Abstract
Physiological studies have shown that Streptomyces rimosus produces the polyketide antibiotic oxytetracycline abundantly when its mycelial growth is limited by phosphate starvation. We show here that transcripts originating from the promoter for one of the biosynthetic genes, otcC (encoding anhydrotetracycline oxygenase), and from a promoter for the divergent otcX genes peak in abundance at the onset of antibiotic production induced by phosphate starvation, indicating that the synthesis of oxytetracycline is controlled, at least in part, at the level of transcription. Furthermore, analysis of the sequences of the promoters for otcC, otcX, and the polyketide synthase (otcY) genes revealed tandem repeats having significant similarity to the DNA-binding sites of ActII-Orf4 and DnrI, which are Streptomyces antibiotic regulatory proteins (SARPs) related to the OmpR family of transcription activators. Together, the above results suggest that oxytetracycline production by S. rimosus requires a SARP-like transcription factor that is either produced or activated or both under conditions of low phosphate concentrations. We also provide evidence consistent with the otrA resistance gene being cotranscribed with otcC as part of a polycistronic message, suggesting a simple mechanism of coordinate regulation which ensures that resistance to the antibiotic increases in proportion to production.
Oxytetracycline (5′-hydroxytetracycline; OTC), a polyketide antibiotic (48) effective against a broad spectrum of microorganisms, is produced by strains of Streptomyces rimosus (for reviews, see references 5 and 27). Environmental factors, such as carbon and nutrient sources, temperature, and method of cultivation can affect the timing and extent of production of this antibiotic, as has been found for many other secondary metabolites (30, 35). However, OTC production is particularly sensitive to phosphate; i.e., OTC can be produced abundantly only in media containing levels of phosphate lower than that required for rapid mycelial growth (3, 4). Moreover, by starving cultures of only phosphate, it is possible to induce abundant OTC production by S. rimosus, including the Pfizer production lineage derived from strain 4018 (42). To date, the molecular basis of phosphate regulation of OTC production by S. rimosus has not been investigated beyond showing that the activity of one of the biosynthetic enzymes, anhydrotetracycline oxygenase, is lower in mycelia grown in excess phosphate (2, 8).
The cellular activity of enzymes associated with the production of other antibiotics has also been found to be regulated by phosphate; these include p-aminobenzoate (PABA) synthase from the candicidin producer S. griseus and deacetoxycephalosporin C synthase from S. clavuligerus and Cephalosporium acremonium (for a review, see reference 31). The structural gene (pabS) and promoter for PABA synthase have been cloned and characterized (18, 19, 41). Comparison of the cellular levels of transcripts originating from the pabS promoter has revealed that they are higher when phosphate is limiting, suggesting that the cellular activity of PABA synthase is controlled, at least in part, at the level of transcription (1). For deacetoxycephalosporin C synthase from S. clavuligerus and C. acremonium, there is evidence that the catalytic activity of this enzyme is inhibited by growth in medium containing high levels of phosphate (55), indicating that transcription may not be the only level at which regulation occurs.
The otc biosynthetic genes are clustered and are flanked by two resistance genes in a 34-kb segment of the S. rimosus chromosome (9) that is sufficient to confer OTC production when introduced on a plasmid into the heterologous hosts S. albus and S. lividans (6). Much of the otc cluster, including a 6.6-kb region (Fig. 1) that contains the gene encoding anhydrotetracycline oxygenase (otcC), has been sequenced (27). Upstream and reading in the direction opposite that of otcC are two genes (otcX-orf1 and orf2) with unknown functions. Given this genetic organization, we considered it likely that any elements controlling the transcriptional initiation of otcC would be located in the intergenic region between otcC and otcX-orf1. Here we report the mapping of promoters within this region and provide evidence that they are activated, when mycelial growth is limited by phosphate starvation, by a transcriptional factor related to ActII-Orf4 and DnrI of the actinorhodin and daunorubicin clusters, respectively (16, 46). This switch may also regulate OTC resistance, as the otcC transcript appears to be polycistronic and to include the otrA gene, whose product, a homologue of elongation factor G (14), protects ribosomes from translational arrest (40). In addition, we show that the three major promoters of the polyketide synthase complex (otcY) share common regulatory motifs with otcC.
FIG. 1.
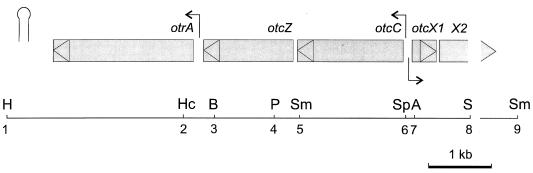
Genetic organization at the otrA end of the otc cluster. Shaded boxes indicate the positions of identified genes. The triangles within each box show the direction of translation. The stem-loop structure indicates the position of a rho-independent terminator (10), and the arrows (→) indicate the positions of the otrAp1 promoter (14) and the otcCp1 and otcXp1 promoters identified here. Only the restriction sites mentioned in the text are shown: H, HindIII; Hc, HincII; B, BamHI; P, PstI; Sm, SmaI; Sp, SphI; A, AvaI; and S, SstI.
MATERIALS AND METHODS
Streptomyces strains, growth conditions, and bacteriophage M13-based constructs.
S. rimosus 4018 (42) and 15883 were derived by Pfizer Ltd. (Sandwich, Kent, United Kingdom) during a strain improvement program. 15883S was derived from 15883 by spontaneous deletion of the OTC gene cluster. To analyze otc transcription during antibiotic fermentation, 15883 was grown in Trusoya Medium 1 (TSM1), a proprietary medium from Pfizer which contains soya flour, starch, vegetable oil, and inorganic salts (7), while 4018 was grown in liquid complete medium (LCM) (42). Control RNA was isolated from 15883S grown in tryptone soya broth (TSB) (25). Cultures (50 ml) were grown in 250-ml Erlenmeyer flasks at 28°C with shaking (250 rpm). All of the constructs used to generate single-stranded DNA (ssDNA) probes for the analysis of transcription were derived from M13mp18 (54). mL6A has, in the SmaI site, a 2.1-kb SmaI fragment containing the intergenic region between otcC and otcX; the insert is oriented such that the otcC gene is nearest the HindIII site in the polylinker. In mL6C, the SmaI insert is in the opposite orientation. mAT12a and mAT12b differ only in the orientation of a 1.4-kb SphI insert that encodes the otcY2 intergenic region, which in mAT12a is oriented such that the orf1 gene is nearest the HindIII site in the polylinker. mKM803 has a 250-bp PstI/SmaI fragment containing the 5′ end of the otcZ gene in the corresponding sites of the polylinker. Similarly, the insert in mKM804 is a 387-bp HincII/BamHI fragment containing the 5′ end of the otrA gene.
Measurement of OTC in cultures.
Culture samples (1 ml) were extracted in 9 ml of HCl (pH 1.7) for 30 min and filtered through Whatman no. 1 papers. Samples (1 ml) were then analyzed for OTC by isocratic, ion-paired, reverse-phase high-performance liquid chromatography with a C18 μBondabank column (Waters) and a mobile phase of acetonitrile-water (3:7) that contained 5% (wt/vol) 1-hexanesulfonic acid and that had a pH adjusted to 1.7 with sulfuric acid.
Dry weight determinations.
Whatman GF/C filters (4.7-cm diameter) were dried for 15 min in a microwave oven set at 150 W and allowed to cool to room temperature in a desiccator for 30 min. The weight of each filter was determined to three decimal places. Mycelia were harvested by passing 5 ml of culture through a dried filter held in a sintered-glass vacuum unit (Millipore). The filter was washed with 15 ml of distilled H2O, dried, and weighed as described above, and the net weight was recorded. At least three independent measurements were used to produce each dry weight value, which had a standard error of the mean of less than 5%.
Isolation of total RNA.
At the appropriate stage of growth, 50 ml of culture was decanted into a 250-ml flask containing 20 g of 0.5-cm3 ice pellets prepared by freezing double-distilled water at −20°C and was mixed by swirling. This step rapidly lowered the temperature of the culture to ca. 0°C. Using the protocol described by Hopwood et al. (25), we harvested the mycelia by centrifugation; the cells were lysed by a modification of the method of Kirby et al. (29), and the lysate was extracted with phenol-chloroform. The aqueous phase of the lysate (3.5 ml) was carefully added to the top of 1.5 ml of 5.7 M CsCl–0.1 M EDTA (density, 1.71 g ml−1) in a 5-ml Polyallomer centrifuge tube (Beckman) and centrifuged at 35,000 rpm in an SW40 rotor for 16 h at 20°C. The RNA in the pellet was redissolved in 400 μl of 0.1 M NaCl, extracted with 0.5 volumes of phenol-chloroform and chloroform, precipitated with 2 volumes of ethanol, reharvested by centrifugation in a Microfuge (12,000 × g for 10 min at 4°C), washed twice with 70% (vol/vol) ethanol, dried in open Microfuge tubes at room temperature, and redissolved in 100 μl of H2O. The concentration of the RNA was determined by measuring the absorbance at 260 nm, the integrity of the RNA was checked by testing in 0.8% (wt/vol) agarose–Tris-borate-EDTA gels, and 10-μg aliquots were stored as ethanol precipitates at −70°C.
Primer extension mapping of RNA 5′ ends.
Oligonucleotide primers were labelled at the 5′ ends as described by Sambrook et al. (43), and 1-ng aliquots were mixed with 10 μg of RNA precipitate. The nucleic acid mixture was harvested by centrifugation, washed with 70% (vol/vol) ethanol, and dried in an open Microfuge tube at room temperature. The oligonucleotide primer was annealed to the RNA and extended with avian myeloblastosis virus reverse transcriptase (Pharmacia) as described previously (17), and the reaction was terminated by the addition of an appropriate amount of sequencing gel loading buffer (43); after heating to 85°C, samples were run in an 8% (wt/vol) polyacrylamide sequencing gel. The sizes of the extension products were determined by comparing their migration with that of sequencing ladders generated from single-stranded M13-derived templates by use of a Sequenase kit as described by the vendor (Amersham). Single-stranded M13 templates were prepared as described by Sambrook et al. (43).
Synthesis of ssDNA probes and nuclease protection assays.
5′-end-labelled or continuously labelled probes were synthesized and used as described by Sambrook et al. (43). The procedure involved extension of oligonucleotide primers annealed to single-stranded templates derived from recombinant bacteriophage M13, digestion of the newly synthesized double-stranded DNA with a suitable restriction enzyme, and separation of the radiolabelled probe from the linearized ssDNA template by electrophoresis through a 6% (wt/vol) polyacrylamide sequencing gel. The position of the probe was determined by autoradiography, and the probe was eluted from a slab of gel by overnight incubation in 0.5 ml of 0.5 M ammonium acetate (pH 8.0)–10 mM magnesium acetate–1 mM EDTA–10% (wt/vol) sodium dodecyl sulfate. The eluate was extracted twice with phenol-chloroform and twice with chloroform, and the probe was precipitated by the addition of 2.5 volumes of ethanol. The precipitate was harvested by centrifugation in a microcentrifuge (12,000 × g for 30 min at 4°C), washed with 70% (vol/vol) ethanol, dried as described above, and redissolved in 50 μl of H2O. The probe (0.2 pmol) was mixed with 10 μg of total RNA precipitate, harvested, washed, dried as described above, and redissolved in 20 μl of hybridization buffer (43). The hybridization mixture was denatured at 85°C for 10 min, allowed to anneal at 55 to 65°C overnight, and treated with S1 nuclease (Sigma) or exonuclease VII (Life Technologies). The S1 nuclease reaction mixture (300 μl) contained 375 U of enzyme, and the reaction was carried out at 37°C for 30 min. The hybridization mixture was treated with exonuclease VII by the addition of 280 μl of 50 mM Tris-Cl (pH 7.8)–50 mM KCl–10 mM EDTA containing 10 U of enzyme and incubation at 37°C for 45 min. The reaction was terminated by extraction with phenol-chloroform and chloroform and processed as described for the S1 nuclease digestion products (43).
Densitometry scanning.
The intensity of autoradiographic images was determined with a PDI scanner and software.
RESULTS
Mapping of promoters within the intergenic region between otcC and otcX-orf1.
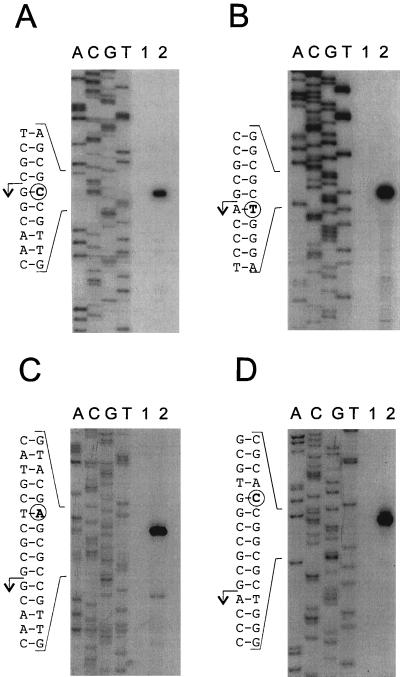
To identify promoters within the intergenic region between the 5′ end of otcC and the 5′ end of otcX-orf1, we first used a primer extension assay to analyze total RNA isolated from S. rimosus 15883 grown under conditions that produce OTC and, as a negative control, from 15883S, which has a deletion of the entire otc cluster. Single extension products were obtained for both otcC and otcX mRNAs (Fig. 2A and B, respectively). Confirmation that both the otcC and the otcX extension products terminated at RNA 5′ ends and not at internal secondary structures, which occur frequently in streptomycete transcripts due to their high (73%) G+C content (15), was obtained by use of a high-resolution exonuclease VII protection assay (Fig. 2C and D, respectively). The protected products for otcC and otcX mRNAs were 5 and 7 nucleotides (nt) longer, respectively, than the corresponding primer extension products (Fig. 2A and B) because exonuclease VII is only able to digest single-stranded probes to within a few nucleotides of protected regions (11). No primer extension or nuclease protection was obtained with RNA from 15883S, indicating that our assays were specific for otc transcripts.
FIG. 2.
Primer extension and nuclease protection analyses of the otcC and otcX transcripts. (A) Lanes 1 and 2 contain cDNA products extended from oligonucleotide C68 hybridized to 10 μg of total RNA from S. rimosus 15883S grown in TSB and from S. rimosus 15883 grown in TSM1 for 46 h (see Fig. 6A), respectively. (B) Like panel A, except that the cDNA products were extended from oligonucleotide X84. (C) Lanes 1 and 2 contain the products of exonuclease VII digestion of otcC transcripts in 10 μg of total RNA from S. rimosus 15883S and 15883, respectively. (D) Like panel C, except that otcX transcripts were analyzed. The probe for otcC transcripts was generated by extending 5′-labelled oligonucleotide C64 annealed to template mL6A and then cutting with AvaI, while the otcX transcript probe was generated by extending oligonucleotide X84 annealed to template mL6C and then cutting with SphI. Both probes were hybridized with S. rimosus RNA at 37°C. See Fig. 3 for the sequences of the primers and the locations of the restriction sites. The sequencing ladders (lanes A, C, G, and T) for analysis of the otcC and otcX transcripts were extended from oligonucleotides C68 and X84 annealed to templates mL6A and mL6C, respectively. Circles indicate the 3′ ends of the products of either primer extension (A and B) or nuclease protection (C and D) analyses. The arrows show the deduced 5′ ends of the otcC and otcX transcripts.
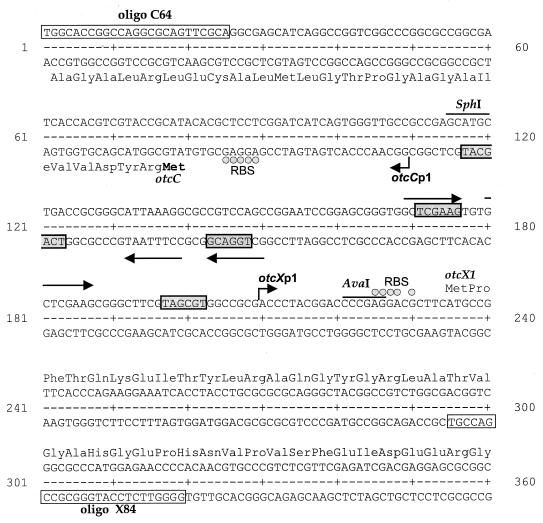
Analysis of the DNA sequence upstream of the positions mapped for the 5′ ends of the otcC and otcX mRNAs revealed hexanucleotide sequences that are centered around positions −10 and −36 (Fig. 3) and that are similar to the consensus sequences for the −10 and −35 regions of the major class of promoters in eubacteria (22) and in streptomycetes (26, 45), suggesting that these 5′ ends are the starts of primary transcripts rather than the 5′ ends of RNA decay intermediates. Accordingly, we designated these conserved hexanucleotide sequences to be the −10 and −35 regions of promoters otcCp1 and otcXp1. Further sequence analysis revealed a perfect tandem repeat between positions −41 and −23 of otcXp1 (Fig. 3). Similarly, a related, but imperfect, tandem repeat was located at precisely the same positions in otcCp1.
FIG. 3.
Annotated sequences of otcCp1 and otcXp1. The positions of the transcription start sites (bent arrows) are indicated, together with the putative −10 and −35 regions of the promoters (shaded boxes), the direct repeats which have sequence similarity to the DNA-binding sites of E. coli PhoB (straight arrows), the primer sequences (unshaded boxes), and restriction enzyme sites used in mapping the 5′ ends of the otcC and otcX transcripts (see Fig. 2). Potential ribosome-binding sites (RBS) are shown by shaded circles.
Tandem repeats also overlap the otcY promoters.
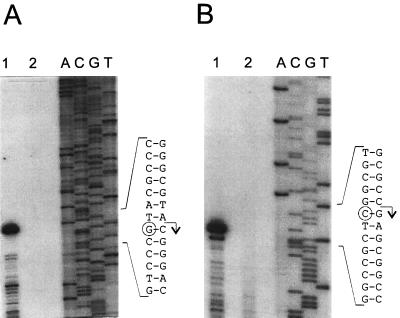
To determine whether tandem repeats overlapping −35 regions are a general feature of the promoters of otc biosynthetic genes, we also mapped and analyzed the sequences of promoters in the intergenic region between the polyketide synthase genes otcY2-orf1 and otcY2-orf2. This is the only other segment where biosynthetic genes diverge in the otc cluster (27). Single major primer extension products were obtained for both of the otcY2 transcripts (Fig. 4); moreover, as was found for otcCp1 and otcXp1, the promoters in the otcY2 intergenic region were overlapped by tandem repeats (Fig. 5). Interestingly, the 11-nt separation (one turn of supercoiled B-form DNA) between the centers of these repeats and the location of the first repeat 10 nt upstream of the −10 regions of the otc promoters is typical of the DNA-binding sites of members of the OmpR family of transcription factors (32–34, 44, 51).
FIG. 4.
Primer extension analysis of transcription from divergent promoters in the intergenic region between otcY2-orf1 and orf2. (A) Lanes 1 and 2 contain cDNA products extended from primer L12A3 (5′-TCACCACGGCGAGCCCGATG) hybridized to 10 μg of total RNA from S. rimosus 4018 grown in TSM1 for 2 days and from 15883S grown in TSB, respectively. (B) Like panel A, except that the cDNA products were extended from primer L12B1 (5′-AGCGCGGTGTCCACGGGGACGC). The sequencing ladders (lanes A, C, G, and T) for analysis of the otcY2-orf1 and orf2 transcripts were extended from L12A3 and L12B1 annealed to templates mAT12a and mAT12b, respectively. As in Fig. 2, circles indicate the 3′ ends of primer extension products, and arrows show the deduced 5′ ends of the transcripts.
FIG. 5.
Alignment of the nucleotide sequences of otc promoter regions. The transcription start sites (designated +1) were determined as described in the legends to Fig. 2 and 4. Underlined sequences are hexanucleotide segments having similarity to consensus sequences TAG[Pu][Pu]T and TTGAC[Pu], which correspond to the −10 and −35 regions, respectively, of the major class of streptomycete promoters (22, 49). The putative −35 regions of the otc promoters are overlapped by tandem repeats indicated by arrows. To allow a consensus sequence to be derived for the repeated sequences, the repeat closest to the transcriptional start site of each promoter was copied, placed within parentheses, and aligned under its partner. In the consensus sequence for the repeated sequences, lowercase, uppercase, bold, and underlined characters represent nucleotides conserved at the same positions in five, six, seven, and eight of the repeats, respectively. The putative sequence of the promoter for the otcY1 genes (28) is also shown but was not used to derive the consensus sequence for the repeated sequences. The otcY1 promoter is overlapped by three related repeats whose centers are separated by 11 nt.
Transcription from otcCp1 and otcXp1 is regulated.
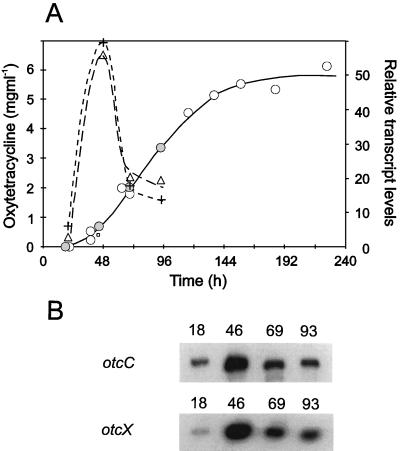
We investigated whether transcription from otc promoters overlapped by tandem repeats is temporally regulated. Primer extension assays were used to compare the levels of transcripts extending from otcCp1 and otcXp1 at different stages during a strain 15883 fermentation that produced significant levels of OTC (ca. 6 mg ml−1) when phosphate became limiting. As shown in Fig. 6, after 18 h, when OTC was barely detectable in the culture (<50 μg ml−1), transcripts originating from otcCp1 and otcXp1 were present at low levels; however by 46 h, when antibiotic production had started, the amount of each transcript had increased 10- to 20-fold. Despite the fact that the amount of OTC in the medium continued to increase at later times (69 and 93 h), the abundance of the otcC transcripts declined steadily. The coincidence of the peak in the abundance of transcripts originating from otcCp1 and otcXp1 with the onset of OTC production suggests that the biosynthesis of this antibiotic is controlled, at least in part, at the level of transcription.
FIG. 6.
Quantification of levels of transcripts originating from otcCp1 and otcXp1 at different stages during the production of OTC by S. rimosus 15883 grown in TSM1. (A) Open circles indicate the amount of OTC in the cultures. The values were obtained from samples taken from 14 cultures (50 ml) that were inoculated and incubated at 28°C on a rotary platform at the same time. Closed circles indicate the concentration of OTC in cultures used to analyze otc transcripts. Crosses and triangles indicate the levels of transcripts originating from otcCp1 and otcXp1, respectively, as measured by densitometry scanning of the autoradiographs of the primer extension products shown in panel B. (B) Primer extension assays were done as described in the legend to Fig. 2 with total RNA isolated from S. rimosus 15883 grown in TSM1 for 18, 46, 69, and 93 h. TSM1 contains a colloidal suspension of starch and soya flour, which prevented the isolation of RNA and prevented the measurement of mycelial growth before 18 h.
Evidence for an otcC-otcC-otrA polycistronic message.
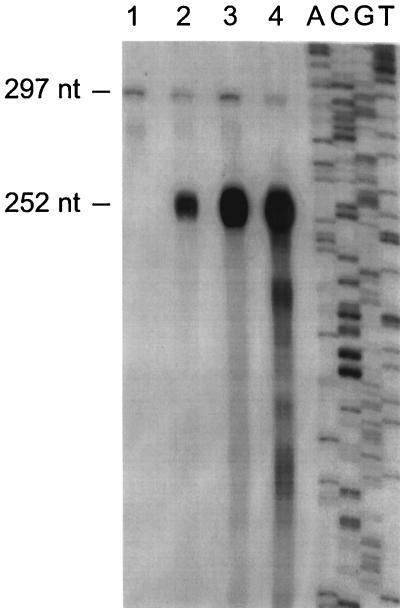
Previously, it had been shown that the otrA resistance gene is transcribed in part by read-through from otcZ, the methyltransferase biosynthetic gene (38, 39) that lies between otrA and otcC (Fig. 1); therefore, if transcription extends from otcC into otcZ, then otcCp1 could participate in the regulation of antibiotic resistance as well as biosynthesis. This possibility was investigated with an S1 nuclease protection assay (Fig. 7). Two protected species of 252 and 297 nt were detected with total RNA isolated from strains 4018 and 15883 (Fig. 7, lanes 2 and 3 and lane 4, respectively); however, the 297-nt species was also detected with total RNA isolated from strain 15883S (lane 1), indicating that only the 252-nt fragment was protected by otc transcripts. The length of the otc-specific fragment corresponded to protection of the probe by transcripts covering the entire otcC-otcZ intergenic region between the PstI and SmaI sites (Fig. 1, positions 4 and 5), indicating that otcZ and thus otrA could be transcribed from otcCp1. Consistent with the results of the protection assay, we were unable to detect any sequence between otcC and otcZ that was predicted to form a rho-independent terminator. The 297-nt species detected in all the samples in Fig. 7 corresponds to the full-length probe protected from S1 nuclease digestion by a small amount of contaminating template DNA in the probe preparation (see Materials and Methods).
FIG. 7.
Analysis of transcription in the intergenic region between otcC and otcZ. High-resolution S1 nuclease protection assays were performed with 10 μg of total RNA from S. rimosus 15883S grown in TSB until the late logarithmic phase, 4018 grown in the same manner as 15883S, 4018 producing OTC (ca. 60 μg ml−1) after 56 h in LCM (see Fig. 9), and 15883 producing OTC (ca. 700 μg ml−1) after growth in TSM1 (see Fig. 6) for 46 h (lanes 1, 2, 3, and 4, respectively). A continuously labelled ssDNA probe was generated from template mKM803 by extension of the −20 universal primer and EcoRI digestion. The sequencing ladder (lanes A, C, G, and T) was produced by extension of the −40 universal primer annealed to template mKM803. The numbers on the left indicate the sizes of the protected probe fragments, as determined by comparison with the sequencing ladder.
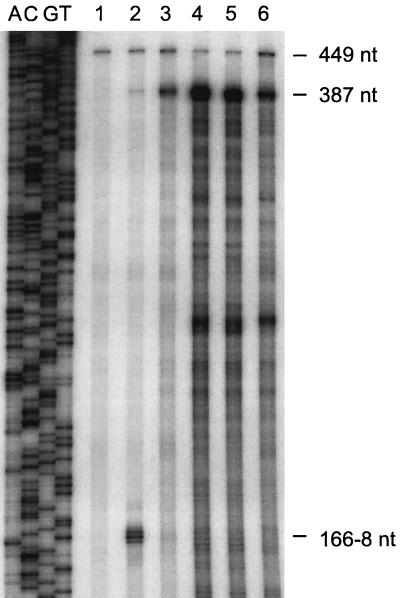
Attempts to confirm the presence of the 6-kb otcC-otcZ-otrA polycistronic message by Northern blot analysis were unsuccessful. Although otc-specific RNA species were detected, they were heterogeneous in size, from 1.5 to 3.0 kb (data not shown). A possible explanation for this result is that decay of the otcC-otcZ-otrA polycistronic message commences before transcription has terminated. In any case, further evidence supporting the notion that otrA is transcribed from otcCp1 was obtained by use of a high-resolution S1 nuclease protection assay to determine the relative levels of transcripts for the segment between otcZ and otrA (Fig. 8) in the RNA samples used to analyze transcription from otcCp1 at different stages during OTC production (Fig. 6). The probe used was complementary to the sense strand between the HincII and BamHI sites (Fig. 1, positions 2 and 3), and total RNA isolated from strain 15883S served as a negative control (Fig. 8, lane 1). The 449-nt species in the 15883S RNA sample (Fig. 8, lane 1) corresponds to the full-length probe, while the 387-nt protected fragment in the RNA samples isolated from 15883 at the stages analyzed during the production of OTC (lanes 3 to 6) corresponds to transcriptional read-through from the otcZ gene. Consistent with otrA being transcribed from otcCp1, the abundance of transcripts encoding the segment between the 3′ end of otcZ and the 5′ end of otrA peaked early in OTC production, at 48 h (compare with Fig. 6). Interestingly, in strain 15883, no transcription was detected from otrAp1 (14), which lies in the intergenic region between otcZ and otrA; however, this strain produced detectable levels of transcripts during growth on TSB (protection of 166 to 167 nt in Fig. 8, lane 2), a medium that does not support antibiotic production.
FIG. 8.
High-resolution S1 nuclease protection analysis of otrA transcription at the onset of OTC production by S. rimosus 15883 grown in TSM1. Lanes 1 and 2 contain probe fragments protected by 10 μg of total RNA from S. rimosus 15883 and 4018 grown in TSB, respectively, while lanes 3, 4, 5, and 6 correspond to 15883 grown in TSM1 for 18, 46, 69, and 93 h, respectively (see Fig. 6). A continuously labelled ssDNA probe was generated by extension of the −20 universal primer annealed to template mKM804 and digestion with EcoRI. The sequencing ladder (lanes A, C, G, and T) was produced by extension of the −40 universal primer annealed to template mKM804 (36). The numbers on the right indicate the sizes of the protected probe fragments. Species shorter that 387 nt in lanes 3, 4, 5, and 6 may represent RNA processing or degradation or an artifact of the S1 nuclease mapping procedure.
The otc genes are transcribed as mycelial growth slows.
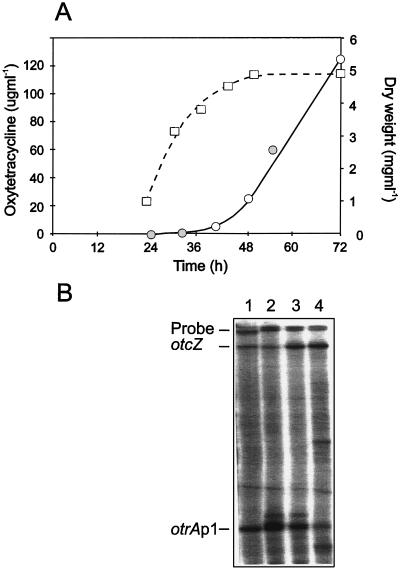
The fermentation conditions for strain 15883 (Fig. 6) made it impossible to correlate the transcription of the otc genes with the phase of mycelial growth or to show that otrA is transcribed from otrAp1 before antibiotic production, because TSM1, the production medium developed for 15883, contains insoluble components that interfered with both the measurement of mycelial growth and the isolation of RNA at early times. We therefore analyzed transcription during the early stages of OTC production by strain 4018 grown in LCM, a completely soluble medium. As shown in Fig. 9, the combined results from the 4018 fermentation indicate that the onset of OTC production coincides not only with increased transcription of otrA by read-through from otcZ but also with the slowing of mycelial growth and decreased transcription of otrA from its own promoter (otrAp1). The transition between rapid growth and the stationary phase is also the point at which increased transcription of the actinorhodin biosynthetic genes in S. coelicolor occurs (20).
FIG. 9.
Analysis of otrA transcription and mycelial growth at the onset of OTC production by S. rimosus 4018 grown in LCM. (A) Open circles indicate the amount of OTC in samples taken from six cultures (50 ml) that were inoculated at the same time and incubated at 28°C on the same rotary platform. Closed circles indicate the concentration of OTC in cultures used to analyze otc transcripts. Squares indicate the mean dry weight of the mycelium at different stages in the fermentation. The determination of dry weight had a variance of less than 5% error for the mean. (B) S1 nuclease protection analysis of otrA transcription. Assay conditions were as described in the legend to Fig. 8. Lane 1, RNA from 4018 grown in TSB; lanes 2, 3, and 4, RNA from 4018 grown in LCM for 24, 32, and 56 h, respectively.
DISCUSSION
Our findings that (i) promoters of otc biosynthetic genes are overlapped by tandem repeats (Fig. 5), whose spacing resembles that of the activation sites of the OmpR family of transcriptional factors, and (ii) transcripts originating from these promoters reach a peak at the onset of antibiotic production that is triggered by limiting phosphate (Fig. 6) together suggest that the expression of some, and possibly all, of the otc biosynthetic genes is regulated, at least in part, at the level of transcription by an activation mechanism. Furthermore, this notion provides a possible explanation for the observation that three nonoverlapping segments of the otc cluster prevent the production of OTC by S. rimosus when cloned with high-copy-number but not low-copy-number plasmids (the switch-off phenomenon [9]). We now know that two of these segments correlate with the intergenic region between otcC and otcX-orf1 and the intergenic region between otcY2-orf1 and otcY2-orf2, which have been shown here to contain divergent promoters overlapped by tandem repeats (Fig. 5). The third corresponds to the likely location of the promoter for the transcription of the otcY1 genes, which encode the minimal polyketide synthase (27). Although the transcription start site of the otcY1 promoter has not yet been determined, tandem repeats resembling those overlapping the otcC-otcX and otcY promoters have been identified (Fig. 5). Given the above correlation, we suggest that the basis of the switch-off phenomenon (9) is the sequestration of a transcriptional activator by an excessive number of plasmid-borne copies of the otc tandem repeats.
Transcriptional activators are also known to regulate the expression of antibiotic gene clusters of other streptomycetes, e.g., ActII-Orf4 and DnrI of the actinorhodin and daunorubicin clusters, respectively (16, 46). Recently, it has been noted (53) that these streptomycete antibiotic regulatory proteins (SARPs) are similar to the DNA-binding domains of the OmpR family (37). Furthermore, an analysis of the promoter regions of the act and dnr biosynthetic genes (53) has revealed tandem repeats which, like the otc repeats identified in this study, are characteristic of the DNA-binding sites of all members of the OmpR family (34). Combined with the knowledge that the dnr tandem repeats were located within footprints of bound DnrI, Wietzorrek and Bibb (53) suggested that the tandem repeats identified in the act and dnr promoter regions are the binding sites of ActII-Orf4 and DnrI, respectively. We find that in addition to having similar spacing, the most highly conserved nucleotide positions (underlined) of the otc tandem repeats (consensus sequence, 5′-GCTCGAA [Fig. 5]) are also the most highly conserved positions in the proposed DNA-binding sites of ActII-Orf4 and DnrI [repeat consensus sequence, 5′-TCGAGC(G/C) (53)]. In the case of the dnr repeats, the most conserved nucleotide positions have been shown to be protected by DnrI from digestion by DNase I (47). On the basis of the above similarity, we suggest that the activator of the otc genes may be a member of the OmpR family, most likely a member of the SARP group. Experiments are currently under way to identify the factor that activates transcription from the otc promoters as, unlike the situation for actinorhodin and daunorubicin, no sarp-like gene has been identified within the otc cluster (28).
The experimental system used to investigate OTC production (shown best in Fig. 9) used limitation by phosphate to arrest the growth of the culture, at which time antibiotic biosynthesis was induced. Thus, transcription from the otc promoters occurred in response to the environmental stress of phosphate limitation. ActII-Orf4 and DnrI, together with their cognate DNA-binding sites, may also be involved in mediating phosphate control of the production of actinorhodin (23) and daunorubicin (12, 47). Consistent with this notion, detailed physiological studies have shown that phosphate starvation can induce actinorhodin production at least in part at the level of transcription of actIII (23, 24), whose promoter (21) is now known to be overlapped by a proposed DNA-binding site of ActII-Orf4 (53).
At present, the best-characterized phosphate-controlled system is the pho regulon of Escherichia coli, which encodes a set of genes required for the uptake of phosphate (for reviews, see references 44 and 51). Under conditions of low phosphate, PhoB, a transcriptional activator that (like SARPs) is a member of the OmpR family, is phosphorylated by PhoR (a sensory kinase) and then binds to promoters to activate the transcription of genes required for the active uptake of phosphate. Whether covalent modification of SARPs plays a role in the regulation of antibiotic production remains to be determined. Interestingly, however, in E. coli cells that lack functional PhoR, the pho regulon is no longer under the control of phosphate but is tightly regulated by carbon and energy sources via controls that were not obvious under conditions of phosphate starvation in the presence of functional PhoR (49, 50). Current evidence suggests that PhoB is activated not only by PhoR but also by other sensory kinases, thus allowing the process of phosphate assimilation to be integrated with central pathways for carbon and energy metabolism (for a review, see reference 51). The notion that the control of antibiotic production in Streptomyces is subject to cross regulation, similar to that described above for the E. coli pho regulon, could explain how factors other than phosphate, for example, carbon and nitrogen sources, can affect the timing and extent of antibiotic production (see references 30 and 36).
Regardless of the actual details of the molecular mechanism controlling antibiotic production, it is important for the survival of the producer that it is resistant at the onset of biosynthesis. The combined results of our transcriptional analyses (Fig. 7 to 9) suggest that antibiotic resistance and production may be coordinated in S. rimosus at least in part via cotranscription of the otrA resistance gene with the otcC and otcZ biosynthetic genes from the otcCp1 promoter.
OTC resistance is also regulated independently of OTC production: preexposure of S. rimosus to sublethal concentrations of OTC during vegetative growth induces higher levels of resistance (40). This response is independent of otcCp1 but probably involves otrAp1, as a DNA segment extending from the 3′ end of otrA to the middle of otcZ is sufficient to confer inducible resistance (13). At the onset of antibiotic production, when otrA is transcribed along with otcZ and otcC, transcription from otrAp1 appears to be superfluous, as there is a significant (>80%) decrease in the level of transcripts originating from this promoter (Fig. 9). The molecular basis of this observation remains to be determined, but it is conceivable that otcCp1 and otrAp1 are transcribed by different RNA polymerase holoenzymes (10, 52) which are active under different physiological conditions.
ACKNOWLEDGMENTS
This work was supported by awards of studentships from the British MRC (to K.J.M.) and the Thai Ministry of Science, Technology and Environment (to A.T.).
We are grateful to Maggie Smith, Craig Binnie, and Mike Butler for useful discussions and to Mel Warren for assistance with oxytetracycline measurements.
REFERENCES
- 1.Asturias J A, Liras P, Martin J F. Phosphate control of pabS gene transcription during candicidin biosynthesis. Gene. 1990;93:79–84. doi: 10.1016/0378-1119(90)90139-i. [DOI] [PubMed] [Google Scholar]
- 2.Behal V, Hostalek Z, Vanek Z. Anhydrotetracycline oxygenase activity and biosynthesis of tetracyclines in Streptomyces aureofaciens. Biotechnol Lett. 1979;1:275–318. [Google Scholar]
- 3.Behal V. Enzymes of secondary metabolism: regulation of their expression and activity. In: Kleinkauf H, von Dohren H, Dornauer H, Nesemann G, editors. Regulation of secondary metabolite formation. Weinheim, Germany: VCH; 1986. pp. 264–281. [Google Scholar]
- 4.Behal V. The tetracycline fermentation and its regulation. Crit Rev Biotechnol. 1987;5:275–318. [Google Scholar]
- 5.Behal V, Hunter I S. Tetracyclines. In: Vining L C, Studdard C, editors. Genetics and biochemistry of antibiotic production. Boston, Mass: Butterworth-Heinemann; 1995. pp. 359–385. [Google Scholar]
- 6.Binnie C, Warren M, Butler M J. Cloning and heterologous expression in Streptomyces lividans of Streptomyces rimosus genes involved in oxytetracycline biosynthesis. J Bacteriol. 1989;171:887–895. doi: 10.1128/jb.171.2.887-895.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butler, M. J. Personal communication.
- 8.Butler, M. J. Unpublished results.
- 9.Butler M J, Friend E J, Hunter I S, Kaczmarek F, Sugden D A, Warren M. Molecular cloning of resistance genes and architecture of a linked gene cluster involved in biosynthesis of oxytetracycline by Streptomyces rimosus. Mol Gen Genet. 1989;215:231–238. doi: 10.1007/BF00339722. [DOI] [PubMed] [Google Scholar]
- 10.Buttner M J, Smith A M, Bibb M J. At least three different RNA polymerase holoenzymes direct transcription of the agarase gene (dagA) of Streptomyces coelicolor A3(2) Cell. 1988;52:599–607. doi: 10.1016/0092-8674(88)90472-2. [DOI] [PubMed] [Google Scholar]
- 11.Calzone F J, Britten F J, Davidson E H. Mapping of transcripts by nuclease protection assays and cDNA primer extension. In: Berger S L, Kimmel A R, editors. Guide to molecular cloning techniques. London, England: Academic Press Ltd.; 1987. pp. 611–632. [Google Scholar]
- 12.Dekeva M L, Titus J A, Strohl W R. Nutrient effects on anthracycline production by Streptomyces peucetius in a defined medium. Can J Microbiol. 1985;31:287–294. doi: 10.1139/m85-053. [DOI] [PubMed] [Google Scholar]
- 13.Doyle, D., and I. S. Hunter. Unpublished results.
- 14.Doyle D, McDowall K J, Butler M J, Hunter I S. Characterization of an oxytetracycline-resistance gene, otrA, of Streptomyces rimosus. Mol Microbiol. 1991;5:2923–2933. doi: 10.1111/j.1365-2958.1991.tb01852.x. [DOI] [PubMed] [Google Scholar]
- 15.Enquist S D, Bradley S G. Characterisation of deoxyribonucleic acid from Streptomyces venezuelae species. Dev Ind Microbiol. 1971;12:2431–2441. [Google Scholar]
- 16.Fernandez-Moreno M A, Caballero J L, Hopwood D A, Malpartida F. The act cluster contains regulatory and antibiotic export genes, direct targets for translational control by the bldA tRNA gene of Streptomyces. Cell. 1991;66:769–780. doi: 10.1016/0092-8674(91)90120-n. [DOI] [PubMed] [Google Scholar]
- 17.Fisher S H, Wray L V. Regulation of glutamine synthase in Streptomyces coelicolor. J Bacteriol. 1989;171:2378–2383. doi: 10.1128/jb.171.5.2378-2383.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gil J A, Hopwood D A. Cloning and expression of a ρ-aminobenzoic acid synthase gene from the candicidin producer Streptomyces griseus. Gene. 1983;25:119–132. doi: 10.1016/0378-1119(83)90174-9. [DOI] [PubMed] [Google Scholar]
- 19.Gil J A, Naharro G, Villanueva J R, Martin J F. Characterisation and regulation of ρ-aminobenzoic acid synthase from Streptomyces griseus. J Gen Microbiol. 1985;131:1279–1287. doi: 10.1099/00221287-131-6-1279. [DOI] [PubMed] [Google Scholar]
- 20.Gramajo H C, Takano E, Bibb M J. Stationary-phase production of the antibiotic actinorhodin in Streptomyces coelicolor A3(2) is transcriptionally regulated. Mol Microbiol. 1993;7:837–845. doi: 10.1111/j.1365-2958.1993.tb01174.x. [DOI] [PubMed] [Google Scholar]
- 21.Hallam S E, Malpartida F, Hopwood D A. Nucleotide sequence, transcription and deduced function of a gene involved in polyketide antibiotic synthesis in Streptomyces coelicolor. Gene. 1988;74:305–320. doi: 10.1016/0378-1119(88)90165-5. [DOI] [PubMed] [Google Scholar]
- 22.Hawley D K, McClure W R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hobbs G, Frazer C M, Gardner D C J, Flett F, Oliver S G. Pigmented antibiotic production by Streptomyces coelicolor A3(2): kinetics and the influence of nutrients. J Gen Microbiol. 1990;136:2291–2296. [Google Scholar]
- 24.Hobbs G, Obanye A I C, Petty J, Mason J C, Barratt E, Gardner D C J, Flett F, Smith C P, Broda P, Oliver S G. An integrated approach to studying regulation of production of the antibiotic methylenomycin by Streptomyces coelicolor A3(2) J Bacteriol. 1992;174:1487–1494. doi: 10.1128/jb.174.5.1487-1494.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, United Kingdom: The John Innes Foundation; 1985. [Google Scholar]
- 26.Hopwood D A, Bibb M J, Chater K F, Janssen G R, Malpartida F, Smith C P. Regulation of gene expression in antibiotic-producing Streptomyces. In: Booth I R, Higgins C F, editors. Regulation of gene expression in antibiotic-producing Streptomyces. Cambridge, United Kingdom: Cambridge University Press; 1986. pp. 251–276. [Google Scholar]
- 27.Hunter I S, Hill R A. Tetracyclines: chemistry and molecular genetics of their formation. In: Strohl W R, editor. Biotechnology of industrial antibiotics. New York, N.Y: Marcel Decker, Inc.; 1997. pp. 659–682. [Google Scholar]
- 28.Kim E S, Bibb M J, Butler M J, Hopwood D A, Sherman D H. Sequences of the oxytetracycline polyketide synthase-encoding otc genes from Streptomyces rimosus. Gene. 1994;141:141–142. doi: 10.1016/0378-1119(94)90144-9. [DOI] [PubMed] [Google Scholar]
- 29.Kirby K S, Fox-Carter E, Guest M. Isolation of deoxyribonucleic acid and ribosomal ribonucleic acid from bacteria. Biochem J. 1967;104:258–262. doi: 10.1042/bj1040258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kleinkauf H, von Dohren H, Dornauer H, Nesemann G, editors. Regulation of secondary metabolite formation. Weinheim, Germany: VCH; 1986. [Google Scholar]
- 31.Liras P, Asturias J A, Martin J F. Phosphate control sequences involved in transcriptional regulation of antibiotic biosynthesis. Trends Biotechnol. 1990;8:184–189. doi: 10.1016/0167-7799(90)90170-3. [DOI] [PubMed] [Google Scholar]
- 32.Makino K, Shinagawa H, Amemura M, Nakata A. Nucleotide sequence of the phoB gene, the positive regulatory gene for the phosphate regulon of Escherichia coli K-12. J Mol Biol. 1986;190:37–44. doi: 10.1016/0022-2836(86)90073-2. [DOI] [PubMed] [Google Scholar]
- 33.Makino K, Shinagawa H, Amemura M, Kimura S, Nakata A, Ishihama A. Regulation of the phosphate regulon of Escherichia coli: activation of pstS transcription by PhoB protein in vitro. J Mol Biol. 1988;203:85–95. doi: 10.1016/0022-2836(88)90093-9. [DOI] [PubMed] [Google Scholar]
- 34.Makino K, Amemura M, Kawamoto T, Kimura S, Shinagawa H, Nakata A, Suzuki M. DNA binding of PhoB and its interaction with RNA polymerase. J Mol Biol. 1996;259:15–26. doi: 10.1006/jmbi.1996.0298. [DOI] [PubMed] [Google Scholar]
- 35.Martin J F, Demain A. Control of antibiotic biosynthesis. Microbiol Rev. 1980;44:230–251. doi: 10.1128/mr.44.2.230-251.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin J F, Daza A, Asturias J A, Gil J A, Liras P. Transcriptional control of antibiotic biosynthesis at phosphate-regulated promoters and cloning of a gene involved in the expression of multiple pathways in Streptomyces. In: Okami Y, editor. Biology of Actinomycetes ’88. Tokyo: Japanese Scientific Societies Press; 1988. pp. 424–430. [Google Scholar]
- 37.Martinez-Hackert E, Stock A M. Structural relationships in the OmpR family of winged-helix transcription factors. J Mol Biol. 1997;269:301–312. doi: 10.1006/jmbi.1997.1065. [DOI] [PubMed] [Google Scholar]
- 38.McDowall K J, Doyle D, Butler M J, Binnie C, Warren M, Hunter I S. Molecular genetics of oxytetracycline production by Streptomyces rimosus. In: Noack D, Krugel H, Baumberg S, editors. Genetics and product formation in Streptomyces. New York, N.Y: Plenum Press; 1991. pp. 105–116. [Google Scholar]
- 39.McDowall K J. Molecular genetics of oxytetracycline production in Streptomyces rimosus. Ph.D. thesis. Glasgow, Scotland: University of Glasgow; 1991. [Google Scholar]
- 40.Ohnuki T, Katoh T, Imanaka T, Aiba S. Molecular cloning of tetracycline resistance genes from Streptomyces rimosus in Streptomyces griseus and characterization of the cloned genes. J Bacteriol. 1985;161:1010–1016. doi: 10.1128/jb.161.3.1010-1016.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rebello A, Gil J A, Asturias J A, Liras P, Martin J F. Cloning and characterization of a phosphate regulated promoter involved in phosphate control of candicidin biosynthesis. Gene. 1989;79:47–58. doi: 10.1016/0378-1119(89)90091-7. [DOI] [PubMed] [Google Scholar]
- 42.Rhodes P M, Winskill N, Friend E J, Warren M. Biochemical and genetic characterisation of Streptomyces rimosus mutants impaired in oxytetracycline biosynthesis. J Gen Microbiol. 1981;124:329–338. [Google Scholar]
- 43.Sambrook J, Fritisch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 44.Stock J B, Ninfa A J, Stock A M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strohl W R. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 1992;20:961–974. doi: 10.1093/nar/20.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stutzman-Engwall K J, Otten S L, Hutchinson C R. Regulation of metabolism in Streptomyces spp. and overproduction in Streptomyces peucetius. J Bacteriol. 1992;174:144–154. doi: 10.1128/jb.174.1.144-154.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang L, Grimm A, Zhang Y X, Hutchinson C R. Purification and characterization of the DNA-binding protein Dnr1, a transcriptional factor of daunorubicin biosynthesis in Streptomyces peucetius. Mol Microbiol. 1996;22:801–813. doi: 10.1046/j.1365-2958.1996.01528.x. [DOI] [PubMed] [Google Scholar]
- 48.Thomas R, Williams D J. Oxytetracycline biosynthesis: mode of incorporation of [1-13C] and [1,2-13C2] acetate. J Chem Soc Chem Commun. 1983;12:677–679. [Google Scholar]
- 49.Wanner B L, Wilmes M R, Young D C. Control of bacterial alkaline phosphatase synthesis and variation in an Escherichia coli K-12 phoR mutant by adenyl cyclase, the cyclic AMP receptor protein, and the phoM operon. J Bacteriol. 1988;170:1092–1102. doi: 10.1128/jb.170.3.1092-1102.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wanner B L, Wilmes-Riesenberg M R. Involvement of phosphotransacetylase, acetate kinase, and acetyl phosphate synthesis in the control of the phosphate regulon in Escherichia coli. J Bacteriol. 1992;174:2124–2130. doi: 10.1128/jb.174.7.2124-2130.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wanner B L. Phosphorus assimilation and control of the phosphate regulon. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Reznikoff W S, Riley M, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1357–1381. [Google Scholar]
- 52.Westpheling J E, Raines M, Losick R. RNA polymerase heterogeneity in Streptomyces coelicolor. Nature. 1985;313:22–27. doi: 10.1038/313022a0. [DOI] [PubMed] [Google Scholar]
- 53.Wietzorrek A, Bibb M J. A novel family of proteins that regulates antibiotic production in Streptomyces appears to contain an OmpR-like DNA-binding fold. Mol Microbiol. 1997;25:1181–1184. doi: 10.1046/j.1365-2958.1997.5421903.x. [DOI] [PubMed] [Google Scholar]
- 54.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 55.Zhang J Y, Demain A L. Regulation of ACV synthetase in penicillin-producing and cephalosporin-producing microorganisms. Biotechnol Adv. 1991;9:623–641. doi: 10.1016/0734-9750(91)90735-e. [DOI] [PubMed] [Google Scholar]