Abstract
Aim: Due to the fact that patients with COVID--19 can have a bacterial co-infection, physicians should be careful when prescribing antibiotics, with rather considering the sensitivity and resistance of these drugs than various bacteria. Therefore, the main purpose of the present study was to evaluate bacterial coinfections and antibiotic resistance in positive COVID-19 patients.
Method:This descriptive cross-sectional study was performed on 450 hospitalized COVID-19 patients who were selected by simple random sampling. Blood culture (BC) and endotracheal aspirate (ETA) were performed for all COVID-19 patients participating in the study. Antibacterial susceptibility was assessed using the standard Kirby-Bauer disk diffusion method on Mueller Hinton agar for all isolated strains in accordance with the Institute of Clinical and Laboratory Standards guidelines. Finally, susceptibility of all identified bacteria to 10 types of antibiotics was assessed.
Results:Based on the results of endotracheal aspirate (ETA) culture, we found that 79 (17.5%) patients had COVID-19 and bacterial co-infection. Among COVID-19 patients with bacterial co-infection, Klebsiella species had the highest frequency (21.6%), followed by Methicillin-sensitive Staphylococcus aureus (MSSA) (19%), Escherichia coli (17.7%), Methicillin-resistant Staphylococcus aureus (MRSA) (15.2%), Enterobacter species (13.9%) and Pseudomonas aeruginosa (12.6%), respectively. Based on the results of the present study, it was found that the level of antibiotic resistance for different bacteria varied from 0-100%.
Conclusion:The results of the present study indicate that patients with COVID-19 are susceptible to bacterial co-infection, which leads to the conclusion that excessive use of antibiotics is an important factor in the development of antimicrobial resistance. Therefore, caution is needed in prescribing different antibiotics to patients with COVID-19. In addition, considering the SARS-CoV-2 co-infection with other pathogens, it is necessary to use an optimal treatment method for this purpose.
Keywords:COVID-19, co-infections, bacteria, antibiotics resistance.
INTRODUCTION
Coronavirus (CoV) is a ribonucleic acid (RNA) virus isolated from the Coronaviridae family and belonging to the Nidovirales order, which commonly causes respiratory and gastrointestinal infections. Complications may be mild to more severe, such as viral pneumonia with systemic dysfunction (1). In the last decade, CoV has been responsible for two major epidemics, acute respiratory syndrome (SARC) and Middle East respiratory syndrome (MERS), which killed 8,098 (mortality rate 10.5%) and 2,519 (mortality rate 34.4%) people (1, 2). Later on, Coronavirus 2019 (COVID-19) was first detected in December 2019 and spread from Wuhan, Hubei Province, China (2). The virus was initially named nCoV-2019 and subsequently renamed SARSCoV- 2, and eventually the associated disease was renamed COVID-19 (3).
Because the disease can cause serious problems in the human respiratory system, some patients need hospitalization and severe cases require intensive care with mechanical ventilation support (4, 5).
Bacterial co-infections are often found in viral infections of the respiratory tract such as the flu and they represent a major cause of death. Therefore, timely diagnosis and antibacterial treatment for such infections is essential (6-8). The frequency, incidence and characteristics of bacterial co-infections in COVID-19 patients are not known, so this is a large knowledge gap in critical situations like these ones (9-12). Although antibiotics are ineffective in treating COVID-19, physicians prescribe them to patients with suspected or confirmed COVID-19 for various reasons (13).
COVID-19 patients with severe bacterial infections during this pandemic received several instructions for the use of experimental antibiotic therapy (13). It is difficult to prevent bacterial coinfection when COVID-19 occurs, but there is also a high risk of secondary bacterial infection during the disease. For these reasons, there are concerns about the potential overuse of antibiotics and subsequent harmful consequences of bacterial resistance. Due to the increase in mortality among patients with severe bacterial infection during the influenza pandemic, various guidelines have been developed to support the use of experimental antibiotics for patients with severe COVID-19 (13).
Although COVID-19 mortality has occurred mainly in the elderly with severe underlying diseases (14), nosocomial pneumonia (NP) is a major risk factor for patients in intensive care units (ICU). In addition, NP may pose a greater risk to patients' health, especially when intubated. The presence of nosocomial infections (NIs) is commonly described as infections that occur during hospitalization within 48 to 72 hours of hospitalization and are spread mainly through contact, personal devices, and tools (15). The most common bacterial pathogens causing Nis include Staphylococcus, Enterococcus, Klebsiella pneumoniae, Enterobacter, Escherichia coli, Acinetobacter and Pseudomonas (16).
Due to the fact that patients with COVID-19 can have a bacterial co-infection, physicians should be careful when prescribing antibiotics, rather considering the sensitivity and resistance of these drugs than various bacteria (11). Therefore, the main purpose of the present study was to evaluate bacterial co-infections and antibiotic resistance in positive COVID-19 patients as a case study in India.
METHOD
This descriptive cross-sectional study was performed on 450 patients admitted to an Indian hospital from April 25, 2020 to December 31, 2020, who were selected by simple random sampling. Participation eligibility was based on meeting the criteria in the questionnaire completed by each patient.
Study plan
Initial laboratory evaluations for study participants included complete blood count (CBC), erythrocyte sedimentation rate (ESR), arterial blood gas (ABG), lactate dehydrogenase (LDH), and C reactive protein (CRP). All patients were positive for SARS-CoV-2 in vitro swab samples using quantitative RT-PCR (qRT-PCR). Clinical laboratory findings were evaluated for all COVID-19 patients and finally all obtained information was recorded in a data collection form.
Isolation and identification of bacteria
Blood culture (BC) and endotracheal aspirate (ETA) were performed for all COVID-19 patients in the study. To identify and separate the bacteria from each other, swabs and blood were cultured on blood agar and McConkey agar plates and then incubated at 37 °C for 24 hours. Standard microbiological methods were used to identify isolated bacteria (17).
Evaluation of bacterial susceptibility to antibiotics
Antibacterial susceptibility was assessed using the standard Kirby-Bauer disk diffusion method on Müller-Hinton agar (Merk Co., Germany) for all isolated strains in accordance with guidelines of the Institute of Clinical and Laboratory Standards (CLSI; 2019, M100-S29). To test for the aforementioned discs from Gentamicin (10 micrograms), Vancomycin (30 mg), Trimethoprim/sulfamethoxazole (25 mg), Amikacin (30 mg), Tobramycin (10 mg), Cephalothin (30 mg), Norfloxacin (50 mg), and Ceftizoxime (30 mg) were used (18-20).
RESULTS
The identified bacterial species isolated from COVID-19 patients as well as the abundance of each of the identified species are shown in Table 1. Among the 79 (17.5%) patients with COVID-19 and bacterial co-infection, the presence of Klebsiella spp., Methicillin-sensitive Staphylococcus aureus (MSSA), Methicillin-resistant Staphylococcus aureus (MRSA), Escherichia coli, Pseudomonas aeruginosa and Enterobacter species was reveled in 10 (12.7%), 9 (11.4%), 5 (6.3%), 9 (11.4%), 5 (6.3%) and 6 (7.6%) subjects, respectively, according to the results of the endotracheal aspirate (ETA) culture, and in 7 (8.9%), 6 (7.6%), 7 (8.9%), 5 (6.3%), 5 (6.3%) and 5 (6.3%) patients, respectively, according to results based on blood culture. Antimicrobial susceptibility patterns of various bacteria isolated from COVID-19 patients were shown in Figures 1 to 6.
DISCUSSION
COVID-19, an unusually prevalent viral pneumonia, is considered a new concern and threat to public health worldwide. Based on the results of some previous studies, it has been determined that 2019-nCoV or SARS-CoV-2 originated from an animal source and later, by crossing the species barrier, eventually caused the infection of humans (20-24). The main purpose of the present study was to evaluate the burden of concurrent infections in 450 patients with COVID-19 in an Indian hospital. A total of 75 (17.5%) patients had secondary bacterial infection among the evaluated patients with COVID-19. The results of the present study showed that the most commonly identified bacteria isolated from endotracheal aspirate culture and blood culture included Klebsiella species, Methicillin-resistant Staphylococcus aureus (MRSA), Escherichia coli, Methicillin-sensitive Staphylococcus aureus (MSSA), Enterobacter and Pseudomonas aeruginosa. Among people with COVID-19 who had a bacterial co-infection, Klebsiella species was the most frequent one (21.6%), followed by MSSA (19%), Escherichia coli (17.7%), MRSA (15.2%), Enterobacter species (13.9%) and Pseudomonas aeruginosa (12.6%), respectively.
According to the results of previous studies, hospitalization for any reason can increase the risk of secondary infections related to health care as well as the risk of transmission of multidrugresistant diseases. In this regard, various microorganisms can increase the use of antibiotics (25, 26). A recent study in ICU across 88 countries found that although only 54% of patients had suspected or confirmed COVID-19, 70% of them had received at least one antibiotic to either treat or prevent bacterial infection (27). In Norton’s study conducted at Montefiore Medical Center (New York), 71% of more than 5,800 COVID-19 patients who had been hospitalized from March to May 2020 received at least one dose of antibiotics (28). Results of studies carried on by Norton (28) and Getahun et al (27) showed that about 72% of hospitalized COVID-19 patients received various antibiotics, while only 8% of them had severe bacterial or fungal infections.
The results of the present study in terms of the amount and type of bacterial co-infection were consistent with those reported by previous studies, but not also with some others. Sharifipour et al (2020) found that COVID-19 patients admitted to ICU had a secondary infection with Acinetobacter baumannii and two strains of Staphylococcus aureus (21). Mahmoudi (2020) also showed that 12.5% of 340 COVID-19 patients had a secondary bacterial infection, most commonly with Klebsiella spp, MRSA, E. coli, MSSA, Enterobacter and Pseudomonas aeruginosa spp (20). In terms of bacterial species, the results of Mahmoudi’s study were very similar to ours, while the amount of secondary bacterial infection in the present study was higher than that reported in the above-mentioned study (17.5% vs. 12.5%). In this regard, the results obtained by Yang et al in their research (2020) showed that 13.5% of patients with COVID-19 had nosocomial infections; among them, one (2%) patient had a pulmonary and blood infection with K. pneumoniae. Other microorganisms identified from respiratory secretions of five (10%) patients with secondary infections included Aspergillus flavus, Aspergillus fumigatus, extended- spectrum β-lactamase (ESBL)-positive K. pneumoniae, ESBL-positive P. aeruginosa, and SBL-negative Serratia marcescens (29). Xavier Lescure et al (2020) reported that patients with evaluated COVID-19 had two pathogens, including antibiotic-susceptible A. baumannii and A. flavus (30).
Chen et al (2019) reported that some COVID-19 patients, especially with severe forms, had a bacterial co-infection. The results of their study revealed that the usual bacterial cultures of patients with secondary infections identified A. baumannii and K. pneumoniae, with the first one being more resistant to antibiotics than the latter (31). Based on the findings of Xing et al (2020), Mycoplasma pneumoniae and Legionella pneumophila were the most commonly identified respiratory pathogens in COVID-19 patients, with prevalence rates of 23.3% and 20%, respectively (32). Based on a systematic review performed by Lansbury et al (2020), about 7% of hospitalized COVID-19 patients had a bacterial co-infection, and the most commonly reported strains included M. pneumoniae, P. aeruginosa and Haemophilus influenza (24). In the UK, Toombs et al (2020) reported that two COVID-19 patients had co-infection with Staphylococcus pneumoniae (33). The results of all the abovementioned studies (20-33) support the hypothesis of secondary infection in COVID-19 patients.
According to data from studies on the use of antibiotics in the treatment of COVID-19 patients, an average of 70% of patients receive antibiotics. However, given that the improper use or overuse of antibiotics is an important factor in the development of antimicrobial resistance, physicians should be very careful when prescribing various antibiotics to COVID-19 patients. Therefore, many efforts have been already made against antimicrobial resistance to reduce inappropriate or excessive use of antibiotics (34). In addition, during the COVID-19 pandemic, soaps and antimicrobial disinfectants have been increasingly used by hospital staff. If these products are based on chlorhexidine digluconate or quater nary ammonium compounds, this can also lead to antibiotic resistance (20).
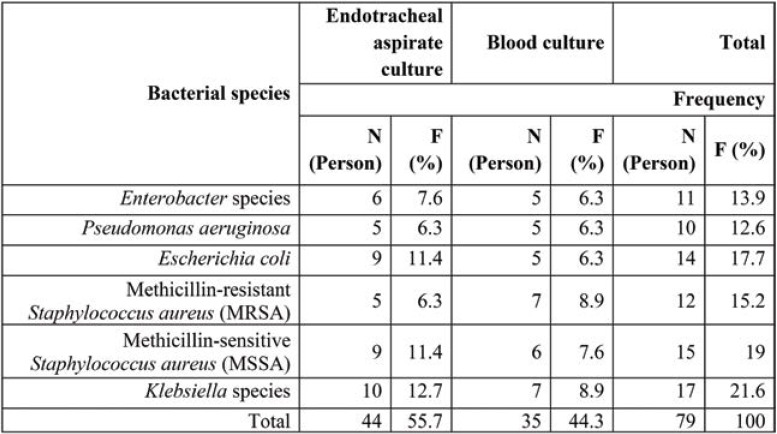
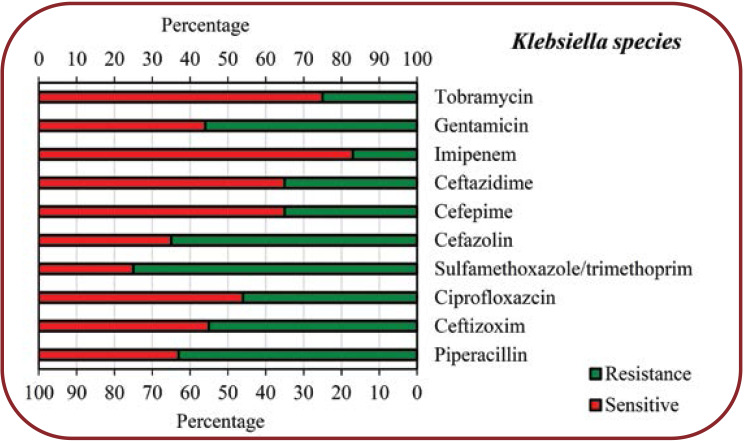
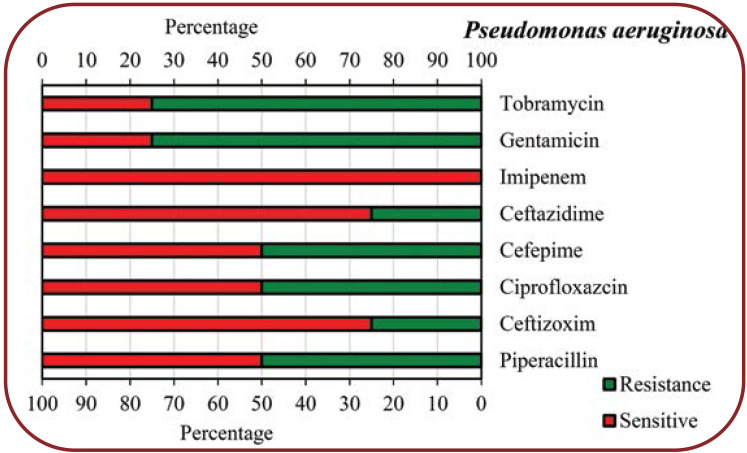
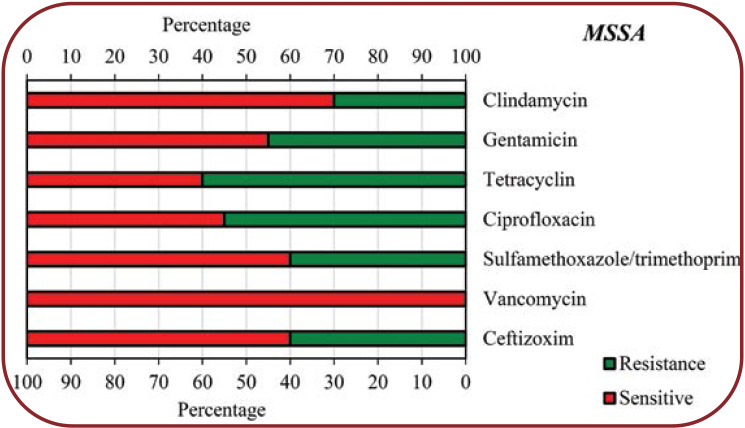
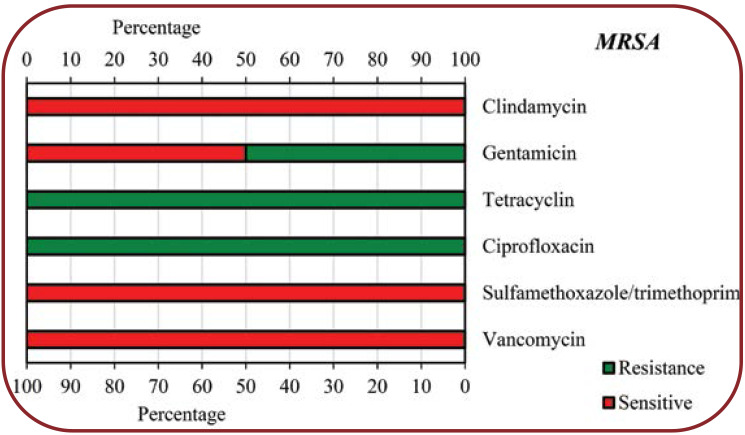
The present study showed that COVID-19 patients with secondary bacterial infections were highly resistant to some common antibiotics for isolated bacteria. According to our findings, Klebsiella species showed the highest resistance to Sulfamethoxazole/trimethoprim (75%), Cefazolin (65%), Piperacillin (63%), Gentamicin (56%) and Ceftizoxim (55%), respectively, while other antibiotics showed less resistance (<50%) to this bacterium. Regarding Escherichia coli, the highest resistance was seen towards Ciprofloxazcin (65%), Cefazolin (54%), Ceftazidime (54%) and Ceftizoxim (54%), while for other antibiotics, the resistance level was less than 50%. Enterobacter species had a high resistance (60%) only to Cefazolin, 10-40% resistance to other antibiotics and no resistance to Imipenem. Pseudomonas aeruginosa showed 75% resistance to Gentamicin and Tobramycin, 50% to Cefepime, Ciprofloxazcin and Piperacillin, 25% to Ceftizoxim and Ceftazidime, but no resistance to Imipenem. MSSA showed a high resistance to Tetracyclin (60%) and Ciprofloxacin (55%), moderate resistance to other antibiotics (30-45%) and no resistance to Vancomycin, while MRSA showed the highest resistance (100%) to Ciprofloxacin and Tetracyclin, 50% resistance to Gentamicin and no resistance to Vancomycin, Sulfamethoxazole/ Trimethoprim, and Clindamycin.
The study of Mahmoudi (2020) found that enterobacteriaceae isolates from COVID-19 patients had the highest resistance to Cotrimoxazole (74%), Piperacillin (67.5%), Ceftazidime (47.5%) and Cefepime (42.5%). In addition, all isolates were susceptible to Amikacin (100%). S. aureus isolates were susceptible to Vancomycin (100%) and the rates of resistance to Oxacillin, Erythromycin and Clindamycin were higher than 90%. P. aeruginosa was susceptible (90%) to Imipenem (20).
Our results together with those reported by Mahmoudi (20) show that the excessive use of antibiotics without following medical instructions as well as the use of various antiseptics, disinfectants and detergents can increase not only antibiotic resistance for different bacteria, but also the health-related risks of COVID-19 patients, the length of hospital stay as well as hospital staff’s workload.
Study limitations
One of the main limitations of the present study was the low number of samples taken for the culture and identification of bacteria. Other limitations include the fact that the study was conducted in a single hospital setting and did not review previous records of bacterial infection.
CONCLUSION
The results of the present study show that hospitalization for any reason, especially COVID-19, can increase the risk of secondary bacterial infections related to health care as well as that of transmission of multidrug-resistant diseases. Also, our findings enable us to conclude that excessive use of antibiotics is an important factor in the development of antimicrobial resistance. Therefore, caution is needed when prescribing antibiotics to COVID-19 patients. In addition, considering the co-infection of SARS-CoV-2 and other pathogens requires the use of an optimal treatment approach.
Conflicts of interests: none declared.
Financial support: none declared.
TABLE 1.
The frequency of bacterial species isolated from positive COVID-19 patients
FIGURE 1.
Antimicrobial susceptibility patterns of Klebsiella species isolated from COVID-19 patients
FIGURE 2.
Antimicrobial susceptibility patterns of Escherichia coli isolated from COVID-19 patients
FIGURE 3.
Antimicrobial susceptibility patterns of Enterobacter species isolated from COVID-19 patients
FIGURE 4.
Antimicrobial susceptibility patterns of Enterobacter species isolated from COVID-19 patients
FIGURE 5.
Antimicrobial susceptibility patterns of Methicillin-sensitive Staphylococcus aureus (MSSA) isolated from COVID-19 patients
FIGURE 6.
Antimicrobial susceptibility patterns of Methicillin-resistant Staphylococcus aureus (MRSA) isolated from COVID-19 patients
Contributor Information
Alpesh M MARUA, Department of Pathology, Dr. N. D. Desai Faculty of Medical Science and Research, Nadiad, India.
Nimisha D. SHETHWALA, Department of Microbiology, GMERS Medical College, Himmatnagar, India
Parth BHATT, Department of Pathology, Dr. N. D. Desai Faculty of Medical Science and Research, Dharmsinh Desai University, Nadiad, India.
Amar SHAH, Department of Pathology, Dr. N. D. Desai Faculty of Medical Science and Research, Dharmsinh Desai University, Nadiad, India.
References
- 1.Di Mascio D, Khalil A, Saccone G, et al. Outcome of coronavirus spectrum infections (SARS, MERS, COVID-19) during pregnancy: a systematic review and meta-analysis. Am J Obstet Gynecol. 2020;2:100107. doi: 10.1016/j.ajogmf.2020.100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen H, Guo J, Wang C, et al. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: a retrospective review of medical records. Lancet. 2020;395:809–815. doi: 10.1016/S0140-6736(20)30360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwartz DA. An analysis of 38 pregnant women with COVID-19, their newborn infants, and maternal-fetal transmission of SARS-CoV-2: maternal coronavirus infections and pregnancy outcomes. Arch Pathol Lab Med. 2020;144:799–805. doi: 10.5858/arpa.2020-0901-SA. [DOI] [PubMed] [Google Scholar]
- 4.Zu ZY, Jiang MD, Xu PP, et al. Coronavirus disease 2019 (COVID-19): a perspective from China. Radiology. 2020;296:E15–E25. doi: 10.1148/radiol.2020200490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Möhlenkamp S, Thiele H. Ventilation of COVID-19 patients in intensive care units. Herz. 2020;45:329–331. doi: 10.1007/s00059-020-04923-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clancy CJ, Nguyen MH. COVID-19, superinfections and antimicrobial development: What can we expect? Clin Infect Dis 2020. [DOI] [PMC free article] [PubMed]
- 7.Morens DM, Taubenberger JK, Fauci AS. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis. 2008;198:962. doi: 10.1086/591708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esper FP, Spahlinger T, Zhou L. Rate and influence of respiratory virus co-infection on pandemic (H1N1) influenza disease. J Infect. 2011;63:260–266. doi: 10.1016/j.jinf.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langford BJ, So M, Raybardhan S, et al. Bacterial co-infection and secondary infection in patients with COVID-19: a living rapid review and meta-analysis. Clin Microbiol Infect. 2020;26:1622–1629. doi: 10.1016/j.cmi.2020.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adeiza SS, Shuaibu AB, Shuaibu GM. Random effects meta-analysis of COVID-19/S. aureus partnership in co-infection. GMS Hyg Infect Control. 2020;15 doi: 10.3205/dgkh000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huttner BD, Catho G, Pano-Pardo JR, et al. COVID-19: don't neglect antimicrobial stewardship principles! Clin Microbiol Infect. 2020;26:808–810. doi: 10.1016/j.cmi.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox MJ, Loman N, Bogaert D, et al. Co-infections: potentially lethal and unexplored in COVID-19. The Lancet Microbe. 2020;1:e11. doi: 10.1016/S2666-5247(20)30009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alhazzani W, Møller MH, Arabi YM, et al. Surviving Sepsis Campaign: guidelines on the management of critically ill adults with Coronavirus Disease 2019 (COVID-19). Intensive Care Med. 2020;46:854–887. doi: 10.1007/s00134-020-06022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Velavan TP, Meyer CG. The COVID‐19 epidemic. Trop Med Int Health. 2020;25:278. doi: 10.1111/tmi.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agaba P, Tumukunde J, Tindimwebwa JV, et al. Nosocomial bacterial infections and their antimicrobial susceptibility patterns among patients in Ugandan intensive care units: a cross sectional study. BMC Res Notes. 2017;10:1–2. doi: 10.1186/s13104-017-2695-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dandagi GL. Nosocomial pneumonia in critically ill patients. Lung India. 2010;27:149. doi: 10.4103/0970-2113.68321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall GS. Bailey & Scott's Diagnostic Microbiology, 13th edition. Lab Med. 2013;44:e138–e139. [Google Scholar]
- 18.Sharifipour E, Shams S, Esmkhani M, et al. Evaluation of bacterial co-infections of the respiratory tract in COVID-19 patients admitted to ICU. BMC Infect Dis. 2020;20:1–7. doi: 10.1186/s12879-020-05374-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahmoudi H. Bacterial co-infections and antibiotic resistance in patients with COVID-19. GMS Hyg Infect Control. 2020;15:1–6. doi: 10.3205/dgkh000370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharifipour E, Shams S, Esmkhani M, et al. Evaluation of bacterial co-infections of the respiratory tract in COVID-19 patients admitted to ICU. BMC Infect Dis. 2020;20:1–7. doi: 10.1186/s12879-020-05374-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singhal T. A review of coronavirus disease-2019 (COVID-19). Indian J Pediatr. 2020;87:281–286. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, Geng M, Peng Y, et al. Molecular immune pathogenesis and diagnosis of COVID-19. J Pharm Anal. 2020;10:102–108. doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lansbury L, Lim B, Baskaran V, et al. Co-infections in people with COVID-19: a systematic review and meta-analysis. Int J Infect. 2020;81:266–275. doi: 10.1016/j.jinf.2020.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rifat M, Milton AH, Hall J, et al. Development of multidrug resistant tuberculosis in Bangladesh: a case-control study on risk factors. PloS One. 2014;9:e105214. doi: 10.1371/journal.pone.0105214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arvelo W, Hinkle CJ, Nguyen TA, et al. Transmission risk factors and treatment of pediatric shigellosis during a large daycare center-associated outbreak of multidrug resistant Shigella sonnei: implications for the management of shigellosis outbreaks among children. J Pediatr Infect Dis. 2009;28:976–980. doi: 10.1097/INF.0b013e3181a76eab. [DOI] [PubMed] [Google Scholar]
- 27.Getahun H, Smith I, Trivedi K, et al. Tackling antimicrobial resistance in the COVID-19 pandemic. Bull World Health Organ. 2020;98:442. doi: 10.2471/BLT.20.268573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X, Yu Y, Xu J, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lescure FX, Bouadma L, Nguyen D, et al. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect Dis. 2020;20:697–706. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xing Q, Li GJ, Xing YH, et al. Precautions are needed for COVID-19 patients with coinfection of common respiratory pathogens. Lancet 2020.
- 33.Toombs JM, Van den Abbeele K, Democratis J, et al. Pneumococcal coinfection in COVID-19 patients. J Med Virol. 2020;93:177–179. doi: 10.1002/jmv.26278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murray AK. The novel coronavirus COVID-19 outbreak: global implications for antimicrobial resistance. Front Microbiol. 2020;11:1020. doi: 10.3389/fmicb.2020.01020. [DOI] [PMC free article] [PubMed] [Google Scholar]