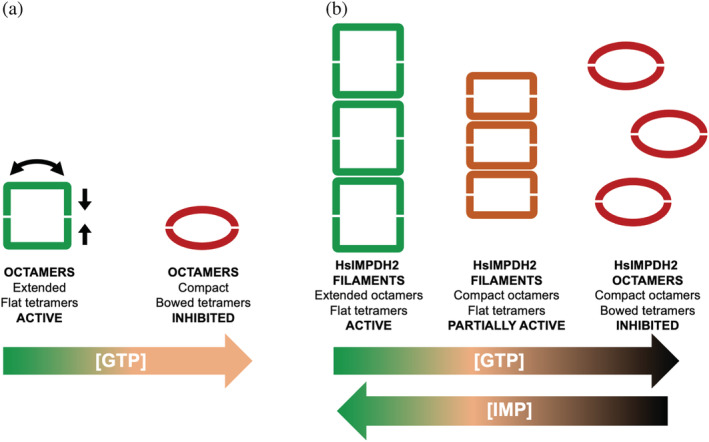
Filamentation of human IMPDH2 tunes allosteric inhibition. (a) Schematic representation of the human IMPDH conformational changes in non‐polymerized IMPDH octamers upon allosteric inhibition induced by moderate GTP concentrations (orange‐colored GTP range): octamer compaction and tetramer bowing. (b) The polymer lattice forces human IMPDH2 tetramers to remain flat within compressed octamers (orange rectangles), remaining partially active at moderate concentrations of GTP (orange‐colored GTP range). Thus, polymerization shifts the sensitivity of human IMPDH2 to GTP. At very high GTP concentrations (dark red‐colored GTP range), tetramers acquire the bowed conformation (red ellipsoids) and disassemble. The substrate IMP, on the other hand, increases resistance to GTP‐induced compaction and depolymerization
Additionally, a substrate dependence of IMPDH2 filament assembly has been reported. When the concentration of IMP is high, IMPDH2 filaments resist both disassembly and the fully compressed inhibited conformation. Thus, at high IMP levels, a portion of IMPDH2 octamers within the filament will remain active, even at elevated GTP concentrations 25 , 26 (Figure 11). These results, obtained in vitro using purified recombinant protein, perfectly agree with those reported in murine embryonic stem cells showing that the intracellular balance IMP/GTP modulate IMPDH filamentation; this is, during rapid cell proliferation, IMP accumulation promotes IMPDH assembly, whereas elevated GTP levels trigger depolymerization. 63
Human IMPDH1 is more sensitive to GTP‐feedback inhibition than IMPDH2. 29 In marked contrast to IMPDH2, filament assembly of IMPDH1 does not impose conformational constraints, that is, IMPDH1 filaments can accommodate catalytically inactive compressed octamers with bowed tetramers 27 (Figure 12). Thereby, there are no significant differences in GTP‐mediated allosteric inhibition between octameric and polymerized IMPDH1. IMPDH1 filamentation does not appear to play a role either in the catalytic activity or in tuning the response to GTP inhibition, which raises the unanswered question about the physiological function of IMPDH1 filaments. 27 Instead, it has been speculated that IMPDH1 filaments might serve as signaling or scaffolding of other enzymes, such as the regulatory protein Ankyrin Repeat Domain 9 68 or CTP synthase. 62
FIGURE 12.
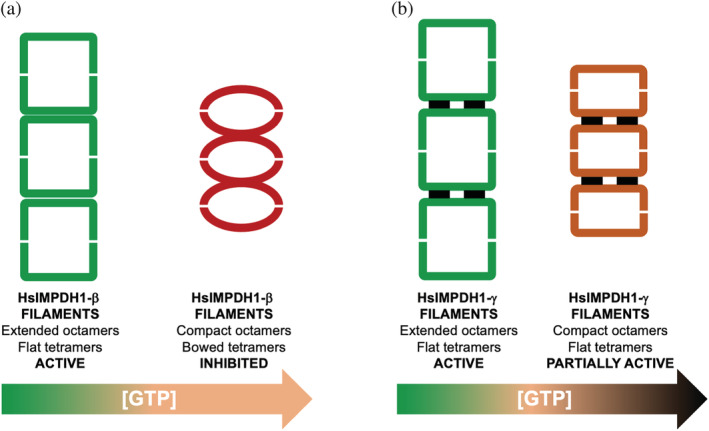
Retinal variants are more resistant to GTP allosteric inhibition than the canonical variant of human IMPDH1. (a) Polymers of the canonical variant β of human IMPDH1 enzyme can accommodate extended active octamers with flat tetramers (green squares), as well as compact inhibited octamers with bowed tetramers (red ellipsoids), which are induced at moderate concentrations of GTP (orange‐colored GTP range). (b) The N‐terminal α‐helical extension (black rectangles) of the retinal variant β of human IMPDH1 forces the tetramers in the compressed octamers to remain flat, partially retaining catalytic activity, even at very high concentrations of GTP (dark red‐colored GTP range)
6.3. Tissue‐specific splicing variants
On top of the differences between the two human IMPDH enzymes described above, tissue‐specific splicing variants add an additional layer of allosteric regulation to finely tune the enzyme allosteric regulation.
In mammalian retinal cells, the IMPDH1 gene is predominantly expressed as two major variants, 47 , 69 where alternative splicing adds additional residues to the C‐terminus (variant α, 546 residues) or both the C‐ and the N‐termini (variant γ, 595 residues) of the minoritarian canonical enzyme (variant β, 514 residues). 47 The majoritarian retinal splice variants α and γ have been reported to be significantly more resistant to GTP feedback inhibition than the canonical variant α, 27 , 70 as they are adapted to the especially high guanine nucleotide demand in retinal cells, especially photoreceptors. 43 , 44
How the apparently disordered C‐terminal extension contributes to increase the resistance to GTP inhibition remains unclear, but the N‐terminal extension in the variant γ forms a short 10‐residue helix that changes filament architecture. This N‐terminal helix sits at the interface between octamers within the polymer and stabilizes the flat conformation of tetramers, even when the octamers in the polymer get compressed upon GTP binding 27 (Figure 12). Therefore, the N‐terminal extension of the retinal variant γ makes polymerized IMPDH more resistant to GTP.
7. MULTILAYER REGULATION
The mechanisms of regulation described above show the diversity and complexity of the physiological allosteric regulation of IMPDH, which is arranged in multiple interconnected layers. In cells, it is expected an interplay among these layers to adapt IMPDH regulation to the metabolic requirements of the different stages of the cell cycle or in response to external stimuli. For instance, the phosphorylation of the residue Ser477 in retinal cells 28 may disrupt filament assembly, because this residue is located at the assembly interface in the filaments of the IMPDH1 retina variant γ. 27 Phosphorylation of Ser477, which occurs preferentially in dark‐adapted retinas, 28 would then increase the sensitivity to GTP‐feedback inhibition to adapt IMPDH allosteric regulation to the low GTP demand of photoreceptors in the dark. 43 , 44
It is expected that IMPDH allosteric regulation might coexist with orthosteric inhibition, mediated by XMP and GMP, although the contribution of these metabolites to IMPDH inhibition remains to be corroborated in vivo. 8 , 13 , 71 , 72
Finally, the simultaneous presence of IMPDH2, IMPDH1, and its different splice variants within cells, all of them with similar tetramerization and filament assembly contacts, suggests that they might co‐assemble into heterogeneous tetramers, octamers, or filaments, adding complexity and versatility to the physiological regulation of IMPDH.
8. ALLOSTERIC DYSREGULATION IN DISEASE
Missense mutations in the two human IMPDH genes associated with pathologies have been reported (reviewed in reference 39). Mutations in IMPDH1 result in autosomal dominant blindness either through retinitis pigmentosa or the more severe and early onset Leber congenital amaurosis. 73 , 74 , 75 , 76 , 77 , 78 On the other hand, mutations in IMPDH2 lead to severe neurodevelopmental disorders, such as dystonia. 79 , 80
Despite IMPDH1 is ubiquitously expressed in most human tissues, mutations in IMPDH1 have only been associated to retinopathies, 47 indicating that retinal cells are exceptionally sensitive to defects in purine biosynthesis. 43 , 44 Because photoreceptors have low expression of IMPDH2 and HPRT, 81 , 82 the major enzyme from the purine salvage biosynthetic pathway, alterations of IMPDH1 function will result in imbalanced purine nucleotide pools that can lead to cell death. 43 Twelve missense mutations in IMPDH1 associated with retinopathies have been identified, although none of them have significant effects on the catalytic activity of IMPDH1 in vitro. 22 , 29 , 74 Six of these mutations map around the second canonical and the third non‐canonical allosteric sites in the Bateman domain, while the rest are scattered in the catalytic domain, but distal from the IMP binding site, the flap region, or the finger domain 8 , 39 (Figure 13). As expected, the six mutations that map into the allosteric sites interfere with nucleotide binding and confer resistance to GTP allosteric inhibition. 27 , 29
FIGURE 13.
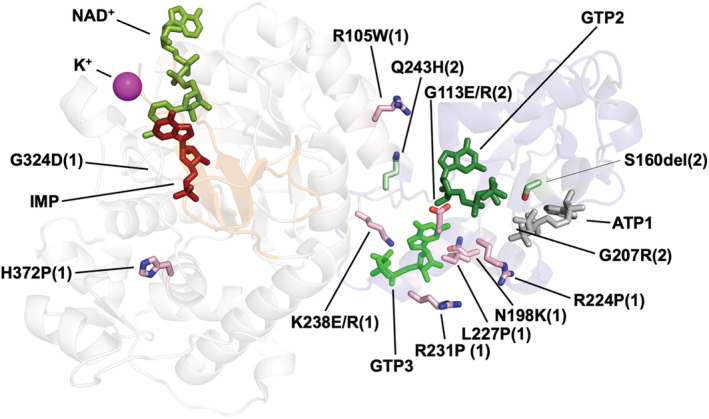
Pathogenic mutations in human IMPDH genes. Ribbon representation of a monomer of human IMPDH with mutations associated to either retinopathies (IMPDH1; indicated with (1) and side‐chain sticks colored in light pink) or neuropathies (IMPDH2; indicated with (2) and side‐chain stick colored in light green). Substrates IMP and NAD+ are shown as red and green sticks, respectively. The K+ ion necessary for catalysis is represented as a pink sphere. ATP and GTP molecules bound to the allosteric sites are also shown. It can be clearly seen that most of these mutations cluster around the allosteric sites, altering nucleotide binding and inhibition
Like IMPDH1, neuropathy‐associated IMPDH2 mutations cluster around the second canonical and the third non‐canonical allosteric sites in the Bateman domain (Figure 13) and, thereby, it is reasonable to propose that they will also disrupt allosteric inhibition. 39 Further experimental work is needed to support this hypothesis, however.
Altogether, a significant number of reported missense mutations in the two human IMPDH genes appear to generate constitutively active mutant enzymes, unable to be allosterically inhibited by GTP, a hypothesis that is supported by the dominant genetic character of the disease. 82 Why mutations in the ubiquitously IMPDH expressed genes lead only to neuronal diseases is an intriguing question that remains open. 47
9. TARGETING ALLOSTERIC REGULATION FOR DRUG DESIGN
Protein allosteric modulation is attracting considerable attention in drug discovery. Ligands that target allosteric sites offer significant advantages over the corresponding orthosteric ligands due to unprecedent selectivity and improved physicochemical properties, while minimizing toxicity and other side effects. Therefore, allosteric ligands offer excellent opportunities for the development of therapeutic strategies spanning a broad spectrum of disease and are at present widely used in drug discovery. 83 , 84 , 85
Several IMPDH inhibitors are widely used at present in the clinic as antivirals and immunosuppressants, all of them binding at the active site as othosteric ligands. 3 , 4 , 9 Given the therapeutic relevance of IMPDH, 4 the identification and development of allosteric modulators might expand the chemical space of inhibitors, and has an obvious pharmacological interest. A clear advantage of targeting the Bateman domain is that this is the most divergent part of IMPDH. Therefore, targeting specifically the allosteric sites exclusive of certain bacteria 20 or kinetoplastids, 40 might lead to molecules with antibiotic 86 or antiprotozoal activity, 87 respectively. We will briefly review next the reported allosteric inhibitors of IMPDH.
In a pioneer study, Alexandre et al. screened large chemical libraries to identify the first‐in‐class allosteric inhibitors of the Pseudomonas aeruginosa IMPDH (Figure 14), which compete with ATP for the allosteric sites in the Bateman domain and block the enzyme in the inhibited compact conformation. 88 Further in vivo experiments are required to decipher the antibiotic potential of these compounds, however.
FIGURE 14.

Allosteric inhibitors of IMPDH. Structures of the reported allosteric inhibitors of Pseudomonas aeruginosa IMPDH (upper row) and human IMPDH2 enzyme (sanglifehrin A and sapannone A). The pendant side chain of sanglifehrin A, which binds specifically to a pocket in the Bateman domain of IMPDH2, is shown inside a rectangle. It should be noted the diversity of chemical scaffolds of the reported allosteric inhibitors that bind to the Bateman domain of IMPDH
The Bateman domain is also the most divergent part between the two human IMPDH enzymes and, to this respect, sappanone A (Figure 14), a small‐molecule probe covalently binding to the conserved Cys140 residue of human IMPDH2, was recently identified. 89 On binding to this residue, the probe exerts an allosteric regulation and induces IMPDH2 inactivation, leading to an effective suppression of neuroinflammatory responses. However, the probe does not bind to IMPDH1, where the corresponding residue is serine, unable to react with sapannone A. Thus, this study shows Cys140 as a druggable site for selective inhibition of IMPDH2. 89
Another example of a small molecule that specifically targets the Bateman domain of the IMPDH2 enzyme is sanglifehrin A (Figure 14), which displays immunosuppressive activity in vitro by blocking T cell proliferation. 90 Sanglifehrin A is a mixed polyketide and non‐ribosomal peptide synthase natural product with sub‐nano‐molar affinity for its receptor cyclophilin A. 91 Sanglifehrin A can be split into two chemical moieties with different target specificities: a macrocyclic core, responsible for binding to cyclophilin A and a pendant side chain, which binds specifically to a pocket in the Bateman domain of IMPDH2 92 (inboxed in Figure 14). The formation of the ternary complex sanglifehrin A–cyclophilin A–IMPDH2 modulates cell growth, despite the activity of IMPDH2 is unaffected. 92
Altogether, these studies demonstrate the versatility of the Bateman domain to accommodate totally unrelated chemical scaffolds (Figure 14) and open the door for the development of IMPDH allosteric inhibitors with potentially have high therapeutic relevance that might provide guidance for clinical trial design.
AUTHOR CONTRIBUTIONS
Rubén M. Buey: Conceptualization (lead); writing – original draft (lead); writing – review and editing (lead). David Fernández‐Justel: Writing – original draft (equal). Alberto Jiménez: Writing – original draft (equal); writing – review and editing (equal). José L. Revuelta: Writing – original draft (equal); writing – review and editing (equal).
ACKNOWLEDGMENTS
This study was funded by the Spanish Ministerio de Ciencia e Innovación‐FEDER‐Fondo Social Europeo (grants PID2019‐109671GB‐I00 to R.M.B. and PID2020‐118200RB‐I00 to J.L.R. and A.J.). D.F.J. was supported by a pre‐doctoral contract from the Junta de Castilla y León. The authors thank Mónica Balsera for her critical comments and helpful suggestions.
Buey RM, Fernández‐Justel D, Jiménez A, Revuelta JL. The gateway to guanine nucleotides: Allosteric regulation of IMP dehydrogenases. Protein Science. 2022;31(9):e4399. 10.1002/pro.4399
Review Editor: John Kuriyan
Funding information Ministerio de Ciencia e Innovación, Grant/Award Numbers: PID2019‐109671GB‐I00, PID2020‐118200RB‐I00; Junta de Castilla y León
REFERENCES
- 1. Allison AC, Eugui EM. Mycophenolate mofetil and its mechanisms of action. Immunopharmacology. 2000;47:85–118. [DOI] [PubMed] [Google Scholar]
- 2. Jayaram H, Cooney DA, Grusch M, Krupitza G. Consequences of IMP dehydrogenase inhibition, and its relationship to cancer and apoptosis. Curr Med Chem. 1999;6:561–574. [PubMed] [Google Scholar]
- 3. Braun‐Sand SB, Peetz M. Inosine monophosphate dehydrogenase as a target for antiviral, anticancer, antimicrobial and immunosuppressive therapeutics. Future Med Chem. 2009;2:81–92. [DOI] [PubMed] [Google Scholar]
- 4. Cuny GD, Suebsuwong C, Ray SS. Inosine‐5′‐monophosphate dehydrogenase (IMPDH) inhibitors: A patent and scientific literature review (2002‐2016). Expert Opin Ther Pat. 2017;27:677–690. [DOI] [PubMed] [Google Scholar]
- 5. Nagano N, Orengo CA, Thornton JM. One fold with many functions: The evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J Mol Biol. 2002;321:741–765. [DOI] [PubMed] [Google Scholar]
- 6. Bateman A. The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem Sci. 1997;22:12–13. [DOI] [PubMed] [Google Scholar]
- 7. Labesse G, Alexandre T, Vaupré L, et al. MgATP regulates allostery and fiber formation in IMPDHs. Structure. 2013;21:975–985. [DOI] [PubMed] [Google Scholar]
- 8. Buey RM, Ledesma‐Amaro R, Velazquez‐Campoy A, et al. Guanine nucleotide binding to the Bateman domain mediates the allosteric inhibition of eukaryotic IMP dehydrogenases. Nat Commun. 2015;6:8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hedstrom L. IMP dehydrogenase: Structure, mechanism, and inhibition. Chem Rev. 2009;109:2903–2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang RG, Evans G, Rotella FJ, et al. Characteristics and crystal structure of bacterial inosine‐5′‐monophosphate dehydrogenase. Biochemistry. 1999;38:4691–4700. [DOI] [PubMed] [Google Scholar]
- 11. Makowska‐Grzyska M, Kim Y, Wu R, et al. Bacillus anthracis inosine 5′‐monophosphate dehydrogenase in action: The first bacterial series of structures of phosphate ion‐, substrate‐, and product‐bound complexes. Biochemistry. 2012;51:6148–6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Buey RM, Fernandez‐Justel D, Marcos‐Alcalde I, et al. A nucleotide‐controlled conformational switch modulates the activity of eukaryotic IMP dehydrogenases. Sci Rep. 2017;7:2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pimkin M, Markham GD. The CBS subdomain of inosine 5′‐monophosphate dehydrogenase regulates purine nucleotide turnover. Mol Microbiol. 2008;68:342–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ledesma‐Amaro R, Buey RM, Revuelta JL. Increased production of inosine and guanosine by means of metabolic engineering of the purine pathway in Ashbya gossypii . Microb Cell Fact. 2015;14:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nimmesgern E, Black J, Futer O, et al. Biochemical analysis of the modular enzyme inosine 5′‐monophosphate dehydrogenase. Protein Expr Purif. 1999;17:282–289. [DOI] [PubMed] [Google Scholar]
- 16. Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem. 1994;140:1–22. [DOI] [PubMed] [Google Scholar]
- 17. Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli . Nat Chem Biol. 2009;58(5):593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alexandre T, Rayna B, Munier‐Lehmann H. Two classes of bacterial IMPDHs according to their quaternary structures and catalytic properties. PLoS One. 2015;10:e0116578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Scott JW, Hawley SA, Green KA, et al. CBS domains form energy‐sensing modules whose binding of adenosine ligands is disrupted by disease mutations. J Clin Invest. 2004;113:274–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fernandez‐Justel D, Marcos‐Alcalde Í, Abascal F, et al. Diversity of mechanisms to control bacterial GTP homeostasis by the mutually exclusive binding of adenine and guanine nucleotides to IMP dehydrogenase. Protein Sci. 2022;31:e4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thomas EC, Gunter JH, Webster JA, et al. Different characteristics and nucleotide binding properties of inosine monophosphate dehydrogenase (IMPDH) isoforms. PLoS One. 2012;7:e51096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mortimer SE, Hedstrom L. Autosomal dominant retinitis pigmentosa mutations in inosine 5′‐monophosphate dehydrogenase type I disrupt nucleic acid binding. Biochem J. 2005;390:41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Duong‐Ly KC, Kuo YM, Johnson MC, et al. T cell activation triggers reversible inosine‐5′‐monophosphate dehydrogenase assembly. J Cell Sci. 2018;131:jcs223289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fernández‐Justel D, Peláez R, Revuelta JL, Buey RM. The Bateman domain of IMP dehydrogenase is a binding target for dinucleoside polyphosphates. J Biol Chem. 2019;294:14768–14775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Johnson MC, Kollman JM. Cryo‐EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation. Elife. 2020;9:e53243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Anthony SA, Burrell AL, Johnson MC, et al. Reconstituted IMPDH polymers accommodate both catalytically active and inactive conformations. Mol Biol Cell. 2017;28:2600–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Burrell AL, Nie C, Said M, et al. IMPDH1 retinal variants control filament architecture to tune allosteric regulation. Nat Struct Mol Biol. 2022;29:47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Plana‐Bonamaiso A, Lopez‐Begines S, Fernondez‐Justel D, et al. Post‐translational regulation of retinal IMPDH1 in vivo to adjust GTP synthesis to illumination conditions. Elife. 2020;9:e56418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fernandez‐Justel D, Nunez R, Martin‐Benito J, et al. A nucleotide‐dependent conformational switch controls the polymerization of human IMP dehydrogenases to modulate their catalytic activity. J Mol Biol. 2019;431:956–969. [DOI] [PubMed] [Google Scholar]
- 30. Koshland DE, Nemethy JG, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5:365–385. [DOI] [PubMed] [Google Scholar]
- 31. Ereno‐Orbea J, Oyenarte I, Martinez‐Cruz LA. CBS domains: Ligand binding sites and conformational variability. Arch Biochem Biophys. 2013;540:70–81. [DOI] [PubMed] [Google Scholar]
- 32. Baykov AA, Tuominen HK, Lahti R. The CBS domain: A protein module with an emerging prominent role in regulation. ACS Chem Biol. 2011;6:1156–1163. [DOI] [PubMed] [Google Scholar]
- 33. Pimkin M, Pimkina J, Markham GD. A regulatory role of the Bateman domain of IMP dehydrogenase in adenylate nucleotide biosynthesis. J Biol Chem. 2009;284:7960–7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McLennan AG, Zamecnik PC. Dinucleoside polyphosphates—An introduction. Ap4A other dinucleoside polyphosphates. Boca Raton, FL: CRC Press, 2020; p. 1–7. [Google Scholar]
- 35. McLennan AG. Dinucleoside polyphosphates—Friend or foe? Pharmacol Ther. 2000;87:73–89. [DOI] [PubMed] [Google Scholar]
- 36. Plateau P, Blanquet S. Dinucleoside oligophosphates in micro‐organisms. Adv Microb Physiol. 1994;36:81–109. [DOI] [PubMed] [Google Scholar]
- 37. Anderson BW, Fung DK, Wang JD. Regulatory themes and variations by the stress‐signaling nucleotide alarmones (p)ppGpp in bacteria. Annu Rev Genet. 2021;55:115–133. [DOI] [PubMed] [Google Scholar]
- 38. Steinchen W, Zegarra V, Bange G. (p)ppGpp: Magic modulators of bacterial physiology and metabolism. Front Microbiol. 2020;11:2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Burrell AL, Kollman JM. IMPDH dysregulation in disease: A mini review. Biochem Soc Trans. 2022;50:71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nass K, Redecke L, Perbandt M, et al. In cellulo crystallization of Trypanosoma brucei IMP dehydrogenase enables the identification of genuine co‐factors. Nat Commun. 2020;11:620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dobie F, Berg A, Boitz JM, Jardim A. Kinetic characterization of inosine monophosphate dehydrogenase of Leishmania donovani . Mol Biochem Parasitol. 2007;152:11–21. [DOI] [PubMed] [Google Scholar]
- 42. Chang CC, Lin WC, Pai LM, et al. Cytoophidium assembly reflects upregulation of IMPDH activity. J Cell Sci. 2015;128:3550–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wong‐Riley M. Energy metabolism of the visual system. Eye Brain. 2010;2:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Country MW. Retinal metabolism: A comparative look at energetics in the retina. Brain Res. 2017;1672:50–57. [DOI] [PubMed] [Google Scholar]
- 45. Lin H, Caroll KS. Introduction: Posttranslational protein modification. Chem Rev. 2018;118:887–888. [DOI] [PubMed] [Google Scholar]
- 46. Kramer RH, Molokanova E. Review modulation of cyclic‐nucleotide‐gated channels and regulation of vertebrate phototransduction cyclic‐nucleotide‐gated channels and phototransduction. J Exp Biol. 2001;204:2921–2931. [DOI] [PubMed] [Google Scholar]
- 47. Bowne SJ, Liu Q, Sullivan LS, et al. Why do mutations in the ubiquitously expressed housekeeping gene IMPDH1 cause retina‐specific photoreceptor degeneration? Investig Ophthalmol Vis Sci. 2006;47:3754–3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xu H, Zhou J, Lin S, Deng W, Zhang Y, Xue Y. PLMD: An updated data resource of protein lysine modifications. J Genet Genomics. 2017;44:243–250. [DOI] [PubMed] [Google Scholar]
- 49. Shi Y, Zhang Y, Lin S, et al. dbPSP 2.0, an updated database of protein phosphorylation sites in prokaryotes. Sci Data. 2020;7:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Castaño‐Cerezo S, Bernal V, Post H, et al. Protein acetylation affects acetate metabolism, motility and acid stress response in Escherichia coli . Mol Syst Biol. 2014;10:762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Weinert BT, Iesmantavicius V, Wagner SA, et al. Acetyl‐phosphate is a critical determinant of lysine acetylation in E. coli . Mol Cell. 2013;51:265–272. [DOI] [PubMed] [Google Scholar]
- 52. Kim D, Yu B, Kim J, et al. The acetylproteome of Gram‐positive model bacterium Bacillus subtilis . Proteomics. 2013;13:1726–1736. [DOI] [PubMed] [Google Scholar]
- 53. Carabetta VJ, Greco TM, Tanner AW, Cristea IM, Dubnau D. Temporal regulation of the Bacillus subtilis acetylome and evidence for a role of MreB acetylation in cell wall growth. mSystems. 2016;1:e00005‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yang Y, Zhang H, Guo Z, et al. Global insights into lysine Acylomes reveal crosstalk between lysine acetylation and succinylation in Streptomyces coelicolor metabolic pathways in brief. Mol Cell Proteomics. 2021;20:100148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mizuno Y, Nagano‐Shoji M, Kubo S, et al. Altered acetylation and succinylation profiles in Corynebacterium glutamicum in response to conditions inducing glutamate overproduction. Microbiology. 2016;5:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Meng Q, Liu P, Wang J, et al. Systematic analysis of the lysine acetylome of the pathogenic bacterium Spiroplasma eriocheiris reveals acetylated proteins related to metabolism and helical structure. J Proteomics. 2016;148:159–169. [DOI] [PubMed] [Google Scholar]
- 57. Liu L, Wang G, Song L, Lv B, Liang W. Acetylome analysis reveals the involvement of lysine acetylation in biosynthesis of antibiotics in Bacillus amyloliquefaciens . Sci Rep. 2016;6:20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schwer B, Eckersdorff M, Li Y, et al. Calorie restriction alters mitochondrial protein acetylation. Aging Cell. 2009;8:604–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Aughey GN, Liu JL. Metabolic regulation via enzyme filamentation. Crit Rev Biochem Mol Biol. 2016;51:282–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. O'Connell JD, Zhao A, Ellington AD, Marcotte EM. Dynamic reorganization of metabolic enzymes into intracellular bodies. Annu Rev Cell Dev Biol. 2012;28:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Liu J. The cytoophidium and its kind: Filamentation and compartmentation of metabolic enzymes. Annu Rev Cell Dev Biol. 2016;32:349–372. [DOI] [PubMed] [Google Scholar]
- 62. Chang C‐C, Keppeke GD, Sung L‐Y, Liu J‐L. Interfilament interaction between IMPDH and CTPS cytoophidia. FEBS J. 2018;285:3753–3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Keppeke GD, Chang CC, Peng M, et al. Liu J‐L (2018) IMP/GTP balance modulates cytoophidium assembly and IMPDH activity. Cell Div. 2018;131(13):1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Senda M, Natsumeda Y. Tissue‐differential expression of two distinct genes for human IMP dehydrogenase (E.C.1.1.1.205). Life Sci. 1994;54:1917–1926. [DOI] [PubMed] [Google Scholar]
- 65. Ji YS, Gu JJ, Makhov AM, Griffith JD, Mitchell BS. Regulation of the interaction of inosine monophosphate dehydrogenase with mycophenolic acid by GTP. J Biol Chem. 2006;281:206–212. [DOI] [PubMed] [Google Scholar]
- 66. Juda P, Šmigová J, Kováčik L, Bártová E, Raška I. Ultrastructure of cytoplasmic and nuclear Inosine‐5’‐monophosphate dehydrogenase 2 “rods and rings” inclusions. J Histochem Cytochem. 2014;62:739–750. [DOI] [PubMed] [Google Scholar]
- 67. Labesse G, Alexandre T, Gelin M, Haouz A, Munier‐Lehmann H. Crystallographic studies of two variants of Pseudomonas aeruginosa IMPDH with impaired allosteric regulation. Acta Crystallogr Sect D Biol Crystallogr. 2015;71:1890–1899. [DOI] [PubMed] [Google Scholar]
- 68. Hayward D, Kouznetsova VL, Pierson HE, et al. ANKRD9 is a metabolically‐controlled regulator of IMPDH2 abundance and macro‐assembly. J Biol Chem. 2019;294:14454–14466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Spellicy CJ, Daiger SP, Sullivan LS, et al. Characterization of retinal inosine monophosphate dehydrogenase 1 in several mammalian species. Mol Vis. 2007;13:1866–1872. [PubMed] [Google Scholar]
- 70. Andashti B, Yazdanparast R, Motahar M, Barzegari E, Galehdari H. Terminal peptide extensions augment the retinal IMPDH1 catalytic activity and attenuate the ATP‐induced fibrillation events. Cell Biochem Biophys. 2021;79:221–229. [DOI] [PubMed] [Google Scholar]
- 71. Gilbert HJ, Lowe CR, Drabble WT. Inosine 5′‐monophosphate dehydrogenase of Escherichia coli. Purification by affinity chromatography, subunit structure and inhibition by guanosine 5′‐monophosphate. Biochem J. 1979;183:481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Buey RM, Ledesma‐Amaro R, Balsera M, de Pereda JM, Revuelta JL. Increased riboflavin production by manipulation of inosine 5′‐monophosphate dehydrogenase in Ashbya gossypii. Appl Microbiol Biotechnol. 2015;99:9577–9589. [DOI] [PubMed] [Google Scholar]
- 73. Bowne SJ, Sullivan LS, Blanton SH, et al. Mutations in the inosine monophosphate dehydrogenase 1 gene (IMPDH1) cause the RP10 form of autosomal dominant retinitis pigmentosa. Hum Mol Genet. 2002;11:559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bowne SJ, Sullivan LS, Mortimer SE, et al. Spectrum and frequency of mutations in IMPDH1 associated with autosomal dominant retinitis pigmentosa and leber congenital amaurosis. Investig Ophthalmol Vis Sci. 2006;47:34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wada Y, Tada A, Itabashi T, Kawamura M, Sato H, Tamai M. Screening for mutations in the IMPDH1 gene in Japanese patients with autosomal dominant retinitis pigmentosa. Am J Ophthalmol. 2005;140:163–165. [DOI] [PubMed] [Google Scholar]
- 76. Wada Y, Sandberg MA, McGee TL, Stillberger MA, Berson EL, Dryja TP. Screen of the IMPDH1 gene among patients with dominant retinitis pigmentosa and clinical features associated with the most common mutation, Asp226Asn. Investig Ophthalmol Vis Sci. 2005;46:1735–1741. [DOI] [PubMed] [Google Scholar]
- 77. Grover S, Fishman GA, Stone EM. A novel IMPDH1 mutation (Arg231Pro) in a family with a severe form of autosomal dominant retinitis pigmentosa. Ophthalmology. 2004;111:1910–1916. [DOI] [PubMed] [Google Scholar]
- 78. Kennan A, Aherne A, Palfi A, et al. Identification of an IMPDH1 mutation in autosomal dominant retinitis pigmentosa (RP10) revealed following comparative microarray analysis of transcripts derived from retinas of wild‐type and Rho(−/−) mice. Hum Mol Genet. 2002;11:547–558. [DOI] [PubMed] [Google Scholar]
- 79. Zech M, Jech R, Boesch S, et al. Monogenic variants in dystonia: An exome‐wide sequencing study. Lancet Neurol. 2020;19:908–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kuukasjärvi A, Landoni JC, Kaukonen J, et al. IMPDH2: A new gene associated with dominant juvenile‐onset dystonia‐tremor disorder. Eur J Hum Genet. 2021;29:1833–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Aherne A, Kennan A, Kenna PF, et al. On the molecular pathology of neurodegeneration in IMPDH1‐based retinitis pigmentosa. Hum Mol Genet. 2004;13:641–650. [DOI] [PubMed] [Google Scholar]
- 82. Kennan A, Aherne A, Bowne SJ, et al. On the role of IMPDH1 in retinal degeneration. Adv Exp Med Biol. 2003;533:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Nussinov R, Tsai CJ. Allostery in disease and in drug discovery. Cell. 2013;153:293–305. [DOI] [PubMed] [Google Scholar]
- 84. Han B, Salituro FG, Blanco MJ. Impact of allosteric modulation in drug discovery:Innovation in emerging chemical modalities. ACS Med Chem Lett. 2020;11:1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wenthur CJ, Gentry PR, Mathews TP, Lindsley CW. Drugs for allosteric sites on receptors. Annu Rev Pharmacol Toxicol. 2014;54:165–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Juvale K, Shaik A, Kirubakaran S. Inhibitors of inosine 5′‐monophosphate dehydrogenase as emerging new generation antimicrobial agents. MedChemComm. 2019;10:1290–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Fotie J. Inosine 5’‐monophosphate dehydrogenase (IMPDH) as a potential target for the development of a new generation of Antiprotozoan agents. Mini Rev Med Chem. 2018;18:656–671. [DOI] [PubMed] [Google Scholar]
- 88. Alexandre T, Lupan A, Helynck O, et al. First‐in‐class allosteric inhibitors of bacterial IMPDHs. Eur J Med Chem. 2019;167:124–132. [DOI] [PubMed] [Google Scholar]
- 89. Liao LX, Song XM, Wang LC, et al. Highly selective inhibition of IMPDH2 provides the basis of antineuroinflammation therapy. Proc Natl Acad Sci U S A. 2017;114:E5986–E5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Sanglier JJ, Quesniaux V, Fehr T, et al. Sanglifehrins A, B, C and D, novel cyclophilin‐binding compounds isolated from Streptomyces sp. A92‐308110. I. Taxonomy, fermentation, isolation and biological activity. J Antibiot. 1999;52:466–473. [DOI] [PubMed] [Google Scholar]
- 91. Zenke G, Strittmatter U, Fuchs S, et al. Sanglifehrin A, a novel Cyclophilin‐binding compound showing immunosuppressive activity with a new mechanism of action. J Immunol. 2001;166:7165–7171. [DOI] [PubMed] [Google Scholar]
- 92. Pua KH, Stiles DT, Sowa ME, Verdine GL. IMPDH2 is an intracellular target of the cyclophilin A and sanglifehrin A complex. Cell Rep. 2017;18:432–442. [DOI] [PubMed] [Google Scholar]


