Abstract
Melanins are the most common and the most enigmatic natural pigments in the nature that found in many different taxa group such as bacteria, yeasts, fungi, insects, plants, reptiles, birds and mammals. These biological macromolecules are highly complex cross-linked, heterogeneous biopolymers and composed of polymerized phenolic and/or indolic compounds. Recently, interest in these ubiquitous biopolymers has been increasing considerably in many different areas such as medicine, pharmacology, cosmetics, organic electronic and optoelectronics because of their versatile properties. In this study, four different extracellular eumelanin pigments (two bacterial eumelanins and two fungal eumelanins) were characterized by different spectrometric techniques such as FT-IR, XRD, NMR and UV-vis. In XRD analyzes, purified fungal and bacterial eumelanin pigments were characterized by giving a wide peak at about 22o with an angle of 2θ. Furthermore, in the 1 H NMR spectra of these biopolymers, it was observed that all pigments have signals in both aromatic and aliphatic regions. In addition to these analyzes, nanostructures of these biopolymers were characterized using atomic force microscopy (AFM) and scanning electron microscopy (SEM). Finally, eumelanin pigment producer microorganisms were molecularly characterized. 16 S rDNA and 18 S rDNA sequence analysis results of these microorganisms (Streptomyces fulvissimus MPPS4, Streptomyces xiamenensis MPPS6, Aspergillus niger MPPF16 and Aspergillus terreus MPPF25) were deposited in NCBI GenBank® database with accession number MT825594, MT973972, MW652652 and MW652653 respectively.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12088-022-01012-1.
Keywords: Eumelanin, AFM, SEM, FT-IR, XRD, 1H NMR
Introduction
Melanins are natural biopolymeric pigments found in many different taxa [1]. These amorphous macromolecular pigments have many different chemical, physical and biological properties such as radioprotective, photoprotective, heavy metal chelating, antioxidant, antimicrobial, and anticancer activity [2–6]. Due to these properties, melanins provide important advantages to living organisms under adverse environmental conditions and in this way, melanins contribute to the protection and survival of living organisms under adverse conditions [7–9].
Melanins are classified under five different sub-headings namely: eumelanin, pheomelanin, allomelanin, neuromelanin and pyomelanin. Each of these melanin pigments are synthesized by different metabolic pathways and many enzymatic or non-enzymatic reactions take place in these process [10].
Eumelanin pigment is dark brown or black in color, while pheomelanin is yellow or red in color. In the process of synthesis of eumelanin pigment, the oxidative polymerization of tyrosine and/or phenylalanine amino acids into L-DOPA takes place initially. Pheomelanin pigments are initially synthesized highly similar to eumelanines but DOPA undergoes cysteinylation and as a result of this cysteinylation reaction, pheomelanins, unlike eumelanins, contain Sulphur [6].
Allomelanines contain different polymer groups such as dihydrofolate, homogentisic acid and catechols and therefore have a highly heterogeneous structure. Additionally, the most distinctive feature of the allomelanin polymers is that these pigments don’t contain nitrogen [11].
Neuromelanin pigment was first observed as granules in the human central nervous system in the 1930s. After that, histological studies have shown pigmentation in the substantia nigra of mammals such as horses and sheep too, in addition to primates such as gibbons, chimpanzees, and baboons. Subsequent investigations have shown that this pigment is insoluble in organic solvents and bleached with hydrogen peroxide. These properties have revealed that this black / brown pigment has similar structure to melanins. However, the metal chelating property and electron paramagnetic resonance (EPR) analysis results confirmed that this granular pigment is melanin. In recent years, obtained evidence, is pointing that neuromelanin accumulation with aging in humans causes Parkinson’s disease [12].
Pyomelanin, one of the another type of melanins, is formed by the catabolism of tyrosine and / or phenylalanine. As a result of the activity of homogentisic acid oxidase (HGA-oxidase) and 4-hydroxyphenylpyruvic acid dioxygenase (4-HPPD) enzymes, tyrosine amino acid is completely breakdown into acetoacetate and fumarate. In case of inhibition of HGA-oxidase enzyme or excessive production of homogentisic acid, this component (HGA) is transferred outside the cell and polymerizes. As a result of these events, extracellular pyomelanine pigment is formed [13].
Due to their physicochemical and bioactive properties, melanins have a widespread use potential in many different industrial areas such as medicine, pharmacology, cosmetics, food packaging, optoelectronic and organic electronics. To date, many articles have been written about the drug carrier, organic semiconductor, metal chelator, radioprotective, photoprotective, organic electronic, optoelectronic, antiproliferative, anticancer, antioxidant, antiviral, antimicrobial etc. properties of these versatile and multifunctional biopolymers [3, 14–21]. However, the fact that melanins are insoluble in water and organic solvents causes these amorphous macromolecular pigments to maintain their mysteries. The obtained results in many different studies conducted so far have revealed that melanin pigments are highly heterogeneous and complex polymers [22, 23]. For this reason, today it appears that the physical, chemical and bioactive properties of these versatile and multifunctional pigments need to be studied in more detail and elucidated in more depth. In this study, the physical and chemical properties of these complex macromolecular pigments purified from four different microorganisms (Streptomyces fulvissimus MPPS4, Streptomyces xiamenensis MPPS6, Aspergillus niger MPPF16 and Aspergillus terreus MPPF25) were tried to be comparatively elucidated with different spectrometric techniques. For this purpose, Fourier transform infrared (FT-IR), X ray diffraction (XRD), nuclear magnetic resonance (NMR) and UV-vis analyzes of the melanin samples were performed and in addition to these analyzes, atomic force microscope (AFM) and scanning electron (SEM) micrographs of these pigment samples were taken. Finally, in order to identify the pigment producer microorganisms, 16 S rDNA and 18 S rDNA gene regions were isolated and sequence analyzes were completed.
Materials and methods
Production and purification of bacterial eumelanin pigments
Eumelanin pigment production from Streptomyces strains was carried out based on the study by Bayram (2021) with small modifications [24]. Streptomyces strains (BSB16 and BSB25), which were found to produce extracellular melanin pigment in ISP 2 medium, were inoculated into nutrient broth (NB) medium (150 mL volume) prepared in 250 mL flasks. These inoculated broths were incubated for one week in a shaking incubator (200 rpm, 35oC). The dark black colored medium obtained at the end of the one-week incubation period was transferred to Falcon tubes (50 mL volume) and centrifuged at 10.000 rpm for 10 min. After centrifugation, the supernatants were carefully transferred to empty Falcon tubes and thus the supernatant was separated from the bacterial biomass (pellet) and these centrifuge processes were repeated two more times. Afterwards, the pH of the obtained pellet-free black suspension was adjusted to 2 using 6 M HCL. After pH adjustment, the resulting solution was left to polymerize at room temperature for 24 h. At the end of this 24-hour polymerization period, the suspension was centrifuged again at same conditions and the supernatant was carefully poured. Obtained brownish-black colored melanin pigment pellet was dried at 60oC for 24 h and kept at 2-8oC until analysis [24].
Production and purification of fungal eumelanin pigments
The processes of the production and purification of fungal eumelanin pigments were carried out by the methods applied by Geib et al. (2020) with small modifications [25]. For this purpose, Aspergillus minimal media (AMM) containing 7.0 mM KCl, 4.3 mM MgSO4, 11.2 mM KH2PO4 was prepared and 100 mM D-glucose as carbon source and 70 mM sodium nitrate as nitrogen source was added to this minimal medium. The prepared Aspergillus minimal medium was dispensed into 250 mL flasks as 100 mL volume and inoculated with melanin producing Aspergillus strains (approximately 106 conidia/ml). After that, these mediums were incubated at 35 °C at 220 rpm for 72 h. At the end of the 72 h incubation period, in order to purify the obtained melanin pigment, medium and mycelium were separated via filtration. For this purpose, the pH of the medium was adjusted to 2 using 6 M HCL and left to stand at room temperature for 24 h for polymerization [26]. At the end of the 24-hour polymerization process, the precipitated melanin pigment was purified by centrifugation at same conditions and kept at 2-8oC until analysis. In addition to these, the processes of the extraction and purification of mycelial eumelanin pigments were carried out by the methods applied by Sava et al. (2001) with minor modifications [27]. For this purpose, firstly, 2 g of mycelial biomass was weighed and transferred into 100 mL of sterile saline, and the pH value was adjusted to 11 using 2 M NaOH. Afterwards, it was kept at 40oC for 36 h. The resulting dark solution was centrifuged at 10.000 rpm for 10 min and the supernatant was carefully transferred to a beaker and acidified with 6 M HCL to pH 2.0. Afterwards, this acidified solution was kept in a Pasteur-oven at 100oC for 2 h. After the 2-hour period, the precipitated residues were purified by centrifugation. This purified dark pigment was dried at 60oC for 24 h and kept at 2-8oC until analysis [26].
Molecular characterization of eumelanin producer microorganisms
Eumelanin pigment producer Streptomyces and Aspergillus strains were molecularly characterized using 16 S rRNA and 18 S rRNA sequencing respectively. In this processes, bacterial 16 S rRNA gene amplicons were obtained using 27 F and 1492R universal primers and 18 S rRNA genes amplicons were obtained using ITS1 and ITS4 primers. Total genomic DNA extraction and 16 S rDNA PCR amplification of Streptomyces strains were performed as described by Bayram (2021) [24] and 18 S rDNA PCR amplification of euelanin pigment producer Aspergillus strains were performed as described by Sezen et al. (2020) [28].
Fourier transform‑infrared (FT‑IR) spectroscopy
Fourier transform infrared spectroscopy (FT-IR) analysis of the purified melanin pigments were performed on a Perkin-Elmer FT-IR spectrometer and melanin samples was scanned at wavenumbers 4000 − 400 cm− 1 and infrared (IR) spectrums were recorded [6].
X Ray diffraction analysis of purified eumelanins
Wide-angle X-ray diffraction (XRD) analysis of purified melanin pigments with Cu Kα radiation (λ = 1.5406 Å) for 2θ angles ranging between 20o and 90o was performed at room temperature by an X-ray diffractometer (Bruker - D8 Discover XRD) [29].
1 H NMR spectroscopy of purified eumelanins
Nuclear magnetic resonance (NMR) spectroscopy of the purified melanin pigments were performed using a 400 MHz NMR Spectrometer (Bruker AVANCE III) at room temperature (9 Tesla) as described before by [6].
Dissolution of purified eumelanin pigments and UV-vis spectroscopy
Purified extracellular eumelanin pigments were dissolved with dimethyl sulfoxide (DMSO) at 35 kHz frequency in an ultrasonic water bath (Kudos). For this purpose, prepared eumelanin pigments (500 µg/mL) were kept in ultrasonic bath at 60oC for 30 min. After these processes, UV-vis absorbance measurements of the obtained eumelanin pigment solutions were carried out. Obtained absorbance values in the wavelength range from 200 to 700 nm were recorded. The blank control was conducted with DMSO [4].
Atomic Force Microscopy (AFM) of purified eumelanins
Two dimensional and three-dimensional AFM micrographs of the eumelanin pigments were captured with tapping mode using Hitachi AFM 5000 II device.
Scanning Electron microscopy (SEM)
Scanning electron microscopy (SEM) image of the eumelanin pigments were captured using Zeiss Gemini Sigma 300 FEG-SEM (Jena, Germany) at a working distance (WD) of 3.1 mm and taken at an electron high tension (EHT) voltage setting of 5.0 kV.
Results
Spectrometric Techniques
When we evaluated obtained Fourier transform‑infrared (FT‑IR) spectra in this study, it was observed that our melanin pigments have broad and strong infrared absorption in the region between 3600 cm− 1 and 2800 cm− 1 (Fig. 1 A). This broad peak region can be attributed to the functional groups found in the indole and pyrrole rings (amine, amide and carboxylic acid). In our study, it was observed that similar spectra were obtained in all purified melanin samples and that these spectra were largely similar to the results in previously published articles [4, 6, 30–32].
Fig. 1.
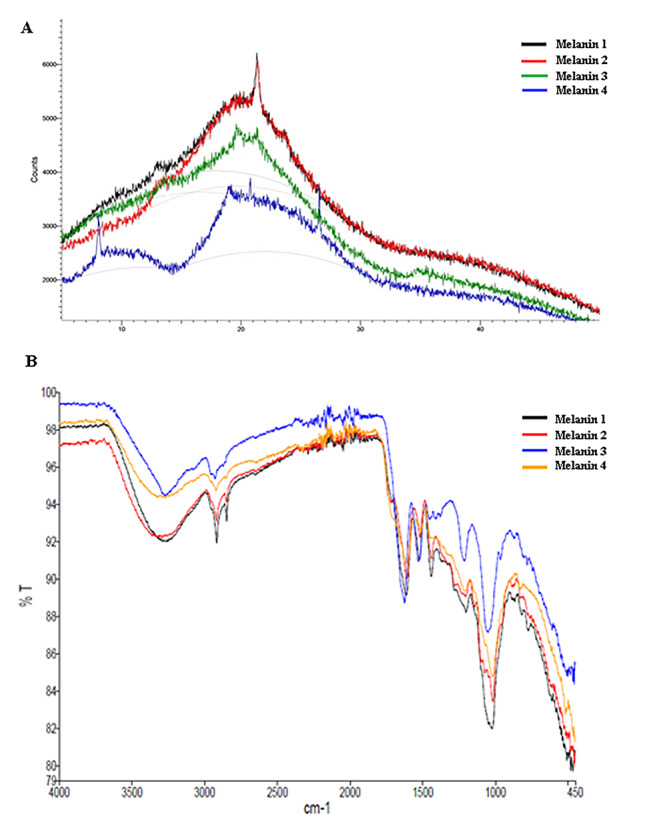
(A): FT-IR spectra of the purified eumelanin pigments (Eumelanin sample 1 purified from Streptomyces fulvissimus MPPS4; eumelanin sample 2 purified from Streptomyces xiamenensis MPPS6; eumelanin sample 3 purified from Aspergillus niger MPPF16; eumelanin sample 4 purified from Aspergillus terreus MPPF25). (B): X Ray diffraction spectra of the purified eumelanin pigments (Eumelanin sample 1 purified from Streptomyces fulvissimus MPPS4; eumelanin sample 2 purified from Streptomyces xiamenensis MPPS6; eumelanin sample 3 purified from Aspergillus niger MPPF16; eumelanin sample 4 purified from Aspergillus terreus MPPF25)
After that, when we used the X-ray diffraction (XRD) technique to examine the atomic and molecular structure of eumelanin pigments it was obtained a wide peak at about 22o with an angle of 2θ (Fig. 1B). The same characteristic peak was observed in all bacterial and fungal melanin pigments. These results indicate that amorphous and heterogeneous structure of melanin pigments. In addition, it was observed that these obtained results were quite similar to the spectrum results previously published in the literature [33–36].
In addition, when we examine the 1 H NMR spectra of the purified bacterial and fungal eumelanin pigments, it was observed that all pigments have signals in both aromatic and aliphatic regions. Similar to previously published articles, in the 1 H NMR spectra, repeating units of indole and pyrrole rings were observed in all examined samples (between 7.00 and 9.00 ppm). Obtained peak between 0.8 and 1.0 ppm in the aliphatic region can be attributed to CH3 groups of alkyl fragments. In addition to these signals, it was observed that residues of solvents (DMSO and H2O) were resonated in the region of 2.5–3.0 ppm. Similar signals were obtained in the 1 H NMR spectra of the examined four different eumelanin samples, and it was observed that all these spectra were quite consistent with the spectra previously published in the literature [4, 6, 24, 32, 36–38].
Furthermore, melanin pigments have the ability to absorb light strongly between 200 and 400 nm wavelengths (in the UV region) and it is seen that the absorbance values decrease as the wavelength increases in the visible light region (between 400 and 700 nm wavelengths). In this study, it was observed that while a high level of absorbance was obtained in the UV region (200–400 nm wavelengths) in all four different eumelanin samples, these absorbance values decreased considerably in the visible region (400–700 nm wavelengths). Obtained UV-vis spectrophotometric measurement results are in line with the results previously published in the literature [33, 39–42].
Atomic Force Microscopy (AFM) and Scanning Electron microscopy (SEM) micrographs
In addition to these spectroscopic techniques, eumelanin pigments were visualized by AFM and SEM. The obtained micrographs are given in Figs. 2 and 3 respectively. In previously published articles in the literature, eumelanin pigments are characterized as spherical shaped particles or aggregates in SEM and AFM images [43–46]. As a result of these microscopic imaging, spherical shaped particles of varying sizes at nanoscale were observed in AFM and SEM micrographs in line with the literature.
Fig. 2.
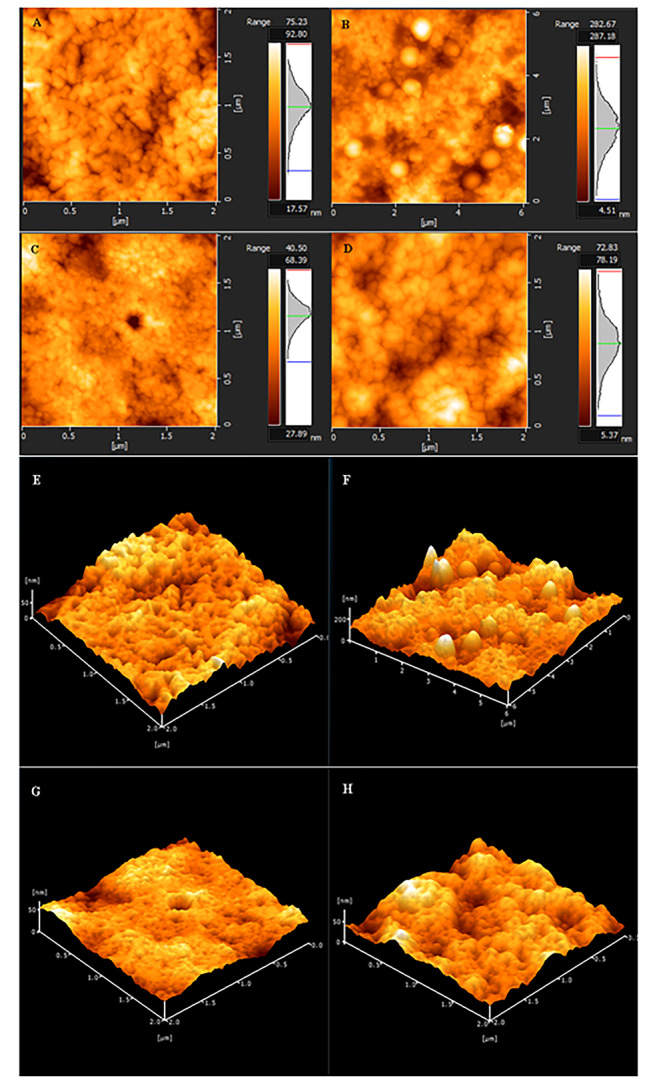
Atomic force electron micrographs of the eumelanin pigments. A, B, C, D two dimensional; E, F, G, H three dimensional images. (A- Eumelanin sample 1 purified from Streptomyces fulvissimus MPPS4, B- Eumelanin sample 2 purified from Streptomyces xiamenensis MPPS6, C- Eumelanin sample 3 purified from Aspergillus niger MPPF16, D- Eumelanin sample 4 purified from Aspergillus terreus MPPF25)
Fig. 3.
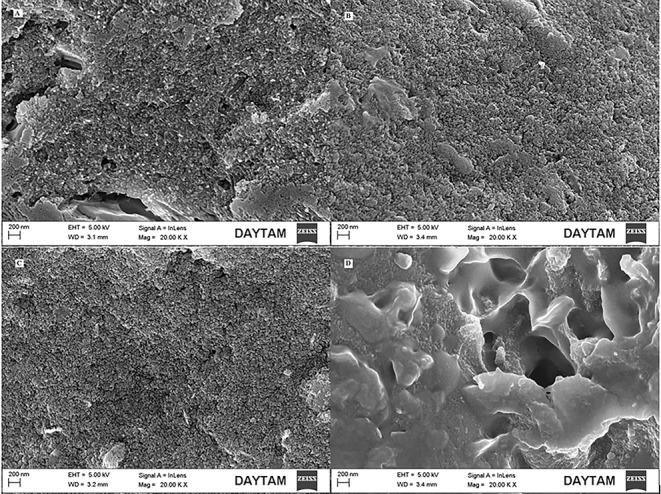
SEM micrographs of the eumelanin pigments. (A- Eumelanin sample 1 purified from Streptomyces fulvissimus MPPS4, B- Eumelanin sample 2 purified from Streptomyces xiamenensis MPPS6, C- Eumelanin sample 3 purified from Aspergillus niger MPPF16, D- Eumelanin sample 4 purified from Aspergillus terreus MPPF25) (20 KX magnification, working distance (WD) of 3.1 mm and taken at an electron high tension (EHT) voltage setting of 5.0 kV.)
Discussion
In this study, it was aimed to characterize four different eumelanin pigments by different spectrometric techniques such as Fourier transform infrared (FTIR) spectroscopy (Fig. 1 A), X-ray diffraction (XRD) spectroscopy (Fig. 1B), 1 H nuclear magnetic resonance (NMR) spectroscopy (Figure S1), and UV-vis spectroscopy (Figuse S2). In addition to these spectrometric techniques atomic force microscopic (AFM) images (Fig. 2) and scanning electron microscopic (SEM) images (Fig. 3) of the eumelanin pigments were taken. Finally, eumelanin-producing bacterial and fungal strains were molecularly characterized. In this processes, bacterial 16 S rRNA gene amplicons were obtained using 27 F and 1492R universal primers and 18 S rRNA genes amplicons were obtained using ITS1 and ITS4 primers. 16 and 18 S rRNA sequence analysis results of these melanin pigment producer microorganisms (Streptomyces fulvissimus MPPS4, Streptomyces xiamenensis MPPS6, Aspergillus niger MPPF16 and Aspergillus terreus MPPF25) were recorded in NCBI GenBank® with accession number MT825594, MT973972, MW652652 and MW652653 respectively.
Are spectrometric techniques sufficient to characterize melanins?
Until now, many different scientific studies have been carried out to determine the chemical structure of melanin pigments. For this purpose, techniques such as FT-IR, NMR, XRD and UV-vis are widely used. In FT-IR analyses, by measuring the absorbance and emission values of eumelanin pigments against infrared rays, the functional groups of these pigments are determined and it is seen that many analysis results in the literature overlap. Melanin pigments have broad and strong infrared absorption in the region between 3600 cm− 1 and 2800 cm− 1 and these signals are attributed to the stretching vibrations of functional groups such as amine, amide and carboxylic acid present in the indole and pyrrole rings [4, 6, 30–32]. When we evaluated obtained FT-IR spectra in this study, it was observed that infrared spectra results were highly consistent and compatible with the previously performed studies (Fig. 1 A). However, these results appear to be insufficient in distinguishing bacterial and fungal eumelanin pigments [4, 6, 30, 31].
In addition to this technique, many different research teams have used the X-ray diffraction analyses to clarify the atomic and molecular structure of these natural pigments. As a result of these studies, consistent sharp peaks, which reflect the characteristic features of melanin samples have been added to the literature with many XRD spectra. Eumelanin pigments are characterized by giving a wide peak at about 22o with an angle of 2θ in XRD analyzes [33–35]. But, these complex biopolymers still remain uncertain due to their amorphous, heterogeneous and insoluble nature. When we evaluated our eumelanin samples in terms of X-ray diffraction (XRD) spectroscopy (Fig. 1B), it was observed that the XRD analysis results of purified bacterial and fungal eumelanin pigments were very compatible with the literature, but no data were obtained for the distinction of bacterial and fungal eumelanin pigments.
In addition, many research teams have studied the 1 H NMR spectra of these natural polymers for the characterization. In previously published research results, it is seen that melanin pigments give peaks in both aliphatic and aromatic regions in 1 H NMR analysis spectra. In this study, when we examine the 1 H NMR spectra of the purified bacterial and fungal eumelanin pigments, signals are seen in both aromatic and aliphatic regions (Please see supplementary data. Figure S1). However, due to the complex and heterogeneous structure of melanin polymers, the resonances in these regions are observed quite uncertainly, similar to previously published articles. In the obtained 1 H NMR spectra, repeating units of indole and pyrrole rings are seen in all examined samples (between 7.00 and 9.00 ppm). Peak between 0.8 and 1.0 ppm in the aliphatic region is attributed to CH3 groups of alkyl fragments in the absorption region. In addition to these signals, residues of solvents (DMSO and H2O) were resonated in the region of 2.5–3.0 ppm. Consequently, it was observed that the 1 H NMR spectra obtained in this study were largely compatible with the previously performed 1 H NMR spectra [4, 6, 24, 32, 36–38]. However, in order to elucidate the structure of eumelanin pigments in detail, more melanin samples need to be analyzed with more techniques such as 13 C, 15 N as well as 1 H NMR analysis.
As the latest spectrometric technique, in all the UV-vis spectrometric measurements, it was observed that bacterial and fungal eumelanin pigments had much higher absorbance values in the UV region (200–400 nm wavelength) compared to the visible region (400–700 nm wavelength) (Please see supplementary data. Figure S2). However, even if this simple characteristic is accepted as a clue for the confirmation of melanin pigments, it does not provide sufficient information to distinguish the bacterial and fungal eumelanin pigments [39–42].
Consequently, melanin pigments are becoming more and more popular in the scientific world and become the subject of many scientific researches because of their radioprotective, photoprotective, drug carrier, antioxidant, organic electronic and optoelectronic properties etc. [3, 14, 20, 47]. However, the fact that these enigmatic pigments are generally insoluble in water and organic solvents makes analysis difficult and this situation prevents us from having detailed information about melanin pigments [23]. For this reason, much more extensive research is required to determine the physical, chemical and bioactive properties of these ubiquitous, versatile and multifunctional pigments. In this study, as a result of the comparative analysis of these pigments, it has been observed that bacterial eumelanin and fungal eumelanin pigments show great similarities physically and chemically. The obtained spectrometric data reveal these similarities (Fig. 1 A-B, Figure S1 and Figure S2). In addition to these processes, as a result of the evaluation of atomic force microscopy (AFM) and scanning electron microscopy (SEM) images, it was seen that eumelanin pigments have spherical and granular structures (Figs. 2 and 3). The obtained results gave positive results for the confirmation of these pigments, however, these results cannot be considered as a distinguishing criterion for the differentiation of fungal and bacterial eumelanin samples.
As a result, in this study, it was aimed to investigate eumelanin pigments obtained from different microorganisms in detail with different spectroscopic and electron microscopic techniques and to determine their distinctive features. For this purpose, purified bacterial and fungal eumelanin samples were analyzed by FT-IR, XRD, NMR and UV-vis spectrometric methods. In addition to these spectrometric techniques, AFM and SEM electron micrographs of eumelanin samples were taken and evaluated comparatively. When all these results are examined, it has been observed that these results are consistent with the literature, but do not provide discriminatory data between bacterial and fungal eumelanin pigments. Based on these results, it can be said that spectrophotometric techniques will be sufficient for the verification of melanin pigments. However, the secrets of these mysterious pigments should be solved as soon as possible and their use in fields such as medicine, cosmetics, textiles, material sciences and biomedical engineering should be further expanded. For this reason, much more extensive research is needed with much more sophisticated techniques such as pyrolysis GC-MS and X-ray photoelectron spectroscopy (XPS) to clarify the structure of these valuable and multifunctional pigments. In this way, it seems likely that these versatile pigments can be used more widely in the fields of medicine, pharmacology and organic electronics, and used in the production of functional bio-composite materials.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
XRD and FT-IR analysis results were obtained from the Bayburt University Central Research Laboratory (BUMER) and NMR analysis reults were obtained from Science Application and Research Center (VAN-MERLAB) in Van Yüzüncü Yıl University. SEM and AFM micrographs were obtained from Center of East Anatolian High-Technology Research and Application (DAYTAM).
Declarations
Conflict of interest
The author declare no conflict of interest.
Human or Animals Participants
This study does not contain any studies with human participants or animals.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Solano F. Melanin and melanin-related polymers as materials with biomedical and biotechnological applications—cuttlefish ink and mussel foot proteins as inspired biomolecules. Int J Mol Sci. 2017;18(7):1561. doi: 10.3390/ijms18071561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dadachova E, Casadevall A. Ionizing radiation: how fungi cope, adapt, and exploit with the help of melanin. Curr Opin Microbiol. 2008;11(6):525–531. doi: 10.1016/j.mib.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schweitzer AD, Revskaya E, Chu P, Pazo V, Friedman M, Nosanchuk JD, Cahill S, Frases S, Casadevall A, Dadachova E. Melanin-covered nanoparticles for protection of bone marrow during radiation therapy of cancer. Int J Radiat Oncol Biol Phys. 2010;78(5):1494–1502. doi: 10.1016/j.ijrobp.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayram S, Dengiz C, Gerçek YC, Cetin I, Topcul MR. Bioproduction, structure elucidation and in vitro antiproliferative effect of eumelanin pigment from Streptomyces parvus BSB49. Arch Microbiol. 2020;202:2401–2409. doi: 10.1007/s00203-020-01956-2. [DOI] [PubMed] [Google Scholar]
- 5.Zerrad A, Anissi J, Ghanam J, Sendide K, Mohammed EH. Antioxidant and antimicrobial activities of melanin produced by a Pseudomonas balearica strain. J Biotechnol Lett. 2014;5(1):87–94. [Google Scholar]
- 6.El-Naggar NA, El-Ewasy SM. Bioproduction, characterization, anticancer and antioxidant activities of extracellular melanin pigment produced by newly isolated microbial cell factories Streptomyces glaucescens NEAE-H. Sci Rep. 2017;7:42129. doi: 10.1038/srep42129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu P, Li Q, Zhang C, Na Y, Xu L. Bcpks12 gene inactivation substantiates biological functions of sclerotium melanization in Botrytis cinerea. Physiol Mol Plant Pathol. 2017;98:80–84. doi: 10.1016/j.pmpp.2017.03.009. [DOI] [Google Scholar]
- 8.Xiao M, Shawkey MD, Dhinojwala A. Bioinspired melanin-based optically active materials. Adv Opt Mater. 2020;8(19):2000932. doi: 10.1002/adom.202000932. [DOI] [Google Scholar]
- 9.Mostert AB. Melanin, the What, the Why and the How: An Introductory Review for Materials Scientists Interested in Flexible and Versatile Polymers. Polymers. 2021;13(10):1670. doi: 10.3390/polym13101670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banerjee A, Supakar S, Banerjee R. Melanin from the nitrogen-fixing bacterium Azotobacter chroococcum: a spectroscopic characterization. PLoS ONE. 2014;9(1):e84574. doi: 10.1371/journal.pone.0084574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCallum NC, Son FA, Clemons TD, Weigand SJ, Gnanasekaran K, Battistella C, Gianneschi NC. Allomelanin: A biopolymer of intrinsic microporosity. J Am Chem Soc. 2021;143(10):4005–4016. doi: 10.1021/jacs.1c00748. [DOI] [PubMed] [Google Scholar]
- 12.Vila M. Neuromelanin, aging, and neuronal vulnerability in Parkinson’s disease. Mov Disord. 2019;34(10):1440–1451. doi: 10.1002/mds.27776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turick CE, Knox AS, Becnel JM, Ekechukwu AA, Milliken CE. Properties and function of pyomelanin. Biopolymers. 2010;449:72. doi: 10.5772/10273. [DOI] [Google Scholar]
- 14.Araújo M, Viveiros R, Correia TR, Correia IJ, Correia VD, Bonifácio T, Casimiro A, Aguiar-Ricardo A. Natural melanin: A potential pH-responsive drug release device. Int J Pharm. 2014;469(1):140–145. doi: 10.1016/j.ijpharm.2014.04.051. [DOI] [PubMed] [Google Scholar]
- 15.Park J, Moon H, Hong S. Recent advances in melanin-like nanomaterials in biomedical applications: a mini review. Biomater Res. 2019;23(1):1–10. doi: 10.1186/s40824-019-0175-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caldas M, Santos AC, Veiga F, Rebelo R, Reis RL, Correlo VM. Melanin nanoparticles as a promising tool for biomedical applications–a review. Acta Biomat. 2020;105:26–43. doi: 10.1016/j.actbio.2020.01.044. [DOI] [PubMed] [Google Scholar]
- 17.Eskandari S, Etemadifar Z. Melanin biopolymers from newly isolated Pseudomonas koreensis strain UIS 19 with potential for cosmetics application, and optimization on molasses waste medium. J Appl Microbiol. 2021;131(3):1331–1343. doi: 10.1111/jam.15046. [DOI] [PubMed] [Google Scholar]
- 18.Roy S, Rhim JW. New insight into melanin for food packaging and biotechnology applications. Crit Rev Food Sci Nutr. 2021;1:1–27. doi: 10.1080/10408398.2021.1878097. [DOI] [PubMed] [Google Scholar]
- 19.Cuzzubbo S, Carpentier AF. Applications of Melanin and Melanin-Like Nanoparticles in Cancer Therapy: A Review of Recent Advances. Cancers. 2021;13(6):1463. doi: 10.3390/cancers13061463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paulin JV, Graeff CF. From nature to organic (bio) electronics: a review on a melanin-inspired material. J Mater Chem C. 2021;9:14514–14531. doi: 10.1039/D1TC03029A. [DOI] [Google Scholar]
- 21.Reali M, Camus A, Beaulieu G, De Angelis J, Pellerin C, Pezzella A, Santato C. Eumelanin: From Molecular State to Film. J Phys Chem C. 2021;125(6):3567–3576. doi: 10.1021/acs.jpcc.0c10063. [DOI] [Google Scholar]
- 22.Yao Z, Qi J, Wang L. Isolation, fractionation and characterization of melanin-like pigments from chestnut (Castanea mollissima) shells. J Food Sci. 2012;77(6):C671–C676. doi: 10.1111/j.1750-3841.2012.02714.x. [DOI] [PubMed] [Google Scholar]
- 23.Ito S, Wakamatsu K, Ozeki H. Chemical analysis of melanins and its application to the study of the regulation of melanogenesis. Pigment Cell Res. 2000;13:103–109. doi: 10.1034/j.1600-0749.13.s8.19.x. [DOI] [PubMed] [Google Scholar]
- 24.Bayram S. Production, purification, and characterization of Streptomyces sp. strain MPPS2 extracellular pyomelanin pigment. Arch Microbiol. 2021;203:4419–4426. doi: 10.1007/s00203-021-02437-w. [DOI] [PubMed] [Google Scholar]
- 25.Geib E, Gressler M, Viediernikova I, Hillmann F, Jacobsen ID, Nietzsche S, Hertweck C, Brock M. A non-canonical melanin biosynthesis pathway protects Aspergillus terreus conidia from environmental stress. Cell Chem Biol. 2016;23(5):587–597. doi: 10.1016/j.chembiol.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 26.Gonçalves RCR, Lisboa HCF, Pombeiro-Sponchiado SR. Characterization of melanin pigment produced by Aspergillus nidulans. World J Microbiol Biotechnol. 2012;28(4):1467–1474. doi: 10.1007/s11274-011-0948-3. [DOI] [PubMed] [Google Scholar]
- 27.Sava VM, Galkin BN, Hong MY, Yang PC, Huang GS. A novel melanin-like pigment derived from black tea leaves with immuno-stimulating activity. Food Res Int. 2001;34(4):337–343. doi: 10.1016/S0963-9969(00)00173-3. [DOI] [Google Scholar]
- 28.Sezen S, Güllüce M, Karadayi M, Alaylar B. First report of fungal strains from Afşin–Elbistan mine for microbial lignite process. Geomicrobiol J. 2020;37(2):143–146. doi: 10.1080/01490451.2019.1668511. [DOI] [Google Scholar]
- 29.Li Q, Liao G, Tian J, Xu Z. Preparation of Novel Fluorinated Copolyimide/Amine-Functionalized Sepia Eumelanin Nanocomposites with Enhanced Mechanical, Thermal, and UV‐Shielding Properties. Macromol Mater Eng. 2018;303(2):1700407. doi: 10.1002/mame.201700407. [DOI] [Google Scholar]
- 30.Ghadge V, Kumar P, Singh S, Mathew DE, Bhattacharya S, Nimse SB, Shinde PB. Natural melanin produced by the endophytic Bacillus subtilis 4NP-BL Associated with the Halophyte Salicornia brachiata. J Agric Food Chem. 2020;68(25):6854–6863. doi: 10.1021/acs.jafc.0c01997. [DOI] [PubMed] [Google Scholar]
- 31.Suwannarach N, Kumla J, Watanabe B, Matsui K, Lumyong S. Characterization of melanin and optimal conditions for pigment production by an endophytic fungus, Spissiomyces endophytica SDBR-CMU319. PLoS ONE. 2019;14(9):e0222187. doi: 10.1371/journal.pone.0222187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Müjdeci GN. Natural Melanin Synthesized by Aureobasidium pullulans Using Food Wastes and its Characterization. Appl Food Biotechnol. 2021;8(4):307–318. doi: 10.22037/afb.v8i4.34599. [DOI] [Google Scholar]
- 33.Madkhali N, Alqahtani HR, Alterary S, Albrithen HA, Laref A, Hassib A. Characterization and electrochemical deposition of natural melanin thin films. Arab J Chem. 2020;13(4):4987–4993. doi: 10.1016/j.arabjc.2020.01.021. [DOI] [Google Scholar]
- 34.Tarangini K, Mishra S (2014) Production of melanin by soil microbial isolate on fruit waste extract: two step optimization of key parameters. Biotechnol Rep 4(2014):139–146. 10.1016/j.btre.2014.10.001 [DOI] [PMC free article] [PubMed]
- 35.Mekala LP, Mohammed M, Chinthalapati S, Chinthalapati VR. Pyomelanin production: Insights into the incomplete aerobic l-phenylalanine catabolism of a photosynthetic bacterium, Rubrivivax benzoatilyticus JA2. Int J Biol Macromol. 2019;126:755–764. doi: 10.1016/j.ijbiomac.2018.12.142. [DOI] [PubMed] [Google Scholar]
- 36.Aime S, Fasano M, Terreno E, Groombridge CJ. NMR studies of melanins: characterization of a soluble melanin free acid from Sepia ink. Pigment Cell Res. 1991;4(5–6):216–221. doi: 10.1111/j.1600-0749.1991.tb00443.x. [DOI] [PubMed] [Google Scholar]
- 37.Hou R, Liu X, Yan J, Xiang K, Wu X, Lin W, Fu J. Characterization of natural melanin from Auricularia auricula and its hepatoprotective effect on acute alcohol liver injury in mice. Food Funct. 2019;10(2):1017–1027. doi: 10.1039/C8FO01624K. [DOI] [PubMed] [Google Scholar]
- 38.Xin C, Ma JH, Tan CJ, Yang Z, Ye F, Long C, Ye S, Hou DB. Preparation of melanin from Catharsius molossus L. and preliminary study on its chemical structure. J Biosci Bioeng. 2015;119(4):446–454. doi: 10.1016/j.jbiosc.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 39.De Souza RA, Kamat NM, Nadkarni VS. Purification and characterisation of a sulphur rich melanin from edible mushroom Termitomyces albuminosus Heim. Mycology. 2018;9(4):296–306. doi: 10.1080/21501203.2018.1494060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.El-Sayyad GS, Mosallam FM, El-Batal AI. One-pot green synthesis of magnesium oxide nanoparticles using Penicillium chrysogenum melanin pigment and gamma rays with antimicrobial activity against multidrug-resistant microbes. Adv Powder Technol. 2018;29(11):2616–2625. doi: 10.1016/j.apt.2018.07.009. [DOI] [Google Scholar]
- 41.Sun S, Zhang X, Sun S, Zhang L, Shan S, Zhu H. Production of natural melanin by Auricularia auricula and study on its molecular structure. Food Chem. 2016;190:801–807. doi: 10.1016/j.foodchem.2015.06.042. [DOI] [PubMed] [Google Scholar]
- 42.Al Khatib M, Harir M, Costa J, Baratto MC, Schiavo I, Trabalzini L, Pogni R. Spectroscopic characterization of natural melanin from a Streptomyces cyaneofuscatus strain and comparison with melanin enzymatically synthesized by tyrosinase and laccase. Molecules. 2018;23(8):1916. doi: 10.3390/molecules23081916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clancy CM, Simon JD. Ultrastructural organization of eumelanin from Sepia officinalis measured by atomic force microscopy. Biochem. 2001;40(44):13353–13360. doi: 10.1021/bi010786t. [DOI] [PubMed] [Google Scholar]
- 44.Bloisi F, Pezzella A, Barra M, Alfè M, Chiarella F, Cassinese A, Vicari L. Effect of substrate temperature on MAPLE deposition of synthetic eumelanin films. Appl Phys A. 2011;105(3):619–627. doi: 10.1007/s00339-011-6603-x. [DOI] [Google Scholar]
- 45.Srisuk P, Correlo VM, Leonor IB, Palladino P, Reis RL. Effect of melanomal proteins on sepia melanin assembly. J Macromol Sci B. 2015;54(12):1532–1540. doi: 10.1080/00222348.2015.1103430. [DOI] [Google Scholar]
- 46.De Trizio A, Srisuk P, Costa RR, Fraga AG, Modena T, Genta I, Dorati R, Pedrosa J, Conti B, Corello VM, Reis RL. Natural based eumelanin nanoparticles functionalization and preliminary evaluation as carrier for gentamicin. React Funct Polym. 2017;114:38–48. doi: 10.1016/j.reactfunctpolym.2017.03.004. [DOI] [Google Scholar]
- 47.Xu R, Santato C, Soavi F. An Electrochemical Study on the Effect of Metal Chelation and Reactive Oxygen Species on a Synthetic Neuromelanin Model. Front Bioeng Biotechnol. 2019;7:227. doi: 10.3389/fbioe.2019.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


