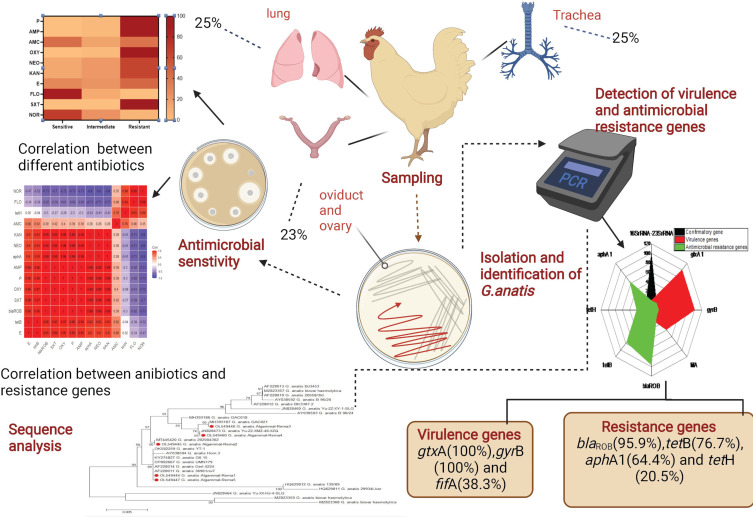
Abstract
Background
Gallibacterium anatis is incriminated frequently in severe economic losses and mortalities in the poultry industry. This study aimed to detect the prevalence of G. anatis in layer chickens, sequence analysis, the antibiogram profiles, and PCR screening of virulence determinants and antibiotic resistance genes.
Methods
Accordingly, 300 samples (tracheal swabs, ovary and oviduct, and lung) were randomly collected from 100 diseased layer chickens from private commercial layer farms at Elsharkia Governorate, Egypt. The bacteriological examination was carried out. The retrieved isolates were tested for 16S rRNA-23S rRNA gene sequencing, antibiogram profiling, PCR screening of virulence (gtxA, fifA, and gyrB), and antibiotic resistance genes (blaROB, aphA1, tetB, and tetH).
Results
The prevalence of G. anatis was 25% in the examined diseased layer chickens. The sequence analyses emphasized that the tested strains derived from a common ancestor and exhibited a notable genetic similarity with other G. anatis strains from USA, China, and Denmark. The isolated G. anatis strains were highly resistant to sulfamethoxazole-trimethoprim, oxytetracycline, penicillin, ampicillin, kanamycin, neomycin, and erythromycin. The PCR revealed that the retrieved G. anatis strains carried gtxA, gyrB, and fifA virulence genes with a prevalence of 100%, 100%, and 38.3%, respectively. Approximately 30.1% of the retrieved G. anatis isolates were XDR to six antimicrobial classes and harbored blaROB, aphA1, and tetB resistance genes. Moreover, 20.5% of the isolated G. anatis strains were MDR to three different classes and carried blaROB and tetH resistance genes.
Conclusion
Briefly, this study emphasized the existence of XDR and MDR G. anatis strains in poultry. Florfenicol and norfloxacin displayed a promising antimicrobial effect against the emerging XDR and MDR G. anatis in poultry. The emergence of XDR and MDR G. anatis is considered a public health alarm.
Keywords: G. anatis, poultry, antibiogram, XDR, sequence analyses, virulence genes, antibiotic resistance genes
Graphical Abstract
Introduction
Globally, there is an increased need for poultry meat and eggs as essential dietary components. Poultry diseases caused by resident microbiota are deleterious diseases that had severe losses in the poultry industry due to the marked decrease in growth and egg production, costs of treatment, and high mortality rates.1,2
Based on 16S rRNA gene sequencing, Genus Gallibacterium is categorized as a member of the Pasteurellaceae family.3 Gallibacterium anatis (G. anatis) is an opportunistic microorganism that normally inhabitant the genital and respiratory tracts of intensively reared chickens and other domestic birds. G. anatis is a Gram-negative coccobacillus frequently incriminated in mortalities in domestic birds, especially chickens, and sporadic human cases.4 Phenotypically, G. anatis is classified into two different biotypes: G. anatis biovar haemolytica (hemolytic biotype) and the G. anatis biovar anatis (non-hemolytic biotype).5 G. anatis is a multidrug-resistant (MDR) pathogen of poultry; causes serious diseases in poultry including salpingitis, decreased egg production, peritonitis, epididymitis, respiratory manifestations, and high mortalities. Moreover, infections caused by G. anatis in humans are rarely accompanied by abscessation of the lung, bacteremia, bronchitis, and mortalities.6
It is crucial to understand the pathogenesis and the virulence determinants of G. anatis to avoid the adverse effects of this pathogen.7 The capability of G. anatis to adhere and invade the host epithelial cells is believed to induce a remarkable role in the pathogenesis of G. anatis infection in poultry.8 The most prevalent virulence determinants accompanying G. anatis are metalloproteases, capsule, fimbriae, hemagglutinin, and innovative elements such as the rtx–like toxin (gtxA). Cell-free filtrates of the hemolytic G. anatis strains are highly toxic against the avian-derived macrophage-like cells (HD11). The leukotoxic and hemolytic activities of G.anatis are attributed mainly to the gtxA toxin which is encoded by the gtxA gene.9 Moreover, the gtxA toxin has a cytotoxic effect on poultry macrophages in-vivo. Similarly, fimbriae are considered one of the vital virulence determinants of G. anatis. The fimbriae of G. anatis are categorized in the F17-like family. Mutant strains of G. anatis that lack the flfA fimbriae were noticed to be mild pathogenic to the experimentally infected birds. Other virulence determinants of G. anatis include the proteolytic proteases and the antiphagocytic capsules that initiate biofilm production.10
Multidrug resistance has noticeably increased globally in the last decade, returning as a public health threat. Various modern surveys demonstrated the emergence of XDR and MDR bacterial pathogens from distinct sources such as poultry, fish, animals, food products, and humans.11–16 Despite that infections induced by G. anatis could be treated with antibiotics, certain unresponsive cases were reported.17,18 Multidrug resistance patterns of G. anatis to various antimicrobial classes (such as β-lactam antibiotics, sulfonamides, aminoglycosides, and tetracyclines) are frequently reported by several previous studies.3,6,19,20
This study aimed to detect the prevalence of G. anatis in layer chickens, sequence analysis, the antibiogram profiles, and PCR screening of virulence determinants (gtxA, fifA, and gyrB) and antibiotic resistance genes (blaROB, aphA1, tetB, and tetH) among the retrieved G. anatis strains.
Methods
Animal Ethics
All procedures were carried out consistent with relevant regulations. All procedures and handling of birds were approved by the Animal Ethics Review Committee, Suez Canal University, Egypt.
Sampling
A total of 300 samples (tracheal swabs, ovary and oviduct, and lung; n=100 for each) were randomly collected from 100 diseased layer chickens (3 types of samples from each bird) with an average age of 25–40 weeks from private commercial layer farms at Elsharkia Governorate, Egypt (from February to May 2020). The examined diseased layer chickens were suffering from depression, respiratory signs, occasionally head swelling, and decreased in egg production (5–10%). The post-mortem inspection of sacrificed and/or freshly dead birds displayed oophoritis, peritonitis, salpingitis, and tracheitis. All samples were gathered in sterile plastic bags, placed in an icebox and quickly transferred to the Microbiology laboratory for bacteriological examination.
Isolation and Identification of G. anatis
Swabs from the obtained samples (tracheal swabs, ovary and oviduct, and lung) were directly streaked out on 5% Columbia blood agar and MacConkey agar plates (Difco, USA) and incubated aerobically at 37 °C for 24 h. The identification of G. anatis was carried out according to Gram’s staining, culture characteristics, motility test, and the biochemical characteristics (oxidase, nitrate reduction, catalase, indole production, methyl red, Voges-Proskauer, citrate utilization, urease test, gelatinase, and sugar fermentation tests).5,21 Moreover, the identification of the retrieved G. anatis isolates was ensured using PCR-based detection of the 16S rRNA-23S rRNA gene according to Bojesen,22 followed by gene sequencing.
G. anatis 16S rRNA-23S rRNA Gene Sequencing
All the retrieved G. anatis isolates displayed harmony in the biochemical and phenotypic features. Consequently, the PCR products of five indiscriminately selected G. anatis strains were purified using the QIAquick PCR-Product extraction kit (QIAGEN GmbH, Hilden, D-40724, Germany), then were tested for sequencing in both directions through the Bigdye Terminator V3.1 cycle sequencing kit (Thermo Fisher Scientific GmbH, Dreieich 63,303, Germany). The recovered sequences were placed in the GenBank with accession numbers: OL549444, OL549445, OL549446, OL549447, and OL549460. The BLAST analysis was carried out to investigate the sequence identity. Moreover, the phylogenetic analysis was performed according to the neighbor-joining in MEGA6 as previously described.23
Antimicrobial Susceptibility Testing of the Recovered G. anatis
The recovered G. anatis strains were examined for the susceptibility to several antimicrobial agents using the disc diffusion technique on Muller-Hinton agar (Oxoid, UK) was implemented consistently with the procedures of CLSI, 2018.24 Ten antimicrobial agents were involved, including; penicillin (PEN, 30μg), ampicillin (AMP, 30μg), amoxicillin-clavulanic acid (AMC, 30 μg), oxytetracycline (OX, 30μg), neomycin (NEO, 10 μg), kanamycin (KAN, 10 μg), erythromycin (E, 15 μg), florfenicol (FFC, 30μg), sulphamethoxazole-trimethoprim (SXT, 30 μg), and norfloxacin (NOR, 10 μg) (ThermoFisher Scientific, USA). Moreover, E. coli-ATCC 25922 was involved as a control strain. The tested G. anatis isolates were categorized according to their resistance patterns into MDR (Multidrug-resistant: resistant to ≥ one antimicrobial agent in ≥ 3 classes) and XDR (Extensively drug-resistant: resistant to ≥ one agent in all tested antimicrobial classes except one or two) according to Magiorakos.25 Moreover, the multiple antibiotic resistance (MAR) index was determined as previously described.26
PCR-Based Screening of Virulence and Antimicrobial Resistance Genes in the Isolated G. anatis
PCR was used to detect the virulence (gtxA: Cytolytic-hemolytic gene, fifA: Flageller gene, and gyrB: The gyrase subunit B gene) and the antimicrobial resistance genes (blaROB: β-lactam resistance gene, aphA1: aminoglycosides resistance gene, tetB and tetH: tetracycline resistance gene) in the retrieved G. anatis strains. DNA of the tested G. anatis strains was extracted following the manufacturer’s instructions of the QIAampDNA Mini Kit (Qiagen, GmbH, Germany/Cat. No.56304). The reaction volume was 25-μL, including 5 μL of extracted DNA, 12.5 μL of 2 × Master Mix, 20 pmol of tested primer, and distilled H2O). Negative control (reactions with no DNA template) and Positive controls (positive strains supplied by A.H.R.I, Egypt) were involved in every reaction. The used primers (Biobasic, Canada) and cycling conditions were demonstrated in Table 1. Lastly, the PCR products were separated using the agar gel electrophoresis (1.5% agarose stained using ethidium bromide 10 mg/mL), and then photographed the gel.
Table 1.
Primers Sequences and PCR Cycling Conditions
| Genes | Oligonucleotides Sequences | Amplicon size (bp) | (35 Cycles) | References | ||
|---|---|---|---|---|---|---|
| Den. | Annealing | Ext. | ||||
| 16SrRNA (1133fgal)-23SrRNA (114r) | F: TATTCTTTGTTACCARCGG R: GGTTTCCCCATTCGG |
1032 | 94°C 30 sec |
55°C 50 sec |
72°C 1 min |
[22] |
|
gtxA (Cytolytic-hemolytic gene) |
F: CAAACCTAATTCAATCGGATG R: TGCTTCAATAATTTTCCATTTTC |
1257 | 94°C 30 sec |
51°C 40 sec |
72°C 1 min |
[1] |
|
fifA (Flageller gene) |
F: CACCATGGGTGCATTTGCGGATGATCC R: TATTCGTATGCGATAGTATAGTTC |
538 | 94°C 30 sec |
55°C 40 sec |
72°C 45 sec |
[27] |
|
gyrB (The gyrase subunit B gene) |
F: TGTGCGTTTCTGGCCAAGTC R: CGCTCACCAACTGCAGATTC |
561 | 94°C 30 sec |
55°C 40 sec |
72°C 45 sec |
[28] |
|
blaROB (Beta lactam resistance gene) |
F: AATAACCCTTGCCCCAATTC R: TCGCTTATCAGGTGTGCTTG |
685 | 94°C 30 sec |
60°C 40 sec |
72°C 45 sec |
[29] |
| aphA1 (Aminoglycosides resistance gene) |
F: TTATGCCTCTTCCGACCATC R: GAGAAAACTCACCGAGGCAG |
489 | 94°C 30 sec |
54°C 40 sec |
72°C 40 sec |
|
|
tetH (Tetracycline resistance gene) |
F: ATACTGCTGATCACCGT R: TCCCAATAAGCGACGCT |
1076 | 94°C 30 sec |
60°C 40 sec |
72°C 1 min |
|
|
tetB (Tetracycline resistance gene) |
F: CCT TAT CATGCC AGT CTTGC R: ACT GCCGTT TTT TTCGCC |
774 | 94°C 30 sec |
50°C 40 sec |
72°C 1 min |
[30] |
Statistical Analyses
The Chi-square test was used in data analyses (SAS software, 9.4 M6, SAS Institute, Cary, NC, USA) (The significance level was (p-value < 0.05). Besides, the correlation coefficient was estimated via the R-software (version 02.1; http://www.r-project.org/) using “corr” and “corrplot”.
Results
Phenotypic Characteristics of the Isolated G. anatis from Diseased Layers
The recovered G. anatis colonies were bright translucent, low convex, fine, circular, smooth-edged with greyish color, and mostly showed β-hemolysis on blood agar. Moreover, the colonies were small (pin point-like) and pink (lactose fermenter) on MacConkey agar. Gram’s staining displayed Gram-negative coccoid to small pleomorphic bacilli. The retrieved G. anatis isolates were positive for oxidase, catalase, nitrate reduction, sucrose, and mannitol fermentation tests. Conversely, the recovered isolates were negative for citrate utilization, indole, urease, gelatinase, motility, methyl red, and Voges-Proskauer tests.
Prevalence of G. anatis in the Examined Samples of Diseased Layer Chickens
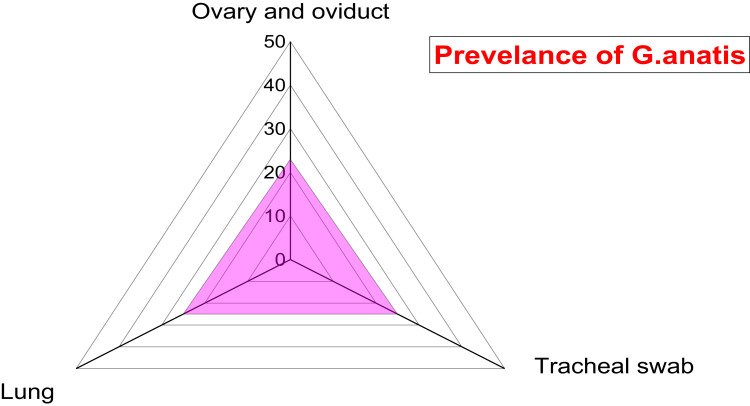
A total of 73 G. anatis isolates were recovered from 300 bacteriologically examined samples obtained from 100 diseased layer chickens. Twenty-five isolates (25%) were obtained from tracheal swabs, 25 isolates (25%) from lung samples, and 23 isolates (23%) from ovary and oviduct samples. From the twenty-five infected birds with G. anatis, the pathogen was isolated from tracheal swab, lung, and ovary and oviduct samples of the same bird in 23 infected birds. Moreover, G. anatis was recovered from the lung and tracheal swabs of the same bird in two infected birds. There is no significant difference in the dissemination of G. anatis among the examined organs obtained from the diseased layer chickens (p > 0.05%), as illustrated in Table 2 and Figure 1. The prevalence of G. anatis was 25% (25/100) in the examined diseased layer chickens.
Table 2.
The Prevalence of G. anatis Isolated from Examined Diseased Layer Chickens (n=73)
| No. of Examined Diseased Birds | Types of Organs | No. of Organs | No. of Positive Samples | Percentage of Positive Samples |
|---|---|---|---|---|
| 100 | Ovary and oviduct | 100 | 23 | 23 |
| Tracheal swab | 100 | 25 | 25 | |
| Lung | 100 | 25 | 25 | |
| Total | 300 | 73 | 24.3 | |
|
Chi square p value |
0.10959 0.9467NS |
|||
Abbreviation: NS, Non-significant.
Figure 1.
Prevalence of G. anatis among different examined samples obtained from diseased layers.
Phylogenetic Analyses of G. anatis 16S rRNA-23S rRNA Gene
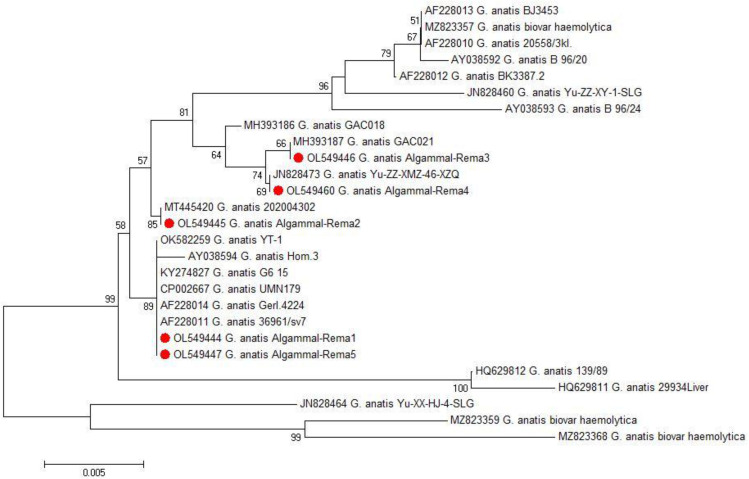
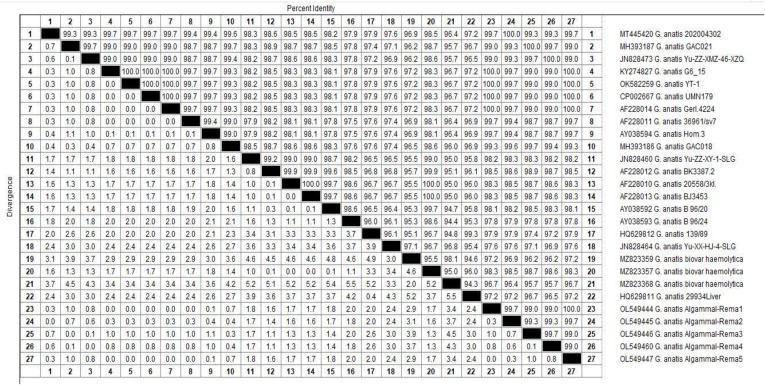
Phylogenetic and sequence analyses of the 16S rRNA-23S rRNA gene emphasized that the tested G. anatis strains (n=5) derived from a common ancestor (Accession Numbers: OL549444, OL549445, OL549446, OL549447, and OL549460). In the present study, the tested G. anatis strains exhibited a notable genetic similarity (96.7–100%) with other G. anatis strains from various origins. For example, G. anatis strain G6_15 (99–100%) was isolated from a tracheal swab of diseased layer chicken in Egypt (Accession No. KY274827). G. anatis strain UMN179 (99–100%) was isolated from an Iowa laying hen with peritonitis in the USA (Accession No. CP002667). G. anatis biovar hemolytica (98.3–98.7%) was isolated from poultry in Pennsylvania, USA (Accession No. MZ823357). G. anatis strain 202,004,302 (99.7–100%) was isolated from layer chicken in China (Accession No. MT445420), G. anatis strain GAC021 (99–100%) was isolated from poultry in China (Accession No. MH393187), G. anatis strain Yu-ZZ-XMZ-46-XZQ (99–100%) isolated from a cloacal sample in China (Accession No. JN828473), G. anatis strain YT-1 (99–100%) isolated from oviduct in China (Accession No. OK582259), G. anatis strain 36,961/sv7 (99.4–99.7%) in Denmark (Accession No. AF228011), and G. anatis strain Gerl.4224 (99–100%) in Denmark (Accession No. AF228014) as shown in Figures 2 and 3.
Figure 2.
The phylogenetic analysis was carried out according to the 16SrRNA-23SrRNA gene sequencing. The phylogenetic tree clarifies the genetic relatedness between the recovered G. anatis strains and other strains deposited in the GenBank. The retrieved G. anatis strains in the present study were emphasized with red circles.
Figure 3.
Illustrates the percentage of G. anatis 16SrRNA-23SrRNA nucleotides sequences identity.
The in-vitro Antimicrobial Susceptibility of G. anatis
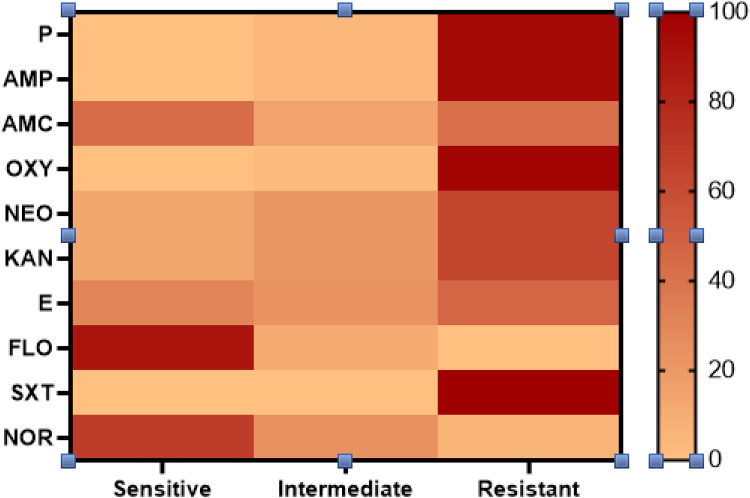
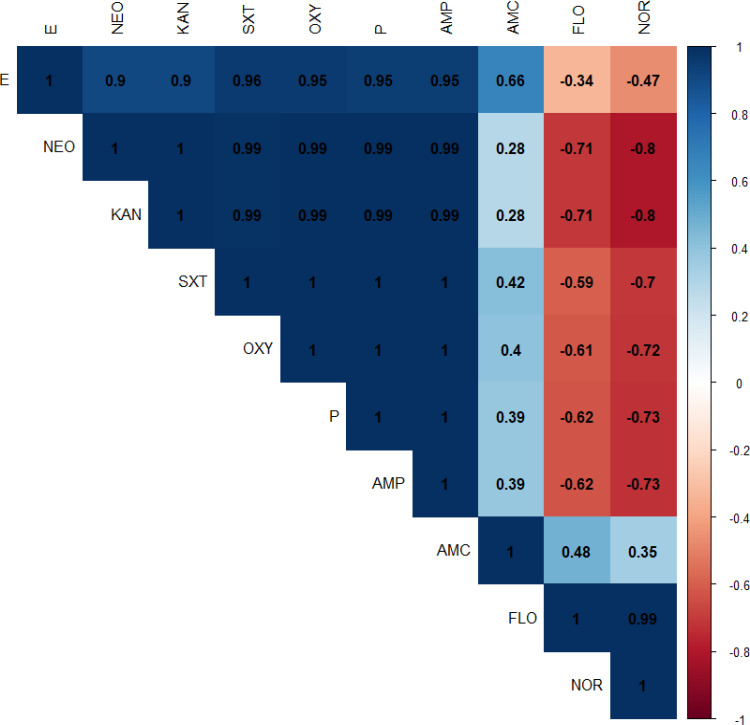
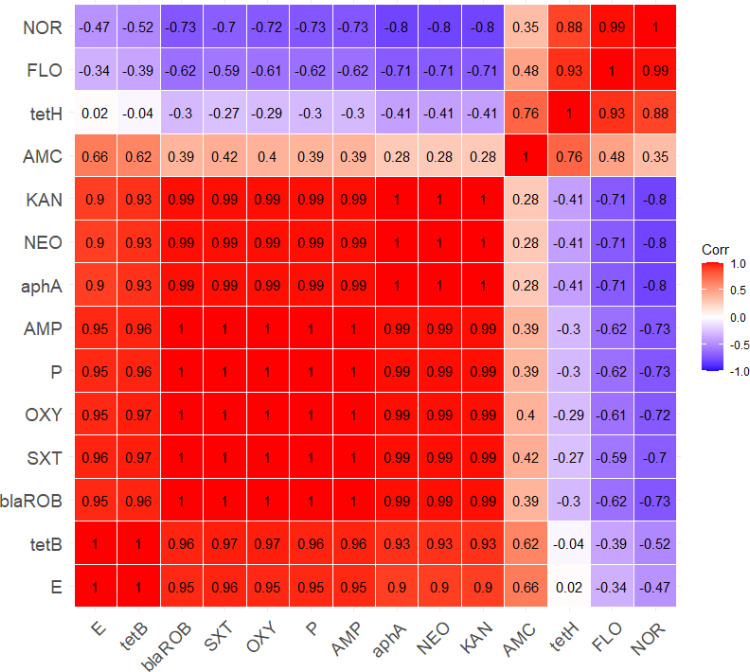
The antimicrobial susceptibility of the retrieved G. anatis isolates showed that the tested strains were resistant to sulfamethoxazole-trimethoprim (100%), oxytetracycline (97.3%), penicillin and ampicillin (95.9%), kanamycin and neomycin (64.4%), and erythromycin (46.6%). Moreover, the recovered isolates were sensitive to florfenicol (89%), norfloxacin (68.5%), and amoxicillin-clavulanic acid (43.8%) (As shown in Table 3 and Figure 4). Statistically, the recovered G. anatis strains displayed a significant difference in their susceptibility patterns to different tested antimicrobial classes (p < 0.05). Besides, the correlation coefficient was detected among the involved antimicrobial agents. Accordingly, strong positive correlations were detected between: KAN, NEO, P, AMP, OXY, and SXT (r=0.99); FLO and NOR(r=0.99); E, P, AMP, OXY, and SXT (r=0.95); E, KAN, and NEO (r=0.90). Moreover, moderate positive correlations were noticed between AMC and E (r=0.66); AMC and FLO (r=0.48); AMC and SXT (r=0.42); AMC and OXY (r=0.40); AMC, P, and AMP (r=0.39) (Figure 5).
Table 3.
The in-vitro Antimicrobial Susceptibility Testing of the Isolated G. anatis (n=73)
| Antimicrobial Class | Antimicrobial Agents | Sensitive | Intermediate | Resistant | |||
|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | ||
| Penicillin | Penicillin | 0 | 0 | 3 | 4.1 | 70 | 95.9 |
| Ampicillin | 0 | 0 | 3 | 4.1 | 70 | 95.9 | |
| β-lactam-β Lactamase inhibitor combination | Amoxicillin -clavulanic acid | 32 | 43.8 | 11 | 15.1 | 30 | 41.1 |
| Tetracyclines | Oxytetracycline | 0 | 0 | 2 | 2.7 | 71 | 97.3 |
| Aminoglycosides | Neomycin | 10 | 13.7 | 16 | 21.9 | 47 | 64.4 |
| Kanamycin | 10 | 13.7 | 16 | 21.9 | 47 | 64.4 | |
| Macrolides | Erythromycin | 22 | 30.1 | 17 | 23.3 | 34 | 46.6 |
| Phenicols | Florfenicol | 65 | 89 | 8 | 11 | 0 | 0 |
| Sulfonamides | Sulfamethoxazole trimethoprim | 0 | 0 | 0 | 0 | 73 | 100 |
| Fluroquinolones | Norfloxacin | 55 | 68.5 | 18 | 24.6 | 5 | 6.8 |
|
Chi-square p value |
267.75 0.0001 |
47.702 0.0001 |
149.62 0.0001 |
||||
Figure 4.
Illustrates the antimicrobial susceptibility of the recovered G. anatis from diseased layer chickens.
Figure 5.
The heat map demonstrates the correlation coefficient (r) between different tested antimicrobial involved in the current study.
Virulence Determinant and Antibiotic Resistance Genes of the Isolated G. anatis Strains
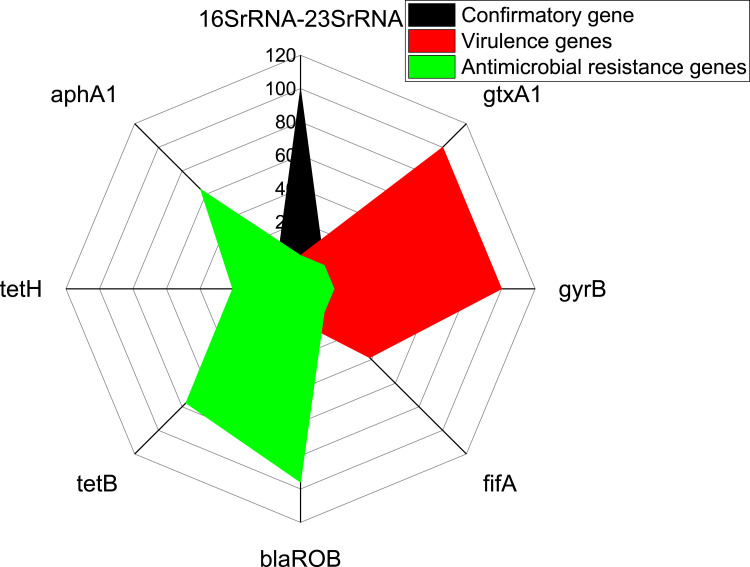
Using PCR, all the retrieved G. anatis isolates (100%) were positive for the confirmatory gene (16SrRNA-23SrRNA). Moreover, the recovered G. anatis strains commonly harbored the virulence genes gtxA1 (cytolytic-hemolytic gene) (100%) and gyrB (gyrase subunit B gene) (100%), followed by fifA gene (fimbrial gene) (38.3%). Furthermore, the tested G. anatis strains harbored the blaROB (β-lactam resistance), tetB (tetracycline resistance), aphA (aminoglycosides resistance gene), and tetH (tetracycline resistance) resistance genes with a prevalence of 95.9%%, 76.7%, 64.4%, and 20.5%, respectively (Table 4 and Figure 6). The statistical analysis showed a significant difference (p < 0.05) in the distribution of antibiotic resistance and virulence genes in the isolated G. anatis strains.
Table 4.
Prevalence of Virulence and Antimicrobial Resistance Genes Among the Recovered G. anatis (n=73)
| Types of Genes | n | % | Chi-Square p value |
|
|---|---|---|---|---|
| Confirmation gene | 16SrRNA-23SrRNA | 73 | 100 | |
| Virulence-determinant genes | gtxA1 | 73 | 100 | 23.276 0.001 |
| gyrB | 73 | 100 | ||
| fifA | 28 | 38.3 | ||
| Antimicrobial resistance genes | blaROB | 70 | 95.9 | 34.766 0.0001 |
| tetB | 56 | 76.7 | ||
| tetH | 15 | 20.5 | ||
| aphA1 | 47 | 64.4 | ||
Figure 6.
The distribution of antimicrobial resistance and virulence determinant genes in the retrieved G. anatis strains from diseased layer chickens.
Multidrug Resistance Patterns of the Retrieved G. anatis Strains
Our results evidenced that 30.1% (22/73) of the retrieved G. anatis isolates were XDR to six antimicrobial classes (Penicillins: PEN and AMP, β-lactam-β-Lactamase-inhibitor combination: AMC, Tetracyclines: OX, Aminoglycosides: NEO and KAN, Macrolides: E, and Sulfonamides: SXT) and harbored blaROB, aphA1, and tetB resistance genes. Moreover, 20.5% (15/73) of the isolated G. anatis strains were MDR to three different classes (Penicillins: PEN and AMP, Tetracyclines: OX, and Sulfonamides: SXT) and carried blaROB and tetH resistance genes. Furthermore, 17.8% (13/73) of the retrieved G. anatis strains were MDR to four different classes (Penicillins: PEN and AMP, Tetracyclines: OX, Aminoglycosides: NEO and KAN, and Sulfonamides: SXT) and possessed blaROB, aphA1, and tetB resistance genes. In addition, 10.9% (8/73) of the isolated G. anatis strains were MDR to four different classes (Penicillins: PEN and AMP, Tetracyclines: OX, β-lactam-β-Lactamase-inhibitor combination: AMC, and Sulfonamides: SXT) and carried blaROB and tetB resistance genes. Besides, 9.6% (7/73) of the isolated G. anatis strains were MDR to five different classes (Penicillins: PEN and AMP, Tetracyclines: OX, Aminoglycosides: NEO and KAN, Macrolides: E, and Sulfonamides: SXT) and carried blaROB, aphA1, and tetB resistance genes as described in Table 5 and Figure 7. In the present study, the MAR index values (0.3–0.6) revealed multiple resistance patterns suggesting that the recovered G. anatis strains are derived from high-risk contamination. The correlation coefficient (r) was calculated among the demonstrated antimicrobial resistance genes in the recovered G. anatis strains and different involved antimicrobial agents. Our findings revealed positive correlations between: blaROB gene, P, and AMP (r=1); aphA1 gene, NEO, and KAN (r=1); tetB and OXY (r=0.97) as illustrated in Figure 8.
Table 5.
The Prevalence of Multidrug-Resistance Profiles and the Resistance Genes Among the Recovered G. anatis Strains (n =73)
| No. of Strains | % | Type of Resistance | Multidrug Resistance Profiles | Antimicrobial-Resistance Genes | MAR-Index |
|---|---|---|---|---|---|
| 22 | 30.1 | XDR |
- Six classes: PEN and AMP AMC OX NEO and KAN E SXT |
blaROB, aphA1,and tetB | 0.6 |
| 15 | 20.5 | MDR |
- Three classes: PEN and AMP OX SXT |
blaROB and tetH | 0.3 |
| 13 | 17.8 | MDR |
- Four classes: PEN and AMP OX NEO and KAN SXT |
blaROB, aphA1, and tetB | 0.4 |
| 8 | 10.9 | MDR |
- Four classes: PEN and AMP AMC OX SXT |
blaROB, and tetB | 0.4 |
| 7 | 9.6 | MDR |
- Five classes: PEN and AMP OX NEO and KAN E SXT |
blaROB, aphA1, and tetB | 0.5 |
| 5 | 6.8 | XDR |
-Six classes: PEN and AMP OX NEO and KAN E SXT NOR |
blaROB, aphA1, and tetB | 0.6 |
Figure 7.
The emergence of XDR and MDR patterns between among the recovered G. anatis strains from diseased layer chickens.
Figure 8.
The heat map illustrates the correlation coefficient (r) between the demonstrated antimicrobial resistance genes in the retrieved G. anatis strains and different involved antimicrobial agents.
Discussion
This study aimed to investigate the prevalence of G. anatis in layer chickens, sequence analysis, the antibiogram profiles, and PCR screening of gtxA, fifA, and gyrB virulence genes and blaROB, aphA1, tetB, and tetH antibiotic resistance genes among the retrieved G. anatis strains.
In the current work, G. anatis was identified in layer chickens that suffered from respiratory manifestations and a drop in egg production. Besides, the PM inspection displayed oophoritis, peritonitis, salpingitis, and tracheitis. In the last decade, G. anatis was recovered from diseased chickens with tracheitis and salpingitis in different geographical areas all over the world. G. anatis is associated mainly with severe economic losses in the poultry industry.20,31,32 G. anatis is frequently retrieved from the genital and respiratory tracts of diseased poultry suffering from severe pathological lesions.33
In the present study, the bacteriological examination proved that the prevalence of G. anatis was 25% in the examined diseased layer chickens, where the lung and trachea are the most predominant infected organs. Moreover, there is no inconsistency in the biochemical and phenotypic features of the isolated G. anatis strains that showed an obvious harmony. The recovered G. anatis colonies were circular, smooth-edged with greyish color, and mostly β-hemolytic on blood agar. Moreover, the colonies were small (poor growth) and pink on MacConkey agar. Furthermore, the retrieved G. anatis isolates were positive for oxidase, catalase, nitrate reduction, sucrose, and mannitol fermentation tests. On the other hand, the recovered isolates were negative for citrate utilization, indole, urease, gelatinase, and Voges-Proskauer tests. Our findings are consistent with those reported by Johnson34 and Van Driessche.4
In the current study, the 16S rRNA-23S rRNA phylogenetic analyses highlighted that the tested G. anatis strains have a common ancestor (Accession Numbers: OL549444, OL549445, OL549446, OL549447, and OL549460). Moreover, the tested G. anatis strains showed high genetic similarity with other G. anatis strains isolated from diseased poultry in different regions, such as G. anatis strain G6_15 isolated by Elbestawy20 from a tracheal swab of diseased layer chicken in Egypt, G. anatis strain UMN179 isolated from an Iowa laying hen with peritonitis in USA.34 G. anatis strain Gerl.4224 was isolated from poultry in Denmark,5 and G. anatis strain GAC021 was isolated from silky chicken in China.35 Our findings disagreed with the results of Paudel,1 who reported that strains of G. anatis are divergent and usually exhibit genetic differences. Besides, these findings illustrated the epidemiological map and ensured the public health importance of G. anatis.
Regarding the antimicrobial resistance patterns, the isolated G. anatis strains showed remarkable resistance patterns to sulfonamides, tetracyclines, β-Lactam antibiotics, aminoglycosides, and macrolides. These findings nearly agreed with Bojesen17 and Elbestawy.36 The extensive uncontrolled use of antibiotics in the poultry farms, as well as the ability of G. anatis to gain antibiotic resistance genes from other resistant bacteria, are the main factors that favor of the existence of these superbugs.18
In the present study, PCR evidenced that the tested G. anatis strains are commonly harbored gtxA and gyrB, virulence genes, followed by the fifA gene. Our results are nearly agreed with those reported by Krishnegowda,2 Sorour,37 and Nassik.38 The rtx-like toxin (gtxA) is one of the main virulence determinants of G. anatis encoded by the gtxA gene.27 The gtxA toxin was demonstrated for the first time in a hemolytic G. anatis strain isolated from diseased chicken in 2010 in Denmark. The toxin is accountable for hemolytic and leukotoxic activities of G. anatis. Lacking the gtxA gene indicates a reduced bacterial virulence.9 Besides, the gyrB gene encodes the ATPase domain of DNA gyrase which is necessary for the replication of DNA in G. anatis. The gyrB gene is used frequently to identify G. anatis infection in poultry.39 Fimbriae are a common virulence determinant of G. anatis that plays a significant role in bacterial adhesion to the glycoprotein receptors in the host mucous membranes. Different F17-like fimbrial genes were recently determined in G. anatis from different origins, where the flfA gene is the most predominant one.27,28
Regarding the multidrug resistance patterns in the isolated G. anatis strains, a high proportion of the retrieved G. anatis isolates were XDR to 6 antimicrobial classes and harbored blaROB, aphA1, and tetB resistance genes. Moreover, a high percentage of the recovered G. anatis strains were MDR to 3–5 different classes and carried blaROB, aphA, and tetB or tetH resistance genes. Antimicrobial resistance is considered one of the main threats to public health globally. It remains occurred due to the abuse of antibiotics in both veterinary and health practices and the bacterial acquirement of antibiotic resistance genes through mobile genetic components.40,41 The resistance to β-lactam antibiotics (such as amoxicillin, penicillin, and ampicillin) is mainly mediated by the blaROB-1 gene. Stimulatingly, the blaROB-1 gene is the most prevalent β-lactamase gene, frequently demonstrated in the members of Pasteurellaceae.42 The tetB gene is the most prevalent tetracycline resistance gene in the present study, followed by the tetH gene. These findings were supported by Bojesen, who reported the determination of the tetB gene in 27 (27/49) tetracycline-resistant G. anatis strains.17 Moreover, the acquired resistance to aminoglycosides antibiotics is mainly attributed to the enzymatic alteration mechanism, resulting in the inactivation of aminoglycosides in several bacterial pathogens, especially G. anatis. The aminoglycosides phosphotransferases, encoded by the aphA gene, are the most common aminoglycosides modifying enzymes.4,43
Briefly, from what we know, this is the first report that emphasized the existence of XDR and MDR G. anatis strains in diseased layers in Egypt, with particular reference to the sequence analyses and multidrug resistance profiles. The retrieved G. anatis strains commonly harbor the gtxA and gyrB virulence genes, followed by the fifA gene. The obtained G. anatis strains are XDR or MDR to several antimicrobial classes (such as sulfonamides, tetracyclines, β-Lactam antibiotics, aminoglycosides, and macrolides) commonly harbored blaROB, aphA, and tetB or tetH resistance genes. Florfenicol and norfloxacin displayed a promising antimicrobial effect against the emerging XDR and MDR G. anatis strains. Combining the conventional and molecular techniques is a consistent epidemiological tool used to diagnose G. anatis infection in poultry. Alarmingly, the emergence of XDR and MDR G. anatis strains establishes a public threat that specifies a lousy prognosis of diseases induced by these superbugs. Besides, it has an adverse impact on the poultry production. Consequently, it encourages the routine application of antimicrobial susceptibility testing along with the appropriate use of antimicrobial agents in the health sector and the veterinary practice.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no conflict of interest.
References
- 1.Paudel S, Alispahic M, Liebhart D, Hess M, Hess C. Assessing pathogenicity of Gallibacterium anatis in a natural infection model: the respiratory and reproductive tracts of chickens are targets for bacterial colonization. Avian Pathol. 2013;42(6):527–535. doi: 10.1080/03079457.2013.843160 [DOI] [PubMed] [Google Scholar]
- 2.Krishnegowda DN, Dhama K, Mariappan AK, et al. Etiology, epidemiology, pathology, and advances in diagnosis, vaccine development, and treatment of Gallibacterium anatis infection in poultry: a review. Vet Q. 2020;40(1):16. doi: 10.1080/01652176.2020.1712495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh SV, Singh B, Sinha D, et al. Gallibacterium anatis: an emerging pathogen of poultry birds and domiciled birds. J Vet Sci Technol. 2016;7(3):324. [Google Scholar]
- 4.Van Driessche L, Vanneste K, Bogaerts B, et al. Isolation of drug-resistant Gallibacterium anatis from calves with unresponsive bronchopneumonia, Belgium. Emerg Infect Dis. 2020;26(4):721. doi: 10.3201/eid2604.190962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christensen H, Bisgaard M, Bojesen AM, Mutters R, Olsen JE. Genetic relationships among avian isolates classified as Pasteurella haemolytica,‘Actinobacillus salpingitidis’ or Pasteurella anatis with proposal of Gallibacterium anatis gen. nov., comb. nov. and description of additional genomospecies within Gallibacterium gen. nov. Int J Syst Evol Microbiol. 2003;53(1):275–287. doi: 10.1099/ijs.0.02330-0 [DOI] [PubMed] [Google Scholar]
- 6.El-Adawy H, Bocklisch H, Neubauer H, Hafez HM, Hotzel H. Identification, differentiation and antibiotic susceptibility of Gallibacterium isolates from diseased poultry. Ir Vet J. 2018;71(1):1–10. doi: 10.1186/s13620-018-0116-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bager RJ, Persson G, Nesta B, et al. Outer membrane vesicles reflect environmental cues in Gallibacterium anatis. Vet Microbiol. 2013;167(3–4):565–572. doi: 10.1016/j.vetmic.2013.09.005 [DOI] [PubMed] [Google Scholar]
- 8.Zhang X-P, Lu C-J, Li Y-T, et al. In vitro adherence and invasion of primary chicken oviduct epithelial cells by Gallibacterium anatis. Vet Microbiol. 2017;203:136–142. doi: 10.1016/j.vetmic.2017.02.009 [DOI] [PubMed] [Google Scholar]
- 9.Kristensen BM, Frees D, Bojesen AM. GtxA from Gallibacterium anatis, a cytolytic RTX-toxin with a novel domain organisation. Vet Res. 2010;41(3):25. doi: 10.1051/vetres/2009073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Persson G, Bojesen AM. Bacterial determinants of importance in the virulence of Gallibacterium anatis in poultry. Vet Res. 2015;46(1):1–11. doi: 10.1186/s13567-015-0206-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Algammal AM, Hashem HR, Alfifi KJ, et al. atpD gene sequencing, multidrug resistance traits, virulence-determinants, and antimicrobial resistance genes of emerging XDR and MDR-Proteus mirabilis. Sci Rep. 2021;11(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Algammal AM, Hashem HR, Al-Otaibi AS, et al. Emerging MDR-Mycobacterium avium subsp. avium in house-reared domestic birds as the first report in Egypt. BMC Microbiol. 2021;21(1):1–11. doi: 10.1186/s12866-021-02287-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hetta HF, Al-Kadmy I, Khazaal SS, et al. Antibiofilm and antivirulence potential of silver nanoparticles against multidrug-resistant Acinetobacter baumannii. Sci Rep. 2021;11(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Algammal AM, Alfifi KJ, Mabrok M, et al. Newly emerging MDR B. cereus in Mugil seheli as the first report commonly harbor nhe, hbl, cytK, and pc-plc virulence genes and bla1, bla2, tetA, and ermA resistance genes. Infect Drug Resist. 2022;15:2167–2185. doi: 10.2147/IDR.S365254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Algammal AM, Mabrok M, Ezzat M, et al. Prevalence, antimicrobial resistance (AMR) pattern, virulence determinant and AMR genes of emerging multi-drug resistant Edwardsiella tarda in Nile tilapia and African catfish. Aquaculture. 2022;548:737643. doi: 10.1016/j.aquaculture.2021.737643 [DOI] [Google Scholar]
- 16.Kareem SM, Al-Kadmy IMS, Kazaal SS, et al. Detection of gyrA and parC Mutations and Prevalence of Plasmid-Mediated Quinolone Resistance Genes in Klebsiella pneumoniae. Infect Drug Resist. 2021; 12;14:555-563. doi: 10.2147/IDR.S275852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bojesen AM, Vazquez ME, Bager RJ, Ifrah D, Gonzalez C, Aarestrup FM. Antimicrobial susceptibility and tetracycline resistance determinant genotyping of Gallibacterium anatis. Vet Microbiol. 2011;148(1):105–110. doi: 10.1016/j.vetmic.2010.08.011 [DOI] [PubMed] [Google Scholar]
- 18.Hess C, Grafl B, Bagheri S, Kaesbohrer A, Zloch A, Hess M. Antimicrobial resistance profiling of Gallibacterium anatis from layers reveals high number of multiresistant strains and substantial variability even between isolates from the same organ. Microbial Drug Resist. 2020;26(2):169–177. doi: 10.1089/mdr.2019.0056 [DOI] [PubMed] [Google Scholar]
- 19.Jones K, Thornton J, Zhang Y, Mauel M. A 5-year retrospective report of Gallibacterium anatis and Pasteurella multocida isolates from chickens in Mississippi. Poult Sci. 2013;92(12):3166–3171. doi: 10.3382/ps.2013-03321 [DOI] [PubMed] [Google Scholar]
- 20.Elbestawy AR, Ellakany HF, Abd El-Hamid HS, et al. Isolation, characterization, and antibiotic sensitivity assessment of Gallibacterium anatis biovar haemolytica, from diseased Egyptian chicken flocks during the years 2013 and 2015. Poult Sci. 2018;97(5):1519–1525. doi: 10.3382/ps/pey007 [DOI] [PubMed] [Google Scholar]
- 21.Kleven S, Dufour-Zavala L, Swayne D, et al. A Laboratory Manual for the Isolation, Identification, and Characterization of Avian Pathogens. American Association of Avian Pathologists. 2008. [Google Scholar]
- 22.Bojesen AM, Vazquez ME, Robles F, et al. Specific identification of Gallibacterium by a PCR using primers targeting the 16S rRNA and 23S rRNA genes. Vet Microbiol. 2007;123(1–3):262–268. doi: 10.1016/j.vetmic.2007.02.013 [DOI] [PubMed] [Google Scholar]
- 23.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals. 4th ed. CLSI supplement VET08. Wayne: Clinical Laboratory Standards Institute; 2018 [Google Scholar]
- 25.Magiorakos A-P, Srinivasan A, Carey R, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- 26.Krumperman PH. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl Environ Microbiol. 1983;46(1):165–170. doi: 10.1128/aem.46.1.165-170.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bager RJ, Nesta B, Pors SE, et al. The fimbrial protein FlfA from Gallibacterium anatis is a virulence factor and vaccine candidate. Infect Immun. 2013;81(6):1964–1973. doi: 10.1128/IAI.00059-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson TJ, Danzeisen JL, Trampel D, et al. Genome analysis and phylogenetic relatedness of Gallibacterium anatis strains from poultry. PLoS One. 2013;8(1):e54844. doi: 10.1371/journal.pone.0054844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klima CL, Alexander TW, Hendrick S, McAllister TA. Characterization of Mannheimia haemolytica isolated from feedlot cattle that were healthy or treated for bovine respiratory disease. Can J Vet Res. 2014;78(1):38–45. [PMC free article] [PubMed] [Google Scholar]
- 30.Petrocchi-Rilo M, Gutiérrez-Martín C-B, Pérez-Fernández E, Vilaró A, Fraile L, Martínez-Martínez SJA. Antimicrobial resistance genes in porcine Pasteurella multocida are not associated with its antimicrobial susceptibility pattern. Antibiotics. 2020;9(9):614. doi: 10.3390/antibiotics9090614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yaman S, Sahan Yapicier O. Diagnosis of Gallibacterium anatis in layers: first report in Turkey. Braz J Poultry Sci. 2019:21. doi: 10.1590/1806-9061-2019-1019 [DOI] [Google Scholar]
- 32.Lozica L, Kazazić SP, Gottstein Ž. High phylogenetic diversity of Gallibacterium anatis is correlated with low biosecurity measures and management practices on poultry farms. Avian Pathol. 2020;49(5):467–475. doi: 10.1080/03079457.2020.1765970 [DOI] [PubMed] [Google Scholar]
- 33.Antenucci F, Arak H, Gao J, Allahgadry T, Thøfner I, Bojesen AM. Hydrostatic filtration enables large-scale production of outer membrane vesicles that effectively protect chickens against Gallibacterium anatis. Vaccines. 2020;8(1):40. doi: 10.3390/vaccines8010040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson TJ, Fernandez-Alarcon C, Bojesen AM, Nolan LK, Trampel DW, Seemann T. Complete genome sequence of Gallibacterium anatis strain UMN179, isolated from a laying hen with peritonitis. J Bacteriol. 2011;193(14):3676–3677. doi: 10.1128/JB.05177-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huangfu H, Dong Q, Wang H, et al.Isolation, identification and phylogenetic analysis of Gallibacterium anatis from silky population in Henan Province. Acta Vet. 2018;49(11):2451–2460. [Google Scholar]
- 36.Elbestawy A Studies on Gallibacterium anatis infection in chickens. PhD Fac Vet Med, Alexandria University, Egypt. 2014. [Google Scholar]
- 37.Sorour HK, Atfeehy NMA, Shalaby AG. Gallibacterium anatis infection in chickens and ducks. Assiut Vet Med J. 2015;61(147):80–86. [Google Scholar]
- 38.Nassik S, Tallouzt S, Karbach N, et al. First report of isolation of Gallibacterium anatis from layer chickens in Morocco with decrease in laying performance. Avian Dis. 2019;63(4):727–730. doi: 10.1637/aviandiseases-D-19-00119 [DOI] [PubMed] [Google Scholar]
- 39.Wang C, Robles F, Ramirez S, Riber AB, Bojesen AM. Culture-independent identification and quantification of Gallibacterium anatis (G. anatis) by real-time quantitative PCR. Avian Pathol. 2016;45(5):538–544. doi: 10.1080/03079457.2016.1184743 [DOI] [PubMed] [Google Scholar]
- 40.Soler N, Forterre P. Vesiduction: the fourth way of HGT. Environ Microbiol. 2020;22(7):2457–2460. doi: 10.1111/1462-2920.15056 [DOI] [PubMed] [Google Scholar]
- 41.Rodríguez-Beltrán J, DelaFuente J, Leon-Sampedro R, MacLean RC, San Millan A. Beyond horizontal gene transfer: the role of plasmids in bacterial evolution. Nat Rev Microbiol. 2021;19(6):347–359. doi: 10.1038/s41579-020-00497-1 [DOI] [PubMed] [Google Scholar]
- 42.Michael GB, Bossé JT, Schwarz S. Antimicrobial resistance in Pasteurellaceae of veterinary origin. Microbiol Spectr. 2018;6(3). doi: 10.1128/microbiolspec.ARBA-0022-2017 [DOI] [PubMed] [Google Scholar]
- 43.Ramirez MS, Tolmasky ME. Aminoglycoside modifying enzymes. Drug Resist Updat. 2010;13(6):151–171. doi: 10.1016/j.drup.2010.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]