Abstract
The SKIV2L RNA exosome is an evolutionarily conserved RNA degradation complex in the eukaryotes. Mutations in the SKIV2L gene are associated with a severe inherited disorder, trichohepatoenteric syndrome (THES), with multi-system involvement but unknown disease mechanism. Here, we reported a THES patient with SKIV2L mutations showing severe primary B-cell immunodeficiency, hypogammaglobulinemia, kappa-restricted plasma cell dyscrasia but normal T-cell and NK cell function. To corroborate these findings, we made B-cell-specific Skiv2l knockout mice (Skiv2lfl/flCd79a-Cre) which lacked both conventional B-2 and innate-like B-1 B cells in the periphery and secondary lymphoid organs. This was linked to a requirement of SKIV2L RNA exosome activity in the bone marrow during early B cell development at the pro-B to large pre-B transition. Mechanistically, Skiv2l-deficient pro-B cells exhibited cell cycle arrest and DNA damage. Furthermore, loss of Skiv2l led to substantial out-of-frame V(D)J rearrangement of immunoglobulin heavy chain and severely reduced surface expression of μH, both of which are crucial for pre-BCR signaling and proliferative burst during early B cells development. Together, our data demonstrated a crucial role for SKIV2L RNA exosome in early B cell development in both human and mice by ensuring proper V(D)J recombination and Igh expression, which serve as the molecular basis for immunodeficiency associated with THES.
One-sentence summary
SKIV2L RNA exosome is essential for V(D)J recombination and the pro-B to large pre-B transition during early B cell development.
Introduction
The RNA exosome is an evolutionarily conserved intracellular RNA degradation complex in eukaryotes, and it is involved in RNA processing, maturation, surveillance and turnover (1). Mammalian RNA exosome is a multi-subunit complex containing three major components: RNA helicases and cofactors that recruit RNA for degradation, a barrel-shaped core for RNA to pass through, and exonucleases such as EXOSC10, DIS3 or DIS3L to degrade RNA (2). The super-killer (SKI) complex is one of several RNA helicase and cofactor complexes that target selective RNA substrates for degradation (3). The SKI complex consists of Ski2 like RNA helicase (SKIV2L), tetratricopeptide repeat domain 37 (TTC37) and WD repeat domain 61 (WDR61) (4). SKIV2L is an RNA helicase that unwinds RNA substrates and threads them through the RNA exosome for degradation, while TTC37 and WDR61 contribute to the structure and activity of the SKI complex.
Studies in yeast have established clear biochemical and structural understanding of the RNA exosome and its cofactors (4). Most RNA exosome genes are evolutionarily conserved from yeast to mammals. However, physiological functions of the RNA exosome in mammals remain largely unexplored. Mutations in either SKIV2L or TTC37 gene are associated with an inherited autosomal recessive disorder, trichohepatoenteric syndrome (THES) (OMIM #222470 THES1 with TTC37 mutation, #614602 THES2 with SKIV2L mutation) (4-6). THES is clinically characterized by intrauterine growth retardation, intractable diarrhea, distinctive hair abnormalities and liver diseases with infancy onset (7). Approximately half of THES patients exhibit primary immunodeficiency, including hypogammaglobulinemia and poor or lack of antibody response to childhood vaccination, which usually complicates the disease with opportunistic infections (7-9). These clinical observations implicate important and potentially diverse physiological functions of the SKIV2L RNA exosome in humans. Molecular mechanism of THES is unknown and no targeted treatment is currently available.
Here we reported a THES2 patient harboring SKIV2L mutations who developed severe primary B cell immunodeficiency. We further generated a B-cell-specific Skiv2l knockout mouse model and uncovered a critical requirement of SKIV2L RNA exosome for early B cell development in the bone marrow. Our findings in mice established a molecular basis for immunodeficiency associated with THES.
Results
Primary B cell immunodeficiency in a THES2 patient with SKIV2L mutations
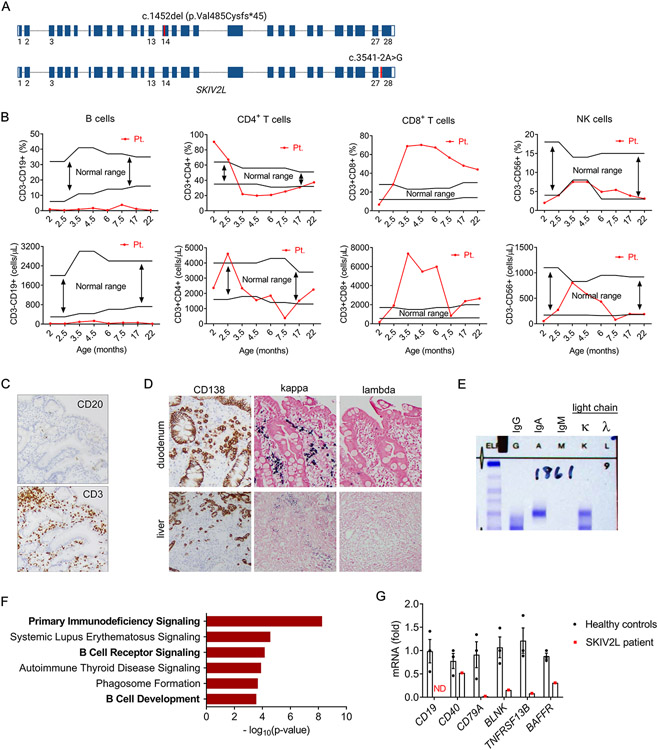
We identified a THES2 patient with biallelic SKIV2L mutations (Fig. 1A). The collection of clinical features (Table 1), including intractable diarrhea in infancy, gastrointestinal and hepatic pathologies, and woolly hair appearance, led to the presumed diagnosis of THES, which was further confirmed with whole-genome sequencing. Immunological parameters of the patient demonstrated hypogammaglobulinemia and elevated IgA (Table 1). Both percentage and absolute number of B cells in peripheral blood were lower than the normal range observed in healthy individuals of similar age (10), while no decrease was observed in CD4+, CD8+ T cells, or NK cells (Fig. 1B). Functional assay revealed normal mitogen responses of lymphocytes and normal NK function (Table 1). The absence of CD20+ B cells was observed in duodenum biopsies, whereas CD3+ T cells were detected (Fig. 1C). A plasma cell dyscrasia with kappa-restricted plasma cells was noted in both duodenum and liver biopsies (Fig. 1D). Serum protein electrophoresis/immunofixation was present for IgA and kappa light chain (Fig. 1E). The patient developed ANCA-associated vasculitis with positive p-ANCA and serine protease 3 at 17-month old and kidney biopsy revealed pauci-immune necrotizing and crescentic glomerulonephritis (Table 1).
Fig. 1. B cell immunodeficiency in a THES2 patient with SKIV2L mutations.
(A) A schematic diagram showing biallelic SKIV2L mutations in the patient, c.1452del (p.Val485Cysfs*45) in exon 14 and c.3541-2A>G in intron 27 (marked with red).
(B) Percentage (upper) and absolute numbers (lower) of B, CD4+ T, CD8+ T, NK cells in patient peripheral blood at indicated ages. Black lines indicate the normal range observed in healthy individuals of different ages (10). The patient was started on glucocorticoid steroid therapy at ~5 months of age.
(C) Immunohistochemistry analysis of B cells (CD20) and T cells (CD3) in duodenum biopsy. Magnification on the images is 100x total magnification (10x ocular x 10x objective).
(D) Immunohistochemistry analysis of plasma cells (CD138) and immunoglobulin light chains (kappa and lambda) in duodenum and liver biopsies. Magnification on the images is 100x total magnification (10x ocular x 10x objective).
(E) Serum protein electrophoresis/immunofixation analysis of immunoglobulin isotypes of the SKIV2L-null patient serum.
(F) Ingenuity pathway analysis of differentially expressed transcripts from RNA-seq analysis of PBMCs of SKIV2L THES patient versus two healthy controls.
(G) qRT-PCR analysis of B cell-related genes in PBMCs of SKIV2L THES patient and three healthy controls. ND, not detected.
Table 1.
Clinical findings of the THES2 patient
| Age | 3 mo | 5 mo* | 6 mo | 7 mo |
|---|---|---|---|---|
| Immunoglobulins (mg/dL) | ||||
| IgA | 37 | 359 | 1120 | 921 |
| IgE | 210 | 106 | 58 | ND |
| IgG | 88 | 1670 | 1020 | 823 |
| IgM | 66 | 24 | 5.3 | 11.2 |
| Functional assay | ||||
| Mitogens (PHA, ConA, PWM) | Normal | |||
| NK Function | Normal | |||
| Clinical phenotypes | ||||
| Intractable diarrhea in infancy | ||||
| Gastrointestinal and hepatic pathologies | ||||
| Woolly hair appearance | ||||
| ANCA-associated vasculitis | ||||
| Positive p-ANCA and serine protease 3 | ||||
| Pauci-immune necrotizing and crescentic glomerulonephritis | ||||
Patient started steroids and IgG replacement therapy at 5 months of age. ND, not done.
We further performed RNA sequencing (RNA-seq) on peripheral blood mononuclear cells (PBMCs) from the patient and healthy controls. Ingenuity pathway analysis of differentially expressed genes revealed primary immunodeficiency and B cell receptor signaling among the top canonical pathways (Fig. 1F), consistent with clinical immunological findings. Quantitative RT-PCR further confirmed that a panel of B-cell related genes were either reduced or not detectable in the patient’s PBMCs (Fig. 1G). Analysis of immunoglobulin heavy chain in RNA-seq dataset showed that IGH V was broadly downregulated while the constant region of only IgA class was detected (Fig. S1A), consistent with serological findings. Expression of the kappa but not lambda light chain was in line with kappa-restricted plasma cell dyscrasia (Fig. S1B). While the overall expression of TCRα and β chains in THES2 patient PBMCs was largely unaffected, the diversity of TCRα and β repertoire was significantly altered compared to the healthy control (Figure S1C). These immunologic and transcriptomic analyses confirmed a B cell immunodeficiency in this THES2 patient carrying SKIV2L mutation.
Loss of SKIV2L leads to B cell immunodeficiency in mice
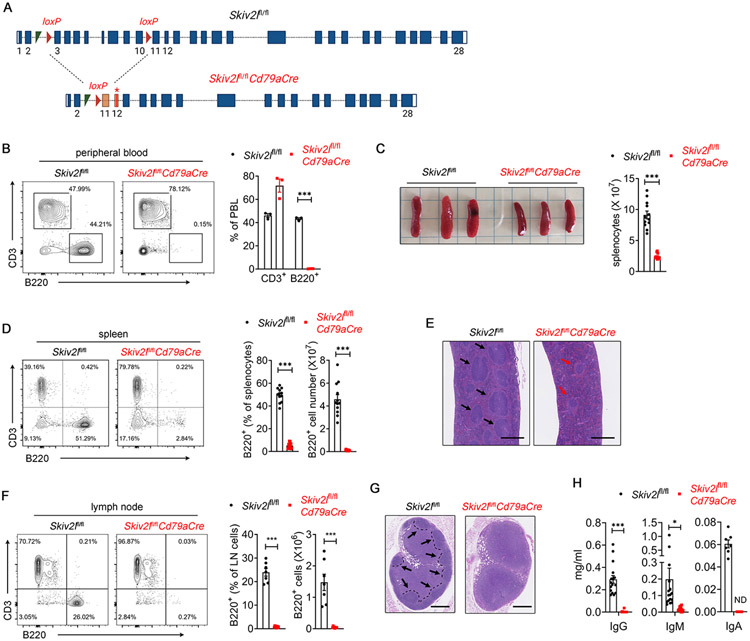
To model the human THES disease in vivo, we generated Skiv2l conditional allele mice and bred Skiv2lfl/fl with Cd79aCre mice to delete Skiv2l specifically at a very early stage of B cell development (Fig. 2A). Skiv2lfl/flCd79aCre mice (one Cd79a-driven Cre allele throughout the study) were born at the expected Mendelian ratio as littermate Skiv2lfl/fl mice. In the peripheral blood, B220+ B cells were completely lost in Skiv2lfl/flCd79aCre mice, whereas CD3+ T cells were relatively unaffected (Fig. 2B). Skiv2lfl/flCd79aCre mice also showed significantly reduced spleen size and splenocyte numbers (Fig. 2C). Flow cytometry analysis of splenocytes revealed a substantial decrease in B220+ B cells in Skiv2lfl/flCd79aCre mice compared with Skiv2lfl/fl controls (Fig. 2D). Histopathological analysis showed that B cell follicles were either absent or architecturally disorganized in Skiv2lfl/flCd79aCre spleen (Fig. 2E). Similarly, B220+ B cells were nearly undetectable in lymph nodes (Fig. 2F) and B cell follicles were absent in lymph node cortex of Skiv2lfl/flCd79aCre mice (Fig. 2G). Total serum immunoglobulin G (IgG), IgM and IgA levels in Skiv2lfl/flCd79aCre mice were undetectable or substantially reduced compared with Skiv2lfl/fl controls (Fig. 2H). These results collectively suggested that Skiv2l deficiency leads to B cell immunodeficiency in a cell-intrinsic manner in mice.
Fig. 2. B cell-specific deletion of Skiv2l leads to primary B cell immunodeficiency in mice.
(A) A schematic diagram showing Skiv2l loci (not to scale) of Skiv2lfl/fl and Skiv2lfl/flCd79aCre mice. Cre-mediated deletion of loxP-flanked exon 3-10 results in a frameshift (orange exon 11) and premature stop codon (asterisk in exon 12).
(B) Flow cytometry analysis of B220+ B cells and CD3+ T cells in peripheral blood of Skiv2lfl/flCd79aCre mice and Skiv2lfl/fl controls (n=3 mice per genotype). Data are shown as means ± SEM and are representative of at least two independent experiments. Unpaired t test, ***P < 0.001.
(C) Spleens of Skiv2lfl/flCd79aCre mice and Skiv2lfl/fl controls. Splenocyte number is shown on the right (n=12 mice per genotype). Data are shown as means ± SEM and are the pool of at least three independent experiments. Unpaired t test, ***P < 0.001.
(D) Flow cytometry analysis of B220+ B cells and CD3+ T cells in the spleen of Skiv2lfl/flCd79aCre mice and Skiv2lfl/fl controls. Percentage and absolute number of B220+ B cells in the spleen are shown on the right (n= 12 mice per genotype). Data are shown as means ± SEM and are the pool of at least three independent experiments. Unpaired t test, ***P < 0.001.
(E) Representative H&E staining of spleen of Skiv2lfl/flCd79aCre mice and Skiv2lfl/fl controls (n=3 mice per genotype). Black arrow, normal lymphoid follicles. Red arrow, disorganized lymphoid follicles. Scale bar, 500 μm.
(F) Flow cytometry analysis of B220+ B cells and CD3+ T cells in the inguinal lymph node of Skiv2lfl/flCd79aCre mice (n=8) and Skiv2lfl/fl controls (n=7). Percentage and the absolute number of B220+ B cells in inguinal lymph nodes are shown on the right. Data are shown as means ± SEM and are the pool of at least three independent experiments. Unpaired t test, ***P < 0.001.
(G) Representative H&E staining of inguinal lymph node of Skiv2lfl/flCd79aCre mice and Skiv2lfl/fl controls (n=3 mice per genotype). Follicles in the cortex are outlined with dashed lines. Black arrow, normal lymphoid follicles. Scale bar, 500 μm.
(H) Serum total IgG, IgM (n=16 mice per genotype) and IgA (n=7 mice per genotype) of 3-month-old Skiv2lfl/flCd79aCre mice and Skiv2lfl/fl controls. IgG, IgM and IgA were undetectable in 12, 2 and 7 samples of Skiv2lfl/flCd79aCre mice respectively, and their values were designated as zero arbitrarily in the graph. ND, not detectable. Data are shown as means ± SEM and are the pool of at least three independent experiments. Unpaired t test, *P < 0.05, ***P < 0.001.
B-1 B cell deficiency in Skiv2l-deficient mice
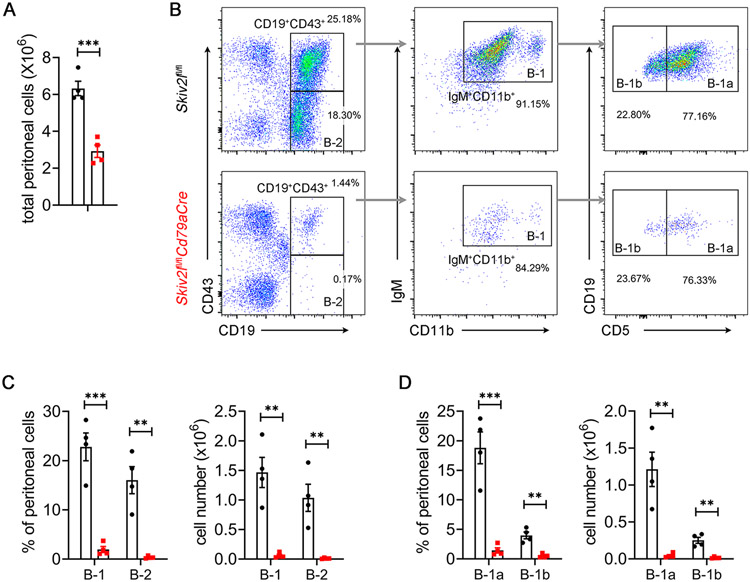
We further examined whether loss of SKIV2L affected long-lived, self-renewing innate-like B-1 B cells that are responsible for steady-state natural antibody production and rapid response to infections (11). We analyzed B-1 B cells in the peritoneal cavity, where the highest frequency of B-1 B cells are found in adult mice. Fewer peritoneal cavity cells were recovered from Skiv2lfl/flCd79aCre mice compared to Skiv2lfl/fl controls (Fig. 3A). Flow cytometry analysis revealed a substantial reduction of B-1 B cells (CD19+IgM+CD43+CD11b+) in the peritoneum of Skiv2lfl/flCd79aCre mice (Fig. 3B and 3C). Similar to the secondary lymphoid organs above, the peritoneal cavity had nearly undetectable conventional B-2 cells (CD19+IgM+CD43−) in Skiv2lfl/flCd79aCre mice (Fig. 3B and 3C). Among B-1 B cells, both CD5+ B-1a and CD5− B-1b subsets were decreased by the loss of SKIV2L (Fig. 3B and 3D). These results suggested that SKIV2L is also essential for the development of B-1 B cell lineage.
Fig. 3. B-1 B cell deficiency in Skiv2l-deficient mice.
(A) Total peritoneal cell number of Skiv2lfl/flCd79aCre mice and Skiv2lfl/fl littermate controls. n=4 mice per genotype. Data are shown as means ± SEM and are the pool of at least two independent experiments. Unpaired t test, ***P < 0.001.
(B) Flow cytometry analysis of B cell subset of peritoneal cells. Representative plot graph showing gating strategy of B cell subsets.
(C, D) Percentage and absolute numbers of different subsets of B cells in peritoneal cells. n = 4 mice per genotype. Data are shown as means ± SEM and are the pool of at least two independent experiments. Unpaired t test, **P < 0.01; ***P < 0.001.
Effect of Skiv2l deficiency on mature B cells
We further examined the effect of SKIV2L deficiency on mature B cells by depleting Skiv2l ex vivo. We bred Skiv2lfl/fl mice to an inducible UBC-Cre/ERT2 allele to generate Skiv2lfl/flUBC-Cre/ERT2 mice. Splenic B cells were isolated from Skiv2lfl/flUBC-Cre/ERT2 and Skiv2lfl/fl littermate mice (Fig. S2A) and treated with 4-OHT to induce Skiv2l gene deletion (Fig. S2B). Treatment with 4-OHT effectively depleted Skiv2l in Skiv2lfl/flUBC-Cre/ERT2 B cells but not in Skiv2lfl/fl cells (Fig. S2C). The proliferation of Skiv2l-deficient mature B cells after LPS stimulation was comparable to that of control cells (Fig. S2D and S2E). Loss of Skiv2l did not affect the viability of mature B cells (Fig. S2F and S2G). We further evaluated the impact of Skiv2l deficiency on IgG1 class switch recombination (CSR) after stimulation with LPS plus IL-4. The IgG1 CSR was indistinguishable between Skiv2l-deficient cells and controls (Fig. S2H and 2I). These results suggested Skiv2l deficiency does not affect proliferation and CSR of mature B cells after in vitro stimulation.
SKIV2L is required for pro-B to large pre-B transition in the bone marrow
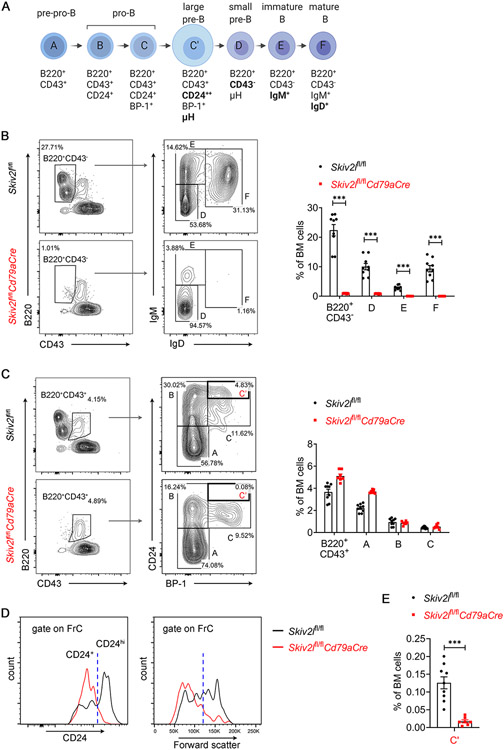
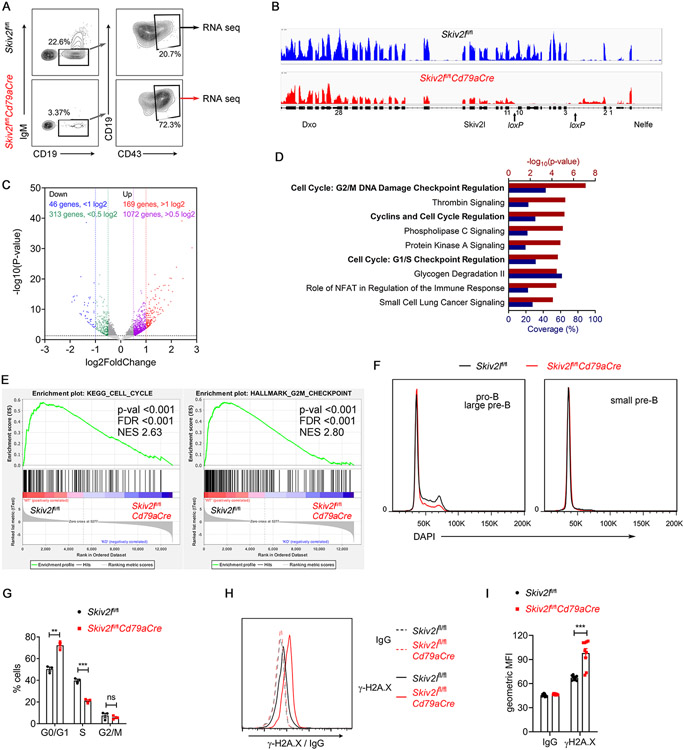
To pinpoint the stage at which conventional B cell development was perturbed by the loss of SKIV2L, we next analyzed early B cells in the bone marrow using Hardy Fraction nomenclature that includes seven discrete developmental stages in B lymphopoiesis (Fig. 4A). Comparing Skiv2lfl/flCd79aCre with Skiv2lfl/fl control bone marrow, we observed a dramatic reduction in B220+CD43− B cells that included small pre-B (B220+CD43−IgM−IgD−, D), immature (B220+CD43−IgM+IgD−, E) and mature (B220+CD43−IgM+IgD+, F) B cells, in Skiv2lfl/flCd79aCre mice (Fig. 4B). In contrast, Skiv2lfl/flCd79aCre mice showed a normal number of B220+CD43− B cells, of which the percentages of pre-pro B (B220+CD43+CD24−BP-1−, A) and pro B (B220+CD43+CD24+BP-1−, B) were also comparable to those of Skiv2lfl/fl counterparts (Fig. 4C). However, further analysis of CD24 expression level in Hardy Fraction C population (B220+CD43+CD24+BP-1+) revealed lack of CD24hi cells that are large pre-B cells (C’), in Skiv2lfl/flCd79aCre mice (Fig. 4D and 4E). Forward scatter that indicates cell size also showed that Skiv2lfl/flCd79aCre had markedly reduced number of Hardy Fraction C cells that were large in size (Fig. 4D). These results suggested a developmental block at the pro-B to large pre-B cell transition in Skiv2l-deficient mouse bone marrow.
Fig. 4. B cell developmental arrest at pro-B cells in Skiv2l-deficient mice.
(A) Diagram showing developmental stages of early B cells, Hardy fractions (A-F) and cell surface markers of each stage in the bone marrow.
(B, C) Flow cytometry analysis of B cell development in the bone marrow of Skiv2lfl/flCd79aCre mice (n=9) and Skiv2lfl/fl controls (n=7). B220+CD43+ cells were further analyzed for Hardy Fractions A-C (B) and B220+CD43− cells for FrD-F (C). Representative contour plots are shown in the left panel and percentages of each developmental stage in the bone marrow are shown in the right bar graphs. Data are shown as means ± SEM and are the pool of at least three independent experiments. Unpaired t test, ***P < 0.001.
(D) Representative histogram of CD24 expression (left panel) and forward scatter (right panel, indicates cell size) of FrC cells from Skiv2lfl/flCd79aCre mice and Skiv2lfl/fl controls.
(E) Percentages of B220+CD43+CD24hiBP-1+ large pre-B cells (FrC’) in the bone marrow of Skiv2lfl/flCd79aCre mice (n=9) and Skiv2lfl/fl controls (n=7). Data are shown as means ± SEM and are the pool of at least three independent experiments. Unpaired t test, ***P < 0.001.
To gain insights into the molecular mechanism by which SKIV2L regulated early B cell development, we performed RNA-seq transcriptomic analysis on pro-/large pre-B cells sorted from Skiv2lfl/flCd79aCre mice and Skiv2lfl/fl controls (Fig. 5A). Deletion of loxP-flanked Skiv2l exons in Skiv2lfl/flCd79aCre cells was confirmed in RNA-seq data (Fig. 5B). Differential gene expression analysis of mRNA (exclude Igh, analyzed separately below) revealed 1072 upregulated and 313 downregulated genes in Skiv2lfl/flCd79aCre cells with fold change greater than half log2, suggesting loss of SKIV2L was less likely to globally impact gene expression (Fig. 5C). Further ingenuity pathway analysis (IPA) of differentially expressed genes demonstrated an enrichment for pathways related to cell cycle regulation (Fig. 5D). Gene set enrichment analysis (GSEA) further revealed that cycle cell-related genes were downregulated in Skiv2lfl/flCd79aCre cells compared to Skiv2lfl/fl controls (Fig. 5E). Additionally, cell cycle analysis by flow cytometry showed increased numbers of Skiv2lfl/flCd79aCre pro-/large pre-B cells in the G0/G1 phase and concomitant decreased cells in S phase compared to Skiv2lfl/fl controls (Fig. 5F and 5G), suggesting a defective proliferation of Skiv2lfl/flCd79aCre early B cells. Moreover, we found genes involved in DNA damage response (Bbc3, Bcl2l2, Atf3 and Phlda3) and cell cycle (E2f2, Cdc25c, Cdca3, Hdac5 and Cks2) were significantly differentially expressed in Skiv2lfl/flCd79aCre early B cells (Fig. S3B), which confirmed the enriched pathways described above (Fig. 5D). We also stained cells with γ-H2A.X to detect DNA damage, and we found increased γ-H2A.X signal in Skiv2lfl/flCd79aCre pro-/large pre-B cells compared to Skiv2lfl/fl controls, consistent with elevated DNA damage response (Fig. 5H and 5I).
Fig. 5. Defective proliferation and DNA damage in Skiv2l-deficient early B cells.
(A) Representative plots showing sorting of pro-/large pre-B cells of Skiv2lfl/flCd79aCre mice versus Skiv2lfl/fl controls for RNA sequencing (n=3 mice per genotype).
(B) RNA sequencing IGV tracks showing Skiv2l deletion (exon 3-10) in Skiv2lfl/flCd79aCre pro/large pre-B cells cells compared to Skiv2lfl/fl control cells.
(C) Volcano plot showing differential mRNA expression (excluding Igh) in Skiv2lfl/flCd79aCre versus Skiv2lfl/fl early B cells (n=3 mice per genotype). Horizontal dashed line, P=0.05.
(D) Ingenuity Pathway Analysis of differentially expressed genes. Top pathways were ranked with −log10(P-value) (red). Coverage (blue) represents the percentage of the differentially expressed transcripts among the gene set of individual pathways.
(E) Gene set enrichment analysis (GSEA) of Skiv2lfl/fl and Skiv2lfl/flCd79aCre pro/large pre-B cells cells RNA-seq dataset. GSEA plots of KEGG cell cycle and hallmark G2M checkpoint are shown. NES, normalized enrichment score. FDR, false discovery rate.
(F, G) Cell cycle analysis of pro-/large pre-B cells in bone marrow of Skiv2lfl/flCd79aCre mice and Skiv2lfl/fl littermate controls (n=3 mice per genotype). Representative histograms are shown in (F). Quantitation of each cell cycle phase is shown in (G). Data are shown as means ± SEM and are representative of at least two independent experiments. Unpaired t test, **P < 0.01; ***P < 0.001; ns, not significant.
(H, I) Flow cytometry analysis of γ-H2A.X in pro-/large pre-B cells of Skiv2lfl/flCd79aCre mice versus Skiv2lfl/fl controls (n=9 mice per genotype). Representative histograms are shown in (H) and geometric mean fluorescence intensity (gMFI) of γ-H2A.X in (I). Data are shown as means ± SEM and are the pool of at least three independent experiments. Unpaired t test, ***P < 0.001.
To rule out the possibility that Cd79a-Cre may contribute to DNA damage either non-specifically or during recombination between loxP sites, we compared WT versus Cd79aCre/+ and Skiv2lfl/+ versus Skiv2lfl/+Cd79aCre mice. Neither Cd79aCre/+ nor Skiv2lfl/+Cd79aCre mice showed B-cell deficiency (Fig. S4A and S4D); we also did not detect increased expression of γ-H2A.X (Fig. S4B and 4E) and DNA damage response genes (Fig. S4C and 4F). Thus, DNA damage in Skiv2lfl/flCd79aCre B cells is due to loss of Skiv2l function and not Cd79a-Cre activity. Together, these data demonstrated that DNA damage and cell cycle arrest likely led to the developmental block observed in Skiv2l-deficient early B cells.
SKIV2L is required for V(D)J recombination and immunoglobulin μ heavy chain expression
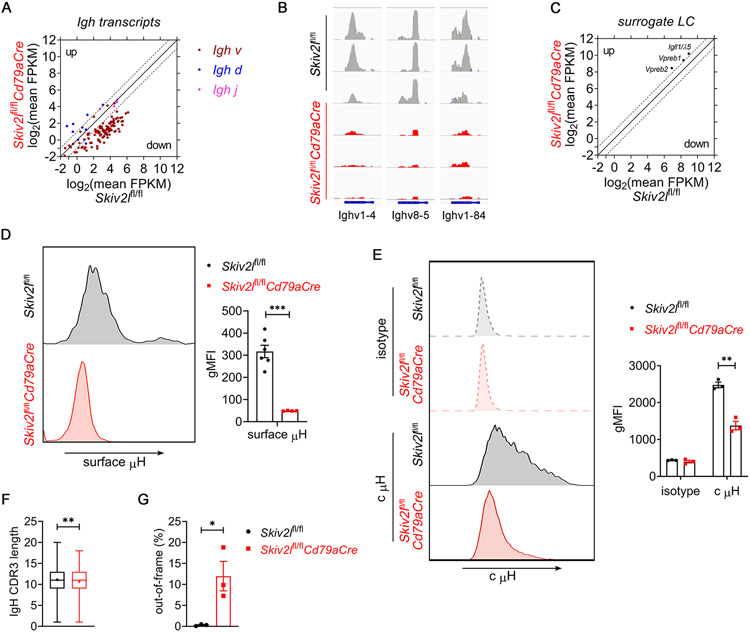
The pre-B cell stage is a critical checkpoint during B cell development that occurs after productive immunoglobulin heavy chain (Igh) rearrangement and precursor B cell receptor (pre-BCR) assembly from μ heavy chain (μH) and surrogate light chains λ5 and VpreB1/2. In the RNA-seq dataset of pro-/large pre-B cells, we found that Skiv2lfl/flCd79aCre pro-/large pre-B cells exhibited a global reduction in Igh expression, particularly V segments, compared to Skiv2lfl/fl counterpart (Fig. 6A and 6B, Fig. S3A). However, expression of surrogate light chains Igll1/λ5, Vpreb1 and Vpreb2 was not reduced in Skiv2lfl/flCd79aCre pro-/large pre-B cells (Fig. 6C). To validate the downregulation of Igh expression, we measured cell surface immunoglobulin μ heavy chain (μH). Flow cytometry analysis revealed that Skiv2lfl/flCd79aCre pro-/large pre-B cells expressed much less surface μH than Skiv2lfl/fl control cells (Fig. 6D). To distinguish whether reduced surface μH is caused by downregulation of μH production or defect in μH trafficking towards cell surface, we further examined cytoplasmic μH level. A significant reduction of cytoplasmic μH was observed in Skiv2lfl/flCd79aCre pro-/large pre-B cells, suggesting that production of μH was indeed decreased in the absence of SKIV2L (Fig. 6E).
Fig. 6. SKIV2L is required for Ig μ heavy chain expression during early B cell development.
(A) Scatter plot showing log2 of average fragment per kilobase per million (FPKM) of Igh transcripts in Skiv2lfl/flCd79aCre versus Skiv2lfl/fl control pro-/large pre-B cells. Data are shown as the mean of 3 biological replicates. Two-fold increase or decrease in gene expression is shown as dashed gray lines.
(B) Representative examples of downregulated Ighv expression in Skiv2lfl/flCd79aCre pro-/large pre-B cells compared to Skiv2lfl/fl control cells. n=3 mice per genotype.
(C) Scatter plot showing log2 of average FPKM of surrogate light chains Igll1/λ5, Vpreb1 and Vpreb2 transcripts in Skiv2lfl/flCd79aCre versus Skiv2lfl/fl control pro-/large pre-B cells. Data are shown as the mean of 3 biological replicates. Two-fold increase or decrease in gene expression is shown as dashed gray lines.
(D) Flow cytometry analysis of surface Ig μ heavy chain (μH) in pro-/large pre-B cells of Skiv2lfl/flCd79aCre mice (n=4) and Skiv2lfl/fl controls (n=6). Geometric mean fluorescence intensity (gMFI) of surface Igμ is shown in the right bar graphs. Data are shown as means ± SEM and are the pool of at least two independent experiments. Unpaired t test, ***P < 0.001.
(E) Flow cytometry analysis of cytoplasmic Ig μ heavy chain (c μH) in CD19+IgM−CD43+ pro-/large pre-B cells of Skiv2lfl/flCd79aCre mice (n=3) and Skiv2lfl/fl controls (n=3). Geometric mean fluorescence intensity (gMFI) of c μH is shown in the right bar graphs. Data are shown as means ± SEM and are representative of at least two independent experiments. Unpaired t test, **P < 0.01.
(F, G) Out-of-frame CDR3 (F) and CDR3 length (G) analysis of Igh in Skiv2lfl/flCd79aCre versus Skiv2lfl/fl pro-B cells. n=3 mice per genotype. “+” indicates mean in (F). Error bars indicate SEM (G). Unpaired t test, *P < 0.05, **P < 0.01.
During early B cell development, μH is expressed only when a productive VHDHJH recombination is completed on one of two Igh alleles in pro-B cells. We next used the TRUST4 algorithm (12) to reconstruct immune repertoire from bulk RNA-seq data. We found that Skiv2lfl/flCd79aCre pro-B cells exhibited a moderate but statistically significant decrease in CDR3 length and higher amount of out-of-frame CDR3 of Igh compared to control cells (Fig. 6F and 6G). These data suggested that SKIV2L RNA exosome activity was required for proper V(D)J recombination and productive μH expression at the pro-B cells stage in the bone marrow (Fig. 7).
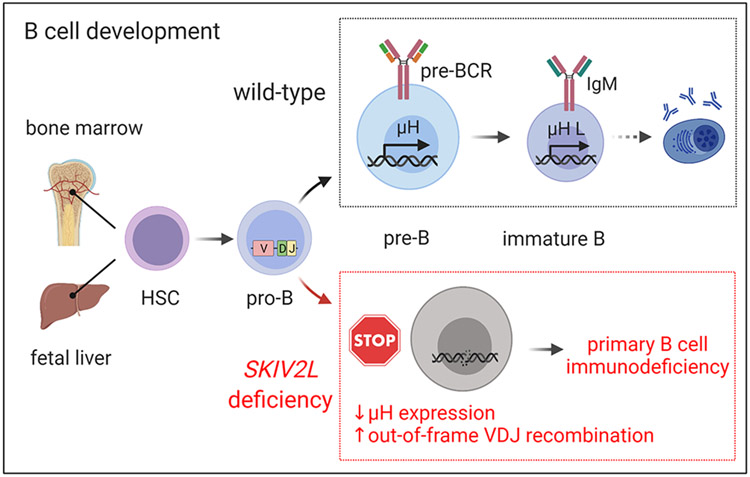
Figure 7. A model of primary B cell immunodeficiency in SKIV2L deficiency (THES2).
B cells develop from hematopoietic stem cells in fetal liver and adult bone marrow. Pro-B cells undergo V(D)J recombination and become large pre-B cells when μ heavy chains are expressed on the membrane and assemble the pre-BCR with surrogate light chains. Following proliferative burst of large pre-B cells, the resulting small pre-B cells rearrange Ig light chains that form IgM with μ heavy chains, and become immature B cells. In the absence of SKIV2L, pro-B cells failed to recombine V(D)J and could not express μ heavy chain expression, which led to B-cell developmental block at the pro-B cell stage and caused B cell deficiency in THES2 patients.
Discussion
A critical checkpoint during early B cell development is V(D)J recombination of the Igh locus and expression of rearranged μH on the cell surface to assemble the pre-BCR with surrogate light chains (13). Pre-BCR signaling is essential for the proliferative burst of large pre-B cells that is needed to expand the immune repertoire before light chain rearrangement. RAG1/2 DNA recombinases and DNA repair machinery are key players in this process. Here, we showed that RNA degradation machinery SKIV2L RNA exosome was also essential for this step of early B cell development, and SKIV2L loss-of-function led to severe B-cell immunodeficiency in human and mice.
Mutations in genes encoding the RNA exosome core complex and its cofactors are associated with human diseases, including pontocerebellar hypoplasia and THES (14). Although the genetic etiology of THES has been identified as mutations in SKI RNA exosome cofactor complex (either SKIV2L or TTC37), the disease mechanism is unknown. Approximately half of THES patients exhibit primary immunodeficiency, including the SKIV2L mutant THES2 patient we reported here. Of note, this THES2 patient still has some B cells in the peripheral blood and plasma cells in certain tissues, likely due to residual SKIV2L expression from c.3541-2A>C allele that locates in intron-exon junction. Early-onset intractable diarrhea and the resulting disruption of intestinal homeostasis may contribute to the elevation of IgA, which is mainly produced in the intestinal mucosa. While it is usually found in B-cell related neoplasm, Ig light chain restriction is reported in several autoimmune disorders, including Sjogren syndrome (15), myasthenia gravis (16), cold agglutinins hemolytic anemia (17), and Russell body gastritis/duodenitis (18). the light chain-restricted plasma cell response is thought to play a crucial role in autoreactive antibody production and the pathogenesis of autoimmune diseases. The kappa light chain restriction in this THES2 patient, who also developed autoimmune phenotypes including ANCA-associated vasculitis, may also represent a similar autoimmune response, which needs further investigation.
We generated aB-cell-specific Skiv2l knockout mouse model that revealed an essential role for SKIV2L RNA exosome activity in early B cell development, specifically at the pro-B to large pre-B cell transition in the bone marrow. Skiv2l-deficiency led to out-of-frame V(D)J recombination, decreased expression of Igh mRNA and surface μH protein, thus failed to pass this critical checkpoint of B cell development. We also observed enriched DNA damage and cell cycle pathways in Skiv2l-deficient early B cells. While defects in pre-BCR signaling could dampen cell proliferation of pre-B cells, DNA damage and p53/p21 activation may also lead to cell cycle arrest and impair B cell development. Consistent with this notion, premature activation of p53 signaling results in similar stage-specific blockage of B cell development (19). The RNA exosome core complex is recruited to programmed DSBs and participate in DNA repair during lymphocyte development, such as class switch recombination and somatic hypermutation of B cells (20-23).
Non-coding RNAs (ncRNAs) are key substrates of RNA exosome (23), and ncRNAs overlapping with DBSs need to be removed to allow accessibility for the DNA repair machinery (21). Although we did not investigate ncRNA in Skiv2l-deficient pro-B cells, the companion manuscript by Laffleur, Basu et al. analyzed pro-B cells lacking core components of the RNA exosome (Exosc3, Exosc10 or Dis3) (24). Loss-of-function of RNA exosome core subunits led to an accumulation of ncRNA from Igh loci, upregulation of p53 pathway, failed V(D)J recombination and arrest of early B cell development at the pro-B to pre-B transition. Previous molecular study of Dis3-deficient mature B cells demonstrates that accumulation of chromatin-associated ncRNA and subsequent Igh chromosomal architectural defects perturbs CTCF/cohesin binding to CTCF-binding elements at the Igh locus, and therefore decreases class-switch recombination (25). Abnormal CTCF/cohesion-mediated chromatin looping is associated with defects in V(D)J recombination (26, 27). It is likely that the accumulated ncRNA in RNA exosome deficient pro-B cells impairs Igh V(D)J recombination through a similar molecular mechanism. We found that knocking out Skiv2l in mature B cells did not affect their functions, including proliferation and class switch recombination. However, deletion of RNA exosome core component Exosc3 impairs class switch recombination through ncRNAs that target activation-induced cytidine deaminase (23). Moreover, MTR4, another helicase of RNA exosome, unwinds RNA exosome-sensitive ncRNAs in mature B cells and regulate class switch recombination of the IgH locus (28). Therefore, the mammalian RNA exosome likely uses different RNA helicases to safeguard recombination events in immature and mature B cells.
This study describes the human clinical pathophysiology of a THES patient and mouse cellular mechanisms of mammalian RNA exosome using cofactor Skiv2l-deficient mice. The companion manuscript by Laffleur, Basu et al. described a detailed molecular mechanism by which RNA exosome regulates V(D)J recombination using catalytic core (Exosc3, Exosc10, or Dis3) deficient mouse models (24). Both studies found the RNA exosome are required for early B cell development at the exact same stage of pro-B to pre-B transition. Collectively, these findings provide a clear and unified molecular mechanism for B cell immunodeficiency associated with THES, which will serve as the scientific base for further therapeutic development.
Materials and Methods
Study design
The objective of this study was to understand the role of mammalian SKIV2L RNA exosome in B cell development. We first characterized the immunological defect of a THES patient carrying SKIV2L mutations. We then generated B-cell-specific Skiv2l knockout mice to investigate the mechanism of B-cell deficiency. We performed multi-color flow cytometry to identify the developmental block of early B cells in the bone marrow. Further RNA sequencing of sorted early B cells and pathway enrichment analysis revealed cell cycle defect and DNA damage response. Last, we analyzed expression of immunoglobulin μ heavy chain in pre-B cells.
Human studies
Genetic, histological and immunological analyses of the affected patient were performed after informed consent was obtained. All human studies were reviewed and approved by the Institutional Review Board at UT Southwestern and have been carried out in accordance with the Declaration of Helsinki. When necessary, peripheral blood samples were collected to assess cell population during hospitalization. Flow cytometry analysis of Extended Lymphocyte Subset and Severe Combined Immunodeficiency (SCID) panels was performed in the clinical laboratory. The normal ranges of different immune cell populations at indicated age are from a previous study (10). Biopsies of the liver and duodenum were obtained and analyzed histologically. Lymphocyte Mitogens Proliferation (phytohemagglutinin, concanavalin A and pokeweed mitogen) was performed in ARUP Laboratories (Salt Lake City, UT). The Natural killer cell (NK) function test using chromium release methodology was performed in the Diagnostic Immunology Laboratory at Cincinnati Children's Hospital Medical Center (Cincinnati, OH).
Mice
Skiv2lfl/fl and Skiv2lfl/flUBC-Cre/ERT2 mice were generated as described previously (29). Skiv2lfl/fl mice were further crossed with B6.C(Cg)-Cd79atm1(cre)Reth/EhobJ mice (CD79a-Cre, Jackson Laboratory, Stock # 020505) to generate B-cell specific (Skiv2lfl/flCd79aCre) knockout mice. Four- to six-week-old Skiv2lfl/flCd79aCre mice and their Skiv2lfl/fl littermate controls were used for experiments. Both male and female animals were used in the study and sex variable did not affect experimental outcomes. All mice were housed in specific pathogen-free barrier facilities of UT Southwestern Medical Center. The animal protocol was approved by the Institutional Animal Care and Use Committee at UT Southwestern Medical Center (APN 2017-101968).
Flow cytometry
Antibodies used for flow cytometry are as following: APC anti-mouse CD19 (Clone 6D5, BioLegend, Cat # 115512), APC anti-mouse CD45R/B220 (Clone RA3-6B2, eBioscience, Cat # 17-0452-82), PE anti-mouse/human CD45R/B220 (Clone RA3-6B2, BioLegend, Cat # 103208), PE anti-mouse CD5 (Clone 53-7.3, eBioscience, Cat # 12-0051-82), PE anti-mouse CD3 (Clone 17A2, BioLegend, Cat # 100206), PE anti-mouse CD43 (Clone S11, BioLegend, Cat # 143205), BV421 anti-Mouse CD43 (Clone S7, BD Bioscience, Cat # 562958), FITC anti-mouse CD249 (BP-1) (Clone 6C3, eBioscience, Cat # 11-5891-81), PerCP/Cy5.5 anti-mouse CD24 (Clone M1/69, BioLegend, Cat # 101823), FITC anti-mouse IgM (Clone RMM-1, BioLegend, Cat # 406505), PE/Cy7 anti-mouse IgD, (Clone 11-26c.2a, BioLegend, Cat # 405719), FITC anti-mouse IgG1 (Clone RMG1-1, BioLegend, Cat # 406606), PerCP/Cy5.5 anti-mouse/human CD11b (Clone M1/70, BioLegend, Cat # 101228), PE Phospho-Histone H2A.X (Ser139) (20E3) Rabbit mAb (Cell Signaling, Cat # 5763), PE Rabbit (DA1E) mAb IgG XP® Isotype Control (Cell Signaling, Cat # 5742), R-Phycoerythrin AffiniPure F(ab')2 Fragment Goat Anti-Mouse IgM, μ chain specific (Jackson ImmunoResearch, Cat # 115-116-075).
Spleen or lymph node was mashed through 70 μm cell strainer into Petri dishes to generate single-cell suspension. Peritoneal cavity cells were collected from peritoneal lavage using ice-cold PBS with 0.5% BSA. Bone marrow (BM) cells were flushed from the femurs and tibias. Red blood cells were removed using ACK (Ammonium-Chloride-Potassium) lysis buffer. Cells were suspended in PBS containing 0.5% BSA (FACS buffer) and filtered through a 70 μm strainer. For cell surface staining, cells were incubated with antibodies in FACS buffer for 30 mins at 4°C and then washed with FACS buffer. Dead cells were excluded by using Zombie Aqua™ Fixable Viability Kit (BioLegend, Cat # 423102). For intracellular staining or cell cycle analysis using DAPI (4′,6-diamidino-2-phenylindole), cells were first stained for surface markers, then fixed in Cytofix/Cytoperm™ solution (BD, Cat # 554722) at 4°C for 30 mins, washed in perm/wash buffer (BD, Cat # 554723) and stained with antibodies for intracellular markers or DAPI (1 μg/ml) at 4°C for 30 mins. Data were collected on an LSRII flow cytometer (BD) and was analyzed using FlowJo software (version 10.8.1). All FACS gating strategies are shown in Fig. S5.
Histopathology
Mouse spleen and inguinal lymph node were fixed in 4% paraformaldehyde. Paraffin embedding, sectioning, and H&E staining were performed at UT Southwestern Medical Center Histo Pathology Core. H&E staining and immunohistochemistry of patient’s biopsies was performed by clinical pathologists in Children’s Medical Center at Dallas. Briefly, paraffin sections were deparaffinized in xylene and serial ethanol and rehydrated in water before hematoxalin staining. Sections were further stained with eosin staining and dehydrated in serial ethanol and xylene before covering. For immunohistochemistry, paraffin sections were deparaffinized in xylene and serial ethanol and rehydrated in water. Endogenous peroxidase activity was blocked using hydrogen peroxide. Epitope retrieval procedure was performed before incubation with primary antibodies. After incubation with biotinylated secondary antibody and HRP-labeled streptavidin, chromogens diaminobenzidine (DAB) was used to visualize the staining.
Splenic B cell isolation and stimulation
Splenic B cells were isolated with the B Cell Isolation Kit (Miltenyi Biotec, cat # 130-090-862) using autoMACS® Pro Separator. Isolated splenic B cells were cultured in RPMI-1640 medium supplemented with 10% FBS, penicillin/streptomycin antibiotics, 2 mM L-glutamine, and β-mercaptoethanol (50 μM). For ex vitro deletion of Skiv2l, isolated B cells were treated with 100 nM 4-OHT (Sigma, cat # H7904) or vehicle (ethanol) for 24 h, then washed to remove 4-OHT for the following stimulation. Cell viability was assessed using Annexin V/PI staining (BioLegend, cat # 640914). For proliferation assay, cells were loaded with 5 μM CellTrace™ Violet (Thermo Fisher, Cat# C34557) and dilution of the dye in B cells after LPS (20 μg/ml) stimulation was analyzed by flow cytometry. For assay of class switch recombination to IgG1, B cells were treated with LPS (20 μg/ml) and IL-4 (20 ng/ml) for 72 h. Cells were analyzed by flow cytometry using B220 and anti-IgG1 antibodies.
RNA sequencing and bioinformatics analysis
Cell sorting of bone marrow pro-/large pre-B cells was performed using a FACS Aria sorter (BD) at Flow Cytometry Core of UT Southwestern Medical Center. Briefly, BM cells were first depleted of red blood cells (Ter119), myeloid cells (Gr-1), T cells (CD3ε, CD4, CD8a), and NK cells (CD49b) using Pan B Cell Isolation Kit II, mouse (Miltenyi, Cat # 130-104-443) on autoMACS Pro separator (Miltenyi), and then stained with fluorophore-labeled APC anti-mouse CD19, FITC anti-mouse IgM, and PE anti-mouse CD43 antibodies. Pro-/large pre-B cells were sorted as CD19+IgM−CD43+ cells and freshly sorted cells were used for RNA isolation and RNA-seq.
Total RNA was isolated from sorted pro/large pre-B cells or peripheral blood mononuclear cells from the patient and healthy individuals using RNeasy Mini Kit (QIAGEN, Cat # 74104). RNA integrity was measured by Bioanalyzer (Agilent). The mRNAseq-Strand specific Library was prepared and sequenced at Genomics Sequencing & Microarray Core of UT Southwestern Medical Center. Raw sequencing data were processed with Astrocyte RNASeq Analysis Workflows at UT Southwestern Bioinformatics Core Facility (https://astrocyte.biohpc.swmed.edu/workflow/12/version/324/docs) to generate Fragments Per Kilobase Million (FPKM) for each transcript. Differential gene expression analysis was performed using the DESeq2 package in Astrocyte RNASeq Analysis Workflows. Volcano plot showing differentially expressed genes was generated using Graphpad Prism. Differentially expressed genes (Skiv2lfl/flCd79aCre/ Skiv2lfl/fl >0.5log2 or <0.5log2) were analyzed using Ingenuity pathway analysis (IPA) software package (QIAGEN Bioinformatics). For Gene Set Enrichment Analysis (GSEA), protein-coding transcripts of total RNA sequencing dataset was run on GSEA 4.1.0 software (Broad Institute) using H: hallmarks gene sets database or C2: KEGG subset of canonical pathways (MSigDB), with 1000 “gene_set” permutations. The gene set size was set between 15 and 500, and t test was used as the metric for ranking genes.
RNA isolation and RT-qPCR
Total RNA was isolated from cells using TRI reagent (Sigma) per manufacturer’s instruction, and cDNA were synthesized with iScript cDNA Synthesis Kit (Bio-Rad). iTaq Universal SYBR Green Supermix (Bio-Rad) was used to quantify mRNA expression with CFX96™ Real-Time PCR Detection System. Gene expression was normalized internal controls. Primer sequences used were as following: hCD19, Fwd 5’- GGCAACCTGACCATGTCATT-3’, Rev 5’-TCACAGCTGAGACCTTCCAG-3’; hCD40, Fwd 5’- TATGGTTCGTCTGCCTCTGC-3’, Rev 5’-GGCACAAAGAACAGCACTGA-3’; hCD79A, Fwd 5’- GAGTCCTCCAAGCTCTGCCT-3’, Rev 5’-ATCAATGATGCTGGGACCTT-3’; hBLNK, Fwd 5’- CCGCCAGTCAGAAGTTGAG-3’, Rev 5’-CTTCGAGGAACACTTGGAGG-3’; hTNFRSF13B, Fwd 5’- TGAGTAATGAGTGGCCTGGG-3’, Rev 5’-GGCAGGATCTCATAGCCACC-3’; mCdkn1a, Fwd 5’- GTTCCTTGCCACTTCTTACCT-3’, Rev 5’-TCATCCTAGCTGGCCTTAGA-’3; mPhlda3, Fwd 5’- TCATCATGGAAGGAGCCAAC-3’, Rev 5’- CGCAAGTCTGAGCTGGATATAA-3'; mBbc3, Fwd 5’- CTGGAGGGTCATGTACAATCTC-3’, Rev 5’- ACCTAGTTGGGCTCCATTTC-3’.
Enzyme-linked immunosorbent assay (ELISA)
Total serum IgG and IgM of 3-month-old Skiv2lfl/flCd79aCre mice and Skiv2lfl/fl littermates were measured by using IgG (Total) Mouse Uncoated ELISA Kit (Thermo Fisher, Cat #88-50400-22) and IgM Mouse Uncoated ELISA Kit (Thermo Fisher, Cat # 88-50470-22) following manufacture’s instruction. The detection limits for IgG and IgM are 1.56 and 0.39 ng/ml, respectively.
Western blotting
Western blotting was performed as previously described (29). Briefly, cultured or sorted cells were lysed in RIPA buffer, and cell lysate was quantified using BCA. Proteins were separated on SDS-PAGE and transferred to nitrocellulose membrane. Blotting membranes were blocked with 5% nonfat milk and incubated with diluted primary antibodies at 4°C overnight per the manufacturers’ instructions. After washing with TBST, membranes were further incubated with HRP-conjugated secondary antibody (Bio-Rad). SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific) was used to develop the blots on film. Primary antibodies used were as follows: SKIV2L Rabbit Polyclonal Antibody (Proteintech, catalog 11462-1-AP, 1:500), Cre Recombinase (D7L7L) XP® Rabbit mAb (Cell Signaling, cat # 15036, 1:1000), Phospho-Histone H2A.X (Ser139) (20E3) Rabbit mAb (Cell Signaling, cat # 9718, 1:1000), Anti-α-Tubulin antibody, Mouse monoclonal (Sigma, catalog T6199, 1:10,000), Anti-β-Actin antibody, Mouse monoclonal (Sigma, cat # A1978, 1:10,000).
Statistical analysis
Graphpad Prism (version 9.3.0) was used for statistical analysis. GSEA 4.1.0 software (Broad Institute) was used for gene set enrichment analysis with H: hallmarks gene sets database or C2: KEGG subset of canonical pathways (MSigDB). DESeq2 was used for differential gene expression analysis. Flow cytometry data were analyzed using FlowJo version 10.8.1 (Tree Star).The number of individual mice analyzed per genotype or condition was indicated in each figure legend. Student’s unpaired two-tailed t test, Chi-square test were performed for comparison as indicated in each figure legend. Numerical data in bar graphs were shown as mean ± SEM. Statistical significance was identified with *P < 0.05, **P < 0.01, ***P < 0.001.
Supplementary Material
Acknowledgments
We thank the THES2 patient and her parents for participating in the study, Wanwei Zhang and Brice Laffleur (Columbia University) for help with bioinformatics analysis of RNA-seq data, members of the Yan lab for helpful discussion.
Funding:
This work was supported by the NIH (AI153576 to N.Y.) and the Burroughs Wellcome Fund (N.Y.).
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The RNA sequencing data in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession numbers GSE186477 and GSE188795. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
Reference and notes
- 1.Houseley J, LaCava J, Tollervey D, RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol 7, 529–539 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Januszyk K, Lima CD, The eukaryotic RNA exosome. Curr. Opin. Struct. Biol 24, 132–140 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kilchert C, Wittmann S, Vasiljeva L, The regulation and functions of the nuclear RNA exosome complex. Nat. Rev. Mol. Cell Biol 17, 227–239 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Halbach F, Reichelt P, Rode M, Conti E, The yeast ski complex: crystal structure and RNA channeling to the exosome complex. Cell 154, 814–826 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Fabre A, Charroux B, Martinez-Vinson C, Roquelaure B, Odul E, Sayar E, Smith H, Colomb V, Andre N, Hugot JP, Goulet O, Lacoste C, Sarles J, Royet J, Levy N, Badens C, SKIV2L mutations cause syndromic diarrhea, or trichohepatoenteric syndrome. Am. J. Hum. Genet 90, 689–692 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartley JL, Zachos NC, Dawood B, Donowitz M, Forman J, Pollitt RJ, Morgan NV, Tee L, Gissen P, Kahr WH, Knisely AS, Watson S, Chitayat D, Booth IW, Protheroe S, Murphy S, de Vries E, Kelly DA, Maher ER, Mutations in TTC37 cause trichohepatoenteric syndrome (phenotypic diarrhea of infancy). Gastroenterology 138, 2388–2398, 2398 e2381-2382 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourgeois P, Esteve C, Chaix C, Beroud C, Levy N, Fabre A, Badens C, Tricho-Hepato-Enteric Syndrome mutation update: Mutations spectrum of TTC37 and SKIV2L, clinical analysis and future prospects. Hum. Mutat 39, 774–789 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Vely F, Barlogis V, Marinier E, Coste ME, Dubern B, Dugelay E, Lemale J, Martinez-Vinson C, Peretti N, Perry A, Bourgeois P, Badens C, Goulet O, Hugot JP, Farnarier C, Fabre A, Combined Immunodeficiency in Patients With Trichohepatoenteric Syndrome. Front. Immunol 9, 1036 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hosking LM, Bannister EG, Cook MC, Choo S, Kumble S, Cole TS, Trichohepatoenteric Syndrome Presenting with Severe Infection and Later Onset Diarrhoea. J. Clin. Immunol 38, 1–3 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Shearer WT, Rosenblatt HM, Gelman RS, Oyomopito R, Plaeger S, Stiehm ER, Wara DW, Douglas SD, Luzuriaga K, McFarland EJ, Yogev R, Rathore MH, Levy W, Graham BL, Spector SA, Pediatric ACTG, Lymphocyte subsets in healthy children from birth through 18 years of age: the Pediatric AIDS Clinical Trials Group P1009 study. J. Allergy Clin. Immunol 112, 973–980 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Smith FL, Baumgarth N, B-1 cell responses to infections. Curr. Opin. Immunol 57, 23–31 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song L, Cohen D, Ouyang Z, Cao Y, Hu X, Liu XS, TRUST4: immune repertoire reconstruction from bulk and single-cell RNA-seq data. Nat. Methods 18, 627–630 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herzog S, Reth M, Jumaa H, Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat. Rev. Immunol 9, 195–205 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Morton DJ, Kuiper EG, Jones SK, Leung SW, Corbett AH, Fasken MB, The RNA exosome and RNA exosome-linked disease. RNA 24, 127–142 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pollard KM, Toumasatos V, Furphy L, Webb J, Immunohistologically definable light chain restriction in autoimmune disease. J. Pathol 156, 349–352 (1988). [DOI] [PubMed] [Google Scholar]
- 16.Knight JG, Laing P, Adams DD, Bray JJ, Ling NR, Autoantibodies to acetylcholine receptor in myasthenia gravis: light chains. Neurology 36, 1531–1533 (1986). [DOI] [PubMed] [Google Scholar]
- 17.Harboe M, Lind K, Light chain types of transiently occurring cold haemagglutinins. Scand. J. Haematol 3, 269–276 (1966). [DOI] [PubMed] [Google Scholar]
- 18.Zhang H, Jin Z, Cui R, Russell body gastritis/duodenitis: a case series and description of immunoglobulin light chain restriction. Clin. Res. Hepatol. Gastroenterol 38, e89–97 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Cantor DJ, King B, Blumenberg L, DiMauro T, Aifantis I, Koralov SB, Skok JA, David G, Impaired Expression of Rearranged Immunoglobulin Genes and Premature p53 Activation Block B Cell Development in BMI1 Null Mice. Cell Rep. 26, 108–118 e104 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marin-Vicente C, Domingo-Prim J, Eberle AB, Visa N, RRP6/EXOSC10 is required for the repair of DNA double-strand breaks by homologous recombination. J. Cell Sci 128, 1097–1107 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Domingo-Prim J, Endara-Coll M, Bonath F, Jimeno S, Prados-Carvajal R, Friedlander MR, Huertas P, Visa N, EXOSC10 is required for RPA assembly and controlled DNA end resection at DNA double-strand breaks. Nat. Commun 10, 2135 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Basu U, Meng FL, Keim C, Grinstein V, Pefanis E, Eccleston J, Zhang T, Myers D, Wasserman CR, Wesemann DR, Januszyk K, Gregory RI, Deng H, Lima CD, Alt FW, The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell 144, 353–363 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pefanis E, Wang J, Rothschild G, Lim J, Chao J, Rabadan R, Economides AN, Basu U, Noncoding RNA transcription targets AID to divergently transcribed loci in B cells. Nature 514, 389–393 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laffleur B, Batista CR, Zhang W, Lim J, Yang B, Rossille D, Wu L, Estrella J, Rothschild G, Pefanis E, Basu U, RNA exosome drives early B cell development via non-coding RNA processing mechanisms. Sci. Immunol xx, xx (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laffleur B, Lim J, Zhang W, Chen Y, Pefanis E, Bizarro J, Batista CR, Wu L, Economides AN, Wang J, Basu U, Noncoding RNA processing by DIS3 regulates chromosomal architecture and somatic hypermutation in B cells. Nat. Genet 53, 230–242 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Zhang X, Ba Z, Liang Z, Dring EW, Hu H, Lou J, Kyritsis N, Zurita J, Shamim MS, Presser Aiden A, Lieberman Aiden E, Alt FW, The fundamental role of chromatin loop extrusion in physiological V(D)J recombination. Nature 573, 600–604 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ba Z, Lou J, Ye AY, Dai HQ, Dring EW, Lin SG, Jain S, Kyritsis N, Kieffer-Kwon KR, Casellas R, Alt FW, CTCF orchestrates long-range cohesin-driven V(D)J recombinational scanning. Nature 586, 305–310 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim J, Giri PK, Kazadi D, Laffleur B, Zhang W, Grinstein V, Pefanis E, Brown LM, Ladewig E, Martin O, Chen Y, Rabadan R, Boyer F, Rothschild G, Cogne M, Pinaud E, Deng H, Basu U, Nuclear Proximity of Mtr4 to RNA Exosome Restricts DNA Mutational Asymmetry. Cell 169, 523–537 e515 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang K, Han J, Asada M, Gill JG, Park JY, Sathe MN, Gattineni J, Wright T, Wysocki CA, de la Morena MT, Garza LA, Yan N, Cytoplasmic RNA quality control failure engages mTORC1-mediated autoinflammatory disease. J. Clin. Invest 132, e146176 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.