Abstract
The region of the Caulobacter crescentus chromosome harboring the genes for the ClpXP protease was isolated and characterized. Comparison of the deduced amino acid sequences of the C. crescentus ClpP and ClpX proteins with those of their homologues from several gram-positive and gram-negative bacteria revealed stronger conservation for the ATPase regulatory subunit (ClpX) than for the peptidase subunit (ClpP). The C. crescentus clpX gene was shown by complementation analysis to be functional in Escherichia coli. However, clpX from E. coli was not able to substitute for the essential nature of the clpX gene in C. crescentus. The clpP and clpX genes are separated on the C. crescentus chromosome by an open reading frame pointing in the opposite direction from the clp genes, and transcription of clpP and clpX was found to be uncoupled. clpP is transcribed as a monocistronic unit with a promoter (PP1) located immediately upstream of the 5′ end of the gene and a terminator structure following its 3′ end. PP1 is under heat shock control and is induced upon entry of the cells into the stationary phase. At least three promoters for clpX (PX1, PX2, and PX3) were mapped in the clpP-clpX intergenic region. In contrast to PP1, the clpX promoters were found to be downregulated after heat shock but were also subject to growth phase control. In addition, the clpP and clpX promoters showed different activity patterns during the cell cycle. Together, these results demonstrate that the genes coding for the peptidase and the regulatory subunits of the ClpXP protease are under independent transcriptional control in C. crescentus. Determination of the numbers of ClpP and ClpX molecules per cell suggested that ClpX is the limiting component compared with ClpP.
In Caulobacter crescentus, cell differentiation is an integral part of each cell cycle (18). Asymmetric cell division gives rise to two distinct progeny, only one of which, the stalked cell, is competent for a new round of DNA replication and cell division. The other daughter cell, the swarmer cell, has to go through an obligatory differentiation step into a stalked cell to regain its replicative ability. Recent studies have revealed that in this organism, specific proteolytic events are key control mechanisms for cell differentiation as well as for proper cell cycle progression (1, 6, 17, 20, 51). For example, CcrM, an essential DNA methyltransferase, is synthesized only in C. crescentus late predivisional cells, where it is required to fully methylate the newly replicated chromosomes (43, 44, 55). To avoid early methylation of the new chromosomes at the beginning of the S phase, DNA methylation activity is restricted to late predivisional cells. This restriction is accomplished by rapid degradation of CcrM, resulting in its presence in cells only when it is actively synthesized. The instability of CcrM and, thus, part of its temporal control are dependent on the C. crescentus Lon protease (51). Several other C. crescentus proteins have been shown to be degraded in a cell cycle-dependent way; these include the flagellar anchor protein FliF (17); the McpA chemoreceptor (1); CtrA, a cell cycle transcriptional regulator (6); and FtsZ, a tubulin-like GTPase required for cytokinesis (20). To understand how the proteolytic turnover of these key regulatory, metabolic, and structural proteins is controlled by the cell cycle, it is necessary to identify the corresponding protease(s) and to understand the regulation of its activity. However, none of the proteases responsible for the turnover of these proteins has been isolated so far.
Short stretches of amino acids at the carboxyl termini of the CtrA, FliF, and McpA proteins have been identified as turnover signals and shown to be strictly required for the cell cycle-dependent degradation of these proteins (1, 6, 17). Similarly, in Escherichia coli, several proteins are tagged for degradation by the ATP-dependent ClpXP proteases by a short C-terminal domain (12, 24, 28). This possible parallel in the mechanism of substrate recognition led us to test the hypothesis that in C. crescentus, ClpXP might be responsible for the degradation of some of the proteins mentioned above. Here we report the isolation and characterization of the C. crescentus chromosomal locus that contains the genes for the peptidase and the ATPase regulatory subunits of the ClpXP protease. In a parallel study, we showed that both the clpP and the clpX genes are essential for growth, viability, and cell cycle progression (16). In addition, ClpXP was shown to be required for cell cycle-dependent degradation of the CtrA response regulator protein. To better understand the physiological role of the ClpXP protease in Caulobacter, the promoters and transcripts of the clpP and clpX genes were mapped and their transcriptional control was investigated. The analysis of heat shock induction as well as the cell cycle- and growth phase-dependent activities of the mapped promoters demonstrated that clpP and clpX are controlled differently at the transcriptional level. The significance of this finding is discussed with respect to the stoichiometry of ClpP and ClpX in cells and the role of the ClpXP protease in Caulobacter cell cycle progression.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli DH10B and S17-1 were used as the host strain for molecular cloning experiments and as the donor strain for conjugation experiments, respectively. E. coli strains were cultured at 37°C in Luria-Bertani (LB) broth (39) supplemented with ampicillin (100 μg/ml), kanamycin (30 μg/ml), or tetracycline (10 μg/ml) as necessary. C. crescentus strains were grown on either PYE complex medium (35) or M2 minimal glucose medium (7) supplemented with nalidixic acid (20 μg/ml), kanamycin (20 μg/ml), or tetracycline (2 μg/ml) as necessary. Synchronizable C. crescentus NA1000 was used as the wild-type strain in all experiments. Cell cycle synchronization was carried out with this strain by Ludox density gradient centrifugation as described previously (8).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH10B | F−mcrA Δ(mrr hsdRMS mcrBC) φ80dlacZΔM15 ΔlacX74 endA1 recA1 deoR Δ(ara-leu)7697 araD139 galU galK nupG rpsL | Gibco BRL |
| S17-1 | M294::RP4-2 (Tet::Mu) (Kan::Tn7) | 42 |
| W3110 | Wild-type K-12 strain; sup0 | 34 |
| SG22080 | MC4100 ΔclpX1::Kan | 11 |
| SSN1 | W3110 × P1 (SG22080); sup0 ΔclpX1::Kan | This study |
| C. crescentus NA1000 | syn-1000; synchronizable derivative of C. crescentus wild-type strain | 8 |
| Plasmids | ||
| pAR33 | pBBR1 derivative containing the C. crescentus rpoH gene | 36 |
| pAS1 | EcoRI-SalI fragment containing the 3′ end of tig in pRKlac290 | This study |
| pAS2 | SalI-XhoI fragment containing the clpP 5′ region in pRKlac290 | This study |
| pAS3 | SalI fragment containing the clpP 5′ region in pRKlac290 | This study |
| pAS4 | SalI-BamHI fragment containing clpP and cicA in pRKlac290 | This study |
| pAS5 | XhoI-BamHI fragment containing the clpP 3′ region and cicA in pRKlac290 | This study |
| pAS6 | BamHI-FspI fragment containing the clpX 5′ region in pRKlac290 | This study |
| pAS7 | MluI-FspI fragment containing the clpX 5′ region in pRKlac290 | This study |
| pAS8 | EcoRI-BamHI fragment containing the 3′ end of tig, clpP, and cicA in pRKlac290 | This study |
| pAS9 | EcoRI-EcoRV fragment containing the 3′ end of tig and clpP in pRKlac290 | This study |
| pAS23 | ApaI-FspI fragment containing the clpX 5′ region in pRKlac290 | This study |
| pAS24 | SalI-FspI fragment containing cicA and the clpX 5′ region in pRKlac290 | This study |
| pAS25 | EcoRV-FspI fragment containing cicA in pRKlac290 | This study |
| pAS26 | ApaI-BamHI fragment containing the cicA-clpX intergenic region in pRKlac290 | This study |
| pAS27 | SalI-BamHI fragment containing cicA in pRKlac290 | This study |
| pAS29 | SalI-MluI fragment containing clpP in pRKlac290 | This study |
| pAS63 | ApaI-FspI fragment containing the cicA 5′ region in pRKlac290 | This study |
| pAS64 | ApaI fragment containing the internal cicA region in pRKlac290 | This study |
| pBluescript SK(+) | Ampr cloning vector | Stratagene |
| pCS225 | NcoI fragment containing PxylX in pRKlac290 | 32 |
| pGB2ts | pSC101-based vector; thermosensitive for replication; Spr Smr | 26 |
| pGB2ts::phd-doc | Derivative of pGB2ts expressing phd and doc; Spr Smr | 26 |
| pMR10 | RK2-based Kanr broad-host-range vector | Rick Roberts and Chris Moor |
| pMR20 | RK2-based Tetr broad-host-range vector | Rick Roberts and Chris Moor |
| pSSN6 | pMR20 containing clpXCc | This study |
| pSSN3 | pMR20 containing clpXEc | This study |
| pRKlac290 | RK2-based lacZ transcriptional fusion vector | 10 |
PxylX, xylX promoter.
DNA manipulations and sequence analysis.
DNA preparation and manipulation techniques used in this study were as previously described (2, 39). Transformations of E. coli were done by electroporation, and plasmids were transferred from E. coli S17-1 to C. crescentus by conjugation as described previously (7). E. coli clones for sequencing were obtained by subcloning specific DNA fragments into vector pBluescript SK(+) (Stratagene, La Jolla, Calif.). The DNA sequence of the C. crescentus chromosomal clp region was determined for both strands by the dideoxy chain termination method (40).
ClpX activity assay with E. coli.
The clpX genes from E. coli and C. crescentus were amplified by PCR with specific primers (5′-CGA CTC TAG AGC ATA TGA CAG ATA AAC GCA AAG ATG GCT C-3′, 5′-CGC GGA TCC CCT TTT TGG TTA ACT TAT TGT ATG GG-3′, 5′-AAA CAT ATG ACG AAA GCC GCG AGC-3′, and 5′-AAA GGA TCC GCT TCG AAA GCA CGC GCT-3′) and cloned as NdeI-BamHI fragments into pET21b to introduce a ribosome-binding site upstream of clpX. The resulting XbaI-BamHI fragments were cloned into the low-copy-number vector pMR20, resulting in plasmids pSSN3 (E. coli) and pSSN6 (C. crescentus) with the clpX gene under the control of the lacZ promoter.
ClpX activity was determined by the cell killing assay based on the bacteriophage P1 plasmid addiction module Phd-Doc (25). The stable cellular toxin Doc is inhibited by the antidote protein Phd, which is unstable in E. coli due to its rapid degradation by the ClpXP protease (26). ClpX activity was determined by monitoring the growth of E. coli cells containing the phd and doc genes on a plasmid with a temperature-sensitive replicon (pGB2ts::phd-doc) after a shift to the nonpermissive temperature. While the progressive loss of the plasmid stops the new synthesis of Phd and Doc, functional ClpX will degrade the antidote protein Phd, allowing the toxic protein Doc to stop cell growth. In the absence of ClpX, both Phd and Doc are stable and growth is not affected.
For the plasmid addiction assay, combinations of plasmid pGB2ts or pGB2ts::phd-doc with plasmid pSSN3 or pSSN6 were introduced into strains W3110 and SSN1. Cultures in LB broth supplemented with the appropriate antibiotics were grown at 30°C and successively diluted so as to maintain logarithmic growth conditions. The assay was started by diluting the cultures into antibiotic-free LB broth at 40°C and monitoring cell growth by measuring the absorbance at 600 nm.
Promoter mapping and transcriptional activity assays.
DNA fragments containing promoters for clpP, clpX, and cicA were identified by cloning restriction fragments spanning the entire chromosomal clp region (see Fig. 4) into vector pRKlac290, generating transcriptional fusions to the lacZ reporter gene. The resulting plasmids were transferred into C. crescentus NA1000 wild-type cells, and overnight cultures of the resulting strains were diluted in fresh PYE medium and grown to an optical density at 660 nm (OD660) of 0.5 to 0.6. β-Galactosidase activity was then determined as described previously (33).
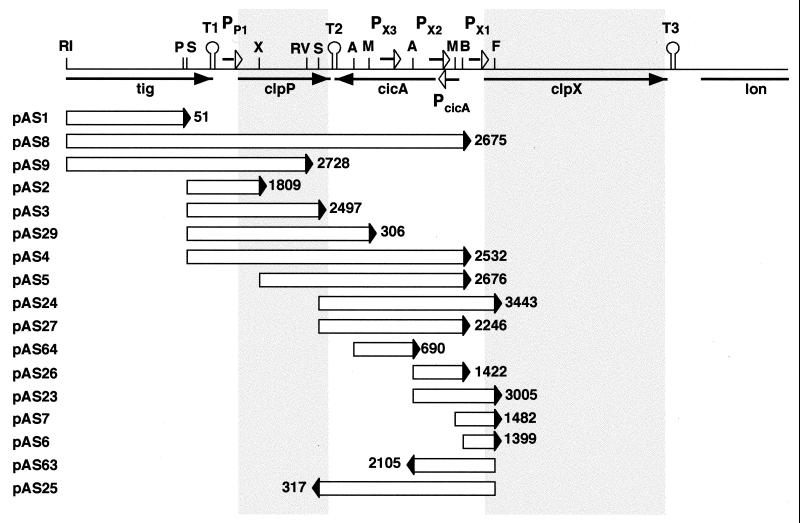
FIG. 4.
Identification of promoter regions in the C. crescentus clp locus. A schematic of the chromosomal clp region is shown at the top. The tig (trigger factor), clpP (ClpP peptidase), cicA (unknown function), clpX (ClpX ATPase), and lon (Lon protease) genes are indicated by black arrows. The approximate locations and orientations of the promoters identified (PP1, PX1, PX2, and PX3) are marked by short open arrows, and putative transcriptional terminator structures (T1, T2, and T3) are indicated as stem-loop outlines. Fragments that were cloned into the lacZ reporter plasmid pRKlac290 (see Materials and Methods) are indicated as open bars below the schematic, with the filled triangles marking the location and orientation of the lacZ reporter gene. The names of the corresponding constructs are on the left, and the number on the right indicates the β-galactosidase activity (Miller units) generated by each fusion construct. All measurements were determined in triplicate, and average numbers are presented. The following abbreviations are used for recognition sites of restriction enzymes: RI, EcoRI; P, PstI; S, SalI; X, XhoI; RV, EcoRV; A, ApaI; M, MluI; B, BamHI; and F, FspI.
For primer extension assays, C. crescentus NA1000 was grown in PYE medium to an OD660 of 0.5, and total RNA was isolated as described previously (38). Residual DNA was removed by precipitating the RNA twice with 3 volumes of sodium acetate (pH 7, 3 M) at −20°C for 6 h before centrifugation for 15 min. Specific primers complementary to the 5′ end of the clpP gene (clpPPE1, 5′-TCG ACC ACC ATC GGC ACC AGG TTC AT-3′; clpPPE2, 5′-ATG ATC CGT TCC TTC AAC AGG CGC GA-3′) and the clpX gene (clpXPE1, 5′-GGC TTT CGT CAT GAT CGC TTC TCA CA-3′; clpXPE2, 5′-GCT TGC GCA CCT CAT GTT GGC TCT TT-3′) were end labeled with [γ-33P]ATP (370 MBq/ml; Amersham) and T4 polynucleotide kinase (NEN BioLabs). Radiolabeled primer (5 × 105 dpm) was annealed to 40 μg of C. crescentus total RNA, and the extension reaction was carried out with a SuperScript II kit (Bethesda Research Laboratories). A 35S-labeled DNA sequencing reaction with the desired primer and pUJ138 DNA as a template was carried out with a ThermoSequenase cycle sequencing kit (Amersham) and served as a standard to identify the transcriptional start site.
The promoter activities of clpP and clpX promoters throughout the cell cycle and following heat shock were determined by pulse-labeling of cultures harboring plasmid-encoded lacZ reporter gene fusions. Pulse labeling and immunoprecipitation of β-galactosidase were carried out as described previously (32).
Immunoblotting.
Immunoblotting was performed as described previously (17). Polyclonal sera against ClpP or ClpX were diluted 1:10,000 and 1:5,000, respectively, before use. Secondary antibody (goat anti-rabbit immunoglobulin G [IgG] coupled to horseradish peroxidase [HRP]) was used at a 1:10,000 dilution. Immunoblots were developed with a Renaissance kit from DuPont NEN by following the manufacturer’s instructions. The cellular levels of ClpP and ClpX were estimated by comparison to a dilution range of purified His-tagged Clp proteins (16). Protein concentrations were determined by the method of Lowry with a Bio-Rad DC protein assay kit and bovine serum albumin as the standard. Signals from the immunoblot analysis were quantitated by use of scanned images and ImageQuant software.
Nucleotide sequence accession number.
The DNA sequence for clp has been deposited in the EMBL nucleotide sequence database under accession no. AJ010321.
RESULTS
Cloning of the genes for the subunits of the Caulobacter ClpXP protease.
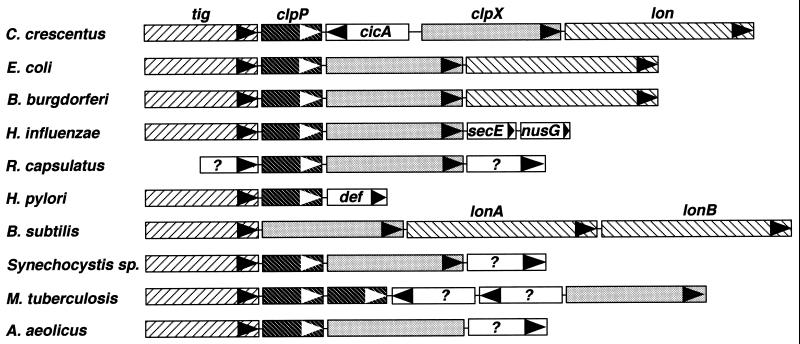
We isolated the C. crescentus clpP and clpX genes on the basis of their proximity to the lon gene, coding for the Lon protease (51). In E. coli, the clpP and clpX genes are located immediately upstream of the lon gene (Fig. 1) (11). Assuming a conserved gene order, we isolated and analyzed the chromosomal region upstream of the C. crescentus lon gene. The genes coding for the ClpP and ClpX homologues were identified together with the tig gene, coding for the homologue of the E. coli trigger factor (Fig. 1). The gene order tig-clpP-clpX is conserved in 7 of 10 bacteria analyzed so far, with the exception of Rhodobacter capsulatus, Helicobacter pylori, and Bacillus subtilis, in which one of the three genes is located elsewhere on the chromosome (Fig. 1). The C. crescentus clp locus contains an additional piece of DNA of about 1 kb between clpP and clpX; this piece is not present in any of the organisms listed in Fig. 1. This region contains an open reading frame, cicA (clp intergenic region in Caulobacter), transcribed in the direction opposite that of the clp genes. Database searches revealed a weak similarity between the deduced amino acid sequence of cicA and that of a family of bacterial proteins with unknown function (data not shown).
FIG. 1.
Schematic diagram of the clpP-clpX regions of gram-positive and gram-negative bacteria. The genes coding for the trigger factor (tig), the ClpP peptidase subunit (clpP), the ClpX ATPase subunit (clpX), and the Lon protease (lon) are indicated. In most organisms analyzed so far, the gene order tig-clpP-clpX is conserved, with the lon gene located immediately downstream. In C. crescentus, the clp genes are separated by cicA, a gene of unknown function; in Haemophilus influenzae, the lon gene is not linked to the clp genes, which are followed by the secE (preprotein translocase subunit) and nusG (transcription antiterminator protein) genes (9); in R. capsulatus, neither tig nor lon is linked to the clp genes, which are flanked by genes of unknown function (?) (35a); in H. pylori, the clpX and lon genes, although linked, are not found at the same location as the tig and clpP genes, which are followed by the def (polypeptide deformylase) gene (45); and in B. subtilis, clpP is not linked to the tig, clpX, and lon genes. The lon gene has undergone duplication in B. subtilis (23); in Synechocystis and Mycobacterium tuberculosis, no gene coding for a homolog of the Lon protease is found. However, two clpP genes are present in M. tuberculosis, and four clpP genes are found in Synechocystis, one of them linked to the tig and clpX genes (19). Two genes are present between the clpP and clpX genes of M. tuberculosis, and the deduced sequences of their products are similar to those of a methyltransferase and a drug efflux protein (4). In Aquifex aeolicus, the lon gene is not linked to the tig, clpP, and clpX genes and is found elsewhere on the chromosome (5).
It is interesting to note that the ClpX ATPase subunit seems significantly more conserved in bacteria than the ClpP peptidase subunit. The C. crescentus ClpP amino acid sequence showed between 31.6 and 65.6% similarity and the ClpX amino acid sequence showed 49.9 to 78.3% similarity with homologues from other bacteria. For both proteins, the weakest similarity was observed with ClpX and ClpP of the spirochete Borrelia burgdorferi, while the strongest match was with ClpX and ClpP of R. capsulatus, like C. crescentus a member of the α-purple group of gram-negative bacteria. The similarity of the Caulobacter ClpP protein with its counterparts extends over the entire protein sequence, and the Ser, His, and Asp residues of the catalytic triad (31, 49) are conserved (Fig. 2A). Similarly, the ClpX sequences are conserved throughout the entire protein length, with the highest homology around the ATP-binding boxes (48) and the C-terminal signature sequences (41) (Fig. 2B). A Zn finger-like motif, CXXC(X18)CXXC, is also conserved in all ClpX sequences shown in Fig. 2B (11). The weakest similarity among all ClpX amino acid sequences is found in the tandem C-terminal PDZ-like (PSD-95, Dlg, and ZO-1 proteins, where the domain was first identified) domains that were proposed to specifically bind target proteins (Fig. 2B) (27).
FIG. 2.
Sequence alignment of ClpP (A) and ClpX (B) from C. crescentus (Ccr) with homologous sequences from R. capsulatus (Rca) (35a), E. coli (Eco) (11, 30), B. subtilis (Bsu) (23), and A. aeolicus (Aae) (5). The amino acid residues identical to those of ClpP or ClpX from C. crescentus are indicated by white letters on a black background. A putative Zn finger-binding site, the ATP-binding (Walker box and ATP-bind.) motifs, and the proposed PDZ-like domains of ClpX (27) are marked. The serine, histidine, and aspartate residues involved in ClpP activity (31, 49) are boxed. Gaps are represented by dashes and were introduced to maximize the alignment. The alignment was generated with the Megalignment program of the DNAstar program package (DNAstar, Madison, Wis.).
ClpXCc is functional in E. coli.
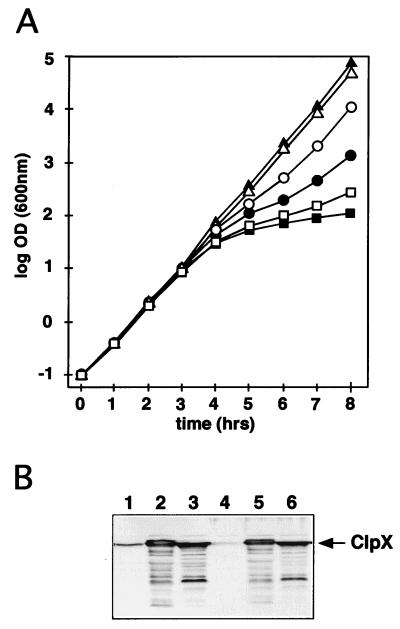
C. crescentus ClpX (ClpXCc) showed 66.8% similarity to E. coli ClpX (ClpXEc). To test if ClpXCc is a functional homologue of the ClpXEc chaperone (50), we tried to complement the E. coli clpX mutant SSN1 with the gene for ClpXCc (clpXCc). For this purpose, the clpXCc coding region was fused to the E. coli Plac promoter with efficient translational start signals (see Materials and Methods), and the fusion was cloned into the low-copy-number vector pMR20. As a test system for functionality, we used the cell killing assay based on the bacteriophage P1 plasmid addiction module Phd-Doc (25) (see Materials and Methods). The growth of E. coli with the temperature-sensitive replicon that does not contain the doc and phd genes (pGB2ts) was unaffected. While wild-type strain W3110 with pGB2ts::phd-doc stopped growing at a nonpermissive temperature (Fig. 3A), the growth of clpX mutant strain SSN1 was not affected by the loss of plasmid pGB2ts::phd-doc (data not shown), demonstrating that the basis of plasmid stabilization by the Phd-Doc module is the degradation of Phd by the ClpXP protease (26). Growth was affected, however, when the clpX mutant strain was complemented with a plasmid-encoded copy of the ClpXEc gene (clpXEc) (pSSN3). Similarly, expressing clpXCc from plasmid pSSN6 in a clpX mutant background resulted in growth inhibition after a temperature shift (Fig. 3A). Growth inhibition in cells expressing ClpXCc was slightly less severe than that in cells expressing the homologous ClpXEc protein (Fig. 3A). This result could be due to reduced activity of the ClpXCc protein in the heterologous host, as immunoblot analysis with an anti-ClpX antibody (16) confirmed that ClpXEc and ClpXCc were present in comparable amounts in the clpX mutant test strain (Fig. 3B). These results suggest that ClpXCc not only is able to recognize the Phd protein but also can interact with the proteolytic ClpP component of E. coli. Interestingly, while a plasmid-encoded copy of clpXCc (pSSN6) was able to support the growth of a C. crescentus mutant with a disrupted chromosomal clpX copy, clpXEc (pSSN3) could not rescue the mutant phenotype (data not shown). This result suggested that ClpXEc is not able to interact with the C. crescentus ClpP subunits, potential substrates, or accessory proteins.
FIG. 3.
clpXCc is able to complement an E. coli clpX mutant. (A) The activity of ClpX was monitored by its ability to degrade, together with ClpP, the antidote protein Phd of the P1 plasmid addiction module Phd-Doc (26). Loss of the phd gene results in Doc-dependent cell killing if the Phd protein is degraded by the ClpXP protease. Growth of cultures containing the phd and doc genes on a plasmid with a temperature-sensitive replicon (pGB2ts::phd-doc) was monitored after a shift to the nonpermissive temperature. Growth is shown as the log OD600. The time after the temperature increase is indicated in hours. Cessation of growth 3 to 5 h after the temperature shift was an indicator of the rapid disappearance of the antidote protein Phd and thus of ClpX activity. The following plasmids were used: plasmid pGB2ts is temperature sensitive for replication; pGB2ts::phd-doc is identical to pGB2ts except that it contains the plasmid addiction genes phd and doc; pSSN6 contains clpXCc; and pSSN3 carries clpXEc. Growth of the following E. coli strains was monitored: W3110/pGB2ts/pSSN6 plus isopropyl-β-d-thiogalactopyranoside (IPTG) (▴; negative control); W3110/pGB2ts::phd-doc/pSSN6 plus IPTG (■; positive control); SSN1/pGB2ts/pSSN6 plus IPTG (▵); SSN1/pGB2ts::phd-doc/pSSN3 plus IPTG (□); SSN1/pGB2ts::phd-doc/pSSN6 plus IPTG (●); and SSN1/pGB2ts::phd-doc/pSSN6 (○). (B) Immunoblot analysis with an anti-ClpX serum and extracts of E. coli strains expressing ClpXCc and ClpXEc. Equal amounts of total protein from the following strains were analyzed: W3110/pMR20 (lane 1); W3110/pSSN3 (lane 2); W3110/pSSN6 (lane 3); SSN1/pMR20 (lane 4); SSN1/pSSN3 (lane 5); and SSN1/pSSN6 (lane 6). The band corresponding to ClpX is marked by an arrow.
The clpP and clpX genes are transcribed from independent promoters.
In E. coli, the clpP and clpX genes are transcribed as an operon from the same promoter, indicating that the syntheses of the regulatory and proteolytic subunits of the ClpXP protease are coupled (11). Three GC-rich inverted repeats followed by several thymidine residues, located downstream of the tig (T1, 5′-GGCGCGGCTCGCGAGGGCCGCGCCTTTTT-3′), clpP (T2, 5′-ACAAAGCCGCCGGCCAGGAGGTCGGCGGCTTTTTT-3′), and clpX (T3, 5′-CGCCATCATCGGATGGCGCGCTTTT-3′) genes, could act as rho-independent transcriptional terminators, implying that these genes are expressed independently from each other (Fig. 4). Also, the large intergenic region between clpP and clpX (Fig. 4) suggested that clpP transcription and clpX transcription are not coupled in C. crescentus.
To analyze the transcription of the clp genes and to localize their promoter regions, we cloned several fragments of the clp locus into vector pRKlac290, generating transcriptional fusions between potential promoter fragments and the lacZ reporter gene. The transcriptional activities of the corresponding fragments were determined by introducing the resulting constructs into C. crescentus NA1000 wild-type cells and measuring β-galactosidase activities. A SalI-XhoI fragment (pAS2; Fig. 4) containing the 5′ end of clpP and its upstream region generated 1,809 Miller units. This fragment most likely contains the promoter for clpP (PP1; see below). The 3′ part of the tig gene generated only background levels of β-galactosidase activity and thus does not carry a promoter (pAS1; Fig. 4). However, we cannot exclude the possibility that some transcriptional activity originating upstream of tig reads through the putative terminator structure T1 located immediately downstream of the tig gene (Fig. 4). Comparison of the Miller units generated by constructs pAS3 and pAS29 (Fig. 4) suggested that the majority of the transcripts that originate at PP1 are terminated by the stem-loop structure T2 downstream of the clpP gene. Thus, PP1 does not contribute significantly to the expression of downstream genes.
The region between the clpP and clpX genes generated a total transcriptional activity corresponding to 3,443 Miller units and reading into the clpX gene (pAS24; Fig. 4). This total activity could be assigned to at least three promoter regions represented by the constructs pAS64 (PX3, 690 Miller units), pAS26 (PX2, 1,422 Miller units), and pAS6 (PX1, 1,399 Miller units). Since the activities of these constructs can be added up to the activity found for the entire clp intergenic region (pAS24), we assume that the transcripts originating in all three promoter regions extend into the clpX gene. In addition, construct pAS23 (3,005 Miller units; Fig. 4) combined the transcriptional activities of PX1 and PX2, and the activity generated by construct pAS27 was equal to the sum of the transcriptional activities of PX2 and PX3 (2,246 Miller units; Fig. 4). The expression of clpX in C. crescentus is therefore controlled by at least three different promoter regions located in the clpPX intergenic region. It is not clear if and to what extent these promoters contribute to the expression of the adjacent lon gene. clpX is followed by a putative rho-independent terminator (T3) that could uncouple the expression of the clpX and lon genes (Fig. 4).
The clpPX intergenic region contains several potential open reading frames, the longest, cicA, pointing in the direction opposite that of clpP and clpX (Fig. 4). To identify a potential promoter region for cicA, we created a lacZ fusion to the ApaI-FspI fragment in the direction opposite that in construct pAS23. While this piece of DNA contains PX1 and PX2, generating about 3,000 Miller units reading into clpX, it also gives rise to about 2,100 Miller units reading in the opposite direction (pAS63; Fig. 4). Speculating that this activity drives the expression of cicA, we call this promoter region PcicA (Fig. 4). In contrast to that generated by construct pAS63, the activity generated by construct pAS25 was only marginally above the background of about 200 Miller units found for vector pRKlac290 alone. This result suggested that the transcripts originating from PcicA do not read into the clpP gene but are terminated at the stem-loop structure T2 (Fig. 4). T2 is localized about 20 bp downstream of the clpP stop codon and 30 bp downstream of the stop codon for the potential cicA open reading frame. This observation and the observation that the T2 inverted repeat is flanked at both ends by several thymidine residues on the transcribed strain indicate that this structure could act as a bidirectional transcriptional terminator.
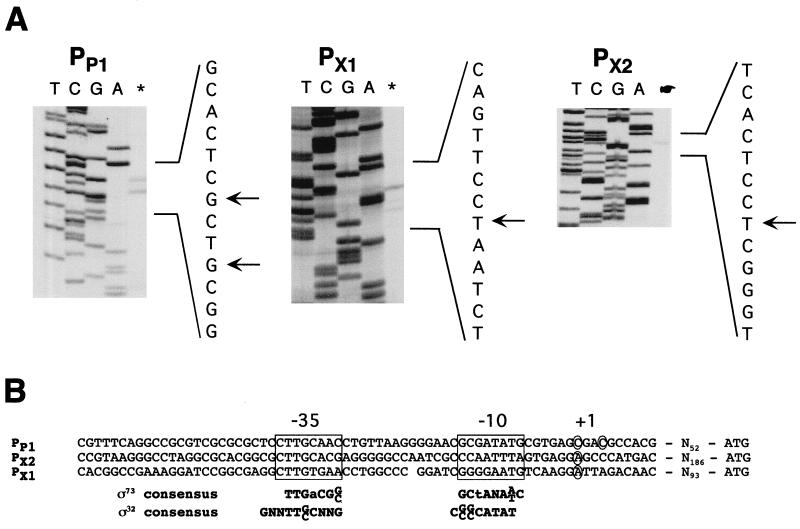
On the basis of the genetic mapping of the promoter regions, we performed primer extension experiments (see Materials and Methods) to precisely localize the transcriptional initiation sites for clpP and clpX. With primers clpPPE1 and clpPPE2 (see Materials and Methods), two close start sites were found to be positioned 59 and 62 bp upstream of the ATG start codon of clpP (Fig. 5). A −10 box and a −35 box designating PP1 were identified upstream of the transcriptional initiation sites for clpP (Fig. 5B). Two separate transcriptional start sites were found directly upstream of clpX by use of two different primers, clpXPE1 and clpXPE2 (see Materials and Methods). One was positioned 103 bp upstream of the proposed translational start codon of clpX and immediately downstream of the BamHI site (Fig. 4 and 5). Two weaker start signals seen in Fig. 5A (PX1) were observed only with the clpXPE1 primer, suggesting that they are artifacts. The second transcriptional start site was located 196 bp upstream of the clpX ATG start codon and immediately upstream of the MluI site (Fig. 4 and 5). Consensus sequences for −10 and −35 promoter boxes were identified upstream of both transcriptional start sites (Fig. 5B). The locations of these two transcriptional start sites downstream of the BamHI site and upstream of the MluI site suggested that the former corresponds to PX1 (pAS6 and pAS7; Fig. 4) and that the latter corresponds to PX2 (pAS26; Fig. 4). It is difficult to unambiguously assign the identified promoter regions to C. crescentus promoter consensus sequences. As depicted in Fig. 5B, the −10 and −35 boxes of PP1, PX1, and PX2 show some similarity to the consensus sequences of both ς32-dependent heat shock promoters (37, 53) and ς73-dependent housekeeping promoters (29).
FIG. 5.
Promoter analysis of the C. crescentus clpP and clpX genes. (A) Primer extension products obtained with the clpX- and clpX-specific primers are shown in the rightmost lanes. Sequencing reactions generated with the same primers are shown in lanes T, C, G, and A. The relevant sequence of the coding strand is shown to the right of each gel, and the positions of the major extension products are indicated by arrows. (B) Sequence alignment of PP1, PX1, and PX2. The −35 and −10 (boxed) and +1 (circled) regions are indicated, and the distance between the transcriptional start site and the presumed translational start codon is shown (Nx). The C. crescentus consensus sequences for ς73-dependent (29) and ς32-dependent (37, 53) promoters are shown in boldface below the three clp promoters.
Heat shock control of clpP and clpX promoters.
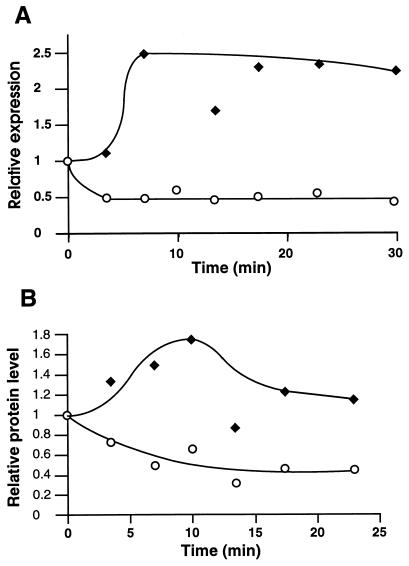
The components of the ClpXP protease in E. coli (22) and the ClpP peptidase in B. subtilis (15, 47) are induced by heat shock, implying a role for the corresponding proteases in the degradation of unfolded and misfolded proteins. To examine if the C. crescentus clpP and clpX genes are also under heat shock control, we assayed the activities of the clpP and clpX promoters as well as the relative concentrations of the ClpP and ClpX proteins after shifting the temperature from 30 to 42°C. Cells of wild-type strain NA1000 carrying plasmid pAS2 (PP1-lacZ) or plasmid pAS24 (PX1-PX2-PX3-lacZ) were grown to the mid-log phase at 30°C, and aliquots of the cultures were shifted to the higher temperature. Promoter activity was assayed by pulse labeling of cells with [35S] Met at different times after the temperature shift and subsequent immunoprecipitation of the β-galactosidase synthesized. The activity of PP1 increased about 2.5 times during the initial 5 to 10 min after heat shock, with transcriptional activity remaining high even after prolonged exposure of the cells to heat (Fig. 6A). In contrast, clpX promoter activity very rapidly decreased by about 50% after heat shock (Fig. 6A). The levels of the ClpP and ClpX proteins followed a similar trend, as the ClpP protein level increased about two times upon heat shock induction, while the ClpX protein level decreased to below 50% its initial value (Fig. 6B). These results suggested that PP1 is under heat shock control and possibly dependent on the C. crescentus sigma factor RpoH (36, 53). Since the clpP gene has recently been shown to be essential for the growth and viability of C. crescentus (16), a requirement for RpoH to activate PP1 could explain the finding that even at low temperatures, the C. crescentus rpoH gene could not be inactivated (36).
FIG. 6.
Heat shock control of ClpP and ClpX expression. (A) The transcription of clpP (⧫) and clpX (○) was determined with cultures of NA1000/pAS2 (clpP::lacZ) and NA1000/pAS24 (clpX::lacZ) by [35S]methionine labeling, β-galactosidase immunoprecipitation, and quantitation at different times after a shift in the temperature from 30 to 42°C. (B) Cellular concentrations of ClpP (⧫) and ClpX (○) after the temperature shift from 30 to 42°C, as determined by immunoblot analysis for the same samples as those analyzed in panel A. The values are relative to the level of transcription or protein at 30°C.
Cell cycle- and growth phase-dependent transcription of clpP and clpX.
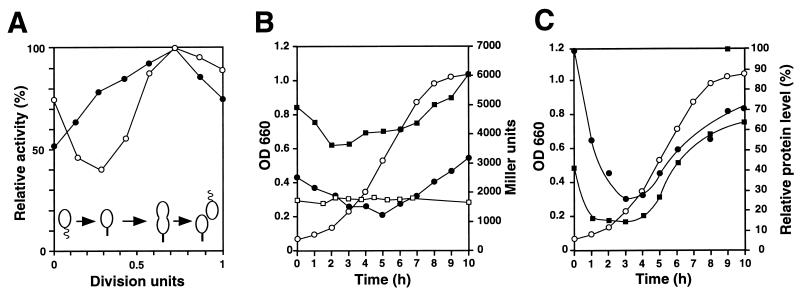
Both clpP and clpX are essential genes in C. crescentus, and their product, the ClpXP protease, is involved in cell cycle control in this organism (16). To examine a possible cell cycle-dependent activity pattern of the clpP and clpX promoters, all four clpP and clpX promoter fragments fused to the lacZ reporter gene (pAS2, pAS6, pAS26, and pAS64; Fig. 4) were assayed by pulse labeling at different short intervals during the cell cycle. Subsequent immunoprecipitation with an anti-β-galactosidase antibody allowed us to quantitate promoter activity at any given time of the cell cycle. Cell cycle fluctuations of promoter activity were observed only for PX1 and PX3 (Fig. 7A). The activity of PP1 and PX2 did not change significantly during the cell cycle. The activity of PX1 and PX3 peaked in predivisional cells (Fig. 7A). However, while PX1 activity was low in stalked cells and high in swarmer cells, the pattern for PX3 activity was the opposite (Fig. 7A). Even though these activity patterns are reproducible, their significance is not clear. Immunoblot analysis with anti-ClpP and anti-ClpX antibodies had revealed that the concentrations of both proteins did not fluctuate significantly during the cell cycle (16).
FIG. 7.
Cell cycle- and growth phase-dependent expression of clpP and clpX. (A) The relative activities of PX1 (○) and PX3 (●) were determined during the cell cycle by pulse labeling of synchronized cultures of strain NA1000 containing plasmids pAS6 (PX1) and pAS64 (PX3), respectively, with [35S]methionine for 5 min at different intervals and determining β-galactosidase synthesis during the pulse time by immunoprecipitation (see Materials and Methods). The promoter activity is shown relative to maximal activity. Progression of the cell cycle is indicated schematically at the bottom. (B and C) Growth phase-dependent expression (B) and cellular levels (C) of ClpP and ClpX. (B) Stationary-phase overnight cultures were diluted in fresh PYE complex medium (NA1000, NA1000/pAS2, and NA1000/pAS24) or PYE complex medium plus 0.2% xylose (NA1000/pCS225), and growth was monitored by determining the OD660 (○). At different intervals, samples were removed from the cultures, and β-galactosidase activity (Miller units) was determined as described in Materials and Methods for the following strains: NA1000/pAS2 (PP1) (●), NA1000/pAS24 (PX1, PX2, and PX3) (■), and NA1000/pCS225 (xylX promoter) (□). (C) Relative cellular levels of ClpP (●) and ClpX (■) in wild-type strain NA1000 were determined as a function of the growth phase by immunoblot analysis. ○, OD660.
During the analysis of the clp promoter fragments, we found that the strength of the clp promoters was dependent on the growth phase of the cells. To determine the influence of the growth phase on the activity of the clpP and clpX promoters, β-galactosidase production from constructs pAS2 and pAS24 (Fig. 4) was monitored through the logarithmic and stationary phases. When cells from an overnight culture were diluted into fresh complex medium, the activity of PP1 was initially high but decreased to about 50% during exponential growth (Fig. 7B). When the cells reached the late logarithmic phase, the activity of PP1 increased coincidently with a decrease in growth rate. Eventually, when the cells entered the stationary phase, the activity of PP1 reached its original value (Fig. 7B). A similar behavior was also observed for the clpX promoters, although the initial decrease occurred more rapidly (Fig. 7B). As a control, the activity of the xylX promoter (32) was assayed as a function of growth phase in the presence of the inducer xylose. In contrast to that of the clp promoters, the activity of the xylX promoter did not change over the course of exponential growth and the stationary phase, demonstrating that promoter induction in the stationary phase is not a general phenomenon in C. crescentus. The levels of the ClpP and ClpX proteins showed a similar, but more pronounced, fluctuation (Fig. 7C). These results suggested that in C. crescentus, the clpP and clpX promoters are induced in the late exponential and stationary phases, resulting in increased ClpP and ClpX levels in cells.
Determination of the number of ClpP and ClpX molecules per cell.
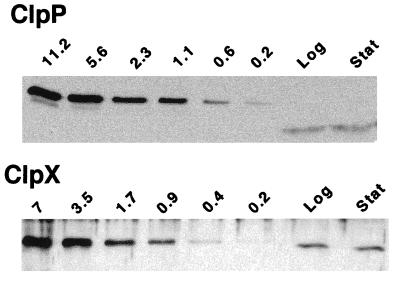
Specific antibodies against ClpP and ClpX were used for an approximate determination of the number of molecules per cell for both components of the protease (see Materials and Methods). Different concentrations of purified His-tagged ClpP and ClpX proteins (16) were analyzed in immunoblot experiments, and the signals obtained were compared quantitatively with the signals from total cellular protein (Fig. 8). On the basis of the assumption that both antibodies react similarly with the full-length His-tagged and native proteins, the numbers of molecules per cell could be calculated from the data. Averages of 23,000 and 41,000 molecules per cell were determined for ClpP in logarithmically growing cells and in the stationary phase, respectively. The corresponding numbers for ClpX were 5,000 molecules per cell in the logarithmic growth phase and 9,000 molecules per cell in the stationary phase. Under the condition that all of the protein pools in the cell are assembled into stable tetradecamers (ClpP) (21, 49) and hexamers (ClpX) (14), the numbers of complexes were about 1,650 ClpP structures and 830 ClpX rings in the logarithmic growth phase and about 3,000 ClpP structures and 1,500 ClpX rings in the stationary phase.
FIG. 8.
Estimation of the number of ClpP and ClpX molecules per cell. Immunoblot analysis was done with anti-ClpP and anti-ClpX sera and with specific amounts (picomoles) of purified His-tagged ClpP and ClpX proteins, respectively, and total proteins from NA1000. The band intensities of total proteins correspond to the amounts of ClpP and ClpX present in 2 × 107 cells and 1 × 108 cells, respectively, during exponential growth (Log) and in 1 × 107 cells and 5 × 107 cells, respectively, during the stationary phase (Stat).
DISCUSSION
We have isolated the genes coding for the ClpXP protease in C. crescentus. ClpXP is a two-component protease with a peptidase subunit (ClpP) and an ATP-dependent regulatory chaperone subunit (ClpX). The peptidase by itself does not have any substrate specificity and requires ClpX for substrate recognition and interaction. Thus, the presence of the regulatory subunit is crucial for the specific activity of the protease. In E. coli, an alternative ATPase, ClpA, is able to interact with and compete for ClpP subunits (14), with ClpXP and ClpAP having distinct substrate spectra (13). However, the molecular basis for substrate selection by the regulatory proteins is not known. In some cases, substrate-specific mediator proteins are required for the interaction between the Clp ATPase and the substrate protein (46, 54). In other cases, a direct substrate interaction is mediated by PDZ-like domains in the C-terminal part of the chaperone (27). In support of this notion, the ClpX C termini are strongly conserved in all bacterial ClpX sequences, indicating a conserved substrate recognition mechanism for ClpX homologues in different organisms. The results of our complementation experiments are in agreement with such a prediction. A copy of the C. crescentus clpX gene can functionally substitute for clpX in E. coli. This fact strongly suggests that the ClpXCc protein is able to interact with at least some of the ClpXEc substrates and with the ClpP peptidase in E. coli. On the other hand, clpXEc was not able to complement a clpXCc mutation. Unlike clpXEc, clpXCc has been shown to be essential for cell cycle progression and thus for growth (16). Thus, one or several substrate proteins for which ClpXP-dependent degradation is indispensable must exist in Caulobacter (16). The failure of ClpXEc to support growth in Caulobacter could be due to its inability to interact with ClpPCc or to inadequate recognition of some or all of the ClpXP substrates. Alternatively, lack of proper control of the ClpXEc-substrate interaction might be the reason for the deficient function of ClpXEc in C. crescentus. Since ClpXP plays a role in controlling the C. crescentus cell cycle, tight temporal control of its activity and thus ClpX substrate accessibility is postulated.
In E. coli, the clpP and clpX genes are coregulated at the transcriptional level (11). In particular, heat shock results in a twofold induction of transcription of the clpPX operon (11), suggesting a role for the ClpXP protease in the stress response in E. coli. In contrast, we found that in C. crescentus, transcription of the clpP and clpX genes is not linked and only the clpP gene is subject to heat shock induction. PP1 was mapped by primer extension analysis and shown to resemble the previously identified ς32-dependent heat shock promoters (3, 36, 37, 52, 53). However, based on the proposed consensus sequences (29, 37, 53), it is difficult to unambiguously assign PP1 to either the ς32-dependent or the ς73-dependent family of promoters. We have observed a twofold increase of PP1 activity in the presence of an additional, plasmid-encoded copy of the rpoH gene (data not shown), indicating that ς32 at least contributes to PP1 activity.
Most transcripts that originated at PP1 terminated immediately downstream of the clpP gene, probably at a transcription terminator-like structure. Thus, heat-inducible PP1 is not responsible for the transcription of the clpX gene. The transcription of clpX is directed by at least three independent promoters in the clpP-clpX intergenic region. This region of about 1 kb was shown to code for an open reading frame (cicA) pointing in the direction opposite that of clpP and clpX. Data obtained recently have confirmed that cicA codes for a protein that, like ClpP and ClpX, is essential for the growth of C. crescentus (50a). Two of the three clpX promoters were localized to the region between the 5′ ends of cicA and clpX. Both show some similarity with the postulated consensus sequence for ς73-dependent promoters (29). The third clpX promoter was located further upstream, in the coding region of cicA. In contrast to the data for clpP, heat shock did not result in an increase but rather resulted in a decrease in total clpX transcription. This finding clearly emphasizes the independent control of the clpP and clpX promoters. Since the change in the concentrations of ClpP and ClpX after heat shock was very similar to the change in the transcription of both genes, we concluded that heat shock control of these genes is exerted primarily at the transcriptional level. It is not clear which mechanism is responsible for the reduction of clpX transcription following heat shock. Since downregulation did not occur for the xylX promoter (15a), the observed reduction of clpX transcription is likely to be the result of a specific regulatory mechanism rather than a general phenomenon. Thus, unlike ClpXEc, ClpXCc does not appear to be involved in the heat shock response. A reduction of ClpX synthesis after heat shock would allow the use of the available ClpP pool more specifically for stress-associated functions. One way to accomplish this would be via an exchange of ClpP-associated ATPases, in which ClpX is replaced by another regulatory ATPase subunit, i.e., ClpA. In vitro, the affinities of the E. coli ClpA and ClpX proteins for ClpP have been shown to be comparable, suggesting that ClpA and ClpX compete for ClpP in vivo (14). The clpACc gene has recently been cloned and shown to be dispensable for growth at a normal temperature (34a). We are currently testing the hypothesis that ClpA is involved in the heat shock response in C. crescentus.
The fact that clpX has three promoters with different cell cycle controls suggested that Caulobacter very carefully regulates the expression of this gene and thereby the cellular concentration of the ClpX protein. While the physiological significance and the individual control of each promoter remain to be elucidated, tight regulation of the clpX gene is consistent with our finding that both the depletion and the overexpression of the ClpX protein are highly toxic for the cell and result in cessation of growth and loss of viability (16). In contrast, the overexpression of the ClpP protein has no obvious physiological consequences for the cell. Assessment of the molar ratio of ClpP to ClpX has revealed that ClpP is present in a molar excess in Caulobacter. Under the assumption that both ClpP and ClpX exist predominantly in their complexed form, about 1,650 assembled ClpP double rings and 830 ClpX rings are present in a cell. Since each ClpX hexamer can complex with two opposite sides of a ClpP tetradecamer in E. coli (14) and assuming that ClpX and ClpP assemble with the same stoichiometry in C. crescentus, the available ClpX pool can saturate at most one-fourth of the theoretically accessible peptidase sites. In comparison, E. coli ClpP has been shown to be limiting compared with ClpX and ClpA combined (14), a finding that is reasonable in view of clpP and clpX being coexpressed and thus most likely present in similar amounts. In Caulobacter, tight control of ClpX expression therefore might be necessary because ClpX is the limiting factor for the degradation of one or several key proteins by the ClpXP protease (16). Alternatively, it is conceivable that ClpX, in addition to its essential role in protein degradation, fulfills critical cellular ClpP-independent functions as a chaperone (50). One of the main future challenges will thus be to characterize the different activities of ClpX in Caulobacter and to identify the proteins with which it is able to interact.
ACKNOWLEDGMENTS
We thank the members of the laboratory for helpful comments and critical reading of the manuscript.
This work was supported by Swiss National Science Foundation fellowship 31-46764.96 to U.J.
REFERENCES
- 1.Alley M R K, Maddock J R, Shapiro L. Requirement of the carboxyl terminus of the bacterial chemoreceptor for its targeted proteolysis. Science. 1993;259:1754–1757. doi: 10.1126/science.8456303. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1992. [Google Scholar]
- 3.Baldini R L, Avedissian M, Gomes S L. The CIRCE element and its putative repressor control cell cycle expression of the Caulobacter crescentus groESL operon. J Bacteriol. 1998;180:1632–1641. doi: 10.1128/jb.180.7.1632-1641.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell B G, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 5.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olsen G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 6.Domian I J, Quon K C, Shapiro L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- 7.Ely B. Genetics of Caulobacter crescentus. Methods Enzymol. 1991;204:372–384. doi: 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
- 8.Evinger M, Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol. 1977;132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 10.Gober J W, Shapiro L. A developmentally regulated Caulobacter flagellar promoter is activated by 3′ enhancer and IHF binding elements. Mol Biol Cell. 1992;3:913–926. doi: 10.1091/mbc.3.8.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottesman S, Clark W P, de Crecy-Lagard V, Maurizi M R. ClpX, an alternative subunit for the ATP-dependent Clp protease of Escherichia coli. Sequence and in vivo activities. J Biol Chem. 1993;268:22618–22626. [PubMed] [Google Scholar]
- 12.Gottesman S, Roche E, Zhou Y, Sauer R T. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottesman S, Wickner S, Maurizi M R. Protein quality control: triage by chaperones and proteases. Genes Dev. 1997;11:815–823. doi: 10.1101/gad.11.7.815. [DOI] [PubMed] [Google Scholar]
- 14.Grimaud R, Kessel M, Beuron F, Steven A C, Maurizi M R. Enzymatic and structural similarities between the Escherichia coli ATP-dependent proteases, ClpXP and ClpAP. J Biol Chem. 1998;273:12476–12481. doi: 10.1074/jbc.273.20.12476. [DOI] [PubMed] [Google Scholar]
- 15.Hecker M, Schumann W, Volker U. Heat-shock and general stress response in Bacillus subtilis. Mol Microbiol. 1996;19:417–428. doi: 10.1046/j.1365-2958.1996.396932.x. [DOI] [PubMed] [Google Scholar]
- 15a.Jenal, U. Unpublished data.
- 16.Jenal U, Fuchs T. An essential protease involved in bacterial cell cycle control. EMBO J. 1998;17:5658–5669. doi: 10.1093/emboj/17.19.5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jenal U, Shapiro L. Cell cycle-controlled proteolysis of a flagellar motor protein that is asymmetrically distributed in the Caulobacter predivisional cell. EMBO J. 1996;15:2393–2406. [PMC free article] [PubMed] [Google Scholar]
- 18.Jenal U, Stephens C, Shapiro L. Regulation of asymmetry and polarity during the Caulobacter cell cycle. Adv Enzymol. 1995;71:1–39. doi: 10.1002/9780470123171.ch1. [DOI] [PubMed] [Google Scholar]
- 19.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 20.Kelly A J, Sackett M J, Din N, Quardokus E, Brun Y V. Cell cycle-dependent transcriptional and proteolytic regulation of FtsZ in Caulobacter. Genes Dev. 1998;12:880–893. doi: 10.1101/gad.12.6.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kessel M, Maurizi M R, Kim B, Kocsis E, Trus B L, Singh S K, Steven A C. Homology in structural organization between E. coli ClpAP protease and the eukaryotic 26 S proteasome. J Mol Biol. 1995;250:587–594. doi: 10.1006/jmbi.1995.0400. [DOI] [PubMed] [Google Scholar]
- 22.Kroh H E, Simon L D. The ClpP component of Clp protease is the ς32-dependent heat shock protein F21.5. J Bacteriol. 1990;172:6026–6034. doi: 10.1128/jb.172.10.6026-6034.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Danchin A, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 24.Laachouch J E, Desmet L, Geuskens V, Grimaud R, Toussaint A. Bacteriophage Mu repressor as a target for the Escherichia coli ATP-dependent Clp protease. EMBO J. 1996;15:437–444. [PMC free article] [PubMed] [Google Scholar]
- 25.Lehnherr H, Maguin E, Jafri S, Yarmolinsky M B. Plasmid addiction genes of bacteriophage P1: doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. J Mol Biol. 1993;233:414–428. doi: 10.1006/jmbi.1993.1521. [DOI] [PubMed] [Google Scholar]
- 26.Lehnherr H, Yarmolinsky M B. Addiction protein Phd of plasmid prophage P1 is a substrate of the ClpXP serine protease of Escherichia coli. Proc Natl Acad Sci USA. 1995;92:3274–3277. doi: 10.1073/pnas.92.8.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levchenko I, Smith C K, Walsh N P, Sauer R T, Baker T A. PDZ-like domains mediate binding specificity in the Clp/Hsp100 family of chaperones and protease regulatory subunits. Cell. 1997;91:939–947. doi: 10.1016/s0092-8674(00)80485-7. [DOI] [PubMed] [Google Scholar]
- 28.Levchenko I, Yamauchi M, Baker T A. ClpX and MuB interact with overlapping regions of Mu transposase: implications for control of the transposition pathway. Genes Dev. 1997;11:1561–1572. doi: 10.1101/gad.11.12.1561. [DOI] [PubMed] [Google Scholar]
- 29.Malakooti J, Wang S P, Ely B. A consensus promoter sequence for Caulobacter crescentus genes involved in biosynthetic and housekeeping functions. J Bacteriol. 1995;177:4372–4376. doi: 10.1128/jb.177.15.4372-4376.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maurizi M R, Clark W P, Katayama Y, Rudikoff S, Pumphrey J, Bowers B, Gottesman S. Sequence and structure of Clp P, the proteolytic component of the ATP-dependent Clp protease of Escherichia coli. J Biol Chem. 1990;265:12536–12545. [PubMed] [Google Scholar]
- 31.Maurizi M R, Clark W P, Kim S H, Gottesman S. Clp P represents a unique family of serine proteases. J Biol Chem. 1990;265:12546–12552. [PubMed] [Google Scholar]
- 32.Meisenzahl A C, Shapiro L, Jenal U. Isolation and characterization of a xylose-dependent promoter from Caulobacter crescentus. J Bacteriol. 1997;179:592–600. doi: 10.1128/jb.179.3.592-600.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 34.Naas T, Blot M, Fitch W M, Arber W. Insertion sequence-related genetic variation in resting Escherichia coli K-12. Genetics. 1994;136:721–730. doi: 10.1093/genetics/136.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34a.Østerås, M., and U. Jenal. Unpublished data.
- 35.Poindexter J S. Biological properties and classification of the Caulobacter group. Bacteriol Rev. 1964;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a.R. capsulatus. 10 May 1998, revision date. Sequences. [Online.] http://pedant.mips.biochem.mpg.de/rodcap/rodcap.html. [31 March 1999, last date accessed.]
- 36.Reisenauer A, Mohr C D, Shapiro L. Regulation of a heat shock ς32 homolog in Caulobacter crescentus. J Bacteriol. 1996;178:1919–1927. doi: 10.1128/jb.178.7.1919-1927.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts R C, Toochinda C, Avedissian M, Baldini R L, Gomes S L, Shapiro L. Identification of a Caulobacter crescentus operon encoding hrcA, involved in negatively regulating heat-inducible transcription, and the chaperone gene grpE. J Bacteriol. 1996;178:1829–1841. doi: 10.1128/jb.178.7.1829-1841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salser W, Gesteland R F, Bolle E. In vitro synthesis of bacteriophage lysozyme. Nature. 1967;215:588–590. doi: 10.1038/215588a0. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schirmer E C, Glover J R, Singer M A, Lindquist S. HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem Sci. 1996;21:289–296. [PubMed] [Google Scholar]
- 42.Simon R, Prieffer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–790. [Google Scholar]
- 43.Stephens C, Reisenauer A, Wright R, Shapiro L. A cell cycle-regulated bacterial DNA methyltransferase is essential for viability. Proc Natl Acad Sci USA. 1996;93:1210–1214. doi: 10.1073/pnas.93.3.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stephens C M, Zweiger G, Shapiro L. Coordinate cell cycle control of a Caulobacter DNA methyltransferase and the flagellar genetic hierarchy. J Bacteriol. 1995;177:1662–1669. doi: 10.1128/jb.177.7.1662-1669.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzgerald L M, Lee N, Adams M D, Venter J C, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 46.Turgay K, Hahn J, Burghoorn J, Dubnau D. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 1998;17:6730–6738. doi: 10.1093/emboj/17.22.6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Volker U, Mach H, Schmid R, Hecker M. Stress proteins and cross-protection by heat shock and salt stress in Bacillus subtilis. J Gen Microbiol. 1992;138:2125–2135. doi: 10.1099/00221287-138-10-2125. [DOI] [PubMed] [Google Scholar]
- 48.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang J, Hartling J A, Flanagan J M. The structure of ClpP at 2.3 Å resolution suggests a model for ATP-dependent proteolysis. Cell. 1997;91:447–456. doi: 10.1016/s0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]
- 50.Wawrzynow A, Wojtkowiak D, Marszalek J, Banecki B, Jonsen M, Graves B, Georgopoulos C, Zylicz M. The ClpX heat-shock protein of Escherichia coli, the ATP-dependent substrate specificity component of the ClpP-ClpX protease, is a novel molecular chaperone. EMBO J. 1995;14:1867–1877. doi: 10.1002/j.1460-2075.1995.tb07179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50a.Wiget, P., T. Fuchs, M. Østerås, and U. Jenal. Unpublished data.
- 51.Wright R, Stephens C, Zweiger G, Shapiro L, Alley M R. Caulobacter Lon protease has a critical role in cell-cycle control of DNA methylation. Genes Dev. 1996;10:1532–1542. doi: 10.1101/gad.10.12.1532. [DOI] [PubMed] [Google Scholar]
- 52.Wu J, Newton A. The Caulobacter heat shock sigma factor gene rpoH is positively autoregulated from a ς32-dependent promoter. J Bacteriol. 1997;179:514–521. doi: 10.1128/jb.179.2.514-521.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu J, Newton A. Isolation, identification, and transcriptional specificity of the heat shock sigma factor ς32 from Caulobacter crescentus. J Bacteriol. 1996;178:2094–2101. doi: 10.1128/jb.178.7.2094-2101.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou Y, Gottesman S. Regulation of proteolysis of the stationary-phase sigma factor RpoS. J Bacteriol. 1998;180:1154–1158. doi: 10.1128/jb.180.5.1154-1158.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zweiger G, Marczynski G, Shapiro L. A Caulobacter DNA methyltransferase that functions only in the predivisional cell. J Mol Biol. 1994;235:472–485. doi: 10.1006/jmbi.1994.1007. [DOI] [PubMed] [Google Scholar]