Abstract
Vici syndrome is a rare, congenital disorder that affects multiple systems and is caused by mutations in the EPG5 gene that encodes for ectopic P-granules autophagy protein 5 (EPG5). The induced pluripotent stem cell (iPSC) line described here was generated from a dermal fibroblast cell line from an 8-year-old male donor with a homozygous recessive c.1007A>G (p.Q336R) mutation in the EPG5 gene. This iPSC model of Vici syndrome provides a unique and valuable resource for investigators to study the pathology of EPG5 mutations and the aetiology of the disease as well as develop therapeutic treatments for those with Vici syndrome.
1. Resource table
| Unique stem cell line identifier | CIMRi001-A |
| Alternative name(s) of stem cell line | GM27291 |
| Institution | Coriell Institute for Medical Research |
| Contact information of distributor | Dr. Matthew W. Mitchell, mmitchell@coriell.org |
| Type of cell line | iPSC |
| Origin | Human |
| Additional origin info | Age: 8 years Sex: Male Ethnicity: Ashkenazi Jewish |
| Cell source | Dermal fibroblast |
| Clonality | Clonal |
| Method of reprogramming | Integration-free Sendai viral vectors containing OCT4, SOX2, KLF4, and c-MYC |
| Genetic modification | Yes |
| Type of modification | Hereditary |
| Associated disease | Vici syndrome |
| Gene/locus | EPG5, c.1007A>G (p.Q336R) |
| Method of modification | N/A |
| Name of transgene or resistance | N/A |
| Inducible/constitutive system | N/A |
| Date archived/stock date | 11/14/2018 |
| Cell line repository/bank | NIGMS Human Genetic Cell Repository (GM27291) |
| Ethical approval | NIGMS Human Genetic Cell Repository Informed Consent Form was obtained from donor at time of sample submission. NIH Confidentiality Certificate: CC-GM-15-004 |
2. Resource utility
The iPSC line CIMRi001-A is a human-derived disease resource for the study of EPG5 mutations and the Vici syndrome disease phenotype and can be used as a cellular model to develop therapeutic treatments for Vici syndrome patients.
3. Resource details
Vici syndrome is an extensive, multisystem human disorder caused by recessive mutations in the EPG5 gene on chromosome 18q12.3 (Cullup et al., 2013). EPG5 encodes for ectopic P-granules autophagy protein 5 (EPG5), a key protein involved in regulating autophagy (Byrne et al., 2016). Autophagy is an evolutionarily conserved cellular pathway that plays a vital role in the elimination of degraded proteins and organelles (Byrne et al., 2016). The five principal diagnostic findings of Vici syndrome (callosal agenesis, cataracts, cardiomyopathy, hypopigmentation and combined immunodeficiency), are often consistently associated with the three additional findings: profound developmental delay, acquired microcephaly and marked failure to thrive (Byrne et al., 2016; Vici et al., 1988). In this study, we generated a human dermal fibroblast cell line from an 8-year-old male donor (GM26636, Coriell Institute) clinically diagnosed with Vici syndrome with a recurrent, homozygous recessive c.1007A>G (p.Q336R) variant (Byrne et al., 2016), which affects the penultimate base of exon 2 in the EPG5 gene (Cullup et al., 2013).
This iPSC line, CIMRi001-A, was reprogrammed from GM26636 fibroblast cells using the CytoTune 2.0 Sendai Reprogramming kit (Thermo Fisher Scientific) containing the pluripotency reprogramming factors OCT4, SOX2, KLF4, and c-MYC. CIMRi001-A cells were analyzed to confirm successful reprogramming and viability (Table 1). The cells exhibited standard iPSC morphology when observed under a phase contrast microscope, showed 95% alkaline phosphatase activity, and expressed pluripotency markers (Fig. 1a). Post-thaw cell viability was measured by placing a frozen vial into culture, and iPSC colony area increased by 32-fold over a five-day observation period (Fig. 1a). Quantitative flow cytometry of the cells indicated a 93.7% expression rate of SSEA-4 (Fig. 1b) and a 84.4% expression rate of SSEA-3 (data not shown). Cytogenomic analysis (G-banded karyotype and Affymetrix Genome-Wide Human SNP Array 6.0 microarray analysis) showed a karyotype of 46,XY[25].arr[hg19]4q12(57,053,403–58,249,858)x3 (Fig. 1c). The 1.1 Mb 4q12 duplication is also present in the parental fibroblast line (GM26636) and is of uncertain clinical significance - this region includes at least 4 OMIM disease genes, but no known phenotype has been established in association with copy number gain of this region. The genetic variant in the EPG5 gene was verified in this iPSC line as well as the parental fibroblast line by whole exome sequencing (Fig. 1d). After passage 14, there was no detection of Sendai virus (SeV) genome or transgenes by qRT-PCR using SeV-specific primers (Table 2). CIMRi001-A cells demonstrated pluripotency via an embryoid body (EB) formation assay (Fig. 1e). This line was negative for mycoplasma contamination (Table 1). The STR profile of CIMRi001-A matched the profile of its parental fibroblast cell line (GM26636) at all six loci tested (Table 1).
Table 1.
Characterization and validation.
| Classification | Test | Result | Data |
|---|---|---|---|
| Morphology | Photography | Normal | Fig. 1 panel A |
| Phenotype | Qualitative analysis (i.e. Alkaline Phosphatase & Immunocytochemistry) | Assess staining/expression of pluripotency markers: SOX2, OCT4, SSEA4, & TRA-1-60 | Fig. 1 panel A |
| Quantitative analysis (i.e. Flow cytometry) | SSEA-4 (93.68%) | Fig. 1 panel B | |
| Genotype | Karyotype (G-banding and microarray) and resolution | 46, XY[25].arr[hg19]4q12(57,053,403-58,249,858)x3 Resolution 400–550 |
Fig. 1 panel C |
| Identity | Microsatellite PCR (mPCR) OR | Not performed | N/A |
| STR analysis | Six sites tested, all sites matched | Submitted in archive with journal | |
| Mutation analysis | Sequencing | Homozygous mutation EPG5, c.1007A>G (p. Q336R) | Fig. 1 panel D |
| Southern Blot OR WGS | N/A | N/A | |
| Microbiology and virology | Mycoplasma | Mycoplasma testing by qRT-PCR. Negative | Submitted in archive with journal |
| Differentiation potential | Embryoid body formation | Embryoid body (EB) with three germ layers formation (ectoderm, mesoderm, and endoderm) | Fig. 1 panel E |
| Donor screening (OPTIONAL) | HIV-1 | Negative | Not shown but available with author |
| Genotype additional info (OPTIONAL) | Blood group genotyping | N/A | N/A |
| HLA tissue typing | N/A | N/A |
Fig. 1.
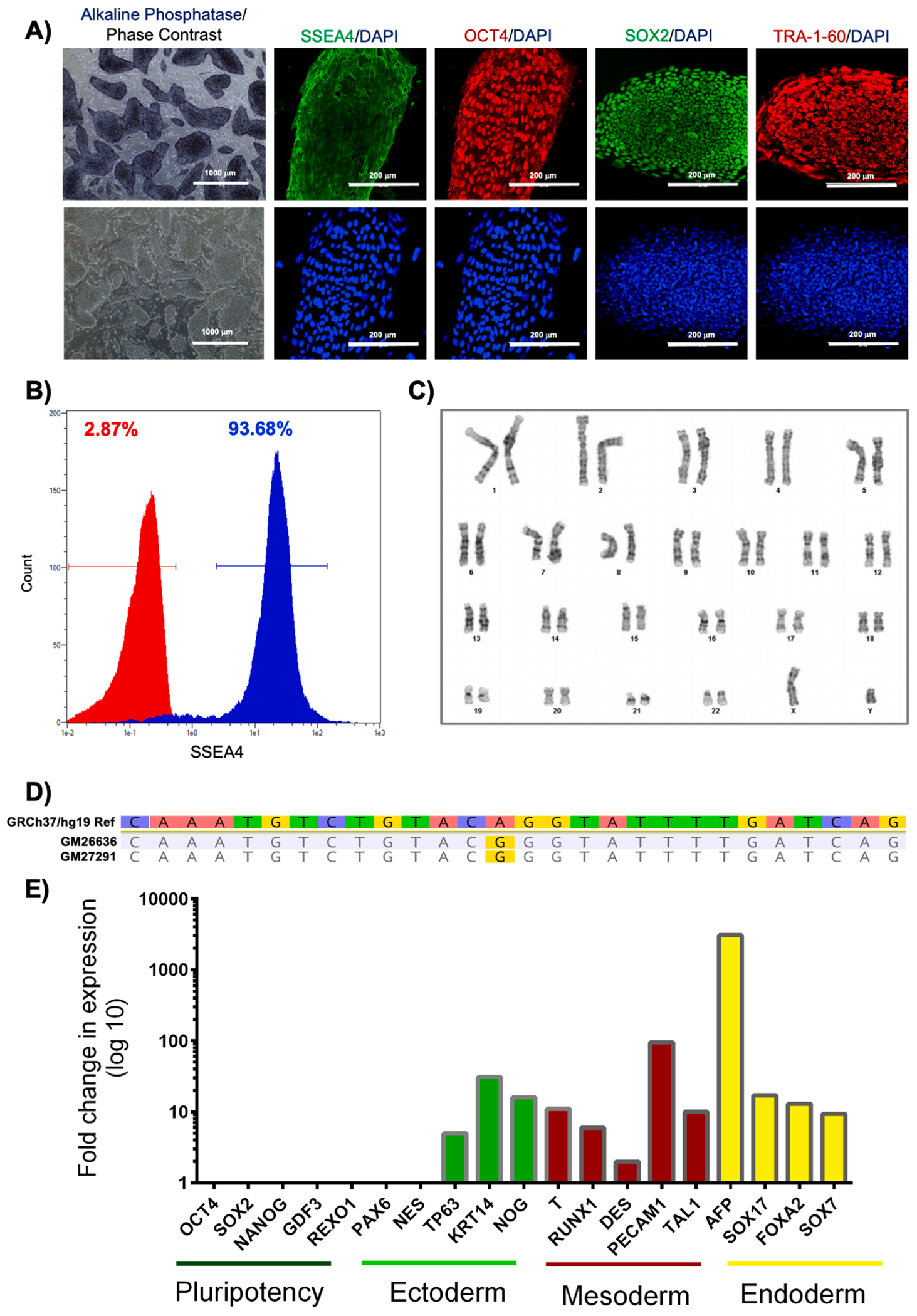
Table 2.
Reagents details.
| Antibodies used for immunocytochemistry/flow-cytometry | |||
|---|---|---|---|
| Antibody | Dilution | Company Cat # and RRID | |
|
| |||
| Pluripotency Markers | Alexa Fluor 647 anti human SSEA4 | 1:200 | BioLegend Cat# 330408, RRID:AB_1089200 |
| Pluripotency Markers | Alexa Fluor 647 Mouse IgG3 κ | 1:300 | BioLegend Cat# 401321, RRID: AB_10683445 |
| Pluripotency Markers | Isotype Control Alexa Fluor 488 anti-human/mouse SSEA3 | 1:200 | BioLegend Cat# 330306, RRID: AB_1279440 |
| Pluripotency Markers | Alexa Fluor 488 Rat IgM κ Isotype Control | 1:300 | BioLegend Cat# 400811, RRID:AB_1659271 |
| Primary Antibody | anti-SOX2 (host: rat) | 1:100 | ThermoFisher Scientific Cat# A24759, RRID:AB_2651000 |
| Primary Antibody | anti-TRA-1–60 (host: mouse IgM | 1:100 | ThermoFisher Scientific Cat# A24868, RRID:AB_2651002 |
| Primary Antibody | anti-OCT4 (host: rabbit) | 1:200 | ThermoFisher Scientific Cat# A24867, RRID:AB_2650999 |
| Primary Antibody | anti-SSEA4 (host: mouse IgG3) | 1:100 | ThermoFisher Scientific Cat# A24866, RRID:AB_2651001 |
| Secondary Antibody | Alexa Fluor 488 donkey anti-rat | 1:100 | Thermo Fisher Scientific Cat# A24876, RRID:AB_2651007 |
| Secondary Antibody | Alexa Fluor 555 goat anti-mouse IgM | 1:100 | Thermo Fisher Scientific Cat# A24871, RRID:AB_2651009 |
| Secondary Antibody | Alexa Fluor 555 donkey anti-rabbit | 1:100 | Thermo Fisher Scientific Cat# A24869, RRID:AB_2651006 |
| Secondary Antibody | Alexa Fluor 488 goat anti-mouse IgG3 | 1:100 | Thermo Fisher Scientific Cat# A24877, RRID:AB_2651008 |
| Primers | |||
| Target | Forward/Reverse primer (5′-3′) | ||
|
| |||
| Sendai virus test (qPCR) | SEV | Mr04269880_mr (TaqMan® probe ID) | |
| Sendai virus test (qPCR) | SEV-KOS | Mr04421257_mr (TaqMan® probe ID) | |
| Sendai virus test (qPCR) | SEV-KLF4 | Mr04421256_mr (TaqMan® probe ID) | |
| Sendai virus test (qPCR) | SEV-CMYC | Mr04269876_mr (TaqMan® probe ID) | |
| Pluripotency Markers (qPCR) | OCT4 | hs00742896_s1 (TaqMan® probe ID) | |
| Pluripotency Markers (qPCR) | SOX2 | hs00602736_s1 (TaqMan® probe ID) | |
| Pluripotency Markers (qPCR) | NANOG | hs02387400_g1 (TaqMan® probe ID) | |
| Pluripotency Markers (qPCR) | GDF3 | hs00220998_m1 (TaqMan® probe ID) | |
| Pluripotency Markers (qPCR) | REXO1 | hs00381890_m1 (TaqMan® probe ID) | |
| House-Keeping Gene (qPCR) | GAPDH | hs00266705_g1 (TaqMan® probe ID) | |
| Differentiation Markers (qPCR) | PAX6 | hs00240871_m1 (TaqMan® probe ID) | |
| Differentiation Markers (qPCR) | NESTIN | hs00707120_s1 (TaqMan® probe ID) | |
| Differentiation Markers (qPCR) | TP63 | hs00978340_m1 (TaqMan® probe ID) | |
| Differentiation Markers (qPCR) | KRT14 | hs00265033_m1 (TaqMan® probe ID) | |
| Differentiation Markers (qPCR) | NOGGIN | hs00271352_s1 (TaqMan® probe ID) | |
| Differentiation Markers (qPCR) | T | hs00610080_m1 (TaqMan® probe ID) | |
| Differentiation Markers (qPCR) | RUNX1 | hs01021970_m1 (TaqMan® probe ID) | |
| Differentiation Markers (qPCR) | DESMIN | hs00157258_m1 (TaqMan® probe ID) | |
| Differentiation Markers (qPCR) | PECAM1 | hs00169777_m1 (TaqMan® probe ID) | |
| Differentiation Markers (qPCR) | TAL1 | hs01097987_m1 (TaqMan® probe ID) | |
| Differentiation Markers (qPCR) | AFP | hs00173490_m1 (TaqMan® probe ID) | |
| Differentiation Markers (qPCR) | SOX17 | hs00751752_s1 (TaqMan® probe ID) | |
| Differentiation Markers (qPCR) | FOXA2 | hs00232764_m1 (TaqMan® probe ID) | |
| Differentiation Markers (qPCR) | SOX7 | hs00846731_s1 (TaqMan® probe ID) | |
4. Materials and methods
4.1. Cell culture and reprogramming
Fibroblast cells derived from the donor’s skin were cultured in MEM supplemented with 15% fetal bovine serum, and 1% Glutamax in a humidified incubator with 5% CO2 at 37 °C. Fibroblast cells were reprogrammed into iPSCs using the CytoTune 2.0 Sendai Reprogramming kit. CIMRi001-A iPSCs were cultured in DMEM/F12 + 20% KOSR + 10 ng/ml bFGF on irradiated CF1 MEFs on 0.1% Gelatin at 37 °C in humidified air with 5% CO2 and 5% O2 up to passage number 13, 20% O2 thereafter. The cells were passaged as cell aggregates (clumps) with TrypLE™ Express (Thermo Fisher Scientific) at a 1:3 ratio without ROCK inhibitor. Post-thaw cell viability was assessed by thawing a frozen vial of cells and placing it in culture. Cultures were observed daily. Colonies were photographed upon first appearance, then 4 days later. The area for 5 colonies was measured using CellSens software on the Olympus 1X50 microscope (Olympus Life Science) at 40x magnification.
4.2. Alkaline phosphatase staining
Cells were stained using the StemTAG™ Alkaline Phosphatase Staining Kit (Cell Biolabs, Inc.).
4.3. Immunocytochemistry
Immunocytochemistry characterization was performed using a 4-marker PSC Immunocytochemistry Kit (ThermoFisher Scientific) at passage 18. There were two combinations of antibodies co-stained: SOX2/TRA-1–60 and SSEA4/OCT4. Cells were fixed, permeabilized, and blocked following kit protocols. The primary antibodies (Table 2) were added to the Blocking Solution at a 1X final dilution and incubated for 3 h at 4 °C. Primary antibodies were removed and 1X DPBS wash buffer was added for 2–3 min and removed, repeating for a total of three times. Secondary antibodies (Table 2) were diluted in Blocking Solution diluted to 1X and incubated at room temperature for 1 h. Secondary antibodies were removed and 1X DPB wash buffer was added for 2–3 min, repeating for a total of three times. NucBlue Fixed Cell Stain (DAPI) was added into the last wash step and incubated for 5 min.
4.4. Flow cytometry
Surface antigen expression of iPSC makers was quantitatively measured by flow cytometry at passage 21. Cells were dissociated by Trypsin (Thermo Fisher Scientific), washed with PBS, and stained with fluorophore-conjugated antibodies (Table 2) for 15 min at room temperature. The cells were analyzed using the MACSQuant Flow Cytometer and MACSQuantify software (Miltyeni Biotec).
4.5. Whole exome sequencing
Whole exome sequencing was performed on the parent fibroblast line (GM26636) prior to submission to the Coriell Institute by GeneDx (Gaithersburg, MD), using the Agilent SureSelectXT2 All Exon V4 Kit. Targeted regions were sequenced using an Illumina HiSeq 2000 with 100 bp paired-end reads, and reads were mapped to the UCSC hg19 reference genome. Whole exome sequencing was performed on the CIMRi001-A iPSC line at the Coriell Institute using the Ion Ampliseq™ Exome RDY Kit and the Ion Gene Studio™ S5 System, and reads were mapped to the UCSC hg19 reference genome.
4.6. G-banded karyotyping & microarray genotyping
The G-banding karyotype analysis was performed using iPSCs at passage 21. Twenty five metaphase cells were counted and analyzed, and five metaphase cells were karyotyped. For the microarray analysis, genomic DNA was extracted from iPSC line CIMRi001-A using a Maxwell RCS 48 (Promega) at passage 21. The Affymetrix Genome-Wide Human SNP 6.0 Array was run using the Core Reagent Kit (Thermo Fisher Scientific). Arrays were scanned on the Affymetrix GenChip Scanner 3000 7G. Array results were analyzed using the Affymetrix Genotyping Console and then the Chromosome Analysis Suite.
4.7. Short tandem repeat (STR) analysis
Six highly polymorphic tetranucleotide microsatellites (STRs) were amplified by PCR and genotyped using the Applied Biosystems™ 3730 DNA Analyzer (Thermo Fisher Scientific). Fragment sizes were measured against GeneScan™ 500 LIZ™ size standard (Themo Fisher Scientific) and allele sizes were analyzed using GeneMapper™ v 4.0 (Thermo Fisher Scientific). This identity screen was used to ensure the iPSC line matched the parental fibroblast line.
4.8. Mycoplasma detection and sterility testing
The MycoSEQ™ Mycoplasma Detection Kit (ThermoFisher Scientific) was used to assess mycoplasma presence according to manufacturer instructions. This real-time PCR assay detects >90 mycoplasma species with proven specificity and demonstrated sensitivity (detecting <10 copies per reaction). Cell culture sterility was tested by growth on trypticase soy agar plates (with 5% sheep blood) and Sabouraud dextrose and Tryptic soy broth.
4.9. Sendai virus detection
Total RNA was extracted using the RNeasy Plus Mini Kit (Qiagen) at passage 13. The cDNA was reverse-transcribed from 1 μg RNA by High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) and gene expression was measured by quantitative RT-PCR. The TaqMan® Gene Expression Master Mix (Applied Biosystems) was used to amplify the target sequence with a PCR program: 50 °C, 2 min, 95 °C, 10 min; 40 cycles of 95 °C, 15 s, and 60 °C, 1 min on QuantStudio 6 Flex (ThermoFisher Scientific) with the specific primers (Table 2). The parental fibroblast cell line (GM26636, Coriell Institute) harvested 7 days after transfection with Sendai virus for 7 days was used as the positive control.
4.10. Differentiation potential
Cells were differentiated by embryoid body (EB) formation to assess pluripotency at passage 21. Embryoid bodies were cultured in DMEM supplemented with 10% fetal bovine serum, 1% L-glutamine, 1% NEAA, 1% sodium pyruvate in ultra-low attachment tissue culture plates and remained in culture for 10 days. RNA was extracted using RNeasy Mini Kit (Qiagen) and quantified using the NanoDrop One spectrophotometer. The cDNA is reverse-transcribed from 1 μg of RNA using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Gene expression is measured by quantitative RT-PCR. Ct values were adjusted to the endogenous housekeeping gene GAPDH. Relative gene expression is shown as the fold difference in expression compared to undifferentiated cells. Calculations were performed using the method (Livak and Schmittgen, 2001).
Supplementary Material
Acknowledgements
The authors would like to thank Laura Scheinfeldt and Swasti Kafle for their assistance for their assistance in reviewing the manuscript. This work was funded by the National Institute of General Medicine Sciences 5U42GM115336 to NT.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Nahid Turan reports financial support was provided by National Institute of General Medical Sciences.
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2022.102833.
Data availability
Data will be made available on request.
References
- Cullup T, Kho AL, Dionisi-Vici C, Brandmeier B, Smith F, Urry Z, Simpson MA, Yau S, Bertini E, McClelland V, Al-Owain M, Koelker S, Koerner C, Hoffmann GF, Wijburg FA, Hoedt A.E.t., Rogers RC, Manchester D, Miyata R, Hayashi M, Said E, Soler D, Kroisel PM, Windpassinger C, Filloux FM, Al-Kaabi S, Hertecant J, Del Campo M, Buk S, Bodi I, Goebel H-H, Sewry CA, Abbs S, Mohammed S, Josifova D, Gautel M, Jungbluth H, 2013. Recessive mutations in EPG5 cause Vici syndrome, a multisystem disorder with defective autophagy. Nat. Genetics 45, 83–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne S, Dionisi-Vici C, Smith L, Gautel M, Jungbluth H, 2016. Vici syndrome: a review. Orphanet J. Rare Diseases 11, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vici CD, Sabetta G, Gambarara M, Vigevano F, Bertini E, Boldrini R, Parisi SG, Quinti I, Aiuti F, Fiorilli M, Optiz JM, Reynolds JF, 1988. Agenesis of the corpus callosum, combined immunodeficiency, bilateral cataract, and hypopigmentation in two brothers. Am. J. Med. Genetics 29, 1–8. [DOI] [PubMed] [Google Scholar]
- Byrne S, Jansen L, U-King-Im J-M, Siddiqui A, Lidov HGW, Bodi I, Smith L, Mein R, Cullup T, Dionisi-Vici C, Al-Gazali L, Al-Owain M, Bruwer Z, Al Thihli K, El-Garhy R, Flanigan KM, Manickam K, Zmuda E, Banks W, Gershoni-Baruch R, Mandel H, Dagan E, Raas-Rothschild A, Barash H, Filloux F, Creel D, Harris M, Hamosh A, Kölker S, Ebrahimi-Fakhari D, Hoffmann GF, Manchester D, Boyer PJ, Manzur AY, Lourenco CM, Pilz DT, Kamath A, Prabhakar P, Rao VK, Rogers RC, Ryan MM, Brown NJ, McLean CA, Said E, Schara U, Stein A, Sewry C, Travan L, Wijburg FA, Zenker M, Mohammed S, Fanto M, Gautel M, Jungbluth H, 2016. EPG5-related Vici syndrome: a paradigm of neurodevelopmental disorders with defective autophagy. Brain 139, 765–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD, 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.


