Abstract
In this study, we elucidate a single cell resolution fate map in the zebrafish in a sub-section of the anterior Lateral Plate Mesoderm (aLPM) at 18 hpf. Our results show that this tissue is not organized into segregated regions but gives rise to intermingled pericardial sac, peritoneum, pharyngeal arch and cardiac precursors. We further report upon asymmetrical contributions of lateral aLPM-derived heart precursors—specifically that twice as many heart precursors arise from the right side versus the left side of the embryo. Cell tracking analyses and large-scale cell labeling of the lateral aLPM corroborate these differences and show that the observed asymmetries are dependent upon Tbx5a expression. Previously, it was shown that cardiac looping was affected in Tbx5a knock-down and knock-out zebrafish (Garrity et al., 2002; Parrie et al., 2013); our present data also implicate tbx5a function in cell specification, establishment and maintenance of cardiac left-right asymmetry.
Keywords: tbx5a, zebrafish, cardiac development, lateral plate mesoderm, left-right asymmetry, fate map
Introduction
Lineage tracing of individual blastomeres in zebrafish show that cardiac precursors within the anterior lateral plate mesoderm (aLPM) can migrate toward the midline to form the primary heart tube (PHT) (Stainier et al., 1993). The cardiac precursors that give rise to the PHT are called the First Heart Field (FHF) and cardiac precursors that later add onto the PHT through the venous and arterial poles are called the Second Heart Field (SHF). While the medially migrating aLPM contributes to the FHF, cells of the lateral aLPM migrate anteriorly to give rise to several fates including parts of the SHF, the pericardial sac, pharyngeal arches, and the peritoneum. The migration and specification of the SHF is essential for the development of arterial pole heart structures such as the bulbous arteriosus and the outflow tract (de Pater et al., 2009; Hami et al., 2011; Lazic and Scott, 2011). We define the anteriorly migrating region of the aLPM to include those cells at 18 hours post fertilization (hpf) that are located lateral to the paraxial and intermediate mesoderm, posterior to the vascular precursors, and anterior to the boundary of the pectoral fin precursor field at somite 1 (Mao et al., 2015). In zebrafish, the aLPM (and pectoral fin field) expresses tbx5a and hand2 at 10 hpf (Ahn et al., 2002; Yelon et al., 2000). Previous fate maps of the aLPM focused on the cardiac precursors and were constructed in midblastula-stage, 40% epiboly and 11 hpf embryos and followed cells to 26 hpf and 44 hpf (Hami et al., 2011; Keegan et al., 2004; Schoenebeck et al., 2007; Stainier et al., 1993). A general fate map of the lateral anteriorly-migrating aLPM was lacking until recently in this study. Additionally, little is known about the migration dynamics of these precursor cells.
The establishment of the left-right axis arises within the Lateral Plate Mesoderm (LPM) by the Nodal-Lefty2-Pitx2 pathway, which results in asymmetric morphogenic events such as cardiac jogging and looping (Long et al., 2003). The addition of the aLPM-derived SHF into the venous and arterial poles of the PHT may be responsible for the asymmetric morphology of the heart, as both SHF addition and morphogenesis occur between 24 and 72 hpf and knockdown of several incompletely-overlapping SHF markers Mef2cb, Ltbp3, Isl2b and Nkx2.5 results in heart looping defects (George et al., 2015; Lazic and Scott, 2011; Witzel et al., 2017; Zhou et al., 2011).
The T-box transcription factor Tbx5 is critically involved in LPM development. Human patients with Holt-Oram Syndrome (HOS, a dominant haploinsufficiency mutation in TBX5) display a range of cardiac septal defects and forelimb abnormalities, which display left-right asymmetries (Basson et al., 1994; Basson et al., 1997; Holt and Oram, 1960; Li et al., 1997). Tbx5 is required in vertebrates after establishment of the heart and forelimb buds for the maintenance of cardiomyocyte fate, septation of the amniote heart, establishment of the conduction system (Moskowitz et al., 2004; Moskowitz et al., 2007), and maintenance of left-right laterality (Sulaiman et al., 2016). Expression of tbx5a is used as an early marker within the LPM to define the anlage of cardiac and pectoral fin precursors in the zebrafish (Ahn et al., 2002). Homozygous zebrafish tbx5a/heartstrings (hst) mutants neither develop pectoral fin buds nor undergo cardiac looping. These mutants subsequently display a gradual slowing of heartbeat and severe pericardial edema, resulting in the eventual deterioration of the heart and death of the zebrafish larva by 6 days post fertilization (dpf) (Garrity et al., 2002). As a transcription factor, Tbx5 directly binds DNA in combination with other cardiomyocyte-specific transcription factors to activate downstream gene activity (Ghosh et al., 2009; Maitra et al., 2009) and to specify cardiomyocyte fate (Small and Krieg, 2003; Takeuchi and Bruneau, 2009). Tbx5a function is required for development of epicardial (Yue et al., 2015) and pectoral fin precursors in zebrafish through directed migration (Mao et al., 2015; Wyngaarden et al., 2010). How Tbx5 mechanistically functions to coordinate fate specification, cellular migration, and laterality of aLPM tissues remains an important unanswered question.
In this study, we describe single-cell resolution fate maps of both wild-type and Tbx5a-deficient aLPM at 18 hpf, a time when cell migration away from the posterior LPM has yet to begin. Analysis of the 18 hpf wild-type fate map shows that the aLPM at this stage is not organized into regionalized, exclusive organ precursor fields, suggesting that final tissue level fates may not be dependent on precursor location at this stage. Of interest, cardiac precursors within the lateral aLPM are asymmetrically distributed across the left-right axis. Knockdown of Tbx5a leads to a disappearance of the observed asymmetries. Our data show that tbx5a has an important role in controlling inherent asymmetries of cardiac precursors with respect to the number of contributing cells, additions to the larval heart, and cellular migration dynamics.
Results
The lateral aLPM exhibits extensive overlap between organ-specific fate map regions
We previously reported a single-cell resolution fate map of the LPM adjacent to somites 1 through 6 (Mao et al., 2015). In that study, the fate mapped region was shown to contribute mainly to the pectoral fin bud. Two other fates were observed to arise in this fate map area: cardiac and peritoneum. In this paper, we sought to assess the fates of the lateral aLPM tissue using similar methods. We injected single blastomeres in 64- to 128- cell stage embryos with Kaede mRNA to create random mosaic expression. We then photoconverted the Kaede from green to red fluorescence in individual aLPM cells that were positioned lateral to the intermediate mesoderm and anterior to somite 1 at 18 hpf. Using the otic vesicles as relational landmarks at all stages of development, we constructed a fate map consisting of 166 labeled single cells and extending posteriorly towards somite 3 to delineate the area that does not give rise to fin mesenchyme (Fig. 1I, Supplemental Table 1). We classified labelled cellular clones into six identity groups according to the location and morphology of the photoconverted cells at 24 and 48 hpf: (1) cardiac (Fig. 1A, E, n=39), cells which gave rise to myocardial and sometimes endocardial tissue (Fig. 2) but did not contribute to the PHT (Fig. 3); (2) pericardial sac (Fig. 1B, F, n=30), cells which gave rise to either the caudal- or rostral-most tissue surrounding the heart; (3) pharyngeal arch (Fig. 1C, G, n=50), cells which gave rise to the mesenchymal cores of the pharyngeal arches; (4) peritoneum (Fig. 1D, H, n=32), cells which gave rise to the tissue laterally positioned over the yolk sac or surrounding the intestinal track; (5) pectoral fin (Fig. 1I green points, n=17), cells which gave rise to the mesenchyme of the pectoral fin bud; (6) other (Fig. 1I black points, n=18), cells which consisted of non-aLPM (somitic or intermediate mesoderm, midline, and eye) clones. The lateral aLPM at 18 hpf therefore gives rise to at least four mesodermal fates: cardiomyocytes; pericardial sac; pharyngeal mesenchyme; and peritoneum.
Figure 1-. Single-cell resolution 18hpf fate map of WT and Tbx5a-deficient aLPM.
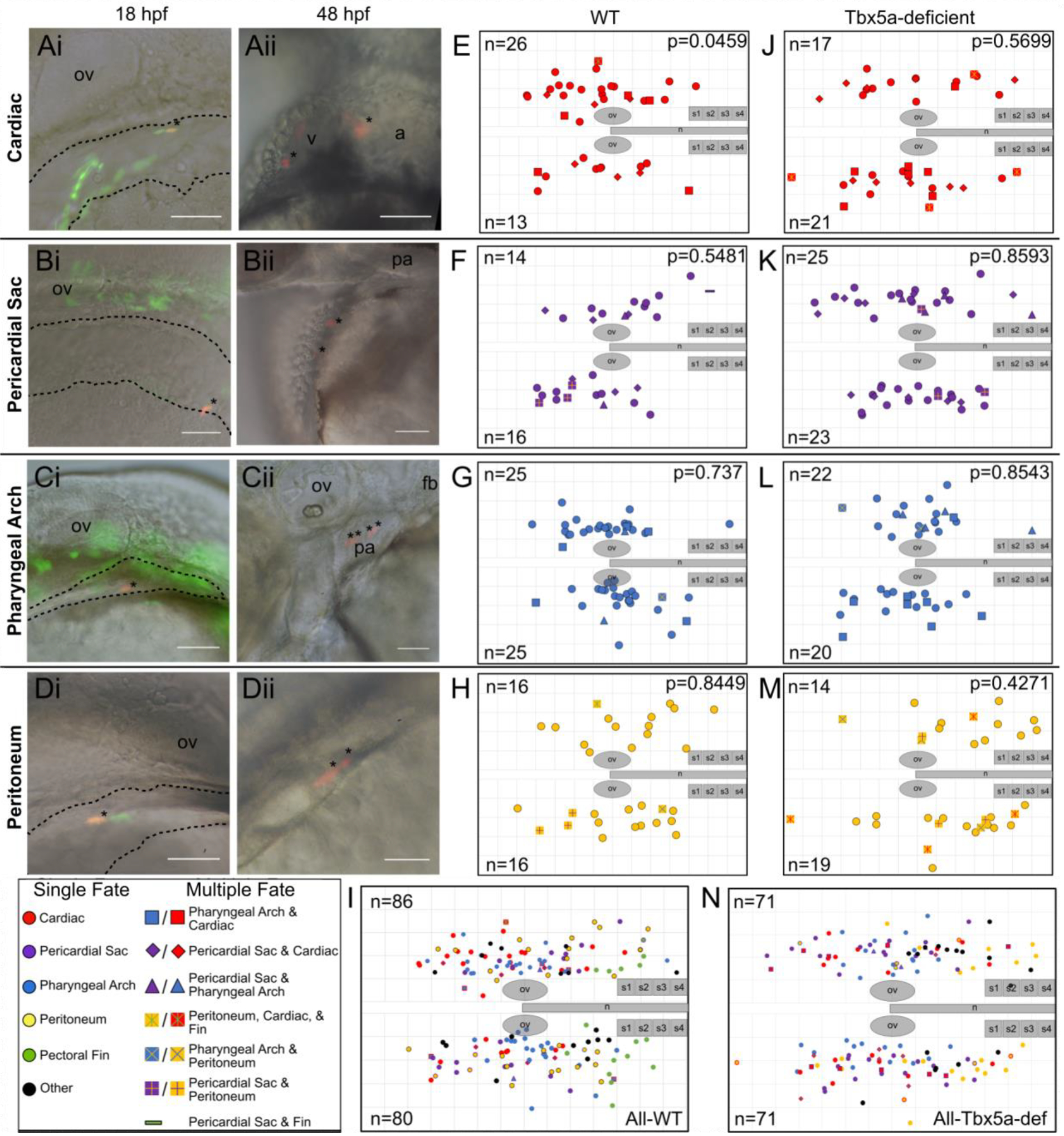
A-D Examples of labeled wild-type single cells within the aLPM, dashed outline. Asterisk marks a single cell photoconverted to express red fluorescent Kaede, whose starting position was measured as the anteroposterior and mediolateral distance from the otic vesicle. Anterior to the left, dorsal to the top; some images taken on the right side of the embryo have been flipped. Ai- Dorso-lateral view of the 18 hpf embryo. Aii- Ventral view of the same embryo at 48 hpf. Asterisks mark resultant clone in the heart. Bi, Bii- Dorso-lateral view of pericardial sac clone. Ci, Cii- Pharyngeal arch clone. Di, Dii- Peritoneum clone.
E-N Combined dorsal-lateral view fate map of aLPM cell fates at 18 hpf from both left and right sides (E-I wild-type, J-N Tbx5a-deficient), showing the four fate categories: E, J-cardiac; F, K-pericardial sac; G, L-pharyngeal arch; and H, M-peritoneum. I, N- Summary fate map of all labeled single cells. Circles denote unipotent clones, other shapes denote multipotent clones, refer to legends for identity. n counts represent the number of precursors from the right or left aLPM that gave rise to that particular fate. p-values determined by comparing the number of left and right precursors using the Fisher’s Exact test. v, ventricle; a, atrium; fb, fin bud; ov, otic vesicle; pa, pharyngeal arches; n, notochord; s1, somite 1. Grid length and scale bar: 100 μm.
Figure 2-. Cardiac precursors within the lateral aLPM give rise to myocardial and infrequent endocardial cells.
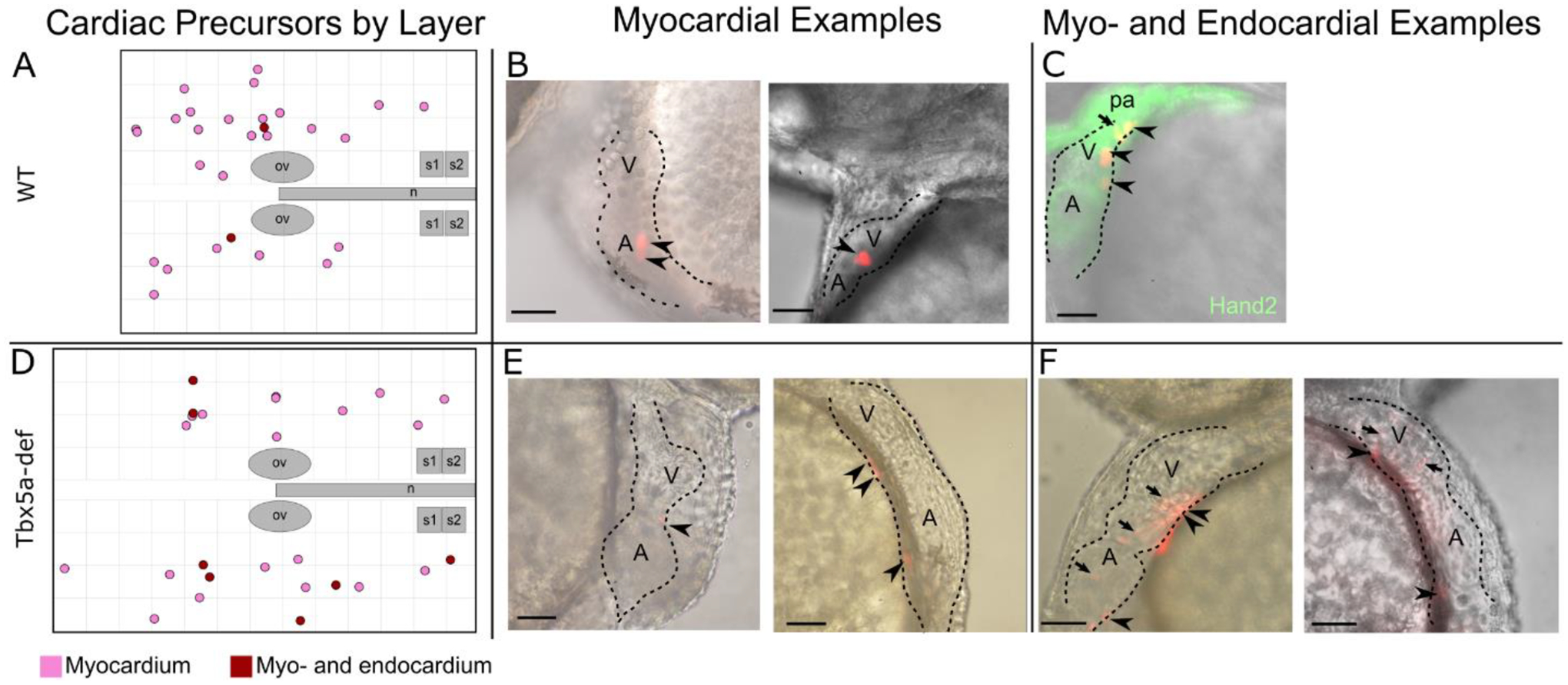
A- Twenty-seven (27) photoconverted wildtype clones gave rise to only myocardium (pink), while two (2) gave rise to myocardium and endocardium (burgundy). B, C, E, F- Examples of larval hearts at 48 hpf, outlined with dashed lines, lateral view. B- WT cardiac precursors that gave rise only to myocardium cells, which display a cuboidal morphology (arrowheads). C- WT cardiac precursor that gave rise to myocardium and as well as endocardium cells, which in general are smaller and more elongated in morphology than myocardial cells (arrow). Hand2 expression marks the larval heart and pharyngeal arches at 48 hpf. D- Nineteen (19) photoconverted Tbx5a-deficient clones gave rise to only myocardium (E arrowheads), while seven (7) gave rise to myocardium and endocardium (F arrows). ov- otic vesicle; n- notochord; s1- somite 1; A- atrium; V- Ventricle, pa- pharyngeal arch. Scale bar 100 μm.
Figure 3-. Photoconverted lateral aLPM cardiac precursor clones do not contribute to the primary heart tube at 24 hpf.
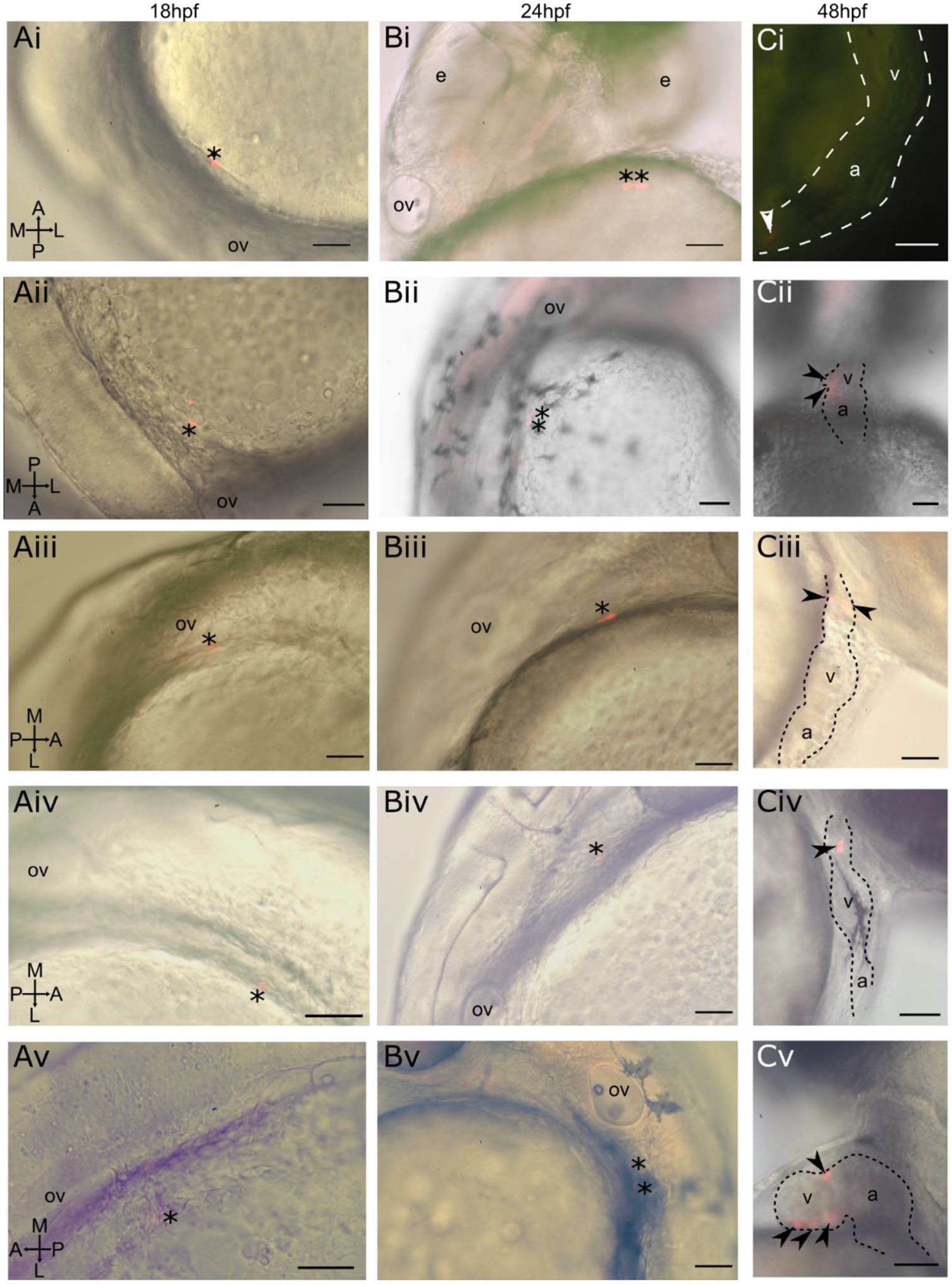
i-v Displaying five of 39 examples of wildtype embryos. A- Dorso-lateral view of 18 hpf embryo. Asterisk marks a single cell photoconverted to express red fluorescent Kaede. B- Lateral view of same embryo at 24 hpf. Asterisk marks photoconverted cardiac precursor still within the lateral ALPM and outside of the primary heart tube, currently out of view on the left side of the embryo. C- Lateral view of same embryo at 48 hpf. Arrowheads mark photoconverted cardiac precursors within the larval heart. Ci- Located in in-flow tract; Cii- located in ventricle and atrioventrical canal; Ciii- located in outflow tract; Civ- located in outflow tract; Cv- located in ventricle. a- atrium; v- ventricle; ov- otic vesicle, e- eye. Scale bar 100 μm.
The total area fate mapped at 18 hpf ranges from 500 μm anterior to the otic vesicle, at the level of the posterior midbrain, to 600 μm posterior to the otic vesicle, adjacent to somite 3. In the mediolateral direction, the aLPM extends from the medial edge of the otic vesicle to about 300 μm lateral of the otic vesicle, a region which was determined by the morphological edge of the aLPM as observed by Nomarski optics (Fig. 1Ai–Di, dashed outline). Most labeled precursors in wildtype embryos gave rise to a single fate, though a small number of labeled precursors gave rise to two fates (Fig. 1I non-circle points, Table 1, Rows 6–15), and one precursor gave rise to three fates. In addition to giving rise to cardiac fate, six precursors also gave rise to pericardial sac fate (Fig. 1E, F diamonds; Table 1 Row 6), five precursors also gave rise to pharyngeal arch fate (Fig. 1E, G squares; Table 1 Row 7), and one precursor also gave rise to peritoneum and pectoral fin fate (Fig. 1E, H red/yellow cross in square; Table 1 Row 16). In addition to giving rise to pericardial sac fate, two precursors also gave rise to pharyngeal arch fate (Fig. 1F, G triangles; Table 1 Row 10), three precursors also gave rise to peritoneum fate (Fig. 1F, H plus in square, Table 1 Row 11), and one precursor also gave rise to pectoral fin fate (Fig. 1F dash; Table 1 Row 12). In addition to giving rise to pharyngeal arch fate, one precursor gave rise to peritoneum fate (Fig. 1G, H x in square; Table 1 Row 13).
Table 1-. ALPM WT clone counts by fate.
Clones designated as ‘other’ are not included.
| Fate at 48hpf | Number of Clones | |||||
|---|---|---|---|---|---|---|
| Clone Fate | Heart | Pericardial Sac | Pharyngeal Arch | Peritoneum | Fin Mesenchyme | |
| Single | x | 27a | ||||
| x | 18 | |||||
| x | 42b | |||||
| x | 27a | |||||
| x | 15 | |||||
| Double | x | x | 6 | |||
| x | x | 5 | ||||
| x | x | 0 | ||||
| x | x | 0 | ||||
| x | x | 2 | ||||
| x | x | 3 | ||||
| x | x | 1 | ||||
| x | x | 1 | ||||
| x | x | 0 | ||||
| x | x | 0 | ||||
| Triple | x | x | x | 1 | ||
| Total Clone # | 39 | 30 | 50 | 32 | 16 | 148 |
| % of all | 26% | 20% | 34% | 22% | 11% | |
| # of multipotent | 12 | 12 | 9 | 5 | 2 | |
| % of total | 8% | 8% | 6% | 3% | 1% | 13% |
| % of fate | 31% | 40% | 18% | 16% | 13% | |
1 also made vasculature.
1 also made blood.
Fates within the 18 hpf lateral aLPM occupy overlapping regions that cannot be distinguished by their center of masses nor by their confidence ellipses (data not shown), except for the pectoral fin-forming region of the fate map, which lies adjacent to somites 1 through 4 (Mao et al., 2015). Despite no statistically significant regionality within any of the fates, slight trends exist along the anteroposterior and mediolateral axes (Fig. 1I). Cardiac precursors arise from positions slightly more anterior than posterior to the otic vesicles (Fig. 1E, J, I, and 4A). However, within the four lateral aLPM organ fields at 48 hpf, in contrast to previous findings for the pectoral fin-forming region (Mao et al., 2015), we found no topological conservation of original antero-posterior (AP) or medio-lateral (ML) position in the aLPM to final position within the separate organ fields. For example, pharyngeal arch precursors that were labeled more anteriorly within the lateral aLPM did not consistently give rise to the most anterior (mandibular or hyoid) arches but instead formed clones that were equally distributed amongst the mesenchymal cores of pharyngeal arches 1–6 (Fig. 1Cii).
Figure 4-. Cardiac precursor asymmetries are affected in Tbx5a-deficiency.
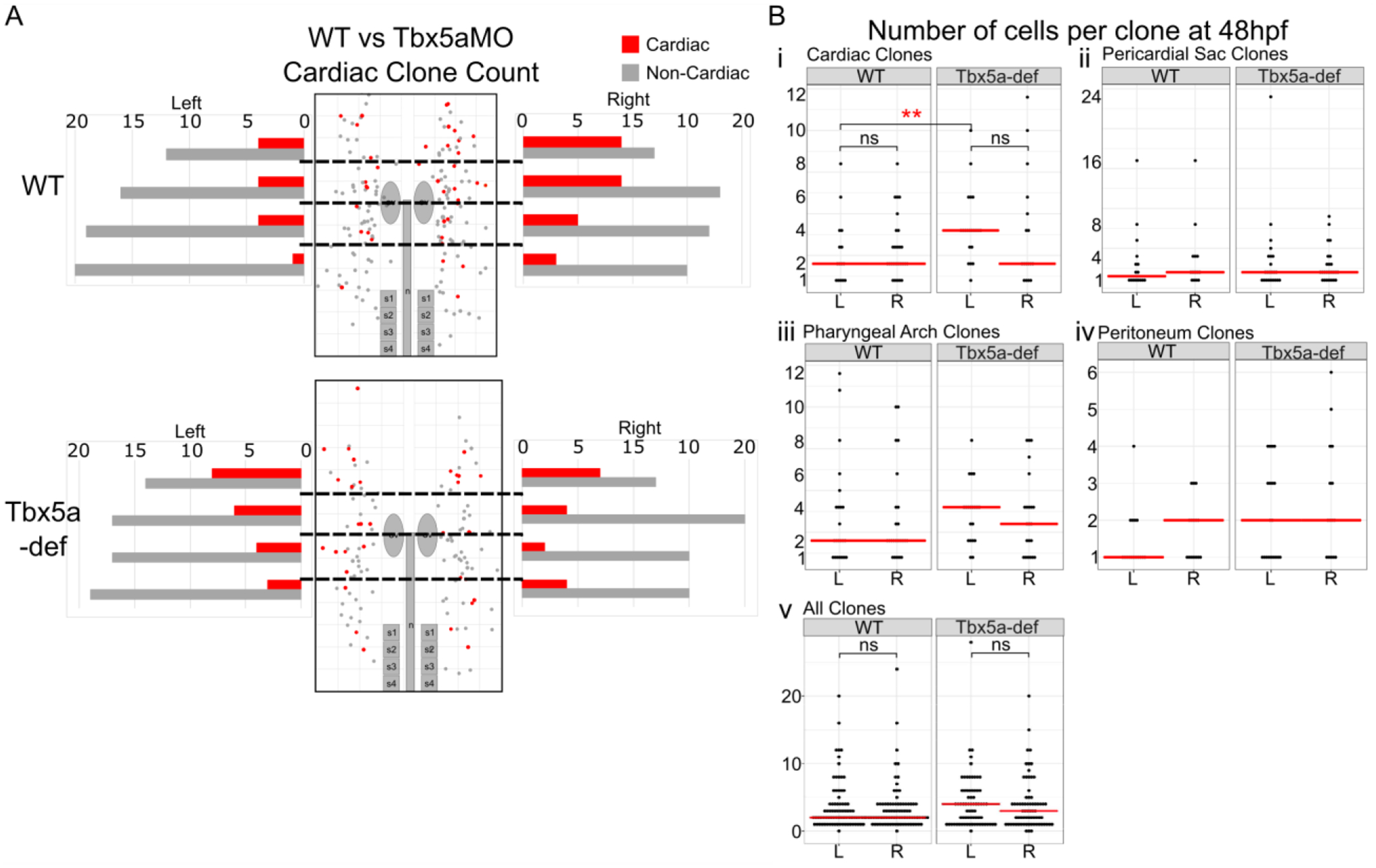
A- Graphical representation of precursors deriving heart versus non-heart fates arising from AP regions of equal bin size of 20 precursors per side. B- Number of cells per clone at 48 hpf reflects the amount of proliferation of aLPM between 18 hpf and 48 hpf. The red bar represents the median. Statistics performed using the Kruskal-Wallis rank sum test.
Left-right asymmetries between cardiac precursors
Within the wildtype aLPM fate map data, we found that twice as many cardiac precursors were distributed on the right side of the embryo than from the left (Fig. 1E, p=0.0459). Importantly, no such asymmetry was observed within any other fates. Furthermore, when the fate map is divided into anteroposterior bins of equal total precursor size, we observed that the anterior bins display a larger left-right cardiac precursor asymmetry than the posterior bins (Fig. 4). To understand better how this right-biased asymmetry contributes to the development of the larval heart, we first assessed how the cardiac precursors proliferated between 18 and 48 hpf. Within individual cardiac clones, there is no significant difference in the average clone size between the left versus right side (Fig. 4B p=0.2806).
Since we observe more cardiac precursors in the right lateral aLPM than left at 18 hpf, we next questioned whether the right lateral aLPM made up a larger proportion of the larval heart than the left lateral aLPM. We photoconverted the left or right side of the lateral aLPM by applying UV-light to a region of interest in 18 hpf embryos that had been injected with Kaede transcripts at the 1-cell stage (Fig. 5A). Overall, the photoconverted (KaedePC-expressing) red cells gave rise to all previously described tissues from our single-cell resolution fate map, which included multiple layers of both heart chambers and the inflow and outflow tracts (Fig. 5B, D-G). Photoconverted lateral aLPM from either the right or left gave rise to equal number of KaedePC-expressing red cells in the pericardial sac, pharyngeal mesenchyme, peritoneum and pectoral fin (data not shown). Additionally, photoconverted cells that gave rise to these tissues retained their laterality between 18 and 48 hpf; i.e., photoconverted cells that originated from the right lateral aLPM were found on the right side of the 48 hpf embryo. However, contributions of photoconverted cells into the heart were not segregated into left versus right sides with respect to the body axis. Instead, heart development proceeds such that medial ALPM precursors undergo a clockwise rotation that positions the left side anteriorly and the right side posteriorly (Baker et al., 2008; Smith et al., 2008). This results in contributions to the heart that are segregated orthogonally/transverse to the body instead of in line with/sagittal to the body (Fig. 5C blue versus orange regions). Recent evidence has shown that later contributions to the heart are also segregated along the AP axis between the right and left sides (Fukui et al., 2018). Our large scale photoconversion experiments also showed that the left side precursors were biased towards populating the anterior heart (Fig. 5C orange region, 5D a–b), whereas cardiomyocytes that originated from the right side aLPM were biased towards populating the posterior heart (Fig. 5C blue region, 5E a–b). Consistent with the left-right asymmetry found within the number of fate-mapped cardiac precursors, photoconverted lateral aLPM from the right side gave rise to more cardiomyocytes than lateral aLPM from the left side (Fig. 5B, D–E, effect size=1.14, p=0.0298, Fig. 6).
Figure 5-. Asymmetric aLPM contributions to the larval heart are affected by Tbx5a knockdown.
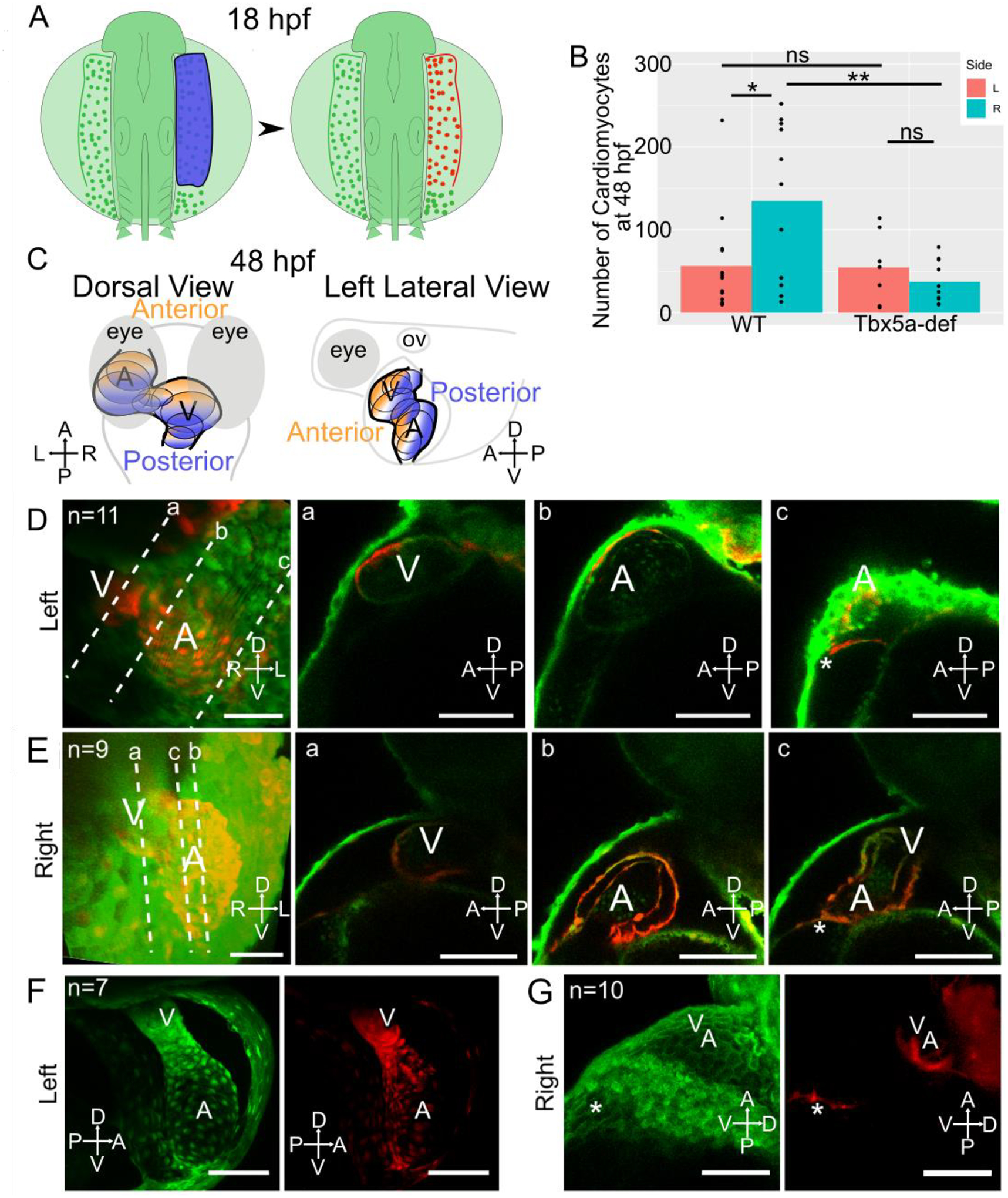
A- Schematic of experimental method with right side lateral aLPM photoconverted using 405 nm light (shown as blue) at 18 hpf. B- Bar plots of mean number of KaedePC cardiomyocytes at 48 hpf in each experimental condition. Points are counts from each embryo (Fig. S5). Statistics performed using the Wilcoxon Rank Sum test. C- Schematic at 48 hpf showing anterior (orange) versus posterior (blue) regions of the heart in dorsal and left lateral views.
D-G- 48 hpf 3D projection of a confocal stack of the larval heart shows the contributions of red fluorescent KaedePC cells among green fluorescent Kaedenon-PC cells, D-E-WT embryos, anterior view. Dotted lines a-c are levels of individual Z-slices through the a-ventricle, b-atrium, and c-connecting region between the venous pole and pericardial sac. D- Heart derived from left side aLPM. E- Heart derived from right side aLPM. F-G- Tbx5a-deficient embryos, F- Heart derived from left side aLPM, anterior view. G- Heart derived from the right side aLPM, left lateral view. A-atrium; V-ventricle; asterisk-venous pole of the heart. Compass abbreviations: A-anterior; P-posterior; D-dorsal; V-ventral; R-right; L-left. Scale bar: 100 μm.
Figure 6-. Photoconverted cardiomyocytes counted manually from a z-stack of the larval heart.
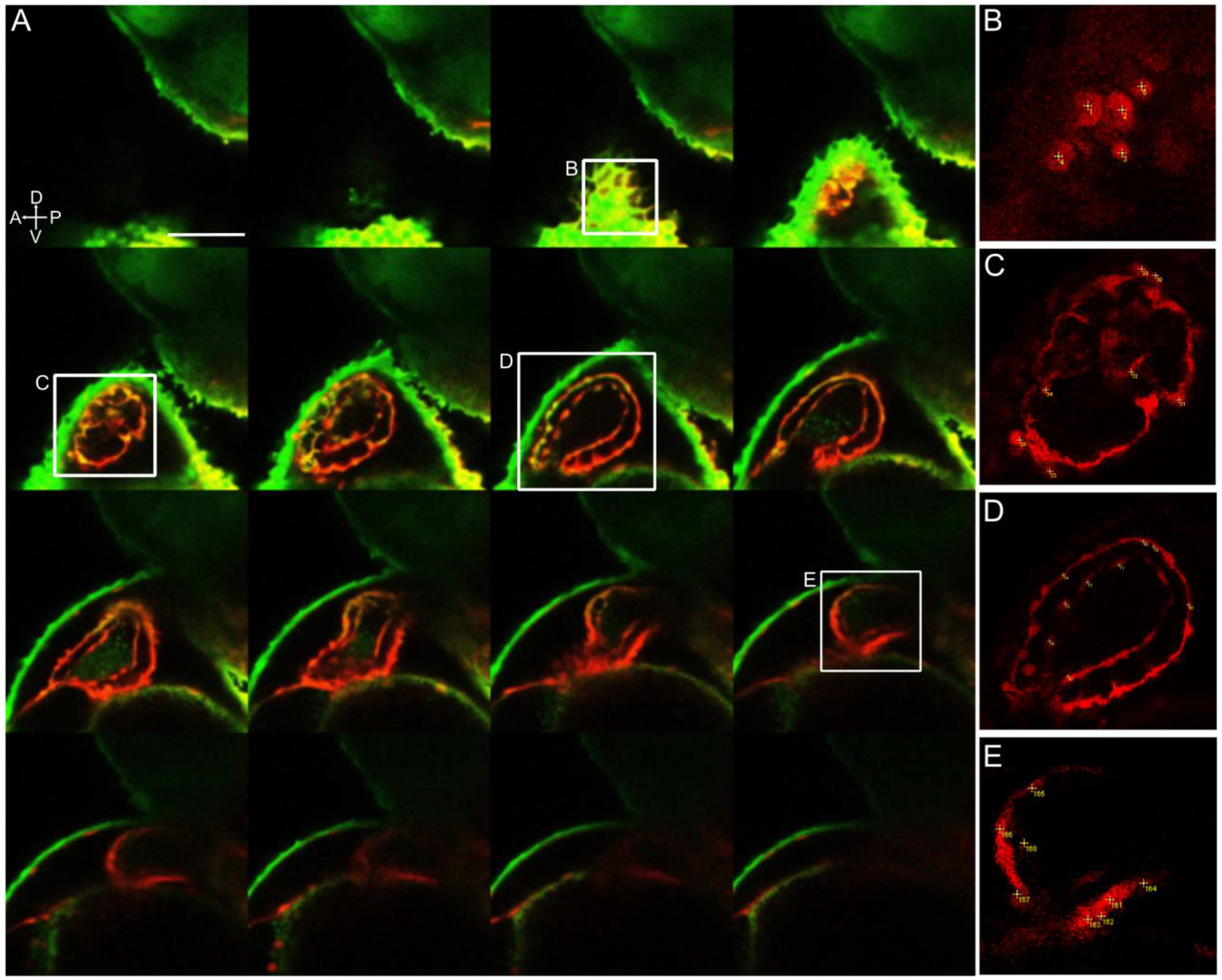
A- 48 hpf left lateral side 3D stack of the heart, displaying every fourth stack of 2.0 μm. Red fluorescence shows right side lateral aLPM-derived Kaede-expressing cells photoconverted at 18 hpf. B-E Insets of the heart showing examples of how cardiomyocytes were counted with numbered points in yellow. This embryo had a total of 185 red fluorescent cardiomyocytes counted, which was verified by the average of three separate manually counted analyses.
tbx5a maintains left-right asymmetry of cardiac precursors in the lateral aLPM
As tbx5a is expressed within the lateral aLPM at 18 hpf and functions in heart and forelimb development, we asked how tbx5a is involved in patterning the lateral aLPM. We constructed a fate map of the lateral aLPM in embryos which had Tbx5a knocked down by MO2-tbx5a morpholino injection, which has been shown to recapitulate cardiac and pectoral fin phenotypes of the hst mutant (Ahn et al., 2002; Garrity et al., 2002). From 142 labeled single cell precursors, we performed similar analyses to the wild-type aLPM fate map (Fig. 1J–N, Supplemental Table 2). As expected, no pectoral fin clones were observed in Tbx5a-deficient embryos, as injections of Tbx5a morpholinos have been shown to faithfully recapitulate the loss of pectoral fin bud formation displayed in the hst mutant. Similar to the wildtype, Tbx5a-deficient aLPM cardiac precursors also were not found in the PHT at 24 and 48 hpf (Fig. 3) and gave rise to multiple layers of all chambers (Fig. 2). In contrast, we found that the Tbx5a-deficient fate map consists of an equal distribution of cardiac precursors on the left and right sides (Fig. 1J). Compared to the right-biased distribution of cardiac precursors in wildtype, the Tbx5a-deficient aLPM does not display left-right asymmetry in the proportions of cardiac precursor numbers (Fig. 4A, p=0.4067). We questioned whether this observation correlates with how precursors from each side proliferated in Tbx5a-deficient embryos. Tbx5a-deficient cardiac precursors that arise from the left lateral aLPM have a median clone size that is twice as high as the wildtype (Fig. 4Bi, red asterisks p=0.006439), suggesting that a tbx5a-dependent mechanism may normally suppress cardiac cell proliferation on the left side. Together, these findings suggest that tbx5a normally functions to maintain differences in the left-right distribution and proliferation of cardiac precursors that contribute to the heart
We questioned whether the absence of asymmetry among cardiac precursor number in the Tbx5a-deficient embryos would affect the composition of the larval heart. We conducted photoconversions of the right or left side of the lateral aLPM (as detailed above for unmanipulated specimens) in Tbx5a-deficient embryos at 18 hpf (Fig. 5F–G). The number of KaedePC-positive red fluorescent cells in all tissue derivatives of the aLPM did not differ based upon their laterality of origin and they retained their laterality between 18 and 48 hpf. For example, photoconverted cardiomyocytes in Tbx5a-deficient embryos from the right and left side lateral aLPM contributed to the larval heart in equal numbers (Fig. 5F–G, B p=0.3302). This result shows that Tbx5a knockdown eliminates the normal left-right difference in gross-level contributions to the larval wildtype heart observed above. Within the Tbx5a-deficient heart, the left lateral aLPM gave rise to the anterior heart and the right lateral aLPM gave rise to the posterior heart, similar to the wildtype (Fig. 5C, F–G). Of note, lateral aLPM from the right side gave rise to fewer cardiomyocytes in the Tbx5a-deficiency than in the wildtype (Fig. 5G, B p=0.01103), while lateral aLPM from the left side gave rise to the same number of cardiomyocytes in the Tbx5a-deficiency compared to the wildtype (Fig. 5F, B p=0.5473). These results further indicate that tbx5a may function to maintain left-right differences in cardiac precursor contribution.
tbx5a functions during specification of lateral aLPM organ-specific fates
tbx5a could maintain the left-right differential cardiac contribution by several non-mutually exclusive mechanisms. First, tbx5a could regulate the specification of cardiac versus other lateral aLPM fates (e.g. pericardial sac, pharyngeal arch, etc.) differently between the right and left sides. Second, tbx5a could differentially affect the migration between the right and left sides into potential ‘organ pools’. To assess the effect of Tbx5a on cardiac cellular specification, we analyzed the degree of multipotency of the fate map precursors at 18 hpf. In the wildtype fate map, most photoconverted precursors among all fates gave rise to a single organ fate. However, a small proportion of wildtype precursors, 13%, gave rise to two fates (Table 1, Row 20). In the Tbx5a-deficient fate map, however, over half (55%) of Tbx5a-deficient cardiac precursors also gave rise to another fate, which was a significant increase (p=0.0194, Fig. 7, Table 2 Row 20). These multipotent precursors include cells that give rise to both pericardial sac and pharyngeal arch fates and an emergence of cardiac precursors that also give rise to peritoneum fates, compared to wildtype. Furthermore, the additional multipotent cardiac precursors in Tbx5a-deficient embryos versus wildtype (p=0.0415, Fig. 7B), arise from the left side lateral aLPM twice as often as from the right side lateral aLPM (Fig. 1J non-circle points). These findings support the hypothesis that tbx5a may regulate the specification of cardiac versus other lateral aLPM fates.
Figure 7-. tbx5a affects the number of multipotent precursors across all fates.
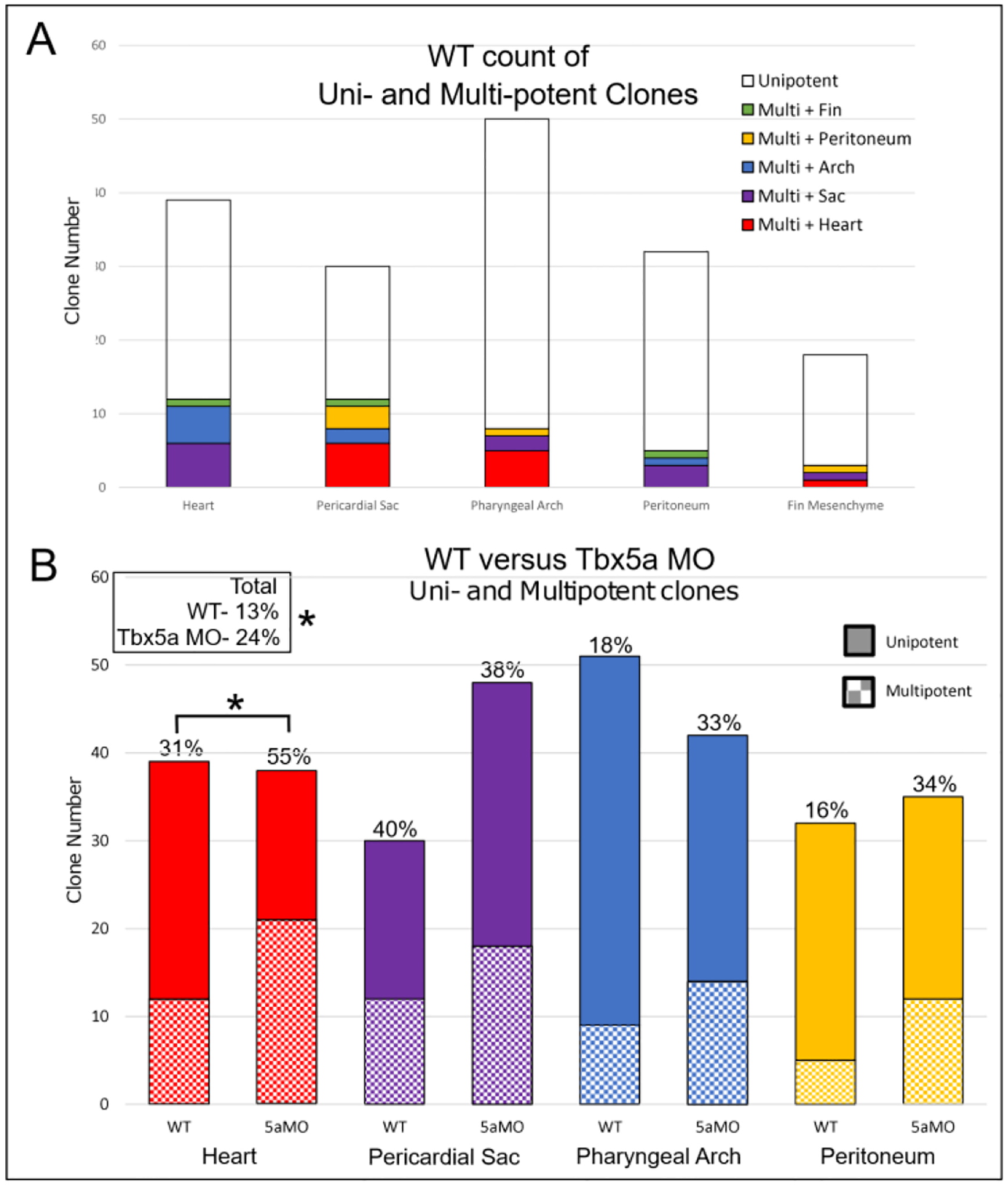
A- Stacked bar graphs of WT aLPM precursor count by fate, including unipotent and multipotent precursors. B-Comparison of WT versus Tbx5a-deficient degree of multipotency. Asterisks denote significant association between multipotency and Tbx5a deficiency of p < 0.05. There was no significant association found among pericardial sac, pharyngeal arch and peritoneum fates.
Table 2-.
ALPM Tbx5a-deficient clone counts by fate.
| Fate at 48hpf | Number of Clones | |||||
|---|---|---|---|---|---|---|
| Clone Fate | Heart | Pericardial Sac | Pharyngeal Arch | Peritoneum | Other | |
| Single | x | 17 | ||||
| x | 30 | |||||
| x | 28 | |||||
| x | 23 | |||||
| x | 10 | |||||
| Double | x | x | 10 | |||
| x | x | 7 | ||||
| x | x | 4 | ||||
| x | x | 0 | ||||
| x | x | 2 | ||||
| x | x | 3 | ||||
| x | x | 1 | ||||
| x | x | 3 | ||||
| x | x | 0 | ||||
| x | x | 2 | ||||
| Triple | x | x | x | 2 | ||
| Total Clone # | 142 | |||||
| % of all | ||||||
| # of multipotent | ||||||
| % of total | 24% | |||||
| % of fate | ||||||
Tbx5a knockdown affects lateral aLPM migration dynamics
We hypothesized that tbx5a could establish and maintain the asymmetric cardiac contribution from the lateral aLPM by differentially affecting the migration dynamics between the right and left sides. To address this issue, we analyzed time-lapse images of the entire aLPM in wildtype and Tbx5a-deficient embryos from 18–34 hpf using Et(hand2:eGFP)ch2; Tg(h2afx:h2afv-mCherry)mw3 fish, which allowed us to visualize mCherry-positive nuclei of individual GFP-expressing cells in the aLPM (Supplemental Movies 1, 2). We observe that GFP-expressing cells migrate over time to contribute to each of the four fates. We first tracked cells backward in time and categorized them based upon which organ field they resided in at 34 hpf and on which side they had arisen from (Fig. 8A, Supplemental Movie 3). We did not track peritoneum precursors as they were not within the field of view. Next, cell tracks between 18 and 24.5 hpf were chosen for analysis to capture individual cell movements into their respective organ fields, while avoiding the later large scale morphogenic movements associated with whole organ formation such as jogging and looping of the PHT, for example. We used the DiPer program to calculate indices of directional migration (Gorelik and Gautreau, 2014), summarized in Fig. 8B. The slope of the log-log plot of mean squared displacement (MSD) over time is the α-value, which represents the degree of ballistic migration (Fig. 9). An α-value of 1 represents random movement and an α-value of 2 represents perfect ballistic movement. The α-value relies on two factors—the speed and persistence, where the persistence is the ratio of net track displacement over total distance traveled, or track length (Fig. 8B).
Figure 8-. Migration dynamics of WT cardiac precursors versus Tbx5a-deficient embryos.
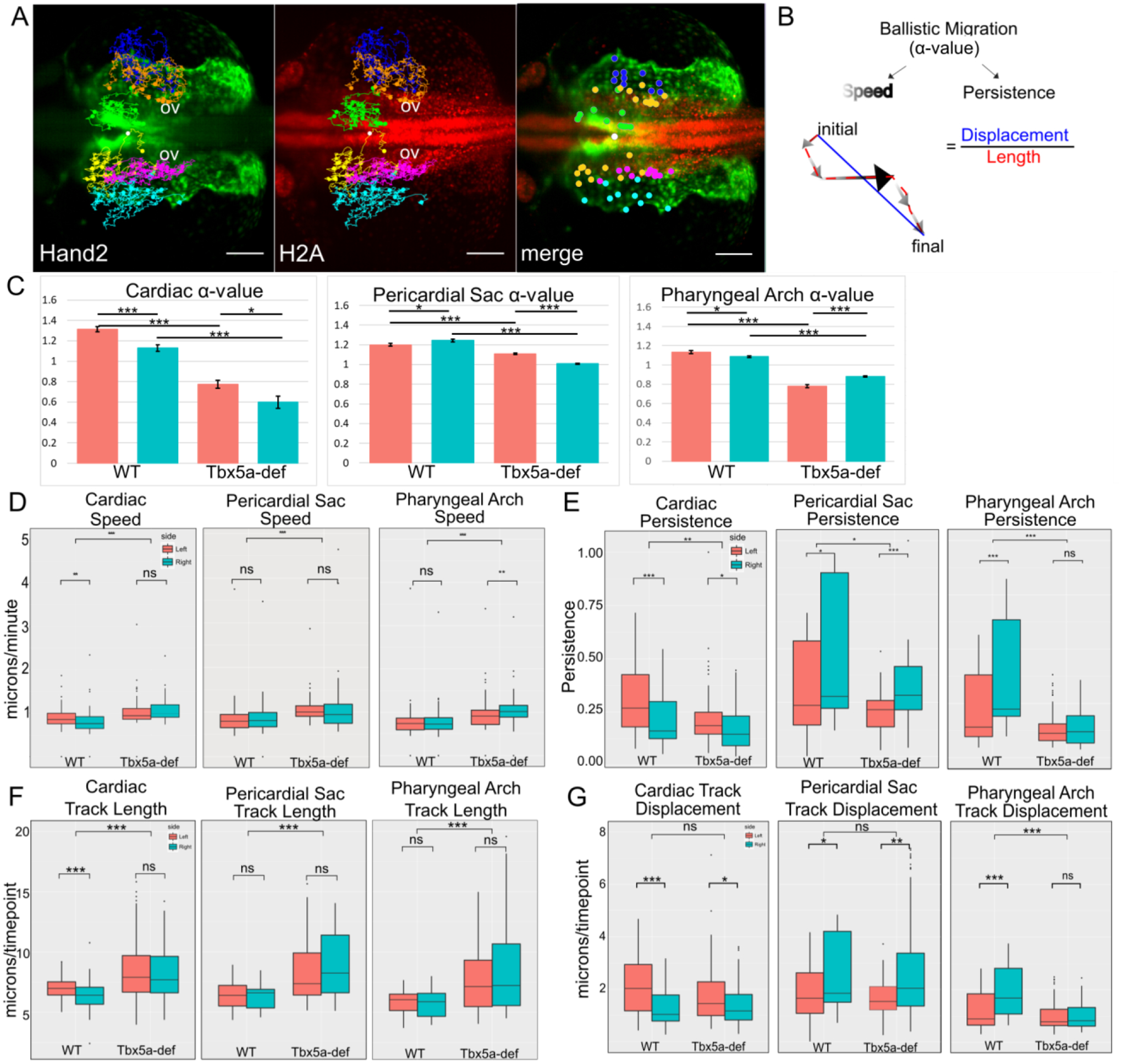
A- Cell tracks of Hand2-expressing aLPM, colored by their fate at 34 hpf and laterality side of origin (White-reference point for drift correction; Green-cardiac precursors from the right; Yellow-cardiac precursors from the left; Orange-pericardial sac precursors from the right; Magenta-pericardial sac precursors from the left; Blue-pharyngeal arch precursors from the right; Cyan-pharyngeal arch precursors from the left). Background maximum-intensity projection image 20x magnification of Hand2-eGFP; H2A-mCherry embryo at 18 hpf, when time-lapse begins (Supplemental Movie 3). Merge projection is overlaid with initial positions of backtracked cells. Dorsal view, anterior to left. Scale bar- 100 μm. B- The degree of ballistic migration can be measured by α-value and is a factor of the speed and persistence of migration. Speed is denoted by the magnitude of individual displacement arrows and the greyscale gradient level. Persistence is the ratio of net displacement (final minus initial position, blue) and total length (distance traveled, red).
C-G- Migration dynamics of cell tracks by organ precursor, comparing WT to Tbx5a MO and progenitors from the left versus the right: C- α-values, and the derivation can be seen in Fig. 9. Statistics performed using a comparison of slopes test; D- Average speed; E- Persistence F-Total track length, normalized by the number of timepoints tracked; G- Net track displacement, normalized by the number of timepoints tracked. Statistics performed using the Kruskal-Wallis rank sum test. 10 cells tracked per group per embryo. n= 6 embryos
Figure 9-. Derivation of α-value from migration dynamics of lateral aLPM cells.
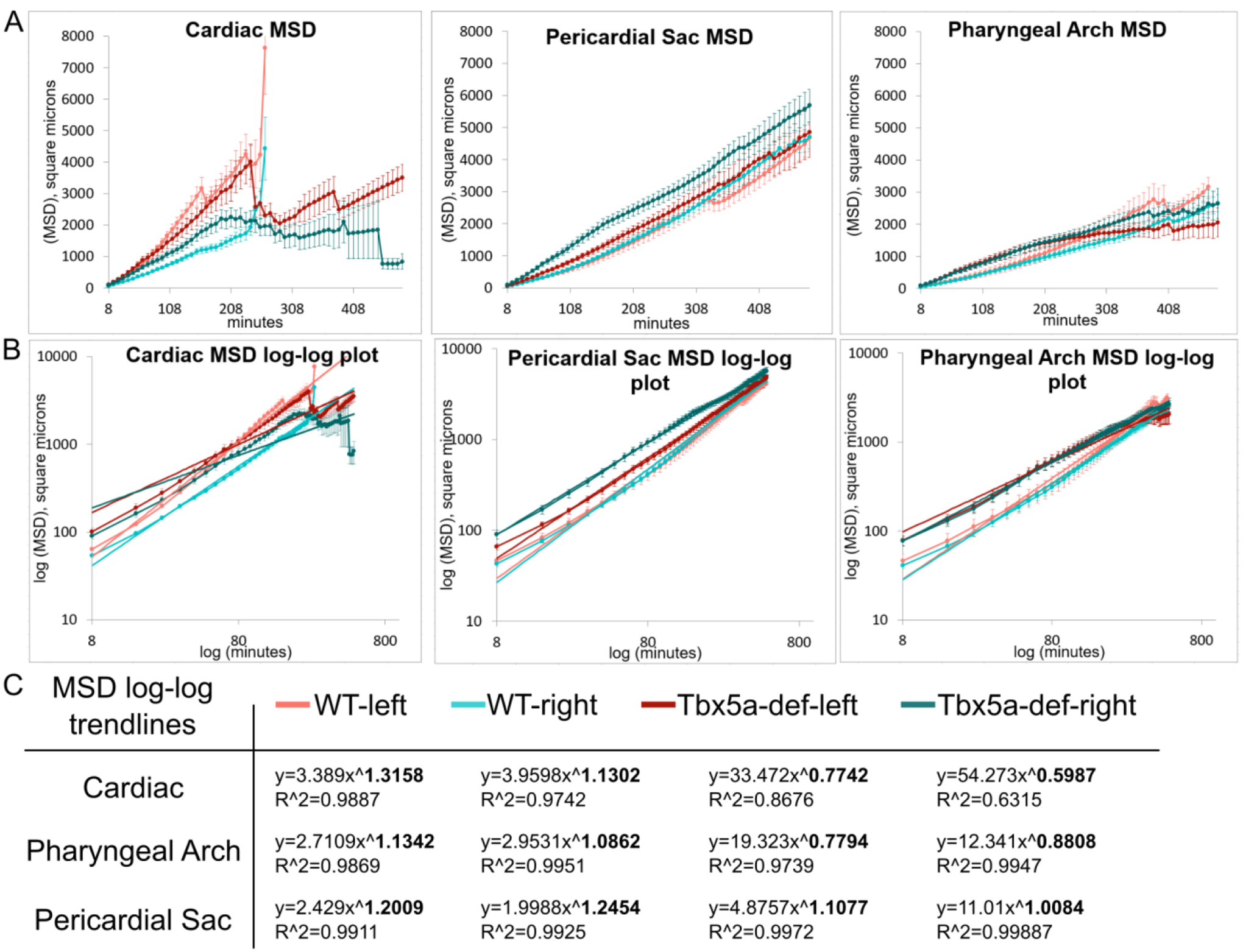
A- Mean squared displacement (MSD) over time, which was calculated using the DiPer excel macro on cell migration data backtracked in 3D. This measure approximates how much area the cells explore over time. B- log-log plot of MSD over time overlaid with power trendlines. C- Table of power trendlines and correlation coefficients, R^2 value. The α-value is the exponent of the trendline, in bold.
The group of wildtype precursors that had the highest degree of ballistic migration was the cardiac precursors that arose from the left side lateral aLPM, as evidenced by their α-value (Fig.8C). As shown in Fig. 8D–G, the migration metrics of speed, persistence, track length, and displacement were higher in cardiac precursors from the left than the right in wildtype embryos. By comparison, the degree of ballistic migration, persistence, and displacement were higher on the right side in the pericardial sac and pharyngeal arch precursors. These results indicate that there are inherent differences in migration dynamics of lateral aLPM precursors correlating with their final fate and laterality of origin.
Tbx5a-deficient precursors have significantly decreased measures of ballistic migration in all fates and laterality of origin (Fig. 8C, p=6.57×10−12). More specifically, the α-values are at or below 1, which signifies that their migration is not ballistic (Fig. 8C). The loss of ballistic migration in Tbx5a-deficient precursors is most likely due to the loss of persistence, since migration speed increases in the knockdown condition (Fig. 8D, p=1.595×10−5). The loss of persistence in Tbx5a-deficient precursors is most likely due to increased track length, as seen by the large effect sizes, compared to the wildtype precursors (Fig. 8F, cardiac effect size=0.91; pericardial sac effect size=0.99; pharyngeal arch effect size=0.88). Increased track length alongside higher speed and fixed displacement indicate that tbx5a-deficient precursors meander more than wildtype precursors. These data suggest that tbx5a functions normally to enable persistent migration of lateral aLPM precursors that give rise to cardiac, pharyngeal arch and pericardial sac.
Cardiac precursors from the left lateral aLPM have higher ballistic migration than precursors from the right in Tbx5a-deficient embryos (Fig. 8C, p=0.0154). The laterality difference in migration is maintained despite the changes in other migration dynamics compared to wildtype: there is no laterality difference in cardiac speed (Fig. 8D, p=0.1095) and in track length (Fig. 8F, p=0.3287). Therefore, the left-right differential migration in the Tbx5a knockdown is most likely due to the observed difference in persistence (Fig. 8E, p=0.01824) and net displacement (Fig. 8G, p=0.01924). Together, these data suggest that tbx5a functions in maintaining left-right asymmetry in cardiac precursor migration speed and track length.
Tbx5a knockdown causes the reversal of laterality difference in the ballistic migration of pericardial sac precursors as measured by the α-values (Fig. 8C, left greater than right, p=1.54×10−21). This difference is most likely due to the changes in speed, since the persistence is higher in precursors from the right (Fig. 8E, p=0.0003778), which is most likely due to the increase in track length, since the displacement is higher in precursors from the right (Fig. 8G, p=0.00281). Tbx5a knockdown results in the disappearance of laterality difference in the ballistic migration of pharyngeal arch precursors displayed in the wildtype (Fig. 8C, p=0.01727), which is most likely due to equal measures of persistence (Fig. 8E, p=0.4766), length (Fig. 8F, p=0.2871) and displacement (Fig. 8G, p=0.3691) between the right and left sides. Together, these data suggest that tbx5a functions normally to maintain left-right asymmetry in the ballistic migration of pericardial sac and pharyngeal arch precursors.
Discussion
Single-cell resolution fate map at 18 hpf identifies aLPM-derived tissues
This paper describes a single-cell fate map analysis of cells within the zebrafish lateral aLPM. The LPM gives rise to many different organs in the early embryo, and previous studies have usually focused upon the development of a particular organ, such as the heart or forelimb (Keegan et al., 2004; Mao et al., 2015; Schoenebeck et al., 2007; Stainier et al., 1993). Our goal was to provide an unbiased description of the aLPM using the photoconversion of single Kaede-containing cells to describe the organization of the organ anlages prior to the laterally- and anteriorly-directed cell migration movements. In a previous fate map study of the posterior LPM, the region between somites 1–4 gave rise to mainly the mesenchymal cells of the pectoral fin bud; furthermore, the migrations of these fin precursors proceeded such that they conserved topological arrangement along the AP body axis (Mao et al., 2015). In contrast to that study, we observe that the lateral aLPM is not organized into organ-specific regions (Fig. 1N, 4), and precursors of four main organs are derived from this area; namely, the heart, pharyngeal arches, peritoneum, and pericardial sac precursors are all interspersed throughout the AP and ML extent of the lateral aLPM region that we examined (Fig. 1N, 4, 5). Cell tracking analyses corroborated that specific organ precursors are derived from varied locations within the pre-migratory aLPM, and that migration pathways were not correlated with the starting position of cells nor location of the progeny within the formed organ by 48 hpf (Fig. 2, 3 for examples of cardiac precursors).
Proliferation rates of the aLPM cells are not high between 18 and 48 hpf as most photoconverted cells divided only once or twice during this period (Fig. 4B). However, a very high proportion of labeled cells gave rise to single-organ lineages with only 13% giving rise to clones that contributed to two or more organs (Fig. 1, 7, Table 1 Row 20). The identification of multipotent precursors at 18 hpf is important for the interpretation of previous studies on this region of the embryo. For example, Schoenebeck et al. studied Hand2-expressing cardiac precursors within the aLPM around 11 hpf and observed that over 90% of embryos additionally generated labeled pharyngeal pouch cells. The authors suggested that the pharyngeal pouch cells could have been derived from the anterior endoderm which overlaps the lateral aLPM at this time in development (Schoenebeck et al., 2007). However, our single-cell labeling technique provides evidence that mesenchymal cores of the pharyngeal arch precursors can arise from within the lateral aLPM rather than solely from endoderm or neural crest (Piotrowski et al., 2003). The study from Hami et al. 2011 observed that a significant proportion of fate mapped aLPM cells at 12 hpf also give rise to the branchial region and pericardial sac at 30 hpf (Hami et al., 2011). Furthermore, time-lapse images of the Tg(Hand2-eGFP)ch2 line allowed us to observe that the lateral aLPM cells migrate into the mesenchymal cores of the pharyngeal arches (Supplemental Movies 1–3). This latter point is important in regards to recent data from Paffett-Lugassy et al. which showed that Nkx2.5-expressing cells first migrate to the pharyngeal arch core regions before proceeding to the heart (Paffett-Lugassy et al., 2017). Therefore, some lateral aLPM-derived clones scored as pharyngeal arch at 48 hpf in our fate map could possibly later migrate to contribute to the heart after 48 hpf.
Our present study also reveals new details on the development of the pericardial sac. At early stages of heart development, the pericardial sac is contiguous with the venous pole (Fig. 5D–G i-ii,c asterisk) (Ocaña et al., 2017). Ocaña et al.’s recent regional-level labeling of the lateral aLPM showed that groups of photoconverted cells can contribute to both the heart and pericardial sac but did not define whether these two fates were derived from the same or different individual precursors. This same study confirmed that these pericardial sac cells also express tbx5a (Ocaña et al., 2017). Our fate map advances these findings by showing that an individual cell within the lateral aLPM at 18 hpf can give rise to both heart and pericardial sac fates (Fig. 1E, F-diamonds), and in fact, this shared fate was the most common multipotent lineage found in our study (Table 1 Row 6).
Left-right asymmetries in wildtype zebrafish lateral aLPM cardiac precursors
Differences between the left and right are not morphologically observed in zebrafish until the formation of the PHT around 22 hpf when medial aLPM rotates clockwise to position left side precursors anteriorly and right side precursors posteriorly with respect to the main body axis (Baker et al., 2008; Fukui et al., 2018; Smith et al., 2008). By characterizing hand2-driven BAC expression at 26 hpf, Fukui et al. 2018 observed that left side lateral aLPM contributes to the anterior heart (Fukui et al., 2018). This observation is aligned with our findings in labeling the entire right versus left lateral aLPM (Fig. 5), specifically that the left lateral aLPM contributes to the anterior heart. Additionally, our use of single cell labeling techniques allowed for the finding of a left-right differential contribution in the aLPM cardiac precursors in wildtype embryos at 18 hpf, specifically that twice as many cardiac precursors arise from the right lateral aLPM compared to the left lateral aLPM (Fig. 1E, p=0.0459).
Therefore, our methods expand the finding of Fukui et al. 2018 to not just cells expressing their BAC transgene, but to the lateral populations of the aLPM. We emphasize that these findings may only hold for the non-PHT precursors of the heart, as our labeling methods do not mark the early, medially migrating population of aLPM cells (Keegan et al., 2004; Schoenebeck et al., 2007; Stainier et al., 1993). However, this finding is notable because left-right differences in the number of precursors were not observed for any of the other ALPM-derived fates (Fig. 1F–H). The photoconversion of larger regions of the aLPM in embryos that had been injected with Kaede mRNA at the one-cell stage showed that the early asymmetry in precursor numbers was retained in the larval heart tissues (Fig. 5). Furthermore, there is left-right asymmetry in ballistic migration between cardiac precursors such that the left lateral aLPM exhibit higher speed and persistence while traveling over a larger displacement than cardiac precursors from the right lateral aLPM (Fig. 8C–G). Corroborating these findings, a study in mouse finds that left-right asymmetries emerge in how cardiac precursors, specifically the posterior SHF, migrate and contribute to different aspects of the mammalian heart (Domínguez et al., 2012).
Proposed mechanisms of tbx5a function in cardiac precursors
The range in our methodologies allows us to assay the development of the lateral aLPM along multiple different developmental timepoints. It is important to distinguish between each of these timepoints, as tbx5a may function differently across time. First, the composition of our fate map is a readout on the specification of the lateral aLPM up until 18 hpf, the pre-migratory stage. Second, our backtracking experiment on migrating lateral aLPM cells addresses the next stage, which is from 18 to 24.5 hpf. Third, our assessment of proliferation and contribution to the larval heart via fate mapping and large-scale photoconversion addresses a wider window of time from 18 to 48 hpf. In the following discussion, we propose that tbx5a has distinct functions on the developing lateral aLPM in each of these time windows.
Currently, the extent of tbx5a function in zebrafish lateral aLPM is unknown. It has also been reported that Tbx5a-knockdown causes a changed ratio in FHF to SHF cells as measured by Draculin expression in the FHF (Mosimann et al., 2015). Mosimann et al. conclude that tbx5a may function ‘by altering migration to the heart tube or by specifying corresponding ALPM fates before the bilateral field migration’ toward the midline. That study aligns with our data and our interpretation of tbx5a function in cardiac development. Although we do not directly see an effect of Tbx5a knockdown on the total number of cardiac precursors in the fate map, Tbx5a knockdown results in fewer photoconverted cardiomyocytes from the right aLPM (Fig. 5B). We provide evidence that tbx5a functions by altering ballistic migration, speed and persistence to the heart (Fig. 8C–E). However, using genetic lineage tracing of BAC transgenes in zebrafish, Sánchez-Iranzo et al’s 2018 report concluded that tbx5a-expressing cells only make minor contributions to the SHF (Sánchez-Iranzo et al., 2018). We show that Tbx5a-deficiency affects the lateral aLPM fate map, migration dynamics, and contributions of precursors to the heart (Fig. 1J–N, 2, 4, 5B, 7B, 8C–G, 9, Table 2). More specifically, tbx5a is required for the left-right asymmetry in cardiac precursors by 18 hpf (Fig. 1J). Additionally, the Tbx5a-deficient fate map has proportionally twice as many multipotent precursors as in the wildtype (Fig. 7B). tbx5a may maintain single-organ lineage of lateral aLPM precursors by individually biasing certain fates over others or by maintaining the progression of development in the tissue by 18 hpf. Our results do not necessarily distinguish between these two possible hypotheses, yet, our results show an overall increase in multipotent precursors across all fates in the Tbx5a-deficient fate map, which supports the latter hypothesis. There is support for the former hypothesis in that tbx5a is specifically involved in the development of cardiomyocyte specification, though not during somitogenesis stages (Bruneau et al., 2001; Ghosh et al., 2009; Plageman and Yutzey, 2006; Small and Krieg, 2003).
Next, tbx5a is required for the ballistic migration of lateral aLPM precursors; more specifically, Tbx5a-deficient cells migrate a longer track length with faster speed, meaning that they are less persistent than wildtype cells (Fig. 8C–G). Both wildtype and Tbx5a-deficient cardiac precursors from the left undergo higher ballistic migration than the right (Fig. 8C). Altogether, tbx5a functions within 18 to 24.5 hpf to maintain ballistic migration on both sides of the lateral aLPM. tbx5a could be affecting the migration by cell-autonomous defects that could include the recognition of the ECM that aLPM cells migrate over, inability to receive migration signals, and/or dysfunction in cellular and cytoskeletal dynamics. Also, we and others have observed evidence for non-cell autonomous effects of Tbx5a and Tbx5b that could affect the coordination, migration and specification of the aLPM (Boyle Anderson and Ho, 2018; Mao et al., 2015; Pi-Roig et al., 2014).
Between 18 and 48 hpf, a left-right asymmetry emerges in the Tbx5a-deficient cardiac precursor proliferation that does not exist in the wildtype (Fig. 4B). Since Tbx5a-deficient cardiac precursors originating from the left lateral aLPM exhibit a larger clone size by 48 hpf than those originating from the right (Fig. 4B), we hypothesize that tbx5a is required normally in limiting the proliferation of lateral aLPM cardiac precursors just on the left side during this time window, perhaps to compensate for the lack of persistence and ballistic migration exhibited by left side wild-type precursors. Within the same time in development, we observe that right side lateral aLPM contributes more final cells than the left side to the larval heart by 48 hpf in wildtype embryos (Fig. 5B). In the Tbx5a-deficiency, the right side lateral aLPM contributes fewer cardiomyocytes such that the contribution is equal to that from the left side (Fig. 5B). At least two hypotheses can explain these observations: tbx5a may have separate functions across the left-right axis and/or tbx5a changes function over time between 18 to 48 hpf. The involvement of TBX5 in left-right asymmetry is supported by multiple previous observations in amniotes and zebrafish. Loss of TBX5 function affects the left side more severely than the right side during mammalian limb development (Sletten and Pierpont, 1996; Sulaiman et al., 2016). As TBX5 is known to be downstream of left-right patterning genes in mice (Nadadur et al., 2006), we propose that tbx5a may also be a downstream effector of the left-right patterning pathway in zebrafish cardiac development. For example, zebrafish prrx1a and chicken PRRX1 are both expressed asymmetrically within the LPM with stronger expression on the right side (Ocaña et al., 2017). Ocaña et al. suggest that prrx1a is responsible for directing the preferential migration of Tbx5a-expressing cells into the posterior pole of the heart, perhaps contributing to proper heart jogging. Other authors have postulated tbx5a’s role in proliferation, cell death, and migration (Hatcher et al., 2001; Hatcher et al., 2004; Lu et al., 2011; Tsai et al., 2018), which may also be able to explain these observed results.
Altogether, we suggest a model of tbx5a function in the lateral aLPM that proceeds as follows in developmental time: First tbx5a acts downstream of BMP and Prrx1a to asymmetrically enable more cardiac precursors within the right side lateral aLPM to undergo cell migration than the left by 18 hpf. Then, tbx5a enables the persistent ballistic cell migration of precursors from both sides into their respective organ fields from 18 to 24.5 hpf. As precursors are migrating between 18 and 48 hpf, when we assessed their fates, tbx5a functions to restrict most lateral aLPM precursors towards unipotential fates. At the same time, tbx5a restricts proliferation of cardiac precursors on the left side once they reach the heart. Lastly in our model, tbx5a allows more cardiomyocytes to contribute to the larval heart from the right side lateral ALPM to maintain proper organ size, perhaps involving a mechanism such as the Hippo pathway (Fukui et al., 2018). Further study must be done to determine on which side tbx5a function is necessary in these mechanisms and to confirm the order of mechanisms we propose.
tbx5a function in pharyngeal arch and pericardial sac precursors
Within the pharyngeal arch precursors, we observe that the displacement is lower in the Tbx5a-deficient condition than in wildtype (Fig. 8G). It is more likely that this change in displacement is due to a change in the initial positions of the precursors at 18 hpf as opposed to a change in the final positions of the calculated tracks at 24.5 hpf. Pharyngeal arch precursors are more likely to arise from medial than lateral bins in the Tbx5a-deficient condition (Fig. 1G, L), and therefore have a shorter distance to travel to medially located pharyngeal arches. Additionally, the entire aLPM tissue has a wider mediolateral width in the Tbx5a-deficient compared to the wildtype condition, which might explain the phenotype (data not shown).
tbx5a functions normally to maintain left-right asymmetry in the ballistic migration of pericardial sac and pharyngeal arch precursors, as knockdown of Tbx5a causes left side pericardial sac precursors to migrate more ballistically than right side precursors and causes pharyngeal arch precursors to show no difference in ballistic migration between the right and left (Fig. 8C). Based upon the variety of effects that Tbx5a-knockdown has on each of the organ progenitor fields, tbx5a possibly functions in different cellular processes depending on the cell-type. This conclusion is in concordance with previous observations of how TBX5 differentially functions between cardiac and non-cardiac cell types in mouse (Waldron et al., 2016).
In summary, our fate map demonstrates that there are no distinct and homogeneous regions defining separate fates within the lateral aLPM at 18 hpf. In wildtype embryos, most aLPM precursors give rise to unipotent lineages. Lateral aLPM precursors exhibit different levels of persistence, directionality, and speed of migration based upon their final fate and laterality of origin. In a specific subset of the lateral aLPM, the higher number of cardiac precursors on the right side lateral aLPM is correlated with a greater contribution from the right side lateral aLPM into the heart. Consequently, the tissue-specific fates of individual lateral aLPM cells might be dependent on a cell’s regional location after migration or on the pattern of the migration itself. tbx5a appears to play a role in this specification by affecting the dynamics of cell migration, thereby affecting fate. The effects of tbx5a knockdown includes the loss of laterality differences in cardiac precursors, changes in the migration movements of cardiac and pharyngeal precursors as well as an increase in the number of multipotent lateral aLPM precursors.
Methods
Zebrafish husbandry and strains
Zebrafish adults and embryos were raised and maintained in standard laboratory conditions (Westerfield, 2007) and approved by the University of Chicago Institutional Animal Care and Use Committee. Et(hand2:eGFP)ch2 was generated by D.G. Ahn and is described by (Mao et al., 2015). Tg(h2afx:h2afv-mCherry)mw3 was a gift from B. Link (McMahon et al., 2009). Morpholino used against tbx5a (5’-CCTGTACGATGTCTACCGTGAGGC-3’) is as described in (Ahn et al., 2002). 1.2 ng per embryo was injected into the single-cell stage. Morphant phenotype was confirmed at 3–4 dpf in all experimental animals. All control and experimental embryos were stage matched based on morphology, including somite count and angle of the tail extension.
Single-cell photoconversion and fate mapping
Single marginal blastomere at the 64- or 128-cell stage were injected with 100 ng/μl Kaede mRNA. 18 hpf embryos were mounted on glass slides under bridged coverslips. A single lateral aLPM precursor at 18 hpf was photoconverted in each individual embryo for 30 s through a pinhole with 20x objective and light from a 100 W HBO mercury bulb through the DAPI filter on a Zeiss Axioplan microscope. Cell identity was assayed by live morphology and final position of the labeled cells over the described timecourse of these experiments. Dorso-lateral views were photographed using a Nikon D5000 camera immediately post-photoconversion and at 24 and 48 hpf. The positions of labeled precursors were measured in ImageJ as medio-lateral and antero-posterial distances from the center of the otic vesicle.
Whole side photoconversion
1-cell stage embryos were injected with 100 ng/μl Kaede mRNA. 18 hpf embryos screened with uniform green fluorescence were embedded into 0.3% low-melting-point agarose (Sigma; A9414) in E3 dorsal-side up. Using an upright Zeiss LSM 710 confocal microscope, a region of interest was drawn around the lateral aLPM tissue on either the right or left sides in a Z-stack, avoiding as much of the medial aLPM as possible. The region of interest was scanned using the diode 405 nm laser at 5% through the 40x water-emersion objective to photoconvert Kaede into red-fluorescent. Since the same 2-dimensional region of interest was drawn based upon the entire extend of lateral aLPM tissue, other ectodermal and endodermal tissue in these regions were also photoconverted. We are confident that these unintended photoconversions did not contribute to the heart as they were not mesodermal in origin nor location.
Confocal stacks were acquired of aLPM-derived tissues after photoconverting the right, left, or both sides lateral aLPM at 48 hpf every 2 μm. Using FIJI, 3D stacks were reconstructed and cell counts were obtained by manually scoring red-fluorescent cardiomyocytes by morphology through both rotating the 3D reconstruction and progressing through the z-slices (Fig. 6). Double-counting of cells was avoided by only counting the first instance of a cardiomyocyte with average depth of 10 μm though the z-slices. Cell counts were therefore corroborated by two different methods of analysis. Image stacks in which cell counts could not be corroborated between methods due to non-uniform Kaede distribution or movements of the heart through the z-stack were not considered in this analysis.
Time-lapse Microscopy and Cell Migration Dynamics
Live imaging was performed using Et(hand2:eGFP)ch2 heterozygous embryos. Selective plane illumination microscopy (SPIM) was performed using multilayer embryo mounting as described in (Kaufmann et al., 2012). Dechorionated embryos were mounted in 0.3% low-melting-point agarose (Sigma; A9414) in E3 containing 200 mg/L tricaine into FEP tubing (Bola, S1815–04, refractive index 1.338, inner diameter 0.8 mm, outer diameter 1.6 mm). FEP tubing was plugged with 1.5% low-melting-point agarose in E3. FEP tubing was suspended into E3 media containing 200 mg/L tricaine, maintained at 28.5°C in the Zeiss Lightsheet Z.1. Z-stacks were collected every 8 min with 20x objectives.
Cell tracking was performed manually using the FIJI MTrackJ plugin backward in time. Ten cell tracks were chosen per organ type per laterality side for analysis in Excel. AP position (x), ML position (y), DV position (z) and time (t) were normalized by a reference track created by following the most postero-dorsal position of the heart cone/primary heart tube at the midline over time. The following metrics were calculated from the original four parameters: Instantaneous Speed, Average Speed across tracks and embryos, Instantaneous Displacement, Net Displacement, Total Length, Persistence, and Mean Squared Displacement. Additionally, DiPer computer program (Gorelik and Gautreau, 2014) was run on 2-dimensional tracks to calculate the α-value and directionality/persistence. Subsequent statistical analyses were performed in R.
Supplementary Material
Highlights.
Fate mapping of anterior lateral plate mesoderm elucidates organ contributions
aLPM precursors are not regionalized by eventual fate in 18 hpf zebrafish embryos
Cardiac precursors are overrepresented on the right side of the embryo
Cell tracking reveals migration variances between left versus right side precursors
Left-Right asymmetry is dependent upon tbx5a function.
Acknowledgements
We thank V. Prince for comments on the manuscript, C. Labno for Lightsheet microscope support and A. Kuuspalu for fish care and confocal microscope maintenance. L.M.F.M was supported by NIH T32 HD055164 and by a Diversifying Faculty in Illinois Fellowship. E.A.T.B.A was supported by NIH T32 GM007197. This work was funded by NIH R01 HD072598 (to R.K.H).
References
- Ahn DG, Kourakis MJ, Rohde LA, Slivert LM and Ho RK (2002). T-box gene Tbx5 is essential for formation of the pectoral limb bud. Nature 417, 754–758. [DOI] [PubMed] [Google Scholar]
- Baker K, Holtzman NG and Burdine RD (2008). Direct and indirect roles for Nodal signaling in two axis conversions during asymmetric morphogenesis of the zebrafish heart. Proc. Natl. Acad. Sci 105, 13924–13929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson CT, Cowley GS, Solomon SD, Weissman B, Poznanski AK, Traill TA, Seidman JG and Seidman CE (1994). The Clinical and Genetic Spectrum of the Holt-Oram Syndrome (Heart-Hand Syndrome). N. Engl. J. Med 330, 885–891. [DOI] [PubMed] [Google Scholar]
- Basson CT, Bachinsky DR, Lin RC, Levi T, Elkins JA, Soults J, Grayzel D, Kroumpouzou E, Traill TA, Leblanc-Straceski J, et al. (1997). Mutations in human cause limb and cardiac malformation in Holt-Oram syndrome. Nat. Genet 15, 30–35. [DOI] [PubMed] [Google Scholar]
- Boyle Anderson EAT and Ho RK (2018). A transcriptomics analysis of the Tbx5 paralogues in zebrafish. PLoS One 13, e0208766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, Conner DA, Gessler M, Nemer M, Seidman CE, et al. (2001). A murine model of Holt-Oram syndrome defines roles of the T-Box transcription factor Tbx5 in cardiogenesis and disease. Cell 106, 709–721. [DOI] [PubMed] [Google Scholar]
- de Pater E, Clijsters L, Marques SR, Lin Y-F, Garavito-Aguilar ZV, Yelon D and Bakkers J (2009). Distinct phases of cardiomyocyte differentiation regulate growth of the zebrafish heart. Development 136, 1633–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez JN, Meilhac SM, Bland YS, Buckingham ME and Brown NA (2012). Asymmetric fate of the posterior part of the second heart field results in unexpected left/right contributions to both poles of the heart. Circ. Res 111, 1323–1335. [DOI] [PubMed] [Google Scholar]
- Fukui H, Miyazaki T, Chow RW-YY, Ishikawa H, Nakajima H, Vermot J, Naoki M and Mochizuki N (2018). Hippo signaling determines the number of venous pole cells that originate from the anterior lateral plate mesoderm. Elife 7, e29106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity DM, Childs S and Fishman MC (2002). The heartstrings mutation in zebrafish causes heart/fin Tbx5 deficiency syndrome. Development 129, 4635–4645. [DOI] [PubMed] [Google Scholar]
- George V, Colombo S and Targoff KL (2015). An early requirement for nkx2.5 ensures the first and second heart field ventricular identity and cardiac function into adulthood. Dev. Biol 400, 10–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh TK, Song FF, Packham EA, Buxton S, Robinson TE, Ronksley J, Self T, Bonser AJ and Brook JD (2009). Physical Interaction between TBX5 and MEF2C Is Required for Early Heart Development. Mol. Cell. Biol 29, 2205–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelik R and Gautreau A (2014). Quantitative and unbiased analysis of directional persistence in cell migration. Nat. Protoc 9, 1931–1943. [DOI] [PubMed] [Google Scholar]
- Hami D, Grimes AC, Tsai H-J and Kirby ML (2011). Zebrafish cardiac development requires a conserved secondary heart field. Development 138, 2389–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatcher CJ, Kim MS, Mah CS, Goldstein MM, Wong B, Mikawa T and Basson CT (2001). TBX5 transcription factor regulates cell proliferation during cardiogenesis. Dev. Biol 230, 177–188. [DOI] [PubMed] [Google Scholar]
- Hatcher CJ, Diman NYS-G, Kim M-S, Pennisi D, Song Y, Goldstein MM, Mikawa T and Basson CT (2004). A role for Tbx5 in proepicardial cell migration during cardiogenesis. Physiol. Genomics 18, 129–140. [DOI] [PubMed] [Google Scholar]
- Holt M and Oram S (1960). Familial heart disease with skeletal malformations. Br. Heart J 22, 236–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann A, Mickoleit M, Weber M and Huisken J (2012). Multilayer mounting enables long-term imaging of zebrafish development in a light sheet microscope. Development 139, 3242–3247. [DOI] [PubMed] [Google Scholar]
- Keegan BR, Meyer D and Yelon D (2004). Organization of cardiac chamber progenitors in the zebrafish blastula. Development 131, 3081–3091. [DOI] [PubMed] [Google Scholar]
- Lazic S and Scott IC (2011). Mef2cb regulates late myocardial cell addition from a second heart field-like population of progenitors in zebrafish. Dev. Biol 354, 123–133. [DOI] [PubMed] [Google Scholar]
- Li QY, Newbury-Ecob RA, Terrett JA, Wilson DI, Curtis ARJ, Yi CH, Gebühr T, Bullen PJ, Robson SC, Strachan T, et al. (1997). Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat. Genet 15, 21–29. [DOI] [PubMed] [Google Scholar]
- Long S, Ahmad N and Rebagliati M (2003). The zebrafish nodal-related gene southpaw is required for visceral and diencephalic left-right asymmetry. Development 130, 2303–2316. [DOI] [PubMed] [Google Scholar]
- Lu J, Tsai T, Choo S, Yeh S, Tang R, Yang A, Lee H and Lu J (2011). Induction of apoptosis and inhibition of cell growth by tbx5 knockdown contribute to dysmorphogenesis in Zebrafish embryos. J. Biomed. Sci 18, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra M, Schluterman MK, Nichols HA, Richardson JA, Lo CW, Srivastava D and Garg V (2009). Interaction of Gata4 and Gata6 with Tbx5 is critical for normal cardiac development. Dev. Biol 326, 368–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Q, Stinnett HK and Ho RK (2015). Asymmetric cell convergence-driven zebrafish fin bud initiation and pre-pattern requires Tbx5a control of a mesenchymal Fgf signal. Development 142, 4329–4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon C, Gestri G, Wilson SW and Link BA (2009). Lmx1b is essential for survival of periocular mesenchymal cells and influences Fgf-mediated retinal patterning in zebrafish. Dev. Biol 332, 287–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann C, Panáková D, Werdich AA, Musso G, Burger A, Lawson KL, Carr LA, Nevis KR, Sabeh MK, Zhou Y, et al. (2015). Chamber identity programs drive early functional partitioning of the heart. Nat. Commun 6, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskowitz IPG, Pizard A, Patel VV, Bruneau BG, Kim JB, Kupershmidt S, Roden D, Berul CI, Seidman CE and Seidman JG (2004). The T-Box transcription factor Tbx5 is required for the patterning and maturation of the murine cardiac conduction system. Development 131, 4107–4116. [DOI] [PubMed] [Google Scholar]
- Moskowitz IPG, Kim JB, Moore ML, Wolf CM, Peterson MA, Shendure J, Nobrega MA, Yokota Y, Berul C, Izumo S, et al. (2007). A Molecular Pathway Including Id2, Tbx5, and Nkx2–5 Required for Cardiac Conduction System Development. Cell 129, 1365–1376. [DOI] [PubMed] [Google Scholar]
- Ocaña OH, Coskun H, Minguillón C, Murawala P, Tanaka EM, Galcerán J, Muñoz-Chápuli R and Nieto MA (2017). A right-handed signalling pathway drives heart looping in vertebrates. Nature 549, 86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paffett-Lugassy N, Novikov N, Jeffrey S, Abrial M, Guner-Ataman B, Sakthivel S, Burns CE and Burns CG (2017). Unique developmental trajectories and genetic regulation of ventricular and outflow tract progenitors in the zebrafish second heart field. Development 144, 4616–4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrie LE, Renfrew EM, Vander Wal A, Mueller RL and Garrity DM (2013). Zebrafish tbx5 paralogs demonstrate independent essential requirements in cardiac and pectoral fin development. Dev. Dyn 242, 485–402. [DOI] [PubMed] [Google Scholar]
- Pi-Roig A, Martin-Blanco E and Minguillon C (2014). Distinct tissue-specific requirements for the zebrafish tbx5 genes during heart, retina and pectoral fin development. Open Biol 4, 140014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski T, Ahn D, Schilling TF, Nair S, Ruvinsky I, Geisler R, Rauch G-J, Haffter P, Zon LI, Zhou Y, et al. (2003). The zebrafish van gogh mutation disrupts tbx1, which is involved in the DiGeorge deletion syndrome in humans. Development 130, 5043–5052. [DOI] [PubMed] [Google Scholar]
- Plageman TF and Yutzey KE (2006). Microarray analysis of Tbx5-induced genes expressed in the developing heart. Dev. Dyn 235, 2868–2880. [DOI] [PubMed] [Google Scholar]
- Sánchez-Iranzo H, Galardi-Castilla M, Minguillón C, Sanz-Morejón A, González-Rosa JM, Felker A, Ernst A, Guzmán-Martínez G, Mosimann C and Mercader N (2018). Tbx5a lineage tracing shows cardiomyocyte plasticity during zebrafish heart regeneration. Nat. Commun 9, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenebeck JJ, Keegan BR and Yelon D (2007). Vessel and Blood Specification Override Cardiac Potential in Anterior Mesoderm. Dev. Cell 13, 254–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sletten LJ and Pierpont MEM (1996). Variation in severity of cardiac disease in Holt-Oram syndrome. Am. J. Med. Genet 65, 128–132. [DOI] [PubMed] [Google Scholar]
- Small EM and Krieg PA (2003). Transgenic analysis of the atrialnatriuretic factor (ANF) promoter: Nkx2–5 and GATA-4 binding sites are required for atrial specific expression of ANF. Dev. Biol 261, 116–131. [DOI] [PubMed] [Google Scholar]
- Smith KA, Chocron S, von der Hardt S, de Pater E, Soufan A, Bussmann J, Schulte-Merker S, Hammerschmidt M and Bakkers J (2008). Rotation and Asymmetric Development of the Zebrafish Heart Requires Directed Migration of Cardiac Progenitor Cells. Dev. Cell 14, 287–297. [DOI] [PubMed] [Google Scholar]
- Stainier DY, Lee RK and Fishman MC (1993). Cardiovascular development in the zebrafish. I. Myocardial fate map and heart tube formation. Development 119, 31–40. [DOI] [PubMed] [Google Scholar]
- Sulaiman FA, Nishimoto S, Murphy GRF, Kucharska A, Butterfield NC, Newbury-Ecob R and Logan MPO (2016). Tbx5 Buffers Inherent Left/Right Asymmetry Ensuring Symmetric Forelimb Formation. PLoS Genet 12, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi JK and Bruneau BG (2009). Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature 459, 708–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai TC, Shih CC, Chien HP, Yang AH, Lu JK and Lu JH (2018). Anti-apoptotic effects of IGF-I on mortality and dysmorphogenesis in tbx5-deficient zebrafish embryos. BMC Dev. Biol 18, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron L, Steimle JD, Greco TM, Gomez NC, Dorr KM, Kweon J, Temple B, Yang XH, Wilczewski CM, Davis IJ, et al. (2016). The Cardiac TBX5 Interactome Reveals a Chromatin Remodeling Network Essential for Cardiac Septation. Dev. Cell 36, 262–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M (2007). The Zebrafish Book. 5th ed. University of Oregon Press. [Google Scholar]
- Witzel HR, Cheedipudi S, Gao R, Stainier DYR and Dobreva GD (2017). Isl2b regulates anterior second heart field development in zebrafish. Sci. Rep 7, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyngaarden LA, Vogeli KM, Ciruna BG, Wells M, Hadjantonakis A-K and Hopyan S (2010). Oriented cell motility and division underlie early limb bud morphogenesis. Development 137, 2551–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelon D, Ticho B, Halpern ME, Ruvinsky I, Ho RK, Silver LM and Stainier DY (2000). The bHLH transcription factor hand2 plays parallel roles in zebrafish heart and pectoral fin development. Development 127, 2573–82. [DOI] [PubMed] [Google Scholar]
- Yue MS, Plavicki JS, Li XY, Peterson RE and Heideman W (2015). A co-culture assay of embryonic zebrafish hearts to assess migration of epicardial cells in vitro Organogenesis. BMC Dev. Biol 15, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Cashman TJ, Nevis KR, Obregon P, Carney SA, Liu Y, Gu A, Mosimann C, Sondalle S, Peterson RE, et al. (2011). Latent TGF-βbinding protein 3 identifies a second heart field in zebrafish. Nature 474, 645–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


