Abstract
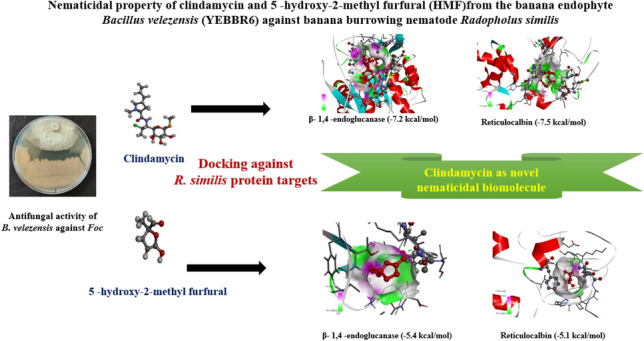
Radopholus similis is a burrowing nematode which causes banana toppling disease and is of major economic threat for the banana production. Bacterial endophyte Bacillus velezensis (YEBBR6) produce biomolecules like 5-hydroxy-2-methyl furfural (HMF) and clindamycin in during interaction with Fusarium oxysporum f.sp. cubense. Molecular modelling and docking studies were performed on Radopholus similis protein targets such as calreticulin, cathepsin S-like cysteine proteinase, β-1,4 -endoglucanase, reticulocalbin, venom allergen-like protein and serine carboxypeptidase to understand the mode of action of HMF and clindamycin against Radopholus similis. Structurally validated protein targets of R. similis were docked with biomolecules through AutoDock Vina module in PyRx 0.8 software to predict the binding energy of ligand and target protein. Among the chosen six targets, docking analysis revealed that clindamycin had the maximum binding affinity for β-1,4-endoglucanase (− 7.2 kcal/mol), reticulocalbin (− 7.5 kcal/mol) and serine carboxypeptidase (− 6.9 kcal/mol) in comparison with HMF and the nematicide, carbofuran 3G. Besides, clindamycin also had the maximum binding energy for the target sites calreticulin and venom allergen-like protein compared to the small molecule HMF. Novel molecule, clindamycin produced by B. velezensis served as a potential inhibitor of the target sites associated in interrupting the functions of β-1,4-endoglucanase, reticulocalbin, serine carboxypeptidase, calreticulin, cathepsin S-like cysteine proteinase, and venom allergen-like proteins. Besides, increased binding affinity of clindamycin with the protein target sites facilitated to explore it as a novel nematicidal molecule for the management of banana burrowing nematode R. similis. Thus, present investigation confirmed that, the small molecules clindamycin can be explored for nematicidal activity.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s12088-022-01011-2.
Keywords: Bacterial endophyte, Nematicidal biomolecule, Bacillus velezensis, Reticulocalbin, Serine carboxypeptidase, Calreticulin, Cathepsin S-like cysteine proteinase, Venom allergen-like proteins and molecular modelling
Introduction
Banana (Musa spp.) is an economically important fruit crop in the world and contributes towards food and livelihood security in developing countries. Though banana contributes to food security, its cultivation is hindered by the outbreak of Fusarium oxysporum f. sp. cubense races (Foc), nematodes and their complex. Banana rhizosphere serves as a repertoire for 132 nematode species representing 54 genera [1]. Among them, burrowing nematode Radopholus, the root-lesion nematode Pratylenchus coffea, spiral nematode Helicotylenchus multicinctus and root knot nematode (Meloidogyne spp.) are the major species threatening banana production in India. Banana cultivation is hampered severely due to the infection of migratory endoparasitic burrowing nematode R. similis accounting for 31 to 41% yield loss [2, 3]. Cultivation of resistant Yengambi Km5 and Pisang Jari Buaya (AAA) suppressed the burrowing nematode infection. However, in Uganda these varieties also become susceptible to R. Similis [4]. But the nematodes infecting bananas are managed mainly through the application of synthetic nematicides. However, several synthetic nematicides are not effective at field level [5]. In this juncture, Trichoderma harzianum, Purpureocillium lilacinum, Pochonia chlamydosporia, Monacrosporium lysipagum, Pseudomonas fluorescens, and Pasteuriapenetrans were explored for the management of nematodes [6–8]. Besides, secondary metabolites from bacterial endophytes suppressed a range of plant parasitic nematodes [9]. Khanna and Ranganathan. [10] reported benzimidazole and piperazine bound to acetylcholine and tubulin β-1 chain receptor from nematodes through in silico approach. They also reported that nitrogen atom in the piperazine ring was found to bind frequently to nematode receptor sites. Furthermore, Kimura et al. [11] reported the nematicidal activity of 5-hydroxymethyl-2-furoic acid against pine wood nematode B. xylophilus and free-living nematode C elegans without affecting plant growth. Similarly, Aissani. [12] reported that HMF from Ailanthus altissima effectively inhibited Meloidogyne Juvenile. Our earlier investigation also emphasized that HMF and clindamycin biomolecules had antifungal activity against Foc [13]. Hence, we attempted to elucidate the mode of action of these molecules towards the suppression of burrowing nematode, which also has chitin as its cell wall component.
However, the details on binding of ligand with the target sites of nematodes has not been explored to discover novel biomolecules with broad spectrum of action. Understanding the interaction of small molecules or ligands to the target sites of the nematodes will help in elucidating the mechanism of action. In this regard, molecular docking of small molecules HMF and clindamycin produced by B. velezensis was analysed to annotate potential therapeutic protein target sequences of the burrowing nematode that play a key role in the nematode survival and parasitism. Earlier research findings emphasized the role of biomolecules binding with different target sites of nematode. But, the study is unique in the sense, the secondary metabolites produced during the interaction of bacterial endophyte with Foc have not been explored for their nematicidal property. It would open avenues for the discovery of novel biomolecules with nematicidal property. Considering this researchable issue, attempts were made to identify the nematicidal property of HMF and clindamycin with antifungal action produced during ditrophic interaction between B. velezensis and Foc. Moreover, to the best of our knowledge, this is the first investigation to explore the nematicidal property of HMF and clindamycin against R. similis through in silico approach.
Materials and Methods
Identification of Protein Targets and Molecular Modelling
The potential protein targets of banana burrowing nematode, R. similis were identified as cathepsin L-like cysteine proteinase [14], calreticulin (CRT) [15, 16], venom allergen proteins (VAP) [17], serine carboxypeptidases (SCPs) [18], β-1,4-endoglucanase [19] and reticulocalbin [20] based on the literature survey. The UniProt database was used to retrieve the protein sequence of chosen protein targets of banana burrowing nematode R. similis as targets based on literature mining. DNA sequence of the target, Venom allergen-like proteins (VAPs) was obtained as described by Li et al. [21]. DNA sequence was translated to a protein sequence using the ExPASY translate tool. (Table S1).
The selected virulent target of burrowing nematode do not have experimentally and computationally solved structures. Hence, molecular modelling was done using SWISS-MODEL (Method: Rigid-body assembly), Phyre2 (Method: Profile based alignment), and ROBETTA (Metaserver). All the target sequences were first developed into a model using SWISS-MODEL server. In the absence of template structure modelling using SWISS-MODEL and Phyre2/ROBETTA was employed based on query coverage performance.
The software, SWISS-MODEL was used to develop homology modelling for the targets calreticulin, cathepsin S-like cysteine proteinase and serine carboxypeptidase. Besides, Phyre2 was used for modelling β-1,4-endoglucanaseand reticulocalbin. Software ROBETTA was used for the modelling of Venom allergen-like protein target (Fig. S1, Table S3).
In the homology modelling protocol, the protein targets calreticulin, cathepsin S-like cysteine proteinase, and serine carboxypeptidase were subjected to BLAST search followed by HHblitsin SWISS-MODEL [22]. Global Mean Quality Estimation (GMQE) score close to 1, sequence identity (30–50 percent safe-zone) and maximum query coverage were used as the parameters to ensure the high quality of modelled structures (Fig. S1 Table S4).
The ROBETTA server (http://robetta.bakerlab.org/)uses RoseTTAFold to model multi-chain complexes using comparative modelling domains).
Validation of Protein Model
Modelled protein targets were validated using the Ramachandran plot of the PROCHECK tool from the Structural Analysis and Verification Server (SAVES, Meta server) (https://saves.mbi.ucla.edu/) so as toensure model quality based on the residues lying in favoured and allowed regions. Swiss PDB Viewer(http://www.expasy.org/spdbv/) was used for energy minimization in modelled proteins and loop building for residues in disallowed regions.
Ligand Preparation and Analysis
Three compounds, HMF, clindamycin and carbofuran were retrieved from the Pubchem database (https://pubchem.ncbi.nlm.nih.gov/) in SDF format. The commercial nematicide carbofuran 3G was used as a reference ligand molecule. The software, Open Babel was used to convert compounds from SDF to PDB file format.
Similarity Analysis of Small Molecules
ChemMine online software was used to calculate the similarity score between the compound pairs. Tanimoto coefficients, such as Atom pair tanimoto (AP), Maximum Common Substructure tanimoto (MCS), MCS size, MCS min/max, and SMILES were assessed to study the similarity in small molecule structure. Tanimoto is defined as c/(a + b + c), where c is the number of features in a compound pair, a denotes features unique to one compound, and b denotes features unique to another compound. The tanimoto coefficient for atom pairs spans from 0 to 1, with a larger coefficient indicating greater similarity (similar structural descriptors). Compounds with large tanimoto differences (> 0.20) and the MCS facilitate to obtaining the most accurate and sensitive similarity measure. A large MCS size (> 9) is likely to be shared by similar molecules and thus the similarity analysis was ascertained.
Virtual Screening and Molecular Docking
The AutoDock vina module in PyRx 0.8 was used to perform molecular docking (Dallakyan and Olson, 2015). Protein preparation was performed using the make macromolecule option in PyRx software. All ligand structures were minimized using conjugate gradient, the first-order derivatives of optimization process with 200 steps and commercial molecular mechanics parameters-Unified Force Field (UFF). The Computed Atlas Topography of Proteins CASTp 3.0 server was used to find binding site pockets for the targets [23]. Grid setting, and docking was carried out using AutoDock4 and autogrid4 parameter files. Ligands were allowed to generate flexible conformations and orientations with a value of 8 exhaustiveness during the execution of docking protocol. Interactions of docked conformations of protein–ligand complexes were imported into BIOVIA Discovery studio client 2021 (https://www.3ds.com/products-services/biovia/) for visualization. Different colours were assigned to the receptor, ligand and interacting atoms to distinguish between each other.
Results
Molecular Modelling
Calreticulin was modelled (SWISS-MODEL) using a template protein (PDB ID-6ENY) that had previously been described by electron microscopy, which had 66.13 percent identity, 93 percent coverage, and a 0.76 GMQE score. Template protein for cathepsin S-like cysteine proteinase (PDB ID-1CS8) had 46.67 percent identity, 91 percent coverage, and a GMQE score of 0.75, while serine carboxypeptidase had 39.14 percent identity, 91 percent coverage, and a GMQE score of 0.72 with template protein (PDB ID-1IVY), which was experimentally solved using X-ray crystallography method. The targets β-1,4-endoglucanase and reticulocalbin had no homologs or template to model with SWISS-MODEL, hence Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2/) was performed. Protein sequence for targets β-1,4-endoglucanase (467 residues) and reticulocalbin (279 residues) were retrieved using the UniProt ID B9U290, A0A6G6CU92. The protein structures such as PDB ID-1WKY and IKFX were used as templates for β-1,4-endoglucanase and reticulocalbin protein models β-1,4-endoglucanase had 100% confidence score and 89 percent coverage. Reticulocalbin had a 100% confidence score with 78 percent coverage (Fig. S1 Table S3, 4,5). ROBETTA was used model a venom allergen-like protein target with 60% confidence score (Fig. S1, Table S5).
Model Validation
The target β-1,4-endoglucanase had 83.1 percent of its residues in the most favoured region or core region, 14.9 percent in additionally allowed regions and 2.0 percent in generously allowed regions, according to Ramachandran plot (Fig. S3). Residues in the allowed, additionally allowed and generously allowed regions of calreticulin target are 87 percent, 11.5 percent and 1.5 percent, respectively (Fig. S4). The target Cathepsin S-like cysteine proteinase had 84.6 percent of its residues in the most favoured region, 14.2 percent in additionally allowed regions, and 1.2 percent in generously allowed regions (Fig. S5). The target reticulocalbin had 68.4 percent residues in the most favoured region, 28.4 percent residues in additionally allowed regions and 3.1 percent residues in the generously allowed region (Fig. S6). Residues in the most favoured region account for 89.1 percent of the target serine carboxypeptidase, 10.4 percent of residues in additionally allowed regions, and 0.5 percent of residues in generously allowed regions (Fig. S7). Residues in allowed, additionally allowed and generously allowed regions for the target, Venom allergen-like protein, were 84.2 percent, 13.7 percent, and 2.2 percent respectively (Fig. S8). This confirmed the acceptability and quality of modelled structures.
Sequence Similarity Analysis
Sequence similarity was performed using the BLASTP tool for the nematode target proteins as a query against the banana genome proteins, in order to find the presence of any similar proteins in the banana genome. There was no single hit or similar sequences that were observed in similarity search. Thus, it indicated the specific binding of HMF and clindamycin to nematode protein targets and no binding was observed with respect to banana proteins.
Small Molecule Analysis
MCS tanimoto coefficient was less than 0.5 for both HMF and clindamycin compared to carbofuran 3G (Fig. S2, Table S2). Further, MCS size was less than 6 in both the above-mentioned compounds, while comparing with nematicide carbofuran 3G. These results imply the diverse structural nature for the HMF and clindamycin with profound nematicidal property in comparison with existing nematicide carbofuran 3G.
Virtual Screening and Molecular Docking
5-hydroxy-2-methylfurfural
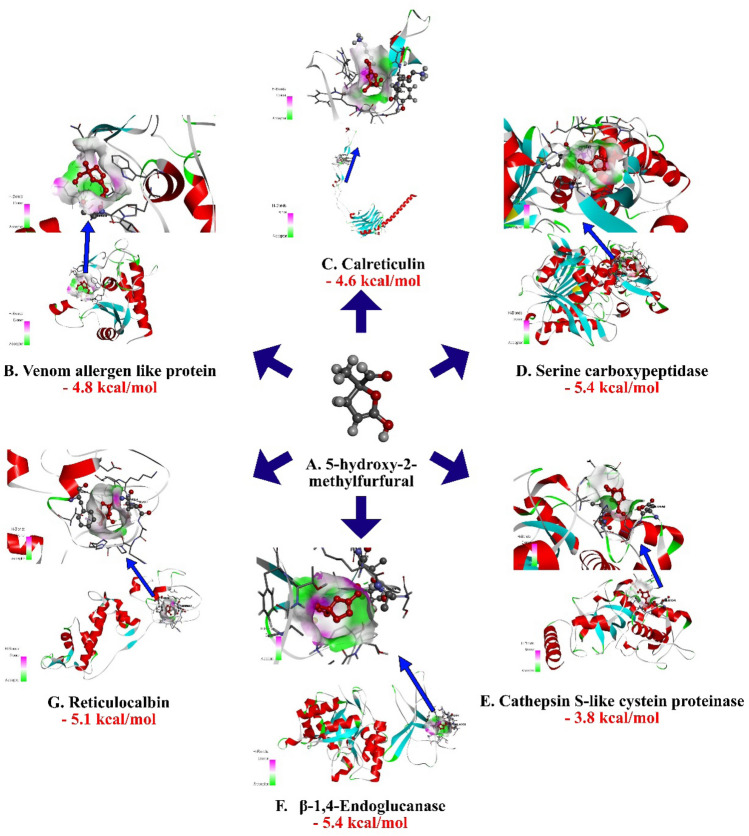
Modelled protein structures were docked with selected compounds to understand their binding mode (Fig. 1, Table 1). HMF had the binding affinity value of − 5.4 kcal/mol with the target β-1,4-endoglucanase (H-bonds: THR 354, ALA 356, THR 359, THR 361), − 4.6 kcal/mol (H-bonds: LYS 237, LYS 271) for calreticulin, binding affinity of 3.8 kcal/mol (H-bonds: GLU 154, LEU 162) for Cathepsin S-like cysteine proteinase, − 5.1 kcal/mol (H-bonds: SER 16) for reticulocalbin, − 5.4 kcal/mol as binding affinity (H-bonds: CYS 342, MET 343) for the target serine carboxypeptidase and was − 4.8 kcal/mol (H-bonds: SER 29) for venom allergen respectively. Hydrogen bonds were reported in all the complexes which denoted the stability and binding strength of HMF towards R. similis targets.
Fig. 1.
Molecular docking interaction of 5-hydroxy-2-methylfurfural with active site residues of Calreticulin, Cathepsin S-like cysteine proteinase, β-1,4-endoglucanaseand reticulocalbin, venom allergen-like protein and Serine carboxypeptidase
Table 1.
Binding affinity values of 5-hydroxy-2-methylfurfural, Clindamycin, Carbofuran with virulent target and H-bond formed
| Targets | Binding affinity (kcal/mol) of small molecules and carbofuran on different targets | H-bonds formed | ||||
|---|---|---|---|---|---|---|
5-hydroxy-2-methylfurfural
|
Clindamycin
|
Carbofuran 3G
|
5-hydroxy-2-methylfurfural
|
Clindamycin
|
Carbofuran 3G
|
|
| β 1,4 endoglucanase | − 5.4 | − 7.2 | − 6.8 |
THR 354, ALA 356, THR 359, THR 361 |
THR 332,THR 334,THR 335,THR 392 | GLU 156, LYS 283 |
| Calreticulin | 4.6 | − 6.3 | − 7.2 |
LYS 237, LYS 271 |
LYS 277 | LYS 237, THR 273 |
| Cathepsin S-like cysteine proteinase | − 3.8 | − 4.3 | − 5.5 |
GLU 154, LEU 162 |
ARG 79, ASN 82 | GLN 313 |
| Reticulocalbin | − 5.1 | − 7.5 | − 6.9 | SER 16 | ASP 53 | PRO 52 |
| Serine carboxypeptidase | − 5.4 | − 6.9 | − 6.2 |
CYS 342, MET 343 |
THR 319 | THR 321 |
| Venom allergen-like protein | − 4.8 | − 5.8 | − 7.7 | SER 329 | MET 283, GLN 333 | TRP 292,THR 294, SER 329 |
Hydrogen bonds with the backbone and side-chain of amino acid residues are the two types of hydrogen bonds formed between docked complexes. Other forms of contacts, such as hydrophobic interactions, van der waals, pi-pi, alkyl, and pi-alkyl, were visible in the docked complex
Clindamycin
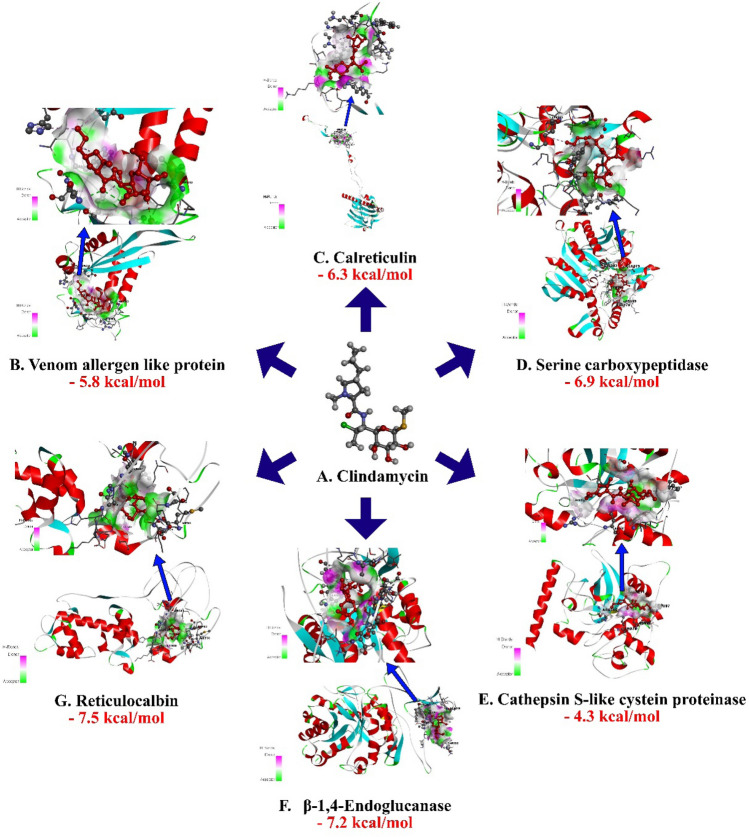
Compound clindamycin had the binding affinity of − 7.2 kcal/mol with target β-1,4 -endoglucanase(H-bonds: THR 332, THR 334, THR 335, THR 392), − 6.3 kcal/mol for calreticulin (H-bonds: LYS 277), − 4.3 kcal/mol for Cathepsin S-like cysteine proteinase (H-bonds: ARG 79, ASN 82), − 7.5 kcal/mol for reticulocalbin (H-bonds: ASP 53), − 6.9 kcal/mol for serine carboxypeptidase (H-bonds: THR 319), − 5.8 kcal/mol for venom allergen (H-bonds: MET 283, GLN 333), respectively (Fig. 2, Table 1).
Fig.2.
Molecular docking interaction of Clindamycin with active site residues of Calreticulin, Cathepsin S-like cysteine proteinase, β-1,4-endoglucanaseand reticulocalbin, venom allergen-like protein and Serine carboxypeptidase
Carbofuran 3G
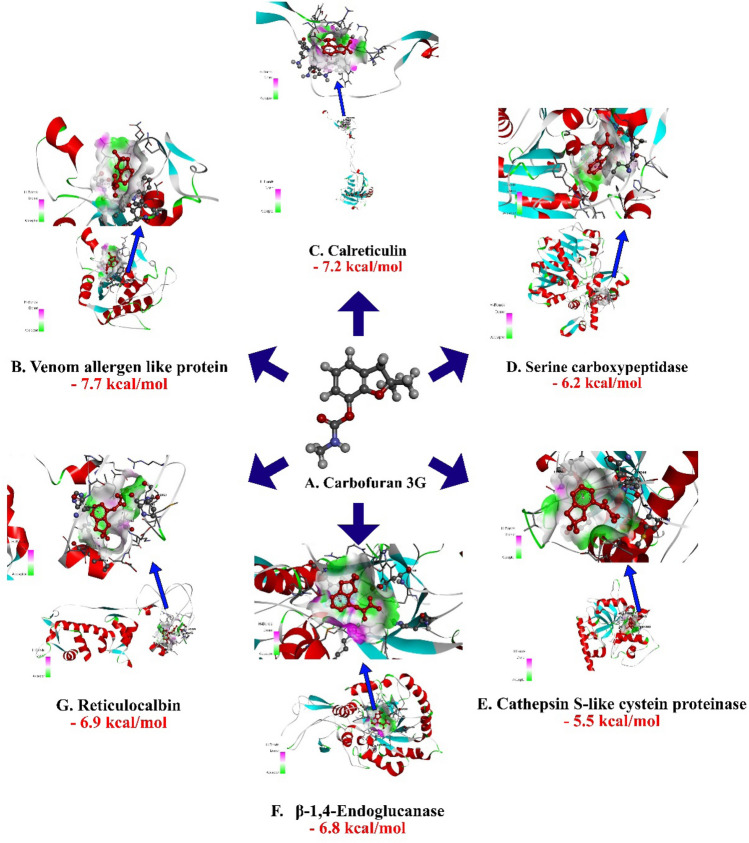
Carbofuran 3G is being used commercially by the farming community for the management of nematode. Hence, it was used as a positive check to compare the binding affinity. The binding affinity value of carbofuran 3G was − 6.8 kcal/mol (H-bonds: GLU 156, LYS 283) with the target β-1,4-endoglucanase, − 7.2 kcal/mol (H-bonds: LYS 237,THR 273) for calreticulin, –5.5 kcal/mol (H-bonds: GLN 313) for Cathepsin S-like cysteine proteinase, − 6.9 kcal/mol (H-bonds: PRO 52) for reticulocalbin, − 6.2 kcal/mol for serine carboxypeptidase (H-bonds: THR 321), − 7.7 kcal/mol (H-bonds: TRP 292, THR 294, SER 329) for venom allergen. The H-bond donor and acceptor groups have been represented by the 3D docked complex for all the targets (Fig. 3, Table 1). Hydrogen bonds are established with the backbone and side-chain of binding site residues in the docked complexes. Other forms of contacts, such as hydrophobic interactions, van der waals, pi-pi stacked, alkyl, and pi-alkyl, were also visible in the docked complex.
Fig.3.
Molecular docking interaction of Carbofuran 3G with active site residues of Calreticulin, Cathepsin S-like cysteine proteinase, β- 1,4 -endoglucanaseand reticulocalbin, venom allergen-like protein and Serine carboxypeptidase
Discussion
The burrowing nematode, R. similis being an endoparasitic migratory nematode is the most economically important nematode parasite of bananas across the globe and was responsible for causing banana toppling disease. The entire life cycle of R. similis is completed within root tissue. Organophosphates or carbamates based nematicides were used for nematode management under field conditions. When compared to untreated controls, nematicide can enhance banana yields by 50% [24]. Nematode management in large commercial banana plantations were predicated on two to four applications of nematicide which are highly dangerous to soil microbial population, and biodiversity [25]. In India, banana growers manage nematode by the application of granular carbofuran 3G to the soil [26]. However, it is only a temporary solution, since the nematode population grows disproportionately after a few months, necessitating frequent applications of nematicides, which eventually become toxic to the environment and not economical [27]. As a direct consequence of the negative impacts of nematicides there is a high desire among scientists for innovating an alternate nematode management strategy. In this juncture, bacterial endophytes are bestowed with the ability to produce VOCs with a broad spectrum of action. They produce flavonoids, peptides, quinones, alkaloids, steroids, phenols, terpenoids, polyketones and lytic enzymes including chitinases, cellulases, hemicellulases, and 1,3-glucanases [28, 29]. Similarly, B. velezensis ZSY-1 produced a phenolic compound, 4-chloro-3-methyl possessing antifungal activity against Alternaria solani and Botrytis cinerea for the first time [30]. Saravanan et al. [13] have also reported the antifungal activity of HMF and clindamycin against Foc. Cuiet al. (2020) also reported the broad spectrum action of B. velezensis 8–4 against Streptomyces galilaeus and fungal pathogens infecting potato [31]. B. velezensis Bvel 1 produced azelaic acid responsible for antifungal action against B. cinereal [32]. However, the black-box of biomolecules produced during the interaction of biocontrol agents and fungal pathogens for nematicidal property remains in the stage of infancy. Hence, there is a need to identify an effective and eco-friendly biomolecule for the management of nematodes of microbial origin. Considering the same, discovery of biomolecules in the black-box produced by bacterial antagonist B. velezensis with Foc confirmed the nematicidal property of HMF and clindamycin for the first time against R. similis.
Virtual screening approaches like molecular docking had a substantial impact in finding a promising novel small molecule with a broad spectrum of action. To harness the advantage of molecular docking for the detection of a molecule with maximum binding affinity on different target sites of R. similis, we explored the possible nematicidal activity of biomolecules HMF and clindamycin against the potential effector and virulent protein targets of R.similis. Among the studied six protein targets of R. similis, the nematicidal activity of clindamycin was predicted to have possible inhibition equivalent to carbofuran 3G. Clindamycin had the maximum binding energy in relation to target site β-1,4 -endoglucanase, reticulocalbin and serine carboxypeptidase than HMF and Carbofuran 3G, which was used as positive control for nematicidal activity. The maximum binding (− 7.2 kcal/mol) of β-1,4-endoglucanase with ligand clindamycin could inhibit the activity of nematode to degrade β 1,4 linkage in cellulose polymer and might have prevented in gaining entry into plant tissues [19]. Likewise, maximum binding energy of clindamycin with reticulocalbin could block the essential protein required for calcium regulation and nematode reproduction [20]. Further, the maximum binding energy with respect to serine carboxypeptidase might have interrupted the hydrolysis of peptides and proteins in banana tissues [18]. Thereby suppress invasion and pathogenicity of burrowing nematode. Thus, the results indicated the chances of clindamycin to possess nematicidal property better than the commercially available nematicide carbofuran 3G. Subsequently clindamycin also had maximum binding energy with protein targets calreticulin (− 6.3 kcal/mol), cathepsin S-like cysteine proteinaseprotease (− 4.3 kcal/mol) and venom allergen-like protein (− 5.8 kcal/mol) compared to HMFand was slightly lower than carbofuran 3G. It indicated that binding of clindamycin with calreticulin can block Ca2+ multifunctional protein responsible for the suppression of immune invasion, reproduction and pathogenesis of burrowing nematode [15, 33]. Besides binding of clindamycin with cathepsin S-like cysteine protein as protease can also suppress the embryonic development of burrowing nematode [14]. Similarly, binding with venom allergen-like protein would inhibit persistent infection of burrowing nematode in banana plants [34]. Thus, the multiple mode of action and maximum binding energy with different target proteins by clindamycin might contribute for the maximum nematicidal action compared to HMF and Carbofuran 3G. Functional groups analysis showed that sulfanyloxan group of clindamycin had interaction with hydrogen bond of all the nematode protein targets. Likewise, aldehyde and hydroxyl group were found to be as interactive centres. But, in the case of HMF this interacting groups might have replace the methyl carbamate present in carbofuran3G.In a similar study, Babu et al. [33]. virtually screened the phenylpropanoids phytochemicals from Piper nigrum L. (black pepper) against various targets including β- 1,4 -endoglucanase, cathepsin β -like cysteine proteinase and glutathione S-transferase from R. similis and reported the increased mortality through in-vitro assay. Sharma et al. [35]. performed the docking interactions of nematicidal compounds with β-tubulin protein from Brugiamalayi and reported albendazol sulfone as best nematicidal (anti-filarial) drug. Taylor et al. [36]. has also reported the common choke points reactions and enzymes in nematodes and prioritized the drug targets and suggested perhexiline as a nematicidal compound and its binding efficacy against carnitine palmitoyl transferase 2 from Caenorhabditis elegans. Further, Babu et al. [33] also performed the comparative docking analysis of plant derived phenyl propanoid compounds and nematicide carbofuran 3G against potential effector proteins of R. similis. They explained the highest binding affinity of carbofuran 3G towards the target calreticulin in comparison with phenyl propanoid compounds. Similarly, in our investigation, carbofuran 3G had the higher binding value against calreticulin, cathepsin S-like cysteine proteinase and venom allergen like protein than HMF and clindamycin. However, there are no reports on the nematicidal activity of clindamycin. Small molecule, clindamycin had the higher binding affinity against different protein targets including β-1,4-endoglucanase, reticulocalbin and serine carboxypeptidases emphasizing the multiple mode of action as like nematicide carbofuran 3G. Thus, the present investigation, is unique in the sense that, clindamycin can be developed as a formulation for the management of banana burrowing nematode. Besides, it was also having antifungal action against Foc infecting banana [13]. Furthermore, the research finding emphasized the avenue for harnessing the potential of clindamycin as a unique small molecule for the management of fungal nematode complex infecting banana. However, further investigation is required to confirm the nematicidal property of clindamycin in wet lab through qRTPCR and transcriptome profiling of clindamycin treated and untreated banana plants challenged with R. similis. The efficacy of clindamycin at field conditions need to be validated as it has both antifungal and nematicidal property. Thus, it would further add more insights towards understanding the mode of action of clindamycin produced by B. velezensis YEBBR6.
Conclusion
The present study explained the multiple mode of action of endophytic bacterial derived biomolecule clindamycin against various protein targets of banana burrowing nematode through in silico approach. Thus, the nematicidal biomolecule clindamycin serve as a potential inhibitor of the target sites associated in interrupting the functions of β-1,4-endoglucanase, reticulocalbin, serine carboxypeptidase, calreticulin, Cathepsin S-like cysteine proteinase, and venom allergen-like proteins. Thus, clindamycin can be explored as a nematicidal molecule for the management of banana burrowing nematode.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by DBT–BTIS facility available at Department of Plant Molecular Biology and Bioinformatics, Centre for Plant Molecular biology and Biotechnology, Tamil Nadu Agricultural University, Coimbatore, Tamil Nadu, India. The authors acknowledge the Department of Plant Pathology, Centre for Plant Protection studies, Tamil Nadu Agricultural University, Coimbatore, Tamil Nadu, India for providing facilities.
Author Contributions
NS, KM and KAS conceptualized the research; SR performed moleculardocking experiments; SN, and RV have supported in the analysis of docking interactions and in the preparation of manuscript; RV illustrated the docking files. The authors read and approved the final manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cruz FSD, Van Den Bergh I, De Waele D, Hautea DM, Molina AB (2005) Towards manage of Musa nematodes in Asia and the Pacifics. INIBAP Montpellier. https://www.bioversityinternational.org/fileadmin/_migrated/uploads/tx_news/Towards_management_of_Musa_nematodes_in_Asia_and_the_Pacific_-_Technical_manual_1099.pdf
- 2.Krishnappa K, Reddy BMR. Integrated management of nematode complex on banana. Indian J Nematol. 1993;23:7–12. [Google Scholar]
- 3.Reddy PR, Khan RM, Rao MS (1992) Crop loss estimation in banana due to burrowing nematode, Radopholussimilis. National Symposium on Productivity and Utilization of Banana, pp 22–23 (National symposium Abstract)
- 4.Hölscher D, Dhakshinamoorthy S, Alexandrov T, Becker M, Bretschneider T, Buerkert A, Crecelius AC, De Waele D, Elsen A, Heckel DG, Heklau H. Phenalenone-type phytoalexins mediate resistance of banana plants (Musa spp.) to the burrowing nematode Radopholussimilis. Proc Natl Acad Sci. 2014;111(1):105–110. doi: 10.1073/pnas.1314168110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.López-Lima D, Sánchez-Nava P, Carrión G, Núñez-Sánchez AE. 89% reduction of a potato cyst nematode population using biological control and rotation. Agron Sustain Dev. 2013;33(2):425–431. doi: 10.1007/s13593-012-0116-7. [DOI] [Google Scholar]
- 6.Davies K, Srivastava A, Kumar K, Mohan S (2015) Understanding nematode suppressive soils: Molecular interactions between Pasteuria endospores and the nematode surface coat. In: 4th Symposium of potato cyst nematode management (including other nematode parasites of potatoes). Asp Appl Biol (National symposium Abstract)
- 7.Li J, Zou C, Xu J, Ji X, Niu X, Yang J, Huang X, Zhang KQ. Molecular mechanisms of nematode-nematophagous microbe interactions: basis for biological control of plant-parasitic nematodes. Annu Rev Phytopathol. 2015;4(53):67–95. doi: 10.1146/annurev-phyto-080614-120336. [DOI] [PubMed] [Google Scholar]
- 8.Silva JC, Medeiros FH, Campos VP. Building soil suppressiveness against plant-parasitic nematodes. Biocontrol Sci Technol. 2018;28(5):423–445. doi: 10.1080/09583157.2018.1460316. [DOI] [Google Scholar]
- 9.Yadav AN. Endophytic fungi for plant growth promotion and adaptation under abiotic stress conditions. Acta Sci Agric. 2019;3(1):91–93. [Google Scholar]
- 10.Khanna V, Ranganathan S. In silico approach to screen compounds active against parasitic nematodes of major socio-economic importance. BMC Bioinform. 2011;12(13):1–12. doi: 10.1186/1471-2105-12-S13-S25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimura Y, Tani S, Hayashi A, Ohtani K, Fujioka S, Kawano T, Shimada A. Nematicidal activity of 5-hydroxymethyl-2-furoic acid against plant-parasitic nematodes. NLM MathSciNet. 2007;62(3–4):234–238. doi: 10.1515/znc-2007-3-413. [DOI] [PubMed] [Google Scholar]
- 12.Aissani N (2014) Nematicidal, antimicrobial and acaricidal activity of plant secondary metabolites. PhD thesis Department of Live and Environmental Sciences, Cagliari Italy. https://core.ac.uk/download/pdf/35315799.pdf
- 13.Saravanan R, Nakkeeran S, Saranya N, Haripriya S, Kavino M, Anandham R, Krishnamoorthy AS, Malathi VG. Differential bacterial endophytome in Foc resistant banana cultivar displays enhanced antagonistic activity against Fusarium oxysporum f. sp. cubense (Foc) Environ Microbiol. 2021 doi: 10.1111/1462-2920.15800. [DOI] [PubMed] [Google Scholar]
- 14.Wang K, Li Y, Huang X, Wang DW, Xu CL, Xie H. The cathepsin S cysteine proteinase of the burrowing nematode Radopholussimilis is essential for the reproduction and invasion. Cell Biosci. 2016;6(1):1–15. doi: 10.1186/s13578-015-0066-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Wang K, Xie H, Wang YT, Wang DW, Xu CL, Huang X, Wang DS. A nematode calreticulin, Rs-CRT, is a key effector in reproduction and pathogenicity of Radopholussimilis. PloS one. 2015;10(6):e0129351. doi: 10.1371/journal.pone.0129351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaouannet M, Magliano M, Arguel MJ, Gourgues M, Evangelisti E, Abad P, Rosso MN. The root-knot nematode calreticulin Mi-CRT is a key effector in plant defense suppression. Mol Plant Microbe Interact. 2013;26(1):97–105. doi: 10.1094/MPMI-05-12-0130-R. [DOI] [PubMed] [Google Scholar]
- 17.Wilbers RH, Schneiter R, Holterman MH, Drurey C, Smant G, Asojo OA, Maizels RM, Lozano-Torres JL. Secreted venom allergen-like proteins of helminths: conserved modulators of host responses in animals and plants. PLoS Pathog. 2018;14(10):e1007300. doi: 10.1371/journal.ppat.1007300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang X, Xu CL, Chen WZ, Chen C, Xie H. Cloning and characterization of the first serine carboxypeptidase from a plant parasitic nematode Radopholus similis. Sci Rep. 2017;7(1):1–9. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamamouch N, Li C, Hewezi T, Baum TJ, Mitchum MG, Hussey RS, Vodkin LO, Davis EL. The interaction of the novel 30C02 cyst nematode effector protein with a plant β-1, 3-endoglucanase may suppress host defence to promote parasitism. J Exp Bot. 2012;63(10):3683–3695. doi: 10.1093/jxb/ers058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi TW, Cho JM, Ahnn J, Song HO. Novel findings of anti-filarial drug target and structure-based virtual screening for drug discovery. Int J Mol Sci. 2018;19(11):3579. doi: 10.3390/ijms19113579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Xu C, Yang S, Chen C, Tang S, Wang J, Xie H. A venom allergen-like protein, RsVAP, the first discovered effector protein of Radopholussimilis that inhibits plant defense and facilitates parasitism. Int J Mol Sci. 2021;22(9):4782. doi: 10.3390/ijms22094782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer RFT, de Beer TA, Rempfer C, Bordoli L, Lepore L. SWISS-MODEL: homology modelling of protein structures and complexes. Nucl Acids Res. 2018;46(W1):W296–303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tai W, He L, Zhang X, Pu J, Voronin D, Jiang S, Zhou Y, Du L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol. 2020;17(6):613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fogain R. Effect of Radopholussimilis on plant growth and yield of plantains (Musa, AAB) Nematology. 2000;2(2):129–133. doi: 10.1163/156854100509015. [DOI] [Google Scholar]
- 25.Chabrier C, Queneherve P. Control of the burrowing nematode (Radopholussimilis Cobb) on banana: impact of the banana field destruction method on the efficiency of the following fallow. Crop Prot. 2003;22(1):121–127. doi: 10.1016/S0261-2194(02)00121-7. [DOI] [Google Scholar]
- 26.Jada MY, Gungula DT, Jacob I. Efficacy of carbofuran in controlling root-knot nematode (Meloidogyne javanica Whitehead, 1949) on cultivars of bambara groundnut (Vigna subterranea (L.) Verdc.) in Yola Nigeria. Int J Agron. 2011 doi: 10.1155/2011/358213. [DOI] [Google Scholar]
- 27.Seenivasan N, Poornima K. Bio-management of root-knot nematode, meloidogyne incognita (Kofoid and White) Chitwood in jasmine (Jasminum sambac L.) Pest Manage Horticecsyst. 2010;16(1):34–40. [Google Scholar]
- 28.Escudero N, Lopez-Llorca LV. Effects on plant growth and root-knot nematode infection of an endophytic GFP transformant of the nematophagous fungus Pochoniachlamydosporia. Symbiosis. 2012;57(1):33–42. doi: 10.1007/s13199-012-0173-3. [DOI] [Google Scholar]
- 29.Schouten A. Mechanisms involved in nematode control by endophytic fungi. Annu Rev Phytopathol. 2016;54:121–142. doi: 10.1146/annurev-phyto-080615-100114. [DOI] [PubMed] [Google Scholar]
- 30.Gao Z, Zhang B, Liu H, Han J, Zhang Y. Identification of endophytic Bacillus velezensis ZSY-1 strain and antifungal activity of its volatile compounds against Alternaria solani and Botrytis cinerea. Biol Control. 2017;105:27–39. doi: 10.1016/j.biocontrol.2016.11.007. [DOI] [Google Scholar]
- 31.Cui L, Yang C, Wei L, Li T, Chen X. Isolation and identification of an endophytic bacteria Bacillus velezensis 8–4 exhibiting biocontrol activity against potato scab. Biol Control. 2020;141:104156. doi: 10.1016/j.biocontrol.2019.104156. [DOI] [Google Scholar]
- 32.Nifakos K, Tsalgatidou PC, Thomloudi EE, Skagia A, Kotopoulis D, Baira E, Delis C, Papadimitriou K, Markellou E, Venieraki A, Katinakis P. Genomic analysis and secondary metabolites production of the endophytic Bacillus velezensis Bvel1: a biocontrol agent against Botrytis cinerea causing bunch rot in post-harvest table grapes. Plants. 2021;10(8):1716. doi: 10.3390/plants10081716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Babu RO, Krishna PB, Eapen SJ. Virtual screening and in vitro assay to explore novel inhibitors from black pepper against potential targets of Radopholussimilis. Int J Comput Appl. 2014;86(14):35–43. [Google Scholar]
- 34.Torres JL (2014) Venom allergen-like proteins in secretions of plant-parasitic nematodes activate and suppress extracellular plant immune receptors. Thesis Wageningen University and Research. https://edepot.wur.nl/304163
- 35.Sharma OP, Pan A, Hoti SL, Jadhav A, Kannan M, Mathur PP. Modeling, docking, simulation, and inhibitory activity of the benzimidazole analogue against β-tubulin protein from Brugiamalayi for treating lymphatic filariasis. Med Chem Res. 2012;21(9):2415–2427. doi: 10.1007/s00044-011-9763-5. [DOI] [Google Scholar]
- 36.Taylor CM, Wang Q, Rosa BA, Huang SC, Powell K, Schedl T, Pearce EJ, Abubucker S, Mitreva M. Discovery of anthelmintic drug targets and drugs using chokepoints in nematode metabolic pathways. PLoS Pathog. 2013;9(8):e1003505. doi: 10.1371/journal.ppat.1003505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.