Abstract
The rhizosphere nitrogen-fixing bacterium Azospirillum irakense KBC1 is able to grow on pectin and β-glucosides such as cellobiose, arbutin, and salicin. Two adjacent genes, salA and salB, conferring β-glucosidase activity to Escherichia coli, have been identified in a cosmid library of A. irakense DNA. The SalA and SalB enzymes preferentially hydrolyzed aryl β-glucosides. A Δ(salA-salB) A. irakense mutant was not able to grow on salicin but could still utilize arbutin, cellobiose, and glucose for growth. This mutant could be complemented by either salA or salB, suggesting functional redundancy of these genes in salicin utilization. In contrast to this functional homology, the SalA and SalB proteins, members of family 3 of the glycosyl hydrolases, show a low degree of amino acid similarity. Unlike SalA, the SalB protein exhibits an atypical truncated C-terminal region. We propose that SalA and SalB are representatives of the AB and AB′ subfamilies, respectively, in glycosyl hydrolase family 3. This is the first genetic implication of this β-glucosidase family in the utilization of β-glucosides for microbial growth.
β-Glucosidases are a heterogeneous group of enzymes, present in eukaryotic and prokaryotic organisms, which catalyze the hydrolysis of cellobiose and chemically related β-glucosides of which the aglycone can be an aromatic compound, such as in arbutin and salicin. Because of the abundance of cellulose in plant cell walls, most microbial β-glucosidases have been investigated in soil and digestive microflora and have been described as the ultimate enzymatic step in the biological conversion of cellulose into glucose. These β-glucosidases were formerly called cellobiases. Nevertheless, some β-glucosidases preferentially hydrolyze aryl β-glucosides, such as arbutin and salicin.
β-Glucosidase activities have been also found in plant growth-promoting rhizobacteria such as Rhizobium (24), Azoarcus (31), and Azospirillum (3) spp., but their corresponding genes have not been characterized. We investigated the genetic determinants of these enzymes in bacteria of the genus Azospirillum which preferentially colonize the rhizosphere of graminae. In the genus Azospirillum, both A. lipoferum and A. brasilense are known for their morphogenic effects on plant roots and for beneficial effects on plant growth (28). The root-colonizing properties and ability to produce phytohormones such as indole-3-acetic acid have been investigated particularly in A. brasilense (5, 6, 7, 29, 30, 38–40). On the other hand, A. irakense, in contrast, is a low indole-3-acetic acid producer (46) and is unique among the Azospirillum species for its ability to grow on pectin (21).
We report on the characterization of two β-glucosidases in the type strain of A. irakense and the genetic analysis of the two corresponding genes. The two purified enzymes, SalA and SalB, show a higher specific affinity for aromatic β-glucosides, such as salicin and arbutin, than for cellobiose. Analysis of the deduced amino acid sequences of salA and salB allowed their unambiguous classification in family 3 of the glycosyl hydrolases. This is the first genetic implication of this β-glucosidase family in the utilization of β-glucosides as carbon sources.
MATERIALS AND METHODS
Strains, vectors, and growth conditions.
The Escherichia coli and A. irakense strains and plasmids used are listed in Table 1. E. coli strains were grown in Luria-Bertani medium (LB) (33) at 37°C. Azospirillum was grown in LB supplemented with 2.5 mM CaCl2 and 2.5 mM MgSO4 (LB*) at 30°C. For solid media, 15 g of agar liter−1 was added. Conjugations were performed on LB* plates, and the A. irakense transconjugants were selected on MMAB minimal medium (41) with 0.5% malate as the C source and 18 mM NH4Cl as the N source. Antibiotics were used at the following concentrations: ampicillin, 50 μg ml−1; kanamycin, 25 μg ml−1; and tetracycline, 10 μg ml−1. Growth rates in liquid minimal medium (MMAB) supplemented with 18 mM NH4Cl and with the appropriate C source (cellobiose, salicin, or arbutin) at 8 mM was measured by monitoring the optical density 595 nm. Malate (0.5%) was used as the C source in all Azospirillum precultures.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| A. irakense | ||
| KBC1 | Wild type, LMG 10653, isolated from rice rhizophere | 20 |
| FAJ0691 | KBC1 (salA salB)::kan Kmr | This work |
| E. coli DH5α | F80 dlacZΔM15 Δ(lacZYA-argF) recA endA hsdR supE | Gibco BRL |
| Plasmids | ||
| pUC19 | Cloning vector, Apr | 45 |
| pBluescript SK(−) | Cloning vector, Apr | Stratagene |
| pSUP202 | Mobilizable plasmid, suicide vector for A. irakense, Cmr Tcr Apr | 35 |
| pRK2013 | Tra+ helper plasmid | 11 |
| pLAFR1 | IncP1 broad-host-range plasmid, Tcr | 12 |
| pLAFR3 | pLAFR1 derivative containing the pUC8 multiple cloning site, Tcr | 36 |
| pHP45Ω-Km | Kmr cassette | 10 |
| pFAJ0650 | pLAFR1 with 24-kb EcoRI genomic fragment from A. irakense KBC1 | This work |
| pFAJ0651 | pLAFR1 with 15-kb EcoRI insert from pFAJ0650 | This work |
| pFAJ0652 | pUC19 with 6.3-kb KpnI-EcoRI insert from pFAJ0651, expressing salA | This work |
| pFAJ0654 | pUC19 with 4.5-kb SphI-EcoRI insert from pFAJ0652, expressing salA | This work |
| pFAJ0655 | pUC19 with 2.7-kb HindIII-EcoRI insert from pFAJ0651 | This work |
| pFAJ0660 | pLAFR3 with 5.5-kb BamHI-HindIII insert from pFAJ0651 | This work |
| pFAJ0662 | pUC19 with 6.5-kb HindIII-SalI insert from pFAJ0650, expressing salB | This work |
| pFAJ0663 | pUC19 with 1.9-kb PstI insert from pFAJ0650 | This work |
| pFAJ0665 | SK− with 2.7-kb HindIII-EcoRI from pFAJ0655 and 0.4-kb EcoRI-PstI insert from pFAJ0663, expressing salB | This work |
| pFAJ0666 | pFAJ0660 with 1.2-kb HindIII fragment containing the 3′ region of salA, expressing salA | This work |
| pFAJ0667 | pFAJ0660 with blunted 2.7-kb HindIII-BamHI insert from pFAJ0665, expressing salB | This work |
DNA techniques and sequence analysis.
Standard methods were used for plasmid isolation, chromosomal DNA preparation, transformation, Southern blotting, and hybridization (33). DNA fragments were recovered from agarose gels by using a Nucleotrap kit (Macherey-Nagel). For Southern hybridization, DNA was transferred to a Hybond-N membrane (Amersham), and DNA probe was labelled with digoxigenin-dUTP by using a random labelling kit (Boehringer Mannheim). The hybridization signal was detected with a chemiluminescence detection kit (Boehringer Mannheim). For sequence analysis, recombinant plasmids were purified with a Qiagen kit. DNA sequencing of pUC18/19 subclones, using the chain-terminating dideoxynucleoside triphosphate method (34), was carried out with an AutoRead sequencing kit (Pharmacia-LKB) on an automated sequencer (ALF; Pharmacia-LKB). DNA sequence analysis was done on both strands. Sequence data were processed and analyzed by using the PCGene software package (Intelligenetics). The prediction of the amino-terminal signal sequence was done with the SignalpWWW server (27). The classification of glycosyl hydrolases is available on the ExPASy server (17).
Enzyme purification and N-terminal amino acid sequence analysis.
Overnight cultures of E. coli clones harboring pFAJ0654 (salA gene) or pFAJ0662 (salB gene) were centrifuged and resuspended in 0.5× phosphate-buffered saline (50 mM sodium phosphate, 75 mM sodium chloride [pH 7.2]) supplemented with DNase I (Sigma) at 200 U/ml. The lysis was performed in FastRNA Tubes-Blues (Bio 101) in a FastPrep machine (Bio 101-Savant) for 30 s at speed 6. The lysate was cleared by two centrifugations (5 min at 13,000 rpm) and filtration through Millex 0.22-μm-pore-size filters. For preparation of SalB enzyme, the lysate was cleared by centrifugation for 2.5 h at 120,000 × g at 4°C and concentrated on Microcon10 concentrators (Amicon). The two enzymatic samples (500 μl of extract per run) were applied to a gel filtration column (Superdex 200 Prep Grade HR16/50; Pharmacia), which was eluted with 0.5× phosphate-buffered saline at a flow rate of 1 ml/min. Positive fractions were pooled and adjusted to pH 5.0 (SalA preparation) or pH 4.6 (SalB preparation) with glacial acetic acid and then filtered through Millex 0.22-μm-pore-size filters. Then cation-exchange chromatography was performed with gradients of buffers A (50 mM sodium acetate) and B (50 mM sodium acetate, 1 M sodium chloride) which were adjusted at pH 5 (SalA purification) or pH 4.6 (SalB purification). The SalA preparation was applied to cation-exchange column (S Sepharose High Load XK16/50; Pharmacia). The column was rinsed with 90% buffer A–10% buffer B and eluted with 75% buffer A–25% buffer B. In the case of SalB purification, the sample was loaded onto MonoS HR5/5 column (Pharmacia) and eluted with a gradient of 0 to 25% buffer B at 0.5 ml/min for 25 min. In both cases, the positive fractions were pooled and purity was estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). N-terminal amino acid sequence analysis of the purified SalA and SalB was accomplished by Edman degradation using an Applied Biosystems 477A protein sequencer (Applied Biosystems/Perkin Elmer, Foster City, Calif.).
Enzyme assays and zymograms.
To detect β-glucosidase or cellobiohydrolase activity in E. coli clones containing A. irakense DNA, cells were grown overnight on LB plates and then incubated with 4-methylumbelliferyl-β-d-glucoside (MUG; Sigma) or 4-methylumbelliferyl-β-d-cellobioside (MUC; Sigma) in an overlay (0.2 mg ml−1 in 0.7% agarose buffer–0.1 M K2HPO4-KH2PO4 [pH 7.0]) for 10 min to 1 h at 37°C. Colony fluorescence was detected with a transilluminator at 302 nm.
p-Nitrophenyl-β-d-glucoside (PNPG; Sigma), p-nitrophenyl-β-d-cellobioside (PNPC; Sigma), and p-nitrophenyl-β-d-xyloside (PNPX; Sigma) were also used as synthetic substrates. Ranges of pH from 5.0 to 8.0 and temperature from 30 to 50°C were first tested with PNPG. Then a sample of SalA was added to 1 ml of 50 mM morpholineethanesulfonic acid buffer (pH 6.5) and allowed to equilibrate to 45°C for 5 min. In the case of SalB, N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES) buffer was used at 50 mM, pH 7.0. The reaction was started by the addition of a preheated substrate solution to give a final concentration of 10 mM of PNPG, PNPX, or PNPC. Enzymatic reaction was terminated by the addition of 1 ml of 1 M Na2CO3, and the absorbance at 420 nm was immediately measured and compared against a p-nitrophenol standard curve. The hydrolysis of cellobiose, gentiobiose, or salicin was measured by glucose production in a total volume of 400 μl. The reaction was terminated by the addition of 400 μl of 0.5 M Na2CO3 and neutralized by 200 μl of 1 M HCl. Glucose assays were performed by monitoring the reduction of NADP+ to NADPH at 340 nm in the presence of hexokinase and glucose-6-phosphate dehydrogenase (G6PDH). The assays were carried out as follows. A 500-μl reaction mixture containing 100 mM TES buffer (pH 7.4), 8 mM MgCl2 · 6H2O, 1.5 mM ATP, 1 mM NADP+, and 10 U of a hexokinase-G6PDH coupled enzymatic preparation (Sigma) was added to 400 μl of each stopped and equilibrated sample. The absorbance values were compared to those obtained for a standard curve of glucose. The hydrolysis of arbutin was measured by the release of hydroquinone. The absorbance at 520 nm was measured in an alkaline solution after the addition of 400 μl of 0.5 Na2CO3 and compared against a hydroquinone standard curve.
For separation of the SalA and SalB enzymes by isoelectric focusing (IEF), PhastGel IEF in the range of pH 5 to 8 (Pharmacia) was used with a PhastSystem electrophoresis chamber (Pharmacia). Electrophoresis was carried out according to the manufacturer’s instructions. Agarose overlays contained 0.7% agarose in 0.1 M K2HPO4-KH2PO4 buffer (pH 7.0) and 0.4 mg of MUG per ml. After IEF migration, the IEF gel was covered by a cooled overlay and incubated at 37°C. MUGase activity was detected under UV light at 302 nm.
Nucleotide sequence accession number.
Sequence data have been submitted to the GenBank database under accession no. AF090429.
RESULTS
Cloning of two adjacent loci encoding β-glucosidase activity.
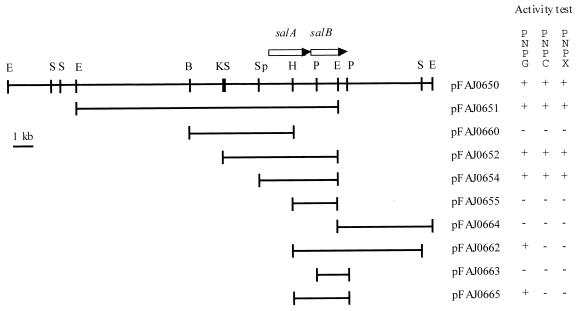
Approximately 3,000 E. coli clones from a genomic cosmid library of A. irakense KBC1 have been tested for β-glucosidase and cellobiohydrolase activities with MUG or MUC, using an overlay technique. Seven clones showed both MUG- and MUC-hydrolyzing activities, and similar EcoRI, BamHI, and HindIII restriction patterns of the corresponding recombinant cosmid DNAs were obtained (data not shown). One of these cosmids, pFAJ0650, was retained for further characterization. The restriction map of the pFAJ0650 cosmid containing a 24-kb insert is shown in Fig. 1. From this cosmid, two groups of MUG/PNPG-positive subclones have been obtained. The first group shared the 4.5-kb SphI-EcoRI fragment (pFAJ0651, pFAJ0652, and pFAJ0654); the second group shared the 3.1-kb HindIII-PstI (pFAJ0662 and pFAJ0665). The 2.7-kb HindIII-EcoRI overlapping fragment (pFAJ0655) from these two subcloning groups was MUG/PNPG negative, suggesting that two loci encoded β-glucosidase activity. Clones of the first group also exhibited PNPX activity. In the E. coli pUC19 subclones, transcription of the salA and salB genes is driven by the lacZ promoter of the pUC19 vector since β-glucosidase activity was observed for only one orientation of the insert DNA.
FIG. 1.
Restriction map of pFAJ0650 cosmid and its derivative subclones harboring the salA or salB gene. The expression of β-glucosidase and β-xylosidase activities in each of the E. coli clones harboring pFAJ0650 or its subclones was tested with the substrates PNPG and PNPX, respectively. B, BamHI; E, EcoRI; H, HindIII; K, KpnI; P, PstI; S, SalI; Sp, SphI.
Sequence analysis of the DNA region encoding β-glucosidase activity.
The region which contained the two putative β-glucosidase encoding loci was sequenced. The deduced amino acid sequence revealed two open reading frames, one from nucleotides 121 to 2319 and the second from nucleotides 2342 to 4291. These open reading frames, named salA and salB, encoded putative proteins of 732 and 649 amino acids, respectively. Sequence analysis did not reveal a Rho-independent transcriptional terminator between salA and salB or downstream of salB. Each putative translation start codon is preceded by a sequence which is similar to the ribosome-binding sequence of gram-negative bacteria.
The N-terminal regions of SalA and SalB displayed characteristics of signal peptides, suggesting transport through the plasma membrane. Cleavage sites predicted by the SignalpWWW server, and confirmed by N-terminal amino acid sequencing of the purified SalA and SalB proteins, occurred between amino acid residues 26 and 27 in SalA and between amino acid residues 21 and 22 in SalB protein. The N-terminal sequences of SalA and SalB are MKVHQLFKAALATSLCLTAFAGGAMA/QAKGAWQNTSL and MRRLPHLSLLALMLYSGTALA/APQQPALPEGQ, respectively (sequenced amino acids of the mature protein are in boldface; slash signs indicate the cleavage site).
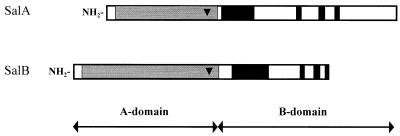
Comparison of the deduced amino acid sequences of SalA and SalB with other known β-glucosidases in data banks revealed that SalA and SalB are not similar to the β-glucosidases of glycosyl hydrolase families 1 and 4. However, a good match was found with β-glucosidases of family 3; 36% identity was observed between SalA and Cbg1 of Agrobacterium tumefaciens, and 52% identity was observed between SalB and BgxA of Erwinia chrysanthemi. Identity between SalA and SalB of A. irakense was only 12%. Consistent with these results, the structures of mature SalA and SalB exhibited all of the main traits of the family 3 β-glucosidases. Nevertheless, the SalB protein is, like BgxA of E. chrysanthemi, an atypical member of this family. As for most members of glycosyl hydrolase family 3, the mature SalA and SalB proteins contain an N-terminal catalytic domain (A domain) and a C-terminal domain (B domain) (Fig. 2). In the C-terminal region of the A domain, a putative catalytic aspartate residue is conserved in all proteins of this family (2), as well as in SalA (position 254) and SalB (position 334). However, the C-terminal region of the B domain of SalB is truncated and is very similar to that of BgxA of E. chrysanthemi. In this shorter B domain, the C-terminal region conserved among family 3 members is also present in SalB but appears more compressed than in the other enzymes (Fig. 2). Moreover, the 45 amino acids of the SalB C-terminal region are required for the expression of β-glucosidase activity in E. coli (E. coli with pFAJ0655 was negative for MUG and PNPG activity [Fig. 1]), suggesting a functional role of this conserved region in SalB.
FIG. 2.
Schematic representation of the AB and AB′ organization of SalA and SalB. The A domain of the glycosyl hydrolase family 3 (in grey) is the largest conserved region and contains the catalytic site (triangle). The B domain contains different highly conserved regions (black bars) which are present in SalA as well as in the atypical truncated B domain of SalB.
Purification and characteristics of the two β-glucosidases.
The two β-glucosidases SalA and SalB were purified from E. coli clones harboring either salA in pFAJ654 or salB in pFAJ662. The two β-glucosidases could be distinguished by molecular weight, pI, and substrate specificity. The apparent molecular weight of the purified SalA protein determined by SDS-PAGE was 78,500, consistent with the deduced value of 75,000. In contrast, the SalB protein exhibited a lower molecular weight 64,600, on SDS-PAGE and a deduced value of 66,600. As previously described, the N-terminal amino acid sequences of SalA and SalB were experimentally determined and compared with obtained DNA sequences. The pI values of SalA (6.0) and SalB (5.7) also exhibited an unambiguous difference which allowed their efficient separation by IEF. The substrate specificity of the two β-glucosidases was evaluated in the presence of synthetic (PNPG, PNPX, and PNPC) or natural (cellobiose, gentiobiose, arbutin, and salicin) substrates. The optimal conditions for PNPG hydrolysis by SalA and SalB extract were 45°C and pH 6.5 or 7.0, respectively. With both SalA and SalB, the Km measurements revealed their greater affinity for natural or synthetic aryl β-glucosides (PNPG, arbutin, and salicin) than for cellobiose or gentiobiose (Table 2). These enzymes are therefore not cellobiase or gentiobiase types of β-glucosidases. SalA also exhibited the capacity to cleave PNPX and PNPC.
TABLE 2.
Biochemical characteristics of SalA and SalB
| Substrate | SalA
|
SalB
|
||||
|---|---|---|---|---|---|---|
| Km | Sp acta | Relative activityb | Km | Sp act | Relative activity | |
| PNPG | 0.11 | 10.80 | 100 | 0.05 | 19.68 | 100 |
| PNPX | 0.40 | 6.16 | 57 | NDc | —d | ND |
| PNPC | NDc | 0.97 | 9 | ND | — | ND |
| Salicin | 0.40 | 10.82 | 100 | 0.06 | 17.10 | 86 |
| Arbutin | 0.25 | 11.28 | 104 | 0.06 | 26.13 | 132 |
| Cellobiose | 16.40 | 0.08 | 0.7 | 5.56 | 1.61 | 9 |
| Gentiobiose | 11.11 | 0.34 | 3 | 1.12 | 0.65 | 3 |
Micromoles of glucose, p-nitrophenol, or hydroquinone (see Materials and Methods for details of experiments) produced per minute per milligram of protein.
Percentage of specific activity in the presence of PNPG.
ND, not determined.
—, no activity detected.
Phenotype and complementation of a salA-salB deletion mutant.
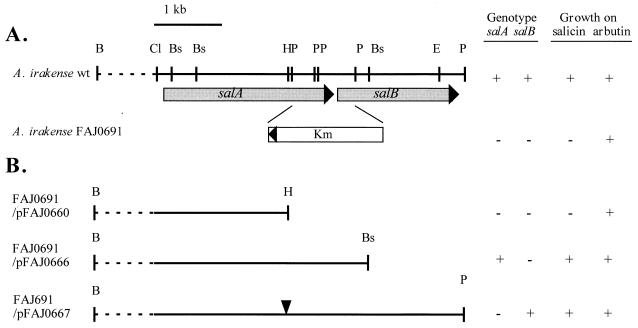
A kanamycin resistance (Kmr) cassette from pHP45Ω-Km was inserted into the blunted ends of pFAJ0654 digested with PstI (Fig. 3). The construction was subcloned into the suicide vector pSUP202 and then transferred into A. irakense KBC1 by a triparental mating. The Kmr transconjugants were purified, and insertion of the Kmr gene in the A. irakense chromosome as well as the absence of pSUP202 were verified by hybridization. The Δ(salA-salB) mutant (strain FAJ0691) was unable to grow on salicin after 24 h of culture at 30°C and showed a slightly delayed growth on arbutin. However, growth on cellobiose was similar to that of the wild-type strain. No effect of this deletion was observed for growth on malic acid, gentiobiose, and glucose (data not shown).
FIG. 3.
Phenotype and complementation of a Δ(salAsalB) A. irakense mutant. (A) Restriction map of the salA salB region of A. irakense and schematic representation of the salA salB deletion constructed by Kmr cassette exchange. (B) Restriction map of A. irakense DNA cloned in the three plasmids: pFAJ0660 (without salA or salB), pFAJ0666 (expressing salA), and pFAJ0667 (expressing salB). The inverted triangle in pFAJ0667 indicates the mutation in salA created by blunted ligation. The genotype and phenotype of each constructed A. irakense strain are also indicated. The dashed line represents the out-of-scale DNA. B, BamHI; Bs, BssHII; Cl, ClaI; E, EcoRI; H, HindIII; P, PstI.
We investigated the ability of each β-glucosidase to restore the Sal− phenotype by complementing FAJ0691 with either salA or salB. First, plasmid pFAJ660, which contains the presumed promoter region of the salA and salB genes (data not shown) and part of SalA, was introduced into FAJ0691. As expected, this plasmid could not restore growth on salicin (Fig. 3). Then plasmid pFAJ0666 was obtained by the addition of the complete 3′ end of salA in pFAJ0660 digested with HindIII. The E. coli clones harboring pFAJ0666 showed both PNPG and PNPX activities, suggesting that SalA was functional. The Δ(salA-salB) mutant which contained pFAJ0666 was able to grow on salicin as the sole carbon source. To test the role of salB, plasmid pFAJ0667 was obtained by the addition of the complete sequence of the salB gene in HindIII-digested and blunted pFAJ0660. By this blunted cloning, a nonsense mutation in salA was created, as indicated in Fig. 3. The presence of salB in pFAJ0667 was first verified by restriction mapping. Second, in situ detection of MUG activity after IEF revealed that the unique β-glucosidase activity of pFAJ0667 comigrated with that of pFAJ0665 (harboring salB) and not with that of pFAJ0654 and pFAJ0666 (harboring salA). Finally, because only SalA exhibited both PNPX and PNPG activities, the lack of PNPX activity in E. coli clones harboring pFAJ0667 was additional evidence that the SalA enzyme is not functional in this construct. Therefore, plasmid pFAJ0667 conferred only SalB β-glucosidase to the host cells. pFAJ0667 was introduced into FAJ0691, and growth on salicin was restored (Fig. 3), suggesting that either salA or salB is required and sufficient for salicin utilization by A. irakense.
DISCUSSION
The rhizosphere nitrogen-fixing bacterium A. irakense KBC1 exhibits pectinolytic activity (21) and is able to grow on several β-glucosides such as cellobiose (20) and, as described in this work, arbutin and salicin. We investigated in this rhizosphere bacterium the genes and encoded enzymes which allow the utilization of such plant-derived β-glucosides. From a cosmid library of A. irakense expressed in E. coli, we identified and characterized at genetic and biochemical levels two β-glucosidases, SalA and SalB, which are required for the growth of A. irakense on salicin.
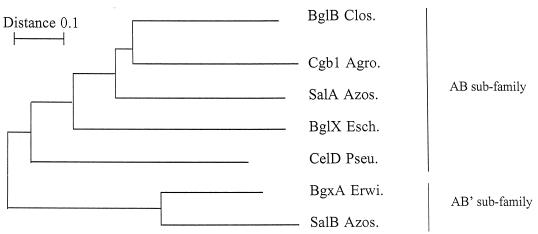
Based on their deduced amino acid sequences, these β-glucosidases were classified as members of family 3 of the glycosyl hydrolases (15–17). All bacterial β-glucosidases of family 3 are organized into two main domains, a catalytic A domain and a noncatalytic B domain (18). In contrast to the AB organization of SalA and most β-glucosidases of this family, the SalB structure is unusual (Fig. 2). SalB shows an atypical truncated B domain and a high degree of similarity with the β-glucosidase BgxA of E. chrysanthemi in which the truncated C-terminal B domain was renamed B′ (42). Because of the surprisingly high degree of similarity between SalB and BgxA, the hypothesis of gene transfer between these two gram-negative bacteria which exhibit the similar host range of monocotyledonous plants can be proposed. However, the GC contents of salB and bgxA, 65 and 57%, respectively, are quite similar to those of their corresponding host genomes, 66% in A. irakense and 57% in E. chrysanthemi. Because this is the first report of two different copies of family 3 β-glucosidases in the same bacterium, the phylogenetic relationship between some of the best-known bacterial β-glucosidases of this family was evaluated. By using the whole amino acid sequence of these enzymes, their most conserved A domain, or their A domain and the most conserved residues of the B domains, similar phylogenetic trees were obtained (Fig. 4). The SalB of A. irakense and the BgxA of E. chrysanthemi constitute an unambiguous AB′ cluster which is separated from SalA of A. irakense and all the other representative AB bacterial β-glucosidases of family 3. It is of interest that the percentage of similarity between SalB and BgxA of these taxonomically unrelated α and γ subclass proteobacteria is higher (52%) than the similarity between SalA and Cbg1 of the two α subclass proteobacteria A. irakense and A. tumefaciens (36%). This feature could suggest that the amino acid sequence of the AB′ group is highly conserved. In conclusion, analysis of their deduced amino acid sequences indicate that the adjacent salA and salB genes are two paralogous genes which encode AB and AB′ β-glucosidases of glycosyl hydrolase family 3.
FIG. 4.
Phylogenetic tree of several bacterial members of the glycosyl hydrolase family 3. The tree was constructed by the neighbor-joining method. SalB and BgxA constitute an unambigous AB′ cluster, while the AB subfamily seems to be a more heterogeneous group within the glycosyl hydrolase family 3. BglB Clos., BglB of Clostridium thermocellum (13); Cbg1 Agro., Cbg1 of A. tumefaciens (4); SalA Azos., SalA of A. irakense; BglX Esch., BglX of E. coli (44); CelD Pseu., CelD of Pseudomonas fluorescens subsp. cellulosa (32); BgxA Erwi., BgxA of E. chrysanthemi (42); SalB Azos., SalB of A. irakense.
In gram-positive and gram-negative bacteria, the β-glucosidases required for the utilization of β-glucosides belong to family 1 or 4 of the glycosyl hydrolases (17, 37). However, no role in β-glucoside assimilation has been assigned to the β-glucosidase BglX of E. coli K-12 (44), the β-glucosidase BgxA of E. chrysanthemi (42), and the other known bacterial β-glucosidases of family 3. The constructed Δ(salA-salB) A. irakense mutant was not able to grown on salicin but could still utilize arbutin, cellobiose, gentiobiose, and other carbon sources such as glucose or malate. This is the first genetic evidence that β-glucosidases of family 3 are involved in the assimilation of aryl β-glucosides. In A. irakense, the β-glucosidases of family 3 could be evolutionary alternatives to the β-glucosidases of families 1 and 4 in the assimilatory pathways of β-glucosides. The structure of the β-glucosidases implicated in the utilization of arbutin or cellobiose in A. irakense is under investigation.
The presence of signal sequences predicts a periplasmic location of SalA and SalB, as has been suggested for BglX of E. coli (44) and BgxA of E. chysanthemi (42). Indeed, supernatants of A. irakense or E. coli, containing the salA and salB genes of A. irakense, did not have PNPG- or MUG-hydrolyzing activity. This raises the question on how β-glucosides are transported through the outer membrane of gram-negative bacteria. Recently Andersen et al. (1) identified the bglH gene of the E. coli bgl operon, encoding a carbohydrate-specific outer membrane porin with high specificity for arbutin, salicin, gentiobiose, and cellobiose. Whether a similar situation exists for A. irakense remains to be determined.
The Sal− phenotype of the Δ(salA-salB) A. irakense mutant is in agreement with the greater affinities of the SalA and SalB enzymes for aromatic β-glucosides than cellobiose (Table 2). SalB has strict specificity for glucose in the aryl β-glucoside, and its active site appears to exclude substrates of a particular size, explaining the finding of no activity with PNPC but weak activity with cellobiose. SalA has a broader substrate spectrum, including xylose in the β linkage. For the best-known bacterial β-glucosidases of family 3, cellobiose is also a less efficient as a substrate than aromatic natural or synthetic β-glucosides or cellodextrins (13, 14, 23, 32, 43). However, because SalA and SalB exhibited similar Kms with arbutin or salicin, and because we observed a slightly delayed growth of the Δ(salA-salB) mutant on arbutin (data not shown), these two proteins could be also involved in arbutin utilization. Nevertheless, it is clear that A. irakense contains a second assimilatory pathway for arbutin. This arbutin pathway could to some extent be involved in the utilization of salicin, because of the slight growth of the Δ(salA-salB) mutant on salicin after 36 h (data not shown).
Since either salA or salB can complement the Δ(salA-salB) A. irakense mutant for growth on salicin (Fig. 3), these two AB and AB′ β-glucosidase encoding genes may be functionally redundant for the utilization of salicin. The salA and salB genes most likely are part of an operon because their coding regions are separated by only 22 nucleotides and because the presumed promoter which controls their expression in A. irakense is located at least 2 kb upstream of salA (data not shown). The latter can be deduced from the minimum length of DNA required 5′ upstream of the salA coding region for complementation of the Δ(salA-salB) A. irakense mutant (Fig. 3). Nevertheless, we cannot at this stage exclude the possibility that SalA and SalB have a specific function for the hydrolysis of still unknown plant-derived aromatic β-glucosides. The ability of SalA to hydrolyze a higher range of substrates than SalB may support this hypothesis. Plant-derived aryl β-glucosides or the aglycones have been found to have a role in signalling or saprophytic relationships between plants and bacteria. Arbutin and salicin, but not their aglycones, induce the production of a cyclic lipodepsinonapeptide toxin, called syringomycin, by phytopathogenic strains of Pseudomonas syringae (25). P. syringae cannot grow on arbutin and salicin as sole carbon sources (19). In the case of some virulent Agrobacterium strains, the aryl β-glucoside coniferin, purified from Pseudotsuga menziesii shoot extract, has been identified as the major inducer of a virE-lacZ fusion introduced in these Agrobacterium strains (26). However, most of these strains exhibit high β-glucosidase activity, and one of them, A. tumefaciens B3/73, produces a β-glucosidase which in vitro hydrolyzes coniferin to the common vir inducer coniferyl alcohol (4). In this case, the aglycone could be the true active compound. Finally, the glucose released after biological cleavage of aryl β-glucosides is a suitable carbon source for many bacteria, and this nutritional capacity could contribute to the competitiveness and survival of bacteria in the rhizosphere. This catabolic property has been well studied in the phytopathogen E. chrysanthemi 3665 (8, 9), in the enteric E. coli K-12 (see reference 22 for a review), and in other gram-negative and gram-positive bacteria (37).
ACKNOWLEDGMENTS
We thank J. M. Penninckx (Laboratoire d’Ecologie Microbienne, ULB, Brussels, Belgium) for helpful discussions and laboratory facilities for the preliminary experiments on the substrate specificity of SalA.
D.F. was recipient of a postdoctoral fellowship from the Katholieke Universiteit Leuven (1996 and 1997). We acknowledge financial support from the Fund for Scientific Research-Flanders, the Flemish government (GOA-Vanderleyden), and the Ministry of Agriculture.
REFERENCES
- 1.Andersen C, Rak B, Benz R. The gene bglH present in the bgl operon of Escherichia coli, responsible for uptake and fermentation of β-glucosides encodes for a carbohydrate-specific outer membrane porin. Mol Microbiol. 1999;31:499–510. doi: 10.1046/j.1365-2958.1999.01191.x. [DOI] [PubMed] [Google Scholar]
- 2.Bause E, Legler G. Isolation and structure of a tryptic glycopeptide from the active site of β-glucosidase A3 from Aspergillus wentii. Biochim Biophys Acta. 1980;626:459–465. doi: 10.1016/0005-2795(80)90142-7. [DOI] [PubMed] [Google Scholar]
- 3.Bekri A. Genetic analysis of pectinolytic and cellulolytic activities of Azospirillum irakense KBC1. Ph.D. thesis. Leuven, Belgium: K.U. Leuven; 1998. [Google Scholar]
- 4.Castle L A, Smith K D, Morris R O. Cloning and sequencing of an Agrobacterium tumefaciens β-glucosidase gene involved in modifying a vir-inducing plant signal molecule. J Bacteriol. 1992;174:1478–1486. doi: 10.1128/jb.174.5.1478-1486.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costacurta A, Keijers V, Vanderleyden J. Molecular cloning and sequence analysis of an Azospirillum brasilense indole-3-pyruvate decarboxylase. Mol Gen Genet. 1994;243:463–472. doi: 10.1007/BF00280477. [DOI] [PubMed] [Google Scholar]
- 6.Croes C, Van Bastelaere E, Declercq E, Eyers M, Vanderleyden J, Michiels C. Identification and mapping of loci involved in motility, adsorption to wheat roots, colony morphology and growth on minimal medium on the Azospirillum brasilense Sp7 90 Mda plasmid. Plasmid. 1991;26:83–93. doi: 10.1016/0147-619x(91)90048-2. [DOI] [PubMed] [Google Scholar]
- 7.Croes C, Moens S, Van Bastelaere E, Vanderleyden J, Michiels C. The polar flagellum mediates Azospirillum brasilense adsorption to wheat roots. J Gen Microbiol. 1993;139:2261–2269. [Google Scholar]
- 8.El Hassouni M, Chippaux M, Barras F. Analysis of the Erwinia chrysanthemi genes, which mediate metabolism of aromatic β-glucosides. J Bacteriol. 1990;172:6261–6267. doi: 10.1128/jb.172.11.6261-6267.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El Hassouni M, Henrissat B, Chippaux M, Barras F. Nucleotide sequence of the arb genes, which control β-glucoside utilization in Erwinia chrysanthemi: comparison with the Escherichia coli sal operon and evidence for a new β-glycohydrolase family including enzymes from eubacteria, archaebacteria, and humans. J Bacteriol. 1992;174:765–777. doi: 10.1128/jb.174.3.765-777.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fellay R, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designated for in vitro inertional mutagenesis of gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 11.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2, dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedman A M, Long S R, Brown S E, Buikema W J, Ausubel F M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982;18:289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- 13.Gräbnitz F, Staudenbauer W. Characterization of two β-glucosidase genes from Clostridium thermocellum. Biotechnol Lett. 1988;10:72–78. [Google Scholar]
- 14.Gräbnitz F, Rücknagel K P, Seib M, Staudenbauer W. Nucleotide sequence of the Clostridium thermocellum bglB gene encoding thermostable β-glucosidase B: homology to fungal β-glucosidases. Mol Gen Genet. 1989;217:70–76. doi: 10.1007/BF00330944. [DOI] [PubMed] [Google Scholar]
- 15.Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henrissat B, Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henrissat B, Bairoch A. Updating the sequence-based classification of glycosyl hydrolases. Biochem J. 1996;316:695–696. doi: 10.1042/bj3160695. http://expasy.hcuge.ch/cgi-bin/lists?glycosid.txt . (The program described can be accessed online at http://expasy.hcuge.ch/cgi-bin/lists?glycosid.txt.) .) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hrmova M, Fincher G B. Barley β-d-glucan hydrolases. Substrate specificity and kinetic properties. Carbohydr Res. 1997;305:209–221. [Google Scholar]
- 19.Joubert J J, Hildebrand D C, Schroth M N. Nonutilization of β-glucosides for growth by fluorescent pseudomonads. Phytopathology. 1970;60:502–505. doi: 10.1094/phyto-60-502. [DOI] [PubMed] [Google Scholar]
- 20.Khammas K M, Ageron E, Grimont P A D, Kaiser P. Azospirillum irakense sp. nov., a nitrogen-fixing bacterium associated with rice roots and rhizosphere soil. Res Microbiol. 1989;140:679–693. doi: 10.1016/0923-2508(89)90199-x. [DOI] [PubMed] [Google Scholar]
- 21.Khammas K M, Kaiser P. Characterization of a pectinolytic activity in Azospirillum irakense. Plant Soil. 1991;137:75–79. [Google Scholar]
- 22.Lin E C C. Dissimilatory pathways for sugars, polyols, and carboxylates. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 307–342. [Google Scholar]
- 23.Lin L L, Rumbak E, Zappe H, Thomson J A, Woods D R. Cloning, sequencing and analysis of expression of a Butyrivibrio fibrisolvens gene encoding a β-glucosidase. J Gen Microbiol. 1990;136:1567–1576. doi: 10.1099/00221287-136-8-1567. [DOI] [PubMed] [Google Scholar]
- 24.Mateos P F, Jiminez-Zurdo J I, Chen J, Squartini A S, Haak S K, Martinez-Molina E, Hubbell D H, Dazzo F B. Cell-associated pectinolytic and cellololytic enzymes in Rhizobium leguminosarum biovar trifolii. Appl Environ Microbiol. 1992;58:1816–1822. doi: 10.1128/aem.58.6.1816-1822.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mo Y-Y, Gross D C. Plant signal molecules activate the syrB gene, which is required for syringomycin production by Peudomonas syringae pv. syringae. J Bacteriol. 1991;173:5784–5792. doi: 10.1128/jb.173.18.5784-5792.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morris J W, Morris R O. Identification of an Agrobacterium tumefaciens virulence gene inducer from the pinaceous gymnosperm Pseudotsuga menziesii. Proc Natl Acad Sci USA. 1990;87:3614–3618. doi: 10.1073/pnas.87.9.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. http://www.cbs.dtu.dk/services/SignalP/ . (The program described can be accessed online at http://www.cbs.dtu.dk/services/SignalP/.) .) [DOI] [PubMed] [Google Scholar]
- 28.Okon Y, Vanderleyden J. Root-associated Azospirillum species can stimilate plants. ASM News. 1997;63:366–370. [Google Scholar]
- 29.Pereg-Gerk L, Paquelin A, Gounon P, Kennedy I R, Elmerich C. A transcriptional regulator of the LuxR-UhpA family, FlcA, controls flocculation and wheat root surface colonization by Azospirillum brasilense Sp7. Mol Plant-Microbe Interact. 1998;11:177–187. doi: 10.1094/MPMI.1998.11.3.177. [DOI] [PubMed] [Google Scholar]
- 30.Prinsen E, Costacurta A, Michiels K, Vanderleyden J, van Onckelen H. Azospirillum brasilense indole-3-acetic acid biosynthesis: evidence for a non-tryptophan dependent pathway. Mol Plant-Microbe Interact. 1993;6:609–615. [Google Scholar]
- 31.Reinhold-Hurek B, Hurek T, Clayssens M, van Montagu M. Cloning, expression in Escherichia coli, and characterization of cellulolytic enzymes of Azoarcus sp., a root-invading diazotroph. J Bacteriol. 1993;175:7056–7065. doi: 10.1128/jb.175.21.7056-7065.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rixon J E, Ferreira L M A, Durrant A J, Laurie J I, Hazlewood G P, Gilbert H J. Characterization of the gene celD and its encoded product 1,4-β-d-glucan hydrolase D from Pseudomonas fluorescens subsp. cellulosa. Biochem J. 1992;285:947–955. doi: 10.1042/bj2850947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 34.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain termination inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simon R, Priefer U, Pühler A. A broad host range mobilisation system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 36.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stülke J, Arnaud M, Rapoport G, Martin-Verstraete I. PRD-a protein domain involved in PTS-dependent induction and carbon catabolite repression of catabolic operons in bacteria. Mol Microbiol. 1998;28:865–874. doi: 10.1046/j.1365-2958.1998.00839.x. [DOI] [PubMed] [Google Scholar]
- 38.Vande Broek A, Lambrecht M, Eggermont K, Vanderleyden J. Auxins upregulate expression of the indole-3-pyruvate decarboxylase gene in Azospirillum brasilense. J Bacteriol. 1999;181:1338–1342. doi: 10.1128/jb.181.4.1338-1342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vande Broek A, Vanderleyden J. Review: genetics of the Azospirillum-plant root association. Crit Rev Plant Sci. 1995;14:445–466. [Google Scholar]
- 40.Vande Broek A, Michiels J, Van Gool A, Vanderleyden J. Spatial-temporal colonization patterns of Azospirillum brasilense on the wheat root surface and expression of the bacterial nifH gene during association. Mol Plant-Microb Interact. 1993;6:592–600. [Google Scholar]
- 41.Vanstockem M, Michiels K, Vanderleyden J, Van Gool A P. Transposon mutagenesis of Azospirillum brasilense and Azospirillum lipoferum: physical analysis of Tn5 and Tn5-Mob insertion mutants. Appl Environ Microbiol. 1987;53:410–415. doi: 10.1128/aem.53.2.410-415.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vroemen S, Heldens J, Boyd C, Henrissat B, Keen N T. Cloning and characterization of the bgxA gene from Erwinia chrysanthemi D1 which encodes a β-glucosidase/xylosidase enzyme. Mol Gen Genet. 1995;246:465–477. doi: 10.1007/BF00290450. [DOI] [PubMed] [Google Scholar]
- 43.Wulff-Strobel C R, Wilson D B. Cloning, sequencing, and characterization of a membrane-associated Prevotella ruminicola B14 β-glucosidase with cellodextrinase and cyanoglycosidase activities. J Bacteriol. 1995;177:5884–5890. doi: 10.1128/jb.177.20.5884-5890.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang M, Luoh S M, Goddard A, Reilly D, Henzel W, Bass S. The bglX gene located at 47.8 min on the Escherichia coli chromosome encodes a periplasmic beta-glucosidase. Microbiology. 1996;142:1659–1665. doi: 10.1099/13500872-142-7-1659. [DOI] [PubMed] [Google Scholar]
- 45.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 46.Zimmer W, Aparicio C, Elmerich C. Relationship between tryptophan biosynthesis and indole-3-acetic acid production in Azospirillum: identification and sequencing of a trpGDC cluster. Mol Gen Genet. 1991;229:41–51. doi: 10.1007/BF00264211. [DOI] [PubMed] [Google Scholar]