Abstract
The Hha protein belongs to a new family of regulators involved in the environmental regulation of virulence factors. The aim of this work was to study the effect of the hha mutation on the overall protein pattern of Escherichia coli cells by two-dimensional polyacrylamide gel electrophoresis. The growth medium osmolarity clearly influenced the effect of the hha mutation. The number of proteins whose expression was altered in hha cells, compared with wild-type cells, was three times larger at a high osmolarity than at a low osmolarity. Among the proteins whose expression was modified by the hha allele, both OmpA and protein IIAGlc of the phosphotransferase system could be identified. As this latter enzyme participates in the regulation of the synthesis of cyclic AMP and hence influences the catabolite repression system, we tested whether the expression of the lacZ gene was also modified in hha mutants. This was the case, suggesting that at least some of the pleiotropic effects of the hha mutation could be caused by its effect on the catabolite repression system.
The chromosomal hha gene of Escherichia coli was identified in a search for mutants of E. coli 5K that overproduced the toxin alpha-hemolysin from a plasmid-encoded hemolytic operon (11). The product of the hha gene is the 8.6-kDa Hha protein (25), which is highly homologous (82%) to the Yersinia enterocolitica YmoA protein (10). The YmoA protein is a temperature-dependent modulator of the expression of some virulence factors in this species (8). Hha participates in the osmolarity-dependent expression of hemolysin (7, 22). Both the ymoA mutation (8) and the hha mutation (11, 22) are pleiotropic. Effects on environmentally dependent modulation of gene expression and pleiotropy are properties of a well-characterized class of mutants, the hns mutants (4, 12, 14, 17; see reference 1 for a review). Because the product of the hns gene is the nucleoid-associated protein H-NS, it was suggested that Hha and YmoA are also nucleoid-associated proteins. With respect to Hha, several lines of experimental evidence support this hypothesis: hha mutants show alterations in the topology of a reporter plasmid (7), overexpression of Hha increases the frequency of transposition of insertion elements in the E. coli chromosome (2, 19), and Hha is a DNA-binding protein (17a).
The aim of this work was to better characterize the pleiotropic effect of the hha mutation with respect to its osmolarity-modulating role. Two-dimensional (2-D) polyacrylamide gel electrophoresis (PAGE) analysis of total cellular extracts obtained from wild-type E. coli 5K and from its hha derivative, strain Hha-3, after growth in media of low or high osmolarity was done. 2-D PAGE protein maps of E. coli K-12 (34) were used to identify some of the proteins whose expression was found to be altered by the mutation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
E. coli 5K, Hha-3 (hha::Tn5) (11), CCB21, and CCB21H (hha::Tn5) (22) have been previously described. Plasmid pCB26R contains a translational hlyC::lacZ fusion that generates an in-frame gene fusion that is transcribed under the control of the hly promoter and gives rise to a hybrid HlyC-LacZ protein. Its construction has already been described (21). Synthetic medium Rich-MOPS (24) was used for the growth of strains. Glucose (0.4%) was included as a carbon source in all the experiments in which Rich-MOPS was used. NaCl was added to the medium for high-osmolarity experiments. Cultures were incubated at 37°C.
Protein labelling.
When the optical density at 600 nm (OD600) of the cultures reached 0.5, a 1-ml sample was removed and pulse-chased for 15 min with [35S]methionine. The procedure was used for both one-dimensional sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and two-dimensional (2-D) PAGE analyses of whole-cell extracts.
Gel electrophoresis and Western blotting.
SDS-PAGE (10% polyacrylamide) was done as described by Laemmli (16). For Western blot analysis, polyclonal rabbit serum raised against OmpA (13) was used at a dilution of 1:5,000 from a stock containing 75 mg ml−1.
2-D SDS-PAGE.
Preparation of cell extracts for 2-D PAGE and the 2-D PAGE itself were performed by the method of O’Farrell (26) with modifications (34). We used an Investigator 2-D Electrophoresis System (Millipore Corporation); all reagents were used as described in the product manual. Carrier ampholytes for the first dimension were in the range of pH 3 to 10. An SDS–12.5% polyacrylamide gel was used for the second dimension. The amounts of extracts loaded into the gels were adjusted after measurement of the radioactivity incorporated by each sample. Comparative studies of protein spot intensities were done by analysis of the gels with a Milligen Bioimage 2-D analysis package.
Measurement of β-galactosidase.
Strain 5K containing plasmid pCB26R was cultured in 0.4 M NaCl–Rich-MOPS and in 0 M NaCl–Rich-MOPS. Strains CCB21 and CCB21H were grown in low-glucose-containing Luria broth (LB) (tryptone yeast broth; Svenska LabFab) either without NaCl or with 0.5 M NaCl and supplemented with 0.2% lactose. When cultures reached an OD600 of 0.5, samples were taken and β-galactosidase activities in permeabilized cells were assayed as described by Miller (20). When needed, cyclic AMP (cAMP) was added at a final concentration of 3 mM.
SDS-PAGE analysis of whole membrane proteins.
Purification of the whole membrane fraction was done as follows. Cultures (400 ml) of strains 5K and Hha-3 grown in LB either without NaCl or with 0.5 M NaCl were harvested at an OD600 of 0.5. Cells were pelleted and resuspended in 10 ml of buffer A (10 mM Tris-HCl, 5 mM MgCl2 [pH 7.6]). This step was done twice. Cells were disrupted by use of a French press (SLM Instruments, Inc.) at 900 lb/in2. This step was also done twice. Cell extracts were then centrifuged at 16,000 rpm for 2 min to eliminate unbroken cells, and the supernatant was centrifuged at 22,300 rpm for 2 h (Beckman L8-M ultracentrifuge, rotor type 70.1 Ti). The pellet was finally resuspended in 2 ml of buffer A. The protein concentration was measured, 5 μg of each sample was mixed with standard protein sample buffer (containing SDS and β-mercaptoethanol), and the mixture was heated at 100°C for 5 min and loaded onto an SDS–10% polyacrylamide gel. This procedure allows the isolation of proteins located in both membranes.
Transport assay.
The transport assay was basically carried out as described by Buhr et al. (6), with slight modifications. Cells were grown by diluting an overnight culture 200-fold in 100 ml of Rich-MOPS. When the culture had reached an OD600 of 0.5, cells from 20 ml of culture were washed in ice-cold Rich-MOPS without glucose and resuspended in 1 ml of the same medium. The cell suspension was preincubated for 10 min at room temperature with aeration. [U-14C]glucose was then added to a final concentration of 400 μM (diluted to 1,000 dpm/nmol; Amersham). Aliquots (100 μl) were removed after 10, 20, 30, 50, 100, and 300 s, quenched by dilution in 8 ml of ice-cold Rich-MOPS complemented with 400 μM unlabelled glucose, and immediately applied to glass fiber filters (GF/F; Whatman) under suction. The filters were washed with 20 ml of ice-cold 0.9% NaCl, and counts were determined with 5 ml of Optiphase (Wallac). The glucose uptake rate was calculated as nanomoles minute−1 milligram−1 (dry weight) from the linear part of the uptake curve.
RESULTS AND DISCUSSION
Growth kinetics analysis.
Our previous work on the influence of growth medium osmolarity on the expression of alpha-hemolysin in strains 5K and Hha-3 was carried out by growing cells in LB without NaCl (low-osmolarity medium) and in LB containing 0.5 M NaCl (high-osmolarity medium) (7, 22). No differences in growth rates were detected between these two strains for a given medium (unpublished data). Pulse-labelling experiments required a defined synthetic medium (Rich-MOPS); therefore, additional growth kinetics comparisons of the two strains were necessary. Standard Rich-MOPS either without NaCl or with 0.1 to 0.5 M NaCl was used to study the growth kinetics. Differences in the growth rates of the two strains were detected only when the cells were cultured at 0.5 M NaCl (data not shown). In accordance with these results, Rich-MOPS with 0.4 M NaCl was used in experiments requiring high-osmolarity conditions, while Rich-MOPS without NaCl was used in experiments requiring low-osmolarity conditions.
Previous work with strain 5K cultured in LB without NaCl and in LB containing 0.5 M NaCl was carried out by us to analyze the influence of the osmolarity of the medium on hemolysin expression. Low osmolarity increased hemolysin expression (21). In that study, transcription of the hemolysin operon was monitored by measuring β-galactosidase activity in cells harboring plasmid pCB26R, which contains an in-frame gene fusion between the hlyC gene of the hemolysin operon and the lacZ gene. In order to confirm that growth in Rich-MOPS was equivalent to growth in LB with respect to hemolysin expression, we measured the β-galactosidase activities of strain 5K carrying plasmid pCB26R in 0.4 M NaCl–Rich-MOPS and in Rich-MOPS without added NaCl (0 M NaCl). The activity at low osmolarity was about twice the activity at high osmolarity (1,592 versus 799 Miller units). These results agreed with those obtained when LB was used (21) and indicated that Rich-MOPS could be used to monitor the effect of different concentrations of NaCl on the pattern of proteins expressed by the strains that we studied.
Electrophoretic analysis.
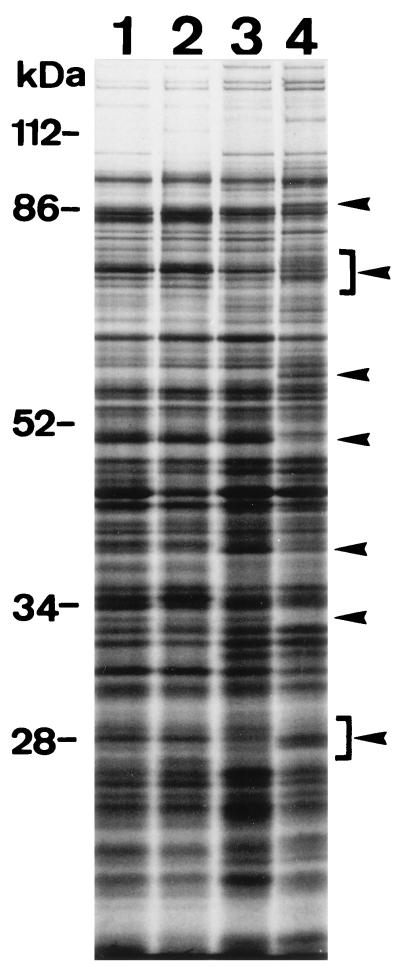
As hha mutants exhibit a pleiotropic phenotype, we tried to observe changes in the overall pattern of protein synthesis due to hha mutations. An initial comparison between the parental and mutant strains was carried out with cultures grown at different osmolarities. Figure 1 shows the autoradiogram obtained from an SDS-PAGE analysis of total cellular proteins after radioactive labelling. Only minor differences between the parental strain 5K and the hha mutant were observed when the cells were grown at low osmolarity. In contrast, several distinct differences were observed when the cells were cultured at high osmolarity.
FIG. 1.
Autoradiogram from SDS–10% PAGE analysis. Total cellular protein extracts from strains 5K (lanes 1 and 3) and Hha-3 (lanes 2 and 4) after growth at low (lanes 1 and 2) and high (lanes 3 and 4) osmolarities were analyzed (see text for details). Arrowheads indicate major differences.
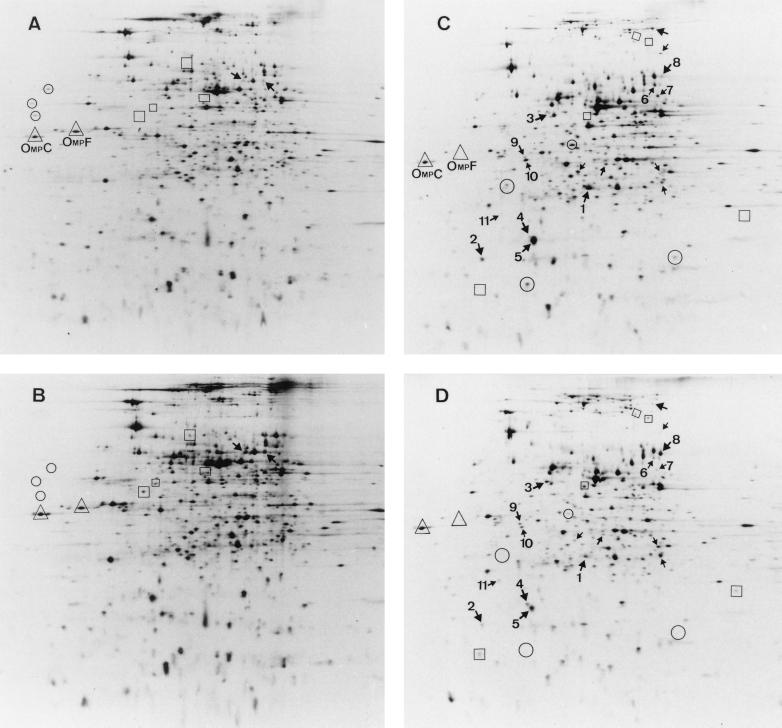
To better characterize such differences in the protein synthesis pattern, we analyzed total cellular extracts by 2-D PAGE and searched for proteins whose expression increased or decreased in strain Hha-3 with respect to the parental strain. Figure 2 shows the results obtained for both strains cultured either at low or at high osmolarity. Although the amount of protein loaded onto the gels was adjusted by the radioactivity incorporated into each sample, the autoradiograms in Fig. 2 are not completely equivalent, and slight background differences are apparent. For this reason and for a given pair of gels, we decided to highlight on Fig. 2 only those spots that were clearly different in both strains. As shown in Fig. 2A and B (low-osmolarity medium), three proteins were present in the wild-type strain, four were present in the mutant, and two were present in both but showed higher expression in the mutant strain. In Fig. 2C and D (high-osmolarity medium), 4 proteins were present in the wild-type strain, 5 were present in the mutant, and 17 were present in both, 11 with higher expression in the wild-type strain and 6 with higher expression in the mutant strain. The lack of the Hha protein not only increased the expression of some genes, as it does with the alpha-hemolysin determinant (11), but also decreased the expression of others.
FIG. 2.
Autoradiograms from 2-D PAGE separation of proteins in total cellular extracts from E. coli 5K and Hha-3 after growth in media of different osmolarities. In each panel, the gel is oriented with the more acidic proteins on the left. (A) Wild-type strain 5K at 0 M NaCl. (B) Mutant strain Hha-3 at 0 M NaCl. (C) Strain 5K at 0.4 M NaCl. (D) Strain Hha-3 at 0.4 M NaCl. For a given osmolarity, proteins in circles are those detected in strain 5K but not in strain Hha-3, while proteins in boxes are those detected in strain Hha-3 but not in strain 5K. Arrows indicate proteins present in both strains but at different intensities. Proteins in triangles are two osmoregulated porins (see text for details).
As the absence of Hha clearly seemed more significant when the cells were cultured at high osmolarity, we further analyzed some of the differences observed in the high-osmolarity gels. Table 1 shows the densitometric values calculated for 11 of the highlighted spots in Fig. 2C and D. By using the gene-protein database for E. coli (34), we were able to locate the alpha-numeric coordinates of five of the affected proteins and to identify the genes for two of them, ompA and crr, which encode proteins OmpA and enzyme IIAGlc of the phosphotransferase system, respectively. A third gene was identified from an Internet database (23a) as ahpC, which encodes the alkyl hydroperoxide reductase that is a member of the superoxide stress regulon.
TABLE 1.
Spot intensity comparison between the protein patterns of strains 5K (wild type) and Hha-3 (hha mutant) from Fig. 2C and D
| Spot | Alpha-numeric coordinatesa | Densitometric valueb for:
|
Genea | Location (min) in E. coli mapc | |
|---|---|---|---|---|---|
| Wild type | hha mutant | ||||
| 1 | F028.0 | 2.19 | 0.90 | ompA | 22.0 |
| 2 | B018.7 | 1.07 | 0.21 | crr | 54.6 |
| 3 | NDd | 0.17 | 0.91 | —e | — |
| 4 | ND | 0.19 | 0.69 | — | — |
| 5 | B020.9 | 5.00 | 2.06 | ahpC | 13.8 |
| 6 | ND | 0.91 | 0.37 | — | — |
| 7 | G057.0 | 0.18 | 0.63 | NIf | — |
| 8 | ND | 0.50 | 1.90 | — | — |
| 9 | ND | 0.19 | 0.42 | — | — |
| 10 | ND | 0.97 | 0.19 | — | — |
| 11 | B024.2 | 0.02 | 0.41 | NId | — |
Comparison of the spots detected by 2-D PAGE analysis with the reference map from Van Bogelen et al. (34) gives only an indication of what certain proteins are; thus, they have to be identified by other means. For that reason and given the relevant role of proteins OmpA and enzyme IIAGlc, we further investigated if the expression of both proteins is indeed affected by hha mutations.
Effect of an hha mutation on the expression of OmpA.
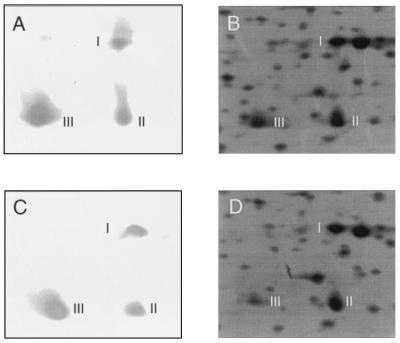
OmpA is one of the most abundant proteins of the E. coli envelope. It maintains the integrity of the E. coli outer membrane (33), it functions as a mediator in F-dependent conjugation (32), it stimulates a strong antibody response (29), and it may play an important role in virulence—an OmpA-deficient mutant of E. coli K-1 showed reduced virulence in a rat model of bacteremia (18). One of the four OmpA isoforms that can be detected by 2-D analysis (34) corresponds to one of the proteins identified as having altered expression in strain Hha-3 grown at high osmolarity. It corresponds to the alpha-numeric coordinates F028.0. Quantification of its expression showed that it decreased by 59% in strain Hha-3 in comparison with parental strain 5K. In order to confirm that the spot was indeed OmpA, we used antibodies against OmpA. Again, 2-D PAGE analysis of the extracts was carried out, proteins being transferred to membranes and immunoblotted. As shown in Fig. 3, the antibodies recognized three of the four OmpA isoforms and did not recognize other proteins in the 2-D gels. The fact that one of the three spots recognized corresponds to a spot highlighted in Fig. 2 confirms it as being OmpA. It is important to note that, when one is looking at the immunodetected spots, differences in intensity seem to be smaller than those detected by autoradiograms, but one must consider that, with the conditions used, immunodetection is qualitative rather than quantitative.
FIG. 3.
Immunodetection of OmpA among the spots obtained by 2-D PAGE. (A and C) Immunodetection of the OmpA isoforms (designated I, II, and III) in cell extracts obtained, respectively, from wild-type and hha strains grown at high osmolarity. (B and D) Magnification of the area from the original 2-D gels used to detect OmpA.
OmpA is an exported protein, like other proteins whose final destination is located outside the cytoplasm. Most of the exported E. coli proteins share a general secretory machinery for leaving the cytoplasm and, for that reason, the effect of the hha mutation on OmpA expression could be the consequence of a more generalized influence on the mechanisms of protein export to the cell envelope. This seemed not to be the case, because two osmoregulated porins, OmpC and OmpF (9), that were identified in Fig. 2 were not affected by the hha mutation.
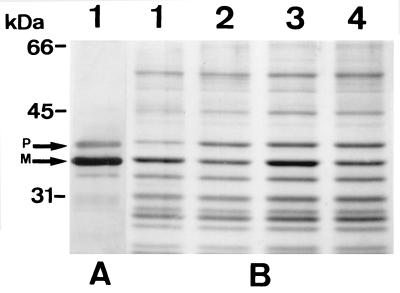
In order to confirm that the effect of the hha mutation was specific for OmpA, envelope proteins from both wild-type and hha mutant strains were analyzed by SDS-PAGE. Immunodetection of OmpA was also carried out to allow for OmpA identification. As shown in Fig. 4, only OmpA was clearly affected by the mutation. At high osmolarity, slight intensity differences were also apparent for a band that was located just above OmpA and that was also recognized by the anti-OmpA antibodies (Fig. 4). As the antibodies previously had been proven to be very specific for detecting OmpA in 2-D PAGE (Fig. 3) and because of the sizes of the immunodetected bands (35.2 and 37.2 kDa), we reasoned that they could be the mature and the precursor forms of OmpA. According to these results, it seems that the effect of the hha allele is specific for OmpA expression and is not a more general effect on a wider range of envelope proteins, at least not those detected by SDS-PAGE. Whether or not this effect on OmpA expression is important for the structural and/or physiological characteristics of the hha mutants is beyond the scope of this work.
FIG. 4.
Analysis of cell envelope proteins in wild-type and hha mutant E. coli strains grown at low and high osmolarities. (A) Western blot analysis with OmpA-specific antibodies of the envelope proteins of strain 5K cultured in standard LB. (B) Coomassie blue-stained gel from SDS-PAGE analysis of envelope proteins of strains 5K (lanes 1 and 3) and Hha-3 (lanes 2 and 4) grown in LB with 0.5 M NaCl (lanes 1 and 2) and in LB without NaCl (lanes 3 and 4). P and M, precursor and mature forms, respectively, of OmpA.
Physiological effects of enzyme IIAGlc levels altered by an hha mutation.
The second identified protein corresponds to the alpha-numeric coordinates B018.7. This protein is enzyme IIAGlc of the phosphoenolpyruvate:glucose phosphotransferase system (PTS) and is encoded by the crr gene. Its expression was decreased by 81% in strain Hha-3 in comparison with the parental strain. This enzyme plays a crucial role in the E. coli carbohydrate phosphotransferase system. It is one of the enzymes involved in the pathway for the uptake of glucose, it regulates the activity of the adenylate cyclase, and it is also responsible for the inducer exclusion of non-PTS carbohydrates (28). It is well known that the phosphorylation state of enzyme IIAGlc regulates the cAMP biosynthetic enzyme, adenylate cyclase (31). cAMP can bind to the cAMP receptor protein (CRP) to form the cAMP-CRP complex, which is a regulatory factor for the transcription of several hundred genes (5, 15). Taking into consideration the important role of the enzyme IIAGlc in E. coli metabolism, we decided to use a physiological approach to confirm that the identified protein was enzyme IIAGlc. We studied the effect of the hha mutation on the expression of a gene that is catabolite repression sensitive, the lacZ gene. We also examined functional implications with respect to effects on inducer exclusion and on glucose uptake.
(i) Effect on cAMP-dependent gene expression.
Variations in the expression of crr influence the activity of adenylate cyclase and therefore modify the level of cAMP (27). This effect could be monitored by measuring the expression of genes regulated by cAMP-CRP. With this aim, we decided to analyze if the expression of the lac operon was affected by the hha mutation. In this experiment, we used the isogenic E. coli Lac+ strains CCB21 and CCB21H (its hha derivative). The expression of the lacZ gene was determined by measuring the β-galactosidase activities of cultures growing in lactose-containing LB either without NaCl or with 0.5 M NaCl. Samples were taken when the cultures reached an OD600 of 0.5. The results obtained and expressed as Miller units (20) for cultures grown at low and high osmolarities were, respectively, as follows: 324 (strain CCB21) and 381 (strain CCB21H) and 564 (strain CCB21) and 329 (strain CCB21H). Differences between the strains were important only under high-osmolarity conditions. Under such conditions, the β-galactosidase activity of the hha mutant strain was only 58% the activity of the parental strain. This result suggests lower levels of cAMP in strain CCB21H caused by the lower expression of enzyme IIAGlc.
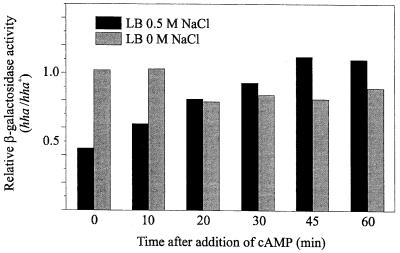
To support this notion, we tested if the addition of cAMP to the culture medium could restore β-galactosidase activity in the hha mutant. With this aim, both strains were grown in the same two culture media already described. At an OD600 of 0.5, samples were taken for β-galactosidase measurements (time zero), and cAMP was added to a final concentration of 3 mM. Thereafter, the β-galactosidase activities of both strains were monitored for 1 h. The results obtained are shown in Fig. 5. cAMP addition allowed the hha mutant strain growing at high osmolarity to gradually increase its β-galactosidase activity to a level similar to that of the parental strain. At low osmolarity, and as expected from the results indicated above, the enzyme activities were already similar in both strains before cAMP was added, so no restoration of β-galactosidase activity was expected.
FIG. 5.
Effect of the addition of cAMP on the expression of β-galactosidase from strains CCB21 and CCB21H. Cells were grown in LB medium with 0.5 M NaCl and in LB medium without NaCl. When the cultures reached an OD600 of 0.5, samples were taken for β-galactosidase measurements (time zero), and cAMP was immediately added to the cultures. Additional samples were then taken for β-galactosidase measurements at the indicated times. Each bar represents the relative activity determined as the ratio of the β-galactosidase activities (Miller units) of the hha (CCB21H) and hha+ (CCB21) strains.
We must point out that the slight decrease in the relative activities that can be observed after 20 min was not due to a decrease in the enzyme activity of the mutant strain but was due to a slight increase in the enzyme activity of the parental strain. This effect was presumably caused by the cAMP addition, although we have no explanation for it. In any case, the results obtained are consistent with the suggestion that the difference detected in the levels of expression of lacZ was due to a difference in the levels of cAMP, which should occur as a consequence of lower activation of adenylate cyclase by enzyme IIAGlc.
(ii) Effect on inducer exclusion.
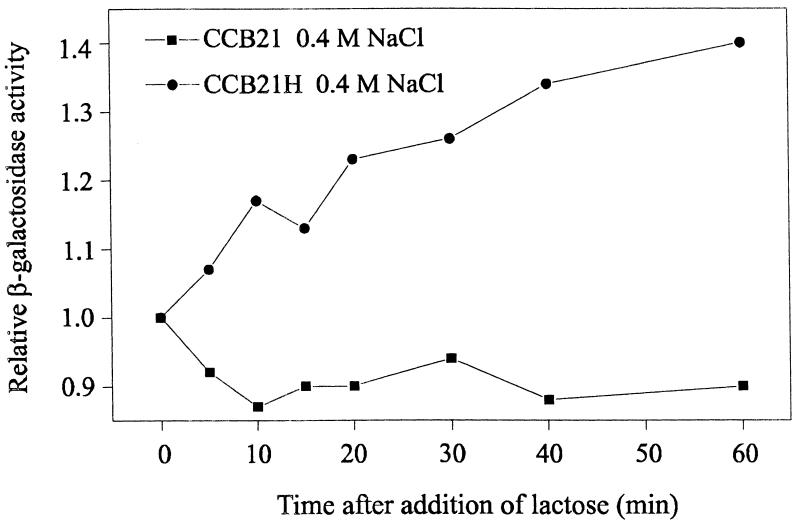
It has been reported that decreasing the amount of enzyme IIAGlc causes cells to escape from the inducer exclusion process (27). An experiment designed to monitor effects at the level of inducer exclusion was performed with strains CCB21 and CCB21H growing in Rich-MOPS containing glucose, a carbohydrate causing this physiological effect. After the wild-type and hha mutant strains were grown at low and high osmolarities to an OD600 of 0.5, lactose was added to a final concentration of 0.2%. Under such conditions, determination of the β-galactosidase activity after the addition of lactose is an indirect quantification of the uptake of lactose and therefore is a measure of the inducer exclusion level. As no differences between the strains were detected under low-osmolarity conditions, Fig. 6 shows only the results obtained at high osmolarity. The addition of lactose caused the β-galactosidase activity of strain CCB21H to increase, while no such effect was detected in strain CCB21. These results clearly indicate that the inducer exclusion process was affected by the hha mutation, as had been anticipated.
FIG. 6.
Effect of the addition of lactose on the β-galactosidase activities of E. coli CCB21 and CCB21H grown in Rich-MOPS containing 0.4 M NaCl. When the cultures reached an OD600 of 0.5 (time zero), lactose was added to a final concentration of 0.2%. Samples were withdrawn at the indicated times. Relative β-galactosidase activity is the ratio of the β-galactosidase activity (Miller units) measured at the indicated times to the β-galactosidase activity (Miller units) measured just before the addition of lactose.
(iii) Effect on the uptake of glucose.
We also examined whether low levels of enzyme IIAGlc might alter the rate of uptake of glucose. The rate of transport of glucose in cultures of strains 5K and Hha-3 growing at low and high osmolarities in Rich-MOPS containing glucose was therefore determined. The results for cultures grown at low and high osmolarities were, respectively, as follows: 2.21 (strain 5K) and 1.20 (strain Hha-3) and 1.10 (strain 5K) and 0.35 (strain Hha-3) nmol of glucose min−1 mg−1. These data show that alterations in osmolarity affect the rates of glucose uptake in both strains. Mutant strain Hha-3 showed lower values than parental strain 5K at both osmolarities, but the largest reduction in the rate of glucose uptake was detected under high-osmolarity conditions, in agreement with the suggestion that the lower enzyme IIAGlc level under such conditions should play a role.
Taken together, all of these results clearly demonstrate that the catabolite repression regulatory system is affected by the hha mutation in the same way as that expected for a decrease in the expression of enzyme IIAGlc. Interestingly, Ryu and Garges (30) found that in vitro transcription from the P1a promoter of the pst operon, which includes the crr gene, is dependent upon a supercoiled DNA template. As the hha mutation has been shown to cause a decrease in the degree of supercoiling of reporter plasmids (7), it is tempting to hypothesize that it is through changes in DNA supercoiling that the hha mutation causes a decrease in the expression of enzyme IIAGlc. Alterations in the expression of protein IIAGlc could, in turn, influence the expression of other genes or operons which belong to the catabolite repression multigene system and which were not considered in this work.
By using as a model the expression of the toxin alpha-hemolysin, we have been able to show that the Hha protein is an osmolarity- and temperature-dependent modulator of hemolysin synthesis (22). In addition, we have shown that DNA supercoiling influences hemolysin expression and that Hha may influence hemolysin expression by modifying the level of DNA supercoiling (7). Here we extend our observations about the modulatory role of Hha and describe new phenotypes for hha mutants. The results provided evidence that many hha-mediated changes in protein expression are dependent on the osmolarity of the growth medium. It is clear that Hha is a global modulator and that osmolarity, probably among other environmental factors, dictates the range of proteins whose expression is affected by Hha. The relationship among osmolarity, Hha, and the proteins OmpA and enzyme IIAGlc remains to be elucidated. However, it is apparent that for E. coli cells, adaptation to high osmolarity and maintenance of adequate levels of certain proteins, such as OmpA and enzyme IIAGlc, require the participation of global modulators, such as Hha. Other well-characterized factors, such as the sigma factor ςs (23) or the nucleoid-associated protein H-NS (see reference 1 for a review), have been described as playing an important role in the adaptation of bacterial cells to the osmolarity of the medium. Hha probably belongs to a cascade of modulators whose combined effects allow E. coli cells to fine-tune the control of the expression of multigene systems in response to environmental stimuli.
ACKNOWLEDGMENTS
This work was supported by the CICYT (grant PB94-0899) and in part by grants from the Swedish Medical Research Council, the Swedish Natural Science Research Council, and the Göran Gustafsson Foundation for Research in Natural Sciences and Medicine. C. Balsalobre was the recipient of an F. I. Fellowship from the Generalitat de Catalunya.
We thank F. C. Neidhardt and the people in his laboratory for providing their 2-D PAGE protocols and Ulf Henning and York Stierhof for the generous gift of antibodies against OmpA.
REFERENCES
- 1.Atlung T, Ingmer H. H-NS: a modulator of environmentally regulated gene expression. Mol Microbiol. 1997;24:7–17. doi: 10.1046/j.1365-2958.1997.3151679.x. [DOI] [PubMed] [Google Scholar]
- 2.Balsalobre C, Juárez A, Madrid C, Mouriño M, Prenafeta A, Muñoa F J. Complementation of the hha mutation in Escherichia coli by the ymoA gene from Yersinia enterocolitica: dependence on the gene dosage. Microbiology. 1996;142:1841–1846. doi: 10.1099/13500872-142-7-1841. [DOI] [PubMed] [Google Scholar]
- 3.Berlyn M K B, Low K B, Rudd K E. Linkage map of Escherichia coli K-12, edition 9. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1715–1902. [Google Scholar]
- 4.Bertin P, Lejeune P, Laurent-Winter C, Danchin A. Mutations in bglY, the structural gene for the DNA-binding protein H1, affect expression of several Escherichia coli genes. Biochimie. 1990;72:889–891. doi: 10.1016/0300-9084(90)90008-5. [DOI] [PubMed] [Google Scholar]
- 5.Botsford J L, Harman J G. Cyclic AMP in prokaryotes. Microbiol Rev. 1992;56:100–122. doi: 10.1128/mr.56.1.100-122.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buhr A, Daniels G A, Erni B. The glucose transporter of Escherichia coli. J Biol Chem. 1992;267:3847–3851. [PubMed] [Google Scholar]
- 7.Carmona M, Balsalobre C, Muñoa F, Mouriño M, Jubete Y, De la Cruz F, Juárez A. Escherichia coli hha mutants, DNA supercoiling and expression of the haemolysin genes from the recombinant plasmid pANN202-312. Mol Microbiol. 1993;9:1011–1018. doi: 10.1111/j.1365-2958.1993.tb01230.x. [DOI] [PubMed] [Google Scholar]
- 8.Cornelis G R, Sluiters C, Delor I, Gelb D, Kaninga K, Lambert de Rouvroit C, Sory M P, Vanooteghem J C, Michaelis T. ymoA, a Yersinia enterocolitica chromosomal gene modulating the expression of virulence functions. Mol Microbiol. 1991;5:1023–1034. doi: 10.1111/j.1365-2958.1991.tb01875.x. [DOI] [PubMed] [Google Scholar]
- 9.Csonka L N. Prokaryotic osmoregulation: genetics and physiology. Annu Rev Microbiol. 1991;45:569–606. doi: 10.1146/annurev.mi.45.100191.003033. [DOI] [PubMed] [Google Scholar]
- 10.De la Cruz F, Carmona M, Juárez A. The Hha protein from Escherichia coli is highly homologous to the YmoA protein from Yersinia enterocolitica. Mol Microbiol. 1992;6:3451–3452. doi: 10.1111/j.1365-2958.1992.tb02214.x. [DOI] [PubMed] [Google Scholar]
- 11.Godessart N, Muñoa F J, Regué M, Juárez A. Chromosomal mutations that increase the production of a plasmid-encoded haemolysin in Escherichia coli. J Gen Microbiol. 1988;134:2779–2787. doi: 10.1099/00221287-134-10-2779. [DOI] [PubMed] [Google Scholar]
- 12.Göransson M, Sondén B, Nilsson P, Dagberg B, Forsman K, Emanuelsson K, Uhlin B E. Transcriptional silencing and thermoregulation of gene expression in Escherichia coli. Nature. 1990;344:682–685. doi: 10.1038/344682a0. [DOI] [PubMed] [Google Scholar]
- 13.Henning U, Schwarz H, Chen R. Radioimmunological screening method for specific membrane proteins. Anal Biochem. 1979;97:153–157. doi: 10.1016/0003-2697(79)90339-7. [DOI] [PubMed] [Google Scholar]
- 14.Hinton J D C, Santos D S, Seirafi A, Hulton C S J, Pavitt G D, Higgins C F. Expression and mutational analysis of the nucleoid-associated protein H-NS of Salmonella typhimurium. Mol Microbiol. 1992;6:2327–2337. doi: 10.1111/j.1365-2958.1992.tb01408.x. [DOI] [PubMed] [Google Scholar]
- 15.Kolb A, Busby S, Buc H, Garges S, Adhya S. Transcriptional regulation by cAMP and its receptor protein. Annu Rev Biochem. 1993;62:749–795. doi: 10.1146/annurev.bi.62.070193.003533. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Laurent-Winter C, Ngo S, Danchin A, Bertin P. Role of the Escherichia coli histone-like nucleoid-structuring protein in bacterial metabolism and stress response. Identification of targets by two-dimensional electrophoresis. Eur J Biochem. 1997;244:767–773. doi: 10.1111/j.1432-1033.1997.00767.x. [DOI] [PubMed] [Google Scholar]
- 17a.Madrid, C., et al. Unpublished data.
- 18.Mahasreshti P J, Murphy G L, Wyckoff III J H, Farmer S, Hancock R E W, Confer A W. Purification and partial characterization of the OmpA family of proteins of Pasteurella haemolytica. Infect Immun. 1997;65:211–218. doi: 10.1128/iai.65.1.211-218.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mikulskis A V, Cornelis G R. A new class of proteins regulating gene expression in enterobacteria. Mol Microbiol. 1994;11:77–86. doi: 10.1111/j.1365-2958.1994.tb00291.x. [DOI] [PubMed] [Google Scholar]
- 20.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 21.Mouriño M, Muñoa F J, Balsalobre C, Díaz P, Madrid C, Juárez A. Environmental regulation of α-haemolysin expression in Escherichia coli. Microb Pathog. 1994;16:249–259. doi: 10.1006/mpat.1994.1026. [DOI] [PubMed] [Google Scholar]
- 22.Mouriño M, Madrid C, Balsalobre C, Prenafeta A, Muñoa F J, Blanco J, Blanco M, Blanco J E, Juárez A. The Hha protein as a modulator of expression of virulence factors in Escherichia coli. Infect Immun. 1996;64:2881–2884. doi: 10.1128/iai.64.7.2881-2884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muffler A, Traulsen D D, Lange R, Hengge-Aronis R. Posttranscriptional osmotic regulation of the ςs subunit of RNA polymerase in Escherichia coli. J Bacteriol. 1996;178:1607–1613. doi: 10.1128/jb.178.6.1607-1613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23a.NCBI website. Sequences. [Online.] http://ECO2DBASE. [July 1997, last date accessed.]
- 24.Neidhardt F C, Bloch P L, Pedersen S, Reeh S. Chemical measurement of steady-state levels of ten aminoacyl-transfer ribonucleic acid synthetases in Escherichia coli. J Bacteriol. 1977;129:378–387. doi: 10.1128/jb.129.1.378-387.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nieto J M, Carmona M, Bolland S, Jubete Y, De la Cruz F, Juárez A. The hha gene modulates haemolysin expression in Escherichia coli. Mol Microbiol. 1991;5:1285–1293. doi: 10.1111/j.1365-2958.1991.tb01902.x. [DOI] [PubMed] [Google Scholar]
- 26.O’Farrell P H. High resolution two dimensional electrophoresis of proteins. J Biol Chem Res. 1975;7:40–46. [PMC free article] [PubMed] [Google Scholar]
- 27.Postma P W, Lengeler J W, Jacobson G R. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Postma P W, Lengeler J W, Jacobson G R. Phosphoenolpyruvate:carbohydrate phosphotransferase systems. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1149–1174. [Google Scholar]
- 29.Puchiniemi R, Karvonen M, Vuopio-Varkila J, Muotiala A, Helander I M, Sarvas M. A strong antibody response to the periplasmic C-terminal domain of the OmpA protein of Escherichia coli is produced by immunization with purified OmpA or with whole E. coli or Salmonella typhimurium bacteria [sic] Infect Immun. 1990;58:1691–1696. doi: 10.1128/iai.58.6.1691-1696.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryu S, Garges S. Promoter switch in the Escherichia coli pts operon. J Biol Chem. 1994;269:4767–4772. [PubMed] [Google Scholar]
- 31.Saier M H, Jr, Ramseier T M, Reizer J. Regulation of carbon utilization. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1325–1343. [Google Scholar]
- 32.Schweizer M, Henning U. Action of major outer cell envelope membrane protein in conjugation of Escherichia coli K-12. J Bacteriol. 1977;129:1651–1652. doi: 10.1128/jb.129.3.1651-1652.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sonntag I, Schwartz H, Hirota Y, Henning U. Cell envelope and shape of Escherichia coli: multiple mutants missing the outer membrane lipoprotein and other major outer membrane proteins. J Bacteriol. 1978;136:280–285. doi: 10.1128/jb.136.1.280-285.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Bogelen R A, Sankar P, Clark R L, Bogan J A, Neidhardt F C. The gene-protein database of Escherichia coli: edition 5. Electrophoresis. 1992;13:1014–1054. doi: 10.1002/elps.11501301203. [DOI] [PubMed] [Google Scholar]