Abstract
Two distinct regions of DNA encode the enzymes needed for phthalate degradation by Burkholderia cepacia DBO1. A gene coding for an enzyme (quinolinate phosphoribosyl transferase) involved in the biosynthesis of NAD+ was identified between these two regions by sequence analysis and functional assays. Southern hybridization experiments indicate that DBO1 and other phthalate-degrading B. cepacia strains have two dissimilar genes for this enzyme, while non-phthalate-degrading B. cepacia strains have only a single gene. The sequenced gene was labeled ophE, due to the fact that it is specifically induced by phthalate as shown by lacZ gene fusions. Insertional knockout mutants lacking ophE grow noticeably slower on phthalate while exhibiting normal rates of growth on other substrates. The fact that elevated levels of quinolinate phosphoribosyl transferase enhance growth on phthalate stems from the structural similarities between phthalate and quinolinate: phthalate is a competitive inhibitor of this enzyme and the phthalate catabolic pathway cometabolizes quinolinate. The recruitment of this gene for growth on phthalate thus gives B. cepacia an advantage over other phthalate-degrading bacteria in the environment.
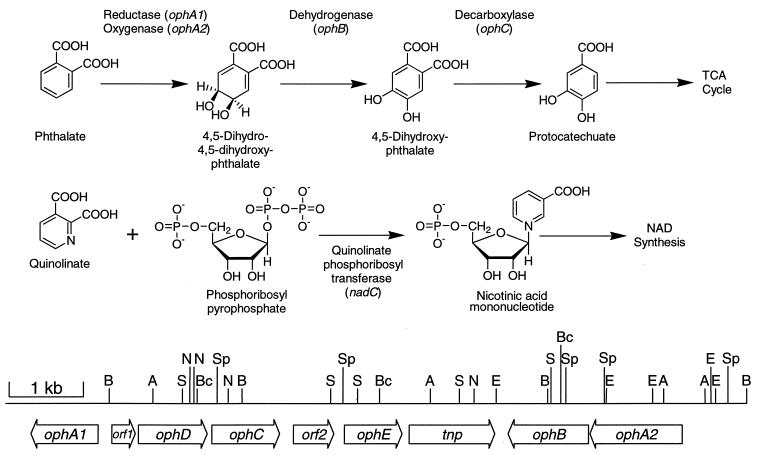
Phthalate is a ubiquitous compound in the environment due to its widespread use not only in the manufacture of plastics and textiles but also as an ingredient in pesticide, munitions, and cosmetic formulations (22, 47). There is some concern over the health effects of phthalate-based compounds, as they have been shown to be both nervous system depressants and stimulators, teratogenic, and estrogen mimics (3, 6, 17, 26, 63, 66). Although many different types of microorganisms have been shown to readily degrade phthalate (31, 44, 50) there have not been many studies on the toxicity of phthalate to microorganisms. Perhaps the best studied phthalate-degrading organism is Burkholderia cepacia DBO1, for which extensive work has been performed on the enzymes and genes involved in the catabolic pathway (2, 10, 31, 48). DBO1 initiates the degradation of phthalate (Fig. 1) through dioxygenase attack to form 4,5-dihydro-4,5-dihydroxyphthalate (cis-phthalate dihydrodiol). The enzyme responsible for this initial step is a two-component enzyme consisting of an oxygenase, which actually catalyzes the addition of oxygen to phthalate, and a reductase, which shuttles electrons from NADH to the oxygenase component (2). Phthalate catabolism continues through the action of a dehydrogenase, which restores the aromatic character of the ring, and a decarboxylase (48), which removes one of the two carboxyl groups. The resulting compound, protocatechuate, then enters the central aromatic catabolic β-ketoadipate pathway (72, 73). The DBO1 genes encoding the enzymes for the conversion of phthalate to protocatechuate have recently been cloned and sequenced (10). Interestingly, the four genes are organized into three operons (Fig. 1) with the two genes for the first catabolic enzymes situated on the two outside ends, approximately 7 kb from each other. In between two of the operons is a 4-kb stretch of DNA, which is not needed for the conversion of phthalate to protocatechuate but is involved in the ability of this strain to grow on phthalate as described in this report. The gene organization that is found in B. cepacia DBO1 is quite different from that described for Pseudomonas putida NMH102-2 (43), in which similar genes for phthalate degradation are present in an operonic structure.
FIG. 1.
Metabolic pathways for the degradation of phthalate and the synthesis of nicotinic acid mononucleotide from quinolinate and phosphoribosyl pyrophosphate. A restriction map and diagram of the cloned and sequenced genes for phthalate degradation (including the nadC analogue ophE reported here) from B. cepacia DBO1 are shown at the bottom. A, AatII; B, BamHI; Bc, BclI; E, EcoRI; N, NotI; S, SalI; Sp, SphI. TCA, tricarboxylic acid.
Aromatic dioxygenases often have a broad substrate range, being able to attack compounds other than the pathway substrate(s). The best-studied examples of this are toluene and naphthalene dioxygenase (49, 71). Phthalate dioxygenase is able to transform other dicarboxylated aromatic compounds to oxygenated products. One example is the transformation of quinolinate (Fig. 1), a dicarboxylated pyridine (42, 59, 60). Since quinolinate is an intermediate in NAD+ synthesis this could be deleterious to the cell since quinolinate pools could be depleted and possibly deleterious dihydroxylated intermediates could form. In addition, because of the similarity in structure between quinolinate and phthalate, quinolinate phosphoribosyl transferase (EC 2.4.2.19) is competitively inhibited by phthalate (4), and thus the pathway for NAD+ synthesis is inhibited. Since phthalate is a negatively charged compound it does not readily diffuse into the bacterial cell, and thus this competitive inhibition is not normally seen in environments where phthalate is present. However, bacteria utilizing phthalate as a carbon source would actively be transporting phthalate into the cell (10), and the combination of effects of phthalate dioxygenase on quinolinate and phthalate on quinolinate phosphoribosyl transferase could result in a loss of competitiveness of the bacterium in the environment. The present work describes one mechanism devised by B. cepacia DBO1 to overcome this potential difficulty.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth of strains.
B. cepacia DBO1 (ATCC 29424) is the wild-type strain capable of utilizing phthalate as the sole carbon and energy source (30). B. cepacia ATCC 17616 (57) is a phthalate-degrading strain isolated by other investigators independently of DBO1. B. cepacia ATCC 17759 (37) and ATCC 25416 (51), clinical isolates 715j (38), K56-2 (39), and K63-3 (39), and field isolates D1 and M53 (34a) cannot utilize phthalate. Escherichia coli DH5α [F− φ80dlacZΔM15 Δ(lacZY-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) supE44 thi-1 gyrA96 relA1] (Gibco-BRL, Gaithersburg, Md.) was used as the recipient strain in the cloning experiments. E. coli WC4546 [nadC8 galT23 IN(rrnD-rrnE)1] (62) was obtained from the E. coli Genetic Stock Center (Yale University, New Haven, Conn.). The pGEM series of vectors (Promega, Madison, Wis.) and pRK415 (29) were used for subcloning DNA fragments. pUC4K with a kanamycin resistance gene cassette was obtained from Pharmacia Biotech (Uppsala, Sweden). pARO180 is a mobilizable narrow-host-range plasmid (46). The promoter probe vector pKRZ-1 has a promoterless lacZ gene cloned into the broad-host-range vector pUCD615 (52). Plasmid pRK2013 was used as a helper strain in mating experiments (19).
L broth (35) was used as the complete medium. Mineral salts basal medium (MSB) (57) was used as the minimal medium. Basal medium (BM) (72) was used for growth curves of B. cepacia DBO1 mutants. Carbon sources were added to the medium at concentrations of 20 mM for succinate, 10 mM for p-hydroxybenzoate, and 10 mM for phthalate. Ampicillin and tetracycline were added at concentrations of 100 μg/ml and 15 μg/ml, respectively, when needed. Nicotinate (10 μg/ml) was added to the minimal medium for culturing E. coli WC4546. Burkholderia strains were grown at 30°C, and E. coli strains were grown at 37°C.
Molecular techniques.
Genomic and plasmid DNA was prepared by established procedures (5, 45). Restriction digests, ligations, transformation, gel electrophoresis, DNA extraction from gels, probe labeling, Southern hybridizations, and automated DNA sequencing were performed following standard procedures as described previously (10, 18, 23, 33, 65). The 0.9-kb probe for the ophE gene was PCR amplified under standard conditions (Perkin-Elmer, Inc., Foster City, Calif.) from pGJZ1331 with the SP6 sequencing primer (Promega) and the internal primer 5′-GCGAAATACGGTCCAC-3′. A modification of the electrotransformation method for Pseudomonas (15) was used to introduce DNA into B. cepacia. Cells (12 ml) were grown to mid- to late-log phase, harvested by centrifugation at 5,000 × g at room temperature, washed in an equal volume of 10% glycerol, and resuspended in 160 μl of the same buffer. DNA (0.2 to 0.5 μg) was added to 40 μl of cells, the solution was incubated at room temperature for 5 min, and an electrical pulse was applied. The electroporator (Gene Pulser; Bio-Rad Laboratories, Rockville Center, N.Y.) was set at 25 μF, 200 Ω, and 1.25 kV for 0.1-cm gap cuvettes. SOC solution (0.5 ml) (53) was added immediately, and the cells were incubated at 30°C for 1 h before being plated on a selective medium.
Construction of an ophE knockout mutant.
The ophE gene was knocked out with a kanamycin resistance cassette. Initially a 4.5-kb BamHI fragment containing ophE from pGJZ1301 (10) was cloned into pGEM7Z-f(−). A 1.3-kb BamHI kanamycin resistance cassette derived from pUC4K was inserted into the unique BclI site in ophE. The BamHI fragment (now 5.8 kb) containing the disrupted ophE gene was moved to the mobilizable suicide vector pARO180. The resulting construct, designated pGJZ1332, was transferred by triparental mating into DBO1 with selection on a minimal medium supplemented with succinate, nicotinate, and kanamycin. Southern hybridization of restriction digested genomic DNA was used to identify clones which had single-crossover (DBO301) or double-crossover (DBO302) recombination events or had become spontaneously kanamycin resistant (DBO303) without the insertion of the resistance gene.
Promoter assays.
The PCR procedure for amplifying the region upstream of the ophE gene involved a step at 94°C for 1 min, 25 repeated cycles of 15 s at 94°C, 30 s at 50°C, and 4 min at 60°C, a 10-min step at 72°C, and a step at 4°C until the tube was removed. Primers HK-A2 (5′-ATTCCGTCGACTCGGGAAG-3′) and HK-B (5′-GCTCTAGAATGTTTCGTCGAACC-3′) were utilized. Primer HK-A2 includes a SalI site (underlined), while a new XbaI site (underlined) was included in primer HK-B. The PCR product was cleaved with SalI and XbaI and cloned into pKRZ-1. The cloned region was sequenced to verify that no changes were introduced by the Taq polymerase. LacZ assays were performed with cells in mid-log phase. Harvested cells were washed twice with phosphate buffer (50 mM sodium/potassium phosphate buffer, pH 7.25) and resuspended at 0.1 times the original volume. The cells were sonicated at 475 W at 4°C for 3 min with intermittent pulsing (1 s on and 3 s off).
A modified β-galactosidase assay (40) was used to measure LacZ activity in cell-free extracts. The crude cell extract (30 μl) was added to 370 μl of buffer Z (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, and 40 mM 2-mercaptoethanol) in a 1.5-ml microcentrifuge tube. o-Nitrophenyl-β-d-galactopyranoside (100 μl of a 4 mg/ml stock) in buffer Z was added to initiate the enzyme reaction, and the mixture was incubated at 37°C for 30 min. The reaction was stopped by adding 500 μl of 1 M sodium carbonate to the mixture. Enzyme activity was calculated as nanomoles of o-nitrophenol formed per minute per milligram of protein. The protein concentration was measured by the Bradford procedure (7) with bovine serum albumin as the standard.
Nucleotide sequence accession number.
The nucleotide sequence has been deposited in the GenBank database under accession no. AF095748.
RESULTS
Identification of a nadC-like gene associated with the genes for phthalate degradation.
The four genes coding for the initial steps in phthalate degradation (Fig. 1) are clustered in two groups approximately 4 kb apart (10). The intervening region was sequenced, and two open reading frames were identified (Fig. 1). One of these open reading frames, designated tnp, shows a high degree of similarity to known transposases (data not shown). The enzyme encoded by the second open reading frame shows a high degree of similarity to quinolinate phosphoribosyl transferase (NadC) from several different sources (Fig. 2). The question as to why a gene involved in the synthesis of NAD+ would be physically associated with the genes for phthalate degradation is the subject of this investigation. This nadC-like gene has been designated ophE, due to its physical association with the oph genes for the degradation of phthalate and its role in phthalate degradation as described below.
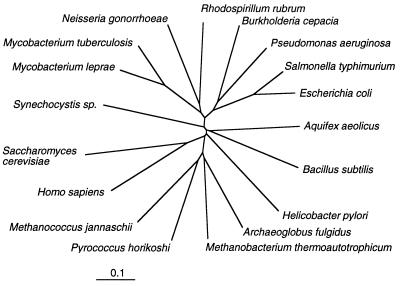
FIG. 2.
Dendrogram showing the phylogenetic relationship of different genes coding for quinolinate phosphoribosyl transferase (nadC or ophE). The nucleotide sequences were aligned with the pileup program of the Genetics Computer Group package (16), the alignment was confirmed by visual inspection and comparison with the deduced amino acid sequence alignments, and the phylogenetic tree was calculated with the PAUP program by using the minimal distance method. The nucleotide sequences were obtained from the following sources: Aquifex aeolicus (13), Archaeoglobus fulgidus (34), Bacillus subtilis, (58), E. coli (68), Helicobacter pylori (61), Homo sapiens, (20), Methanococcus jannaschii (8), Mycobacterium leprae (unpublished data; GenBank accession no. U00010), Methanobacterium thermoautotrophicum (56), Mycobacterium tuberculosis (12), N. gonorrhoeae (51a), P. aeruginosa (47a), Pyrococcus horikoshi (28), Rhodospirillum rubrum, (55), Saccharomyces cerevisiae (41), S. typhimurium (24), and Synechocystis sp. (27).
In order to prove that the identified ophE gene encodes a protein with NadC activity, mutant complementation experiments were performed. A 1.2-kb SphI-AatII fragment containing the entire ophE gene was subcloned into pGEM5Z-f(−). The resulting plasmid, designated pGJZ1331, was introduced into the nadC E. coli strain WC4546. This strain requires nicotinate in order to grow on a minimal medium due to the block in its NAD+ synthesis pathway. However, WC4546(pGJZ1331) is able to grow on MSB medium containing succinate without nicotinate, demonstrating that ophE is able to complement the nadC mutation and thus that the encoded enzyme has quinolinate phosphoribosyl transferase activity. Control experiments with WC4546[pGEM5Z-f(−)] showed no growth on the minimal medium without nicotinate.
Two dissimilar copies of nadC-like genes.
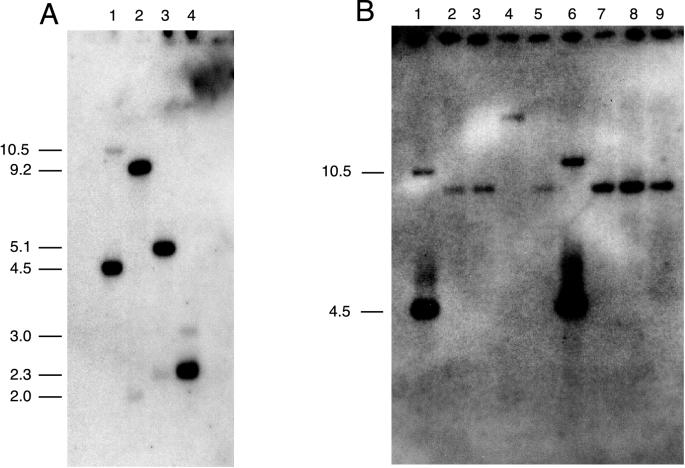
The nadC genes of Salmonella typhimurium and E. coli are not physically linked to other genes for the de novo synthesis of NAD+ (25, 68). The ophE/nadC gene identified here is also located in an isolated area not linked to other genes involved in NAD+ synthesis. It is possible that ophE is really nadC and that its presence near the genes for phthalate degradation is a mere coincidence or that there are two nadC-like genes, one associated with the genes for phthalate degradation and the other located somewhere else. These alternative hypotheses were first tested by performing Southern hybridizations with portions of the ophE gene as probes. Initially, a 0.4-kb SphI-HindIII fragment from pGJZ1331 containing a portion of the ophE gene coding for the first 116 amino acids of OphE was used as a probe against B. cepacia DBO1 genomic DNA digested separately with either EcoRI, BamHI, NotI, or SphI. The results (not shown) indicate that a single restriction fragment hybridized in each case: 9.2 kb for EcoRI, 4.5 kb for BamHI, 3.8 kb for NotI, and 3.3 kb for SphI.
This result suggested that a single ophE/nadC gene exists in B. cepacia DBO1. However, a detailed analysis of the amino acid sequences of all NadC enzymes in the GenBank database indicates that the C-terminal half of the protein is more conserved across genus and species lines, while the N-terminal half of the protein shows more evolutionary divergence. This being the case, a second probe was prepared by PCR in order to include most of the ophE gene. A 0.9-kb fragment of DNA was amplified from pGJZ1331 with primers hybridizing to the SP6 promoter region of the vector and to a position near the end of the ophE gene (see Materials and Methods). A Southern blot with this fragment as a probe against genomic DNA digested with either BamHI, EcoRI, PstI, or XhoI is shown in Fig. 3. In every case one strongly hybridizing band and one weakly hybridizing band can be seen. The size of the strongly hybridizing band corresponds to that predicted for the ophE gene. The weakly hybridizing band is most likely the nadC gene of B. cepacia DBO1. The fact that the bands are not of equal intensity suggests that the ophE gene was not recruited by a simple gene duplication event but actually was obtained from an evolutionarily distinct source. In order to investigate this further Southern hybridizations were carried out with BamHI-digested genomic DNA prepared from phthalate-degrading and non-phthalate-degrading B. cepacia strains and the 0.9-kb ophE gene probe. In every case (Fig. 3) genomic DNA from non-phthalate-degrading B. cepacia strains showed a single faintly hybridizing fragment, while phthalate-degrading B. cepacia DBO1 and ATCC 17616 showed two disparately hybridizing bands. The ophE gene is thus only associated with phthalate-degrading B. cepacia strains and may be somehow related to the ability to metabolize phthalate.
FIG. 3.
Southern hybridization of total genomic DNA from B. cepacia DBO1 (A) and from different B. cepacia strains (B) with the 0.9-kb PCR-amplified ophE gene probe from DBO1. (A) Total DNA from B. cepacia was digested with BamHI (lane 1), EcoRI (lane 2), PstI (lane 3), and XhoI (lane 4). The migration distances of the size standards are indicated on the left. (B) Total DNA from different B. cepacia strains was digested with BamHI. Lanes 1 through 9 contain DNA from strains DBO1, 382, D1, M53, ATCC 25416, ATCC 17616, k56-2, k63-3, and 715j, respectively. Only DBO1 and ATCC 17616 are phthalate degraders. The deduced size of each band is indicated.
OphE enhances growth on phthalate.
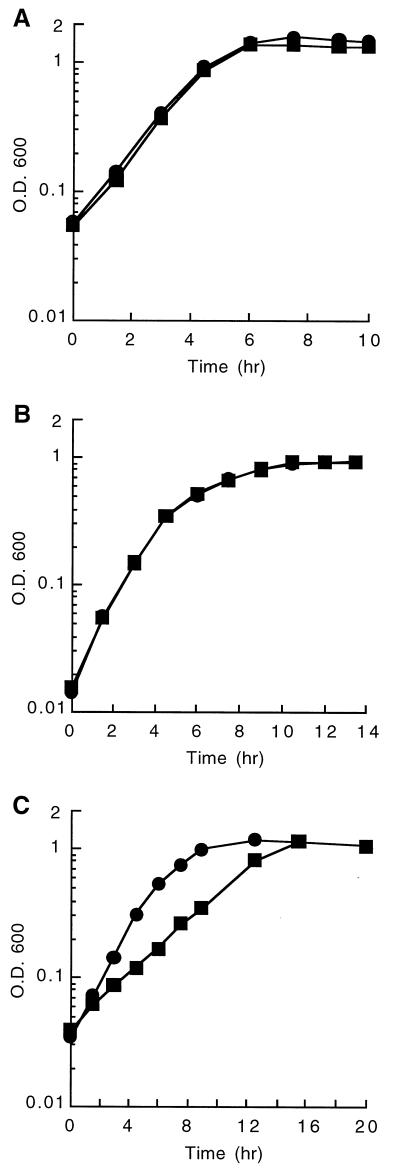
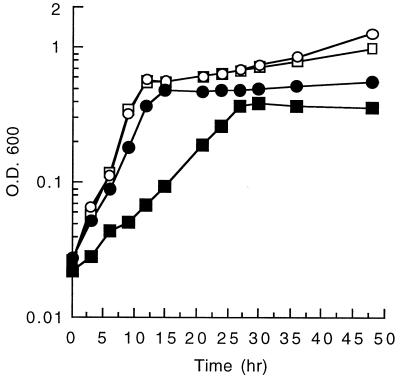
In order to verify that there are two genes (ophE and nadC) in B. cepacia DBO1 coding for quinolinate phosphoribosyl transferase, gene knockout experiments were performed. A mutant strain, designated DBO302, which has a kanamycin resistance cassette inserted into the BclI site of the ophE gene, was constructed by double reciprocal recombination as described in Materials and Methods. DBO302 is able to grow on MSB medium containing succinate, indicating that a lack of ophE does not impair the ability of the strain to synthesize NAD+. This verifies that there must be a functional nadC gene somewhere else in the genome. Additionally, DBO302 is still able to grow on MSB medium containing phthalate, indicating that ophE is not absolutely required for the degradation of phthalate. However, the close physical proximity of ophE to the genes coding for the enzymes involved in phthalate degradation led us to believe that it must play some role in the metabolism of phthalate. This being the case, growth rates of the ophE knockout mutant DBO302 and the spontaneously kanamycin-resistant mutant DBO303 were compared on various substrates (Fig. 4). DBO302 and DBO303 have the same growth characteristics when cultured in BM broth with either succinate or p-hydroxybenzoate as the sole carbon source. The doubling time in all of these cases was approximately 1.1 h. This indicates that the loss of the ophE gene has no discernible effect on central metabolism (succinate-grown cells) or on aromatic metabolism (p-hydroxybenzoate-grown cells). However, DBO302 and DBO303 show much different growth patterns when cultured in BM broth with phthalate as the sole carbon source. In this case DBO303 has a doubling time of 1.4 h, while DBO302 grows at less than half of this rate with a doubling time of 3.1 h. In order to test whether this deficiency of growth rate is due to the missing ophE gene, mutant complementation experiments were performed. The ophE gene was cloned into a broad-host-range vector by first subcloning a 2.4-kb XhoI fragment containing the ophE gene into pGEM11Z-f(−), then by utilizing restriction sites that cleave in the vector, a BamHI to EcoRI fragment was transferred into pRK415, and the resulting clone was designated pGJZ1333. The growth of DBO302 carrying either pRK415 or pGJZ1333 was compared on BM broth containing phthalate (Fig. 5). The cloned ophE gene restored the ability of DBO302 to grow on phthalate, increasing the growth rate above wild-type levels. In fact, the wild-type DBO1 strain grew slightly faster on phthalate when it carried the cloned ophE gene (pGJZ1333) than when it carried the vector alone. These experiments clearly demonstrate that OphE enhances the ability of DBO1 to grow on phthalate while not being required for the actual metabolism of phthalate.
FIG. 4.
Growth of DBO302 (ophE knockout) (■) and DBO303 (spontaneously kanamycin-resistant mutant of DBO1) (●) in a minimal medium with succinate (A), p-hydroxybenzoate (B), or phthalate (C) as the sole carbon source. OD600, optical density at 600 nm.
FIG. 5.
Growth of wild-type DBO1 (circles) and the ophE knockout mutant DBO302 (squares) in a minimal medium containing phthalate with either the vector (pRK415) (black) or the cloned ophE gene (pGJZ1333) (white). Growth is slower than that shown in Fig. 4 due to the incorporation of tetracycline in the medium.
ophE is induced by phthalate.
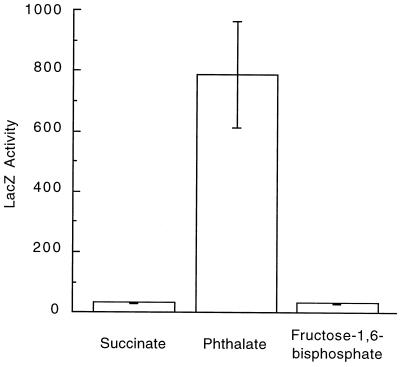
If ophE is indeed involved in phthalate degradation by B. cepacia DBO1 then it stands to reason that the gene should be induced when the strain is grown in the presence of phthalate. The putative promoter region (384 bases from within orf2 to within ophE) was amplified by PCR with primers HK-A2 and HK-B and cloned into the lacZ promoter-probe plasmid pKRZ-1. The resulting plasmid, designated pGJZ1334, was introduced into B. cepacia DBO1 by electroporation. β-Galactosidase activity was measured in crude cell extracts following growth in the presence of various compounds (Fig. 6). DBO1(pGJZ1334) had minimal LacZ activity when grown in MSB broth with succinate (<50 nmol/min/mg). However, growth on phthalate as the sole carbon source resulted in a >16-fold increase in LacZ activity (∼800 nmol/min/mg). In order to verify that this increase in LacZ activity is the result of induction of the ophE gene by phthalate and not just a generalized stress response due to the inhibition of quinolinate phosphoribosyl transferase (OphE or NadC) a second experiment was performed. DBO1(pGJZ1334) was grown on MSB broth with succinate and 20 mM fructose 1,6-bisphosphate, a known inhibitor of NadC. In this case no increase of LacZ activity above basal levels was detected. These experiments clearly indicate that the ophE gene is induced by phthalate and thus is specific for growth in the presence of phthalate.
FIG. 6.
Induction of ophE by phthalate. Comparison of the β-galactosidase activities produced by B. cepacia DBO1 containing the promoter-probe construct pGJZ1334 following growth on succinate, phthalate, or succinate plus fructose 1,6-bisphosphate. LacZ activity is reported in nanomoles per minute per milligram of protein in crude cell extracts. The standard deviation (error bar) is the average of three independent assays.
DISCUSSION
Aromatic hydrocarbons are widely found in the environment. It is not surprising therefore that microorganisms have evolved mechanisms to utilize these compounds as carbon and energy sources for growth. In fact, microorganisms are constantly evolving such abilities, and much literature has been devoted not only to an analysis of the catabolic pathways but also to the mechanisms by which microorganisms evolve such capabilities (64). Catabolic genes are often present as operonic segments that can be recruited or recombined to make metabolic pathways for new substrates. However, only a few mechanisms have been reported by which microorganisms overcome the potential toxic effects of the compounds they are metabolizing. The best known of these mechanisms is perhaps that of solvent resistance, protecting the cell against the deleterious effects of hydrophobic solvents on the cell membrane (32, 67). In the case of growth on phthalate the cell is exposed to two potentially deleterious effects that act on the same step in NAD+ synthesis: the conversion of quinolinate and phosphoribosyl pyrophosphate to nicotinic acid mononucleotide by quinolinate phosphoribosyl transferase. Growth on phthalate results in high levels of phthalate dioxygenase that not only has the ability to oxidize phthalate to a cis-dihydrodiol (Fig. 1) but also is able to convert quinolinate to oxidized products. This effectively depletes the intracellular pool of quinolinate and decreases the rate of synthesis of NAD+. Growth on phthalate also results in high intracellular levels of phthalate due to the transport of this negatively charged molecule into the cell (10). Since phthalate is a known competitive inhibitor of quinolinate phosphoribosyl transferase (4) this also results in a negative effect on the same step in NAD+ synthesis. B. cepacia DBO1 has overcome this potentially deleterious effect of growth on phthalate by recruiting a gene for quinolinate phosphoribosyl transferase and placing it not only under the inducible control of phthalate but also moving it within the cluster of genes responsible for phthalate degradation. To our knowledge, this is the first example of the recruitment of a gene for a biosynthetic activity into a catabolic operon to overcome the possible deleterious effects of a growth substrate on the cell. The fact that the phthalate-inducible ophE-encoded quinolinate phosphoribosyl transferase confers an advantage on the cell is evident by the data presented in Fig. 5. Insertional knockout mutants lacking this gene grow slower on phthalate than does the wild type. The lack of ophE has no discernable effect on the growth of DBO1 on other substrates such as succinate or the related aromatic compound p-hydroxybenzoate.
B. cepacia is one of the more versatile pseudomonads in terms of its ability to utilize a wide variety of carbon sources found in the environment (57). Part of this ability is due to the large size of its genome (11, 36), but the species’ ability to recruit and reorganize DNA also plays a role. For instance, certain insertion sequences in B. cepacia are known to activate the expression of foreign genes under selective pressure (9, 21, 54, 69, 70). In these cases gene expression is often constitutive, driven from a promoter in the insertion sequence. Interestingly, an insertion sequence is adjacent to the ophE gene in DBO1 (Fig. 1). This insertion sequence may have been involved in the recruitment of ophE and its association with the ophA1A2BCD genes for phthalate degradation. However, ophE is specifically inducible by phthalate and thus has evolved its own promoter, contrary to other insertion sequence-recruited genes in B. cepacia where expression is constitutive. It is interesting to note that the ophA1A2BCD structural genes for phthalate degradation are arranged into three operons, on both sides of ophE. ophD codes for a frame-shifted nonfunctional phthalate permease (10), while the real phthalate transporter is encoded elsewhere on the DBO1 genome (14). This means that there are at least five phthalate-inducible operons in B. cepacia DBO1 [ophA1, ophDC, ophE, ophA2B, and the transport gene(s)]. This gene organization is not unique to DBO1, as B. cepacia 249, another phthalate degrader, has a restriction fragment length polymorphism pattern identical to that of DBO1 for this locus. This is contrary to that found in P. putida NMH102-2, in which the genes for phthalate degradation are adjacent to one another and are transcribed in the same direction (43). No ophE analogue has been found clustered with the genes for phthalate degradation in P. putida.
A comparative analysis of ophE and other quinolinate phosphoribosyl transferase genes (Fig. 2) shows that it is most similar to those from Pseudomonas aeruginosa (59.9% identity with zero gaps), S. typhimurium (55.0% identity with two gaps), E. coli (53.0% identity with two gaps), and Neisseria gonorrhoeae (51.0% identity with three gaps). The G+C content of ophE (63%) is less than the 68% reported for the B. cepacia species (1) and the 67% reported for the genes for protocatechuate dioxygenase (73) from DBO1. However, it is in line with the 62 to 63% G+C content of the other oph genes. This suggests that ophE and the other oph genes were recruited from outside of the B. cepacia species. This is backed up by the fact that Southern hybridizations (Fig. 3) with ophE show a strongly hybridizing band (for itself) and a weakly hybridizing band. The latter is presumably due to the hybridization to the housekeeping gene nadC. The fact that there is not a strongly hybridizing second band suggests that ophE was not recruited by a duplication of existing nadC followed by evolution to become phthalate inducible. The similarity of ophE to nadC genes from related gram-negative bacteria indicates that the recruitment was not from an evolutionarily distant species.
ACKNOWLEDGMENTS
This work was supported by cooperative agreement CR822634 from the U.S. Environmental Protection Agency Gulf Breeze Environmental Research Laboratory and a National Science Foundation Young Investigator Award to G.J.Z.
REFERENCES
- 1.Ballard R W, Palleroni N J, Doudoroff M, Stainer R Y, Mandel M. Taxonomy of the aerobic pseudomonads: P. cepacia, P. marginata, P. alliicola and P. caryophylli. J Gen Microbiol. 1970;60:199–214. doi: 10.1099/00221287-60-2-199. [DOI] [PubMed] [Google Scholar]
- 2.Batie C J, LaHaie E, Ballou D P. Purification and characterization of phthalate oxygenase and phthalate oxygenase reductase from Pseudomonas cepacia. J Biol Chem. 1987;262:1510–1518. [PubMed] [Google Scholar]
- 3.Beliles R, Salinas J A, Kluwe W M. A review of di(2-ethylhexyl)phthalate (DEHP) risk assessments. Drug archives of environmental contamination and toxicology. NY Metab Rev. 1989;21:3–12. doi: 10.3109/03602538909029952. [DOI] [PubMed] [Google Scholar]
- 4.Bhatia R, Calvo K C. The sequencing, expression, purification, and steady-state kinetic analysis of quinolinate phosphoribosyl transferase from Escherichia coli. Arch Biochem Biophys. 1996;325:270–278. doi: 10.1006/abbi.1996.0034. [DOI] [PubMed] [Google Scholar]
- 5.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blom A, Ekman E, Johannisson A, Norrgren L, Pesonen M. Effects of xenoestrogenic environmental pollutants on the proliferation of a human breast cancer cell line (MCF-7) Arch Environ Contam Toxicol. 1998;34:306–310. doi: 10.1007/s002449900322. [DOI] [PubMed] [Google Scholar]
- 7.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 8.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J-F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Presley E A, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Hurst M A, Roberts K M, Kaine B P, Borodovsky M, Klenk H-P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 9.Byrne A M, Lessie T G. Characteristics of IS401, a new member of the IS3 family implicated in plasmid rearrangements in Pseudomonas cepacia. Plasmid. 1994;31:138–147. doi: 10.1006/plas.1994.1015. [DOI] [PubMed] [Google Scholar]
- 10.Chang H-K, Zylstra G J. Novel organization of the genes for phthalate degradation from Burkholderia cepacia DBO1. J Bacteriol. 1999;180:6529–6537. doi: 10.1128/jb.180.24.6529-6537.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng H P, Lessie T G. Multiple replicons constituting the genome of Pseudomonas cepacia 17616. J Bacteriol. 1994;176:4034–4042. doi: 10.1128/jb.176.13.4034-4042.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell B G, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–44. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 13.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olson G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 14.Dennis, J. J., H.-K. Chang, and G. J. Zylstra. Unpublished data.
- 15.Dennis J J, Sokol P A. Electrotransformation of Pseudomonas. In: Nickoloff J A, editor. Methods in molecular biology. 47. Electroporation protocols for microorganisms. Totowa, N.J: Humana Press Inc.; 1995. pp. 125–133. [DOI] [PubMed] [Google Scholar]
- 16.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ema M, Miyawaki E, Kawashima K. Reproductive effects of butyl benzyl phthalate in pregnant and pseudopregnant rats. Reprod Toxicol. 1998;12:127–132. doi: 10.1016/s0890-6238(97)00127-5. [DOI] [PubMed] [Google Scholar]
- 18.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 19.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukuoka S I, Nyaruhucha C M, Shibata K. Characterization and functional expression of the cDNA encoding human brain quinolinate phosphoribosyltransferase. Biochim Biophys Acta. 1998;1395:192–201. doi: 10.1016/s0167-4781(97)00143-7. [DOI] [PubMed] [Google Scholar]
- 21.Gaffney T D, Lessie T G. Insertion-sequence-dependent rearrangements of Pseudomonas cepacia plasmid pTGL1. J Bacteriol. 1987;169:224–230. doi: 10.1128/jb.169.1.224-230.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graham P R. Phthalate ester plasticizers—why and how they are used. Environ Health Perspect. 1973;3:3–12. doi: 10.1289/ehp.73033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanahan D. Studies on transformation of E. coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 24.Hughes K T, Dessen A, Gray J P, Grubmeyer C. The Salmonella typhimurium nadC gene: sequence determination by use of Mud-P22 and purification of quinolinate phosphoribosyl transferase. J Bacteriol. 1993;175:479–486. doi: 10.1128/jb.175.2.479-486.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hughes K T, Roth J R, Olivera B M. A genetic characterization of the nadC gene of Salmonella typhimurium. Genetics. 1991;127:657–670. doi: 10.1093/genetics/127.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamrin M A, Mayor G H. Diethyl phthalate: a perspective. J Clin Pharmacol. 1991;31:484–489. doi: 10.1002/j.1552-4604.1991.tb01908.x. [DOI] [PubMed] [Google Scholar]
- 27.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 28.Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, Nagai Y, Sakai M, Ogura K, Otsuka R, Nakazawa H, Takamiya M, Ohfuku Y, Funahashi T, Tanaka T, Kudoh Y, Yamazaki J, Kushida N, Oguchi A, Aoki K, Kikuchi H. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- 29.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 30.Keyser P. Aerobic metabolism of the phthalates by selected pseudomonads. M.S. dissertation. Miami, Fla: University of Miami; 1974. [Google Scholar]
- 31.Keyser P, Pujar R W, Eaton R W, Ribbons D W. Biodegradation of phthalates and their esters by bacteria. Environ Health Perspect. 1976;18:159–166. doi: 10.1289/ehp.7618159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kieboom J, Dennis J J, de Bont J A, Zylstra G J. Identification and molecular characterization of an efflux pump involved in Pseudomonas putida S12 solvent tolerance. J Biol Chem. 1998;273:85–91. doi: 10.1074/jbc.273.1.85. [DOI] [PubMed] [Google Scholar]
- 33.Kim E, Zylstra G J. Molecular and biochemical characterization of two meta-cleavage dioxygenases involved in biphenyl and m-xylene degradation by Beijerinckia sp. strain B1. J Bacteriol. 1995;177:3095–3103. doi: 10.1128/jb.177.11.3095-3103.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klenk H P, Clayton R A, Tomb J, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Peterson S, Reich C I, McNeil L K, Badger J H, Glodek A, Zhou L, Overbeek R, Gocayne J D, Weidman J F, McDonald L, Utterback T, Cotton M D, Spriggs T, Artiach P, Kaine B P, Sykes S M, Sadow P W, D’Andrea K P, Bowman C, Fujii C, Garland S A, Mason T M, Olsen G J, Fraser C M, Smith H O, Woese C R, Venter J C. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 34a.Kobayashi, D. Personal communication.
- 35.Lennox E S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955;1:190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- 36.Lessie T G, Hendrickson W, Manning B D, Devereux R. Genomic complexity and plasticity of Burkholderia cepacia. FEMS Microbiol Lett. 1996;144:117–128. doi: 10.1111/j.1574-6968.1996.tb08517.x. [DOI] [PubMed] [Google Scholar]
- 37.McKenney D, Brown K E, Allison D G. Influence of Pseudomonas aeruginosa exoproducts on virulence factor production in Burkholderia cepacia: evidence of interspecies communication. J Bacteriol. 1995;177:6989–6992. doi: 10.1128/jb.177.23.6989-6992.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKevitt A I, Bajaksouzian S, Klinger J D, Woods D E. Purification and characterization of an extracellular protease from Pseudomonas cepacia. Infect Immun. 1989;41:1099–1104. doi: 10.1128/iai.57.3.771-778.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKevitt A I, Retzer M D, Woods D E. Development and use of a serotyping scheme for Pseudomonas cepacia. Serodiagn Immunother. 1987;1:177–184. [Google Scholar]
- 40.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. Experiment 48: assay of β-galactosidase; pp. 352–355. [Google Scholar]
- 41.Murakami Y, Naitou M, Hagiwara H, Shibata T, Ozawa M, Sasanuma S-I, Sasanuma M, Tsuchiya Y, Soeda E, Yokoyama K, Yamazaki M, Tashiro H, Eki T. Analysis of the nucleotide sequence of chromosome VI from Saccharomyces cerevisiae. Nat Genet. 1995;10:261–268. doi: 10.1038/ng0795-261. [DOI] [PubMed] [Google Scholar]
- 42.Nomura Y, Harashima S, Oshima Y. A simple method for detection of enzyme activities involved in the initial step of phthalate degradation in microorganisms. J Ferment Bioeng. 1989;67:291–296. [Google Scholar]
- 43.Nomura Y, Nakagawa M, Ogawa N, Harashima S, Oshima Y. Genes in PHT plasmid encoding the initial degradation pathway of phthalate in Pseudomonas putida. J Ferment Bioeng. 1992;74:333–344. [Google Scholar]
- 44.Nomura Y, Takada N, Oshima Y. Isolation and identification of phthalate-utilizing bacteria. J Ferment Bioeng. 1989;67:297–299. [Google Scholar]
- 45.Olsen R H, DeBusscher G, McCombie W R. Development of broad-host-range vectors and gene banks: self-cloning of the Pseudomonas aeruginosa PAO chromosome. J Bacteriol. 1982;150:60–69. doi: 10.1128/jb.150.1.60-69.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parke D. Construction of mobilizable vectors derived from plasmids RP4, pUC18, and pUC19. Gene. 1990;93:135–137. doi: 10.1016/0378-1119(90)90147-j. [DOI] [PubMed] [Google Scholar]
- 47.Peakall D B. Phthalate esters: occurrence and biological effects. Residue Rev. 1975;54:1–41. doi: 10.1007/978-1-4612-9857-1_1. [DOI] [PubMed] [Google Scholar]
- 47a.Pseudomonas Genome Project. 15 March 1999, revision date. [Online.] http://www.pseudomonas.com. [5 April 1999, last date accessed.]
- 48.Pujar B G, Ribbons D W. Phthalate metabolism in Pseudomonas fluorescens PHK: purification and properties of 4,5-dihydroxyphthalate decarboxylase. Appl Environ Microbiol. 1985;49:374–376. doi: 10.1128/aem.49.2.374-376.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Resnick S M, Lee K, Gibson D T. Diverse reactions catalyzed by naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816. J Ind Microbiol Biotechnol. 1996;17:438–457. [Google Scholar]
- 50.Ribbons D W, Keyser P, Kunz D A, Taylor B F, Eaton R W, Anderson B N. Microbial degradation of phthalate. In: Gibson D T, editor. Microbial degradation of organic compounds. New York, N.Y: Marcel Dekker, Inc.; 1984. pp. 371–397. [Google Scholar]
- 51.Rodley P D, Romling U, Tummler B. A physical genome map of the Burkholderia cepacia type strain. Mol Microbiol. 1995;17:57–67. doi: 10.1111/j.1365-2958.1995.mmi_17010057.x. [DOI] [PubMed] [Google Scholar]
- 51a.Roe, B. A., S. P. Lin, L. Song, X. Yuan, S. Clifton, T. Ducey, L. Lewis, and D. W. Dyer. 3 April 1999, revision date. Gonococcal genome sequencing project. University of Oklahoma. [Online.] http://www.genome.ou.edu/gono.html. [5 April 1999, last date accessed.]
- 52.Rothmel R K, Shinabarger D L, Parsek M R, Aldrich T L, Chakrabarty A M. Functional analysis of the Pseudomonas putida regulatory protein CatR: transcriptional studies and determination of the CatR DNA-binding site by hydroxyl-radical footprinting. J Bacteriol. 1991;173:4717–4724. doi: 10.1128/jb.173.15.4717-4724.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 54.Scordilis G E, Ree H, Lessie T G. Identification of transposable elements which activate gene expression in Pseudomonas cepacia. J Bacteriol. 1987;169:8–13. doi: 10.1128/jb.169.1.8-13.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shelver D, Kerby R L, He Y, Roberts G P. Carbon monoxide-induced activation of gene expression in Rhodospirillum rubrum requires the product of cooA, a member of the cyclic AMP receptor protein family of transcriptional regulators. J Bacteriol. 1995;177:2157–2163. doi: 10.1128/jb.177.8.2157-2163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrovski S, Church G M, Daniels C J, Mao J-I, Rice P, Nölling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stanier R Y, Palleroni N J, Duodoroff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966;43:159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- 58.Sun D, Setlow P. Cloning, nucleotide sequence, and regulation of the Bacillus subtilis nadB gene and a nifS-like gene, both of which are essential for NAD biosynthesis. J Bacteriol. 1993;175:1423–1432. doi: 10.1128/jb.175.5.1423-1432.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taylor B F, Amador J A. Metabolism of pyridine compounds by phthalate-degrading bacteria. Appl Environ Microbiol. 1988;54:2342–2344. doi: 10.1128/aem.54.10.2342-2344.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taylor B F, King C A. Phthalic acid and pyridine dicarboxylic acids as catabolic analogs. FEMS Microbiol Lett. 1987;44:401–405. [Google Scholar]
- 61.Tomb, J.-F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539–547. [DOI] [PubMed]
- 62.Tritz G J, Matney T S, Chandler J L, Gholson R K. Chromosomal location of the C gene involved in the biosynthesis of nicotinamide adenine dinucleotide in Escherichia coli K-12. J Bacteriol. 1970;104:45–49. doi: 10.1128/jb.104.1.45-49.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Turner K J, Sharpe R M. Environmental oestrogens—present understanding. Rev Reprod. 1997;2:69–73. doi: 10.1530/ror.0.0020069. [DOI] [PubMed] [Google Scholar]
- 64.van der Meer J R, De Vos W M, Harayama S, Zehnder A J B. Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol Rev. 1992;56:677–694. doi: 10.1128/mr.56.4.677-694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vogelstein B, Gillespie D. Preparation and analytical purification of DNA from agarose. Proc Natl Acad Sci USA. 1979;76:615–619. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wams T J. Diethylhexylphthalate as an environmental contaminant—a review. Sci Total Environ. 1987;66:1–16. doi: 10.1016/0048-9697(87)90072-6. [DOI] [PubMed] [Google Scholar]
- 67.Weber F J, de Bont J A M. Adaptation mechanisms of microorganisms to the toxic effects of organic solvents on membranes. Biochim Biophys Acta. 1996;1286:225–245. doi: 10.1016/s0304-4157(96)00010-x. [DOI] [PubMed] [Google Scholar]
- 68.Whitchurch C B, Mattick J S. Escherichia coli contains a set of genes homologous to those involved in protein secretion, DNA uptake and the assembly of type-4 fimbriae in other bacteria. Gene. 1994;150:9–15. doi: 10.1016/0378-1119(94)90851-6. [DOI] [PubMed] [Google Scholar]
- 69.Wood M S, Byrne A, Lessie T G. IS406 and IS407, two gene-activating insertion sequences for Pseudomonas cepacia. Gene. 1991;105:101–105. doi: 10.1016/0378-1119(91)90519-h. [DOI] [PubMed] [Google Scholar]
- 70.Wood M S, Lory C, Lessie T G. Activation of the lac genes of Tn951 by insertion sequences from Pseudomonas cepacia. J Bacteriol. 1990;172:1719–1724. doi: 10.1128/jb.172.4.1719-1724.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zylstra G J, Gibson D T. Aromatic hydrocarbon degradation: a molecular approach. In: Setlow J K, editor. Genetic engineering: principles and methods. New York, N.Y: Plenum Press; 1991. pp. 183–203. [DOI] [PubMed] [Google Scholar]
- 72.Zylstra G J, Olsen R H, Ballou D P. Cloning, expression, and regulation of the Pseudomonas cepacia protocatechuate 3,4-dioxygenase genes. J Bacteriol. 1989;171:5907–5914. doi: 10.1128/jb.171.11.5907-5914.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zylstra G J, Olsen R H, Ballou D P. Genetic organization and sequence of the Pseudomonas cepacia genes for the alpha and beta subunits of protocatechuate 3,4-dioxygenase. J Bacteriol. 1989;171:5915–5921. doi: 10.1128/jb.171.11.5915-5921.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]