Abstract
To study changes in the intestinal flora associated with inflammatory bowel disease (IBD) in the Han population of southwest China, 48 participants were enrolled, 18 of whom had been diagnosed with IBD. Stool samples were collected from the participants. Sequencing of 16S rRNA gene was used to measure and identify the components of the intestinal flora. Diversity analysis and multivariate statistical analysis were conducted to study differences in intestinal flora between patients with IBD and healthy controls. The goods coverage, observed species, Shannon, and Simpson indices of alpha diversity were different (p < 0.05). Beta diversity analysis yielded significant differences between groups (R = 0.5668, p = 0.001 < 0.05). Compared with the composition of the intestinal flora in healthy controls, the relative abundances of Proteobacteria (18.56% vs. 3.56%, p = 0.001) and Fusobacterium (2.08% vs. 0.35%, p = 0.005) were higher in patients with IBD. Therefore, this study provides insight into the role of the microbiome in IBD.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12088-022-01014-z.
Keywords: Inflammatory bowel disease, Intestinal flora, 16S rRNA gene, Southwest China, Alpha diversity, Beta diversity
Introduction
Inflammatory bowel disease (IBD) is a chronic inflammation of the intestinal mucosa that includes Crohn's disease (CD) and ulcerative colitis (UC) [1]. The precise etiology of IBD is unknown, however, the leading hypothesis is that IBD is caused by an excessive immune response to environmental factors and pathogenic microorganisms in hosts with altered intestinal flora or genetic predispositions [2].
More than 50 diseases, including IBD, have been linked to imbalances of intestinal flora. Therefore, intestinal microorganisms are a major focus of current research. The correct and diverse gut microbiome is the factory and direct source of many of the basic building blocks of the human body [3]. There is a wide spectrum of data on bacterial diversity and density changes in patients with inflammation of the bowel mucous membrane. These studies focus on specific bacterial changes, such as oxidative stress and the regulation of nutrition [4, 5]. However, no single microbe with a consistent association to IBD has been identified. It is not clear whether the changes reported to date are the primary drivers of IBD or secondary to underlying IBD.
IBD is diagnosed at any age, but most new diagnoses occur in adolescents and young adults. The prevalence of IBD is expected to rise steadily in Western countries in the future, while the incidence and number of cases are also rising rapidly in Asia. By 2025, China is expected to be on a par with Western countries [6]. As an incurable chronic disease with low mortality, if unattended, IBD can presage a predictable and huge economic and medical burden. Therefore, augmenting our mechanistic understanding of the pathology of IBD and the relationships between intestinal flora and the body's immune system is conducive to the development of personalized therapy and new treatments for IBD.
In this study, we used the Illumina MiSeq sequencing platform to analyze the intestinal flora of patients with IBD and healthy individuals of Han ancestry in southwest China, focusing on structural changes in flora of patients with IBD. Analysis of the structural differences between the intestinal communities of patients with IBD and healthy individuals lays a foundation for understanding their functions and interactions with the immune system.
Materials and Methods
Sample Collection and DNA Extraction
We conducted a case–control study from September 2018 to April 2019, involving 48 subjects who visited the Department of Gastroenterology at the West China Hospital, Sichuan University. This included 18 patients with untreated IBD at the onset (within 1 year) and 30 healthy individuals as controls.
There is no unified diagnostic standard for IBD; the following form the basis for its potential diagnosis: typical clinical manifestations, colonoscopy or (and) radiological features, histopathology (biopsy/surgical specimens), and comprehensive professional opinions of clinicians. A questionnaire was designed to address the following queries: family history; history of rheumatoid arthritis, systemic lupus erythematosus, and other autoimmune diseases; long-term residence of the patients; history of present illness; and past medical history, especially of autoimmune diseases.
Thirty age- and sex-matched healthy controls were selected from the Health Examination Center of West China Hospital. No history or family history of IBD was apparent. Individuals with ankylosing spondylitis, hypertension, and diabetes were excluded. Participants were in good health, based on physical examination; liver and kidney functional indices; blood glucose level; blood lipid level; electrolytes; blood uric acid level; hemocytometry; routine urine/stool analysis; and fecal occult blood analysis.
Members of the two groups were not related. The exclusion criteria for the study participants were: antibiotic treatment within the last 1 month; receiving hormones, immune enhancers/inhibitors, or biological preparations within the last 3 months; and currently taking any medication; as well as hypertension, diabetes, basic metabolic disorders, and other systemic chronic diseases.
Fresh fecal samples from participants (minimum 6 g) were collected in a clean environment in an aseptic sampling tube and kept at − 80 °C for inspection in a specialized laboratory. Samples were collected from patients with untreated IBD (n = 18) and a healthy control group (n = 30). During the collection process, the samples were not contaminated by urine or sewage. The entire case inclusion process is shown in the flowchart of the study (Fig. S1).
DNA extraction was taking by a QIAamp DNA Stool Mini Kit (catalog #, USA), concentration and integrity measurement by NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and agarose gel electrophoresis, respectively.. The V3-V4 hypervariable region of bacterial 16S rRNA gene was amplified by PCR using a universal primer pair (343F: 5′-TACGGRAGGCAGCAG-3′; 798R: 5′-AGGGTATCTAATCCT-3′).
Analysis of Microbiota Composition
To obtain dynamic change trends of the index at different sequencing depths, exponential rarefaction curves were analyzed. Sequencing was performed on an Illumina MiSeq with two paired-end read cycles of 300 base pairs (bp) each (Illumina Inc., San Diego, CA; OE Biotech Company; Shanghai, China). Paired-end reads were preprocessed using Trimmomatic software to detect and cut off ambiguous bases (N); low-quality sequences with an average quality score of < 20 were also cut off using a sliding window trimming approach. After trimming, paired-end reads were assembled using FLASH software. The assembly parameters were: 10 bp of minimal overlap, 200 bp of maximum overlap, and 20% maximum mismatch. Further denoising of the sequences was performed as follows: reads with ambiguous, homologous sequences or less than 200 bp were abandoned; reads with 75% of bases above Q20 were retained using QIIME software (version 1.8.0); and chimeric reads were detected and removed using VSEARCH.
Clean reads were subjected to primer sequence removal and clustered to generate operational taxonomic units (OTUs) using Vsearch software with a 97% similarity cutoff. The representative read of each OTU was selected using the QIIME package. Alpha diversity (Chao1, Shannon, Simpson and Observed Species indices) was used to estimate microbial diversity. Differences in species complexity between samples was evaluating by beta diversity analysis, using QIIME software. The 16S rRNA gene amplicon sequencing and analysis were conducted by OE Biotech Co., Ltd. (Shanghai, China).
Species Annotation
Tag numbers of each classification rank (Phylum, Class, Order, Family, Genus, and Species) or OTU in different samples were summarized in a spectrum table or histogram. Based on the Greengene database (V201305), the RDP classifier 2.2 software Bayesian algorithm was used to carry out similarity and species annotation for representative sequences of each OTU, calculate community composition of each sample at multiple classification levels, and then use histogram for visualization.
Statistical Analysis
Count data are expressed as mean ± standard deviation (x ± SD) (normal distribution), or median and quartile spacing (M; p25–p75) (non-normal distribution). Box diagrams depict minimum, maximum, median, and quartile ranges. SPSS software 24.0 was used for t-tests, variance tests, and the Kruskal–Wallis rank sum test. For distributions that were not normal, the non-parametric test of multiple books was used for inter-group comparisons. R software (v. 3.4.0) was used for inter-group difference analysis and graph drawing. All statistics were tested using a two-sided probability test. The results are considered statistically significant at p < 0.05.
Results
Baseline and Clinical Characteristics
Patients newly diagnosed as IBD within 1 year with typical clinical symptoms and colonoscopic manifestations were included in Group-IBD. The demographic and clinical characteristics were shown in Table 1: age and sex composition are matched (p > 0.05). Results of routine laboratory tests showed significant difference in glucose and cholesterol (p < 0.05).
Table 1.
Demographic and clinical characteristics of the study participants
| Index | IBD | HC | p |
|---|---|---|---|
| n | 18 | 30 | – |
| Age(year) | 34.61 ± 10.3 | 40.03 ± 6.8 | 0.058 |
| Gender(Male/Female) | 11/7 | 17/13 | 0.762 |
| RBC (× 1012/L) | 4.89 ± 0.39 | 4.93 ± 0.61 | 0.875 |
| Hb (g/L) | 136.25 ± 14.32 | 144.83 ± 16.27 | 0.099 |
| PLT (× 109/L) | 263.88 ± 75.16 | 218.93 ± 65.01 | 0.067 |
| WBC (× 109/L) | 6.56 ± 1.54 | 6.12 ± 1.55 | 0.329 |
| ALT (IU/L) | 17.31 ± 9.88 | 29.20 ± 24.25 | 0.055 |
| AST (IU/L) | 20.90 ± 9.34 | 23.70 ± 9.22 | 0.293 |
| GLU (mmol/L) | 4.45 ± 0.51 | 4.99 ± 0.52 | 0.004* |
| CHOL (mmol/L) | 3.91 ± 1.25 | 5.06 ± 0.87 | 0.001* |
| HDL-C (mmol/L) | 1.33 ± 0.46 | 1.52 ± 0.46 | 0.216 |
| LDL-C (mmol/L) | 2.55 ± 0.66 | 3.02 ± 0.79 | 0.055 |
| GGT (IU/L) | 17.69 ± 8.73 | 30.03 ± 27.42 | 0.067 |
*means the difference is statistically significant
OTU Analysis
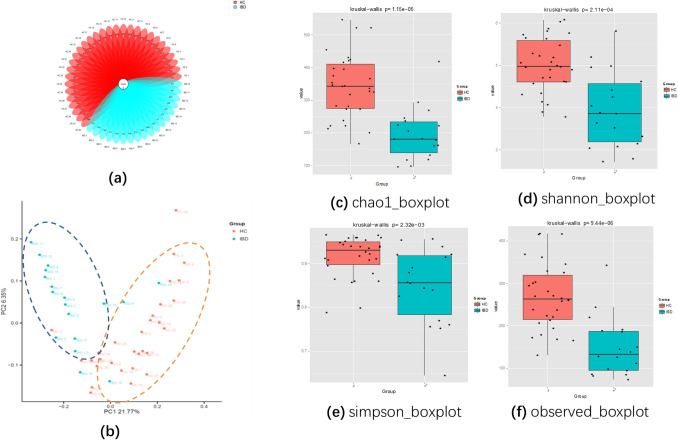
The total number of valid tags was distributed between 20,908 and 38,350 and the number of OTUs was distributed from 79 to 416, with an average of 145 and 270 OTU in Group-IBD and Group-HC (healthy control), respectively. A flower diagram of the OTUs shows that the two groups share two OTUs. The non-overlapping portion represents the group-specific OTUs. Compared with Group-IBD, Group-HC had more unique OTU numbers (Fig. 1).
Fig. 1.
a Flower diagram of operational taxonomic units (OTUs). b Principal coordinates analysis (PCoA) between Group-HC and Group-IBD. Each dot represents a sample and each color represents a group: red for Group-HC and blue for Group-IBD. c-f Alpha diversity boxplot for Group-HC and Group-IBD: c Chao1 index; d Shannon index; e Simpson index; f observed index (color figure online)
Alpha Diversity
The Chao1, Shannon, Simpson, and observed species indices of alpha diversity differed between groups (p < 0.05). The curve of Good’s coverage index flattened, indicating sufficient sequencing depth, which is the rarefaction curve of the alpha diversity index.
The Chao1 richness, Shannon, Simpson and Observed Species diversity indices, calculated based on OTU species and abundance, were used to describe diversity features. Chao1 indices and Observed Species reflect species richness or the numbers of OTU, while Shannon and Simpson indices assess community diversity, including species richness and evenness. The Chao1 indices and Observed Species diversity indices of Group-IBD were significantly lower than those of Group-HC, indicating that IBD decreases species richness. Additionally, the Shannon index and the Simpson index of Group-IBD was both lower than controls (p < 0.05, Fig. 1, Table 2), indicating reduced species diversity.
Table 2.
Richness and diversity analysis
| Index | IBD | HC | p |
|---|---|---|---|
| Chao1 | 180.201 [138.318–418.107] | 342.905 [274.985–546.428] | 0.0000022* |
| Shannon | 3.869[3.192–5.804] | 4.977[4.611–5.804] | 0.000021* |
| Simpson | 0.856[0.784–0.956] | 0.931[0.898–0.965] | 0.001021* |
| Observed | 111.6[85.1–143.5] | 249.8[175.2–312.1] | 0.0003* |
*means the difference is statistically significant
Beta Diversity
Beta diversity significantly differed between the groups (p < 0.05). According to PCoA, species composition of two samples is similar if they are close, and vice versa. PCoA revealed a significant separation in the bacterial community composition between groups, using the first two principal component scores: PC1, the principal coordinate component causing the largest difference, had an explanatory value of 21.77%, while PC2 had 6.35%. We also found clustering among some samples, indicating that the intestinal flora had changed to some extent (Fig. 1).
Structural Microbiota Changes Associated with IBD
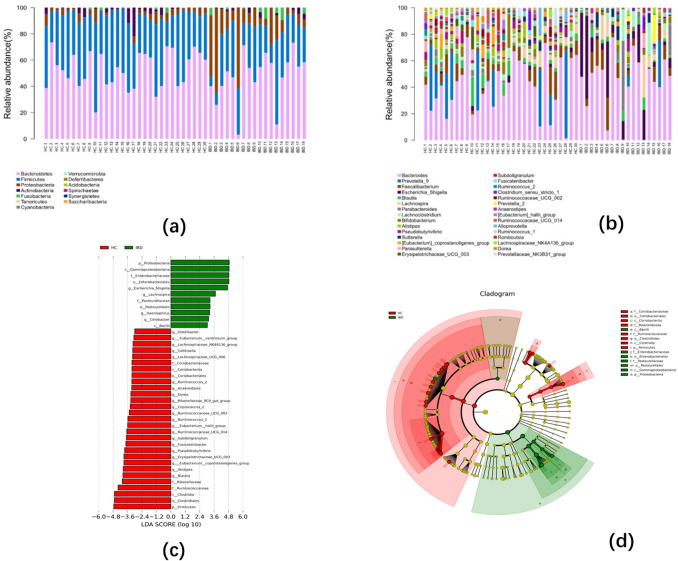
The relative abundance of Proteobacteria (18.56% vs. 3.56%, p = 0.001) and Fusobacterium (2.08% vs. 0.35%, p = 0.005) was higher in patients with IBD than in healthy participants.
Microbial Composition at Different Taxonomic Levels
We detected 13 phyla and 30 genera in our subjects. There were significant differences between groups: Proteobacteria accounted for 3.56–18.56% of the two groups; the proportion of Fusobacteria in the two groups ranged from 0.35 to 2.08% (Fig. 2a). In the patients with IBD, the dominant genera were Bacteroides, Escherichia, Lachnospira, Parabacteroides, and Faecalibacterium in the proportions 46.19, 14.21, 3.77, 5.79, and 7.73% respectively. In healthy control patients, the dominant genera were Bacteroides, Prevotella, Blautia, Bifidobacterium, and Eubacterium in the proportions 41.33, 11.55, 4.39, 2.40, and 2.37%, respectively (Fig. 2b).
Fig. 2.
Taxonomic comparisons of the relative abundances of intestinal microbiota at the phylum (a), and genus (b) level (c) The Group-HC-enriched taxa are indicated with a negative LDA score (red), and Group-IBD-enriched taxa a positive score (green). (d) The circular cladogram was derived from the LEfSe analysis and showed the relationship between the most differentially abundant taxa between Group-IBD (green) and Group-HC (red) (color figure online)
Relative Species Abundance Differences
The two groups differed in microbial richness. In Group-IBD, the proportion of Firmicutes decreased (30.12% vs. 40.88%, p = 0.007), while the proportion of Fusobacteria (2.08% vs. 0.35%, p = 0.005) and Proteobacteria (18.56% vs. 3.56%, p = 0.0001) increased at the phylum level. However, in the 10 most abundant genera, the abundances of eight species were lower in Group-IBD, including Eubacterium coprostanoligenes, Eubacterium hallii group, Alistipes, Erysipelotrichaceae_UCG_003, Fusicatenibacter, Pseudobutyrivibrio, Ruminococcaceae_UCG_013, and Subdoligranulum. Only the abundance of Escherichia_Shigella (14.21% vs. 0.89%, p = 0.0005) and Fusobacteria (1.84% vs. 0.22%, p = 0.026) in Group-IBD was increased. A LEfSe (lineardiscriminant analysis effect size) analysis identified the differentially abundant taxa between Group-IBD and Group-HC. Only taxa with a LDA (linear discriminant analysis) threshold greater than 3.0 are displayed (Fig. 2c, d).
Differences in Metabolic Pathways
Based on the microbial community characteristics derived from 16S rRNA gene sequencing, we inferred the functional content of metagenomes using PICRUST software. There were differences in amino-acid metabolism pathways between groups. In general, microbial communities could be distinguished by their function. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways significantly enriched in Group-IBD included those of shigellosis, bladder cancer, bacterial invasion of epithelial cells, and pathogenic Escherichia coli infection. In the metabolism-related pathways, the KEGG pathways enriched in Group-IBD were those of ether lipid metabolism, transcription-related proteins, glycan biosynthesis and metabolism, and nucleotide metabolism. Hierarchical clustering also revealed differences between groups by the intensities detected in each metabolic pathway (Fig. S2).
Discussion
IBD, including CD and UC, is a complex condition in which the host's genetic susceptibility, environment, intestinal microbes, and other factors are intertwined, leading to an abnormal mucosal immune response and impaired epithelial barrier function. The microbiome may play a larger role in IBD than it does in other diseases [6, 7]. The intestinal flora can alter the severity of intestinal inflammation while being altered itself by the introduction of microbes or their effectors, including lipids, small molecules, proteins, or sugars [8]. Each gastrointestinal disorder has a specific microbial signature; however, they may share common pathophysiological pathways that constitute a "core dysbiosis". Assessment of these microbial markers may assist with diagnostic assessment and choice of therapeutic targets, contributing to the promotion of intestinal health. [9]. In the model of IBD and ileitis, in addition to the bacterial spectrum in the lumen and mucosa, the composition and function of the fungal and viral spectra are changed repeatedly [10].
The intestinal flora plays a major role in the maturation of host immune responses, protection against enteric pathogen proliferation, and response to specific drugs. Since IBD is likely caused by changes in the immune response to intestinal flora and microecology disorders, the relationship between the intestinal immune system and microflora is extremely important [2]. Their interaction is primarily mediated by toll-like receptors (TLRs). In the gastrointestinal mucosa lamina propria, after TLR activation, macrophages and dendritic cells migrate to the intestinal Peyer's patch, activate T cells, promote the activation and differentiation of regulatory T cells (Treg), and generate inhibitory cytokines (such as IL-10 and transforming growth factor-β) that induce tolerance of intestinal flora. Multiple factors, such as diet and antibiotic abuse, can cause intestinal flora imbalance. Dysbacteriosis causes invasive bacterial overgrowth; the production of large quantities of antigens causes an increase in intestinal mucosa permeability and a pathological immune response. Due to the runaway effect of the immune response, intestinal antigen presenting cells and mucous membrane epithelial cells produce many immunoregulatory factors, leading to differentiation and activation of Th1 and Th17 cells, reducing the function of Treg cells, ultimately resulting in intestinal inflammation [11]. Bacteria can alter the differentiation of Th17 and Treg cells, microbial metabolites, and bacteria themselves; they can also affect the immune response, including the alteration of metabolites, such as short-chain fatty acids (SCFAs) and sphingolipids [12]. SCFAs in the intestine are an important energy source for intestinal epithelial cells, and disorders often lead to a decrease in their levels, leading to increased intestinal permeability and inflammation [13]. IBD is hypothesized to result from an aberrant immune response to commensal flora of genetically susceptible hosts with an unbalanced host-microbial relationship. Based on this study, fecal microbiota transplantation to restore the intestinal homeostasis may be an efficient strategy for IBD treatment. Studies indicate that targeting specific microbiota such as Lactobacillus transplantation, offers new treatment strategies for IBD [14].
Most intestinal bacteria cannot be isolated, cultured, and identified by traditional selective culture methods, limiting research on intestinal flora. Recently, however, the rapid development and extensive application of high-throughput sequencing technology has promoted research on microbial community and species, community structure, microbial genomes, transcriptomes, and metabolomics. Studies of intestinal microbiota in other chronic immune-mediated inflammatory diseases can be relevant to the analysis of IBD. Intestinal microbial changes that are not evident in other chronic immune-mediated inflammatory diseases are more likely to be IBD-specific [15]. A study that evaluated the gut microbes of three patients with gastrointestinal disease (diverticulitis, irritable bowel syndrome, and IBD) found that intestinal disease has a specific microbial mode [9]. This microecological disorder promotes the invasive mucosal immune response and long-term damage, characteristic of the disease.
High levels of nitrifying and oxidative stress in the intestinal tract of patients with IBD lead to decreased abundance of obligate anaerobes Bacteroidetes, Firmicutes, and aerobic bacteria actinomycetes, increased abundance of denatured facultative anaerobe Bacteroides, and decreased microbial diversity [12], resulting in intestinal mucosal damage and intestinal inflammation. Inflammation stimulates the production of IFN-γ and produces reactive oxygen species through phagocytosis by innate immune cells, which eventually form anaerobic respiration products. Facultative anaerobes use these products for growth, leading to low bacterial diversity. Changes in the abundance of intestinal flora found in our study were consistent with such statement. Compared with the healthy group, the richness and evenness of intestinal flora in the disease group significantly decreased, like previous studies [16, 17]. In our study, the intestinal flora composition in the IBD group changed significantly. The general trend involves an increase in Proteobacteria and a decrease in Firmicutes. Studies showed that Proteobacteria abundance can be used to characterize unstable intestinal microbial communities and metabolic disorders [18]. Selection pressure caused by microecological imbalance seems to interfere with the stability of the microbial community, and Proteobacteria adaptability will be enhanced to become the dominant bacteria. Mucosal immune system is responsible for removing pathogens. Improper immune responses during this process can disrupt intestinal homeostasis resulting in microdysbiosis, and lead to local or systemic inflammation and metabolic dysfunction. Therefore, some studies have proposed the idea that Proteobacteria increase can be used as a marker for diagnosing diseases and potential diseases [19]. Among the bacteria present in the gut microbiome, Firmicutes and Bacteroidetes are the most beneficial [20]. A decrease in Firmicutes leads to a reduction in the abundance of Clostridium, which releases butyric acid to lower the levels of pro-inflammatory cytokines. Clostridium also enhances Treg cell numbers and function in the colon, and aids resistance to colitis by inducing a TGF-β-rich environment [21]. Moreover, the degree of intestinal flora disorder varies among patients with IBD [22]. Invasive Escherichia coli (AIEC) is associated with patients with CD, while diffusely adherent E. coli (DAEC) is linked to UC. Certain pathogenic features of these E. coli pathobionts suggest that IBD-associated E. coli strains play a catalytic role during IBD flares [23]. These differences in microbial composition can be used to identify biomarkers in patients with IBD. As changes in intestinal microecological structure can lead to intestinal mucosal damage, can these changes inhibit inflammatory responses? Probiotics are known to help in maintaining the gut homeostasis through a wide range of functions, and sometimes play valuable roles in serious pathological conditions such as IBD [24]. In addition, FMT is a prominent research topic in the treatment of IBD. Treatments in some studies maintain intestinal microecological balance and reduce intestinal inflammation by increasing obligate anaerobic bacteria and decreasing facultative anaerobic bacteria, which is also a major strategy for the treatment of intestinal microbiome disruptions [12]. In contrast to FMT, application of a single bacterium (Akkermansia muciniphila) has also proved beneficial in treating various disorders [20].
Interaction of the intestinal flora with the host generally occurs through the intermediate or end products of microbial metabolism. These metabolites, derived from different sources, influence a series of host intestinal functions, including immune maturation, immune homeostasis, and maintenance of mucosal integrity [25]. Therefore, to determine how the microbiome affects gastrointestinal health, we need to transition from censuses to functional studies. A study on the relationship between food additives and human intestinal flora showed that the microbiota can be directly impacted by commonly used food additives, in a manner that subsequently drives intestinal inflammation. [26]. Unlike in healthy individuals, for whom diets rich in fermentable fiber provide an array of health benefits, in patients with IBD, prebiotic fibers can lead to gut dysbiosis and surfeit colonic butyrate, which may exacerbate IBD illness [27]. Psychological stress is also an important inducer of IBD, enhancing intestinal autophagy by regulating intestinal flora and inflammation. [28]. In addition, metabolites can be used as therapeutic targets for patients with IBD, including epigallocatechin gallate which is a potential modulator for gut microbiota to prevent and treat IBD, and rhein which can modulate gut microbiota, indirectly changing purine metabolism in the intestine and subsequently alleviating colitis [13, 29]. Based on the prediction of function in our research work, we found several statistically differences in KEGG metabolic pathways, and these differential findings provide a foundation for future functional research.
Most studies on the relationship between intestinal flora and specific changes in intestinal disease have reported associations, not cause-and-effect relationships. To our knowledge, this is the first study to investigate intestinal flora changes in incipient Han patients with untreated IBD in southwest China, to understand the structure and characteristics of the disease. The inclusion criteria were stringent (excluding individuals with ethnic, diet, drug, disease, and other factors that may affect the gastrointestinal tract) and provided basic data for the study of IBD diagnosis and treatment in the Chinese Han population. However, there were a few limitations. First, the sample size was small and our target of at least 30 patients with diseases was not achieved, mainly because of limited time and strict inclusion criteria. Thus, only an overview of the relative abundance distribution of the intestinal flora at a single point in time could be obtained, failing to capture the complex dynamics of the microbial ecosystem in the intestines of patients. Based on our differential results, we are conducting further metabonomic and other multi-omics studies, and we are also trying to create conditions to further expand the study sample size. With this knowledge, we may be able to develop novel personalized treatments for patients with IBD.
Conclusion
The interaction between specific microbiota and a dysfunctional immune response strongly supports selective targeting of the intestinal flora as a diagnostic and therapeutic approach for IBD. Our study aimed to identify possible targets for IBD diagnosis and treatment and provides basic data for the study of IBD showing specific classes of metabolites, notably bile acids, SCFAs, and tryptophan, in the southwest Chinese Han population. Metagenomic and proteomic analyses identified metabolites that have implicated in the pathogenesis of IBD, which should be addressed in more detailed studies with larger sample sizes.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We would also like to thank Zhonghao Wang and Zhongyi Zhao for their excellent assistance in conducting the language revision.
Authors' Contribution
YB conceived of the study. YRL collected patient samples and basic information. WTT analyzed and interpreted all genomic data and was a major contributor in writing the manuscript. ZLL and WXA performed literature collation and manuscript calibration. All authors read and approved the final manuscript.
Funding
This study was funded by the National Natural Science Foundation of China (No. 81772258) and Science and Technology Agency of Sichuan Province (2019YFS0310, 2018FZ0106).
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Tingting Wang, Email: tingting2003.1@163.com.
Bin Yang, Email: 19728243@qq.com.
References
- 1.Seyedian SS, Nokhostin F, Malamir MD. A review of the diagnosis, prevention, and treatment methods of inflammatory bowel disease. J Med Life. 2019;12:113–122. doi: 10.25122/jml-2018-0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Souza HS, Fiocchi C. Immunopathogenesis of IBD: current state of the art. Nat Rev Gastroenterol Hepatol. 2016;13:13–27. doi: 10.1038/nrgastro.2015.186. [DOI] [PubMed] [Google Scholar]
- 3.Yu B, Yu B, Yu L. Commentary: reconciling hygiene and cleanliness: a new perspective from human microbiome. Indian J Microbiol. 2020;60:259–261. doi: 10.1007/s12088-020-00863-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayers MD, Moon C, Stupp GS, et al. Quantitative metaproteomics and activity-based probe enrichment reveals significant alterations in protein expression from a mouse model of inflammatory bowel disease. J Proteome Res. 2017;16:1014–1026. doi: 10.1021/acs.jproteome.6b00938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu YL, Chen DW, Zheng P, et al. The bidirectional interactions between resveratrol and gut microbiota: an insight into oxidative stress and inflammatory bowel disease therapy. BioMed Res Int. 2019;2019:5403761. doi: 10.1155/2019/5403761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishida A, Inoue R, Inatomi O, et al. Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin J Gastroenterol. 2018;11:1–10. doi: 10.1007/s12328-017-0813-5. [DOI] [PubMed] [Google Scholar]
- 7.Llewellyn SR, Britton GJ, Contijoch EJ, et al. Interactions between diet and the intestinal microbiota alter intestinal permeability and colitis severity in mice. Gastroenterology. 2018;154:1037–1046.e2. doi: 10.1053/j.gastro.2017.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen LJ, Cho JH, Gevers D, et al. Genetic factors and the intestinal microbiome guide development of microbe-based therapies for inflammatory bowel diseases. Gastroenterology. 2019;156:2174–2189. doi: 10.1053/j.gastro.2019.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopetuso LR, Petito V, Graziani C, et al. Gut microbiota in health, diverticular disease, irritable bowel syndrome, and inflammatory bowel diseases: time for microbial marker of gastrointestinal disorders. Dig Dis. 2018;36:56–65. doi: 10.1159/000477205. [DOI] [PubMed] [Google Scholar]
- 10.Sartor RB, Wu GD. Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology. 2017;152:327–339.e4. doi: 10.1053/j.gastro.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Praveen P, Jordan F, Priami C, et al. The role of breast-feeding in infant immune system: A systems perspective on the intestinal microbiome. Microbiome. 2015;3:41. doi: 10.1186/s40168-015-0104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schirmer M, Garner A, Vlamakis H, et al. Microbial genes and pathways in inflammatory bowel disease. Nat Rev Microbiol. 2019;17:497–511. doi: 10.1038/s41579-019-0213-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu J, Wei Z, Cheng P, et al. Rhein modulates host purine metabolism in intestine through gut microbiota and ameliorates experimental colitis. Theranostics. 2020;10:10665–10679. doi: 10.7150/thno.43528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weingarden AR, Vaughn BP. Intestinal microbiota, fecal microbiota transplantation, and inflammatory bowel disease. Gut Microbes. 2017;8:238–252. doi: 10.1080/19490976.2017.1290757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernstein CN, Forbes JD. Gut microbiome in inflammatory bowel disease and other chronic immune-mediated inflammatory diseases. Inflamm Intest Dis. 2017;2:116–123. doi: 10.1159/000481401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang C, Lin H. Dysbiosis in gastrointestinal disorders. Best Pract Res Clin Gastroenterol. 2016;30:3–15. doi: 10.1016/j.bpg.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 17.Josephine N, Gary DW, Lindsey A, et al. Gut microbiota and IBD: causation or correlation? Nat Rev Gastroenterol Hepatol. 2017;14:573–584. doi: 10.1038/nrgastro.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Y, Xu ZZ, He Y, et al. Gut microbiota offers universal biomarkers across ethnicity in inflammatory bowel disease diagnosis and infliximab response prediction. mSystems. 2018;3:e00188–17. doi: 10.1128/mSystems.00188-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altomare A, Putignani L, Chierico FD, et al. Gut mucosal-associated microbiota better discloses inflammatory bowel disease differential patterns than faecal microbiota. Dig Liver Dis. 2019;51:648–656. doi: 10.1016/j.dld.2018.11.021. [DOI] [PubMed] [Google Scholar]
- 20.Kalia VC, Gong C, Shanmugam R, et al. The emerging biotherapeutic agent: Akkermansia. Indian J Microbiol. 2022;62:1–10. doi: 10.1007/s12088-021-00993-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richard ML, Sokol H. The gut mycobiota: insights into analysis, environmental interactions and role in gastrointestinal diseases. Nat Rev Gastroenterol Hepatol. 2019;16:331–345. doi: 10.1038/s41575-019-0121-2. [DOI] [PubMed] [Google Scholar]
- 22.Borren NZ, Conway G, Garber JJ, et al. Differences in clinical course, genetics, and the microbiome between familial and sporadic inflammatory bowel diseases. J Crohns Colitis. 2018;12:525–531. doi: 10.1093/ecco-jcc/jjx154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mirsepasi-Lauridsen HC, Vallance BA, Krogfelt KA, et al. Escherichia coli pathobionts associated with inflammatory bowel disease. Clin Microbiol Rev. 2019;32:e00060–18. doi: 10.1128/CMR.00060-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar R, Sood U, Gupta V, et al. Recent advancements in the development of modern probiotics for restoring human gut microbiome dysbiosis. Indian J Microbiol. 2020;60:12–25. doi: 10.1007/s12088-019-00808-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lavelle A, Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2020;17:223–237. doi: 10.1038/s41575-019-0258-z. [DOI] [PubMed] [Google Scholar]
- 26.Chassaing B, Wiele TV, Bodt JD, et al. Dietary emulsifiers directly alter human microbiota composition and gene expression ex vivo potentiating intestinal inflammation. Gut. 2017;66:1414–1427. doi: 10.1136/gutjnl-2016-313099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh V, Yeoh BS, Walker RE, et al. Microbiota fermentation-NLRP3 axis shapes the impact of dietary fibres on intestinal inflammation. Gut. 2019;68:1801–1812. doi: 10.1136/gutjnl-2018-316250. [DOI] [PubMed] [Google Scholar]
- 28.Wang SL, Shao BZ, Zhao SB, et al. Intestinal autophagy links psychosocial stress with gut microbiota to promote inflammatory bowel disease. Cell Death Dis. 2019;10:391. doi: 10.1038/s41419-019-1634-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu ZH, Huang SM, Li TT, et al. Gut microbiota from green tea polyphenol-dosed mice improves intestinal epithelial homeostasis and ameliorates experimental colitis. Microbiome. 2021;9:184. doi: 10.1186/s40168-021-01115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.