Abstract
Background:
Primary amoebic meningoencephalitis (PAM) is an acute and fulminant CNS infection caused by Naegleria fowleri. Recreational activities and ritual ablution with contaminated warm fresh water are the main reason of PAM. Pakistan ranked the second most affected country, where most of the PAM incidences were reported from Karachi, Pakistan.
Methods:
In May, 2019, a 28-yr-old suspected PAM patient came to the Imam Zain-Ul-Abdin Hospital, Karachi. Biochemical and cytological investigations of patient‘s CSF were carried out at Karachi Diagnostic Center and Molecular Biology Lab. Sequencing of Naegleria sp. specific (ITS) primer-based amplicons was performed from both patient‘s CSF and water samples followed by multiple sequence alignment and phylogenetic studies.
Results:
Biochemical and cytological investigations of patient‘s CSF showed 5 mg/dl glucose, 240 mg/dl total protein and 2260/mm3 TLC suggesting acute meningoencephalitis. PCR-based analyses of patient‘s CSF and his residential tap water samples using Naegleria sp. specific (ITS) and N. fowleri specific primers revealed the presence of N. fowleri DNA. Nucleotide sequences of ITS primer-based amplicons from both patient‘s CSF and water samples were submitted in GenBank under the accession numbers MT726981.1 and MT726226.1, respectively. According to phylogenetic analysis, N. fowleri isolate from Pakistan has shown the least node age of seven.
Conclusion:
Here, for the very first time in Pakistan, N. fowleri genotype has been identified as type-2. Phylogenetic analysis showed that N. fowleri isolate from Pakistan is among the latest descendants, i.e., evolved later in life.
Keywords: Naegleria fowleri, Genotyping, Primary amoebic meningoencephalitis
Introduction
Naegleria fowleri is the causative agent of primary amoebic meningoencephalitis (PAM). It is a habitant of warm lakes, streams, spas, pools, domestic water reservoir and domestic water supplies (1–3). PAM cases are mostly observed in hot summers as N. fowleri proliferates rapidly at higher temperatures, i.e. 40–46 °C (4–6). Hundreds of PAM cases have been reported in last five decades; most probably due to global warming-related environmental changes (7). The WHO has also declared PAM as the second major cause of morbidity and mortality worldwide (8).
PAM incidences have been reported in many countries including America, Australia, Hong Kong, Thailand, Taiwan and China associated with a recent history of swimming in warm fresh water or direct exposure to contaminated tap water (1, 9–11). The clinical manifestations of PAM are quite similar to that of acute bacterial meningitis which makes PAM really hard to get differentiated from other bacterial meningitis. The resulting delayed diagnosis of PAM is one of main reasons of high mortality (1, 12–15). However, encephalopathic patients showing a triad of symptoms, i.e. fever, nausea and a low ESR should be urgently referred to lumbar puncture for confirmed diagnosis (16).
The first PAM case was reported in 2008 from Karachi, Pakistan (1). An obvious increase in PAM incidences has been reported during the last few years. In USA, N. fowleri infection has been identified generally in children of less than 14 years of age. In contrast, most of the PAM patients in Pakistan were adults having 26–45 years of age. This prominent difference indicates somewhat unique N. fowleri strain in Pakistan which needs to be characterized in detail (17–19).
Despite of morphological similarities among N. fowleri isolates, eight distinct N. fowleri genotypes have been characterized on the basis of differences in ribosomal internal transcribed spacers, including 5.8S rDNA. The ITS and 5.8S rDNA sequences will be of additional help in describing new Naegleria spp. in future (20). According to the previous studies, N. fowleri genotypes are unequally dispersed in different continents where genotypes 1, 2 and 3 are found in America, genotypes 2, 3, 4, 5, 6, 7 and 8 in Europe and genotypes 2 and 3 occur in Asia. Out of these eight genotypes, only types 1, 2, 3 and 5 are clinically significant (1, 21). Annotation of particular pathogenic genotype is likely to assist the development of a potential genotype-specific vaccine or drug against it. Additionally, different geographical distribution of diverse genotypes made them important epidemiologic marker that can trace the source of infection in a particular population (22, 23). N. fowleri type-2 genotype has been reported from many Asian countries but has not been testified yet in Pakistan (15, 24, 25).
The main aim of the present study was to perform genotyping of Pakistani N. fowleri isolate and reveal its phylogenetic relationship with other N. fowleri isolates.
Materials and Methods
A 28 yr-old suspected PAM patient came to the Imam Zain-Ul-Abdin Hospital, Karachi in 2019. Biochemical and cytological investigations of patient‘s CSF were carried out at Karachi Diagnostic Center and Molecular Biology Lab.
Ethical approval and consent to participate
This study was approved by the Ethics Committee of Karachi Diagnostic and Molecular Biology Lab (EC Ref No: RECNF02). The patient was unconscious that is why the informed consent was acquired in written form from his elder brother.
Biochemical, cytological and microscopic analysis of patient’s CSF sample
Cytological analysis of fresh CSF sample was carried out using Sysmix KX 21N Hematology analyzer. For further analysis, the CSF sample was centrifuged at 2000×g for 15 min at room temperature (26, 27). The supernatant was analyzed for the estimation of Glucose and overall protein content. The pellet was resuspended in 200 μl of supernatant followed by 30 min incubation at 37 °C. Resuspension was used both for the N. fowleri detection by direct microscopy and N. fowleri cultivation on attenuated E. coli (ATCC number 25922) containing non-nutrient agar plate (28–30).
Detection of N. fowleri DNA in patient’s CSF and residential tap water
The CSF sample was centrifuged at 10,000×g for 10 min. The sediment was subjected to DNA extraction using the QIAamp DNA Mini Kit (Qiagen Inc., USA). Tap water sample (1000 ml) was collected in a sterile bottle from patient’s residency and processed within 12 hours. The water sample was filtered through nitrocellulose membrane (MS®MCE Gridded Membrane Filter) having 0.45 μm pore-size to trap N. fowleri. The nitro-cellulose membrane was aseptically removed from filtration apparatus, cut into small pieces and placed in 50-ml sterile falcon tube. Wash buffer was prepared by adding 10 μl Tween 20 to 5 ml of AE buffer (QIAamp DNA Mini Kit) and immediately transferred in membrane containing falcon tube followed by vortex for 3 minutes. Rest of the protocol was same as recommended in QIAamp DNA Mini Kit (Qiagen Inc., USA). Genomic DNA was stored at −20 °C.
N. fowleri PCR-based detection
Identification of N. fowleri among other Naegleria spp. based on cellular morphology is difficult so PCR-based detection was performed. As reported previously (31–34), amplification assays were performed in a total volume of 25 μl, containing 9.5 μl of ddH2O, 0.5μl of each primer (10μM), 10 μl Green Master Mix (Promega, USA), and 5.0μl of template genomic DNA extracted from the CSF samples. For detection of Naegleria spp. NfITS1-F and NfITS1–R primer set was used. Whereas, for specific detection of pathogenic N. fowleri in the CSF samples, NaeglF1925 and NaeglR344 primer set was used.
Nf-ITS1-F 5′ GAACCTGCGTAGGGATCATTT 3′
Nf-ITS2-R 5′ TTTCTTTTCCTCCCCTTATTA 3′ (35)
NaeglF1925 5′GTGCTGAAACCTAGCTATTGTAACT CAGT 3′
NaeglR344 5′CACTAGAAAAAGCAAACCTGAAAGG 3′ (36)
Followed by a prolonged denaturation i.e. 95 °C for 5 min, amplification reactions with primer pairs were performed in 40 cycles of denaturation at 95 °C for 3 seconds, annealing at 53 °C for 30 seconds and extension at 72 °C for 30 seconds. A final extension at 72 °C for 5 min was included. Amplified product was visualized in a 2 % agarose gel (31).
Sequence Analysis and Genotyping
The purified PCR product of internal transcribed spacer-1 (ITS1), 5.8S rRNA gene, internal transcribed spacer-2 (ITS2) region was sequenced by both ITS-1 and ITS-2 primers using ABI PRISM 3730 Genetic Analyzer (Applied Biosystems Inc., USA). The reaction mixture was prepared by mixing 7 μL of purified PCR product, 0.5 μL primer (10 μM), 1.25 μL buffer and 1.5 μL Big Dye Terminator (Applied Biosystem Inc., USA). The sequencing reaction was subjected to 25 cycles of 96 °C for 10 seconds, 50 °C for 5 seconds and 60 °C for 4 minutes in thermal cycler. Ethanol precipitation was carried out to remove the left over fluorescent dye followed by washing of pellet with 70% ethanol. Each pellet was resuspended in 10 μL of HiDi Formamide (Applied Biosystem Inc., USA), heat denatured for 5 min at 95 ° C and quickly chilled on ice for 3 minutes followed by DNA sequencing. The nucleotide sequences of Nf-ITS regions from patient‘s CSF and tap-water acquired in this study has been submitted to GenBank with the accession numbers MT726981.1 and MT726226.1, respectively.
Genotyping was performed using previously reported method (10). Briefly, reference sequences of eight existing genotypes consisting of ITS-1, 5.8S rDNA and ITS-2 regions of Naegleria spp. including AY376149, X96564, X96562, AJ132030, AJ132028, FR875287, X96563, and FR875288 were retrieved from GenBank and aligned with MT726981.1 and MT726226.1 sequences using MEGA-X program (37). Furthermore, phylogenetic analysis was performed to delineate the evolutionary relationship of N. fowleri isolate from Pakistan with other N. fowleri strains isolated so-far using Maximum Likelihood Tree construction method on Mega-X program (37–39).
Results
Biochemical and cytological investigations of PAM patient’s CSF
The patient was the resident of Liaquat-abad town and had no previous history of any recreational activity. He might have used contaminated water during ritual ablution or bath. His CSF sample was received in Karachi Diagnostic Center and Molecular Biology Lab for the detection of PAM. CSF analysis showed a white blood cell (WBC) count of 2260/mm3including 75% neutrophils and 25% lymphocytes. Analysis of the fresh CSF indicated glucose at a concentration of less than 5 mg/dl and proteins at a concentration of more than 240 mg/dl.
Direct microscopy of patients’ fresh CSF sample
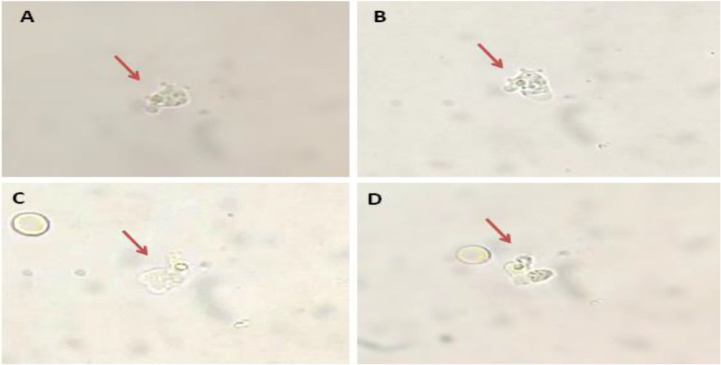
Direct microscopy of fresh CSF smear found alive motile amoebic cells with pseudo-podia in the CSF sample (Fig. 1). The continuous change in cell morphology and formation of pseudopods suggested the trophozoite state of amoeba. The trophozoites were approximately 12–15 μm in size. The crawling amoeba was observed to move rapidly with ∼1 μm/s speed using eruptive pseudopods.
Fig. 1:
Direct microscopy of patients fresh CSF sample showing the presence of motile amoeba (depicted by arrows) using 40X
PCR-based detection of N. fowleri
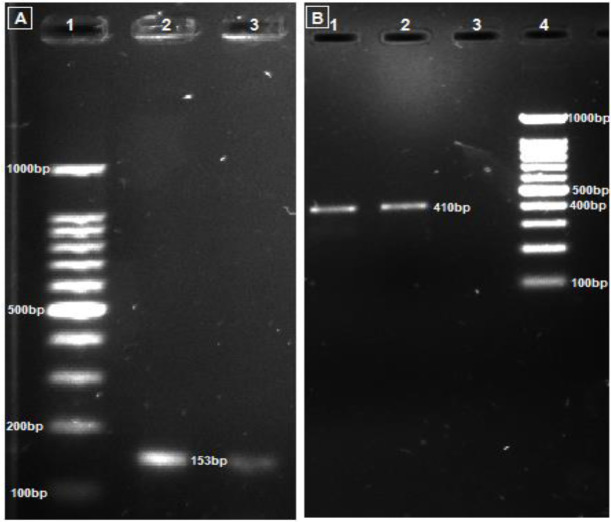
PCR using Nf-ITS1-F_Nf-ITS2-R primer pair amplified a 410bp fragment of Naegleria specie ITS (including ITS1, 5.8S rDNA and ITS2) region which confirmed the presence of Naegleria species in general (Figure 2A). However, the presence of pathogenic N. fowleri in the CSF was confirmed using NaeglF1925_NaeglR344 set of primers; which showed a 153bp amplicon consisting of a region of 18S rDNA in pathogenic N. fowleri (Fig. 2B).
Fig. 2:
PCR amplicons obtained using N. fowleri specific (NaeglF1925: NaeglR344) (A) and Naegleria spp. specific (Nf-ITS1-F: Nf-ITS1-R) primer pairs (B). In part A, lane 1–3: Marker, CSF, Tap water. In part B, lane 1–4: Tap water, CSF, blank, Marker
Isolation of N. fowleri on non-nutrient agar (NNA)
Alternate day examinations of the NNA culture plate was done up to 10 days using a light microscope with 10X magnification (OPTIKA, B-382 PLi, Italy). The trophozoite stage was observed after third day of culture in the CSF sample.
Sequence Analysis and Genotyping
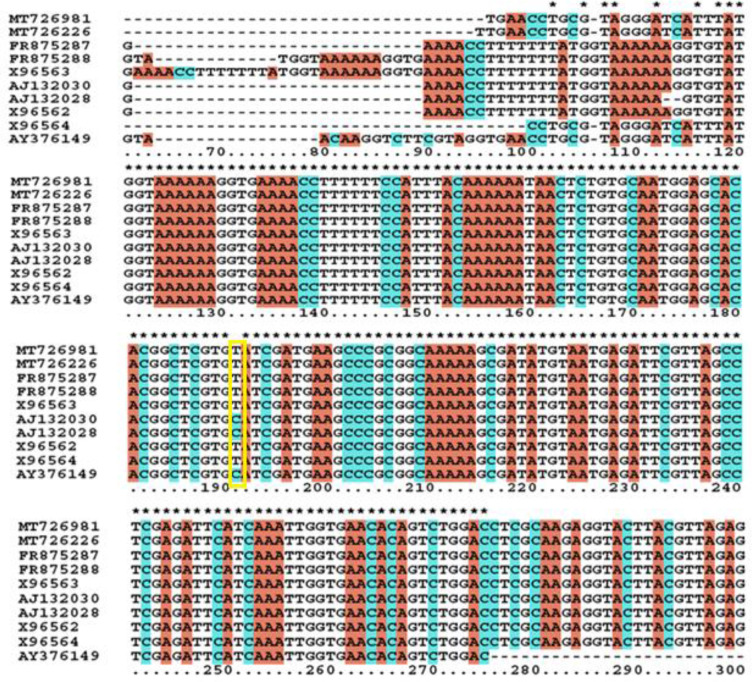
The genotyping of the isolated N. fowleri was elucidated using ITS-1 and 5.8S rRNA gene region. The ITS-1 region is comprised of 85bp which is divided into R1 (16bp), R2 (28bp), M1 (15bp) and M2 (27bp) regions. Sequence analysis showed that repeats R1 and R2 were absent in ITS-1 region of both patient‘s CSF and water samples, i.e. MT726981.1 and MT726226.1, respectively. However, presence of two main regions i.e. M1 and M2 having 42bp length and C>T transition at position 31 in 5.8S rRNA gene sequence was observed (Fig. 3).
Fig. 3:
Multiple sequence alignment of the nucleotide sequences of ITS and 5.8S rDNA from eight existing N. fowleri genotypes including same region from Pakistani N. fowleri. One copy of each M1 and M2 region of ITS-1 is apparent while C>T transition at location 31 of 5.8S rDNA is also shown (yellow box)
Evolutionary relationships with other N. Fowleri genes
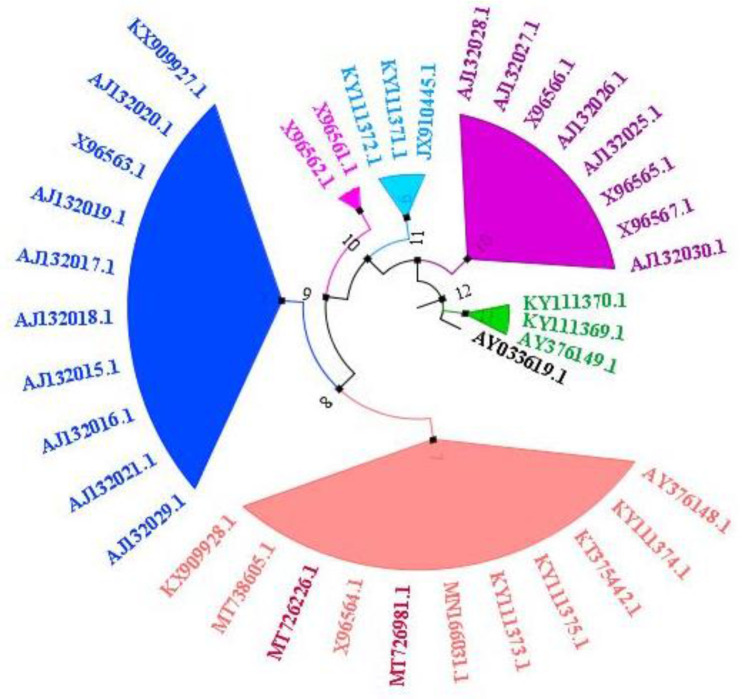
Phylogenetic analysis of MT726981.1 and MT726226.1 sequences was performed by aligning them with other 36 available N. Fowleri ITS-1, 5.8S, ITS-2 region-based sequences. Pakistani N. fowleri isolate showed closed homology with eleven other N. Fowleri isolates including nine from Asia and three from USA (Fig. 4).
Fig. 4:
Phylogenetic analysis of 38 N. Fowleri ITS-1, 5.8S, ITS-2 region-based sequences including those from PAM patient‘s CSF and water samples using Maximum Likelihood Tree construction method (39)
Discussion
In Karachi, Pakistan, PAM remains an overwhelming CNS infection connected with warm freshwater exposure since last decade. Having a population size of ∼27 million, Karachi is the largest city, industrial hub, financial capital and commercial center of Pakistan. The geographical changes especially recurrent heat shocks seem to be the cause of N. fowleri outbreaks in this city. Considering the rapid onset and high mortality rate of this deadly disease, potential vaccine, early diagnosis and effective treatment is very crucial. PAM can be confirmedly diagnosed by direct microscopy, amoebic cultivation on NNA plated with E. coli and PCR. Selective isolation of N. fowleri requires culture temperature between 42–45 °C at which the growth of other amoebae is strictly suppressed. Although N. lovaniensis is exceptional Naegleria species that can grow at 42–45 °C but being non-pathogenic it cannot be present in CSF. Generally, confirmed diagnosis of PAM is carried out from patients‘ CSF using Naegl and ITS primer-based PCR assays (2, 21, 40–42).
The present study was focused on detection and isolation of Pakistani N. fowleri isolate from patient‘s CSF and his residential tap water in order to reveal its yet unknown genotype as this could help in designing genotype-specific vaccine and drugs in future. The biochemical and cytological analyses showed an overall low concentration of glucose, high concentration of total protein and a high value of total leucocyte count with increased percentage of neutrophils. Similar findings have also been reported in other types of meningitis suggesting inadequacy of these analyses in discriminating PAM from other meningitis. However, direct microscopy of the CSF sample detected motile amoeba. Additionally, PCR analysis of both samples using ITS- and Naegl-primers also showed the presence of N. fowleri DNA.
Our primary finding was that the genotype of Pakistani N. fowleri isolate is type-2; the predominant type in Asia. Additionally, Pakistani N. fowleri isolate is among those which originated later in life. According to the calculated node ages, N. fowleri isolate from Phillipine, USA is thought to be the ancestral isolate. Whereas, N. fowleri isolate from Pakistan is among the latest descendants showing a node age of seven.
Conclusion
Molecular genotyping studies revealed that Pakistani N. fowleri isolate belongs to type-2. The identification of N. fowleri in patient‘s residential tap water confirmed the source of infection. Parallel tap water sample analysis showed the presence of coliform along with low chlorine level (data not shown). The presence of coliform strongly indicates sewage contamination in tap water. The Karachi citizens are at high risk as they use the same tap water in raw form for routine ritual ablution. Preventive measures including proper chlorination and water/sewerage pipeline fixture should be taken by Karachi Water and Sewer-age Board (KWSB) on urgent basis.
Acknowledgments
We are thankful to the team of Karachi Diagnostic Center and Molecular Biology Lab for providing us patient‘s CSF sample as well as for conducting biochemical and cytological analyses of CSF sample. The project was not funded.
Footnotes
Conflict of interest
There is no conflict of interest.
References
- 1.Shakoor S, Beg MA, Mahmood SF, et al. Primary amebic meningoencephalitis caused by Naegleria fowleri, Karachi, Pakistan. Emerg Infect Dis. 2011; 17(2):258–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Jonckheere JF. The impact of man on the occurrence of the pathogenic free-living amoeboflagellate Naegleria fowleri. Future Microbiol. 2012; 7(1):5–7 [DOI] [PubMed] [Google Scholar]
- 3.Marciano-Cabral F, MacLean R, Mensah A, et al. Identification of Naegleria fowleri in domestic water sources by nested PCR. Appl Environ Microbiol. 2003; 69(10):5864–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martinez AJ, Visvesvara GS. Free-living, amphizoic and opportunistic amebas. Brain Pathol. 1997; 7(1):583–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Visvesvara GS. Free-living amebae as opportunistic agents of human disease. J Neuroparasitol. 2010; 1:1–3. [Google Scholar]
- 6.Griffin JL. Temperature tolerance of pathogenic and nonpathogenic free-living amoebas. Science. 1972. ;178(4063):869–70 [DOI] [PubMed] [Google Scholar]
- 7.Martínez-Castillo M, Cárdenas-Zúñiga R, Coronado-Velázquez D, et al. Naegleria fowleri after 50 years: is it a neglected pathogen? J Med Microbiol. 2016. ;65(9):885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mishra BB, Gundra UM, Teale JM. Toll-like receptors in CNS parasitic infections. Curr Top Microbiol Immunol. 2009; 336:83–104. [DOI] [PubMed] [Google Scholar]
- 9.De Jonckheere JF. Origin and evolution of the worldwide distributed pathogenic amoeboflagellate Naegleria fowleri. Infect Genet Evol. 2011. ;11(7):1520–8. [DOI] [PubMed] [Google Scholar]
- 10.Wang Q, Li J, Ji J, et al. A case of Naegleria fowleri related primary amoebic meningoencephalitis in China diagnosed by next-generation sequencing. BMC Infect Dis. 2018; 18(1):349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chomba M, Mucheleng’anga LA, Fwoloshi S, et al. A case report: primary amoebic meningoencephalitis in a young Zambian adult. BMC Infect Dis. 2017; 17:532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panda A, Khalil S, Mirdha BR, et al. Prevalence of Naegleria fowleri in Environmental Samples from Northern Part of India. PLoS One. 2015; 10(10):e0137736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petit F, Vilchez V, Torres G, Molina O, et al. [Primary amebic meningoencephalitis: two new cases report from Venezuela]. Arq Neuropsiquiatr. 2006; 64:1043–6. [DOI] [PubMed] [Google Scholar]
- 14.Craun GF, Calderon RL, Craun MF. Outbreaks associated with recreational water in the United States. Int J Environ Health Res. 2005; 15(4):243–62. [DOI] [PubMed] [Google Scholar]
- 15.Tung MC, Hsu BM, Tao CW, et al. Identification and significance of Naegleria fowleri isolated from the hot spring which related to the first primary amebic meningoencephalitis (PAM) patient in Taiwan. Int J Parasitol. 2013; 43(9):691–6. [DOI] [PubMed] [Google Scholar]
- 16.Quist-Paulsen E, Kran AB, Lindland ES, et al. To what extent can clinical characteristics be used to distinguish encephalitis from encephalopathy of other causes? Results from a prospective observational study. BMC Infect Dis. 2019; 19(1):80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naqvi AA, Yazdani N, Ahmad R, et al. Epidemiology of primary amoebic meningoencephalitis-related deaths due to Naegleria fowleri infections from freshwater in Pakistan: An analysis of 8-year dataset. Arch Pharm Pract. 2016; 7(4):119–29. [Google Scholar]
- 18.Ali M, Jamal SB, Farhat SM. Naegleria fowleri in Pakistan. Lancet Infect Dis. 2020; 20(1):27–8. [DOI] [PubMed] [Google Scholar]
- 19.Khalid M, Saif UR, Shahzain K. Suspected Case of Naegleria fowleri (Primary Amebic Meningo-encephalitis). Pak Pediatr J. 2014; 38(3):196–8. [Google Scholar]
- 20.De Jonckheere JF. Sequence Variation in the Ribosomal Internal Transcribed Spacers, Including the 5.8S rDNA, of Naegleria spp. Protist. 1998. ;149(3):221–8. [DOI] [PubMed] [Google Scholar]
- 21.De Jonckheere JF. What do we know by now about the genus Naegleria? Exp Parasitol. 2014; 145 Suppl:S2–9. [DOI] [PubMed] [Google Scholar]
- 22.Escrig JI, Hahn HJ, Debnath A. Activity of Auranofin against Multiple Genotypes of Naegleria fowleri and Its Synergistic Effect with Amphotericin B In Vitro. ACS Chem Neurosci. 2020. ;11(16):2464–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zein NN. Clinical significance of hepatitis C virus genotypes. Clin Microbiol Rev. 2000;13(2):223–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang LL, Wu M, Hu BC, et al. Identification and molecular typing of Naegleria fowleri from a patient with primary amebic meningoencephalitis in China. Int J Infect Dis. 2018. ;72:28–33. [DOI] [PubMed] [Google Scholar]
- 25.Sazzad HMS, Luby SP, Sejvar J, et al. A case of primary amebic meningoencephalitis caused by Naegleria fowleri in Bangladesh. Parasitol Res. 2020; 119(1):339–344. [DOI] [PubMed] [Google Scholar]
- 26.Deisenhammer F, Bartos A, Egg R, et al. Guidelines on routine cerebrospinal fluid analysis. Report from an EFNS task force. Eur J Neurol. 2006; 13(9):913–22. [DOI] [PubMed] [Google Scholar]
- 27.Teunissen CE, Petzold A, Bennett JL, et al. A consensus protocol for the standardization of cerebrospinal fluid collection and biobanking. Neurology. 2009; 73(22):1914–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ithoi I, Ahmad AF, Nissapatorn V, et al. Detection of Naegleria species in environmental samples from Peninsular Malaysia. PLoS One. 2011; 6(9):e24327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barnett ND, Kaplan AM, Hopkin RJ, et al. Primary amoebic meningoencephalitis with Naegleria fowleri: clinical review. Pediatr Neurol. 1996; 15(3):230–4. [DOI] [PubMed] [Google Scholar]
- 30.Moussa M, De Jonckheere JF, Guerlotté J, et al. Survey of Naegleria fowleri in geothermal recreational waters of Guadeloupe (French West Indies). PLoS One. 2013; 8(1):e54414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang H, Seong GS, Sohn HJ, et al. Effective PCR-based detection of Naegleria fowleri from cultured sample and PAM-developed mouse. Eur J Protistol. 2015; 51(5):401–8. [DOI] [PubMed] [Google Scholar]
- 32.Wittwer CT, Ririe KM, Andrew RV, et al. The Light Cycler: a microvolume multisample fluorimeter with rapid temperature control. Biotechniques. 1997; 22(1):176–81. [DOI] [PubMed] [Google Scholar]
- 33.Fotedar R, Stark D, Beebe N, et al. Laboratory diagnostic techniques for Entamoeba species. Clin Microbiol Rev. 2007; 20(3):511–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Behera HS, Satpathy G, Tripathi M. Isolation and genotyping of Acanthamoeba spp. from Acanthamoeba meningitis/meningoencephalitis (AME) patients in India. Parasit Vectors. 2016; 9:442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pélandakis M, Serre S, Pernin P. Analysis of the 5.8S rRNA gene and the internal transcribed spacers in Naegleria spp. and in N. fowleri. J Eukaryot Microbiol. 2000. ;47(2):116–21. [DOI] [PubMed] [Google Scholar]
- 36.Dobrowsky PH, Khan S, Cloete TE, et al. Molecular detection of Acanthamoeba spp., Naegleria fowleri and Vermamoeba (Hartmannella) vermiformis as vectors for Legionella spp. in untreated and solar pasteurized harvested rainwater. Parasit Vectors. 2016. ;9(1):539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar S, Stecher G, Li M, et al. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 2018; 35(6):1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 2004; 101(30):11030–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.U.o.E . Institute of Evolutionary Biology, Edinburgh, FigTree v1.3.1, (2010). [Google Scholar]
- 40.De Jonckheere JF. Century of research on the amoeboflagellate genus Naegleria. Acta Protozool. 2002;41:309–42. [Google Scholar]
- 41.Phu NH, Hoang Mai NT, Nghia HD, et al. Fatal consequences of freshwater pearl diving. Lancet. 2013;381(9861):176. [DOI] [PubMed] [Google Scholar]
- 42.Nicolas M, De Jonckheere JF, Pernin P, et al. [Molecular diagnosis of a fatal primary amoebic meningoencephalitis in Guadeloupe (French West Indies)]. Bull Soc Pathol Exot. 2010;103(1):14–8. [DOI] [PubMed] [Google Scholar]