Abstract
The work is devoted to the study of the structural characteristics of the myeloperoxidase–ceruloplasmin–thrombin complex using small-angle neutron scattering methods in combination with computer modeling, as well as surface plasmon resonance and solid-phase enzyme assay. We have previously shown that the functioning of active myeloperoxidase during inflammation, despite the presence in the blood of an excess of ceruloplasmin which inhibits its activity, is possible due to the partial proteolysis of ceruloplasmin by thrombin. In this study, the myeloperoxidase–ceruloplasmin–thrombin heterohexamer was obtained in vitro. The building of a heterohexamer full-atomic model in silico, considering the glycosylation of the constituent proteins, confirmed the absence of steric barriers for the formation of protein–protein contacts. It was shown that the partial proteolysis of ceruloplasmin does not affect its ability to bind to myeloperoxidase, and a structural model of the heterohexamer was obtained using the small-angle neutron scattering method.
Keywords: Myeloperoxidase, Halogenative stress, Ceruloplasmin, Thrombin, Molecular modeling, Small-angle neutron scattering, Inflammation, Reactive halogen species
Introduction
Myeloperoxidase (MPO) is a dimeric, heme-containing enzyme found in neutrophils and monocytes which takes part in reactions of innate immunity. In the process of neutrophil degranulation, MPO is exposed resulting in the formation of neutrophil extracellular traps (NETs) (Brinkmann and Zychlinsky 2012) and membrane tubulovesicular extensions (or cytomenes) (Galkina et al. 2013). Specific binding of MPO with red blood cells, platelets, neutrophils, endothelial cells, and a number of extracellular matrix components has been extensively studied in the last decade (Kargapolova et al. 2021; Hawkins and Davies 2021). Use of heparin as a soluble competitor significantly increased release of MPO from the extracellular matrix and cell surface (Kargapolova et al. 2021). MPO catalyzes the formation of hypochlorous acid (HOCl) and other reactive halogen species with cytotoxic properties.
In chronic inflammation, reactive halogen species deplete antioxidant reserves and modify cell components, leading to the release of biomarkers of halogenative stress (Hawkins and Davies 2021). Ceruloplasmin (CP), a copper-containing glycoprotein of blood plasma forms a complex with MPO, especially during the acute phase of inflammation; it can inhibit the peroxidase and chlorinating activities of MPO (Park et al. 2000; Segelmark et al. 2003; Sokolov et al. 2015a). Obtaining the 2CP-MPO complex three-dimensional structure and the observation that CP after limited proteolysis loses its ability to inhibit MPO, led to the conclusion that the suppression of MPO activity occurs when the peptide loop connecting the 5th and 6th domains of the CP polypeptide chain blocks the aperture of the MPO heme pocket (Samygina et al. 2013).
The active site of CP does not participate in inhibition of MPO activity, e.g. the apo-form of CP does not interact with MPO, nor inhibit MPO activity. Indeed, removal of copper ions from CP (realized only by strong chelators as cyanide) results in irreversible transition of the protein to a “molten globule” structure (Vachette et al. 2002). Unexpectedly, a synthetic peptide, RPYLKVFNPR, corresponding to 883–892 in CP sequence (loop connecting 5th and 6th domains) is enough for MPO inhibition with IC50 ~ 160 nM, as well as for suppression of neutrophil respiratory burst. We found that this loop of CP is cleaved by thrombin (FIIa) at a noncanonical site, K887-V888 (Sokolov et al. 2015b). Cleavage at this alone resulted not only in loss of CP's inhibitory effect on MPO, but also loss of its suppressive effect on neutrophil respiratory burst (Golenkina et al. 2018). However, FIIa-catalyzed cleavage of R481-S482 (loop connecting 3th and 4th domains) and K887-V888 (loop connecting 5th and 6th domains) can occur, yet without resultant dissociation of fragments from the CP globule. The cleaved fragments are connected by disulphide bridges and hydrophobic interactions. Currently, there is a lack of detailed density map resolution for structural clarification of these loops in published human CP X-ray structures (Samygina et al. 2013).
Considering that all three proteins are glycosylated, and that the type of MPO glycation can affect the degree of its inhibition upon contact with CP (Tjondro et al. 2021), we used full-atomic models accounting for glycosylation to describe protein–protein contacts in this study. We earlier showed a direct interaction between CP and FIIa exosite 2 (Sokolov et al. 2015b). In control experiments, CP did not alter the activity of FIIa. Moreover, FIIa treated by irreversible inhibitors, PMSF or PPACK (so-called suicide inhibitors) did not lose its affinity for CP. In the synovial fluid of patients with rheumatoid arthritis, we found proteolyzed CP (197–1341 nM), active MPO (10.1–36.4 nM), and FIIa (6.7–25.8 nM). However, upon suppression of thrombin activity by hirudin, uncleaved CP was observed in patient synovial fluid, while MPO activity was practically not detected (Sokolov et al. 2015b). Human MPO injected intravenously into rats was detected in complex with rat CP in blood samples 30 min after injection (Sokolov et al. 2007). Complexes including CP and MPO have been detected in plasma and serum samples obtained from patients with acute inflammation and atherosclerosis (Sokolov et al. 2007, 2010). Resistance of the CP-MPO complex to heparin allows purification of the complex from serum obtained from patients with acute inflammation using aminoethyl-agarose and heparin-Sepharose chromatography, followed by mass-spectrometry of tryptic fragments for identification of CP and MPO (Sokolov et al. 2007).
The dissociation constant of the CP-MPO complex was initially estimated to be 7.5 μM (Griffin et al. 1999). However, later study of the inhibition constant for CP as inhibitor of MPO chlorinating activity estimated the value to be 0.46 μM (Sokolov et al. 2015a). In healthy donor plasma, MPO concentrations (about 0.5–1.5 nM) were approximately 1000-fold lower than CP concentrations; with atherosclerosis, however, MPO concentrations were higher, reaching up to 14 nM (Sokolov et al. 2010). Taking into account the high affinity of CP for MPO, and the fact that CP concentration significantly increases (from 3 to 10 µM) during acute inflammation (Gitlin 1988), it remains unclear how MPO can produce markers of halogenative stress with such a large excess of CP. It should be noted that MPO may also exist on surfaces on endothelium, blood cells, or neutrophilic extracellular traps. However, the possibility of interaction of such MPO with CP has not been studied, and we cannot exclude the formation of CP-MPO complexes there during inflammation. It is known that the three proteins (CP, MPO, FIIa) are involved in inflammation and affect each other, hence, the possibility of the existence of their triple complex was indirectly indicated by the presence of pairwise interactions between its components, as described (Sokolov et al. 2015b). Taken together, these data required structural evidence for the possibility of the existence of the complex in solution and clarification of its structural characteristics, if it exists. To study the possibility of complex formation between FIIa, CP, and MPO in solution (considering that FIIa can proteolyze CP), in this study we: compared the affinities of thrombin-proteolyzed CP and intact CP for MPO; compared the binding of FIIa with immobilized partners (CP, MPO, 2CP-MPO complex); and established the structural characteristics of such complexes.
Materials and methods
Materials
The following reagents were used: UNO-Sphere Q (Bio-Rad, USA); sodium chloride (Helicon, Russia); benzamidine-agarose; and Boc-Val-Pro-Arg-MCA (Sigma Aldrich, USA).
Myeloperoxidase (MPO)
Native MPO was obtained from human neutrophil extract by successive chromatography on heparin-Sepharose and phenyl-agarose, followed by gel-filtration on SephacrylS-200 HR, as previously described (Sokolov et al. 2015a). The protein obtained had an A430/A280 ratio of 0.85.
Ceruloplasmin (CP)
A stable preparation of CP (A610/A280 > 0.045) was obtained from healthy donor blood plasma, stabilized by heparin, using chromatography on UNO-Sphere Q and neomycin–agarose, as previously reported (Sokolov et al. 2015b). Protein integrity and purity were assessed by denaturing SDS-PAGE indicating: the CP preparation yielded a 132 kDa fragment with at least 98% purity.
Alpha-thrombin (FIIa)
FIIa was activated from prothrombin by filtration through a column with agarose gel with immobilized ecarin. Prothrombin was isolated from blood plasma during purification of CP by ion exchange chromatography on UNO-Sphere Q and affinity chromatography on neomycin–agarose. After activation of prothrombin by ecarin, FIIa was purified by affinity chromatography on benzamidine-agarose. According to SDS-PAGE data, the resulting FIIa consisted of 31 and 6 kDa chains. It was characterized by a Michaelis constant of 37.5 nM for the specific fluorogenic substrate Boc-Val-Pro-Arg-MCA (100 mM NaCl, 10 mM CaCl2, 50 mM Tris–HCl, pH 7.4, 0.66% PEG-6000, 0.16–320 nM Boc-Val-Pro-Arg-MCA, 1 mg/L FIIa), as described previously.
Proteolysis of ceruloplasmin with thrombin
CP proteolysis was performed in PBS by incubation of the protein with FIIa (50:1, w/w) at 37 °C for 24 h. Traces of FIIa were removed by affinity chromatography on benzamidine-agarose. Cleavage of CP was monitored by SDS-PAGE.
Surface plasmon resonance
Surface plasmon resonance (SPR) experiments were carried out at 25 °C using a Biacore-X100 (GE Healthcare) instrument with two flow cells equipped with a research-grade carboxymethylated dextran chip (CM5). MPO was immobilized using amine coupling chemistry at pH 5.0 (BR-1000-50, GE Healthcare). The sensor chip was first activated with a 1:1 mixture of 0.2 M N-ethyl-N′-dimethylaminopropyl carbodiimide and 0.2 M N-hydroxysuccinimide. It was then treated with an MPO solution (10 μg/mL in 10 mM sodium acetate buffer, pH 5.0) injected at a flow rate of 5 μL/min for 18 min. Unreacted groups on the sensor chip were blocked by reaction with 1 M ethanolamine at pH 8.5. Final immobilization levels of 10,985 resonance units (RU) were obtained, corresponding to approximately 11 ng of bound MPO per mm2. Thereafter, increasing concentrations of CP were injected over the sensor chip coated with MPO at a flow rate of 10 μL/min in HBS-P + buffer, pH 7.5, containing 10 mM HEPES, 150 mM NaCl and 0.05% polyoxyethylene sorbitan. Each reaction cycle consisted of four steps: (1) HBS-P + injection for 1 min; (2) CP (125, 250, 500, 1000, or 2000 nM) injection for 1 min; (3) dissociation for 5 min in HBS-P+; and (4) 1-min regeneration with 1 M NaCl, 10 mM ethanolamine at pH 8.5. The reference flow cell, with no MPO immobilized, was used as a control to evaluate non-specific binding (< 1% of Rmax) of analyte to the sensor surface. The resulting control curves were subtracted from the corresponding binding curves recorded at each analyte concentration. Data analysis was performed using Biacore X100 Evaluation Software (Version 2.0.1).
Binding of FIIa with immobilized partner proteins (CP, MPO, CP-MPO-MPO-CP)
For immobilization on solid phase, 100 µL of each protein solution was dropped into its corresponding well in high-binding polystyrene plates: 400 nM CP; 200 nM MPO; 400 nM CP and 200 nM MPO (complex); or 600 nM BSA in PBS. Immobilization was incubated for 2 h at 37 °C with continuous shaking at 350 rpm. Next, three washes were performed with PBS containing 0.05% Tween-20 (PBST), and 100 µL of 1% BSA in PBST was added to the wells. After 1 h of blocking and three PBST plate washes, 100 µL of FIIa at various concentrations (0, 0.78, 1.56, 3.12, 6.25, 12.5, 25, or 50 nM) was added into wells corresponding to each immobilized protein experimental variant (CP, MPO, CP-MPO-MPO-CP). The plate was again incubated for 1 h at 37 °C, followed by three PBST washes. Next, 100 µL of fluorogenic substrate solution was added (12 µM Boc-Val-Pro-Arg-MCA, Sigma-Aldrich, 100 mM NaCl, 10 mM CaCl2, 50 mM Tris–HCl, pH 7.4). The plate again was incubated for 1 h at 37 °C with continuous shaking at 350 rpm. The fluorescent intensity of released MCA was measured at 455–465 nm (excitation at 375–385 nm) in the CLARIOstar plate reader (BMG LABTECH, Germany) using fluorescent intensity mode. Wells with immobilized BSA were used as controls of non-specific binding of FIIa to solid phase.
To characterize the binding sites on CP, dimeric MPO, and CP-MPO-MPO-CP complex, we used a model in which the effect of the cooperative binding is taken into account. Curves were processed using OriginPro software and were fitted by the Hill equation:
| 1 |
where I is the intensity, arb. u., corresponding to FIIa activity; C is the FIIa concentration, nM; Vmax is the max velocity (maximal intensity), arb. u.; k is the Michaelis constant (concentration of FIIa causing the filling of half of the binding sites), nM; n is the number of cooperative sites. The equilibrium dissociation constant was calculated as Kd = kn, nM.
Small angle scattering (SANS)
For obtaining small-angle neutron scattering spectra, measurements were carried out on an IBR-2 high-flux reactor with a YuMO spectrometer (JINR, Dubna, Russia) using two detectors and the time-of-flight method (Kuklin et al. 2005, 2011) in the range of from 0.006 to 0.3 Å−1. Primary processing of experimental data was carried out in the SAS program (Soloviev et al. 2003), allowing normalization of the obtained spectrum to an independent vanadium scatterer, subtracting the background sample data (Ostanevich 1988). The sample/detector distance was 12.96 m and 5.3 m for two detectors. Sample solutions in buffer prepared in D2O were measured at 20 °C in a Hellma quartz cuvette (2 mm). Temperature was controlled by a Lauda thermostat. The spectrometer was controlled by software (Kirilov et al. 2004) during the experiment. Measurements were carried out for 60 min for all systems under study.
Protein solutions
All proteins and the triple complex (FIIa-CP-dimeric MPO = 2:2:1) were subjected to gel filtration in HBS on Sephacryl S-300 HR (1 × 150 cm). Eluted proteins were concentrated using VivaSpin-20 (Sartorius) with simultaneous substitution of water for D2O. We studied solutions of CP (12.3 mg/mL), MPO (2.8 mg/mL), and FIIa (2 mg/mL). All buffers were prepared using D2O. Judging by their resolution in PAGE, the concentrations of proteins in the complex were 1.9 mg/mL, 1.1 mg/mL, and 0.5 mg/mL, respectively. In addition, the A280/A430/A610 ratio measured was in agreement with the stoichiometry (FIIa-CP-dimeric MPO = 2:2:1).
Processing of SANS spectral data
SANS spectra were processed using OriginPro software and the ATSAS package to estimate the molecular weights of proteins and the complex, as described (Svergun and Koch 2003). Molecular weight M, Da was calculated using the formula:
| 2 |
where I0 is the zero angle intensity, cm−1, QI is the Porod volume, cm−1 Å−3, – volume of 1 Da of protein, Å3Da−1.
At first, the incoherent scattering was determined for each spectrum: the data were plotted in coordinates Iq4 vs q4 and approximated by a linear plot ; the obtained A constants were subtracted from the corresponding spectra. Next, the I0 and radius of gyration Rg, Å were found using the GNOM program (ATSAS package) (Svergun 1992) for a monodisperse system. The maximum diameters of the particles were estimated as 130, 145, 65, and 255 Å for CP, MPO, FIIa, and as a complex, respectively. Porod volume QI was calculated using OriginPro as the area under the corresponding curve, presented in the coordinates Iq2 vs q.
Building a hexameric model
In view that mature myeloperoxidase molecule consists of two identical MPO monomers bridged by a disulfide bond we regarded the overall structure of the complex “FIIa-CP-MPO-MPO-CP-FIIa” as a hexamer. Our models for this hexameric structure were proposed on the basis of 3F9P structure (MPO-MPO) (Carpena et al. 2008) from the PDB database (http://rcsb.org) and published structures (Samygina et al. 2013; Sokolov et al. 2015b) using PyMOL (Schrödinger LLC 2020), MODELLER (Eswar et al. 2008), and GROMACS (Abraham et al. 2015) software. Glycosylation was constructed using the Glycamgmml software toolkit. Docking was performed using hdocklite, while only FIIa were considered as mobile molecules. Further, from all possible FIIa positions, only those that are sterically compatible with the glycosylation of CP-MPO were selected; it was also assumed that the binding sites of both FIIa molecules are symmetric in the heterohexamer structure. The resulting structures were used for the theoretical calculation of SANS spectra. Theoretical SANS spectra were calculated using the CRYSON programs from the ATSAS package (Franke et al. 2017).
Ab initio modeling
Ab initio modeling based on the SANS spectrum of the complex was performed according to the ATSAS manual using DAMMIN (Svergun 1999). The DAMMIN input file was obtained using GNOM as described earlier (Sect. 2.5). Twenty independent structures were obtained with the following parameters: (P2) dimer symmetry with unknown anisometry. The resultant set of structures possessed mirror symmetry. For that reason, only 10 structures with symmetries matching the full-atomic structure were chosen for further analysis. In the next step, the set of structures was processed with DAMAVER (Volkov and Svergun 2003) as follows. We used damsel to find the most probable model, then damsup aligned all the structures to the most probable one. Next, damaver averaged all models and computed the probability map. Finally, we used damstart to create a model, based on the damaver one, for further use in DAMMIN as an initial approximation. We repeated DAMMIN ab initio modeling using the damstart structure as the starting point and received 20 new structures. It should be noted that all of them possess the same mirror symmetry as the full-atomic structure, and they fit the SANS experimental data. We processed these models with DAMAVER as described earlier to receive the probability map using damaver.
Results and discussion
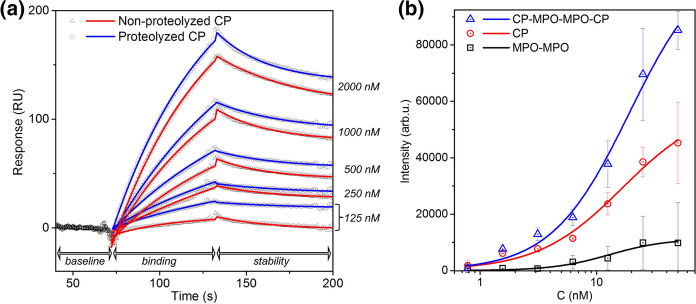
To test the hypothesis that cleavage of CP may affect CP/MPO interaction, we analyzed parameters which might affect the formation of a stable complex between MPO and thrombin-proteolyzed CP. We also used surface plasmon resonance (SPR) to study the binding of both the intact and proteolyzed CP to immobilized MPO. The SPR data are presented in Fig. 1a and Table 1. The calculated equilibrium association rate constants (kon = 12,890 ± 122 M−1 s−1 for proteolyzed CP-MPO and 11,520 ± 410 M−1 s−1 for non-proteolyzed CP-MPO) are within the range of experimental error. This indicates that proteolysis of CP does not change its ability to bind MPO.
Fig. 1.
a Sensograms of non-proteolyzed CP (red) and proteolyzed CP (blue) interaction with immobilized MPO. b Dependence of FIIa activity on FIIa concentration after binding solid-phase immobilized CP, dimeric MPO or CP-MPO-MPO-CP complex
Table 1.
Kinetic parameters characterizing MPO interaction (immobilized on CM5-chip) with intact or proteolyzed CP, calculated from SPR data by Biacore X-100 software
| Analyte | kon, M−1 s−1 | koff, 10–3 s−1 | Kd, nM | Rmax, RU |
|---|---|---|---|---|
| Intact (non-proteolyzed) CP | 11,520 ± 410 | 503 ± 15 | 436 ± 37 | 246 ± 3.5 |
| CP protolyzed by FIIa | 12,890 ± 122 | 422 ± 77 | 327 ± 63 | 245 ± 1.5 |
kon: equilibrium association rate constant, koff: dissociation rate constant, Kd: equilibrium dissociation constant, Rmax: analyte binding capacity
To compare FIIa binding with solid phase immobilized CP, MPO, or 2 CP-dimeric MPO complex (“CP-MPO-MPO-CP”), they were immobilized on a polystyrene plate using BSA as a non-specific binding control. Plates were then incubated with different concentrations of FIIa. After washing off unbound FIIa, activity was detected using a specific fluorogenic substrate. The dependence of substrate hydrolysis product (fluorescence) on FIIa concentration (using different immobilized proteins) is shown in Fig. 1b. It is seen that FIIa binding to MPO does not differ from nonspecific binding to BSA. Therefore, we compared FIIa binding to CP and to the CP-MPO-MPO-CP complex. The Hill equation (1) was used for curve fitting to take into account the possibility of cooperative binding (Table 2). The obtained n values are close to 1 in both cases, while the k and Kd values are quite close. This supports the hypothesis that FIIa can bind to CP in complex with dimeric MPO; it also shows that the FIIa active site remains available for catalysis.
Table 2.
Parameters characterizing cooperative binding of thrombin (FIIa) to solid-phase immobilized analytes, obtained using the Hill equation
| Analyte (immobilized) | Vmax, arb. u | k, nM | n | Kd, nM |
|---|---|---|---|---|
| CP-MPO-MPO-CP | 111,580 ± 11,920 | 19 ± 4 | 1.3 ± 0.2 | 46 ± 17 |
| CP | 58,960 ± 11,280 | 17 ± 8 | 1.2 ± 0.3 | 30 ± 20 |
| MPO-MPO** | 11,260 ± 5090 | 13 ± 10 | 1.8 ± 1.6 | 101 ± 228 |
Vmax: max velocity (maximal intensity); k: Michaelis constant; n: number of cooperative sites; and Kd: equilibrium dissociation constant
MPO in solution exists as a dimer; mature MPO consists of two protomers connected by a disulfide bond
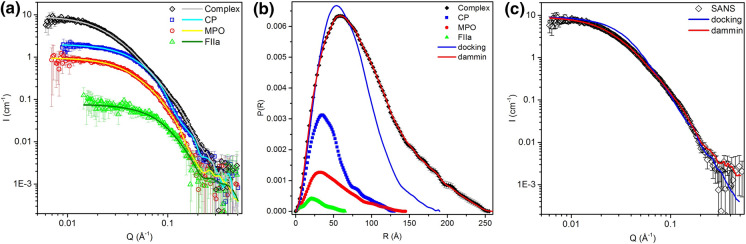
Complex of FIIa-CP-MPO-MPO-CP-FIIa was prepared as described in the Materials and Methods, and small-angle neutron scattering (SANS) spectra of the FIIa-CP-MPO-MPO-CP-FIIa complex in solution were obtained. Individual spectra for pure protein solutions (CP, MPO-MPO, FIIa), as well as for their complex, are presented in Fig. 2a (scatter plots). The scattering spectrum of the complex does not correspond to the arithmetic sum of its components’ spectra. This indicates the emergence of a new structure and interaction among its components.
Fig. 2.
Small-angle neutron scattering spectra (a) and calculated pairwise distributions P(R) (b) of solutions of ceruloplasmin (CP, squares), myeloperoxidase (MPO-MPO, circles), thrombin (FIIa, triangles), and FIIa-CP-MPO-MPO-CP-FIIa complex (diamonds). Solid lines on a show the approximation based on P(R) distribution. Solid lines on (b) show P(R) functions obtained based on theoretical docking spectrum (blue line, ‘docking’), and ab initio modeling spectrum (red line, ‘dammin’). c Comparison of the theoretical neutron scattering spectrum for the FIIa-CP-MPO-MPO-CP-FIIa model, obtained as a result of docking taking into account complete glycosylation (‘docking’, blue line), with the experimental spectrum (black circles, ‘SANS’). The results of ab initio modeling using the DAMMIN program (ATSAS package) are shown as a red line (‘dammin’). As a result of ab initio modeling, twenty structures were obtained; all of them had nearly identical spectra shown by the red line. I (cm−1): scattering intensity, Q (Å): magnitude of the momentum transfer, R (Å): distance
To calculate the molecular weight M of the complex from SANS data, formula (2) was used. The values for I0 ang Rg were found using the GNOM program in ATSAS (Table 3) (Svergun 1992). The resulting pairwise distribution P(R) of each sample is presented in Fig. 2b. The theoretical spectra, calculated based on P(R) functions, are presented in Fig. 2a (solid lines). In every case, the solutions were rated by GNOM as 'good'. The obtained gyration radii Rg for CP (36 Å) and MPO (40 Å) corresponded to the theoretical values calculated from the available spatial structure models (Carpena et al. 2008; Samygina et al. 2013). The FIIa-CP-MPO-MPO-CP-FIIa complex gyration radius according to SANS data was about 72 Å.
Table 3.
Parameters of scattering objects, calculated based on SANS spectra
| Sample | Rg, Å | I0, cm−1 | QI × 10–5, cm−1 Å−3 |
M, kDa | Mtheor, kDa |
|---|---|---|---|---|---|
| CP | 36.1 ± 0.5 | 2.00 ± 0.02 | 21.31 ± 2.00 | 146 ± 19 | 132 |
| MPO-MPO | 40.2 ± 0.7 | 0.96 ± 0.01 | 9.91 ± 1.69 | 153 ± 26 | 150 |
| FIIa | 20.7 ± 0.6 | 0.077 ± 0.003 | 3.51 ± 1.74 | 35 ± 17 | 37 |
| FIIa-CP-MPO-MPO-CP-FIIa | 72.0 ± 1.0 | 9.17 ± 0.14 | 30.22 ± 1.98 | 479 ± 32 | 488 |
Rg: radius of gyration, I0: zero angle intensity, QI: Porod volume, M: experimental molecular weight, Mtheor: theoretical molecular weight, calculated based on amino acid sequence
The calculated molecular weight of the complex was about 480 kDa. This corresponds to the sum of the molecular weights of its monomeric components: kDa. The observed difference (about 10 kDa) is within experimental error and corresponds to the molecular weight of the FIIa-CP-MPO-MPO-CP-FIIa heterohexamer. It should be noted that the molecular weights of the complex's individual components (CP, MPO, FIIa), as calculated by this method from the corresponding scattering spectra, also correspond to the expected ones.
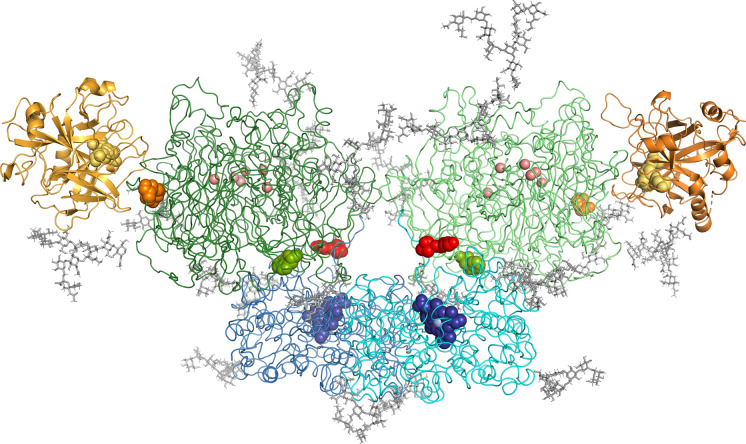
The results make it possible to confidently conclude that the considered complex exists in solution, with good correspondence between the complex's observed molecular weight in solution and the theoretically predicted one. Small-angle neutron scattering also enables low resolution determination of the complex's spatial structure. As a starting point, we used a full-atomic model obtained as described in the Materials and Methods. In brief, using protein database structures (MPO-MPO, CP-MPO complex) and literature data (glycosylation of CP and MPO), models of the CP-MPO-MPO-CP heterotetrameric complex were obtained. Next, using computational docking, a model supplemented with two FIIa molecules was obtained (Fig. 3). It is seen that two CP molecules contact the heme pockets in MPO-MPO (hemes presented as blue spheres). The p-phenylene diamine-binding site in CP, the affinity towards which is modified upon MPO binding, is shown as green spheres. Such a model is in agreement with the data obtained previously that showed a reciprocal effect of CP and MPO on each other's enzymatic activities (Sokolov et al. 2015a). It was suggested that CP does not inhibit the proteolytic activity of FIIa (Sokolov et al. 2015b). Our model supports that notion, showing no steric interaction of CP with the catalytic sites (yellow spheres) of either FIIa molecule (Fig. 3).
Fig. 3.
Structural model of the FIIa-CP-MPO-MPO-CP-FIIa complex obtained as a result of docking, taking into account glycosylation. Key: MPO: blue; CP: green; FIIa: gold; and carbohydrates: grey. Heme is shown as blue spheres in MPO molecules (iron atoms are presented as light blue spheres). The active center of FIIa (a.a.r. S525, H363, D419) is shown as yellow spheres. Copper atoms of CP are shown as pink spheres. Substrate-binding in CP (W669) is shown as green spheres. Amino acid residue flanking loops that contain FIIa-cleavage sites R481-S482 and K887-V888 of CP are shown as orange and red spheres, respectively
In acute inflammation, MPO can be exposed on the surfaces of blood cells, endothelial cells, and NETs. The pathophysiological role of the latter is connected with procoagulant activity and immunothrombosis that characterize severe cases of COVID-19 (Ackermann et al. 2021). Several future research approaches are planned: elucidation of the possibility of formation of complexes involving interaction of CP and FIIa with MPO exposed on NETs; and estimation of the activities of such complexes.
Figure 2c is a comparison of the corresponding neutron scattering spectral data (black circles, experimental) and theoretical data (blue line, model-based). Even though the theoretical spectrum (‘docking’) was close to the experimental one, complete coincidence was not observed. At the same time, the gyration radius of the full-atomic model turned out to be less than the gyration radius calculated based on SANS spectra (61 Å vs 72 Å). To analyze the reasons for the discrepancy, the theoretical and experimental pairwise distribution P(R) curves were compared (Fig. 2b, solid lines) using black circles and a blue line, respectively.
The discrepancy in the right side of the P(R) function curve (R > ~ 75 Å) may be associated with differences between natural/theoretical protein glycosylation or differences in the spatial position of carbohydrate chains in solution. It could be asserted that the complex's real structure in solution possesses a more extended structure than that obtained by computer modeling (docking).
To visualize differences between theoretical SANS spectra (calculated on the basis of the full-atomic structure) and experimental SANS spectra, we performed ab initio modeling with dummy atoms using DAMMIN (Svergun 1999) from the ATSAS package. We obtained 20 structures which were processed using DAMAVER (Volkov and Svergun 2003) (ATSAS package) to combine them. We then used the resulting structure as the starting point to perform ab initio modeling with DAMMIN one more time. All 20 structures in the second set were highly similar to each other and possessed the theoretical spectra shown in Fig. 2c (red line), with the P(R) function as shown (Fig. 2b, red line). They can be considered a good fit. A sample of such an ab initio structure is shown in Fig. 4.
Fig. 4.
The results of ab initio modeling performed using DAMMIN followed by DAMAVER processing (‘Ab initio structure’ and ‘Probability map’) in comparison with full-atomic structure. The right and left sides of the figure represent different projections
After the second ab initio modeling stage, all twenty models were also aligned and averaged in order to compute the probability map (Fig. 4). Based on P(R) analysis (Fig. 2b), it can be concluded that the ‘dummy’ model possesses a more elongated structure (better matching the experimental data), while the full-atomic model is more compact, which is consistent with the observed discrepancy in their Rg. An overlay of the ‘dummy’ and full-atomic models makes vivid such differences (Fig. 4).
Considering that the final, full-atomic structure was obtained using a docking procedure, it is not surprising that the natural hexamer in solution (i.e. in dynamics) may feature a more expanded or extended conformation, even though the complex's molecular weight and stoichiometry (FIIa-CP-MPO-MPO-CP-FIIa) remain unchanged.
It should be noted that two CP molecules in the full-atomic hexameric model do not contact each other, in contrast to the data obtained (Samygina et al. 2013) for CP-MPO complex. Such discrepancy may arise from the noticeable differences between the studies by (X-ray crystallography versus SANS), which necessitates subsequent modeling (Lebedev et al. 2021).
Conclusion
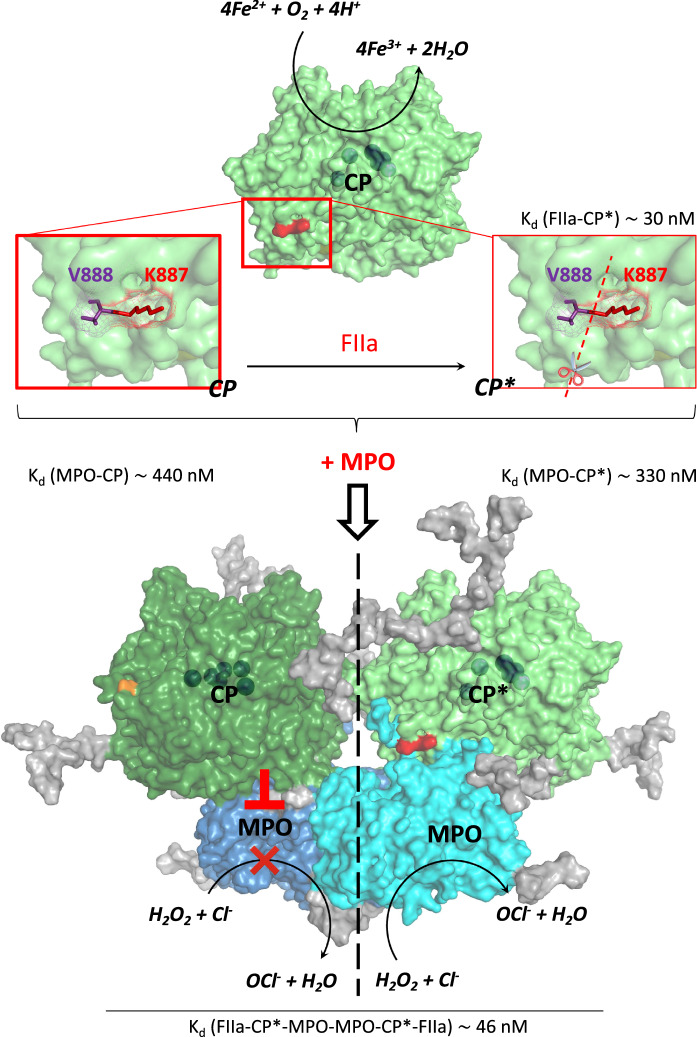
Small-angle neutron scattering of the proteins under study proved to be the method of choice, enabling examination of their complex under quasi-natural conditions while achieving sufficient spatial resolution. Our data show that FIIa after exposure to ceruloplasmin does not dissociate from the stable heterohexameric complex FIIa-CP-MPO-MPO-CP-FIIa; this may cause partial FIIa-induced proteolysis of CP. This, in turn, prevents CP from inhibiting MPO, but does not significantly affect CP/MPO affinity (Fig. 5). Finding such a complex in solution enables a better specific understanding of the mechanism behind the involvement of MPO and CP in inflammation, including the link between inflammation and blood coagulation. Since both active FIIa and active MPO can form a complex with CP, testing for MPO activity and CP intactness seems a promising approach to assessing the severity of inflammation.
Fig. 5.
Scheme of heterohexameric complex components showing affinities and regulation of MPO chlorinating activity by intact or FIIa-proteolzyed ceruloplasmin (CP and CP*, respectively). A loop in CP containing a non-canonical thrombin (FIIa) cleavage site (K887-V888, marked red) interacts with the entrance at MPO's heme pocket (Samygina et al. 2013). After cleavage, CP* loses the ability to inhibit MPO's chlorinating activity (Sokolov et al. 2008), yet it is still able to interact with MPO with an affinity close to that of intact CP. The affinity of FIIa for complex (CP*-MPO-MPO-CP*) and already proteolzyed ceruloplasmin (CP*) are similar
Acknowledgements
This work was supported by Russian Foundation for Basic Research (No 18-015-00241). The authors thank Dr. Vladimir V. Volkov and Dr. Dmitry V. Lebedev for fruitful discussion.
Author contributions
YAZ: Investigation, Formal analysis, Writing—original draft, Visualization; VVE: Investigation, Writing—original draft, Supervision; AVS—Investigation, Writing—original draft, Funding acquisition; AVS—Formal analysis, Resources; OII, and YEG—Investigation, Formal analysis; VAK, NPG, NDF, and ABB—Investigation; ESR: Writing—original draft, Writing—review & editing; VBV: Funding acquisition, Writing—review & editing.
Funding
This work was supported by Russian Foundation for Basic Research (No 18-015-00241).
Declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abraham MJ, Murtola T, Schulz R, et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. doi: 10.1016/J.SOFTX.2015.06.001. [DOI] [Google Scholar]
- Ackermann M, Anders H-J, Bilyy R, et al. Patients with COVID-19: in the dark-NETs of neutrophils. Cell Death Differ. 2021;28:3125–3139. doi: 10.1038/s41418-021-00805-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann V, Zychlinsky A. Neutrophil extracellular traps: Is immunity the second function of chromatin? J Cell Biol. 2012;198:773–783. doi: 10.1083/jcb.201203170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpena X, Fita I, Obinger C (2008) 3F9P: crystal structure of myeloperoxidase from human leukocytes. https://www.wwpdb.org/pdb?id=pdb_00003f9p. Accessed 20 Jan 2022
- Eswar N, Eramian D, Webb B, et al (2008) Protein structure modeling with MODELLER. In: Methods in molecular biology (Clifton, N.J.). pp 145–159 [DOI] [PubMed]
- Franke D, Petoukhov MV, Konarev PV, et al. ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J Appl Crystallogr. 2017;50:1212–1225. doi: 10.1107/S1600576717007786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkina SI, Fedorova NV, Stadnichuk VI, Sud’ina GF Membrane tubulovesicular extensions (cytonemes) Cell Adh Migr. 2013;7:174–186. doi: 10.4161/cam.23130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin JD. Transcriptional regulation of ceruloplasmin gene expression during inflammation. J Biol Chem. 1988;263:6281–6287. doi: 10.1016/S0021-9258(18)68783-6. [DOI] [PubMed] [Google Scholar]
- Golenkina E, Viryasova G, Galkina S, et al. Fine regulation of neutrophil oxidative status and apoptosis by ceruloplasmin and its derivatives. Cells. 2018;7:8. doi: 10.3390/cells7010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin SV, Chapman PT, Lianos EA, Lockwood CM. The inhibition of myeloperoxidase by ceruloplasmin can be reversed by anti-myeloperoxidase antibodies. Kidney Int. 1999;55:917–925. doi: 10.1046/j.1523-1755.1999.055003917.x. [DOI] [PubMed] [Google Scholar]
- Hawkins CL, Davies MJ. Role of myeloperoxidase and oxidant formation in the extracellular environment in inflammation-induced tissue damage. Free Radic Biol Med. 2021;172:633–651. doi: 10.1016/j.freeradbiomed.2021.07.007. [DOI] [PubMed] [Google Scholar]
- Kargapolova Y, Geißen S, Zheng R, et al. The enzymatic and non-enzymatic function of myeloperoxidase (MPO) in inflammatory communication. Antioxidants. 2021;10:562. doi: 10.3390/antiox10040562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirilov AS, Litvinenko EI, Astakhova NV, et al. Evolution of the SONIX software package for the YuMO spectrometer at the IBR-2 reactor. Instruments Exp Tech. 2004;47:334–345. doi: 10.1023/B:INET.0000032899.51622.1e. [DOI] [Google Scholar]
- Kuklin AI, Islamov AK, Gordeliy VI. Scientific reviews: two-detector system for small-angle neutron scattering instrument. Neutron News. 2005;16:16–18. doi: 10.1080/10448630500454361. [DOI] [Google Scholar]
- Kuklin AI, Soloviov DV, Rogachev AV, et al. New opportunities provided by modernized small-angle neutron scattering two-detector system instrument (YuMO) J Phys Conf Ser. 2011;291:012013. doi: 10.1088/1742-6596/291/1/012013. [DOI] [Google Scholar]
- Lebedev DV, Egorov VV, Shvetsov AV, et al. Neutron scattering techniques and complementary methods for structural and functional studies of biological macromolecules and large macromolecular complexes. Crystallogr Rep. 2021;66:242–253. doi: 10.1134/S1063774521020103. [DOI] [Google Scholar]
- Ostanevich YM. Time-of-flight small-angle scattering spectrometers on pulsed neutron sources. Makromol Chemie Macromol Symp. 1988;15:91–103. doi: 10.1002/masy.19880150107. [DOI] [Google Scholar]
- Park YS, Suzuki K, Mumby S, et al. Antioxidant binding of caeruloplasmin to myeloperoxidase: myeloperoxidase is inhibited, but oxidase, peroxidase and immunoreactive properties of caeruloplasmin remain intact. Free Radic Res. 2000;33:261–265. doi: 10.1080/10715760000301421. [DOI] [PubMed] [Google Scholar]
- Samygina VR, Sokolov AV, Bourenkov G, et al. Ceruloplasmin: macromolecular assemblies with iron-containing acute phase proteins. PLoS ONE. 2013 doi: 10.1371/journal.pone.0067145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger LLC (2020) The PyMOL molecular graphics system, Version 2.4. https://pymol.org/2/
- Segelmark M, Persson B, Hellmark T, Wieslander J. Binding and inhibition of myeloperoxidase (MPO): a major function of ceruloplasmin? Clin Exp Immunol. 2003;108:167–174. doi: 10.1046/j.1365-2249.1997.d01-992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolov AV, Pulina MO, Ageeva KV, et al. Interaction of ceruloplasmin, lactoferrin, and myeloperoxidase. Biochem. 2007;72:409–415. doi: 10.1134/S0006297907040074. [DOI] [PubMed] [Google Scholar]
- Sokolov AV, Ageeva KV, Pulina MO, et al. Ceruloplasmin and myeloperoxidase in complex affect the enzymatic properties of each other. Free Radic Res. 2008;42:989–998. doi: 10.1080/10715760802566574. [DOI] [PubMed] [Google Scholar]
- Sokolov AV, Ageeva KV, Cherkalina OS, et al. Identification and properties of complexes formed by myeloperoxidase with lipoproteins and ceruloplasmin. Chem Phys Lipids. 2010;163:347–355. doi: 10.1016/j.chemphyslip.2010.02.002. [DOI] [PubMed] [Google Scholar]
- Sokolov AV, Kostevich VA, Zakharova ET, et al. Interaction of ceruloplasmin with eosinophil peroxidase as compared to its interplay with myeloperoxidase: reciprocal effect on enzymatic properties. Free Radic Res. 2015;49:800–811. doi: 10.3109/10715762.2015.1005615. [DOI] [PubMed] [Google Scholar]
- Sokolov AV, Acquasaliente L, Kostevich VA, et al. Thrombin inhibits the anti-myeloperoxidase and ferroxidase functions of ceruloplasmin: relevance in rheumatoid arthritis. Free Radic Biol Med. 2015;86:279–294. doi: 10.1016/j.freeradbiomed.2015.05.016. [DOI] [PubMed] [Google Scholar]
- Soloviev AG, Stadnik A V, Kuklin AI, et al (2003) SAS. The package for small-angle neutron scattering data treatment. Version 2.4. Long Write-Up and User’s Guide. 22
- Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr. 1992;25:495–503. doi: 10.1107/S0021889892001663. [DOI] [Google Scholar]
- Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI, Koch MHJ. Small-angle scattering studies of biological macromolecules in solution. Reports Prog Phys. 2003;66:1735–1782. doi: 10.1088/0034-4885/66/10/R05. [DOI] [Google Scholar]
- Tjondro HC, Ugonotti J, Kawahara R, et al. Hyper-truncated Asn355- and Asn391-glycans modulate the activity of neutrophil granule myeloperoxidase. J Biol Chem. 2021 doi: 10.1074/jbc.RA120.016342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vachette P, Dainese E, Vasyliev VB, et al. A key structural role for active site type 3 copper ions in human ceruloplasmin. J Biol Chem. 2002;277:40823–40831. doi: 10.1074/jbc.M207188200. [DOI] [PubMed] [Google Scholar]
- Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889803000268. [DOI] [PMC free article] [PubMed] [Google Scholar]