Abstract
Cohesin mediates chromatin loop formation across the genome by extruding chromatin between convergently oriented CTCF-binding elements. Recent studies indicate that cohesin-mediated loop extrusion in developing B cells presents immunoglobulin heavy chain (Igh) variable (V), diversity (D) and joining (J) gene segments to RAG endonuclease through a process referred to as RAG chromatin scanning. RAG initiates V(D)J recombinational joining of these gene segments to generate the large number of different Igh variable region exons that are required for immune responses to diverse pathogens. Antigen-activated mature B cells also use chromatin loop extrusion to mediate the synapsis, breakage and end-joining of switch regions flanking Igh constant region exons during class-switch recombination, which allows for the expression of different antibody constant region isotypes that optimize the functions of antigen-specific antibodies to eliminate pathogens. Here, we review recent advances in our understanding of chromatin loop extrusion during V(D)J recombination and class-switch recombination at the Igh locus.
Introduction
Mammalian genomes are folded into topologically associated domains characterized by increased contact frequency within the domains1,2. These domains have been implicated in regulating various biological activities, including transcription3-9, replication10,11, recombination12,13 and DNA repair14-16, in part by promoting functional interactions between regulatory sequences and other sequences within the domains17. Many such domains are based on chromatin loops with well-defined boundaries anchored by CTCF-binding elements [G] (referred to in the remainder of the text as CTCF sites that are bound by the structural protein CTCF (CCCTC-binding factor) and by the cohesin ring protein complex [G]1,18 (Fig. 1). Cohesin-mediated chromatin loop extrusion modulates chromosome architecture by extruding chromatin between two CTCF sites of convergent orientation [G] to generate loop-shaped contact domains of up to millions of bases in size17,19-24. Recent studies from our lab and others have advanced our mechanistic understanding of the genomic rearrangement processes in B cells that diversify antibody repertoires. These studies have shown that cohesin-mediated chromatin loop extrusion has fundamental roles in both V(D)J recombination [G]25-29 and class-switch recombination [G] (CSR) of the immunoglobulin heavy chain locus (Igh)30,31.
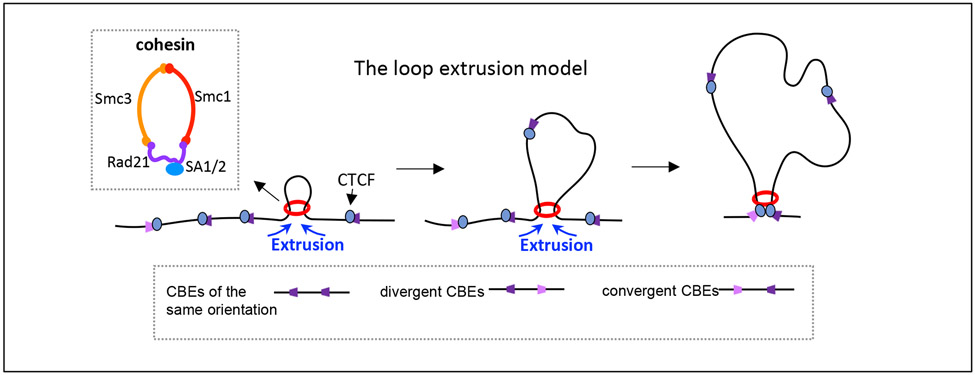
Figure 1 ∣. The loop extrusion model.
A simplified version of the loop extrusion model is outlined (adapted with permission from REF. 53). The extrusion model suggests that progressive extrusion of chromatin by the cohesin complex (red ring) leads to the formation of chromatin loops anchored by the structural protein CTCF (CCCTC-binding factor) bound to convergent CTCF sites. CTCF sites can be found in the genome in three orientations relative to each other (same, divergent and convergent). The inset box shows a schematic of the highly conserved cohesin ring-shaped protein complex, which is composed of the core subunits SMC1, SMC3 and RAD21 associated with either SA1 or SA2 in somatic cells.
Antibody subunits consist of pairs of identical immunoglobulin heavy chains and light chains. Highly diverse amino-terminal variable regions of the heavy and light chains provide antigen-binding specificity, and the carboxy-terminal heavy chain constant regions provide effector functions32. V(D)J recombination in developing B cells assembles germline gene segments into immensely diverse exons that encode the heavy and light chain variable regions32. In antigen-activated mature B cells, CSR replaces IgM-encoding constant region exons with a set of different downstream exons. CSR thus provides antigen-specific antibodies with a heavy chain constant region that is best suited for antigen elimination33. The very different processes of V(D)J recombination and CSR both use chromatin loop extrusion to juxtapose cis-regulatory elements, substrate DNA sequences and initiating enzymes, as well as to promote the proper orientation of joined sequences26,30. Here, we review recent advances in our understanding of the functions of chromatin loop extrusion in V(D)J recombination and CSR of the Igh locus.
Cohesin-mediated loop extrusion
CTCF sites are widely dispersed in the genome34. However they are also highly enriched in certain loci, including the long locus that encodes IgH variable region gene segments35,36. A substantial proportion of chromatin loops occur via cohesin binding to two widely separated CTCF-bound sites1,18. CTCF binds these sites in an orientation-dependent manner37,38. Thus, genomic CTCF sites can occur in the same orientation, divergent orientation or convergent orientation relative to each other18 (Fig. 1). Many major genomic contact loops have endpoints at convergently oriented CTCF sites bound by CTCF and, indirectly, by cohesin18,39,40. Chromatin loops between convergent CTCF sites can be megabases long, and their orientation-specific bias cannot be readily explained by diffusion-based random collisions, which would be expected to promote the interaction of two CTCF sites independently of their chromosomal orientation21,22,24. Thus, models of chromatin loop extrusion have posited that cohesin, upon loading onto chromatin primarily at enhancers and active promoters41-43, actively drives the formation of contact loop domains by linearly extruding chromatin until extrusion is terminated in each direction upon reaching convergently oriented CTCF sites17,21-24 (Fig. 1).
Convergently oriented CTCF sites provide the strongest anchors for chromatin loops, largely owing to stabilization of cohesin through its interaction with the N-terminus of CTCF44-47. However, many studies indicate that these anchors are dynamic48, in part owing to the continuous unloading and loading of CTCF at loop anchors49,50. In addition, CTCF sites that are not in convergent orientation can function as weaker loop anchors51. Loop extrusion also can be impeded by other ‘obstacles’, for example transcription factors bound to enhancers and promoters17,51-53. Therefore, loop boundaries formed by active enhancers and promoters may cooperate with CTCF sites to provide developmentally regulated sub-loops within larger loops based on CTCF sites25,26,30,54-56. Deletion or mutation of convergent CTCF sites results in longer chromatin loop domains4,5,7,21. Moreover, depletion of CTCF57,58 or cohesin58-61, as well as inhibition of cohesin ATPase activity62, eliminates loop domains across the genome. Furthermore, depletion of WAPL (a protein that unloads cohesin) leads to increased retention of cohesin on chromatin63,64 and, thereby, extends chromatin loop length58,59,65. This may reflect that the increased density of cohesin on chromatin provides more opportunities to extrude past dynamic CTCF-site anchors48,50. Finally, elegant in vitro studies have shown that cohesin extrudes naked and nucleosome-bound DNA into loops66-68. Thus, the cohesin-mediated loop extrusion mechanism is now well accepted by the field48,51,52.
V(D)J recombination
V(D)J recombination occurs during B cell development in the bone marrow. Progenitor B cells (pro-B cells) first assemble exons that encode Igh chain variable regions from VH, D and JH gene segments by V(D)J recombination. Subsequent IgH chain expression promotes development to the precursor B cell stage, during which immunoglobulin light chain (Igl) variable region exons are assembled from VL and JL segments. IgH and IgL chains associate to form the B cell receptor on immature B cells, which migrate to peripheral lymphoid organs to become mature B cells36,69. Upon antigen activation, mature B cells can ultimately secrete their B cell receptor as antibodies. Studies of the role of chromatin loop extrusion in V(D)J recombination have mainly focused on the mouse 2.8 Mb Igh locus25-29 (Fig. 2a), and we refer to that locus here unless otherwise specified.
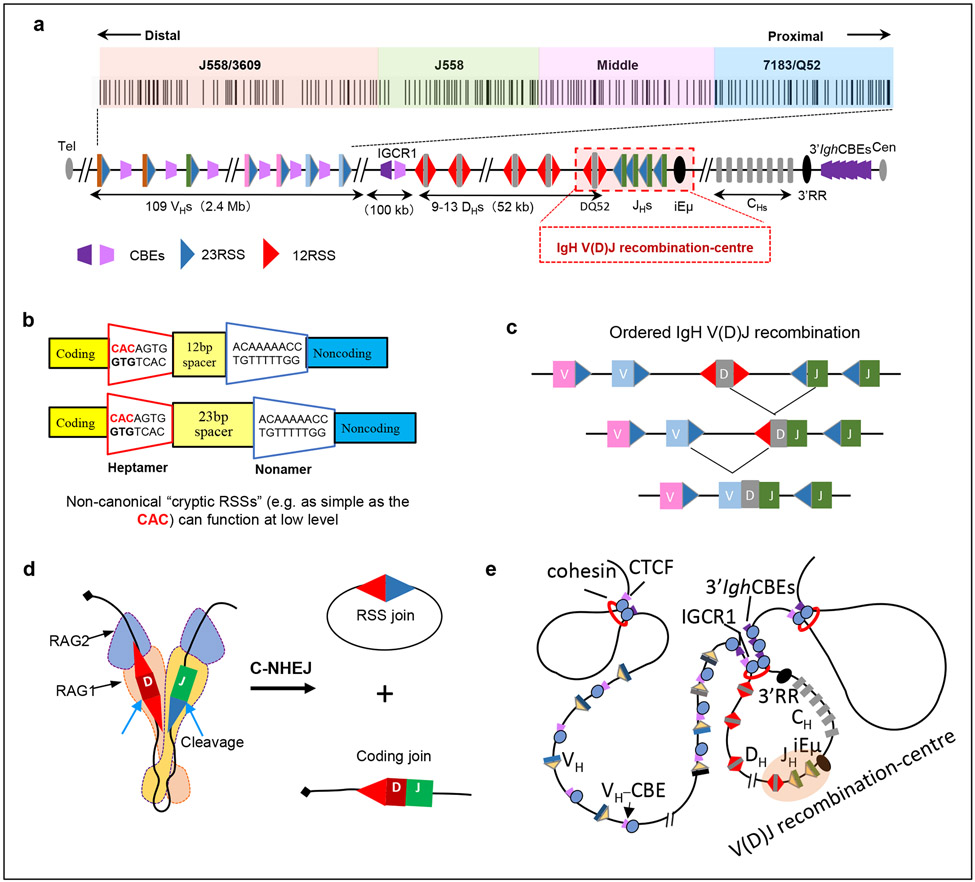
Figure 2 ∣. The Igh locus initiates V(D)J recombination at the V(D)J recombination-centre.
a ∣ The mouse 2.8 Mb immunoglobulin heavy chain (Igh) locus. Upstream of the Igh constant region exon (CH)-containing domain, there is a 3 kb region containing the intronic Igh enhancer (iEμ), 4 JH segments (JHs) and the D segment DQ52, which binds RAG to form the JH-based V(D)J recombination-centre. Upstream of the V(D)J recombination-centre is a 52 kb region containing 9–14 D segments, an approximately 100 kb intervening region containing intergenic control region 1 (IGCR1), and the 2.4 Mb VH-containing telomeric region (locus not drawn to scale). The VH portion of the Igh locus consists of approximately 100 VH segments clustered into four general groups that are specified by VH family type and position; from proximal to distal location these are VH7183/VHQ52, middle VHs without a specific family type, VHJ558 and VHJ558/VH3609. The four VH groups are colour-coded, which is maintained in subsequent figures. The position and orientation of CTCF-binding sites, as well as of recombination signal sequences (12-RSSs and 23-RSSs) are shown. 12-RSSs are located upstream and downstream of D segments, 23-RSSs are located upstream of JHs and downstream of VHs. 3′IghRR, 3′ IgH regulatory region b ∣ Sequence organization of bona fide 12-RSSs and 23-RSSs. The first three nucleotides (CAC) of the heptamer define the cleavage site for RAG endonuclease (RAG1–RAG2) and are crucial for RSS functionality. Cryptic RSSs as simple as CAC can be cleaved by RAG at low levels. c ∣ Igh recombination occurs in an ordered manner, with D-to-JH joining preceding VH-to-DJH joining. d ∣ RAG is recruited to the V(D)J recombination-centre, where it binds a JH 23-RSS paired with a DH 12-RSS and aligns them for 12/23-restricted cleavage, with the ends being joined by classical non-homologous end joining (C-NHEJ). e ∣ Illustration of Igh domain organization showing that DHs, the V(D)J recombination-centre and CHs are contained within a 3′ loop domain bounded by IGCR1 and 3′ Igh CTCF sites. The VHs are located within the upstream 5′ region and are excluded from the D-JH domain loop at the D-to-JH recombination stage. The structural protein CTCF binds CTCF sites and cohesin accumulates at these sites.
V(D)J recombination is initiated by RAG1–RAG2 endonuclease (collectively referred to here as RAG), which cleaves at the junctions between V, D and J coding segments and flanking recombination signal sequences (RSSs)70. RSSs consist of a conserved heptamer (closely related to the sequence CACAGTG) and an AT-rich nonamer that are separated by a 12 base-pair spacer or a 23 base-pair spacer. These RSSs are referred to, respectively, as 12-RSSs and 23-RSSs (Fig. 2b). Such ‘bona fide’ RSSs flank gene segments in all antigen receptor loci that undergo V(D)J recombination, including immunoglobulin and T cell receptor loci. The first three nucleotides (CAC) of the heptamer define the RAG cleavage site and are crucial for RSS functionality70. RAG also can target, at much lower levels, ‘cryptic’ RSSs that are not associated with V, D and J segments71. Such cryptic RSSs range from sequences similar to bona fide RSSs to sequences as simple as CAC12,71. Robust RAG-mediated cleavage is restricted to paired gene segments that are flanked, respectively, by a bona fide 12-RSS or a 23-RSS70. VH segments are flanked by downstream 23-RSSs, D segments are flanked on both sides by 12-RSSs, and JH segments are flanked by upstream 23-RSSs. This organization allows for V(D)J exon formation by the ordered joining of a D segment to a JH segment and then a VH segment to the DJH complex72 (Fig. 2c). Structural studies in the past decade provided a break-through in our understanding of V(D)J recombination. This work showed that RAG functions as a Y-shaped heterodimer in which active sites in the two RAG1 subunits bind a 12-RSS and a 23-RSS and align them for 12/23-restricted cleavage73-77 (Fig. 2d). RAG is not a classic ‘recombinase’ as the RSS and coding-segment ends cleaved by RAG are fused by classical non-homologous end-joining [G]78,79.
Another advance in understanding V(D)J recombination was the discovery of the V(D)J recombination-centre [G]80,81. In pro-B cells, an Igh V(D)J recombination-centre occurs in a 5-kb region of active chromatin spanning the JH-proximal D segment DQ52, the 4 JH segments and the intronic Igh enhancer iEμ81 (Fig. 2a). RAG binds the nascent V(D)J recombination-centre and upon acquisition of a JH 23-RSS forms an activated JH-based V(D)J recombination-centre for D-to-JH joining25,53,71. As DQ52 is located within the V(D)J recombination-centre, RAG can also initiate V(D)J recombination upon acquiring the DQ52 12-RSS26. The intergenic control region 1 (IGCR1), which contains two CTCF sites, lies just upstream of the distal D segment DFL16.182,83, and a cluster of 10 CTCF sites lies just downstream (3′) of the Igh locus84-87. Interactions between the IGCR1 CTCF sites and the 3′ Igh CTCF sites were implicated in anchoring a chromatin loop that sequesters D segments and the recombination-centre away from VH segments82,83 (Fig. 2e). The DJH intermediate then forms a new DJH-based V(D)J recombination-centre with its upstream D segment 12-RSS poised for VH-to-DJH joining25,53. Seminal 3D DNA fluorescence in situ hybridization (FISH) studies88-92, augmented by chromatin conformation capture (3C)-based studies86,93-95, discovered that the VH locus undergoes physical contraction in primary pro-B cells. This process was implicated in bringing distant upstream VH segments into close proximity to the DJH-based V(D)J recombination-centre to enable VH-to-DJH joining36,96-99. As discussed later in this Review, recent studies implicate a mechanistic role for chromatin loop extrusion in contraction of the VH locus27-29.
Discovery of RAG chromatin scanning.
To test the potential of RAG-generated double-strand breaks to generate oncogenic translocations, a cassette containing 12/23-matched bona fide RSSs from D and J segments was inserted into the mouse Myc oncogene100. This ‘Myc–DJ’ cassette was bred into an ATM-deficient background, which allows double-strand breaks to escape from normal C-NHEJ complexes and translocate101,102. Strikingly, these mice developed B cell lymphomas with oncogenic chromosomal translocations that fused RAG-generated DNA breaks in the V(D)J recombination-centre with sequences far downstream of Myc103. Notably, in mice heterozygous for the Myc–DJ cassette allele, translocations occurred exclusively on the cassette-containing allele, but did not involve the cassette103. This finding suggested that RAG activity at bona fide D and J segment RSSs in Myc promoted ‘off-target’ activity far downstream in cis103.
The mechanism of this long-range, off-target activity of RAG was elucidated in v-Abl-transformed pro-B cell lines derived from ATM-deficient mice carrying the Myc–DJ cassette allele12. Such v-Abl cell lines can be viably arrested in G1 stage of the cell cycle, leading to the induction of RAG expression and V(D)J recombination at endogenous and chromosomally integrated cassette targets104. These studies used high-throughput genome-wide translocation sequencing of V(D)J recombination outcomes (HTGTS-V(D)J-Seq [G]), which provides unprecedented sensitivity in elucidating RAG activity at both bona fide RSSs and cryptic RSSs12,53. In addition to the great majority of joining products involving bona fide RSSs in the Myc–DJ cassette, a small subset of break ends in the cassette RSSs were joined to hundreds of cryptic RSSs within the 1.8 Mb downstream portion of the Myc loop domain12 (Fig. 3a). Remarkably, these distant cryptic RSSs required as little as 3 base pairs (CAC) of the canonical RSS heptamer for paired cleavage and joining with a bona fide cassette RSS. In addition, cryptic RSSs were used predominantly when in convergent orientation with the cassette RSS to which they were joined12. The dependence of this process on a convergent orientation suggested the involvement of active linear pairing, rather than diffusion-based random collisions12,17. These findings were confirmed in primary pro-B cells12.
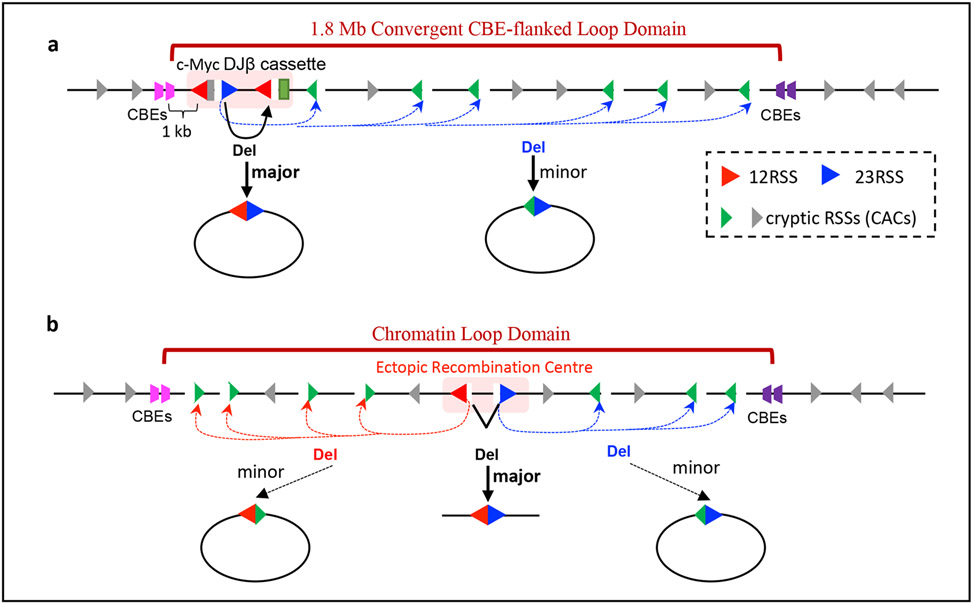
Figure 3 ∣. Discovery of RAG long-range chromatin scanning.
a ∣ Illustration of RAG activity initiated from bona fide D and Jβ recombination signal sequences (RSSs) in the Myc–DJ cassette, which, in addition to normal deletional D-to-Jβ joining within the cassette, also results in low-level joining of bona fide RSSs to hundreds of convergent cryptic RSSs across the 1.8 Mb loop domain of Myc12. Cryptic RSSs in the same orientation as the Myc–DJ cassette generally are not used for such cryptic V(D)J recombination events. b ∣ Ectopic insertion of pairs of bona fide RSSs randomly into 12 other chromatin loop domains across the genome that are known to be based on convergent CTCF sites activates RAG scanning across these loop domains12. The orientation of the initial RAG-binding RSS prescribes the direction of RAG scanning (upstream or downstream), which continues until terminated by a loop anchor determined by convergent CTCF sites.
Ectopic introduction of paired bona fide RSSs into 12 other chromatin loop domains across the genome that are known to be based on convergent CTCF sites showed that the orientation of each of the two initiating bona fide RSSs, respectively, programmed linear, directional chromatin exploration by RAG over long distances until the convergent CTCF sites-based loop boundary was reached12 (Fig. 3b). In each of these chromatin loop domains, RAG predominantly targeted distant cryptic RSSs in convergent orientation with the initiating bona fide RSS12. This process was originally referred to as RAG tracking, and cohesin was proposed as a driver12 owing to similarities with proposed cohesin activities22. Contemporaneously with the RAG tracking study12, two studies provided strong evidence in support of a proposed cohesin-driven chromatin loop extrusion model21,24. Potential relationships between RAG tracking and cohesin-mediated loop extrusion were noted17. In this regard, RAG tracking and loop extrusion both are linear processes, both preferentially use RSSs or CTCF sites in convergent orientation, and both are restricted by the same loop anchors based on CTCF sites12,21,24,25,53. Indeed, additional studies indicated that RAG tracking is more accurately described as RAG chromatin scanning, during which recombination-centre-bound RAG linearly scans chromatin presented by cohesin-mediated loop extrusion25,53. Cohesin-depletion studies in G1-arrested v-Abl cell lines provided direct evidence for the role of cohesin in this process27.
RAG scanning mediates physiological D-to-JH recombination.
Linear tracking had been long ago proposed to explain the highly preferential use of RSSs downstream, rather than upstream, of D segments to mediate D-to-JH joining105. However, extra-chromosomal plasmid studies dismissed tracking105 or scanning106 models for RAG activity in favour of diffusion-based models that proposed D segment downstream 12-RSSs had specialized properties for joining to JH segment 23-RSSs107. These models were revisited in a chromosomal setting in G1-arrested v-Abl cell lines, which activate robust D-to-JH joining; studies analyzed the effects of targeted mutagenesis of the endogenous D locus, including inversion of individual D segments and/or of upstream or downstream 12-RSSs26. The findings implicated a linear RAG scanning mechanism in promoting deletional D-to-JH joining for all D segments except for DQ52 (see below), which is physically proximal to JH segments in the V(D)J recombination-centre26. (Fig. 4a). This process often may be initiated by cohesin loading in active chromatin in the region of the V(D)J recombination-centre (Fig. 4a, parts ii-iv). However, it also could be initiated by cohesin binding upstream of the recombination-centre with the same end result (Fig. 4a, parts v and vi). The V(D)J recombination-centre functions as a dynamic subdomain anchor that strongly impedes, but does not fully block, extrusion of downstream chromatin25. Upstream chromatin extrusion is nearly fully blocked at the strong anchor mediated by the two IGCR1 CTCF sites25,26. Overall, this process allows for upstream D-containing chromatin to be linearly presented to the open RAG1 active site opposite the bound JH-RSS in the recombination-centre25,26. During RAG scanning of upstream chromatin, convergent 12-RSSs downstream of D segments are captured, whereas 12-RSSs upstream of D segments in the same orientation as JH segment 23-RSSs are almost invisible to RAG26. These, and prior studies, showed that the preferential use of RSSs in convergent orientation is an intrinsic property of RAG scanning12,26. To facilitate visualization of this process, we provide an animated model (Supplementary Video 1).
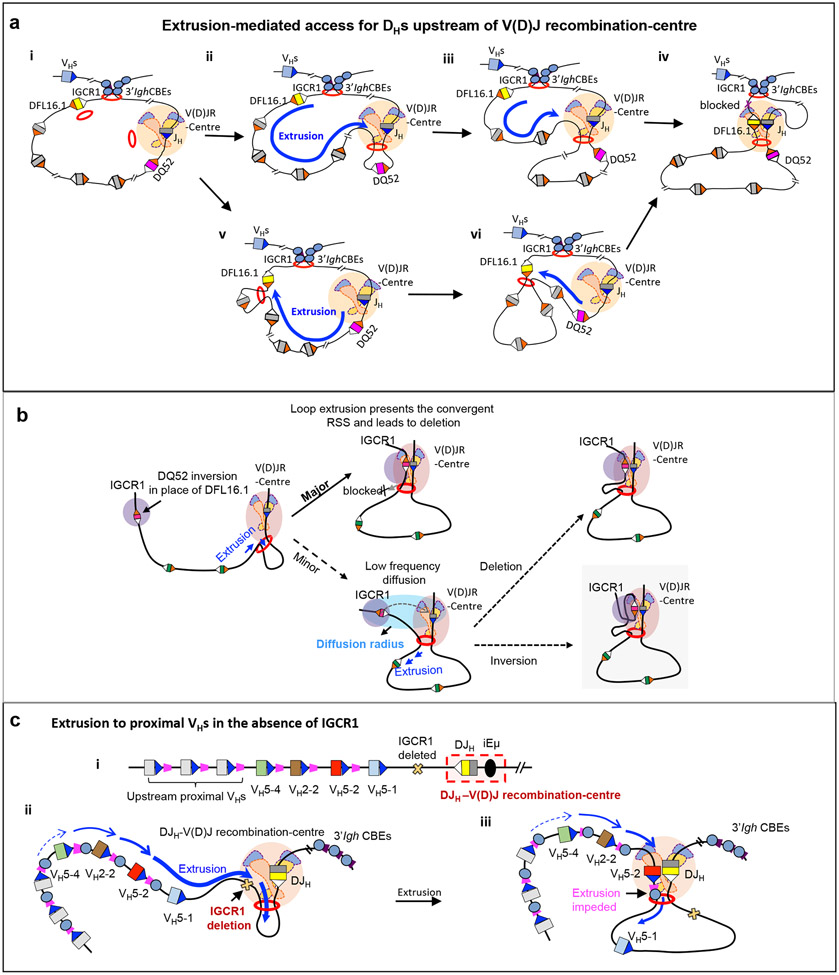
Figure 4 ∣. Loop extrusion-mediated RAG scanning drives D-to-JH and proximal VH-to-DJH recombination.
a ∣ Extrusion-mediated D-to-JH recombination for D segments upstream of DQ52 and the V(D)J recombination-centre. D-to-JH recombination occurs within the 3′ Igh loop domain that is anchored by IGCR1 and the 3′ Igh CTCF sites. Cohesin is known to load near the V(D)J recombination-centre26 to initiate loop extrusion, but likely also at low levels at other sites such as within the D region (part i). The V(D)J recombination-centre functions as a broad and dynamic sub-loop anchor25 (within the 3′ Igh loop domain) that promotes cohesin-mediated loop extrusion of upstream chromatin past a RAG-bound JH recombination signal sequence (RSS) in the V(D)J recombination-centre (parts ii,iii). This linear process aligns 12-RSSs downstream of D segments with the convergent RAG-bound JH 23-RSS for deletional D-to-JH recombination. 12-RSSs upstream of D segments are not used for recombination owing to being in the same orientation as the JH RSS. D segments upstream of DQ52 are frequently passed without being used for recombination, allowing loop extrusion-mediated RAG scanning to continue in many pro-B cells. Loop extrusion is strongly impeded by the IGCR1 anchor of the 3′ Igh loop, which lies just upstream of the distal D segment DFL16.1 (part iv). See also Supplementary Video 1. Upon cohesin loading near DFL16.1, continuous linear extrusion brings the downstream 12-RSS of DFL16.1 to the RAG-bound JH 23-RSS in the recombination centre. The dynamic anchor of the V(D)J recombination-centre impedes further extrusion and promotes DFL16.1 recombination (parts v,vi). b ∣ Model of short-range diffusion-mediated inversional joining of distal D segments. Whereas loop extrusion-mediated RAG scanning drives dominant deletional joining of distal D segments, low-level inversional D joining occurs via short-range diffusion after loop extrusion brings the D segment into a ‘diffusion radius’ of the RAG-bound V(D)J recombination-centre. Such inversional joining is augmented by strong upstream 12-RSSs of D segments, as exemplified here by the normally dominant downstream RSS of DQ52 being placed upstream as a result of inversion of DQ52 in the DFL16.1 position. Reproduced with permission from REF. 26. c ∣ Schematic of the DJH-V(D)J recombination-centre including its 12-RSS (white), and proximal VH segments including their 23-RSSs and associated CTCF sites (part i). Upon deletion of IGCR1 or mutation of its CTCF sites, robust RAG scanning activity is extended into the proximal VH region (part ii). VH5-2 is dominantly rearranged owing to it being the first VH segment that is associated with a CTCF site (which impedes further loop extrusion) to be encountered during RAG scanning; the most D-proximal VH segment VH5-1 lacks a functional CTCF site and is mostly bypassed without rearrangement (part iii). Mutation (to inactivate CTCF binding) or deletion of the VH5-2 CTCF site abolishes VH-5-2 usage, rendering the next VH segment that is flanked by a CTCF site (VH2-2) as the most frequently used. Restoring the functionality of the inactive CTCF site of VH5-1 makes it the most frequently used VH segment. In each case, the first several proximal VH segments flanked by CTCF sites are used for VH-to-DJH joining but at progressively reduced levels, with RAG scanning further upstream ultimately being terminated in this proximal VH region. Parts a and c are adapted with permission from REF. 53.
Given the very close proximity of DQ52 in the V(D)J recombination-centre to the JH segments, both the upstream and downstream 12-RSSs of DQ52 should in theory access V(D)J recombination-centre-bound RAG by short-range diffusive motion, resulting in deletions and inversions during D-to-JH joining 12,26. However, DQ52 joins to JH segments predominantly by deletion. To resolve this potential paradox, DQ52 and its RSSs were inverted in the V(D)J recombination-centre location, leading to predominantly inversional DQ52-to-JH joining mediated by the 12-RSS that is normally downstream of DQ52 but now placed in a divergent orientation upstream of DQ52. Thus, DQ52 seems to have evolved a dominant downstream 12-RSS107. This dominant downstream 12-RSS in its normal location in the V(D)J recombination-centre promotes deletional joining to JH and obviates diffusion-mediated inversional joining by the weaker upstream 12-RSS26. When DQ52 was inserted, in its normal orientation, far upstream in place of DFL16.1, it also was highly used for deletional joins mediated by its robust downstream 12-RSS. However, when DQ52 was inverted in this far upstream location, the weaker 12-RSS normally upstream of DQ52, now in a downstream orientation convergent to JH, became predominantly used to mediate deletional JH joining26 (Fig. 4b). This set of experiments provided clear evidence that linear RAG scanning enforces the deletional joining of D segments that lie distal to the V(D)J recombination-centre26. Finally, when DQ52 was inverted in the DFL16.1 position, its dominant 12-RSS, now in the upstream position, mediated low-level inversional joining to JH (Fig. 4b). A model to explain this latter finding proposed that the strong RSS in upstream position gains low-level access to V(D)J recombination-centre-bound RAG via short-range diffusional motion when brought into proximity by loop extrusion26 (Fig. 4b). Related diffusional mechanisms might contribute more prominantly to programmed inversional V(D)J recombination at other antigen receptor loci, including long-range inversional joining within the Igκ light chain locus (Box 1).
Box 1: Programmed inversional V(D)J recombination.
RSSs and cryptic RSSs located in close linear proximity to V(D)J recombination-centres can gain inversional access to RAG by a short-range diffusional mechanism12,26. This mechanism also may explain inversional V joining at T cell receptor-β150 and T cell receptor-δ151 loci. Both loci mainly undergo deletional V(D)J recombination, but contain a single V segment in close proximity to the V(D)J recombination-centre that undergoes inversional joining. Similar mechanisms could explain inversional joining in the highly compact teleost T cell receptor-α/δ locus151,152. However, long-range inversional V(D)J recombination in the Igκ locus must involve distinct mechanisms. The 3.2 Mb Igκ locus, which is subject to locus contraction91,94,153-156, undergoes Vκ-to-Jκ rearrangement between 92 functional Vκ segments and 4 Jκ segments36,69. Whereas the proximal and distal Vκ segments are oriented for deletional joining to Jκ, the middle Vκ segments are mostly oriented for inversional joining69. The short Vκ–Jκ intergenic region contains Cer157 and Sis158 elements containing CTCF sites. Deletion of either Cer or Sis increases the use of proximal Vκ segments and decreases the use of distal Vκ segments157,159, which is reminiscent of the effects of IGCR1 deletion in the Igh locus82. However, long-range inversional joining of distant middle Vκ segments to Jκ segments is not consistent with direct linear RAG scanning. In theory, Igκ may be structurally optimized for the mechanisms described above for low-level inverted D-to-JH joining26 or incorporate proximity mediated by phase separation [G]110. Such mechanisms could, in theory, bring Vκ RSSs close enough to the Jκ-V(D)J recombination-centre for a short-range diffusion-mediated joining. Although a role for cohesin-mediated loop extrusion in Igκ locus contraction seems likely, it has not been tested. Targeted deletions and inversions of Igκ domains and the use of high-resolution approaches to address cryptic RSS use across Igκ could, as shown for Igh, provide more mechanistic insights.
VH accessibility during RAG chromatin scanning.
All VH segments and their associated 23-RSSs lie in convergent orientation with the upstream 12-RSSs of D segments of the DJH-based V(D)J recombination-centre36. In this regard, early findings that the D-proximal VH segments are the most frequently rearranged suggested a role for linear scanning106. However, that study pointed out the complicating factor that the D-proximal VH5-1 barely rearranges, despite its consensus bona fide 23-RSS106. Remarkably, all of the D-proximal VH segments, except for VH5-1, have CTCF sites directly downstream of their 23-RSSs25,36,85,108,109. To test the potential roles of these CTCF sites in promoting rearrangement, the CTCF site of the most highly rearranging VH5-2 (also known as VH81X), which lies just upstream of VH5-1, was mutationally inactivated, which nearly abrogated use of VH5-2 in primary pro-B cells, despite its bona fide 23-RSS25. For further analysis, G1-arrested v-Abl cells were used, which do not undergo VH locus-contraction or distal VH-to-DJH rearrangement25. Indeed, in v-Abl cells, linear scanning upstream of the DJH-based V(D)J recombination-centre is largely blocked at the IGCR1 anchor12,25. However, mutational inactivation of IGCR1 CTCF sites in v-Abl cells extended RAG scanning through the 100kb intervening region containing IGCR1 to the most proximal VH segments12,25. Moreover, IGCR1 inactivation greatly increased interactions between VH5-2 and the DJH-based V(D)J recombination-centre and, correspondingly, increased its rearrangement frequency about 50-fold12,25,82,83 (Fig. 4c). Remarkably, inactivation of the VH5-2-associated CTCF site in the context of IGCR1 inactivation still nearly abrogated interaction of VH5-2 with the V(D)J recombination-centre and its high-level rearrangement. Moreover, inactivation of the VH5-2 CTCF site promoted robust scanning upstream to the next VH segment VH2-2, which became highly used25. These results solved the mystery of low-level VH5-1 rearrangement, owing to its lack of a downstream CTCF site. Conversion of a non-functional sequence just downstream of VH5-1 into a functional CTCF site promoted robust interaction of VH5-1 with the V(D)J recombination-centre and rendered it, by far, the most frequently rearranged VH25.
These studies showed that the convergent RSSs of proximal VH segments alone do not promote robust rearrangement during linear RAG scanning and that proximal VH CTCF sites are required to enhance chromatin accessibility25. This function, which is not absolutely dependent on orientation of the CTCF site, is based on the ability of VH-associated CTCF sites, but not RSSs, to impede loop extrusion and, thereby, prolong interaction with the V(D)J recombination-centre25 (Fig. 4c). Although proximal VH-associated CTCF sites do not create absolute boundaries for loop extrusion and RAG scanning, the first few proximal VH segments that are encountered dominate recombination-centre interactions and rearrangements in v-Abl cells, albeit at progressively decreasing levels as expected for a linear process (Fig. 4c).
Loop extrusion promotes VH locus contraction and scanning.
Many of the 109 VH segments in the mouse genome lie in a 2.4 Mb region upstream of the proximal VH segments that harbours numerous CTCF-bound sites. Unlike the proximal VH-associated CTCF sites, upstream CTCF sites are not directly adjacent to VH segments25,36,85,108,109, which suggests that they might have different or additional roles. The CTCF sites of the VH locus are in convergent orientation with the ten 3′ CTCF sites downstream of the Igh locus 36 and have substantial interactions with them86,93. In this regard, loop extrusion has been considered as a mechanism for locus contraction27-29,110. However, CTCF-bound sites across the VH locus would be impediments to linear loop extrusion25, unless their inhibitory activity as observed in v-Abl cells is somehow reduced in primary pro-B cells27. Notably, CTCF depletion in non-lymphoid cells suppresses most CTCF site-based chromatin interactions across the genome57,58. To test the potential negative effects of CTCF-bound sites on locus contraction and long-range RAG scanning, CTCF was depleted in G1-arrested v-Abl cells27, which reduced interactions of the recombination-centre with IGCR1 CTCF sites and the CTCF sites of the most proximal VH segments. By contrast, CTCF depletion promoted robust recombination-centre interactions across the distal VH locus, with hallmarks of locus contraction essentially identical to those in locus-contracted primary pro-B cells27. Correspondingly, CTCF depletion activated robust distal VH-to-DJH joining in v-Abl cell lines. In these studies, free CTCF was eliminated but chromatin-bound CTCF remained associated at lower levels with certain CTCF sites27. Thus, it is proposed that bound CTCF was sufficiently depleted to allow for loop extrusion-mediated RAG scanning to frequently pass IGCR1 and VH locus CTCF sites, but that residual CTCF at some sites promoted recombination-centre interactions and use during scanning27. These studies provided proof-of-principle in cell lines that downmodulation of CTCF site impediments activates locus contraction and long-range RAG scanning. However, addressing the mechanism that operates in primary pro-B cells required different approaches.
Cryptic RSS use supports long-range VH locus scanning.
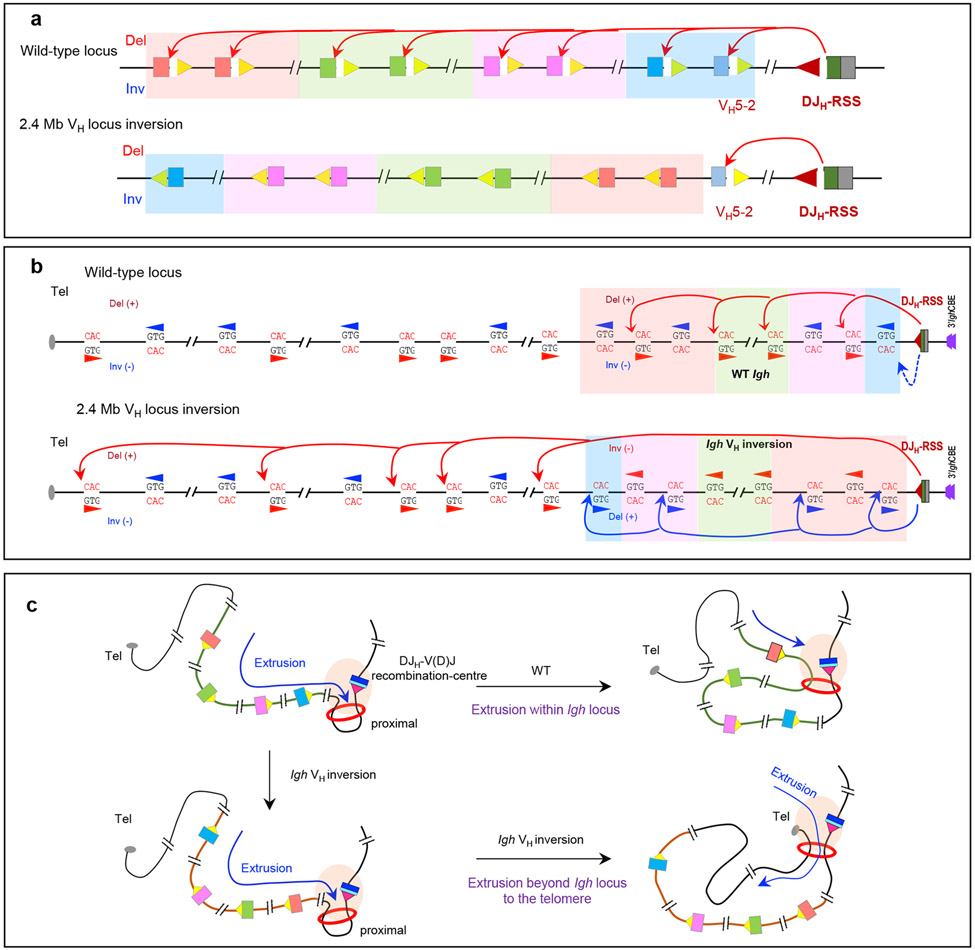
A diffusion-based model of access to distal VH segments, in its simplest form, predicts little impact of VH RSS orientation on access to the DJH-V(D)J recombination-centre. By contrast, a linear scanning model predicts that inverting the normally convergent VH RSS to the same orientation as the RSS of the DJH-V(D)J recombination-centre would markedly decrease its rearrangement. To distinguish between these two models, one study inverted an 890 kb portion of the distal VH locus28, and a second study inverted a 2.4 Mb portion of the VH locus, leaving D-proximal VH5-2 in its normal position and orientation29. VH rearrangements were essentially abrogated in both inverted regions, whereas VH segments left in their normal orientation were still rearranged28,29 (Fig. 5a).
Figure 5 ∣. Long-range RAG scanning mediates VH use across the Igh locus.
a ∣ VH recombination signal sequences (RSSs) are all convergently oriented with respect to the DJH-V(D)J recombination-centre RSS, which prescribes that VH-to-DJH recombination is exclusively deletional in wild-type pro-B cells. A 2.4 Mb inversion of the VH locus spanning all VH segments except for proximal VH5-2 abolished the use of all inverted VH segments in bone-marrow pro-B cells and WAPL-downregulated v-Abl cell lines. b ∣ RAG scanning through the VH locus uses hundreds of cryptic RSSs (which can be as simple as the sequence CAC) in convergent orientation with the RSS of the DJH-V(D)J recombination-centre, with scanning terminated within the VH locus. RAG scanning across the wild-type VH locus would be expected often to be terminated within the VH locus by dominant deletional rearrangements mediated by convergent VH RSSs and DJH-V(D)J recombination-centre RSSs29. Upon inversion of the 2.4 Mb VH locus in primary pro-B cells, use of cryptic RSSs normally in the convergent orientation is abrogated, whereas use of cryptic RSSs normally in the opposite orientation is activated. Inversion of the locus would eliminate the dominant deletional rearrangements in the wild-type locus, which could allow for some RAG scanning to proceed through the VH locus. Remarkably, RAG scanning from the DJH-V(D)J recombination-centre continued upstream of the VH locus all the way to the telomere, targeting normally oriented cryptic RSSs within multiple, consecutive convergent loop domains beyond the Igh domain. This finding indicated that loop anchor impediments to scanning are likely broadly dampened in pro-B cells29. Adapted with permission from REF. 29. c ∣ Model for RAG scanning activity initiated by the DJH-V(D)J recombination-centre RSS with a wild-type VH locus or inverted VH locus, which illustrates the findings described in panels a and b.
To assess effects of the 2.4 Mb inversion on Igh chromatin interactions, a LAM-HTGTS-3C-Seq [G] approach was used25. Such chromatin interaction assays must be carried out in RAG-deficient v-Abl or primary pro-B cells to obviate confounding changes in genome organization associated with V(D)J recombination25-27,29. In primary pro-B cells, the recombination-centre interacts robustly with about 15 highly focused regions29, many of which have been speculated to be involved in locus contraction93. Among the most robust are the PAX5-activated intergenic repeat (PAIR) elements93,97,111,112 in the distal VH locus. Remarkably, all major recombination centre–VH locus interactions were maintained within the inverted VH locus as mirror images29. Transcription profiles were precisely maintained across the inverted locus, albeit in the opposite direction relative to the recombination-centre. Nearly all CTCF sites spread over a 1 Mb distal region of the locus maintained similar levels of interactions with the 3′ CTCF sites downstream of the Igh locus when they were inverted and moved into a proximal position, but the 37 functional VH segments inverted in this region still did not rearrange29.
HTGTS-V(D)J-Seq analyses revealed that in normal VH loci, RAG uses, in addition to convergently oriented bona fide VH RSSs, hundreds of convergent cryptic RSSs embedded across the VH locus29 (Fig. 5b). Use of these normally convergent cryptic RSSs was abrogated in the inverted VH locus, which is fully consistent with a linear scanning mechanism for RAG. Strikingly, however, cryptic VH-locus RSSs normally in the same orientation as the RSS of the DJH-V(D)J recombination-centre became robustly used when placed in a convergent orientation upon locus inversion29 (Fig. 5b). These studies show that RAG scans the entire inverted VH locus from its DJH-recombination centre location, although not necessarily from the same starting point in the VH locus (as described later). Correspondingly, the inability of inverted bona fide VH RSSs to be rearranged during RAG linear scanning of the VH locus, similarly to the RSSs upstream of D segments, predominantly reflects their inability to be properly paired28,29 (Fig. 5c)
WAPL downregulation promotes long-range scanning of the VH locus.
Inversion of the 2.4 Mb VH locus in primary pro-B cells extended the scanning activity of V(D)J recombination-centre-bound RAG to cryptic RSSs beyond the VH locus by extending loop extrusion through multiple convergently oriented CTCF sites to the telomere (Fig. 5b, c). This finding indicated that CTCF site-mediated impediments to loop extrusion are broadly deregulated in primary pro-B cells, both in the Igh locus and beyond29. An interesting unsolved question is why RAG scanning proceeds beyond the VH locus in pro-B cells with an inverted Igh locus but not in those with normally oriented VH segments. One possibility could be decreased impedance of loop extrusion by normally proximal CTCF sites when they are inverted in a distal position29.
Strikingly, 3C-based Hi-C studies of chromatin interactions indicated that chromatin loops are extended across the genome in primary pro-B cells28. To identify factors associated with the cohesin complex that might be deregulated to allow for such extended chromatin looping and scanning in primary pro-B cells, both of the VH locus inversion studies looked for factors that are differentially expressed in primary pro-B cells compared with earlier or later developmental stages and/or v-Abl cell lines28,29. Both studies identified the WAPL cohesin-unloading protein as one such factor28,29. As mentioned above, WAPL depletion in non-lymphoid cells increases the size of CTCF-anchored loops genome wide58,59,65. Thus, WAPL downregulation in pro-B cells was a strong candidate for promoting enhanced loop extrusion past CTCF site-based VH locus impediments.
The transcription factor PAX5 has long been known to be required for locus contraction and recombination of distal VH segments89,113, with one postulated mechanism involving transcription of PAIR elements in the distal VH locus97,111,112. However, an elegant recent study discovered that PAX5 achieves this function by mediating developmental reduction of Wapl expression in pro-B cells28. PAX5 suppresses Wapl transcription by engaging a PAX5-binding element in the Wapl promoter and then recruiting the polycomb repressive complex 2 (PRC2) to induce repressive H3K27me3 histone methylation28. Remarkably, deletion of this PAX5-binding element resulted in loss of VH locus contraction and distal VH rearrangements in pro-B cells28. These studies further revealed that an approximately four-fold reduction in Wapl expression is sufficient for locus contraction and distal VH rearrangement28. Although this reduction in Wapl expression also altered genome-wide chromatin architecture, it did not impact gene expression required for pro-B cell survival28. A contemporaneous study showed that depleting WAPL in G1-arrested v-Abl cells activated VH locus contraction and V(D)J recombination across the entire upstream VH locus29. Moreover, WAPL-depleted v-Abl lines with the 2.4 Mb VH locus inversion had the same phenotype as primary pro-B cells with this inversion, with respect to both effects on the use of bona fide and cryptic RSSs and the activation of RAG scanning upstream of Igh to the telomere29. In support of increased cohesin processivity in WAPL-depleted v-Abl lines, cohesin was redistributed to distal VH regions and to the 3′ Igh CTCF sites, similar to the distribution observed in primary pro-B cells29. Overall, this work showed that WAPL downregulation neutralizes the impediments to VH locus contraction and distal VH rearrangement that would otherwise be mediated by CTCF sites in pro-B cells28,29.
Substrate accessibility during V(D)J recombination.
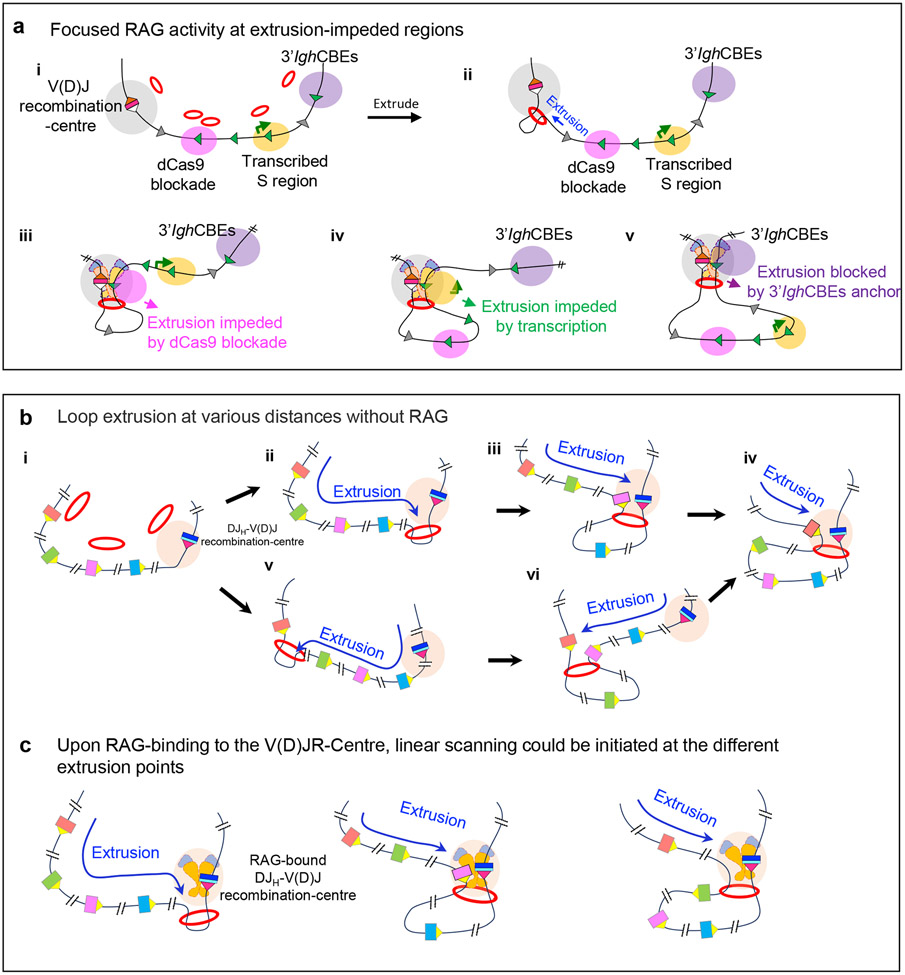
V(D)J recombination is regulated epigenetically by modulating the chromatin accessibility of substrate V, D and J segments to RAG activity32. Factors implicated in determining accessibility include sense and antisense transcription and active chromatin32,96,114-117. New insights into regulating accessibility have recently come from studies of v-Abl cell lines in which downstream-oriented RSSs in the V(D)J recombination-centre programme robust RAG scanning across the downstream CH-containing domain to the 3′ CTCF sites26 (Fig. 6a, parts i-v). This downstream Igh domain lacks dominant bona fide RSSs or CTCF sites that impede scanning, which allows for accessibility mechanisms to be more readily revealed.
Figure 6 ∣. Potential substrate accessibility and distal RAG scanning mechanisms in the VH locus.
a ∣ Mechanisms that promote the targeting of cryptic recombination signal sequences (RSSs) during RAG scanning. Cohesin loaded to chromatin (multiple potential loading sites are shown) extrudes chromatin to promote interaction of the Igh V(D)J recombination-centre with downstream chromatin regions (part i, ii). The RAG-bound V(D)J recombination-centre initiates RAG scanning that can be impeded by various impediments on the scanning path, including targeted blockade by binding of catalytically ‘dead’ Cas9 (part iii), active transcription (part iv) and CTCF site-based loop anchors (part v). b ∣ Model of loop extrusion-mediated locus contraction in the absence of RAG binding. Multiple potential cohesin loading sites are indicated. Loop extrusion theoretically could initiate from any of these sites. Loop extrusion from a non-RAG bound, nascent DJH V(D)J recombination-centre promotes long-range interactions with the V(D)J recombination-centre to mediate Igh locus contraction. Extrusion is proposed to proceed over varied upstream chromatin distances in different pro-B cells prior to RAG binding. Loop extrusion might also initiate from cohesin loaded within the VH locus. c ∣ Model for use of distal VH segments promoted by RAG binding to nascent V(D)J recombination-centres. RAG binding to the V(D)J recombination-centre subsequent to loop extrusion forms an active DJH-V(D)J recombination-centre, which initiates extrusion-mediated scanning at multiple different extrusion points initiated by multiple cohesin loading points across the VH locus. Such scanning from a variety of extrusion intermediates avoids biasing VH recombination to the very proximal segments and thus helps to balance the primary VH repertoire. Part a is adapted with permission from REF. 26. Parts b and c are adapted with permission from REF. 29.
In these v-Abl cells, a 7kb repetitive switch region [G] (S region) that is constitutively transcribed functions as a dynamic loop extrusion anchor and, thereby, targets downstream RAG scanning to cryptic RSSs located all across it26. Indeed, deletion of the S region promoter abrogated transcription, V(D)J recombination-centre interactions and the ability to target RAG scanning, all in association with allowing loop extrusion to proceed through this S region unimpeded26. This work indicated that transcription, similarly to CTCF-bound sites, can target a sequence for accessibility during RAG scanning by increasing its interaction with the recombination centre26 (Fig. 6a, part iv). A related study revealed that binding of catalytically ‘dead’ Cas9 (without endonuclease activity but which retains specific DNA-binding activity) to 16 repeated target sites in a different, untranscribed S region also impeded RAG scanning and focused RAG activity to convergent cryptic RSSs within it26 (Fig. 6a, part iii). This latter study suggested that binding of ‘dead’ Cas9 may sterically hinder RAG scanning. However, a caveat was that binding of ‘dead’ Cas9 promoted accumulation of the cohesin loader NIPBL and of cohesin across the region, which raises the possibility of an indirect inhibition mechanism26.
Analyses of various parameters for individual VH segments in primary pro-B cells and in WAPL-depleted v-Abl lines implicated CTCF site activity and/or transcription in enhancing the accessibility of their RSSs25,27,29. Proximal VH segments lack detectable transcription and their use correlates with impediments to further scanning mediated by CTCF sites25. By contrast, all highly rearranged distal VH segments are transcribed; although not all transcribed segments are highly rearranged27,29. Most of the highly rearranging, transcribed distal VH segments also have CTCF-bound sites within 10 kb that could conceivably function with transcription to enhance interaction of the region containing them with the V(D)J recombination-centre during RAG scanning29. Finally, some frequently rearranged VH segments in the middle of the VH locus lack closely associated CTCF-bound sites and are not transcribed, which suggests that other accessibility factors remain to be elucidated27,29.
Summary of loop extrusion-based models for V(D)J recombination at the Igh locus.
Igh locus V(D)J recombination is predominantly initiated from the V(D)J recombination-centre, which harbours the vast majority of bound RAG80,81. Current findings indicate that the V(D)J recombination-centre also functions as a major downstream anchor for loop extrusion-mediated RAG scanning of upstream chromatin25-27,29. The 3′ CTCF sites downstream of the Igh locus have also been proposed to have roles in anchoring loop extrusion during long-range Igh V(D)J recombination25,28,110. Similarly to other CTCF sites genome wide, the 3′ Igh CTCF sites impede RAG scanning26. Thus, the 3′ CTCF sites might reinforce the loop anchor provided by the V(D)J recombination-centre upon their extrusion-mediated juxtaposition. Surprisingly, however, recent studies have shown that complete deletion of the 3′ Igh CTCF sites has little impact on Igh V(D)J recombination29. But, as described for CSR below, other CTCF sites further downstream may be extruded to the recombination centre and compensate for the putative functions of the 3′ CTCF sites in their absence29,30.
The initial D-to-JH V(D)J recombination step incorporates both short-range diffusion and long-range RAG scanning. Owing to its location in the V(D)J recombination-centre, the frequently used DQ52 accesses RAG mainly by short-range diffusional motion26. Indeed, loop extrusion is thought to counteract even more dominant use of DQ52 by quickly isolating it from recombination centre-bound RAG26,118. All D segments upstream of the V(D)J recombination-centre are accessed primarily by linear RAG scanning26 (Fig. 4a). The very frequent use of distal DFL16.1 may reflect prolonged interaction with the V(D)J recombination-centre owing to scanning being impeded by the IGCR1 CTCF sites26,119. Robust transcription of the IGCR1–DFL16.1 region may also contribute to its high-level targeting during RAG scanning26. Lower-level targeting of intervening D segments also may be mediated by their antisense transcription26,117,120.
IGCR1 prevents direct joining of VH segments to D segments82,83. In pro-B cells, Wapl downregulation suppresses IGCR1-based and other VH locus impediments to RAG scanning sufficiently to promote VH locus contraction and the use of VH segments across the locus29. In theory, D-to-JH joining might activate Wapl downregulation to enable locus contraction. However, locus contraction also occurs in RAG-deficient pro-B cells in the absence of V(D)J recombination88,89,93. Thus, PAX5-mediated downregulation of Wapl expression may be an early event associated with differentiation to the pro-B cell stage. If so, D-to-JH rearrangements may occur first owing to their proximal position to the V(D)J recombination-centre during scanning. Understanding how the persisting activity of CTCF sites following a four-fold reduction in WAPL levels, together with transcription and other factors, contribute to the rearrangement levels of each VH segment across the loop extrusion path remains an interesting challenge.
How RAG scanning promotes diverse distal VH use in the face of downstream scanning impediments and proximal VH rearrangements after Wapl downregulation is another challenging question. One model suggests that pro-B cells often initiate loop extrusion of upstream chromatin past nascent V(D)J recombination-centres that progresses to various distances across the VH locus in individual cells29 (Fig. 6b). Subsequent RAG binding to extruded nascent V(D)J recombination-centres could then generate active V(D)J recombination-centres at various points across the VH locus29 (Fig. 6c). Extrusion of downstream chromatin from cohesin bound to distal VH domains with high transcriptional activity could also mediate this process. Such a mechanism could minimize the potential competitive advantages of D-proximal VH segments during scanning to ensure the generation of diverse antibody repertoires29.
Another recent report proposes a model in which loop extrusion and diffusional access cooperate to promote distal VH use28. This model suggests that 3′ Igh CTCF sites and convergent VH locus CTCF sites anchor ‘stable’ chromatin loops that allow for VH segments within the loop to access the V(D)J recombination-centre by diffusion28. As currently proposed, this diffusion-based model does not readily explain the long-range orientation-specific use of cryptic RSSs across normal and inverted VH loci. However, this report proposes a second model in which orientation specificity is mediated by the alignment of convergent VH region RSSs and V(D)J recombination-centre RSSs for deletional joining via linear loop extrusion28. This second model is very similar to the linear RAG scanning model with one exception27,29 in that it proposes no role for the well-documented ability of the V(D)J recombination-centre to function as an extrusion anchor that increases the frequency of relevant interactions25-27,29.
Class-switch recombination
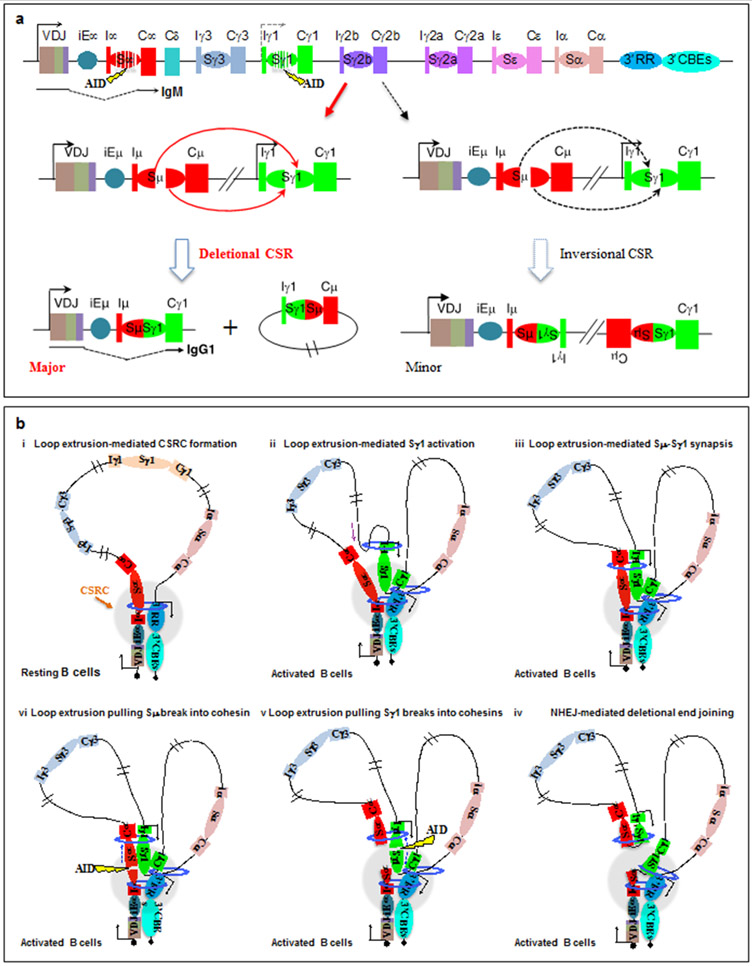
Exons encoding the immunoglobulin heavy-chain constant (CH) regions lie within a downstream 250kb Igh subdomain that lacks CTCF sites. Transcription from an assembled V(D)J exon runs through Cμ exons encoding the C-terminal CH region that specifies IgM. Upon activation, mature B cells undergo CSR to replace the Cμ exons with one of 6 sets of CH exons in the 100–200kb downstream subdomain33 (Fig. 7a). Each set of these CH exons (Cγ3, Cγ1, Cγ2a, Cγ2b, Cε and Cα) specifies an antibody class with different effector properties and functions121. Long, repetitive S regions (up to 12kb) precede Cμ and each downstream CH121. Activation-induced cytidine deaminase (AID)122 initiates CSR by generating deamination lesions at short target motifs within donor Sμ and a downstream acceptor S region33,123. These lesions are converted into double-strand breaks by co-opted general DNA repair pathways33,123,124. Deletional end-joining of the upstream end of an Sμ double-strand break to the downstream end of an acceptor S region double-strand break completes productive CSR in mice and humans13 (Fig. 7a).
Figure 7 ∣. Loop extrusion mediates physiological, deletional class-switch recombination.
a ∣ Schematic of the 200 kb CH region of the immunoglobulin heavy chain (Igh) locus, including the V(D)J exon, the intronic Igh enhancer iEμ, the various CH exons with inducible (I) exon and promoter upstream of each CH segment, S regions, the 3′ Igh regulatory region (3′IghRR) super-enhancer and the 3′ Igh CTCF sites. AID-initiated breaks within Sμ and an activated target Sγ1 are illustrated, which can lead to deletional or inversional class-switch recombination (CSR) outcomes for Sμ to Sγ1 joining. Deletional CSR is the major physiological event, whereas inversional CSR occurs at a lower frequency. Part a is adapted with permission from REF. 13. b ∣ Loop extrusion mediates formation of a class-switch recombination-centre (CSR-centre) and promotes the synapsis of S region double-strand break ends for deletional joining. In resting B cells (part i), cohesin is loaded at 3′ IghRR or iEμ–Sμ and then extrudes the 3′IghRR and iEμ–Sμ into proximity to generate a dynamic CSR-centre. In activated B cells (part ii), a primed S–CH unit (Sγ1–Cγ1 is shown as an example) is extruded to the CSR-centre for transcriptional activation and cohesin loading, which leads to further extrusion to align acceptor and donor S regions (part iii). Tension pulls S region double-strand break ends initiated by AID into opposing cohesin rings (parts iv and v), stalling extrusion and aligning them for deletional joining mediated by non-homologous end joining (NHEJ) (part vi). Part b is adapted with permission from REF. 30. See also Supplementary Video 2.
Enhancers at either end of the CH-containing Igh subdomain have important roles in CSR. Deletion of iEμ variably reduces CSR125-128. The 27kb 3′ Igh regulatory region (3′IghRR) super-enhancer at the downstream end of this subdomain contains closely associated enhancers that are essential for transcription from S region promoters (I-promoters) to induce transcriptionally mediated CSR129-132. Early 3C studies detailed the interaction patterns of the 3′IghRR and iEμ with different CH regions in resting and CSR-stimulated wild-type B cells that corresponded to I-promoter transcription and associated patterns of CSR132. These seminal studies suggested that interactions of 3′IghRR with upstream I-promoters form a ‘synaptosome’ that determines the potential for germline transcription and CSR to associated S regions132. The mechanism by which synaptosome structures are formed between widely separated Igh sequences remained unclear. However, other early studies provided a mechanistic hint with the finding that I-promoters located across a 100kb upstream region undergo linear competition for 3′IghRR-mediated activation to promote CSR133.
Because S regions are up to 12kb long and repetitive, prior CSR assays yielded few CSR junctions, all from S region borders, and gave limited mechanistic insights. Based on studies in transformed cell lines, DNA breaks were proposed to join at similar levels by inversional and deletional joining during CSR, with cellular selection required to sort out productive deletional rearrangements134. However, a recently developed high-throughput, highly sensitive LAM-HTGTS-Seq CSR assay has identified tens of thousands of CSR junctions across the entire length of acceptor S regions in CSR-activated normal B cells13. This assay revealed that AID-initiated double-strand breaks during CSR arise from most of the hundreds of deamination motifs across S regions and confirmed that donor and acceptor S region breaks can be generated independently of S region synapsis13. Moreover, this assay revealed that the great majority of CSR joining events between donor Sμ and acceptor S regions occur in deletional orientation by an unknown mechanism involving Igh organizational features in cis13 (Fig. 7a). This finding was striking as, other than V(D)J recombination-associated double-strand breaks, most breaks ends genome wide join to the ends of other separate double-strand breaks without orientation bias13,135-137. The orientation specificity of CSR was puzzling as there was no known factor, similar to RAG, that could enforce the orientation-specific AID-initiated joining of double-strand breaks. These findings pointed to an active synapsis mechanism during CSR30.
Chromatin loop extrusion-mediated CSR.
Indeed, recent studies have shown in three ways that cohesin-mediated loop extrusion is the fundamental mechanism of CSR30 (Fig. 7b; Supplementary Video 2).
First, in resting B cells, the iEμ–Sμ region and the 3′IghRR load cohesin and also function as dynamic loop extrusion anchors (Fig. 7b, part i). Cohesin-mediated extrusion of chromatin between these two dynamic loop anchors leads to the formation of basal, dynamic CSR loops and a class-switch recombination-centre [G] (CSR-centre) that is dynamic and likely to be deformed and reformed in resting B cells30.
Second, in activated B cells, the 3′IghRR, donor Sμ and acceptor S region are actively aligned in the CSR-centre by an activation-induced secondary loop extrusion process that is not dependent on expression of AID30. Although downstream S regions in the basal CSR loop are constantly extruded through the CSR-centre, they do not impede further extrusion as they are not transcribed and are inert in this regard. However, priming of a particular I region promoter through B cell activation and/or cytokine stimulation leads to transcriptional activation when it is brought proximal to the 3′IghRR in the CSR-centre (Fig. 7b, part ii). Induced I-region transcription leads to binding of the cohesin loader NIPBL and of cohesin at the activated I region promoter and the extrusion of a new sub-loop that aligns the activated downstream S region and Sμ in the CSR-centre30 (Fig. 7b, part iii). B cell activation also induces AID expression, which targets actively aligned S region double-strand breaks138.
Third, physical synapsis of individual double-strand break ends in each synapsed S region for deletional CSR is mediated post-cleavage by a loop extrusion-related mechanism30. Thereby, double-strand breaks on Sμ and the target S region can occur at different times and locations along the S region, as one or both ends of a given S region double-strand break is extruded into an associated cohesin ring, thereby stalling extrusion (Fig. 7b, part iv). The ends of a double-strand break in the second synapsed S region would then be extruded into the same cohesin ring(s), which aligns donor and acceptor break ends for productive, deletional joining30 (Fig. 7b, parts v and vi). Notably, this loop extrusion-mediated alignment process can be impaired by the insertion of CTCF sites within the extrusion path, leading to incomplete alignment and increased inversional joining. Such inversional CSR probably occurs because misaligned S regions are brought into close enough proximity for double-strand break ends in the donor and acceptor S regions to access each other by short-range diffusional motion30. Structural variations in chicken139 and duck140 CH loci may similarly promote inversional joining between oppositely oriented donor and acceptor S regions, with cellular selection for productive inversional events during CSR.
The above findings in support of a loop extrusion-mediated model of CSR indicate an unappreciated role for Igh enhancers and associated sequences in CSR. Namely, the enhancers function as cohesin loading sites and dynamic impediments to loop extrusion that are crucial for S region synapsis30. Similarly to V(D)J recombination25, impeding extrusion past a CSR-centre promotes the accessibility of impeded acceptor S regions for AID-mediated breakage and subsequent joining to Sμ30. Notably, insertion of CTCF sites in the CSR scanning path leads to misalignment of adjacent downstream non-S region sequences with Sμ, causing them to become surrogate S regions that undergo AID-initiated double-strand breaks and joining to Sμ30. Finally, loop extrusion-mediated CSR provided a mechanism for the enigmatic "sequential" CSR (from Sμ to Sγ1 and then Sμ/Sγ1 to Sε)141,142 and "downstream" CSR (between Sγ1 and Sε)143 events that contribute to IgE antibody generation30 (ref 30; supplemental discussion).
3′ Igh CTCF sites prevent ectopic CSR-centre formation.
The 3′ Igh CTCF sites focus chromatin loop extrusion-mediated activities within the upstream CH-containing domain and prevent 3′IghRR-mediated transcription and CSR downstream of the Igh locus31,144. Thus, deleting all ten 3′ Igh CTCF sites in mice significantly decreases the transcription, synapsis and switching of upstream S regions to varying degrees31. Moreover, deletion of the 3′ CTCF sites leads to active transcription of non-Igh downstream sequences, which allows for these sequences to be synapsed with Sμ to become ectopic S regions31.
Potential related roles of loop extrusion in double-strand break repair.
A recent study revealed that double-strand breaks may stall loop extrusion on one side of each break end, allowing extrusion to continue on the other sides15. This mechanism may enable the DNA-damage response protein ATM to scan the chromatin that is extruded past cohesin-bound double-strand break ends and phosphorylate other damage response proteins until arrested by loop anchors to form characteristic foci15. This mechanism is potentially related to the proposed mechanism of loop extrusion in tethering double-strand break ends for proper joining during CSR30 and provides a mechanism for early chromatin compaction models for the tethering of S region breaks for CSR145. This new study, together with related findings146,147, implicates an important role for cohesin-mediated loop extrusion in the maintenance of genome stability through double-strand break repair148.
Concluding remarks
Although tremendous progress has been made in understanding the roles of chromatin loop extrusion in RAG scanning and CSR, further studies are required of these dynamic processes, which are likely far more complex than outlined in current models. Similarly, the detailed mechanisms by which various factors promote the accessibility and pairing of target sequences within V(D)J recombination-centres and CSR-centres is another important area for further investigation. A long-standing question has been whether locus contraction involves an active mechanism in G1-arrested pro-B cells or whether the process requires cell-cycle progression32. The implicated role of cohesin-mediated loop extrusion in locus contraction may provide an insight. Specifically, cohesin-mediated chromatin loops are disassembled during mitosis and reassembled in G148. As V(D)J recombination is shut down upon G1 exit149, the prime opportunity for loop extrusion-mediated locus contraction to intersect with V(D)J recombination would seem to be in G1. Extension of current population-based studies to single-cell resolution may be required to fully elucidate these mechanisms.
Supplementary Material
Supplementary Video 1 ∣ Loop extrusion-mediated RAG chromatin scanning during Igh D-to-JH recombination. Reproduced with permission from REF. 26.
Supplementary Video 2 ∣ Chromatin loop extrusion has a fundamental mechanistic role in antibody class-switch recombination. Reproduced with permission from REF. 30
Acknowledgements
This work was supported by US National Institutes of Health grants (R01AI020047 and R01AI077595 to F.W.A.; R01AI155775 to Y.Z.). H.-Q.D. was a fellow of the Cancer Research Institute (CRI) of New York. H.H is a fellow of CRI. F.W.A. is an investigator of the Howard Hughes Medical Institute.
Glossary
- CTCF-binding elements
(CTCF sites). DNA sequence motifs recognized by the structural protein CTCF (CCCTC-binding factor) that mostly are characterized by a highly conserved 20 base pair core consensus motif but a subset of which are also flanked by upstream and/or downstream regulatory motifs.
- Cohesin ring protein complex
A highly conserved ring-shaped multi-protein complex composed of three core subunits, SMC1, SMC3 and RAD21, that associate with SA1 or SA2 in somatic cells. The cohesin complex has ATPase activity and has essential functions in chromosome segregation and chromatin loop formation, as well as in various fundamental processes including transcription and DNA repair.
- Convergent orientation
Referring to two DNA sequence motifs of opposite orientation that point towards each other in cis.
- V(D)J recombination
A programmed somatic recombination process initiated by RAG1–RAG2 endonuclease in early developing lymphocytes that assembles V, D and J gene segments into exons that encode the variable regions of antibody and T cell receptor chains.
- Class-switch recombination
(CSR). A programmed somatic recombination process initiated by activation-induced cytidine deaminase in antigen-activated mature B cells that replaces the upstream IgM-encoding Cμ exon with a different downstream constant region exon to change the class of antibody expressed. This allows for the antigen-specific effector functions of an antibody with a given variable region specificity to be optimized.
- Classical non-homologous end-joining
A major cellular repair pathway for DNA double-strand breaks that joins DNA ends without requiring a homologous template. This pathway is commonly referred to as ‘classical’ to distinguish it from less robust alternative end-joining pathways that become more obvious in the absence of the classical pathway.
- V(D)J recombination-centre
In the Igh locus, this is the site where RAG1–RAG2 endonuclease binds active chromatin harboring a strong enhancer to generate a hub for the capture of substrate V, D and J segments to allow for their cleavage and joining to create V(D)J variable region exons. Similar recombination-centres occur in the other antigen receptor loci that undergo V(D)J recombination.
- HTGTS-V(D)J-Seq
This assay maps RAG-initiated V(D)J recombination events across the genome and is adapted from the linear amplification-mediated high-throughput genome-wide translocation sequencing method (LAM-HTGTS). HTGTS-V(D)J-Seq has a unique ability to detect thousands of low-level independent cryptic joining events within a large population of cells, that when viewed together provide ‘tracks’ of RAG activity within a domain. These tracks clearly show the chromatin regions that recombination-centre-bound RAG explores, the directionality of the exploration process and the sequences RAG does or does not ‘see’ during exploration.
- LAM-HTGTS-3C-Seq
A high-resolution chromatin conformation capture (3C)-based genomic interaction assay that uses linear amplification-mediated high-throughput genome-wide translocation sequencing (LAM-HTGTS) for its downstream steps. This method detects sequences within chromatin loop domains that interact with a sequence of interest at high resolution.
- Switch region
(S region). A 1–12 kb repetitive DNA sequence that precedes each set of Igh constant region exons. S regions undergo DNA double-strand breaks mediated by activation-induced cytidine deaminase that are then joined to effect IgH class-switch recombination.
- Phase separation
A process that forms condensates of loci with similar chromatin states through homotypic attraction of bridging proteins.
- Class-switch recombination-centre
(CSR-centre). The CSR-centre is an Igh locus site at which activated B cells juxtapose distant cis-regulatory elements with donor and acceptor DNA switch regions, which are then cleaved and joined to carry out the process of antibody isotype switching.
Footnotes
Competing interests
The authors declare no competing interests.
Supplementary information
Supplementary information is available for this paper at https://doi.org/10.1038/s415XX-XXX-XXXX-X
References
- 1.Dixon JR et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nora EP et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dowen JM et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell 159, 374–387 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lupianez DG et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161, 1012–1025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narendra V et al. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science 347, 1017–1021 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hnisz D, Day DS & Young RA Insulated Neighborhoods: Structural and Functional Units of Mammalian Gene Control. Cell 167, 1188–1200 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merkenschlager M & Nora EP CTCF and Cohesin in Genome Folding and Transcriptional Gene Regulation. Annu. Rev. Genomics Hum. Genet 17, 17–43 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Stadhouders R, Filion GJ & Graf T Transcription factors and 3D genome conformation in cell-fate decisions. Nature 569, 345–354 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Guo Y et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell 162, 900–910 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pope BD et al. Topologically associating domains are stable units of replication-timing regulation. Nature 515, 402–405 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moindrot B et al. 3D chromatin conformation correlates with replication timing and is conserved in resting cells. Nucleic Acids Res. 40, 9470–948 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hu J et al. Chromosomal Loop Domains Direct the Recombination of Antigen Receptor Genes. Cell 163, 947–959 (2015). This study was the first to show that RAG, upon binding a bona fide recombination signal sequence (RSS), can then linearly explore mega-base distances within convergent CTCF sites-based chromatin loop domains to identify convergently-oriented cryptic RSS targets for V(D)J recombination-based cleavage and joining, thereby providing the basis for subsequent linear RAG chromatin scanning models.
- 13.Dong J et al. Orientation-specific joining of AID-initiated DNA breaks promotes antibody class switching. Nature 525, 134–139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins PL et al. DNA double-strand breaks induce H2Ax phosphorylation domains in a contact-dependent manner. Nat. Commun 11, 3158 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Arnould C et al. Loop extrusion as a mechanism for formation of DNA damage repair foci. Nature 590, 660–665 (2021). This study provided evidence for a key role of cohesin-mediated loop extrusion in general double-strand break repair which involves facilitating the formation of long double-strand break response foci critical for double-strand break end synapsis and joining.
- 16.Ochs F et al. Stabilization of chromatin topology safeguards genome integrity. Nature 574, 571–574 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Dekker J & Mirny L The 3D Genome as Moderator of Chromosomal Communication. Cell 164, 1110–1121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rao SS et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). This study is the first to report kilobase high-resolution Hi-C maps which revealed a genome-wide prevalence of convergent CTCF sites at the chromatin loop anchors, providing a key observation for the cohesin-mediated loop-extrusion model.
- 19.Nasmyth K Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet 35, 673–745 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Alipour E & Marko JF Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Res. 40, 11202–11212 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sanborn AL et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl Acad. Sci. USA 112, E6456–6465 (2015). This paper, together with the study by Fudenberg et al., (ref 24), provided strong evidence to support a chromatin loop extrusion process, likely driven by cohesin, as the underlying mechanism for the formation of convergent CTCF sites-based contact loop domains.
- 22.Nichols MH & Corces VG A CTCF Code for 3D Genome Architecture. Cell 162, 703–705 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bouwman BA & de Laat W Getting the genome in shape: the formation of loops, domains and compartments. Genome Biol. 16, 154 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fudenberg G et al. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 15, 2038–2049 (2016). This paper, together with the study by Sanborn et al (ref 21), provided strong evidence to support a chromatin loop extrusion process, likely driven by cohesin, as the underlying mechanism for the formation of convergent CTCF sites-based contact loop domains.
- 25. Jain S, Ba Z, Zhang Y, Dai HQ & Alt FW CTCF-Binding Elements Mediate Accessibility of RAG Substrates During Chromatin Scanning. Cell 174, 102–116 e114 (2018). This paper provided evidence that supports a model in which the Igh V(D)J recombination-centre serves as a dynamic loop anchor to allow cohesin-mediated loop extrusion to present upstream chromatin for scanning by RAG endonuclease and also demonstrated that CTCF sites promote accessibility of associated proximal VHs by increasing their interaction with the V(D)J recombination-centre during scanning.
- 26. Zhang Y et al. The fundamental role of chromatin loop extrusion in physiological V(D)J recombination. Nature 573, 600–604 (2019). This paper provided evidence that loop extrusion-mediated RAG scanning is the major mechanism for physiological, deletional D-to-JH recombination and mechanisms beyond CTCF sites-based loop anchors, including active transcription and nuclease-dead Cas9 protein binding can impede loop extrusion-mediated scanning and promote RAG targeting activity.
- 27. Ba Z et al. CTCF orchestrates long-range cohesin-driven V(D)J recombinational scanning. Nature 586, 305–310 (2020). Through a targeted depletion of cohesin or CTCF in v-Abl cell lines, this paper provided evidence that cohesin drives loop-extrusion-mediated RAG chromatin scanning and provided proof-of-principle that dampening of CTCF sites-based anchors can promote loop extrusion, locus contraction, and long-range RAG scanning.
- 28. Hill L et al. Wapl repression by Pax5 promotes V gene recombination by Igh loop extrusion. Nature 584, 142–147 (2020). This study showed that PAX5-mediated suppression of Wapl transcription in pro-B cells results in global extension of chromosomal loops, which implicated loop extrusion as the long-sought mechanism for VH locus contraction and distal VH utilization.
- 29. Dai HQ et al. Loop extrusion mediates physiological Igh locus contraction for RAG scanning. Nature 590, 338–343 (2021). By studying utilization of cryptic RSSs across normal and inverted VH loci, this study demonstrated linear RAG scanning across the VH locus in normal pro-B cells and also implicated WAPL down-regulation in promoting loop extrusion-mediated locus contraction and long-range RAG scanning.
- 30. Zhang X et al. Fundamental roles of chromatin loop extrusion in antibody class switching. Nature 575, 385–389 (2019). This paper showed that cohesin-mediated loop extrusion plays a fundamental role in the physiological, deletional Igh class switch recombination (CSR) mechanism, by establishing a CSR-centre that orchestrates substrate S region activation and synapsis, as well as post-cleavage deletional joining of S region double-strand breaks.
- 31. Zhang X, Yoon HS, Chapdelaine-Williams AM, Kyritsis N & Alt FW Physiological role of the 3'IgH CBEs super-anchor in antibody class switching. Proc. Natl Acad. Sci. USA 118 (2021). This paper showed that the 3′ Igh CTCF sites serve as an insulator to focus chromatin loop extrusion-mediated transcriptional and CSR activities within the upstream CH-containing domain.
- 32.Alt FW, Zhang Y, Meng FL, Guo C & Schwer B Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell 152, 417–429 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Methot SP & Di Noia JM Molecular Mechanisms of Somatic Hypermutation and Class Switch Recombination. Adv. Immunol 133, 37–87 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Ong CT & Corces VG CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet 15, 234–246 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Degner SC, Wong TP, Jankevicius G & Feeney AJ Cutting edge: developmental stage-specific recruitment of cohesin to CTCF sites throughout immunoglobulin loci during B lymphocyte development. J Immunol 182, 44–48 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Proudhon C, Hao B, Raviram R, Chaumeil J & Skok JA Long-Range Regulation of V(D)J Recombination. Adv Immunol 128, 123–182 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacPherson MJ & Sadowski PD The CTCF insulator protein forms an unusual DNA structure. BMC Mol. Biol 11, 101 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakahashi H et al. A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep. 3, 1678–1689 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vietri Rudan M et al. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 10, 1297–1309 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Wit E et al. CTCF Binding Polarity Determines Chromatin Looping. Mol. Cell 60, 676–684 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Kagey MH et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuin J et al. A cohesin-independent role for NIPBL at promoters provides insights in CdLS. PLoS Genet. 10, e1004153 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dorsett D & Merkenschlager M Cohesin at active genes: a unifying theme for cohesin and gene expression from model organisms to humans. Curr. Opin. Cell Biol 25, 327–333 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y et al. The structural basis for cohesin-CTCF-anchored loops. Nature 578, 472–476 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nishana M et al. Defining the relative and combined contribution of CTCF and CTCFL to genomic regulation. Genome Biol. 21, 108 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pugacheva EM et al. CTCF mediates chromatin looping via N-terminal domain-dependent cohesin retention. Proc. Natl Acad. Sci. USA 117, 2020–2031 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nora EP et al. Molecular basis of CTCF binding polarity in genome folding. Nat. Commun 11, 5612 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mirny L & Dekker J Mechanisms of Chromosome Folding and Nuclear Organization: Their Interplay and Open Questions. Cold Spring Harb. Perspect. Biol 10.1101/cshperspect.a040147 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hansen AS, Pustova I, Cattoglio C, Tjian R & Darzacq X CTCF and cohesin regulate chromatin loop stability with distinct dynamics. Elife 6, e25776 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hansen AS, Cattoglio C, Darzacq X & Tjian R Recent evidence that TADs and chromatin loops are dynamic structures. Nucleus 9, 20–32 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rowley MJ & Corces VG Organizational principles of 3D genome architecture. Nat. Rev. Genet 19, 789–800 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davidson IF & Peters JM Genome folding through loop extrusion by SMC complexes. Nat. Rev. Mol. Cell Biol 22, 445–464 (2021). [DOI] [PubMed] [Google Scholar]
- 53.Lin SG, Ba Z, Alt FW & Zhang Y RAG Chromatin Scanning During V(D)J Recombination and Chromatin Loop Extrusion are Related Processes. Adv. Immunol 139, 93–135 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Kraft K et al. Serial genomic inversions induce tissue-specific architectural stripes, gene misexpression and congenital malformations. Nat. Cell Biol 21, 305–310 (2019). [DOI] [PubMed] [Google Scholar]
- 55.Hsieh TS et al. Resolving the 3D Landscape of Transcription-Linked Mammalian Chromatin Folding. Mol. Cell 78, 539–553 e538 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thiecke MJ et al. Cohesin-Dependent and -Independent Mechanisms Mediate Chromosomal Contacts between Promoters and Enhancers. Cell Rep. 32, 107929 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nora EP et al. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell 169, 930–944 e922 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wutz G et al. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 36, 3573–3599 (2017). This study, together with ref 59 and ref 65, showed that depletion of WAPL, the cohesin unloading factor, increases cohesin chromatin retention time and leads to extension of chromatin loop sizes genome-wide.
- 59. Gassler J et al. A mechanism of cohesin-dependent loop extrusion organizes zygotic genome architecture. EMBO J. 36, 3600–3618 (2017). This study, together with ref 58 and ref 65, showed that depletion of WAPL, the cohesin unloading factor, increases cohesin chromatin retention time and leads to extension of chromatin loop sizes genome-wide.
- 60.Rao SSP et al. Cohesin Loss Eliminates All Loop Domains. Cell 171, 305–320 e324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schwarzer W et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature 551, 51–56 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vian L et al. The Energetics and Physiological Impact of Cohesin Extrusion. Cell 173, 1165–1178 e1120 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gandhi R, Gillespie PJ & Hirano T Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr. Biol 16, 2406–2417 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kueng S et al. Wapl controls the dynamic association of cohesin with chromatin. Cell 127, 955–967 (2006). [DOI] [PubMed] [Google Scholar]
- 65. Haarhuis JHI et al. The Cohesin Release Factor WAPL Restricts Chromatin Loop Extension. Cell 169, 693–707 e614 (2017). This study, together with ref 58 and ref 59, showed that depletion of WAPL, the cohesin unloading factor, increases cohesin chromatin retention time and leads to extension of chromatin loop sizes genome-wide.
- 66.Davidson IF et al. DNA loop extrusion by human cohesin. Science 366, 1338–1345 (2019). [DOI] [PubMed] [Google Scholar]
- 67.Kim Y, Shi Z, Zhang H, Finkelstein IJ & Yu H Human cohesin compacts DNA by loop extrusion. Science 366, 1345–1349 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Golfier S, Quail T, Kimura H & Brugues J Cohesin and condensin extrude DNA loops in a cell cycle-dependent manner. Elife 9, e53885 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ribeiro de Almeida C, Hendriks RW & Stadhouders R Dynamic Control of Long-Range Genomic Interactions at the Immunoglobulin kappa Light-Chain Locus. Adv. Immunol 128, 183–271 (2015). [DOI] [PubMed] [Google Scholar]
- 70.Schatz DG & Swanson PC V(D)J recombination: mechanisms of initiation. Annu. Rev. Genet 45, 167–202 (2011). [DOI] [PubMed] [Google Scholar]
- 71.Teng G & Schatz DG Regulation and Evolution of the RAG Recombinase. Adv Immunol 128, 1–39 (2015). [DOI] [PubMed] [Google Scholar]
- 72.Jung D, Giallourakis C, Mostoslavsky R & Alt FW Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu. Rev. Immunol 24, 541–570 (2006). [DOI] [PubMed] [Google Scholar]
- 73.Kim MS, Lapkouski M, Yang W & Gellert M Crystal structure of the V(D)J recombinase RAG1-RAG2. Nature 518, 507–511 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ru H et al. Molecular Mechanism of V(D)J Recombination from Synaptic RAG1-RAG2 Complex Structures. Cell 163, 1138–1152 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim MS et al. Cracking the DNA Code for V(D)J Recombination. Mol. Cell 70, 358–370 e354 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ru H et al. DNA melting initiates the RAG catalytic pathway. Nat. Struct. Mol. Biol 25, 732–742 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grundy GJ et al. Initial stages of V(D)J recombination: the organization of RAG1/2 and RSS DNA in the postcleavage complex. Mol. Cell 35, 217–227 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang XS, Lee BJ & Zha S The recent advances in non-homologous end-joining through the lens of lymphocyte development. DNA Repair (Amst) 94, 102874 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhao B, Rothenberg E, Ramsden DA & Lieber MR The molecular basis and disease relevance of non-homologous DNA end joining. Nat. Rev. Mol. Cell Biol 21, 765–781 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ji Y et al. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell 141, 419–431 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Teng G et al. RAG Represents a Widespread Threat to the Lymphocyte Genome. Cell 162 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guo C et al. CTCF-binding elements mediate control of V(D)J recombination. Nature 477, 424–430 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lin SG, Guo C, Su A, Zhang Y & Alt FW CTCF-binding elements 1 and 2 in the Igh intergenic control region cooperatively regulate V(D)J recombination. Proc. Natl Acad. Sci. USA 112, 1815–1820 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Garrett FE et al. Chromatin architecture near a potential 3' end of the igh locus involves modular regulation of histone modifications during B-Cell development and in vivo occupancy at CTCF sites. Mol. Cell Biol 25, 1511–1525 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Degner SC et al. CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells. Proc. Natl Acad. Sci. USA 108, 9566–9571 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Benner C, Isoda T & Murre C New roles for DNA cytosine modification, eRNA, anchors, and superanchors in developing B cell progenitors. Proc. Natl Acad. Sci. USA 112, 12776–12781 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Aiden EL & Casellas R Somatic Rearrangement in B Cells: It's (Mostly) Nuclear Physics. Cell 162, 708–711 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kosak ST et al. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science 296, 158–162 (2002). [DOI] [PubMed] [Google Scholar]
- 89.Fuxa M et al. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev 18, 411–422 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sayegh CE, Jhunjhunwala S, Riblet R & Murre C Visualization of looping involving the immunoglobulin heavy-chain locus in developing B cells. Genes Dev. 19, 322–327 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Roldan E et al. Locus 'decontraction' and centromeric recruitment contribute to allelic exclusion of the immunoglobulin heavy-chain gene. Nat. Immunol 6, 31–41 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jhunjhunwala S et al. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell 133, 265–279 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Medvedovic J et al. Flexible long-range loops in the VH gene region of the Igh locus facilitate the generation of a diverse antibody repertoire. Immunity 39, 229–244 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rother MB et al. Nuclear positioning rather than contraction controls ordered rearrangements of immunoglobulin loci. Nucleic Acids Res. 44, 175–186 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Montefiori L et al. Extremely Long-Range Chromatin Loops Link Topological Domains to Facilitate a Diverse Antibody Repertoire. Cell Rep. 14, 896–906 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bossen C, Mansson R & Murre C Chromatin topology and the regulation of antigen receptor assembly. Annu. Rev. Immunol 30, 337–356 (2012). [DOI] [PubMed] [Google Scholar]
- 97.Ebert A, Hill L & Busslinger M Spatial Regulation of V-(D)J Recombination at Antigen Receptor Loci. Adv. Immunol 128, 93–121 (2015). [DOI] [PubMed] [Google Scholar]
- 98.Rogers CH, Mielczarek O & Corcoran AE Dynamic 3D Locus Organization and Its Drivers Underpin Immunoglobulin Recombination. Front. Immunol 11, 633705 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lucas JS, Zhang Y, Dudko OK & Murre C 3D trajectories adopted by coding and regulatory DNA elements: first-passage times for genomic interactions. Cell 158, 339–352 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ranganath S et al. Productive coupling of accessible Vbeta14 segments and DJbeta complexes determines the frequency of Vbeta14 rearrangement. J. Immunol 180, 2339–2346 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Helmink BA & Sleckman BP The response to and repair of RAG-mediated DNA double-strand breaks. Annu. Rev Immunol. 30, 175–202 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nussenzweig A & Nussenzweig MC Origin of chromosomal translocations in lymphoid cancer. Cell 141, 27–38 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tepsuporn S, Hu J, Gostissa M & Alt FW Mechanisms that can promote peripheral B-cell lymphoma in ATM-deficient mice. Cancer Immunol. Res 2, 857–866 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bredemeyer AL et al. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature 442, 466–470 (2006). [DOI] [PubMed] [Google Scholar]
- 105.Wood C & Tonegawa S Diversity and joining segments of mouse immunoglobulin heavy chain genes are closely linked and in the same orientation: implications for the joining mechanism. Proc Natl Acad Sci U S A 80, 3030–3034, doi: 10.1073/pnas.80.10.3030 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yancopoulos GD et al. Preferential utilization of the most JH-proximal VH gene segments in pre-B-cell lines. Nature 311, 727–733 (1984). [DOI] [PubMed] [Google Scholar]
- 107.Gauss GH & Lieber MR The basis for the mechanistic bias for deletional over inversional V(D)J recombination. Genes Dev. 6, 1553–1561 (1992). [DOI] [PubMed] [Google Scholar]
- 108.Choi NM et al. Deep sequencing of the murine IgH repertoire reveals complex regulation of nonrandom V gene rearrangement frequencies. J. Immunol 191, 2393–2402 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bolland DJ et al. Two Mutually Exclusive Local Chromatin States Drive Efficient V(D)J Recombination. Cell Rep. 15, 2475–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Khanna N, Zhang Y, Lucas JS, Dudko OK & Murre C Chromosome dynamics near the sol-gel phase transition dictate the timing of remote genomic interactions. Nat. Commun 10, 2771 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ebert A et al. The distal V(H) gene cluster of the Igh locus contains distinct regulatory elements with Pax5 transcription factor-dependent activity in pro-B cells. Immunity 34, 175–187 (2011). [DOI] [PubMed] [Google Scholar]
- 112.Verma-Gaur J et al. Noncoding transcription within the Igh distal V(H) region at PAIR elements affects the 3D structure of the Igh locus in pro-B cells. Proc. Natl Acad. Sci. USA 109, 17004–17009 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hesslein DG et al. Pax5 is required for recombination of transcribed, acetylated, 5' IgH V gene segments. Genes Dev. 17, 37–42 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yancopoulos GD & Alt FW Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell 40, 271–281 (1985). [DOI] [PubMed] [Google Scholar]
- 115.Yancopoulos GD, Blackwell TK, Suh H, Hood L & Alt FW Introduced T cell receptor variable region gene segments recombine in pre-B cells: evidence that B and T cells use a common recombinase. Cell 44 251–259 (1986). [DOI] [PubMed] [Google Scholar]
- 116.Bolland DJ et al. Antisense intergenic transcription in V(D)J recombination. Nat. Immunol 5, 630–637 (2004). [DOI] [PubMed] [Google Scholar]
- 117.Bolland DJ et al. Antisense intergenic transcription precedes Igh D-to-J recombination and is controlled by the intronic enhancer Emu. Mol. Cell Biol 27, 5523–5533 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Beilinson HA et al. The RAG1 N-terminal region regulates the efficiency and pathways of synapsis for V(D)J recombination. J. Exp. Med 218 e20210250 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Qiu X et al. Sequential Enhancer Sequestration Dysregulates Recombination Center Formation at the IgH Locus. Mol Cell 70, 21–33 e26, doi: 10.1016/j.molcel.2018.02.020 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chakraborty T et al. Repeat organization and epigenetic regulation of the DH-Cmu domain of the immunoglobulin heavy-chain gene locus. Mol. Cell 27, 842–850 (2007). [DOI] [PubMed] [Google Scholar]
- 121.Hwang JK, Alt FW & Yeap LS Related Mechanisms of Antibody Somatic Hypermutation and Class Switch Recombination. Microbiol. Spectr 3, MDNA3-0037-2014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Muramatsu M et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102, 553–563 (2000). [DOI] [PubMed] [Google Scholar]
- 123.Yeap LS & Meng FL Cis- and trans-factors affecting AID targeting and mutagenic outcomes in antibody diversification. Adv. Immunol 141, 51–103 (2019). [DOI] [PubMed] [Google Scholar]
- 124.Cascalho M, Wong J, Steinberg C & Wabl M Mismatch repair co-opted by hypermutation. Science 279, 1207–1210 (1998). [DOI] [PubMed] [Google Scholar]
- 125.Bottaro A et al. Deletion of the IgH intronic enhancer and associated matrix-attachment regions decreases, but does not abolish, class switching at the mu locus. Int. Immunol 10, 799–806 (1998). [DOI] [PubMed] [Google Scholar]
- 126.Sakai E, Bottaro A & Alt FW The Ig heavy chain intronic enhancer core region is necessary and sufficient to promote efficient class switch recombination. Int. Immunol 11, 1709–1713 (1999). [DOI] [PubMed] [Google Scholar]
- 127.Perlot T, Alt FW, Bassing CH, Suh H & Pinaud E Elucidation of IgH intronic enhancer functions via germ-line deletion. Proc. Natl Acad. Sci. USA 102, 14362–14367 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li F, Yan Y, Pieretti J, Feldman DA & Eckhardt LA Comparison of identical and functional Igh alleles reveals a nonessential role for Emu in somatic hypermutation and class-switch recombination. J. Immunol 185, 6049–6057 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Vincent-Fabert C et al. Genomic deletion of the whole IgH 3' regulatory region (hs3a, hs1,2, hs3b, and hs4) dramatically affects class switch recombination and Ig secretion to all isotypes. Blood 116, 1895–1898 (2010). [DOI] [PubMed] [Google Scholar]
- 130.Saintamand A et al. Deciphering the importance of the palindromic architecture of the immunoglobulin heavy-chain 3' regulatory region. Nat. Commun 7, 10730 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pinaud E et al. Localization of the 3' IgH locus elements that effect long-distance regulation of class switch recombination. Immunity 15, 187–199 (2001). [DOI] [PubMed] [Google Scholar]
- 132.Wuerffel R et al. S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity 27, 711–722 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Seidl KJ et al. Position-dependent inhibition of class-switch recombination by PGK-neor cassettes inserted into the immunoglobulin heavy chain constant region locus. Proc. Natl Acad. Sci. USA 96, 3000–3005 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Harriman W, Volk H, Defranoux N & Wabl M Immunoglobulin class switch recombination. Annu. Rev. Immunol 11, 361–384 (1993). [DOI] [PubMed] [Google Scholar]
- 135.Chiarle R et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell 147, 107–119 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Frock RL et al. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol 33, 179–186 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wei PC et al. Long Neural Genes Harbor Recurrent DNA Break Clusters in Neural Stem/Progenitor Cells. Cell 164, 644–655 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dudley DD et al. Internal IgH class switch region deletions are position-independent and enhanced by AID expression. Proc. Natl Acad. Sci. USA 99, 9984–9989 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhao Y, Rabbani H, Shimizu A & Hammarstrom L Mapping of the chicken immunoglobulin heavy-chain constant region gene locus reveals an inverted alpha gene upstream of a condensed upsilon gene. Immunology 101, 348–353 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lundqvist ML, Middleton DL, Hazard S & Warr GW The immunoglobulin heavy chain locus of the duck. Genomic organization and expression of D, J, and C region genes. J. Biol. Chem 276, 46729–46736 (2001). [DOI] [PubMed] [Google Scholar]
- 141.Xiong H, Dolpady J, Wabl M, Curotto de Lafaille MA & Lafaille JJ Sequential class switching is required for the generation of high affinity IgE antibodies. J. Exp. Med 209, 353–364 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mandler R, Finkelman FD, Levine AD & Snapper CM IL-4 induction of IgE class switching by lipopolysaccharide-activated murine B cells occurs predominantly through sequential switching. J. Immunol 150, 407–418 (1993). [PubMed] [Google Scholar]
- 143.Zhang T et al. Downstream class switching leads to IgE antibody production by B lymphocytes lacking IgM switch regions. Proc. Natl Acad. Sci. USA 107, 3040–3045 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yu K An insulator that regulates chromatin extrusion and class switch recombination. Proc. Natl Acad. Sci. USA 118, e2026399118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bassing CH & Alt FW The cellular response to general and programmed DNA double strand breaks. DNA Repair (Amst) 3, 781–796, doi: 10.1016/j.dnarep.2004.06.001 (2004). [DOI] [PubMed] [Google Scholar]
- 146.Liu Y et al. Very fast CRISPR on demand. Science 368, 1265–1269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Li K, Bronk G, Kondev J & Haber JE Yeast ATM and ATR kinases use different mechanisms to spread histone H2A phosphorylation around a DNA double-strand break. Proc. Natl Acad. Sci. USA 117, 21354–21363 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mirny LA Cells use loop extrusion to weave and tie the genome. Nature 590, 554–555 (2021). [DOI] [PubMed] [Google Scholar]
- 149.Desiderio S Temporal and spatial regulatory functions of the V(D)J recombinase. Semin. Immunol 22, 362–369 (2010). [DOI] [PubMed] [Google Scholar]
- 150.Majumder K, Bassing CH & Oltz EM Regulation of Tcrb Gene Assembly by Genetic, Epigenetic, and Topological Mechanisms. Adv. Immunol 128, 273–306 (2015). [DOI] [PubMed] [Google Scholar]
- 151.Carico Z & Krangel MS Chromatin Dynamics and the Development of the TCRalpha and TCRdelta Repertoires. Adv. Immunol 128, 307–361 (2015). [DOI] [PubMed] [Google Scholar]
- 152.Fischer C et al. Conservation of the T-cell receptor alpha/delta linkage in the teleost fish Tetraodon nigroviridis. Genomics 79, 241–248 (2002). [DOI] [PubMed] [Google Scholar]
- 153.Fitzsimmons SP, Bernstein RM, Max EE, Skok JA & Shapiro MA Dynamic changes in accessibility, nuclear positioning, recombination, and transcription at the Ig kappa locus. J. Immunol 179, 5264–5273 (2007). [DOI] [PubMed] [Google Scholar]
- 154.Ribeiro de Almeida C et al. The DNA-binding protein CTCF limits proximal Vkappa recombination and restricts kappa enhancer interactions to the immunoglobulin kappa light chain locus. Immunity 35, 501–513 (2011). [DOI] [PubMed] [Google Scholar]
- 155.Stadhouders R et al. Pre-B cell receptor signaling induces immunoglobulin kappa locus accessibility by functional redistribution of enhancer-mediated chromatin interactions. PLoS Biol. 12, e1001791 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lin YC et al. Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat. Immunol 13, 1196–1204 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Xiang Y, Park SK & Garrard WT Vkappa gene repertoire and locus contraction are specified by critical DNase I hypersensitive sites within the Vkappa-Jkappa intervening region. J. Immunol 190, 1819–1826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Liu Z et al. A recombination silencer that specifies heterochromatin positioning and ikaros association in the immunoglobulin kappa locus. Immunity 24, 405–415 (2006). [DOI] [PubMed] [Google Scholar]
- 159.Xiang Y, Zhou X, Hewitt SL, Skok JA & Garrard WT A multifunctional element in the mouse Igkappa locus that specifies repertoire and Ig loci subnuclear location. J. Immunol 186, 5356–5366 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Video 1 ∣ Loop extrusion-mediated RAG chromatin scanning during Igh D-to-JH recombination. Reproduced with permission from REF. 26.
Supplementary Video 2 ∣ Chromatin loop extrusion has a fundamental mechanistic role in antibody class-switch recombination. Reproduced with permission from REF. 30