Abstract
Melanoma is a type of skin lesion that is less common than other types of skin lesions, but it is fast growing and spreading. Therefore, it is classified as a serious disease that directly threatens human health and life. Recently, the number of deaths due to this disease has increased significantly. Thus, researchers are interested in creating computer-aided diagnostic systems that aid in the proper diagnosis and detection of these lesions from dermoscopy images. Relying on manual diagnosis is time consuming in addition to requiring enough experience from dermatologists. Current skin lesion segmentation systems use deep convolutional neural networks to detect skin lesions from RGB dermoscopy images. However, relying on RGB color model is not always the optimal choice to train such networks because some fine details of lesion parts in the dermoscopy images can not clearly appear using RGB color model. Other color models exhibit invariant features of the dermoscopy images so that they can improve the performance of deep neural networks. In the proposed Color Invariant U-Net (CIU-Net) model, a color mixture block is added at the beginning of the contracting path of U-Net. The color mixture block acts as a mixer to learn the fusion of various input color models and create a new one with three channels. Furthermore, a new channel-attention module is included in the connection path between encoder and decoder paths. This channel attention module is developed to enrich the extracted color features. From the experimental result, we found that the proposed CIU-Net works in harmony with the new proposed hybrid loss function to enhance skin segmentation results. The performance of the proposed CIU-Net architecture is evaluated using ISIC 2018 dataset and the results are compared with other recent approaches. Our proposed method outperformed other recent approaches and achieved the best Dice and Jaccard coefficient with values 92.56% and 91.40%, respectively.
Keywords: Channel-wise attention, Hybrid loss Function, Color-invariant Skin lesion segmentation, Color mixture block
Introduction
The computer-aided diagnosis (CAD) system for melanoma detection requires an automatic discrimination between skin lesions and surrounding tissues. The study of dermoscopy images is an important support for clinical decision-making and for image-based diagnosis to detect diseases such as melanoma. Using dermoscopy image is a non-invasive technique which is mainly used for the study of pigmented skin lesions and serve observers to check lesions with a little coloring [1]. It is performed with an instrument called a dermatoscope which requires a high-quality magnifying lens and a powerful lighting system to let identification of morphological features such as globules, lines, blue and white areas, and spots [2] more easier. This arrangement leads to a significant reduction of errors and gives significant differentiation of lesions from normal skin.
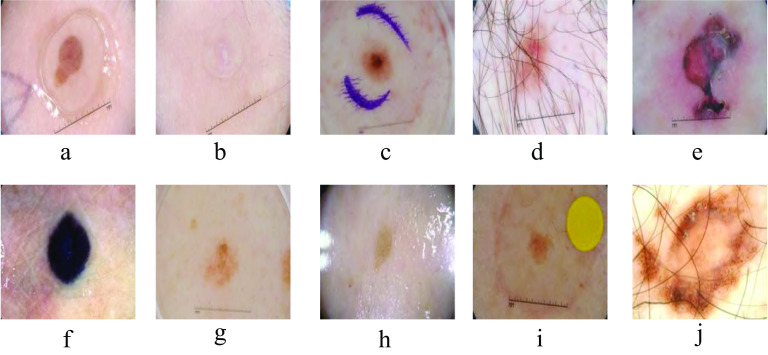
Automatic skin lesion segmentation from dermoscopy images determines lesion parts from the surrounding healthy parts by classifying each pixel as lesion or normal. This task is challenging for many reasons: (1) poor contrast between the lesion parts (foreground) and the surrounding skin area (background); (2) existence of some obstacles such as skin lines, air bubbles and hairs; and (3) color diversification at the borders of the lesion in the dermoscopy images. Figure 1 displays some challenges of skin lesion images from ISIC 2018 dataset. Among these challenges, the color divergence of skin images produces distracting features which deteriorate the performance of skin lesion segmentation methods. Precise color representation of lesion provides important information for an accurate diagnosis. In addition, the extraction of any strange objects on the image such as (hair, pen marker, oil, gel,...etc) can be done easily if the network learned appropriate color representation of the input images.
Fig. 1.
Dataset obstacles in ISIC 2018 dataset a jel, b unclear lesion, c pen markers, d hair markers, e lesion with different colors, f dark-colored lesion area irregular borders, g light-colored lesion area, h oil , i yellow mark, j larg lesion area
Recently, many research works [3–10] tried to overcome the segmentation task challenges by proposing skin lesion segmentation methods based on color invariant models to investigate the color variation in the images. These methods are usually supported by deep neural networks and exploits color model transformation-based approaches to extract features and recognize the lesion areas efficiently. Transformation of the RGB image to another color model was the common strategy between all these methods. The converted color models are used to train already existing deep convolutional neural networks (CNNs) with some modifications [11–17]. However, most transformed color models depends on a fixed transformation operation which ignore the specific characteristics of medical images.
The motivation of this work is to overcome the color variations and low contrast problems exhibited in skin lesion images. Therefore, a new color mixture convolutional block is devised to learn the fusion of multiple color spaces. This work reviews the effect of fusing various color models using U-Net as a baseline network structure. U-Net is considered as one of the effective deep convolutional neural network (CNN) structures in biomedical image segmentation field. A new U-Net variant model called color invariant U-Net (CIU-Net) is proposed by adding a color mixture block to learn the fusion of multiple color spaces. The encoder branch of the proposed network receives multiple color models of the input skin image and apply a convolutional operation to fuse these multiple color channels. In addition, a new channel wise attention units is employed to interconnect the two U-Net paths to direct the network to learn important color features. The proposed model passes the learned features from the encoder levels through an attention unit before sending them to the corresponding decoder’s level to pick up invariant lesions color features to overwhelm color diversity issues. The resulted feature maps from encoder path are sent to a channel-wise attention unit, then it is received by the decoder branch to fuse each feature contribution into the concluding segmentation map. Due to the significant impact of the loss function used in the network training, a combined binary-weighted loss function is employed to optimize the parameters of the network. The new proposed loss function includes cross-entropy, generalized dice, and sensitivity-specificity losses. Different evaluation metrics are used to estimate the segmentation results. The following is a summary of our contributions:
Studying the effect of fusing multiple color models on the behavior of the new color invariant U-Net (CIU-Net) model.
Employing a color mixture block to learn the fusion of multiple color spaces.
Using channel-wise attention in the connection path between encoder and decoder branches.
Employing a hybrid binary-weighted loss function to optimize the network parameters.
A benchmark skin lesion database is used in the experiments to validate the effectiveness of the proposed model and to compare it with other methods.
The organization of the rest of this paper is as follows: Section 2 provides background on some related works in the same field. Section 3 presents the image pre-preparing and the structure of the new skin lesion segmentation model. The details of the obtained results from the implementation of our proposed architecture and composite loss function is explained in Sect. 4. Discussion is viewed in Sects. 5 and 6 includes conclusion of the paper.
Related work
Many research works have been presented to implement fully automated skin lesion image segmentation. These works aimed to overcome the existing obstacles and fulfill the segmentation process with high accuracy. Therefore, in this section, we explain the most recent works which are concerned with the effect of fusing color models in skin lesion segmentation and we will focus on those approaches that are based on applying U-net architecture to obtain features using deep learning Convolutional Neural Network (CNN) architecture.
Color space conversion is utilized in many works to improve skin lesion segmentation results. Ma et al. [18] converted the RGB dermoscopic images to L*u*v and L*a*b color spaces and employed the contained color information to distinguish between the infected and non-infected parts. They defined a speed function and a stopping criterion for the deformable model using the differences in the combined color channels. Their model was robust against the noise and provides an effective and flexible segmentation, but the initial curves of the deformable model need to be defined manually to avoid negative influence from the complicated imaging background.
A region growing method for automatic skin lesion segmentation and classification was developed by Sumithra et al. [19]. They derived various statistical measures such as: mean, standard deviation, variation, and skewness for each channel of HSV, RGB, NTSC, YCbCr, CIE L*u*v, and CIE L*a*b color models to evaluate the color presence in skin lesions. Their proposed method could be used as a supplementary tool for the experts to diagnose skin diseases. However, its performance was decreased for some classes that led to a degradation of the overall system performance. Pour et al. [3] used a limited data size with no augmentation or pre-processing to train their proposed network. They used efficient feature maps concatenation from CIELAB color space with RGB color channels. Their proposed model needs no data augmentation nor any pre-processing and improved the performance of a not very deep and complex CNN. Although they achieved high performance using small dataset, the training of convolution neural network was longer and more complicated than traditional methods. In [20], Khan et al. extracted a discriminative deep features using fully automated method. They used color-controlled histogram intensity values (LCcHIV) and deep saliency segmentation method to enhance the training images before feeding them to a ten-layers custom convolutional neural network (CNN). Finally, the Kernel extreme learning machine (KELM) classifier is used to classify the generated features. Employing the improved moth-flame optimization (IMFO) algorithm improved the accuracy by removing irrelevant and redundant deep features, but on the other hand, it required a high computational time.
A deep fully Convolutional Deconvolutional Neural Network (CDNN) to handle images under varying conditions proposed by Yuan et al. [4]. They combined RGB, HSV color model, and they choose luminance (L) channel from Lab color model to train their suggested model. Their approach resulted in a fast segmentation process. De Angelo et al. [5] developed an application to collect skin lesion images using smartphone. They presented an investigation regarding the color spaces and the post-processing that raise some important remarks about the ground truth of skin lesion images. They associated conditional random fields techniques, deep learning, and color models to segment skin lesions. Their methodology achieved a noteworthy performance to handle ink marks and brightness in the images. Although some color space combinations slightly improved the Jaccard index, they did not report a strong effect in the results. In [21], we proposed three variants of the U-Net model with single, dual, and triple inputs, namely, single input color U-Net (SICU-Net), dual input color U-Net (DICU-Net) and triple input color U-Net (TICU-Net). Each encoder sub-network is fed with different color space of the input image. Later, this work was extended to combine gradient and color information using a new dual gradient-Color U-Net (DGCU-Net) [22] architecture. The DGCU-Net model integrated features extracted from an invariant color image representation with the gradient information of the input image to strengthen the borders of skin lesion.
In the same context, Azad et al. [23] relied on U-Net architecture in their proposed approach. They proposed a frequency re-calibration U-Net (FRCU-Net) for medical image segmentation and used a channel-wise attention mechanism. The FRCU-Net used to segment the medical images in order to generalize a low data regime and decrease the texture bias effect. They represented objects in terms of frequency to reduce the effect of texture bias and using the Laplacian pyramid to represent object in different frequency domains. Re-calibration of these frequency representations resulted in a more distinct representation to describe the object of interest. Tang et al. [24] applied stochastic weight averaging separable-U-Net to introduce a skin lesion segmentation approach for higher semantic feature information with ideal boundaries. The Separable-Unet framework takes advantage of the separable convolutional block and U-Net architectures in capturing the context feature channel correlation with higher semantic feature information. A Recurrent Convolutional Neural Network (RU-Net) and a Recurrent Residual Convolutional Neural Network (R2U-Net) based on U-Net are proposed by Alom et al. [25] for improved representation of skin lesion. The residual unit helps to speed up the training process of deep architecture. Feature accumulation with recurrent residual convolutional layers ensures better feature representation for segmentation tasks. They designed better U-Net architecture using same number of network parameters.
Asadi et al. [26] presented Multi-level Context Gating based on U-Net (MCGU-Net) which utilize multi-level bi-directional ConvLSTM (BConvLSTM) in the skip connection. In addition, densely connected convolutional blocks and squeeze-excitation (SE) blocks are utilized in the decoding path. Batch normalization (BN) operation is employed after the up-convolutional layer for getting more precise results and capturing more discriminative information. The included BN after each up-convolutional layer speed up the network learning process while dense blocks helped the network to increase the representational power of deeper models. Using multi-level BConvLSTMs led to efficiently combine encoded and decoded features. However, this modified architecture increases the complexity of the network and need more computational time. Table 1 summarizes the important related works with advantages and disadvantages of each work.
Table 1.
A summary of crucial related works with their advantages and limitations
| Refs. | Color model | Method | Advantages/limitations |
|---|---|---|---|
| [18] | L*a*b* | Deformable model with speed function. | Robust against the noise |
| L*u*v* | Provides an effective and flexible segmentation | ||
| The initial curves of the deformable model need to be defined manually | |||
| [19] | HSV | Region growing | Used as a supplementary tool for the experts in diagnose |
| RGB | K-NN classifier. | Decreasing in the performance for some classes. | |
| YCbCr | |||
| L*u*v | |||
| L*a*b | |||
| [3] | L*a*b | A deep CNN trained from scratch | No data augmentation or pre-processing |
| RGB | Use multiple input color channels | High performance on small dataset | |
| Training is longer and more complicated. | |||
| [4] |
RGB HSV L*a*b |
A deeper network with smaller kernels | Boosting in the segmentation performance |
| [5] | RGB | Conditional random field | Achieved a noteworthy performance to handle ink marks and brightness |
| HSV | Deep CNN | Slightly improving Jaccard index | |
| L*a*b | Color models | Did not viewed a strong effect in the results | |
| [23] | RGB | A frequency re-calibration U-Net. | Reduces the texture bias effect |
| Channel-wise attention mechanism. | Representation of the object in different frequency domains | ||
| Increased computational cost and complexity. | |||
| [24] | RGB | Separable-Unet | Speed up the network learning |
| Stochastic weight averaging . | Using limited datasets for evaluation | ||
| [25] | RGB | U-Net model | Ensuring better feature representation |
| Designing a network with the same number of parameters | |||
| Increased segmentation time due to recurrence operation. | |||
| [26] | RGB | Multi-level bi-directional ConvLSTM | Speed up the network learning |
| Using dense convolutional blocks | Increasing the representational power of deeper models | ||
| Squeeze excitation (SE) blocks. | Combining paths features | ||
| Need more computational time |
Proposed method
Image pre-processing
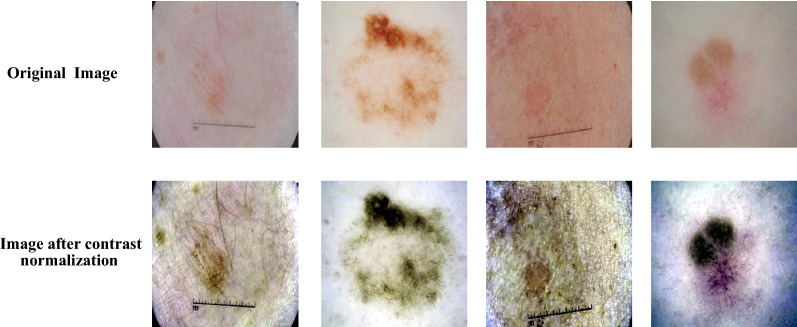
Color space representation of input skin images is significantly affect the performance of segmentation results. In order to obtain the optimum feature representation, recent works using U-Net model [21] reported that the combination of RGB and XYZ color models produced the best results. Thus, this work fuses the RGB-XYZ color spaces with other color models to fed a new modified U-Net architecture. In the proposed method, a simple image contrast normalization is applied on each component of the combined color models to enhance the contrast between lesion and normal skin pixels [21]. Applying the pre-processing operation on each color channel of the fused color models helps to improve the performance of the proposed network. In addition, using contrast normalized channel image for skin segmentation enhances the appearance of skin lesion in the input image. Figure 2 shows some examples of low contrast skin images with the effect of applying contrast normalization on these images.
Fig. 2.
The effect of applying contrast normalization on low contrast skin lesion images.
Color invariant U-Net architecture
Semantic segmentation not only require discriminative features at pixel level but also need to reconstruct a labeled image from the resulted feature maps. This task can not be accomplished using traditional CNN architectures that are prevailed the image classification tasks. Therefore, there is a need for an alternative structure that is capable of doing image segmentation task using encoder-decoder architecture such as U-Net deep model. The original U-Net model introduced by Ronneberger et al. [27] is utilized to segment various biomedical images and label each pixel in these image. It has a simple structure consisting of a repetition of basic building blocks so it is easy to implement. Moreover, it shows good performance with a little amount of training data and it can be fitted to solve any semantic segmentation problem. The U-net structure keeps the segmented image to be in the same size as the input image. This work proposes a new modified U-Net model to tackle color variations problem in the skin lesion segmentation task.
The segmentation results of skin lesion images using classical U-Net model are usually unsatisfactory due to the existing obstacles in the image dataset. Especially, the low contrast of infected skin areas and the color blurring of skin lesions. To address these problems, we propose a new Color Invariant U-Net (CIU-Net) structure which learn the optimum mixture of various color models to represent invariant color information of skin lesion images. The proposed network includes a color mixture block along with the encoder path of U-Net structure. To further improve the feature representation, the encoder and decoder paths are interconnected with an attention units. The input image of the network is constructed by fusing multiple color models or some discriminative channels from multiple color models. The multi-color input image is fed into the color mixture convolutional block to learn the optimum three-channel color representation of the input image for subsequent processing.
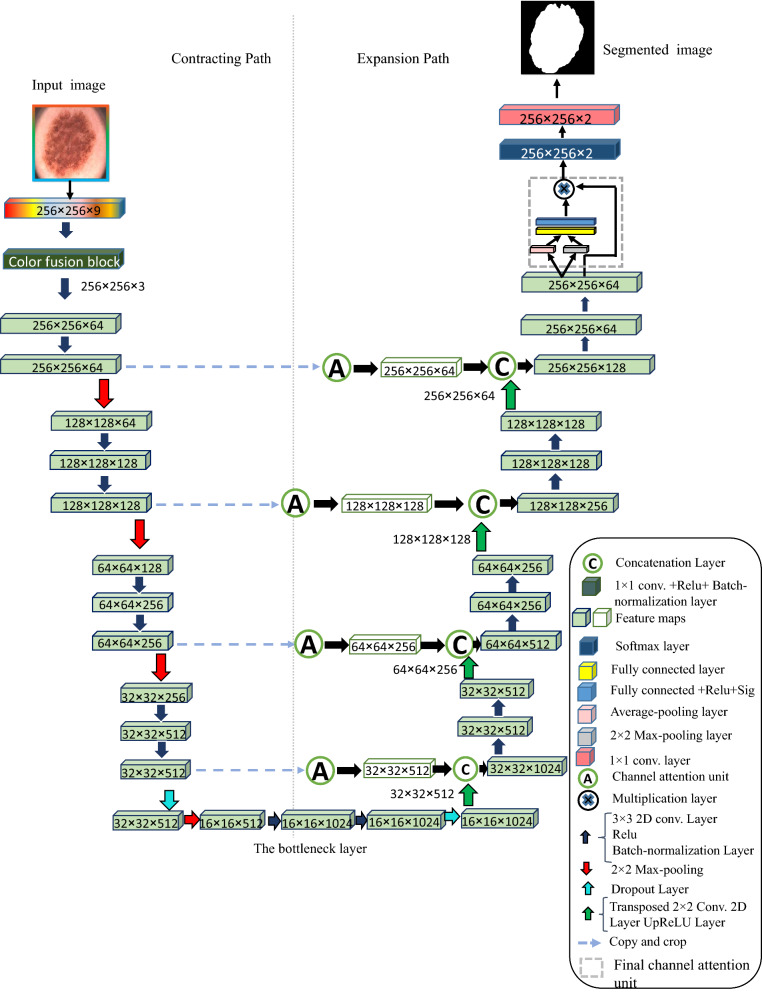
The architecture of proposed CIU-Net model resembles a ‘U’ letter shape that is why it is called by this name. Figure 3 displays the structure of proposed CIU-Net model. The CIU-Net architecture includes (1) The color mixture convolutional block (2) the contracting/encoder path, (3) bridge, and (4) the expanding/decoder path. In the proposed CIU-Net structure, a new color mixture convolutional block is added at the beginning of the contracting/encoder path. It contains a convolution layer with 3 kernels followed by batch-normalization layer and ReLU activation function. This convolution layer acts as a mixer to learn the fusion of the input color models and create a new image with three channels. The contracting path consists of four blocks, each of them has two 3 × 3 convolution layers with ReLU activation function. Then, a max pooling layer with 2 × 2 size and stride 2 is followed to reduce the spatial resolution of the input feature maps by half. The number of feature maps is doubled after each max pooling layer. The encoder path starts with 64 feature maps in the initial stage, then it is increased by 2 until it reach 1024 in the bridge stage. The encoder path tries to pick up the contextual information of the image and outputting feature maps that will be sent to the corresponding decoder block via a channel-wise attention unit. The third part is the bridge, which is placed between the ending of the encoder path and the beginning of the decoder path and consists of two 3 × 3 convolution layers followed by 2 × 2 up-convolution layer followed by dropout operation. The final part is the expansion path which includes 4 blocks each with stride 2 deconvolution layer concatenated with the corresponding attentioned feature maps from the corresponding contracting path. It also contains a pair of 3 × 3 convolution layers with ReLU activation function. At the end of each decoder block, the number of feature maps shrinks by half to maintain similarity. In the final output stage, there is another attention unit attached with a convolutional layer with the number of features equal to the total number of classes in the segmentation task. In the CIU-Net structure, the color model/channels of the image can be selected according to the type of medical image. The segmentation can be enhanced by determining the appropriate color model which appropriately describe the classes of the dataset. CIU-Net model takes whole powers of original U-Net and adds the context and localization information required to predict segmentation map.
Fig. 3.
Color invariant U-Net (CIU-Net) architecture
Channel-wise attention unit
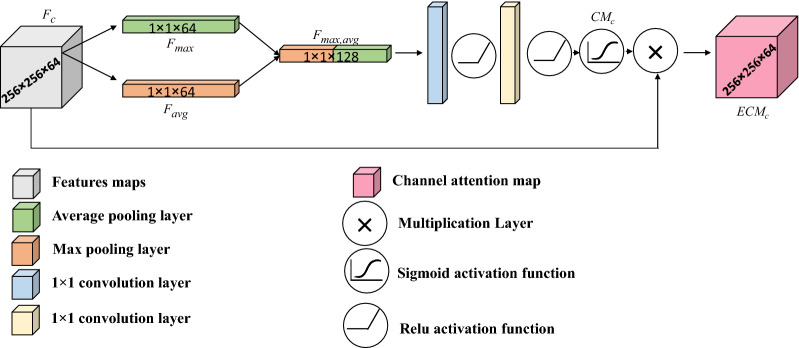
The proposed CIU-Net learns a set of feature maps after each stage of the encoder path. It is favorable to direct the attention of the network on vital feature maps to assert descriptive features while discard meaningless features. A channel-wise attention unit is employed to focus on efficient feature components. Channel-wise attention units are commonly based on understanding the correlation between feature channels. The channel-wise attention unit accepts a set of features obtained by applying a series of convolutional and pooling processes in the contracting path, where r, c, and n are the number of rows, columns, and channels, respectively. The structure of channel attention unit adopted here is driven from [28] by employing average and max pooling process to obtain and , respectively. Next, the derived pooled features are concatenated to get as written in Eq. 1 and delivered into two convolution layers with appropriate number of filters. Each convolution layer is followed by ReLU, then a Sigmoid activation function is appended to get the channel weights in Eq. 2. The input feature maps is magnified by the learned weights to obtain the attentioned feature maps in Eq. 3. Figure 4 illustrates the channel-wise attention unit structure. The following equations describe the channel attention operation steps:
| 1 |
| 2 |
where is sigmoid activation function. The attentioned feature maps are obtained from:
| 3 |
where is the element-wise multiplication and .
Fig. 4.
The structure of channel-wise attention module
Proposed loss function
In the learning process of deep convolution neural networks, it is essential to choose an appropriate loss function to enhance the learning behavior of semantic segmentation problems. Loss function is utilized to check out the network performance by reducing the resulted training error. In our work, we use a hybrid loss function which comprise three fused binary weighted terms. The proposed binary weighted loss function (CE-Di-SS) comprises Sensitivity-Specificity (SS), Dice (Di), and Cross-entropy (CE) loss functions. This hybrid loss function is very suitable to handle imbalance skin lesion datasets. Mathematically, it can be formulated as follows:
| 4 |
The cross-entropy loss measures the performance of model with output probability ranged between 0 and 1 and it increases as the predicted probability (P) diverges from the actual ground truth label (G). Weighted cross-entropy loss is used to tackle the imbalanced data in background/foreground segmentation. The weighed cross-entropy (CE) loss function is defined as.
| 5 |
Dice loss function is based on the Dice coefficient, which measure the overlap between two samples and ranged from 0 to 1 where a Dice coefficient of 1 denotes perfect overlap and 0 value for loss. It can be calculated as:
| 6 |
Sensitivity-specificity loss is defined to address imbalanced semantic segmentation problems, where sensitivity is the first term and sensitivity is the second term. It can be formulated as:
| 7 |
where and define pairs of i pixel predicted and ground-truth values at a distinct class k, respectively.
Experimental results
This section explains the details of results obtained from experiments that were conducted using proposed CIU-Net architecture and loss function. ISIC 2018 benchmark dataset is utilized for evaluation. We also compare our results with other recent state-of-the-art methods. ISIC 2018 is a large-scale dermoscopy image dataset published by International Skin Imaging Collaboration (ISIC)1. It has 2594 training RGB dermoscopic images with resolution ranged from () to (). In this work, the training images are divided into 2076 (80% ) images used for model training and 518 (20% ) images for testing with size of () pixels.
Implementation details
The experiments are conducted using Matlab 2021a running on PC with Windows 10, 32 GB RAM, Intel Core i7 processor and NVIDIA GeForce RTX 2080 Ti. The SGDM optimizer is employed to optimize the network parameters using following hyper-parameter values: The number of epochs equals 30, the learning rate is set to 0.05 and the mini-batch size equals 4. Two techniques are implemented to avoid overfitting problem, the first one is using L2 regularization in the loss function and the second is using dropout layers.
Evaluation Metrics
To check the quality of the segmentation results, it is essential to consider various evaluation metrics to compare the obtained performance with recent methods. The commonly used evaluation metrics are: true positive rate (SEN) calculated from Eq. 8, true negative rate (SPE) in Eq. 9, False positive rate (FPR) in Eq. 10, Dice coefficient (DIC) in Eq. 11, Jaccard index (JAC) in Eq. 12, Accuracy (ACC) detailed in Eq. 13, Area Under Curve score (AUC) in Eq. 14 and Precision (PRE) in Eq. 15.
| 8 |
| 9 |
| 10 |
| 11 |
| 12 |
| 13 |
| 14 |
| 15 |
where, FN, FP, TP, and TN are False Negative, False Positive, True Positive, and True Negative, respectively.
Studying the effect of using various color models on CIU-Net model
In this experiment, the proposed CIU-Net is trained and tested using ISIC 2018 data set with different color models to illustrate the effect of varying the input color model on the (CIU-Net) performance. Results in Table 2 illustrate the superiority of using a fused input color model that consists of a combination of 7 channels (RGB-XYZ-Gray) in most evaluation metrics. The fused input color model achieved 96.95% for sensitivity, 98.88% for specificity, 92.58% for dice coefficient, 91.40% for jaccard coefficient,95.44% for accuracy, and 97.91% for AUC score. While the best false positive rate of 0.16% and the best precision value 98.93% are achieved using the combination of (RGB-XYZ-LUV) channels.
Table 2.
The effect of using various combination of image color models on the CIU-Net for ISIC 2018 dataset
| Input color model | SEN% | SPE% | DICE% | JAC% | ACC% | FPR% | AUC% | PRE% |
|---|---|---|---|---|---|---|---|---|
| RGB-XYZ-Gray | 96.95 | 98.88 | 92.56 | 91.40 | 95.44 | 4.49 | 97.91 | 90.01 |
| RGB-XYZ-YCbCr | 93.73 | 96.20 | 91.73 | 90.61 | 95.02 | 3.98 | 94.97 | 90.81 |
| RGB-XYZ-Lab | 51.13 | 99.50 | 65.10 | 71.65 | 84.29 | 0.80 | 75.32 | 96.31 |
| RGB-XYZ-YIQ | 77.15 | 98.99 | 83.67 | 83.82 | 91.34 | 1.48 | 88.07 | 95.52 |
| RGB-XYZ-HSV | 66.97 | 99.89 | 78.06 | 79.80 | 89.11 | 0.64 | 83.43 | 97.75 |
| RGB-XYZ-LUV | 39.41 | 99.88 | 52.16 | 65.51 | 80.64 | 0.16 | 69.65 | 98.93 |
Bold indicates the best-achieved results
State-of-the-art comparison
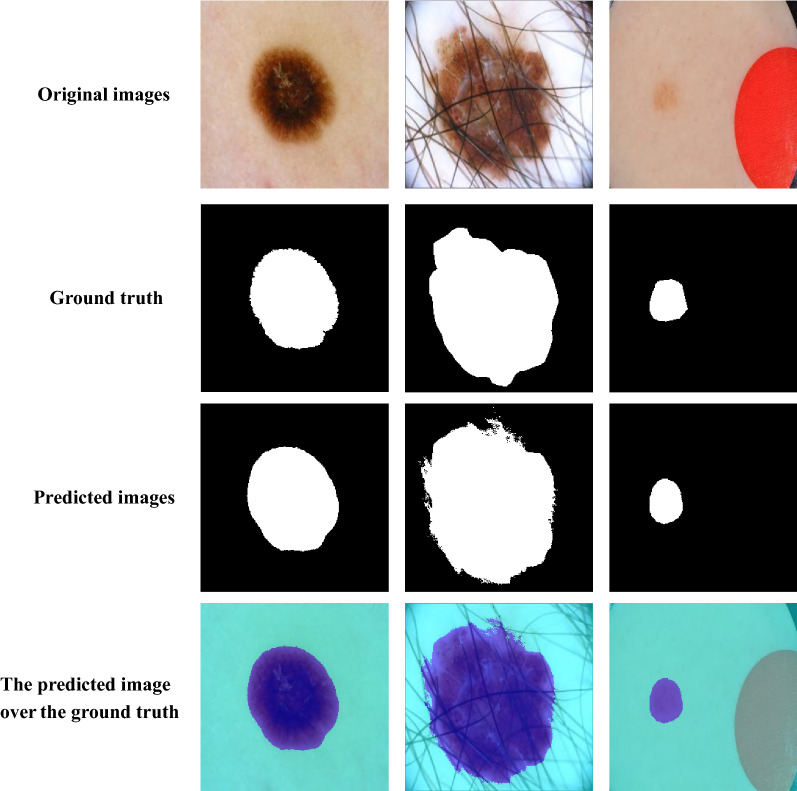
This experiment presents a comparison between our proposed model and other recent approaches which employ ISIC 2018 dataset. The results of the state-of-the-art approaches were obtained from [12, 25, 29, 30]. Table 3 shows the evaluation metric values in comparison with our proposed model and other recent approaches. The proposed model achieves the best values for Sen, Spe, AUC, Dice and Jaccard coefficients with values of 96.95%, 98.88%, 97.91%, 92.56% and 91.40%, respectively while the value of accuracy is close to the best approach. Figure 5 shows sample of segmentation results from CIU-Net model using images from ISIC 2018 dataset. The performance of the proposed method is superior even for low contrast skin images.
Table 3.
Comparison between our proposed method and other recent state-of-the-art methods using ISIC 2018 dataset
| Method | Year | SEN% | SPE% | DIC% | JAC% | ACC% | AUC% |
|---|---|---|---|---|---|---|---|
| U-Net [27] | 2015 | 91.40 | 94.20 | 85.10 | 76.10 | 91.80 | – |
| Deeplab [31] | 2018 | 94.40 | 91.30 | 85.80 | 76.80 | 92.40 | – |
| CENet [32] | 2019 | – | – | 87.50 | 80.00 | 95.50 | – |
| Multi-ResUnet [33] | 2019 | – | – | 80.30 | – | – | – |
| CPFNet [34] | 2019 | – | – | 89.90 | 82.90 | 96.30 | – |
| BCDU-Net [35] | 2019 | 78.50 | 98.20 | 85.10 | 93.70 | 93.70 | 97.88 |
| R2U-Net [25] | 2019 | 89.40 | 96.70 | 86.10 | 78.20 | 92.00 | – |
| Ensemble-A [29] | 2020 | 89.90 | 95.10 | 87.10 | 79.30 | 94.10 | – |
| AFLN-DGCL [12] | 2021 | – | – | 90.00 | 83.50 | 96.30 | – |
| AFLN-DGCL-FUSION [12] | 2021 | – | – | 92.00 | 85.60 | 96.60 | – |
| CKD-Net [36] | 2021 | 96.70 | 90.40 | 87.70 | 79.40 | 93.40 | 97.50 |
| CIU-Net [RGB-XYZ-Gray] | 2022 | 96.95 | 98.88 | 92.56 | 91.40 | 95.44 | 97.91 |
Bold indicates the best-achieved results
Fig. 5.
Segmentation results of CIU-Net using chosen images from ISIC 2018 dataset
Discussion
The proposed color invariant U-Net model (CIU-Net) utilizes a fusion of multi-color models to overcome the color variation and low contrast problems of skin lesion image segmentation. A new color mixture block with single convolution layer is added to learn invariant color representation which is subsequently fed to the modified U-Net model. This work analyzes the efficiency of fusing multiple color models for skin lesion segmentation. The proposed CIU-Net is provided with distinctive color models of the image to capture particular features which show up in some channels of different color models. The proposed model succeeds to enhance the behavior of original U-Net semantic segmentation deep model. The inter-connections between the two network encoder and decoder paths using channel-wise attention unit improves the segmentation results by focusing on the meaningful features while suppressing irrelevant ones. However, one weakness of the introduced approach is increasing computational time due to the addition of the attention units. There are some involved remarks, which can be outlined as follows:
The combination of RGB-XYZ-Gray color model is the best among others.
Applying channel attention unit enhanced the results of some performance metrics.
Utilizing hybrid loss function outperforms other simple loss functions.
Conclusion and future work
The variation of dermoscopy images’ colors contrast is an obstruction for distinguishing the infected regions from the surrounding un-infected ones. As a step toward solving this problem, this paper introduced a novel model for skin lesion segmentation based on U-Net deep learning model. The introduced CIU-Net model is able to achieve comparable results with other state-of-the-art techniques even for low contrast skin lesion images. The presented model is fed with a composite color spaces. The optimum choice of the fused color models significantly affects the behavior of the proposed CIU-Net segmentation results. In addition, the inclusion of the channel-wise attention units between the two network paths help to concentrate on the interesting features derived from the encoder part which lead to a significant improvement of segmentation results. The benchmark ISIC 2018 dataset is used to verify the robustness of the presented model. Additional advancements can be produced by using other attention network units to learn more descriminative features of skin images.
Acknowledgements
Saleh Aly would like to thank the Deanship of Scientific Research at Majmaah University for supporting this work under Project No. R-2022-229.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Rania Ramadan, Email: rania_abdrabou@scince.sohag.edu.eg.
Saleh Aly, Email: s.haridy@mu.edu.sa.
Mahmoud Abdel-Atty, Email: mahmoud.abdelatty@science.sohag.edu.eg.
References
- 1.Sinz C, Tschandl P, Rosendahl C, Akay BN, Argenziano G, Blum A, Braun RP, Cabo H, Gourhant J-Y, Kreusch J. Accuracy of dermatoscopy for the diagnosis of nonpigmented cancers of the skin. J Am Acad Dermatol. 2017;77(6):1100–1109. doi: 10.1016/j.jaad.2017.07.022. [DOI] [PubMed] [Google Scholar]
- 2.Martini M. An Atlas of Surface Microscopy of Pigmented Skin Lesions: Dermoscopy. Arch Dermatol. 2004 doi: 10.1001/archderm.140.2.248-a. [DOI] [Google Scholar]
- 3.Pour MP, Seker H. Transform domain representation-driven convolutional neural networks for skin lesion segmentation. Expert Syst Appl. 2020;144:113129. doi: 10.1016/j.eswa.2019.113129. [DOI] [Google Scholar]
- 4.Yuan Y, Lo YC. Improving Dermoscopic Image Segmentation With Enhanced Convolutional-Deconvolutional Networks. IEEE J Biomed Health Inf. 2019;23(2):519–526. doi: 10.1109/JBHI.2017.2787487. [DOI] [PubMed] [Google Scholar]
- 5.De Angelo GG, Pacheco AGC, Krohling RA. Skin lesion segmentation using deep learning for images acquired from smartphones. In: Proceedings of the International Joint Conference on Neural Networks 2019-July(July); 2019. p. 1–8. 10.1109/IJCNN.2019.8851803.
- 6.Abbas AA, Guo X, Tan WH, Jalab HA. Combined spline and B-spline for an improved automatic skin lesion segmentation in dermoscopic images using optimal color channel systems-level quality improvement. J Med Syst. 2014;38(8):1–8. doi: 10.1007/s10916-014-0080-7. [DOI] [PubMed] [Google Scholar]
- 7.Schaefer G, Rajab MI, Emre Celebi M, Iyatomi H. Colour and contrast enhancement for improved skin lesion segmentation. Comput Med Imaging Gr. 2011;35(2):99–104. doi: 10.1016/j.compmedimag.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Hajabdollahi M, Esfandiarpoor R, Khadivi P, Soroushmehr SMR, Karimi N, Samavi S. Simplification of neural networks for skin lesion image segmentation using color channel pruning. Comput Med Imaging Gr. 2020;82:101729. doi: 10.1016/j.compmedimag.2020.101729. [DOI] [PubMed] [Google Scholar]
- 9.Nisar H, Ch’ng YK, Chew TY, Yap VV, Yeap KH, Tang JJ. A color space study for skin lesion segmentation. In: ICCAS 2013–2013 IEEE International Conference on Circuits and Systems: ”Advanced Circuits and Systems for Sustainability”, 2013. p. 172–6 . 10.1109/CircuitsAndSystems.2013.6671629.
- 10.Ganesan P, Sathish BS, Leo Joseph LMI. HSL Color Space Based Skin Lesion Segmentation Using Fuzzy-Based Techniques. In: Pradhan GS, Morris K, editors. Advances in Electrical Control and Signal Systems. Lecture Notes in Electrical Engineering. Berlin: Springer; 2020. pp. 903–911. [Google Scholar]
- 11.Shan P, Wang Y, Fu C, Song W, Chen J. Automatic skin lesion segmentation based on fc-dpn. Comput Biol Med. 2020;123:103762. doi: 10.1016/j.compbiomed.2020.103762. [DOI] [PubMed] [Google Scholar]
- 12.Tang P, Yan X, Liang Q, Zhang D. Afln-dgcl:adaptive feature learning network with difficulty-guided curriculum learning for skin lesion segmentation. Appl Soft Comput. 2021;110:107656. doi: 10.1016/j.asoc.2021.107656. [DOI] [Google Scholar]
- 13.Zhang L, Yang G, Ye X. Automatic skin lesion segmentation by coupling deep fully convolutional networks and shallow network with textons. J Med Imaging. 2019;6(2):024001. doi: 10.1117/1.JMI.6.2.024001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Jiang X, Ding H, Zhao Y, Liu J. Knowledge-aware deep framework for collaborative skin lesion segmentation and melanoma recognition. Pattern Recogn. 2021;120:108075. doi: 10.1016/j.patcog.2021.108075. [DOI] [Google Scholar]
- 15.Sarker MMK, Rashwan HA, Akram F, Singh VK, Banu SF, Chowdhury FU, Choudhury KA, Chambon S, Radeva P, Puig D, et al. Slsnet: skin lesion segmentation using a lightweight generative adversarial network. Expert Syst Appl. 2021;183:115433. doi: 10.1016/j.eswa.2021.115433. [DOI] [Google Scholar]
- 16.Li W, Raj ANJ, Tjahjadi T, Zhuang Z. Digital hair removal by deep learning for skin lesion segmentation. Pattern Recogn. 2021;117:107994. doi: 10.1016/j.patcog.2021.107994. [DOI] [Google Scholar]
- 17.Tong X, Wei J, Sun B, Su S, Zuo Z, Wu P. Ascu-net: attention gate, spatial and channel attention u-net for skin lesion segmentation. Diagnostics. 2021;11(3):501. doi: 10.3390/diagnostics11030501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma Z, Tavares JMR. A novel approach to segment skin lesions in dermoscopic images based on a deformable model. IEEE J Biomed Health Inform. 2015;20(2):615–623. doi: 10.1109/JBHI.2015.2390032. [DOI] [PubMed] [Google Scholar]
- 19.Sumithra R, Suhil M, Guru DS. Segmentation and classification of skin lesions for disease diagnosis. Procedia Comput Sci. 2015;45:76–85. doi: 10.1016/j.procs.2015.03.090. [DOI] [Google Scholar]
- 20.Khan MA, Sharif M, Akram T, Damaševičius R, Maskeliunas R. Skin Lesion Segmentation and Multiclass Classification Using Deep Learning Features and Improved Moth Flame Optimization. Diagnostics. 2021;11(5):811. doi: 10.3390/diagnostics11050811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramadan R, Aly S. Cu-net: a new improved multi-input color u-net model for skin lesion semantic segmentation. IEEE Access. 2022;10:15539–15564. doi: 10.1109/ACCESS.2022.3148402. [DOI] [Google Scholar]
- 22.Ramadan R, Aly S. Dgcu-net: A new dual gradient-color deep convolutional neural network for efficient skin lesion segmentation. Biomed Signal Process Control. 2022;77:103829. doi: 10.1016/j.bspc.2022.103829. [DOI] [Google Scholar]
- 23.Azad R, Bozorgpour A, Asadi-Aghbolaghi M, Merhof D, Escalera S. Deep frequency re-calibration u-net for medical image segmentation. In: Proceedings of the IEEE/CVF International Conference on Computer Vision; 2021. p. 3274–83.
- 24.Tang P, Liang Q, Yan X, Xiang S, Sun W, Zhang D, Coppola G. Efficient skin lesion segmentation using separable-Unet with stochastic weight averaging. Comput Methods Prog Biomed. 2019;178:289–301. doi: 10.1016/j.cmpb.2019.07.005. [DOI] [PubMed] [Google Scholar]
- 25.Alom MZ, Yakopcic C, Hasan M, Taha TM, Asari VK. Recurrent residual u-net for medical image segmentation. J Med Imaging. 2019;6(1):014006. doi: 10.1117/1.JMI.6.1.014006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asadi-Aghbolaghi M, Azad R, Fathy M, Escalera S. Multi-level context gating of embedded collective knowledge for medical image segmentation. http://arxiv.org/abs/2003.05056 (2020).
- 27.Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) 2015;9351:234–241. doi: 10.1007/978-3-319-24574-4_28. [DOI] [Google Scholar]
- 28.Chen L, Zhang H, Xiao J, Nie L, Shao J, Liu W, Chua TS. SCA-CNN: Spatial and channel-wise attention in convolutional networks for image captioning. In: Proceedings—30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017 2017-Janua; 2017. p. 6298–6306 arXiv:1611.05594. 10.1109/CVPR.2017.667.
- 29.Goyal M, Oakley A, Bansal P, Dancey D, Yap MH. Skin Lesion Segmentation in Dermoscopic Images with Ensemble Deep Learning Methods. IEEE Access. 2020;8:4171–4181. doi: 10.1109/ACCESS.2019.2960504. [DOI] [Google Scholar]
- 30.Al-masni MA, Al-antari MA, Choi MT, Han SM, Kim TS. Skin lesion segmentation in dermoscopy images via deep full resolution convolutional networks. Comput Methods Prog Biomed. 2018;162:221–231. doi: 10.1016/j.cmpb.2018.05.027. [DOI] [PubMed] [Google Scholar]
- 31.Chen LC, Papandreou G, Kokkinos I, Murphy K, Yuille AL. DeepLab: Semantic Image Segmentation with Deep Convolutional Nets, Atrous Convolution, and Fully Connected CRFs. IEEE Trans Pattern Anal Mach Intell. 2018;40(4):834–848. doi: 10.1109/TPAMI.2017.2699184. [DOI] [PubMed] [Google Scholar]
- 32.Gu Z, Cheng J, Fu H, Zhou K, Hao H, Zhao Y, Zhang T, Gao S, Liu J. Ce-net: Context encoder network for 2d medical image segmentation. IEEE Trans Med imaging. 2019;38(10):2281–2292. doi: 10.1109/TMI.2019.2903562. [DOI] [PubMed] [Google Scholar]
- 33.Ibtehaz N, Rahman MS. Multiresunet: Rethinking the u-net architecture for multimodal biomedical image segmentation. Neural Netw. 2020;121:74–87. doi: 10.1016/j.neunet.2019.08.025. [DOI] [PubMed] [Google Scholar]
- 34.Feng S, Zhao H, Shi F, Cheng X, Wang M, Ma Y, Xiang D, Zhu W, Chen X. Cpfnet: Context pyramid fusion network for medical image segmentation. IEEE Trans Med Imaging. 2020;39(10):3008–3018. doi: 10.1109/TMI.2020.2983721. [DOI] [PubMed] [Google Scholar]
- 35.Reza A, Maryam A-A, Mahmood Fathy SE. Bi-Directional ConvLSTM U-Net with densley connected convolutions. http://arxiv.org/abs/1909.00166v1
- 36.Jin Q, Cui H, Sun C, Meng Z, Su R. Cascade knowledge diffusion network for skin lesion diagnosis and segmentation. Appl Soft Comput J. 2021;99:106881. doi: 10.1016/j.asoc.2020.106881. [DOI] [Google Scholar]