Abstract
Background:
Background parenchymal enhancement (BPE) is assessed on breast MRI reports as mandated by the Breast Imaging Reporting and Data System (BI-RADS) but is prone to inter- and intra-reader variation. Semi- and fully-automated BPE assessment tools have been developed but none has surpassed radiologist BPE designations.
Purpose:
To develop a deep learning model for automated BPE classification and to compare its performance with current standard-of-care radiology report BPE designations.
Study Type:
Retrospective.
Population:
Consecutive high-risk patients (i.e., >20% lifetime risk of breast cancer) who underwent contrast-enhanced screening breast MRI from October 2013–January 2019. The study included 5,224 breast MRIs, divided into 3,998 training, 444 validation, and 782 testing exams. On radiology reports, 1,286 exams were categorized as high BPE (i.e., Marked or Moderate) and 3,938 as low BPE (i.e., Mild or Minimal).
Field Strength/Sequence:
1.5T and 3T; 1 pre-contrast and 3 post-contrast phases of fat-saturated T1-weighted dynamic contrast-enhanced imaging.
Assessment:
Breast MRIs were used to develop two deep learning models (Slab Artificial Intelligence (AI); Maximum Intensity Projection (MIP) AI) for BPE categorization using radiology report BPE labels. Models were tested on a held-out test sets using radiology report BPE and three-reader averaged consensus as the reference standards.
Statistical tests:
Model performance was assessed using receiver operating characteristic (ROC) curve analysis. Associations between high BPE and BI-RADS assessments were evaluated using McNemar’s Chi-square test (α*=0.025).
Results:
The Slab AI model significantly outperformed the MIP AI model across the full test set (area under the curve (AUC) of 0.84 vs. 0.79) using the radiology report reference standard. Using three-reader consensus BPE labels reference standard, our AI model significantly outperformed radiology report BPE labels. Finally, the AI model was significantly more likely than the radiologist to assign “high BPE” to suspicious breast MRIs and significantly less likely than the radiologist to assign “high BPE” to negative breast MRIs.
Data Conclusion:
Fully automated BPE assessments for breast MRIs could be more accurate than BPE assessments from radiology reports.
Keywords: Artificial Intelligence, Deep Learning, Background Parenchymal Enhancement, Breast MRI, Cancer Risk Assessment
INTRODUCTION
Artificial intelligence (AI) has exciting potential to revolutionize the field of diagnostic radiology in many ways, but as a first step, it should be more widely applied to automate the radiologist’s most simple, repetitive tasks. In breast MRI, for example, automating the categorization of background parenchymal enhancement (BPE) would not only improve clinical efficiency, but eliminate the high interreader variability of current BPE assessments, which hinders its use as a predictive imaging biomarker(1–3).
BPE refers to the physiologic enhancement of normal breast tissue after intravenous contrast injection(4, 5). BPE depends on tissue vascularity and vascularity permeability, and is influenced by the underlying hormonal milieu (6). The American College of Radiology MRI Breast Imaging Reporting and Data System (BI-RADS) atlas divides BPE into four categories depending on the volume and intensity of post-contrast enhancement: marked, moderate, mild, and minimal (7). It is recommended that breast radiologists include this BPE categorization on every breast MRI report. This categorization is useful because: (i) BPE may be a risk factor for the presence of cancer) (8–12), and (ii) BPE may also give information about a radiologist’s risk of missing that cancer (i.e., high BPE increases abnormal breast MRI interpretation rates) (7, 13).
However, there is wide variability in BPE assessment among radiologists (1, 3). Automated BPE categorization has the potential to standardize the breast MRI interpretation process. Quantitative tools for three-dimensional (3D) assessment of BPE have been developed using a variety of semi- and fully-automated methods (14–17), including more recent work using deep learning (18–21). Ha et al. developed a fully automated model for quantification of fibroglandular tissue (FGT) and BPE using their previously published quantification methods as the ground truth (16, 19). Nam et al. developed a similar model using qualitative FGT and BPE scoring by radiologists as the ground truth. Most recently, Borkowski et al developed a fully automated BPE classification modal using sequential “breast slice detection” and a “BPE classification” neural networks, achieving non-inferior performance to experienced radiologists(21). However, no BPE tool to date has been shown to surpass current standard-of-care radiologist BPE designations, which is the standard clinical practice today.
Herein, we aimed to develop two different AI models to categorize BPE — a two-dimensional (2D) maximum intensity projection (MIP) AI Model, and a Slab AI Model — and to assess the diagnostic performance of of each model using radiology report BPEs as the reference standard. We also generated BPE labels based on consensus reading on a subset of our data and used this reference standard to evaluate whether our top-performing AI model would outperform the radiology report BPE labels. Additionally, we analyzed BPE trends to glean whether our top-performing AI model might better capture breast cancer risk compared with the current practice of radiologist-designated qualitative BPE assessments.
MATERIALS AND METHODS
Patients
The Institutional Review Board, at a tertiary cancer care center, approved this Health Insurance Portability and Accountability Act–compliant retrospective study, and the need for informed consent was waived.
Consecutive contrast-enhanced screening breast MRI exams performed on high-risk patients at our institution from October 2013–January 2019 were reviewed for this study. Exams were included if the radiology report contained a BPE assessment using BI-RADS verbiage (i.e. “Marked”, “Moderate”, “Mild”, or “Minimal”). Exams were excluded if the Digital Imaging and Communications in Medicine (DICOM) header contained non-standardized MRI series description names, if the dynamic post-contrast sequences were obtained in sagittal orientation, if ultrafast imaging was performed, or if image pre-processing errors occurred (e.g. matrix discrepancies between pre- and post-contrast images). Additionally, the following exams were also excluded: (i) BI-RADS 1/2/3 exams that lacked two-year negative breast MRI follow-up, and (ii) BI-RADS 4/5 exams in which histopathologic sampling was recommended but not performed. For all cases with BI-RADS 4/5 designations, biopsy pathology reports from image-guided core needle biopsy were parsed using an in-house natural language processing (NLP) algorithm and labeled as malignant (invasive or in-situ cancer), high-risk benign, or benign. Cases were excluded if the NLP was unable to parse the pathology report.
Breast MRI Acquisition
All breast MRI exams were performed on a 1.5 or 3.0 Tesla system (Discovery 750; GE Medical Systems, Waukesha, WI, USA) using a dedicated 8- or 16-channel breast coil. The gadolinium-based contrast agent was administered at a concentration of 0.1 mmol gadobutrol per kg body weight (Gadavist; Bayer Healthcare Pharmaceuticals, Inc., Whippany, NJ, USA), at a rate of 2 ml/s. One pre-contrast phase and three post-contrast phases of fat-saturated T1-weighted dynamic contrast-enhanced images were acquired (post-contrast imaging began ~30 seconds after contrast injection with each phase lasting ~90 seconds). Additional acquisition parameters are listed in Supplementary Table 1.
Deep Learning Model Development
Data Preprocessing
A deep learning model was developed to assign a BPE label to breast MRI exams. An automated data pipeline was built to extract the axial pre-contrast fat saturated T1-weighted series and the axial first post-contrast series of the exam (obtained at approximately 90 seconds after injection of the gadolinium-based contrast agent). Axial subtraction images were created. The breast was segmented using k-means clustering and central slides were extracted. Axial slices were pooled into three maximum intensity slabs (i.e., three axial subtraction MIPs generated from the upper, middle, and lower breast respectively, to serve as an input for the Slab AI model), as well as into a since standard axial subtraction MIP (to serve as an input for the MIP AI model). Figure 1 provides an illustration of the creation of Slab and MIP images. Each image was then split into “left breast” and “right breast” images. Images were converted to jpg file format and signal intensity was normalized using zero mean, with a standard deviation of 1. Data was augmented with left/right flips. Exams were excluded if this automated algorithm was unable to identify and/or process the pre- and post-contrast series.
Figure 1:
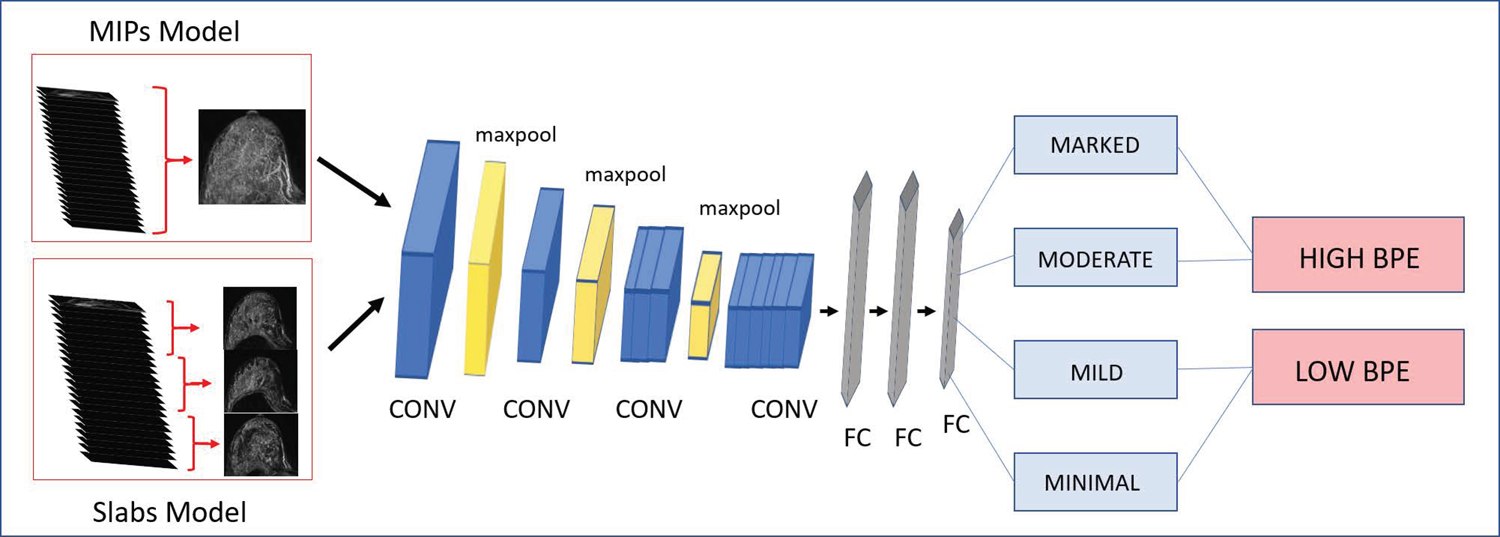
AI model architecture schematic. A VGG-19 architecture was trained to classify images into four BPE categories, which were then pooled into “high BPE” and “low BPE” categories. In the MIP AI model, axial MIPs generated from the first subtraction phase were used as the model input. In the Slab AI model, axial slices from the first subtraction phase were pooled into three maximum intensity slabs, which were each used as an independent model input. CONV = convolutional layer, FC = fully connected layer.
NLP was used to extract BPE labels (marked, moderate, mild, or minimal) from the original radiology reports. Marked and moderate BPE were considered “high BPE”, while mild and minimal BPE were considered “low BPE”. These labels served as Reference Standard #1.
Convolutional Neural Network (CNN) Architecture and Training
A VGG19 architecture (Python 3.7.0, Python Software Foundation, Beaverton, Oregon, USA, with TensorFlow 1.11.0, Google, Mountain View, California, USA) run using a NVIDIA-GTX-1080ti GPU was trained to classify images into the four BPE categories using MRI data alone as input and four BPE labels from the radiology report as the ground truth (i.e., Marked, Moderate, Mild, Minimal). ImageNet weights were used for initialization, and training was run for 20 epochs with a learning rate of 1e-5, a batch size of 32, and a momentum of 0.9. Dropout of 75% was used after the first fully connected layer. The stochastic gradient decent optimizer and a categorical cross entropy loss function were used. The code is publicly available online: https://github.com/eskreis/BPE_project.git.
Two CNN models were trained: (i) a MIP AI model, using a single standard axial 2D subtraction MIP, generated from the whole breast, as the model input, and (ii) a Slab AI model, using three axial separate 2D subtraction MIPs, generated from the upper, middle, and lower breast respectively, as the model input. Slab AI model results were evaluated by averaging the classification scores across all slabs to generate a final BPE ouput on a per breast basis. While models were initially trained at 4-way classification, models were tested using a pooled binary classification of high BPE versus low BPE (i.e., Reference Standard #1) to maximize clinical relevance. Figure 1 shows a schematic illustration of the AI model architecture.
Breast MRI exams were then divided into training, validation, and testing sets. First, each patient was randomly assigned to the training or validation subgroup, using a 9:1 split. Then, two test sets were created with an eye towards clinical relevance: (i) a BI-RADS 4/5 test set, which included all available BI-RADS 4/5 exams extracted by our NLP, and (ii) a reader study subgroup, which consisted of 100 randomly selected BI-RADS 1 exams. Patients with exams in either test set were completely removed from training and validation sets and were not used during model training or validation. In this way, there was no patient crossover between training, validation, and testing sets.
Reader Study BPE Labels
The 100 randomly selected BI-RADS 1 exams were independently reviewed by three breast fellowship-trained radiologists (**, **, **, with 16, 7, and 4 years of experience) who recorded BPE designations based on the first post-contrast phase. Readers were blinded to patient information, radiology report BPE designations, and prior imaging. BPE designations assigned independently by the three radiologists were averaged to generate a combined reading per breast, hereafter referred to as the “consensus reading.” To maximize clinical relevance, BPE designations were pooled into high BPE and low BPE categories. Pooled consensus readings served as Reference Standard #2. Ninety-four exams were included in the reader study since six of the 100 exams in this test set were excluded due to failure of image pre-processing.
Statistical Analysis
Diagnostic performance metrics, i.e., accuracy, sensitivity, and specificity, were calculated over the test sets, with 95% exact confidence intervals (CIs) for both the MIP AI model and the Slab AI model, using Reference Standard #1. Receiver operating characteristic (ROC) curves were generated for the MIP AI and Slab AI models across the full testing set, using reference standard #1. The areas under the curve (AUCs) were compared using DeLong’s test for correlated ROCs. Mulitple comparison correction was done using Bonferroni adjustment (α* = 0.025). Using the top-performing AI model over the consensus reading for BI-RADS 1, performance metrics were calculated using both Reference Standard #1 (i.e., the reference standard used during model training) and Reference Standard #2. ROC curves were generated and AUCs for the different reference standards were also compared using DeLong’s test.
Using the top-performing AI model and Reference Standard #2, diagnostic accuracy of the AI model was compared with that of the radiology report over the consensus reading for BI-RADS 1. This comparison was performed using a one-tailed McNemar’s Chi-square test with a level of statistical significance of 0.005.
To assess the clinical relevance of BPE using the top-performing AI model versus the radiology report, the frequency of high BPE labeling by the top-performing AI model was compared to the radiology report for the BI-RADS 1 and BI-RADS 4/5 subgroups. Comparisons were made using one-tailed McNemar’s Chi-square test with Bonferroni correction for multiple comparisons (α* = 0.025). All statistical analyses were performed using SAS software, version 9.4 (SAS Institute, Cary, NC, USA) and MATLAB 2017b (Mathworks, Natick, Massachusetts, USA).
RESULTS
Patient and Exam Characteristics
Of the 16,235 screening breast MRIs performed at our institution between 2013 and 2019, 1,573 were excluded due to lack of standardized BPE labeling, 6,775 were excluded due to missing pathology information or lack of two-year negative imaging follow-up, and 2,663 were excluded due to non-standardized MRI series descriptions, sagittal acquisition, ultrafast imaging, or image pre-processing errors. Figure 2 provides a flow diagram for patient selection. After these exclusions, the study included 5,224 breast MRI exams from 3,705 female patients (mean age ± standard deviation, 52 years ± 11). The breast MRI data were divided into 3,998 training exams, 444 validation exams, and 782 testing exams, without patient overlap.
Figure 2:
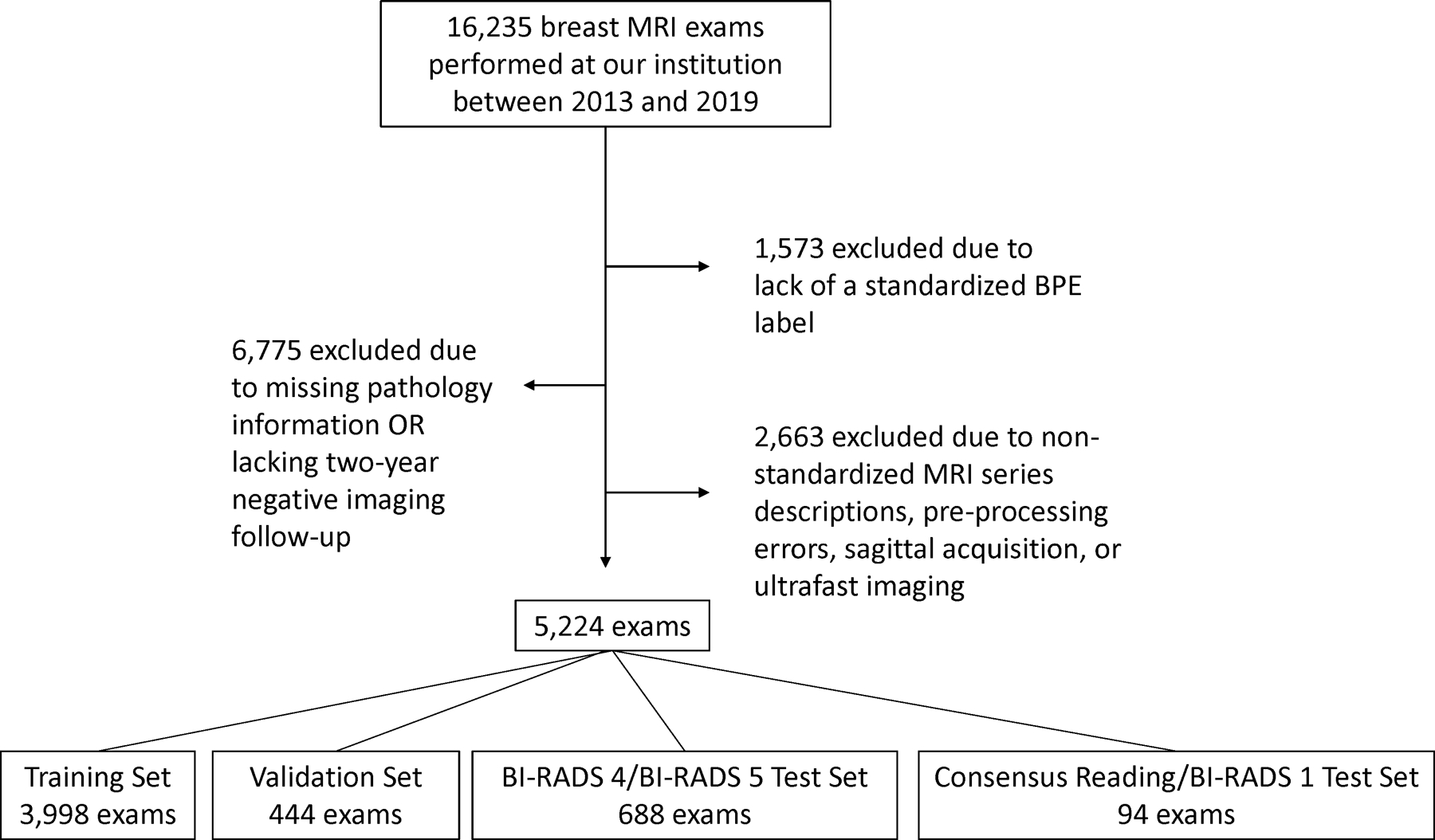
Patient selection flow diagram.
According to the radiology report BPE labels, high BPE was present in 1030/4442 (23.2%) of the training and validation data, in 236/688 (34.3%) of the BI-RADS 4/5 test set subgroup, and in 20/94 (21.3%) of the BI-RADS 1 reader study test set subgroup. Of the BI-RADS 4/5 cases, 228/684 (33.1%) yielded malignancy on subsequent core needle biopsy. Table 1 provides further information on demographics and patient characteristics.
Table 1.
MRI Exam Characteristics
| Training & Validation Sets | Testing Sets | ||
|---|---|---|---|
| BI-RADS 4/BI-RADS 5 Subgroup | Consensus Reading (BI-RADS 1) Subgroup | ||
| Number of MRI Exams | 4442 | 688 | 94 |
|
| |||
| Number of Patients | 2956 | 655 | 94 |
|
| |||
| Patient Age | 53 ± 11 | 49 ± 12 | 51 ± 13 |
|
| |||
| BPE Category | |||
| Marked | 282 (6.3%) | 73 (10.6%) | 8 (8.5%) |
| Moderate | 748 (16.9%) | 163 (23.7%) | 12 (12.8%) |
| Mild | 1535 (34.6%) | 268 (39.0%) | 25 (26.6%) |
| Minimal | 1877 (42.3%) | 184 (26.7%) | 49 (52.1%) |
|
| |||
| BPE Overall Category | |||
| High | 1030 (23.2%) | 236 (34.3%) | 20 (21.3%) |
| Low | 3412 (76.8%) | 452 (65.7%) | 74 (78.7%) |
|
| |||
| BI-RADS Categories | |||
| 1 | 841 (18.9%) | 0 (0%) | 94 (100.0%) |
| 2 | 2451 (55.2%) | 0 (0%) | 0 (0%) |
| 3 | 9 (0.2%) | 0 (0%) | 0 (0%) |
| 4 | 46 (1.0%) | 668 (97.1%) | 0 (0%) |
| 5 | 2 (0.04%) | 20 (2.9%) | 0 (0%) |
| 6 | 1093 (24.6%) | 0 (0%) | 0 (0%) |
|
| |||
| Biopsy Pathology | |||
| Malignant | n/a | 228 (33.1%) | n/a |
| High Risk | 148 (21.5%) | ||
| Benign | 296 (43.0%) | ||
Abbreviations: BI-RADS, Breast Imaging Reporting and Data System; BPE, background parenchymal enhancement
Diagnostic Performance
Using Reference Standard #1, the Slab AI model’s AUC was significantly higher than the MIP AI model’s AUC over the full test set (Slab AI model AUC, 0.84; 95% CI: 0.82, 0.86; MIP AI model AUC, 0.79; 95% CI: 0.76, 0.81). ROC curves and AUCs are displayed in Figure 3 and descriptive statistics are reported in Table 2. See Supplementary Table 2 for the test set results of 4-way classification (i.e. Marked/Moderate/Mild/Minimal).
Figure 3:
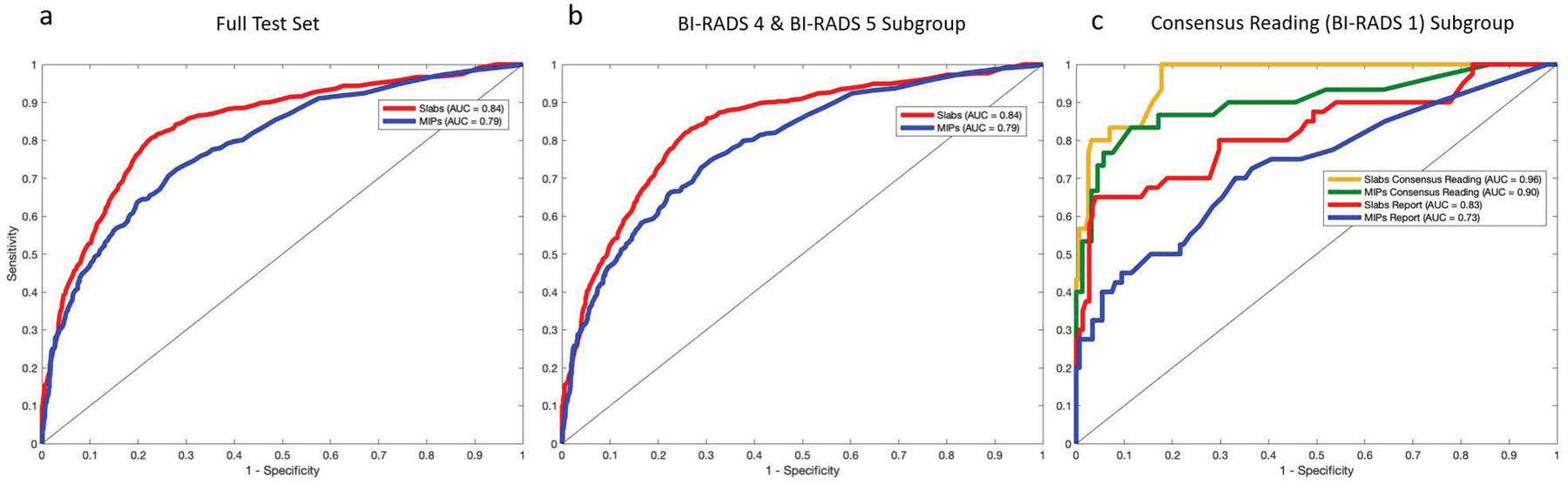
AI model test set results. Receiver operating characteristic curves and areas under the curve for the Slab AI and MIP AI models, over the full test set (a), over the BI-RADS 4/5 subgroup (b) and over the reader study subgroup (c). In (c) results are displayed using both Reference Standard #1 (Radiology Report BPE labels) and Reference Standard #2 (Consensus Reading BPE labels).
Table 2.
AI Model Performance Metrics
| A | B | C | D | E | F | |
|---|---|---|---|---|---|---|
| BPE Classification Method | AI Model (MIPs) | AI Model (Slabs) | AI Model (Slabs) | AI Model (Slabs) | AI Model (Slabs) | Radiology Report |
| Reference Standard | Reference Standard #1 Radiology Report | Reference Standard #1 Radiology Report | Reference Standard #1 Radiology Report | Reference Standard #1 Radiology Report | Reference Standard #2 Consensus Reading | Reference Standard #2 Consensus Reading |
| Test Set | Full Testing Set | Full Testing Set | BI-RADS 4/BI-RADS 5 Subgroup | Reader Study Subgroup | Reader Study Subgroup | Reader Study Subgroup |
| Accuracy | 76% (74%, 79%) | 79% (77%, 81%) | 77% (75%, 80%) | 89% (84%, 93%) | 94% (91%, 98%) | 88% (84%, 93%) |
| Sensitivity | 57% (50%, 63%) | 75% (71%, 79%) | 76% (73%, 80%) | 57% (42%, 73%) | 77% (62%, 92%) | 80% (65%, 94%) |
| Specificity | 86% (83%, 89%) | 81% (79%, 83%) | 78% (76%, 81%) | 97% (95%, 100%) | 97% (95%, 100%) | 90% (85%, 95%) |
| AUC | 0.79 (0.76, 0.81) | 0.84 (0.82, 0.86) | 0.84 (0.81, 0.86) | 0.83 (0.74, 0.91) | 0.96 (0.94, 0.99) | ------ |
Abbreviations: AI, artificial intelligence; AUC, area under the curve; BI-RADS, Breast Imaging Reporting and Data System; BPE, background parenchymal enhancement; MIP, maximum intensity projection
Over the consensus reading for BI-RADS 1, the AUC of the Slab AI model (the top-performing model) was significantly higher when Reference Standard #2, not Reference Standard #1, was used (Ref #2 AUC, 0.96; 95% CI: 0.94, 0.99; Ref #1 AUC, 0.83; 95% CI: 0.74, 0.91). Notably, when Reference Standard #2 was used, the diagnostic accuracy of the Slab AI model was significantly higher than the diagnostic accuracy of the radiology reports (Slab AI model accuracy, 177/188 (94.2%); 95% CI: 90.8%, 97.5%; radiology report accuracy, 166/188 (88.3%); 95% CI: 83.7%, 92.9%). Figure 3 shows ROC curves and Table 2 provides additional performance metrics. In Figure 4, example cases are shown where AI model BPE categorizations agree and disagree with the radiology report BPE assessments.
Figure 4:
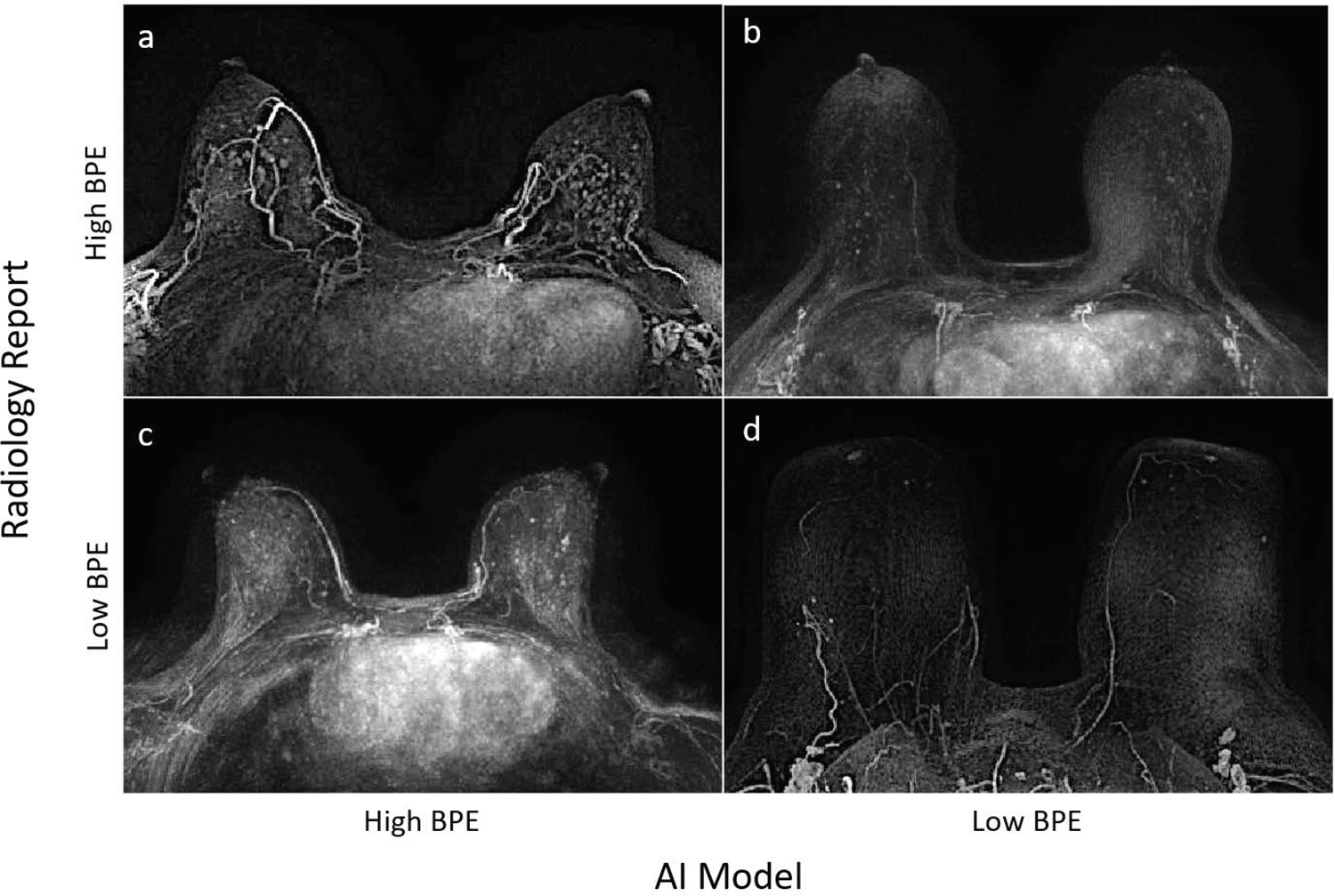
Case examples illustrating (a) the Slab AI model and Radiology Report (Reference Standard #1) both classifying as High BPE, (b) the Slab AI Model classifying as Low BPE and the Radiology Report classifying as High BPE, (c) the Slab AI Model classifying as High BPE and the Radiology Report classifying as Low BPE, and (d) both the Slab AI Model and Radiology Report classifying as Low BPE.
High BPE as a Risk Factor for the Presence of Malignancy
Finally, the AI model was significantly more likely than the radiologist to assign “high BPE” to suspicious breast MRIs (558/1376 (40.1%) vs. 472/1376 (34.3%)) and significantly less likely than the radiologist to assign “high BPE” to negative breast MRIs (27/188 (14.4%) vs. 40/188 (21.3%)), highlighting the AI model’s clinical relevance (see Figure 5).
Figure 5:
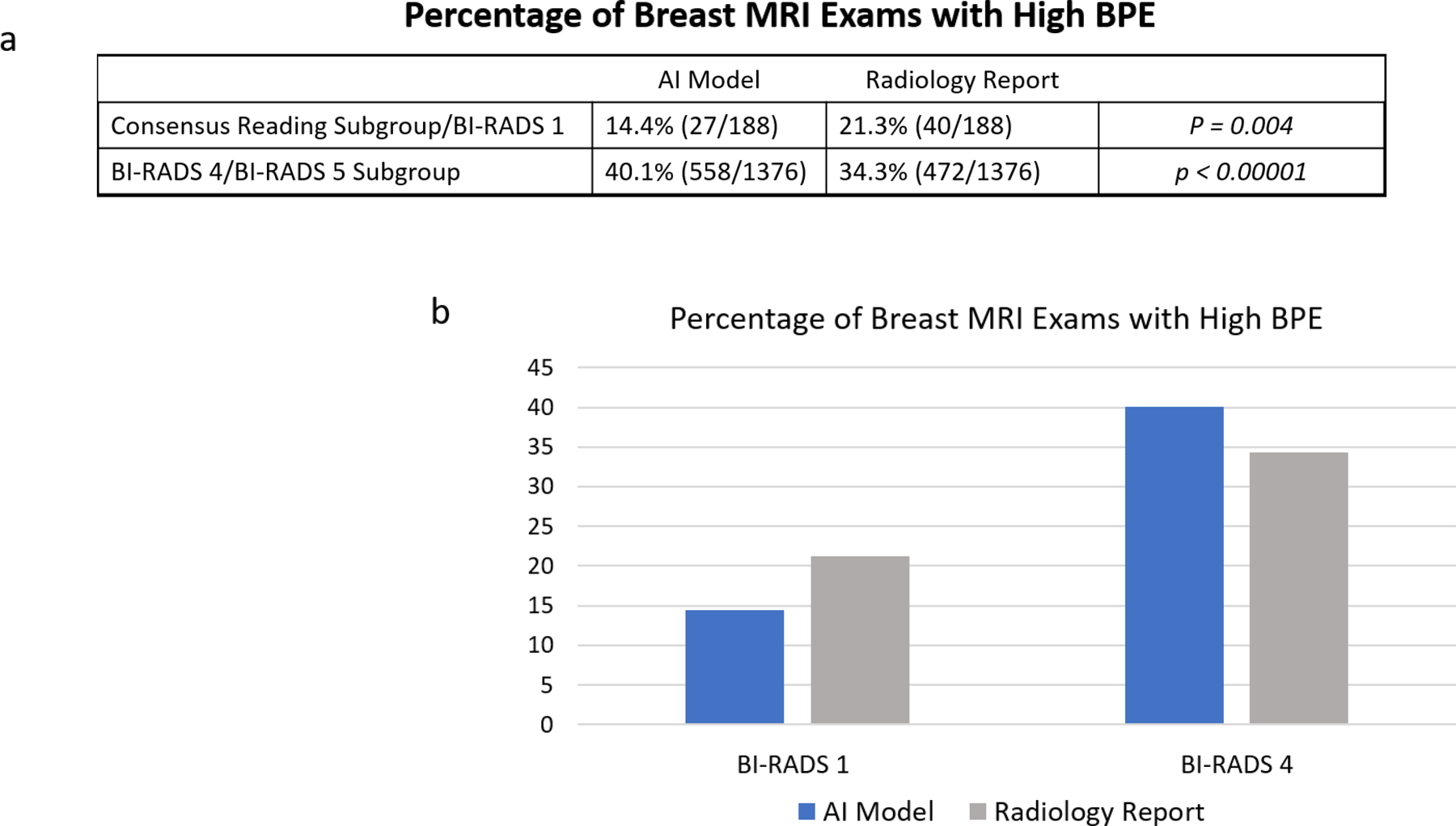
Trends in AI model BPE designations. (a) In the Consensus Reading BI-RADS 1 Subgroup, the Slab AI Model was less likely than the Radiology Report to classify cases as High BPE (p = 0.004). In the BI-RADS 4/5 Subgroup, the trend was reversed (p < 0.0001). (b) Percentages of Breast MRI Exams with High BPE, according to the Slab AI Model versus the Radiology Report.
DISCUSSION
In this study, we developed a fully automated, easy-to-use AI model for BPE categorization on breast MRI. Our top-performing AI model, i.e., Slab AI model, provides significantly improved BPE assessments compared with the radiology reports. It performed similarly well across both normal breast MRI exams (i.e., the BI-RADS 1 test set) and abnormal breast MRI exams (i.e., the BI-RADS 4/5 test set), indicating its robustness across different types of breast MRIs.
Additionally, we showed that our Slab AI model was both more likely than the radiology report to assign “high BPE” to suspicious exams and less likely than the radiology report to assign “high BPE” to negative exams, indicating a potential clinical relevance of the AI model BPE designations.
BPE labeling in radiology reports is subjective and prone to inter- and intra-reader variation, which stymies efforts to use BPE as a clinical tool or biomarker(1–3). Fellowship-trained breast radiologists achieve only fair inter-reader agreement on BPE categorization (κ = 0.36), and show only modest improvement after dedicated training in BPE standardization (κ = 0.45) (1, 3). In prior work, BPE quantification methods included an automated fibroglandular tissue segmentation step and a calculation of percent enhancement (14, 15), but these methods show only slight to fair agreement with radiologists (κ = 0.20–0.36) and are more discordant at higher levels of BPE, precisely when they might be more clinically useful (2). More recently, supervised deep learning models have been used to approximate radiology report BPE designations, with test set accuracies ranging from 0.70 to 0.93 (18, 19, 21, 22). In contrast to prior work, we show that an AI model not just approximates the radiology report BPE classification, but can surpass it.
Our AI model was trained with radiology report BPE labels, but learned to become even more accurate than those labels when we did a head-to-head comparison using consensus readings as a reference standard (i.e., Reference Standard #2). We hypothesize that the large size of our training dataset compensated for noisy labels during training, which allowed our deep learning model to generalize. In fact, mathematical relationships between noisy training examples, dataset size, and deep learning model performance have been well-studied in the deep learning literature (23).
The large size of a four-dimensional (4D) breast MRI dataset makes it necessary to distill the clinically relevant spatial and temporal information into a more computationally feasible form. A popular approach is to create MIPs of the axial subtraction images, thus incorporating both spatial and temporal information into a single 2D image (referred to in our study as the MIP AI model). We used our domain knowledge of the expected spatial and temporal distributions of BPE in the breast to design an improved AI model that harnesses quasi-3D spatial and temporal information from the MRI exam, while still permitting use of a 2D model architecture. Specifically, since BPE is a global property present throughout the fibroglandular tissue, we pooled axial subtraction images into three slabs, from which three MIPs were generated (referred to in our study as the Slab AI model). Our Slab AI model outperformed the standard 2D MIP model, underscoring the importance of incorporating more granular, clinically relevant data into model development.
With an eye towards clinical relevance, we show that, compared with the radiology report, our AI model is more likely to assign “high BPE” to suspicious breast MRI exams and less likely to assign “high BPE” to negative breast MRI exams, suggesting that it might serve as a better surrogate for assessing breast cancer risk. This is in line with two recent meta-analyses, which report that high BPE is associated with the presence of breast cancer (3, 12). However, the meta-analyses conflict regarding certain subcategories of patients, and certain BPE categories. We suspect that these inconsistencies are due in part to the noisy BPE labels from radiology reports and are hopeful that our automated AI tool will standardize BPE labeling and enable us to identify both stronger and more nuanced relationships about BPE and breast cancer.
Our AI tool may be seamlessly incorporated into clinical workflow, potentially resulting in improved clinical efficiency. We are currently working to integrate our BPE model into our hospital’s radiology report dictation system so that reports can be autopopulated with our model’s BPE assessment, eliminating the need for radiologists to manually input this information. This could streamline and automatize the BPE assessment process, saving the radiologist time, reducing inter- and intra-reader variability, and resulting in reports with more accurate and robust BPE labeling.
Limitations
First, all breast MRIs were performed on single-institution and single-vendor scanners; in future work we will externally validate our model with multi-institutional datasets. Additionally, the patients in this study were overwhelmingly white women, and in future work we will also work to secure a more racially diverse test dataset to ensure that our algorithms work equally well on minority groups. In future work we also plan to extend our BPE tool to automate the classification of fibroglandular tissue, which has only moderate intra-/inter-observer agreement (24), and also to further explore whether high BPE exams are associated not just with abnormal breast MRIs but with the presence of current and/or future breast cancer.
Conclusion
Our fully automated, reproducible AI model, i.e., Slab AI model, for BPE categorization provides more accurate BPE assessments than our MIP-based AI model and demonstrates improved diagnostic accuracy compared to standard-of-care radiology report BPE categorization. Our AI tool may be configured to autopopulate breast MRI reports with BPE assessment information, thus improving the accuracy of BPE designations while simultaneously alleviating the radiologist’s workload.
Supplementary Material
ACKNOWLEDGMENTS:
The computation for this study was performed on the Lilac cluster hosted by the Sloan Kettering Institute, New York. The authors thank Joanne Chin, MFA, ELS, for manuscript editing.
Grant support:
The project described was supported by RSNA Research and Education Foundation, through grant number RF1905. The content is solely the responsibility of the authors and does not necessarily represent the official views of the RSNA R&E Foundation. This work was also supported in part through the NIH/NCI Cancer Center Support Grant P30 CA008748.
REFERENCES
- 1.Melsaether A, McDermott M, Gupta D, Pysarenko K, Shaylor SD, Moy L. Inter- and intrareader agreement for categorization of background parenchymal enhancement at baseline and after training. AJR Am J Roentgenol. 2014;203(1):209–15. Epub 2014/06/22. doi: 10.2214/AJR.13.10952. [DOI] [PubMed] [Google Scholar]
- 2.Pujara AC, Mikheev A, Rusinek H, Gao Y, Chhor C, Pysarenko K, et al. Comparison between qualitative and quantitative assessment of background parenchymal enhancement on breast MRI. J Magn Reson Imaging. 2018;47(6):1685–91. Epub 2017/11/16. doi: 10.1002/jmri.25895. [DOI] [PubMed] [Google Scholar]
- 3.Bignotti B, Signori A, Valdora F, Rossi F, Calabrese M, Durando M, et al. Evaluation of background parenchymal enhancement on breast MRI: a systematic review. Br J Radiol. 2017;90(1070):20160542. Epub 2016/12/08. doi: 10.1259/bjr.20160542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris EA CC, Lee CH, et al. ACR BI-RADS ® Magnetic Resonance Imaging. In: ACR BI-RADS ® Atlas, Breast Imaging Reporting and Data System. 2013. Reston, VA: American College of Radiology. [Google Scholar]
- 5.Giess CS, Yeh ED, Raza S, Birdwell RL. Background Parenchymal Enhancement at Breast MR Imaging: Normal Patterns, Diagnostic Challenges, and Potential for False-Positive and False-Negative Interpretation. Radiographics. 2014;34(1):234–U93. doi: 10.1148/rg.341135034. [DOI] [PubMed] [Google Scholar]
- 6.Kuhl CK, Bieling HB, Gieseke J, Kreft BP, Sommer T, Lutterbey G, et al. Healthy premenopausal breast parenchyma in dynamic contrast-enhanced MR imaging of the breast: normal contrast medium enhancement and cyclical-phase dependency. Radiology. 1997;203(1):137–44. Epub 1997/04/01. doi: 10.1148/radiology.203.1.9122382. [DOI] [PubMed] [Google Scholar]
- 7.D’Orsi CJSE, Mendelson EB, Morris EA. ACR BI-RADS® Atlas, Breast Imaging Reporting and Data System. Reston, VA: American College of Radiology; 2013. [Google Scholar]
- 8.King V, Brooks JD, Bernstein JL, Reiner AS, Pike MC, Morris EA. Background parenchymal enhancement at breast MR imaging and breast cancer risk. Radiology. 2011;260(1):50–60. Epub 2011/04/16. doi: 10.1148/radiol.11102156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson CM, Mallawaarachchi I, Dwivedi DK, Ayyappan AP, Shokar NK, Lakshmanaswamy R, et al. The Association of Background Parenchymal Enhancement at Breast MRI with Breast Cancer: A Systematic Review and Meta-Analysis. Radiology. 2019;292(3):552–61. Epub 2019/06/27. doi: 10.1148/radiol.2019182441. [DOI] [PubMed] [Google Scholar]
- 10.Dontchos BN, Rahbar H, Partridge SC, Korde LA, Lam DL, Scheel JR, et al. Are Qualitative Assessments of Background Parenchymal Enhancement, Amount of Fibroglandular Tissue on MR Images, and Mammographic Density Associated with Breast Cancer Risk? Radiology. 2015;276(2):371–80. Epub 2015/05/13. doi: 10.1148/radiol.2015142304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu N, Zhao J, Li Y, Fu Q, Zhao L, Chen H, et al. Breast cancer and background parenchymal enhancement at breast magnetic resonance imaging: a meta-analysis. BMC Med Imaging. 2021;21(1):32. Epub 2021/02/21. doi: 10.1186/s12880-021-00566-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H, Guo L, Tao W, Zhang J, Zhu Y, Abdelrahim MEA, et al. Possible Breast Cancer Risk Related to Background Parenchymal Enhancement at Breast MRI: A Meta-Analysis Study. Nutr Cancer. 2021;73(8):1371–7. Epub 2020/07/24. doi: 10.1080/01635581.2020.1795211. [DOI] [PubMed] [Google Scholar]
- 13.Giess CS, Yeh ED, Raza S, Birdwell RL. Background parenchymal enhancement at breast MR imaging: normal patterns, diagnostic challenges, and potential for false-positive and false-negative interpretation. Radiographics. 2014;34(1):234–47. Epub 2014/01/17. doi: 10.1148/rg.341135034. [DOI] [PubMed] [Google Scholar]
- 14.Klifa C, Suzuki S, Aliu S, Singer L, Wilmes L, Newitt D, et al. Quantification of background enhancement in breast magnetic resonance imaging. J Magn Reson Imaging. 2011;33(5):1229–34. Epub 2011/04/22. doi: 10.1002/jmri.22545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ha R, Mema E, Guo X, Mango V, Desperito E, Ha J, et al. Quantitative 3D breast magnetic resonance imaging fibroglandular tissue analysis and correlation with qualitative assessments: a feasibility study. Quant Imaging Med Surg. 2016;6(2):144–50. Epub 2016/05/18. doi: 10.21037/qims.2016.03.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ha R, Mema E, Guo X, Mango V, Desperito E, Ha J, et al. Three-Dimensional Quantitative Validation of Breast Magnetic Resonance Imaging Background Parenchymal Enhancement Assessments. Curr Probl Diagn Radiol. 2016;45(5):297–303. Epub 2016/04/04. doi: 10.1067/j.cpradiol.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 17.Saha A, Grimm LJ, Ghate SV, Kim CE, Soo MS, Yoon SC, et al. Machine learning-based prediction of future breast cancer using algorithmically measured background parenchymal enhancement on high-risk screening MRI. J Magn Reson Imaging. 2019;50(2):456–64. Epub 2019/01/17. doi: 10.1002/jmri.26636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nam Y, Park GE, Kang J, Kim SH. Fully Automatic Assessment of Background Parenchymal Enhancement on Breast MRI Using Machine-Learning Models. J Magn Reson Imaging. 2021;53(3):818–26. Epub 2020/11/22. doi: 10.1002/jmri.27429. [DOI] [PubMed] [Google Scholar]
- 19.Ha R, Chang P, Mema E, Mutasa S, Karcich J, Wynn RT, et al. Fully Automated Convolutional Neural Network Method for Quantification of Breast MRI Fibroglandular Tissue and Background Parenchymal Enhancement. J Digit Imaging. 2019;32(1):141–7. Epub 2018/08/05. doi: 10.1007/s10278-018-0114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vreemann S, Dalmis MU, Bult P, Karssemeijer N, Broeders MJM, Gubern-Mérida A, et al. Amount of fibroglandular tissue FGT and background parenchymal enhancement BPE in relation to breast cancer risk and false positives in a breast MRI screening program : A retrospective cohort study. Eur Radiol. 2019;29(9):4678–90. Epub 2019/02/24. doi: 10.1007/s00330-019-06020-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borkowski K, Rossi C, Ciritsis A, Marcon M, Hejduk P, Stieb S, et al. Fully automatic classification of breast MRI background parenchymal enhancement using a transfer learning approach. Medicine (Baltimore). 2020;99(29):e21243. Epub 2020/07/25. doi: 10.1097/MD.0000000000021243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ha R, Chang P, Mema E, Mutasa S, Karcich J, Wynn RT, et al. Fully Automated Convolutional Neural Network Method for Quantification of Breast MRI Fibroglandular Tissue and Background Parenchymal Enhancement. J Digit Imaging. 2018. Epub 2018/08/03. doi: 10.1007/s10278-018-0114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rolnick D VA, Belongie S, Shavit N. Deep Learning is Robust to Massive Label Noise. 2018.
- 24.Wengert GJ, Helbich TH, Woitek R, Kapetas P, Clauser P, Baltzer PA, et al. Inter- and intra-observer agreement of BI-RADS-based subjective visual estimation of amount of fibroglandular breast tissue with magnetic resonance imaging: comparison to automated quantitative assessment. Eur Radiol. 2016;26(11):3917–22. Epub 2016/04/25. doi: 10.1007/s00330-016-4274-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


