Abstract
The industrially important polysaccharide alginate is composed of the two sugar monomers β-d-mannuronic acid (M) and its epimer α-l-guluronic acid (G). In the bacterium Azotobacter vinelandii, the G residues originate from a polymer-level reaction catalyzed by one periplasmic and at least five secreted mannuronan C-5-epimerases. The secreted enzymes are composed of repeats of two protein modules designated A (385 amino acids) and R (153 amino acids). The modular structure of one of the epimerases, AlgE1, is A1R1R2R3A2R4. This enzyme has two catalytic sites for epimerization, each site introducing a different G distribution pattern, and in this article we report the DNA-level construction of a variety of truncated forms of the enzyme. Analyses of the properties of the corresponding proteins showed that an A module alone is sufficient for epimerization and that A1 catalyzed the formation of contiguous stretches of G residues in the polymer, while A2 introduces single G residues. These differences are predicted to strongly affect the physical and immunological properties of the reaction product. The epimerization reaction is Ca2+ dependent, and direct binding studies showed that both the A and R modules bind this cation. The R modules appeared to reduce the Ca2+ concentration needed for full activity and also stimulated the reaction rate when positioned both N and C terminally.
Alginate is an industrially important polysaccharide which is manufactured from brown algae (30). It is also produced by some species of the bacterial genera Azotobacter and Pseudomonas (6, 12–14, 18). Its biosynthesis has been most extensively studied in Pseudomonas aeruginosa due to the detrimental infections by alginate-producing strains of this species in the lungs of patients suffering from cystic fibrosis (21). However, most of the Azotobacter vinelandii biosynthetic genes have now been cloned and sequenced (4, 9, 19, 22, 23, 26).
The polysaccharide is composed of 1-4-linked β-d-mannuronic acid (M) and α-l-guluronic acid (G), both of which are distributed nonrandomly. An alginate molecule can be described as a mixture of blocks of different lengths of consecutive M residues (M blocks) or G residues (G blocks) or of alternating M and G residues (MG blocks). The amount and distribution of G residues determine the gel-forming, water-binding, and immunogenic properties of an alginate and also its solubility in acid (28, 30). The polymer is first synthesized as mannuronan, and the G residues are introduced by the action of mannuronan C-5-epimerases (17). This group of enzymes thus determines most of the important properties of the alginate, and they can also be used to tailor alginate in vitro. Alginate is now being evaluated as a gel-forming agent for encapsulation of cells for transplantation into humans (31). This usage requires a homogeneous alginate with good gelling properties (long G blocks) and without long stretches of M blocks, which could stimulate the immune system (28). These demands may be most easily met by using alginate epimerized in vitro.
A. vinelandii encodes both a periplasmic epimerase (AlgG) (26) and a family of secreted epimerases (AlgE) (7, 10, 32). The AlgE epimerases are all composed of one or two copies of a 385-amino-acid module (A module) and one to seven copies of a 153-amino-acid module (R module). Even though the homology within each group of modules is quite high, different epimerases introduce different G distribution patterns (10, 11, 32). All the AlgE epimerases are dependent on Ca2+ for activity, although the Ca2+ concentration needed for optimal activity is not the same for different epimerases (9). Based on sequence similarities to other enzymes, it has been proposed that the A modules are responsible for binding of the alginate and thus probably contain the catalytic site as well (15). The R modules are homologous to the C-terminal part of a group of secreted proteins which are exported by a C-terminal signal sequence and contain four to six repeats of a nine-amino-acid motif shown to bind Ca2+ in other proteins (2, 7). Thus the R modules probably bind Ca2+ and perhaps also participate in the secretion of the enzymes. Since all epimerases contain at least one R module C terminal to each A module, it has been thought that an A module with a downstream R module constitutes the minimal epimerase.
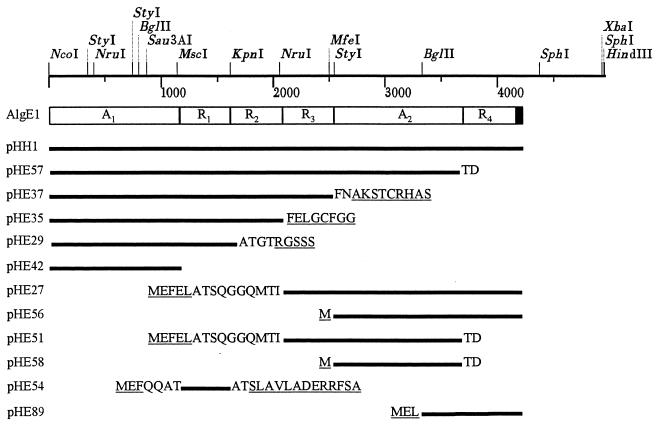
AlgE1 is the second most complex epimerase in that it contains two A modules and four R modules (Fig. 1). We have previously reported that this epimerase can be divided into two catalytically active parts: AlgE1-1, comprising the amino-terminal A module and the following three R modules, and AlgE1-2, comprising the second A module and the C-terminal R module (11). This earlier study further showed that AlgE1-1 predominantly introduced G blocks while AlgE1-2 introduced mainly MG blocks. The reaction rate of AlgE1-1 was found to be low compared to those of AlgE1-2 and the native enzyme. In order to study the contribution of A and R modules to the epimerization rate, epimerization pattern, and calcium dependence of the epimerase, we have expressed several new truncated forms of algE1.
FIG. 1.
The modular structure of AlgE1. The restriction map of the A. vinelandii DNA used in this study is shown at the top. Only the Sau3AI site actually used is shown. The three 3′ restriction sites originate from the vector. The epimerases used in the study are shown below as solid lines with the plasmids encoding them indicated at the left. The amino acids shown are those encoded by vector DNA (underlined) or the preceding or succeeding module.
MATERIALS AND METHODS
Standard techniques.
Escherichia coli JM109 (33) was grown at 37°C in L broth or L agar. Plasmid isolation, enzymatic manipulations of DNA, and gel electrophoresis were performed according to the methods of Sambrook et al. (27). Transformations were performed as described by Chung et al. (5). The construction of the plasmids is described in Table 1 and shown in Fig. 1. The primers used for creating an in-frame stop codon at the 3′ end of the sequence encoding the A2 module were 5′ AGCGGATAACAATTTCACACAGGA 3′, which binds to the vector upstream of algE1, and 5′ CTCAAGCTTAGTCGGTCCCCTGCGG 3′ (the HindIII site is shown in boldface, and the bases complementary to the UAA stop are underlined). Protein concentrations were measured by the Bio-Rad Coomassie brilliant blue-based assay, using bovine serum albumin as a standard.
TABLE 1.
Plasmids used in the study
| Plasmid | Descriptiona | Reference |
|---|---|---|
| pUC18 | Apr, ColE1 replicon | 24 |
| pTrc99A | Apr, ColE1 replicon; trc promoter for expression of cloned genes | 1 |
| pJB658cop271C/celB | Apr, RK2 replicon; contains celB from Acetobacter xylinum | 3 |
| pHH1 | AlgE1 cloned into pTrc99A | 11 |
| pHH14 | Derivative of pTrc99A encoding the first 1.6-kb (AR) of algE1 (NcoI-KpnI) | This study |
| pHE1 | A 4.1-kb partially restricted Sau3AI DNA fragment inserted into the BamHI site of pUC18 | This study |
| pHE21 | Derivative of pTrc99A with the insert of pHE1 | This study |
| pHE26 | Derivative of pHE21 with a 0.6-kb SphI-SphI DNA fragment deleted | This study |
| pHE27 | Derivative of pHE26 with a 1.2-kb KpnI-KpnI-NruI DNA fragment deleted | This study |
| pHE29 | Derivative of pHE26 in which the XbaI site was filled in | This study |
| pHE31 | Derivative of pHE1 from which a 2.4-kb StyI-SalI DNA region was deleted | This study |
| pHE33 | Derivative of pHE26 from which a 0.3-kb EcoRI-MscI DNA fragment was deleted | This study |
| pHE35 | Derivative of pHH1 from which a 2.3-kb partially restricted NruI-HindIII DNA fragment was deleted | This study |
| pHE36 | Derivative of pHH14 in which a 35-bp KpnI-HindIII DNA fragment was replaced by a 0.9-kb KpnI-HindIII DNA fragment from pHE31 | This study |
| pHE37 | ARRR cloned into pTrc99A | 11 |
| pHE42 | Derivative of pHH14 from which a 0.5-kb MscI-XbaI DNA fragment was deleted | This study |
| pHE51 | Derivative of pHE27 where the 1.0-kb BglI-HindIII DNA fragment was replaced with a 0.4-kb BglI-HindIII DNA fragment generated by PCR | This study |
| pHE54 | Derivative of pHE33 from which a 2.8-kb KpnI-HindIII DNA fragment was deleted; encodes R1 | This study |
| pHE56 | A2R cloned into pTrc99A | 11 |
| pHE57 | Derivative of pHE51 in which a 0.45-kb NcoI-MfeI DNA fragment was replaced with a 2.5-kb NcoI-MfeI DNA fragment from pHE37 | This study |
| pHE72 | Derivative of pJB658cop251CcelB where a 0.9-kb NruI-NruI-SalI DNA fragment was replaced with a 0.5-kb NruI-SalI DNA fragment from pHE36 encoding R3 | This study |
| pHE89 | Derivative of pHE21 from which a 2.5-kb EcoRI-BglII DNA fragment was deleted | This study |
Vector-encoded restriction sites are underlined.
Preparation of enzyme extracts.
E. coli JM109 containing the plasmid of interest was grown overnight in 3× L broth (30 g of tryptone, 15 g of yeast extract, and 5 g of NaCl per liter) supplemented with 100 μg of ampicillin/ml. The culture was diluted 1:100 in the same, prewarmed medium and grown for 3 h before being induced by IPTG (isopropyl-β-d-thiogalactopyranoside; final concentration, 0.5 mM). The cells were harvested by centrifugation after another 4 h, resuspended in one-tenth volume in 50 mM MOPS (3-[N-morpholino]propanesulfonic acid [pH 6.9]) containing 5 mM CaCl2, and disrupted by sonication. The cell debris was removed by centrifugation at 10,000 × g for 30 min and filtration of the supernatant through a 0.2-μm-pore-size filter. The epimerases were then partially purified by using a Pharmacia HiTrapQ ion-exchange column with 50 mM MOPS (pH 6.9) containing 1 mM CaCl2 and a 0 to 1 M NaCl gradient.
Measurements of epimerase activity by radioisotope assay.
Epimerase activities were quantified by measuring the liberation of tritium from [5-3H]alginate to water as described previously (29). Epimerase, 50 mM MOPS (pH 6.9), and CaCl2 (final concentration, 3 mM unless otherwise stated) were mixed in a total volume of 550 μl and prewarmed at 37°C for 30 min. Then, 50 μl of prewarmed [5-3H]alginate from P. aeruginosa (2 mg/ml in H2O; specific activity, 100,000 dpm/mg) (26) was added, and the mixtures were incubated at 37°C. The alginate was precipitated by adding 15 μl of NaCl and 800 μl of isopropanol and was incubated at −50°C for at least 15 min. After centrifugation for 30 min, the activity in 1 ml of the supernatant was determined in a liquid scintillation counter. All measurements were performed in duplicate, and the results were confirmed by at least one independent experiment. Since the activities of AlgE1 and truncated forms of AlgE1 are constant for more than 30 h at 37°C (8), the low activity of some of the enzymes was compensated for by increasing the incubation time in order to obtain counts between 1,000 and 2,000 dpm. When the Ca2+ requirements were measured, the analyses were complicated by the fact that as the amounts of G and especially G blocks increase, the substrate will bind more divalent cations, thus possibly diminishing the amount of Ca2+ available to the enzyme. To minimize this effect the reactions were stopped when the degree of epimerization was less than 20%.
Measurements of G distribution pattern by NMR spectroscopy.
Epimerase, 50 mM MOPS (pH 6.9), CaCl2 (final concentration, 3 mM), and 7.5 mg of alginate (degree of epimerization [FG], <0.04) (prepared from P. aeruginosa as described previously [26]) were mixed in a total volume of 6 ml. The amount of enzyme and the incubation time were adjusted for each enzyme to obtain an FG of between 0.3 and 0.5. After incubation at 37°C, the reactions were stopped by adding Na2-EDTA (pH 8.0) to 10 mM, and the mixture was dialyzed extensively against Milli-Q water. The pH was adjusted to 6.9, and the alginate was freeze-dried and subsequently dissolved in D2O. Nuclear magnetic resonance (NMR) spectra were obtained by using a 300-MHz Bruker spectrometer. The spectra were integrated, and FG, FGG, and FMG,GM were calculated as described previously (15) by using the equations FG + FM = 1, FGG + FGM + FMG + FMM = 1, and FGM ≈ FMG.
Binding of 45Ca2+.
The presence of calcium-binding proteins was detected by 45Ca2+ autoradiography as described by Maruyama et al. (20). The proteins were separated on a sodium dodecyl sulfate (SDS)–8% polyacrylamide gel and blotted onto nitrocellulose. After washing four times with 10 mM imidazol (pH 6.8) containing 60 mM KCl, 5 mM MgCl2, and 20 mM Na2-EDTA (first wash only), the filter was incubated in 20 ml of the same buffer (lacking Na2-EDTA) containing 60 μCi of 45CaCl2. The filter was washed in deionized water, air dried, and autoradiographed on Hyperfilm β-max (Amersham). The proteins on the filter were stained with 0.1% amido black.
RESULTS AND DISCUSSION
Construction of the expression plasmids.
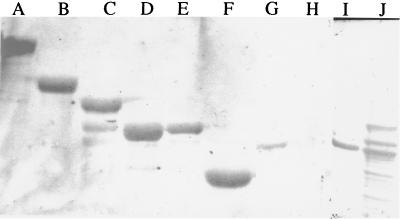
To further analyze the functional role of the different modules in AlgE1, we constructed new truncated forms of the enzyme by removing one or more of the modules at the DNA level (Fig. 1). E. coli cells containing the different plasmids were induced with IPTG, and the corresponding partially purified proteins were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) (Fig. 2). The dominant band in each lane has an apparent molecular mass corresponding to that expected for the recombinant proteins, taking into account that the AlgE epimerases migrate as if their molecular masses are somewhat larger than those calculated from their amino acid sequences (7, 11).
FIG. 2.
Denaturing gel electrophoresis of proteins expressed by the different plasmids. The proteins were stained with Coomassie brilliant blue. The structures of the epimerases were as follows: lane A, R3A2 (pHE51); lane B, A2R4 (pHE56); lane C, R3A2R4 (pHE27); lane D, A2 (pHE58); lane E, A1 (pHE42); lane F, A1R1 (pHE29); lane G, A1R1R2 (pHE35); lane H, A1R1R2R3 (pHE37); lane I, A1R1R2R3A2 (pHE57); lane J, A1R1R2R3A2R4 (pHH1). Six micrograms of protein was loaded in each lane. The proteins were partially purified by ion-exchange chromatography, except for the epimerase in lane B, which was further purified by gel filtration (11). The numbers indicate molecular masses (in kilodaltons) of a molecular mass standard.
The A modules are sufficient for epimerization.
The proteins were then analyzed for epimerase activity, and as expected (11), all enzymes containing an A module with a C-terminal R module were catalytically active (Table 2). The activity may also be measured in the crude extract (11), but removal of most of the contaminating proteins made it possible to compare the activities of the different epimerases. Interestingly, the protein containing an A module with an N-terminal R module but no C-terminal R module (R3A2) was also active, although the activity was somewhat lower than that of the corresponding enzyme with the R module positioned C terminally (A2R4). Finally, all R modules were removed and each A module was expressed alone. Even though the reaction rates were much lower, it was clear that the A modules alone are sufficient for epimerization. A construct containing the R1 module only was also made (pHE54), and crude extracts from cells containing this plasmid did not display any measurable epimerase activity, indicating that the A modules are both necessary and sufficient for epimerization. A construct encoding R4 fused to the last 118 amino acids of A2 was also made (pHE89). This plasmid did not encode any active epimerase either, confirming that the R modules are not sufficient for activity. The result also showed that the C-terminal third of the A module is not sufficient for activity.
TABLE 2.
Specific activities of partially purified epimerases
| Plasmid | Protein | Specific activitya
|
|
|---|---|---|---|
| Ab | Bb | ||
| pHH1 | A1R1R2R3A2R4 | 10,700 | 5,800 |
| pHE57 | A1R1R2R3A2 | 10,600 | 7,700 |
| pHE37 | A1R1R2R3 | 131 | 82 |
| pHE35 | A1R1R2 | 105 | 53 |
| pHE29 | A1R1 | 240 | 151 |
| pHE27 | R3A2R4 | 9,800 | 8,700 |
| pHE56 | A2R4 | 10,200 | 6,100 |
| pHE51 | R3A2 | 7,400 | 3,600 |
| pHE42 | A1 | 19 | 13 |
| pHE58 | A2 | 880 | 620 |
| pHE54c | R1 | 0 | 0 |
| pHE89c | R4 | 0 | 0 |
The specific activity is given in dpm per hour per microgram of protein.
A and B refer to two independent experiments.
Crude extract.
All strains encoding the active epimerases were grown, and the proteins were partially purified in the same experiment. The experiment was repeated in order to see if the differences were caused by occasional variations in enzyme purity for one or more of the enzymes. Even though the specific activities in the two experiments vary (Fig. 2), the figures in Table 2 indicate a significant difference between the two A modules, with the A2 module displaying a reaction rate much higher than that of A1. A C-terminal R module increased the reaction rate about 10-fold for each A module. An N-terminal R module also increased the reaction rate, although not to the same extent. This shows that both an N-terminal and a C-terminal R module influence the reaction rate of an A module.
The A modules determine the structure of the reaction product.
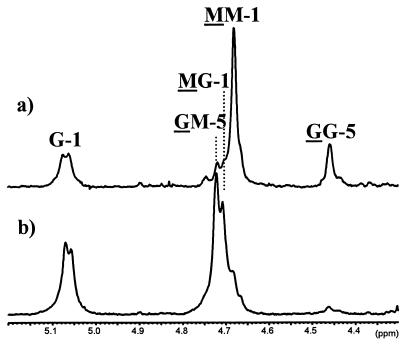
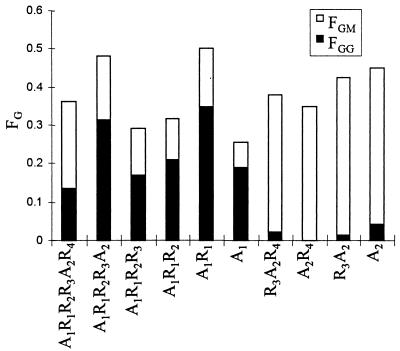
Alginate containing less than 6% G was epimerized by the A1 and A2 modules and analyzed by NMR spectroscopy. The spectra (Fig. 3) show that the A1 module predominantly introduces G residues into G blocks, since the GG peak is much more dominant than the GM peak. The A2 module, on the other hand, predominantly introduces single G residues (as indicated by a dominant GM signal and very small GG peak). Similar analyses were performed for all the truncated epimerases to see if the number of R modules somehow influenced the structure of the epimerized alginate. The experiment was performed at least twice for each enzyme. The results from the experiments having FG closest to 0.3 are summarized in Fig. 4, which shows the fractions of G, G blocks, and MG blocks present in the alginate. All the enzymes which contain only the A1 module introduced more G blocks than MG blocks, while all the enzymes containing only the A2 module made MG blocks and almost no G blocks. It has been found that AlgE4 is also able to make G blocks, although at a much lower rate than it makes MG blocks (16). As shown previously (11), the FGG/FGM ratio increases with increasing FG, and the differences in this ratio among the enzymes containing only the first A module are thus not unexpected. Since there did not seem to be any correlation between the amount of G blocks produced and the number or position of R modules for any of these truncated enzymes, it must be concluded that the A modules are sufficient not only for catalysis but also for determining the structure of the epimerized alginate.
FIG. 3.
1H NMR (300 MHz) spectra of alginate epimerized by A1 (a) and A2 (b). The residues causing the signal are underlined, the numbers denote which H is causing the signal, and the nonunderlined residues refer to the neighboring residue.
FIG. 4.
NMR analyses of alginate epimerized by the truncated epimerases. The lengths of the bars show the G content, the filled parts of the bars show the fraction of G blocks, and the open parts show the fraction of MG blocks.
The removal of only the R4 module from whole AlgE1 had a significant effect on the epimerization pattern of the enzyme. This may, however, be explained by assuming that the R4 module has some effect on the activity of the A2 module relative to that of the A1, such that the reaction rate of A2 becomes lower when R4 is not present. The significant difference in specific activity between the proteins R3A2R4 and R3A2 (Table 2) seems to support this hypothesis.
So far, 10 different A modules in AlgE epimerases from A. vinelandii have been described (10, 32). Since all of these modules are highly homologous, it should be possible to determine which parts of them are responsible for the G distribution pattern and the differences in reaction rates. This might be done by exchanging parts of the modules between different epimerase genes, and such experiments have now been initiated in our laboratory.
Both the A and the R modules bind calcium.
The repeated motifs in the R modules are homologous to a calcium-binding motif found in a metalloprotease from P. aeruginosa (2), suggesting a possible role for the R modules in the binding of this cation. To analyze this, the proteins were blotted on membranes from SDS-polyacrylamide gels and incubated with 45Ca2+ (Fig. 5). Binding of the radioisotope was easily visualized for whole AlgE1 (lane A) and several truncated forms containing at least one A module and one to three of the R modules (lanes B to E). Interestingly, expression of only A1 showed that this module alone binds Ca2+ (lane F). A similar result was obtained for A2 alone (not shown). Exposure of the membrane to Na2-EDTA prior to the binding of calcium was found to stimulate the binding of the radioisotope (not shown), presumably because this pretreatment leads to the removal of already-bound nonradioactive Ca2+.
FIG. 5.
45Ca2+ binding by different truncated enzymes. Lanes: A, A1R1R2R3A2R4; B, A1R1R2R3; C, A1R1R2; D, A1R1; E, R3A2; F, A1; G, CelB-R3; H, CelB; I and J (underlined), amido-black-stained filter after removal of the bound 45Ca2+ of CelB and CelB-R3 (lanes H and G), respectively. The enzymes in lanes A to F were partially purified, whereas crude extracts were loaded in lanes G and H.
Based on these experiments, we concluded that either the R modules are not involved in binding of the cation or both modules are capable of binding. To distinguish between these two possibilities, we have also studied the binding of 45Ca2+ to the R module only. Expression from pHE54 was not sufficiently high to allow visualization of the protein after SDS-PAGE directly from crude extracts, and since this module apparently did not display any epimerase activity, it was difficult to make an R-module preparation containing protein concentrations comparable to those of the other truncated forms of AlgE1. To overcome this problem, we fused (at the DNA level) the R3 module to the CelB protein (phosphoglucomutase) from Acetobacter xylinum. This fusion partner was chosen because we have previously shown that it could be expressed at very high levels in E. coli (3). Lane H shows that CelB itself does not bind the radioisotope, although it is clearly visible on the amido-black-stained membrane (lane I). The crude extract of the fusion protein contains many contaminating proteins (lane J), but only the fusion protein binds radioisotope. It could therefore be concluded that both the A and the R modules bind Ca2+.
In some of the lanes, more than one protein appeared to bind the radioisotope. These signals vary from one extract to another and are believed to result from a tendency of the epimerases to form several distinct bands when separated on denaturing polyacrylamide gels (16).
The R modules modulate the enzymes’ requirements for calcium.
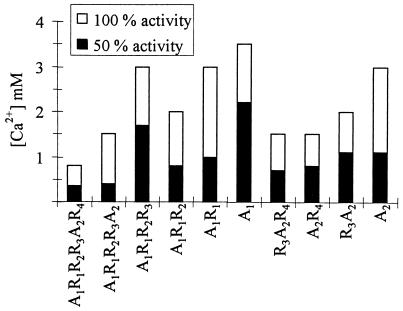
To study the role of the R modules further, we determined the optimal concentrations of Ca2+ for activity of AlgE1 and its truncated forms. We observed that the shapes of the curves varied somewhat among the different enzymes, as, for instance, those lacking A1 displayed a very slight slope around their optimal values. It therefore seemed more meaningful to also compare the values at which each enzyme displayed 50% activity, and both these values and those that were obtained for optimal activity are shown in Fig. 6. The A1 module seems to need slightly more Ca2+ for 50% activity than the A2 module. The R modules appear to lower the requirements for Ca2+, such that the more R modules are present, the less of the cation is needed for full activity. This is, however, not true for A1R1R2R3, which needs more Ca2+ than A1R1R2. The explanations for these observations are not clear, partly because the exact role of the R modules has not been determined. In particular, interpretations are complicated by the fact that Ca2+ is bound by alginate as well as by both the A and the R modules. It could be that the R modules function as a source of Ca2+ for the catalytic part (the A module) and that the positioning of the R module relative to the A module affects its efficiency in donating the cation to the catalytic part. Alternatively, one might imagine that the R-module Ca2+ complex stimulates binding of the epimerase to the substrate, thus indirectly stimulating the epimerization process.
FIG. 6.
Ca2+ requirements of the truncated epimerases. The lengths of the bars show the concentrations of calcium needed for 100% activity, while the filled parts of the bars show the concentrations needed for 50% activity. The concentrations of Ca2+ used were 0.4 to 1.2 mM in 0.2 mM increments and 1.5 to 5.0 mM in 0.5 mM increments. The results for A1R1R2R3A2R4, A1R1R2R3, and A2R4 are from Ertesvåg et al. (11).
The modular structure of AlgE1, where the A modules alone are sufficient for the reaction, may suggest that an ancestral epimerase contained only this module and that the R modules were added to the gene at a later stage. It has been proposed that the Ca2+-binding motifs found in the R modules participate in the folding of the protein and thus also facilitate its secretion (25). In the case of AlgE1, it seems that the binding of Ca2+ by the R modules also increases the epimerization rate, possibly by increasing the amount of this cation available for the catalytic part.
ACKNOWLEDGMENTS
This work was supported by the Norwegian Research Council and by Pronova Biopolymers AS.
We thank Gudmund Skjåk-Bræk and Wenche Iren Strand for providing the alginate and analyzing the NMR samples.
REFERENCES
- 1.Amann I, Ochs B, Abel K-J. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988;69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 2.Baumann U, Wu S, Flaherty K M, McKay D B. Three-dimensional structure of the alkaline protease of Pseudomonas aeruginosa: a two domain protein with a calcium binding parallel roll motif. EMBO J. 1993;12:3357–3364. doi: 10.1002/j.1460-2075.1993.tb06009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blatny J M, Brautaset T, Winther-Larsen H, Karunakaran P, Valla S. Improved broad-host-range RK2 vectors useful for high and low regulated gene expression levels in gram-negative bacteria. Plasmid. 1997;38:35–51. doi: 10.1006/plas.1997.1294. [DOI] [PubMed] [Google Scholar]
- 4.Campos M-E, Martinez-Salazar J M, Lloret L, Moreno S, Nunez C, Espin G, Soberon-Chavez G. Characterization of the gene coding for GDP-mannose dehydrogenase (algD) from Azotobacter vinelandii. J Bacteriol. 1996;178:1793–1799. doi: 10.1128/jb.178.7.1793-1799.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung C T, Niemela S L, Miller R H. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci USA. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cote G L, Krull L H. Characterization of the exocellular polysaccharides from Azotobacter chroococcum. Carbohydr Res. 1988;181:143–152. [Google Scholar]
- 7.Ertesvåg H, Doseth B, Larsen B, Skjåk-Bræk G, Valla S. Cloning and expression of an Azotobacter vinelandii mannuronan C-5-epimerase gene. J Bacteriol. 1994;176:2846–2853. doi: 10.1128/jb.176.10.2846-2853.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ertesvåg H. Ph.D. thesis. Trondheim, Norway: University of Trondheim; 1994. [Google Scholar]
- 9.Ertesvåg H, Erlien F, Skjåk-Bræk G, Rehm B H A, Valla S. Biochemical properties and substrate specificities of a recombinantly produced Azotobacter vinelandii alginate lyase. J Bacteriol. 1998;180:3779–3784. doi: 10.1128/jb.180.15.3779-3784.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ertesvåg H, Høidal H K, Hals I K, Rian A, Doseth B, Valla S. A family of modular type mannuronan C-5-epimerase genes controls alginate structure in Azotobacter vinelandii. Mol Microbiol. 1995;16:719–731. doi: 10.1111/j.1365-2958.1995.tb02433.x. [DOI] [PubMed] [Google Scholar]
- 11.Ertesvåg H, Høidal H K, Skjåk-Bræk G, Valla S. The Azotobacter vinelandii mannuronan C-5-epimerase AlgE1 consists of two separate catalytic domains. J Biol Chem. 1998;273:30927–30932. doi: 10.1074/jbc.273.47.30927. [DOI] [PubMed] [Google Scholar]
- 12.Fett W F, Cescutti P, Wijey C. Exopolysaccharides of the plant pathogens Pseudomonas corrugata and Ps. flavescens and the saprophyte Ps. chlororaphis. J Appl Bacteriol. 1996;81:181–187. [Google Scholar]
- 13.Gorin P A J, Spencer J F T. Exocellular alginic acid from Azotobacter vinelandii. Can J Chem. 1966;44:993–998. [Google Scholar]
- 14.Govan J R W, Fyfe J A M, Jarman T R. Isolation of alginate-producing mutants of Pseudomonas fluorescens, Pseudomonas putida and Pseudomonas mendocina. J Gen Microbiol. 1981;125:217–220. doi: 10.1099/00221287-125-1-217. [DOI] [PubMed] [Google Scholar]
- 15.Grasdalen H, Larsen B, Smidsrød O. A p.m.r. study of the composition and sequence of uronate residues in alginates. Carbohydr Res. 1979;68:23–31. [Google Scholar]
- 16.Høidal, H. K., H. Ertesvåg, G. Skjåk-Bræk, B. T. Stokke, and S. Valla. The recombinant Azotobacter vinelandii mannuronan C-5-epimerase AlgE4 epimerizes alginate by a non-random attack mechanism. J. Biol. Chem., in press. [DOI] [PubMed]
- 17.Larsen B, Haug A. Biosynthesis of alginate. 3. Tritium incorporation with polymannuronic acid 5-epimerase from Azotobacter vinelandii. Carbohydr Res. 1971;20:225–232. doi: 10.1016/s0008-6215(00)81375-0. [DOI] [PubMed] [Google Scholar]
- 18.Linker A, Jones R S. A polysaccharide resembling alginic acid from a Pseudomonas microorganism. Nature. 1964;204:187–188. doi: 10.1038/204187a0. [DOI] [PubMed] [Google Scholar]
- 19.Lloret L, Barreto R, Leon R, Moreno S, Martinez-Salazar J, Espin G, Soberon-Chavez G. Genetic analysis of the transcriptional arrangement of Azotobacter vinelandii alginate biosynthetic genes: identification of two independent promoters. Mol Microbiol. 1996;21:449–457. doi: 10.1111/j.1365-2958.1996.tb02554.x. [DOI] [PubMed] [Google Scholar]
- 20.Maruyama K, Mikawa T, Ebashi S. Detection of calcium binding proteins by 45Ca autoradiography on nitrocellulose membrane after sodium dodecyl sulfate gel electrophoresis. J Biochem (Tokyo) 1984;95:511–519. doi: 10.1093/oxfordjournals.jbchem.a134633. [DOI] [PubMed] [Google Scholar]
- 21.May T B, Chakrabarty A M. Pseudomonas aeruginosa: genes and enzymes of alginate synthesis. Trends Microbiol. 1994;2:151–157. doi: 10.1016/0966-842x(94)90664-5. [DOI] [PubMed] [Google Scholar]
- 22.Mejia-Ruiz H, Guzman J, Moreno S, Soberon-Chavez G, Espin G. The Azotobacter vinelandii alg8 and alg44 genes are essential for alginate synthesis and can be transcribed from an algD-independent promoter. Gene. 1997;199:271–277. doi: 10.1016/s0378-1119(97)00380-6. [DOI] [PubMed] [Google Scholar]
- 23.Mejia-Ruiz H, Moreno S, Guzman J, Najera R, Leon R, Soberon-Chavez G, Espin G. Isolation and characterization of an Azotobacter vinelandii algK mutant. FEMS Microbiol Lett. 1997;156:101–106. doi: 10.1111/j.1574-6968.1997.tb12712.x. [DOI] [PubMed] [Google Scholar]
- 24.Norrander J, Kempe T, Messing J. Improved M13 vectors using oligonucleotide-directed mutagenesis. Gene. 1983;26:101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- 25.Ofstad R, Larsen B. The effect of calcium-ion concentration on poly-d-mannuronate C-5-epimerase. In: Levring T, editor. Proceedings of the 10th International Seaweed Symposium. Berlin, Germany: Walter de Gruyter & Co.; 1981. pp. 485–493. [Google Scholar]
- 26.Rehm B H, Ertesvåg H, Valla S. A new Azotobacter vinelandii mannuronan C-5-epimerase gene (algG) is part of an alg gene cluster physically organized in a manner similar to that in Pseudomonas aeruginosa. J Bacteriol. 1996;178:5884–5889. doi: 10.1128/jb.178.20.5884-5889.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Skjåk-Bræk G, Espevik T. Application of alginate gels in biotechnology and biomedicine. Carbohydr Eur. 1996;14:19–25. [Google Scholar]
- 29.Skjåk-Bræk G, Larsen B. A new assay for mannuronan C-5-epimerase activity. Carbohydr Res. 1982;103:133–136. [Google Scholar]
- 30.Smidsrød O, Draget K I. Chemistry and physical properties of alginates. Carbohydr Eur. 1996;14:6–12. [Google Scholar]
- 31.Soon-Shiong P, Heintz R E, Merideth N, Yao Q X, Yao Z, Zheng T, Murphy M, Moloney M K, Schmehl M, Harris M, Mendez R, Mendez R, Sandford P A. Insulin dependence in a type 1 diabetic patient after encapsulated islet transplantation. Lancet. 1994;343:950–951. doi: 10.1016/s0140-6736(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 32.Svanem B I G, Skjåk-Bræk G, Ertesvåg H, Valla S. Cloning and expression of three new Azotobacter vinelandii genes closely related to a previously described gene family encoding mannuronan C-5-epimerases. J Bacteriol. 1999;181:68–77. doi: 10.1128/jb.181.1.68-77.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]