Abstract
The incorporation of radioactive orthophosphate into the cell walls of Saccharomyces cerevisiae was studied. 33P-labeled cell walls were extensively extracted with hot sodium dodecyl sulfate (SDS). Of the remaining insoluble radioactivity more than 90% could be released by laminarinase. This radioactive material stayed in the stacking gel during SDS-polyacrylamide gel electrophoresis but entered the separating gel upon treatment with N-glycosidase F, indicating that phosphate was linked directly or indirectly to N-mannosylated glycoproteins. The phosphate was bound to covalently linked cell wall proteins as mannose-6-phosphate, the same type of linkage shown previously for soluble mannoproteins (L. Ballou, L. M. Hernandez, E. Alvarado, and C. E. Ballou, Proc. Natl. Acad. Sci. USA 87:3368–3372, 1990). From the phosphate-labeled glycoprotein fraction released by laminarinase, three cell wall mannoproteins, Ccw12p, Ccw13p and Ccw14p, were isolated and identified by N-terminal sequencing. For Ccw13p (encoded by DAN1 [also called TIR3]) and Ccw12p the association with the cell wall has not been described before; Ccw14p is identical with cell wall protein Icwp (I. Moukadiri, J. Armero, A. Abad, R. Sentandreu, and J. Zueco, J. Bacteriol. 179:2154–2162, 1997). In ccw12, ccw13, or ccw14 single or double mutants neither the amount of radioactive phosphate incorporated into cell wall proteins nor its position in the stacking gel was changed. However, the triple mutant brought about a shift of the 33P-labeled glycoprotein components from the stacking gel into the separating gel. The disruption of CCW12 results in a pronounced sensitivity of the cells to calcofluor white and Congo red. In addition, the ccw12 mutant shows a decrease in mating efficiency and a defect in agglutination.
The walls of fungal cells can be considered vital extracellular organelles that have to withstand turgor pressures greater than 15 × 105 Pa (6). Baker’s yeast, Saccharomyces cerevisiae, invests about 20% of its total dry weight into building up this organelle, which consists of approximately equal parts of glucans and mannoproteins and less than 2% chitin (7). The protein moieties of the mannoproteins amount to about 10% of the cell wall weight, and more than 20 individual cell wall proteins have been identified so far (3, 4, 8, 17, 18, 21, 24, 25, 27, 33, 37, 42, 43, 48).
The extraordinary stability of the cell wall against tension due to high internal hydrostatic pressure can be explained, if it is assumed that the various components of the wall are interlinked and form a highly branched meshwork. Partial evidence for such a structure consisting of covalently linked β-1,3- and β-1,6-glucan, chitin, and mannoproteins has been reported (19, 23), whereby a stable phosphodiester linkage has been proposed to connect the mannoproteins to the rest of the meshwork (16).
On the other hand, the small oligosaccharides O-linked to mannoproteins also seem to contribute to the stability of the cell wall, since mutants defective in protein O-mannosylation are osmolabile (9). This phenomenon could be explained by assuming that there are phosphodiester bridges between the O-linked saccharides of mannoproteins and, for example, glucans of the cell wall. Such a type of linkage exists, for example, in the cell wall of Volvox, for which it was shown that O-linked chains may link extracellular matrix components by Ara-5-phospho-5-Ara bridges (44).
To identify such phosphodiester bridges, we studied the incorporation of radioactive phosphate into the cell wall of S. cerevisiae. During the course of this investigation three new covalently linked cell wall proteins (Ccw12p, Ccw13p, and Ccw14p) have been identified, whose electrophoretic behavior correlated with the most highly phosphorylated cell wall fraction. Ccw14p has been identified in the meantime also by Moukadiri et al. (26) as inner cell wall protein Icwp. These cell wall proteins and their knockout phenotypes, as well as the structure of the phosphate-containing saccharides, are described in this paper.
MATERIALS AND METHODS
Yeast strains.
The strains used in this study are shown in Table 1.
TABLE 1.
Yeast strains
| Strain | Genotype | Reference |
|---|---|---|
| SEY6210 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 GAL | 30 |
| SEY6211 | MATa ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 GAL | 30 |
| LB347-1C | MATα mmn9 | 41 |
| MEY12A | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 GAL ccw12::ura3 | This work |
| MEY12B | MATa ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 GAL ccw12::ura3 | This work |
| MEY13 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 GAL ccw13::trp1 | This work |
| MEY14 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 GAL ccw14::ura3 | This work |
| MEY1213 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 GAL ccw12::ura3 ccw13::trp1 | This work |
| MEY234 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 GAL ccw12::ura3 ccw13::trp1 ccw14::his3 | This work |
Phosphate labeling of covalently linked cell wall proteins.
A total of 1.4 × 107 cells from an overnight culture of S. cerevisiae were pelleted and resuspended in 10 ml of a phosphate-free minimal medium (39). After the culture was shaken at 29°C for 2.5 h, 20 μl of 0.1 M K phosphate and 400 μCi of [33P]orthophosphate (specific activity, 3,000 Ci/mmol) were added, and the cells were incubated for another 3.5 h. Cells were spun down, an aliquot of the medium was counted, and the cells were broken by vortexing the suspension four times for 1 min with 0.5-mm-diameter glass beads in 300 μl of an extraction buffer (50 mM Tris-HCl [pH 7.5], 1 mM 2-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, 1 μg of antipain per ml, 5 μg of pepstatin per ml, 20 μg of leupeptin per ml). The extract was removed from the glass beads, and the latter was washed with 300 μl of the same buffer. The pooled extracts (600 μl) were centrifuged at 1,000 × g for 3 min. The supernatant (cytosol) was removed, and the pellet was extracted twice with 1 ml of sodium dodecyl sulfate (SDS)-Laemmli buffer (22) at 95°C. The pellet was then washed with 1 ml of 0.5 M NaCl and with 1 ml of H2O. Subsequently, the pellet was incubated in a total volume of 200 μl of a solution containing 50 mM Tris-HCl [pH 7.5], 50 mM MgCl2, DNase I (110 U), and RNase (50 μg) for 8 h at 30°C. The radioactivity staying in the pellet was considered to be associated with insoluble cell wall material. Most (90%) of this radioactivity was released when the pellet was treated for 4 h with laminarinase (100 μl of a 0.1 M Na acetate buffer [pH 5.5] containing 0.05 U of laminarinase [Sigma, St. Louis, Mo.]). The material released was separated by SDS-polyacrylamide gel electrophoresis according to the method described by Laemmli (22).
Biotinylation and isolation of cell wall proteins.
Labeling and isolation of wall proteins was done essentially as described previously (27). In brief, cells were grown in 1 liter of YPD medium (1% yeast extract, 2% Bacto Peptone, 2% dextrose; Difco Laboratories) to an optical density at 578 nm of 4 to 5 (1 unit corresponds to 2 × 107 cells), then centrifuged, and washed twice with a 50 mM K phosphate buffer (pH 8.0). Cells were resuspended in 20 ml of the same buffer to which 10 mg of Sulfo-NHS-LC-Biotin reagent (Pierce, Rockford, Ill.) was added and labeled for 90 min on ice. Cells were then washed twice with a 50 mM Tris-HCl buffer (pH 7.5) containing 50 mM MgCl2 and once with a K phosphate buffer (pH 8.0). Cells were disrupted mechanically in a glass bead-cell homogenizer (MKS Braun, Melsungen, Germany). Cell walls were centrifuged (15 min at 5,000 × g) and washed five times with the same buffer. SDS-extractable proteins were obtained by a 10-min extraction in 2 to 3 ml of Laemmli sample buffer (22) at 95°C. To release covalently attached proteins, cell walls were further washed five times with Na acetate buffer (pH 5.5) and then digested for 2 h at 37°C in 6 ml of Na acetate buffer to which 0.5 U of laminarinase from mollusks (Sigma) was added.
Purification of Ccw12p, Ccw13p, and Ccw14p proteins.
In order to purify Ccw12p, the laminarinase extract was lyophilized and dissolved in 1 ml of Laemmli sample buffer. Subsequently, proteins were applied to an SDS–10% polyacrylamide gel (100 by 80 by 1.5 mm) with about 5 mm of 4.5% stacking gel. After electrophoresis, the stacking gel was cut off and crushed by passing it through a Teflon net. Gel pieces were extracted overnight in 1 ml of phosphate-buffered saline (10 mM Na phosphate [pH 7.4], 150 mM NaCl). Aliquots (0.2 ml) of the extract were then applied to a Superose 12 column and eluted with a 0.1 M Tris-HCl buffer, pH 6.8, at a flow rate of 0.1 ml/min. The peak eluted in the void volume was collected and dialyzed exhaustively against 5% acetic acid. The protein preparation obtained in this way was used for the N-terminal sequencing of Ccw12p. To purify Ccw13p, the same procedure was followed, except that strain MEY12A was used. Ccw14p was purified from the mnn9 mutant strain (41).
Electrophoresis and blotting.
Protein electrophoresis was performed by the method of Laemmli (22). Blots of biotin-labeled cell wall proteins were obtained by electroblotting proteins onto nitrocellulose membranes, which were then incubated for 1 h in 10 mM Tris-HCl, pH 7.5, containing 0.1% Triton X-100 and 4% bovine serum albumin (BSA), followed by 1 h in the same buffer with 1% BSA and streptavidin-horseradish peroxidase conjugate (Sigma), at a dilution of 1:5,000. Blots were subsequently washed three times with the same buffer and developed with an ECL kit (Amersham, Little Chalfont, United Kingdom). Protein standards used for the estimation of molecular masses (in daltons) of proteins were as follows: myosin (205,000), β-galactosidase (116,000), phosphorylase b (97,400), BSA (67,000), ovalbumin (45,000), and carboanhydrase (29,000).
Disruption of genes coding for identified cell wall proteins.
Standard procedures were used for all DNA manipulations (32). For all PCRs, genomic DNA isolated from strain SEY6210 (28) was used as a template. To construct strains MEY12A and MEY12B, the open reading frame (ORF) YLR110c was amplified by PCR with oligonucleotides 5′-TGGGAGTCTTACTTC-3′ and 5′-CTCCGCGAGGTTCAG-3′ and cloned by blunt-end ligation into the SmaI site of pUC19, resulting in plasmid pME12. The URA3 gene was isolated as a 1.1-kb HindIII fragment from the plasmid YEp24 (14) and cloned into pME12 (digested with Tth111I).
To construct the strain MEY13, the ORF YJR150c was amplified by PCR with oligonucleotides 5′-CTTCCGTAGACGCTCCTCTG-3′ and 5′-GGACCGGAAATAGTTGGAGCAC-3′ and cloned by blunt-end ligation into the SmaI restriction site of pUC19, resulting in plasmid pME13. The TRP1 gene was cloned as a 1.3-kb BspHI-SspI fragment from the plasmid pRS414 (38) into pME13 (digested with StyI). In the case of strains MEY14 and MEY234, N- and C-terminal fragments of YLR391w were amplified with oligonucleotides 5′-ATAAGAATGCGGCCGCCAGAATACGACGAGGACG-3′ and 5′-CTATCCCGGGGATCCAAGCTTGGTGCTGTGAGCTTTGACC-3′ and oligonucleotides 5′-CAAGCTTGGATCCCCGGGATAGGGCCTGTGTTGCGCAAGT-3′ and 5′-TGACACGCGTCGACGGAGGAAGCGCTACTGGA-3′, respectively. Both new PCR fragments of YLR39w1 with a 22-base overlap were fused in a second PCR, creating a new HindIII site and a new BamHI site. These sites were used subsequently for the insertion of the markers URA3 (1.1-kb HindIII fragment from the plasmid YEp24) and HIS3 (1.8-kb BamHI fragment from the plasmid JCW102; kindly supplied by J. Wan, San Diego, Calif.). Single disruptions were performed in strains SEY6210 and SEY6211. Yeast transformation was performed according to the method of Gietz et al. (10). Correct disruptions were analyzed by PCR.
To construct the ccw12 ccw13 double mutant, the direct disruption of CCW13 in the ccw12 mutant (MEY12A) was unsuccessful. Therefore, MEY12A was crossed with SEY6211, and the obtained diploids were transformed with the ccw13::TRP1 construct. The strain MEY1213 resulted from the sporulation of the diploid transformants. In order to construct MEY234 (ccw12 ccw13 ccw14), MEY1213 was transformed with ccw14::HIS3.
Sensitivity of yeast cells to calcofluor white and Congo red.
Cells were grown to a density of 2 × 108 cells/ml, and 2 × 107 cells were suspended in 100 μl of water. Serial 1:10 dilutions thereof were prepared. A total of 5 μl of each dilution was spotted on YPD medium, to which either 10 μg of calcofluor white per ml or 10 or 20 μg of Congo red per ml was added.
Quantitation of mating efficiency.
Cells were grown to a density of 1 × 107 to 4 × 107 cells/ml. Equal numbers (106 cells) of strains of opposite mating types were mixed in 150 μl of YPD medium. The suspension was incubated for 6 h at 30°C, centrifuged, and resuspended in 100 μl of H2O. Serial dilutions (1:10) were made, and 5 μl of the 10−1 to 10−4 dilutions was spotted either on YPD medium or on a minimal medium lacking adenine and lysine, thus allowing only the growth of diploid cells.
Agglutination assay.
Equal numbers (2 × 107 cells) of logarithmically growing cells of opposite mating types were mixed in 50 μl of YPD medium in microtiter plates. Subsequently 150 μl of a 0.1 M Na phosphate buffer (pH 6.3) was added, and the suspension was shaken for 1 h and checked for agglutination.
Chromatography of phosphorylated cell wall components. (i) TLC.
Thin-layer chromatography (TLC) was performed on polyethyleneimine cellulose plates (Schleicher & Schüll) with 1 M ammonium formate, pH 3.4, as the solvent system (44).
(ii) Bio-Gel P 2 chromatography.
Oligosaccharides were separated on a column (0.9 by 104 cm) equilibrated in 50 mM pyridine acetate, pH 5.2. Fractions of 1 ml were collected and analyzed for radioactivity or total carbohydrate by the phenol-sulfuric acid method (5). Molecular size standards were stachyose, raffinose, and mannose.
(iii) High-performance anionic exchange chromatography (HPAEC).
To determine the sugar composition, samples were hydrolyzed with 4 M trifluoroacetic acid (TFA) at 100°C for 4 h and separated on a CarboPac PA1 column (4 by 250 mm) with 16.5 mM NaOH as an eluent at a flow rate of 1 ml/min. For the analysis of sugar-phosphates the same column was used by applying an isocratic elution of 150 mM Na acetate in 54 mM NaOH for 20 min, followed by an increase to 200 mM Na acetate in 39 mM NaOH within 10 min and at a flow rate of 1 ml/min; for detection a PAD (Gold) detector was used. With a mannose-6-phosphate standard the column was qualitatively and quantitatively calibrated.
Preparation and characterization of phosphorylated cell wall oligosaccharides.
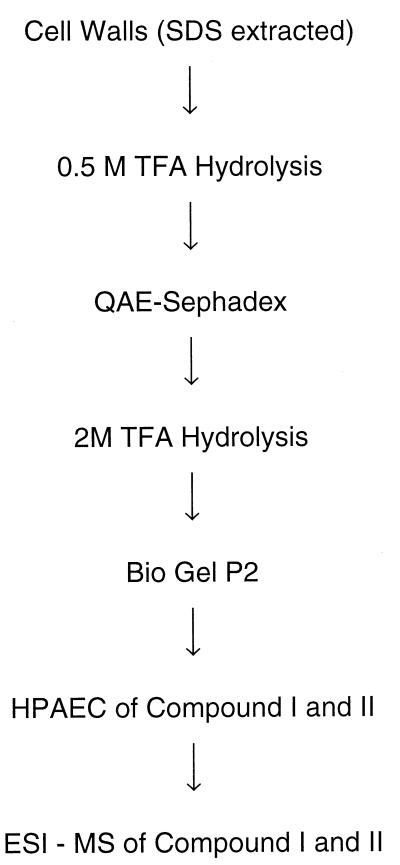
Commercially obtained baker’s yeast (wet weight, 150 g) was suspended in 150 ml of 10 mM Tris-HCl (pH 7.4) containing 1 mM phenylmethylsulfonyl fluoride and broken with a Bio-Spec bead beater (150 g of 0.5-mm-diameter glass beads) four times for 30 s. The disruption of cells was monitored by microscopy. The homogenate was centrifuged for 10 min at 5,000 × g, and the cell wall pellet was washed three times with a homogenization buffer, five times with 1 M NaCl, and five times with water. Cell walls were extracted twice with hot 50 mM Tris-HCl containing 2% SDS, 100 mM EDTA, and 40 mM 2-mercaptoethanol (pH 8) for 15 min at 100°C, followed by three washes with water (final yield was 60 g [wet weight]). The purified cell wall fraction was hydrolyzed in portions of 15 g with 50 ml of 0.5 M TFA in boiling water for 2 h and then extensively lyophilized to remove the acid (final yield was 840 mg).
(i) QAE-Sephadex A50 chromatography.
The lyophilisate (400 mg) was dissolved in 15 ml of 2 mM Tris-base, applied to a column of 2.5 by 15 cm, and eluted at a flow rate of 1 ml/min with a step gradient of 2 mM Tris-base, 2 mM Tris-base containing 100 mM NaCl, and 2 mM Tris-base containing 200 mM NaCl (see Fig. 4). Fractions of 5 ml were collected, pooled, and dialyzed (2,000-molecular-weight cutoff; Spectropor 6) against water.
FIG. 4.
QAE-Sephadex chromatography of 0.5 M TFA hydrolysate. The cell wall fraction released by 0.5 M TFA was loaded and eluted as described in Materials and Methods. Aliquots were analyzed for total carbohydrate by the phenol-sulfuric acid method. The arrows indicate the addition of 100 mM and 200 mM NaCl.
(ii) Bio-Gel P2 chromatography.
Quaternary aminoethyl (QAE) fractions 48 to 60 (see Fig. 4) were pooled and hydrolyzed with 2 M TFA for 2.5 h at 100°C, dried under nitrogen, dissolved in 0.3 ml of water, and chromatographed on the same column, and identical conditions were used for the radiolabeled material.
(iii) HPAEC.
Pooled P2 fractions 54 to 57 and 58 to 62 were lyophilized and purified further in several batches on a PA1 column with the sugar-phosphate gradient. The sugar-phosphate peaks (compound I from the pool of fractions 54 to 57 and compound II from the pool of fractions 58 to 62) were desalted on Dowex 50W × 8 (H+) and used for electrospray ionization tandem mass spectrometry (ESI-MS) and gas chromatography-mass spectrometry analysis.
(iv) ESI-MS.
A Finnigan MAT TSQ 700 triple quadrupole mass spectrometer equipped with a Finnigan electrospray ion source (Finnigan MAT Corp., San Jose, Calif.) was used for ESI-MS. Native oligosaccharides were dissolved in methanol containing 0.3% NH3 to a concentration of approximately 10 pmol/μl, infused at a flow rate of 1 μl/min into the electrospray chamber, and subjected to negative-ion-mode ESI-MS. A voltage of −4.5 kV was applied to the electrospray needle. For collision-induced dissociation (CID) experiments, parent ions were selectively transmitted by the first mass analyzer and directed into the collision cell (argon was used as the collision gas) with the kinetic energy set around 33 eV. The reduced and permethylated samples were dissolved in acetonitrile, saturated with NaCl (concentration, approximately 10 pmol/μl), and detected in the positive-ion mode with inversed voltages on the mass spectrometer.
(v) Methylation analysis.
For methylation analysis, oligosaccharides were permethylated according to the method of Hakomori (12), purified, hydrolyzed, reduced, and peracetylated as described previously (29). The separation and identification of partially methylated alditol acetates were performed on a Finnigan gas chromatograph (Finnigan MAT Corp.), equipped with a 30-m DB5 capillary column, connected to a Finnigan GCQ ion trap mass spectrometer.
RESULTS
Characterization of the phosphate-labeled glycan fraction covalently linked to the cell wall.
In order to address the question whether glucan-phosphate or mannan-phosphate linkages occur in the wall and in particular whether mild acid-stable phophodiester bridges of the type found in Volvox (44) can be detected as a structural element, yeast cells were metabolically labeled with [33P]orthophosphate. Isolated cell walls were washed, and soluble, noncovalently linked glycoproteins were removed by extraction with SDS under reducing conditions. The residual wall was treated with mild acid to release labile phosphodiester mannose of the Man-1-phospho-6-Man type (31) and finally with DNase-RNase to remove radiolabeled nucleic acid contaminants. The cell wall purified and treated in this way contained 0.5 to 0.9% of the total 33P incorporated (three experiments).
(i) TLC analysis.
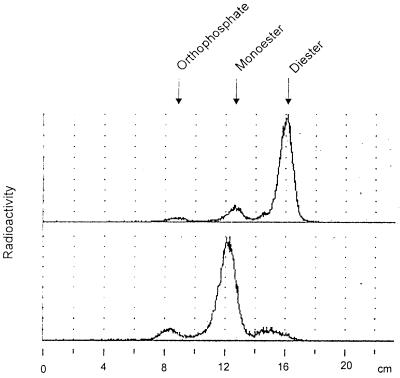
This purified radiolabeled cell wall was partially hydrolyzed with 0.5 M TFA (2 h at 100°C); the total radioactivity became soluble under this condition. Chromatography of the 33P-labeled cell wall hydrolysate on polyethyleneimine cellulose thin-layer plates resulted in the detection of a radioactive product that was hardly retained by the ion-exchange layer, as is typical for the mobility of sugar phosphodiester compounds (Fig. 1). This chromatographic system has previously been used to identify a phosphodiester of arabinose of an extracellular matrix glycoprotein from Volvox carteri (11, 44). It separates sugar phosphodiester from monoester and inorganic phosphate, in that order of mobility. Acid hydrolysis of the fastest running radioactive spot (at 16 cm) by 2 M TFA for 2 h converted the material to a major product comigrating with hexose phosphates (at 12 cm) and to a minor peak comigrating with inorganic phosphate (at 8.5 cm). Chromatography of this same material on a Dionex HPAEC instrument showed that the hexose phosphate has the mobility of mannose-6-phosphate (data not shown; see below).
FIG. 1.
Chromatography of radiolabeled (0.5 M TFA) cell wall hydrolysate. Purified cell walls were hydrolyzed and analyzed by polyethyleneimine TLC. Shown are a 0.5 M TFA extract (top) and a 0.5 M TFA extract upon further treatment with 4 M TFA for 2.5 h (bottom). Arrows indicate the migration of inorganic phosphate, phosphomonoester (mannose-6-phosphate), and phosphodiester (as reported in references 11 and 44).
(ii) Bio-Gel P2 analysis.
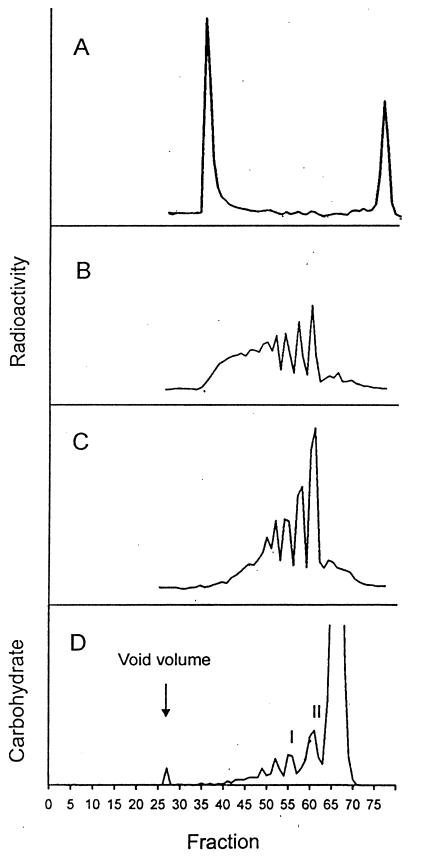
Analysis of the 0.5 M TFA hydrolysate on a Bio-Gel P2 column revealed that the material elutes in the void volume (Fig. 2A), indicating that it is larger than 1.8 kDa. The isolated, excluded fraction was treated with stronger acid and rechromatographed on the same column. As shown in Fig. 2B and C the radiolabeled material shown in Fig. 2A was converted to 33P-labeled compounds of smaller size. Fractions 59 to 62, eluting between a di- and tetrasaccharide reference, as well as fractions 56 to 58 and fractions 53 to 55 (Fig. 2B) were pooled and analyzed by TLC. All three fractions migrated in the phosphodiester region, in accordance with the expected behavior of the ion-exchange layer, which does not separate by size. After further acid hydrolysis these compounds could be converted to radioactive material, running like a hexose monophosphate (data not shown).
FIG. 2.
Bio-Gel P2 profiles. Shown are the analysis of radiolabeled 0.5 M TFA extract (A), the void volume fraction shown in panel A further treated with 2 M TFA for 2 h (B) or 3 h (C), and the profile of the nonradioactive cell wall hydrolysate (D). QAE fractions 48 to 60 (Fig. 4) were pooled, dialyzed, and hydrolyzed with 2 M TFA for 2.5 h and applied to Bio-Gel P2. Fractions 54 to 57 (peak I) and 58 to 62 (peak II) were further purified by HPAEC (Fig. 5). The large carbohydrate peak, fractions 63 to 70, represents the position of monosaccharides.
Structural analysis of the cell wall-bound sugar-phosphate. (i) Isolation of phosphorylated oligosaccharides.
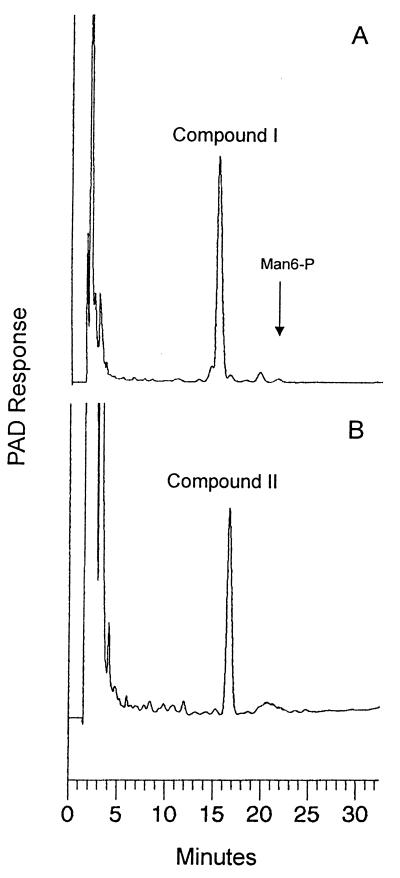
To identify and characterize the nature of the cell wall-bound sugar-phosphate described above, nonradioactive bulk cell wall material was prepared. The procedure followed to obtain SDS-extracted purified cell walls was identical to that described above. A large-scale purification of the 0.5 M TFA-released cell wall fraction was carried out as described in Materials and Methods (Fig. 3). Determination of the carbohydrate composition of this material after complete hydrolysis (4 M TFA at 100°C for 4 h) yielded 57% mannose, 41% glucose, and 2% mannose-6-phosphate. Before the 0.5 M TFA hydrolysate was applied to Bio-Gel P2, it was first fractionated on a QAE-Sephadex column to remove neutral material, according to the procedure of Varki and Kornfeld (45) (Fig. 4). Three fractions were obtained: a neutral, nonretarded one (fractions 12 to 28); fractions 48 to 60, eluted with 100 mM NaCl; and fractions 81 to 105, eluted with 200 mM NaCl. Both charged pools after complete hydrolysis had a similar carbohydrate composition, consisting of 96% mannose, 3.5% mannose-6-phosphate, and hardly any glucose. Only the pool of fractions 48 to 60 was further analyzed; due to the almost identical overall composition, it is assumed that the compounds in the two charged pools differ only by the charge density, i.e., the number of charges per molecule. The Bio-Gel P2 profile obtained after 2 M TFA hydrolysis of the pool of fractions 48 to 60 is depicted in Fig. 2D. Fractions 54 to 57 and 58 to 62 were pooled, and the phosphorylated compounds were purified to homogeneity by HPAEC. Both fractions contained phosphorylated saccharides and in addition still-neutral manno-oligosaccharides eluting in the flowthrough fraction. High-pressure liquid chromatography profiles of the phosphorylated compounds designated I and II are shown in Fig. 5A and B. The retention times of both peaks were shorter than that of mannose-6-phosphate, which was expected for a phosphodiester due to a reduced charge. Also analysis by TLC on polyethyleneimine cellulose revealed that they comigrate with the radiolabeled material (data not shown). Strong acid hydrolysis of compound I resulted in a mannose-6-phosphate (Man-6-P):mannose ratio of 1:2, indicating that the assumption of a diester (e.g., of the type Man-6-P-6-Man) may be wrong, since after the release of either mannosyl residue, the remaining Man-6-P should be resistant to the acid hydrolysis conditions used (Fig. 1).
FIG. 3.
Flow sheet showing the isolation of phosphorylated saccharides from cell walls.
FIG. 5.
Purification of compounds I and II by HPAEC. Pooled fractions 54 to 57 (peak I) and 58 to 62 (peak II) from the Bio-Gel columns (Fig. 2D) were applied to a PA1 column for final purification with the sugar-phosphate gradient. The arrow indicates the elution of the mannose-6-phosphate standard.
(ii) Structural analysis of the phosphorylated oligosaccharides.
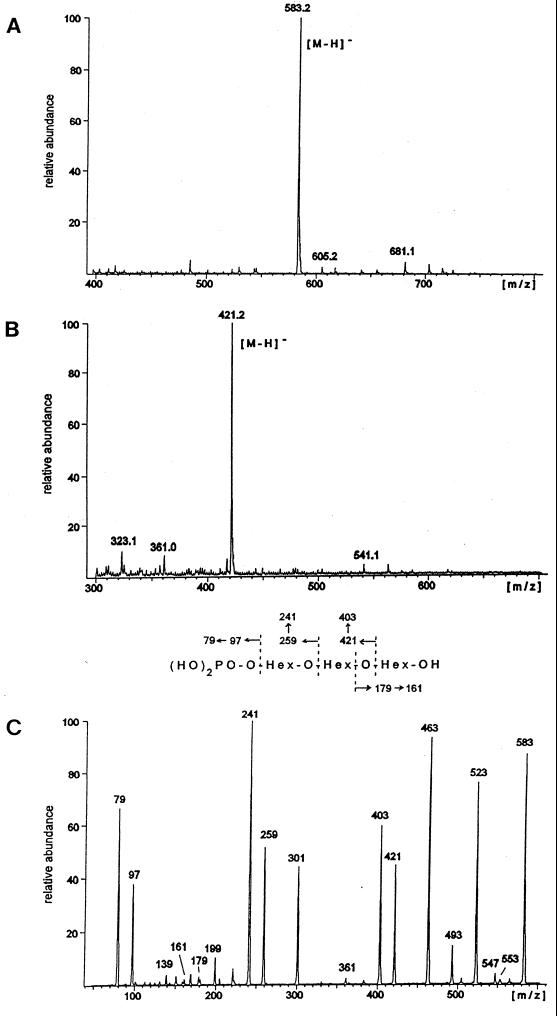
Electrospray mass spectrometry of the native compounds (negative-ion mode) yielded deprotonated molecular ions at m/z 583 (compound I) and m/z 421 (compound II), consistent with the masses of a monophosphorylated tri- and dimannosyl oligosaccharide, respectively (Fig. 6A and B). In Fig. 6C the daughter ion spectrum of the trisaccharide is depicted, obtained after CID. The intense fragment ion series generated from the nonreducing end of the trisaccharide (Fig. 6, fragmentation scheme) clearly indicates a linear structure with the phosphate linked to the terminal mannose residue. The loss of up to four hydroxylated carbon units from the reducing end of the molecule limits the linkage point of the middle mannose to position 6 of the reducing mannose residue. The daughter ion spectrum of the phosphorylated disaccharide (compound II; data not shown) yielded an analogous pattern. In particular, the loss of up to four hydroxylated carbon units from the reducing mannose also suggests a substitution of this mannose at O-6 by the phosphorylated sugar residue. After the reduction and permethylation of the oligosaccharides, molecular ions (sodium adducts) at m/z 791 and 587, respectively, were obtained by positive-ion-mode ESI-MS, indicating a complete methylation of both the carbohydrate part and the phosphate group. The MS-MS spectra (not shown) were compatible with linear tri- and disaccharide structures bearing a terminal dimethylphosphate.
FIG. 6.
ESI-MS spectra of compounds I and II. Negative-ion ESI-MS spectra of compound I (A) and compound II (B). (C) The detected deprotonated molecular ion suggests the presence of a monophosphorylated trisaccharide. The daughter ion spectrum of this molecular ion obtained after CID. The fragmentation pattern is explained in the scheme between panels B and C. The indicated secondary fragment ions are all due to the elimination of water. Additional fragment ions at m/z 553, 523, 493, and 463 are generated by the loss of one to four CH(OH) units from the reducing hexose, excluding a substitution at positions 2 to 4 of this residue. The intense fragment ion at m/z 301 can be assigned to the terminal phosphorylated hexose linked to an inner ring fragment incorporating two hydroxylated carbon units from the middle hexose residue.
Methylation analysis of the trisaccharide yielded 6-O-acetyl-1,2,3,4,5-penta-O-methyl-mannitol and 1,2,5-tri-O-acetyl-3,4,6-O-methyl-mannitol in a ratio of approximately 1:1, confirming the substitution of the reduced mannose residue at O-6 and indicating a linkage of the terminal, phosphorylated sugar residue to O-2 of the inner mannose. We failed to detect a partially methylated alditol acetate characteristic for the phosphorylated mannose residue, suggesting its complete degradation during the derivatization procedure. From the disaccharide in an analogous fashion, exclusively 6-O-acetyl-1,2,3,4,5-penta-O-methyl-mannitol was obtained, characteristic for a 6-substituted mannose at the reducing end.
The combination of these complementary sets of data suggests the structures P-6-Man-1→2-Man-1→6-Man, for the trisaccharide (compound I), and P-6-Man-1→6-Man, for the disaccharide (compound II). Assuming an α-configuration of the mannose residues, which is observed exclusively in mannoproteins isolated from yeast, the structures described are identical to the ones described for phosphorylated manno-oligosaccharides, which were isolated, however, from soluble glycoproteins (1, 15).
Isolation, purification, and identification of new cell wall proteins.
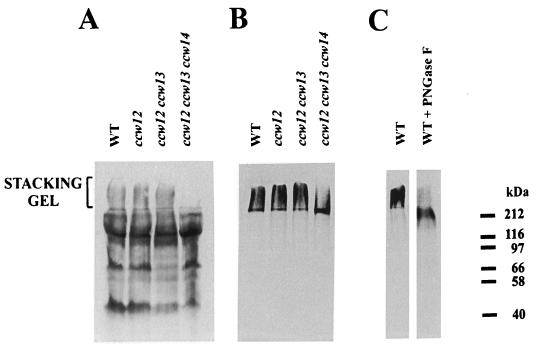
To find out which protein components are associated with the phosphate-labeled material, 33P-labeled, insoluble cell wall material of the strain SEY6210 was treated with a β-glucanase. A total of 90% of the 33P-labeled material was released by laminarinase. Electrophoresis of this material revealed that the radioactivity remained in the stacking gel (Fig. 7B and C, WT lanes). Digestion with PNGase F resulted in the shift of the 33P-labeled material from the stacking gel to the separating gel (Fig. 7C), suggesting that 33P is associated with mannoproteins.
FIG. 7.
Cell wall proteins of different ccw mutants released by digestion with laminarinase. Cell wall proteins of strains SEY6210 (wild type [WT]), MEY12A (ccw12), MEY 213 (ccw12 ccw13), and MEY234 (ccw12 ccw13 ccw14) were labeled with biotin (A) or 33P (B and C). SDS-extracted cell walls were digested with laminarinase (1 mU/μl), and the released material was subjected to electrophoresis and blotted. The blots were visualized either by the reaction with streptavidin-peroxidase conjugates (A) or by autoradiography (B and C).
The results obtained motivated the analysis of proteins contained in the stacking portion of the gel. Mrsa et al. (27) described a method for labeling S. cerevisiae cell wall proteins by biotinylation. By using this method, 20 cell wall proteins were labeled, and 11 of them were identified (4, 27). When proteins extracted from the wall by laminarinase were analyzed previously, the material remaining in the stacking gel was not included. Therefore, the biotinylation procedure was applied to analyze also that part of the gel which contains the phosphate. As shown in Fig. 7A, in addition to the proteins in the separating gel, the stacking gel also contained biotinylated material.
To identify these proteins, they were extracted from the stacking gel and purified on a Superose 12 column as described in Materials and Methods. N-terminal sequencing of the material obtained revealed one major protein sequence (Table 2) corresponding to the ORF assigned the Yeast Protein Database code YLR110c. Consistent with the previous designation of identified cell wall proteins (27), we named this gene CCW12 (because it encodes a covalently linked cell wall protein). CCW12 was found to be identical to the α0.6 gene, which was identified in a screen for genes in which transcription is switched off by α-factor (34). The corresponding protein, however, had not been identified so far. To test whether Ccw12p is the major protein component detected in the stacking gel, a ccw12 mutant was constructed. In the ccw12 mutant there is no obvious decrease in the amount of the biotin- or 33P-labeled material in the stacking gel (Fig. 7A and B).
TABLE 2.
N-terminal sequences of newly identified cell wall proteins
| Protein | N-terminal sequencea | Identical to: | Reference | Yeast ORF |
|---|---|---|---|---|
| Ccw12p | AAXVXXAXVXQEXXXLVXIXXXEDHVXXEXVXP | α0.6p | 34 | YLR110c |
| (AANVTTATVSQESTTLVTITSCEDHVCSETVSP) | ||||
| Ccw13p | SGVXTTLS | Dan1p | 35 | YJR150c |
| (ASVTTTLS) | ||||
| Ccw14p | XPPAXLLAXVAQV | Icwp1p | 26 | YLR391w |
| (TPPACLLACVAQV) |
The experimentally determined sequence obtained (shown first) and the peptide derived from the gene sequence thought to be coding for the experimentally derived peptide (shown in parentheses) are shown for each protein.
To identify further protein components, preparative amounts of cell wall proteins of a ccw12 null mutant were extracted with laminarinase and isolated from the stacking gel as before. This revealed another protein sequence (YJR150c), and the gene was named CCW13 (Table 2). This gene was found to be allelic to the DNA1 gene, which was identified in a screen for genes induced during the anaerobic growth of yeast cells (35). The Dan1 protein had not been identified so far.
The 33P incorporation into the covalently bound cell wall material was checked in the ccw13 and the ccw12 ccw13 null mutants. In a ccw13 mutant no effect on the amount or on the running behavior of the biotinylated or radiolabeled cell wall proteins was found (data not shown). The same was true for the ccw12 ccw13 double mutant, for which the radioactivity was still concentrated in the stacking gel (Fig. 7A and B).
Therefore, we decided to test whether the Ccw14p protein, which we purified in a different context from a mnn9 mutant, might affect the running behavior of the 33P-labeled material. In the zymolyase extract of cell walls of the mnn9 mutant a double band could be seen on Coomassie blue-stained gels, with one band hardly migrating into the separating gel and the other with a molecular mass of about 150 kDa (data not shown). N-terminal sequencing of both bands revealed the same protein, here named Ccw14p (Table 2). The amino acid sequence showed that the protein was identical to the recently described inner cell wall protein (Icwp) (26). The disruption of CCW14 had no effect on the phosphate-labeled material in the stacking gel (data not shown). Only in the ccw12 ccw13 ccw14 triple knockout strain did the radioactivity incorporated into the SDS-extracted cell wall decrease slightly (data not shown) and the radioactively labeled material shifted from the stacking gel into the separating gel (Fig. 7B). In addition, no cell surface biotinylated material could be detected in the stacking gel anymore (Fig. 7A).
Characterization of the newly identified cell wall proteins. (i) Analysis of the sequences of Ccw12p, Ccw13p, and Ccw14p.
Ccw12p is a small, acidic protein of 133 amino acids and has all the properties characteristic of covalently linked cell wall proteins. It contains a typical signal sequence for directing the protein to the secretory pathway (amino acids 1 to 18), a large number of hydroxy amino acids (nearly 40% of the total amino acids), and a C-terminal sequence required for the potential attachment of a glycosylphosphatidylinositol (GPI) anchor. N-terminal sequencing of the protein predicted a mature protein of 93 amino acids and revealed that all serine and threonine residues between amino acid 19 and amino acid 51 were modified, indicating an exhaustive O-glycosylation of the protein. Besides, Ccw12p contains three potential N-glycosylation sites. The pronounced difference in the size of the calculated protein compared to the actual measured one suggests a very high degree of glycosylation. In the C-terminal part of the protein the amino acid motif TTEAPKNGTSTAAP is repeated twice. A similar motif is also present in the cell wall protein Sed1 (13, 36), where it is repeated four times with slight variations within the N-terminal part of the protein. Furthermore, the N-terminal part of Ccw12p (amino acid 28 to amino acid 73) is 72% identical to the region from amino acid 1229 to amino acid 1275 of the flocculation protein Flo1p (47). The same region has similar homology to three hypothetical FLO1 homologues (YKR102w, YHR211w, and YAL063c). It should also be mentioned that the S. cerevisiae genome contains another homologue of the CCW12 gene (YD9302.09c/YD9302.10c), which is 83% identical to CCW12 but contains a stop codon instead of the Q at position 67. The existence of this stop codon in our yeast strain has been confirmed by sequencing the PCR-amplified fragment (data not shown).
The second identified protein, Ccw13p, also conforms to the general properties of covalently linked cell wall proteins. It contains 298 amino acids (263 in the mature form) and a signal sequence (amino acids 1 to 19), about 40% of its total amino acids are hydroxy amino acids clustered in the C-terminal half of the gene, and finally, it has a putative C-terminal GPI-anchoring signal. CCW13 has significant homology with another not characterized yeast gene (ORF YJR151c). In addition, CCW13 is homologous to members of the PAU gene family (46). Altogether 21 additional ORFs, containing sequences homologous to the first 120 to 165 amino acids of Ccw13p, have been found scattered all over the genome.
Ccw14p also contains all sequence elements typical for covalently linked cell wall proteins, including the serine-rich region in the C-terminal half of the protein and the putative GPI-anchoring signal.
(ii) Phenotypic characterization of ccw12, ccw13, and ccw14 null mutants.
To assess possible functions of the three cell wall proteins, deletion mutants were constructed as described in Materials and Methods (Table 1). The ccw12, ccw13, and ccw14 single mutants showed no significant morphological phenotypes. The ccw13 and ccw14 mutants grew as well as the wild type, but the ccw12 mutant showed an increased generation time of about 40% compared to the wild type. In order to test whether Ccw12p, Ccw13p or Ccw14p are involved in stabilizing the cell wall, the effects of calcofluor white and Congo red, compounds known to interfere with cell wall biogenesis (18), on the growth of ccw12, ccw13, and ccw14 mutants were analyzed. As shown in Fig. 8, the ccw12 mutant is at least 100 times more sensitive to both dyes than the wild type. In the case of the ccw14 mutant, an increase in cell wall lability, as already reported by Moukadiri et al. (26), has been confirmed (data not shown). The mutation in the CCW13 gene had no influence on the cell wall sensitivity towards the two agents (data not shown).
FIG. 8.
Sensitivity of the ccw12 mutant to calcofluor white and Congo red. Sequential dilutions of 2 × 107 cells/0.1 ml for strain SEY6210 (wild type [WT]) and MEY12A (ccw12) were prepared, and 5 μl of the suspension was spotted on YPD medium containing 10 μg of either calcofluor white or Congo red per ml.
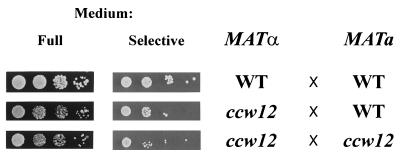
The ability of all three ccw disruptants to mate has also been tested. ccw mutants were mated with the wild-type SEY6211 (MATa) strain and tested for the growth of diploids. No mating defect was found for ccw13 and ccw14 mutants (data not shown). The ccw12 mutant, however, showed an apparent decrease in the mating efficiency (Fig. 9). To check whether the ccw12 mutant was also affected in agglutination, a standard agglutination assay was performed. Fig. 10 shows that the agglutination was indeed impaired, if one or both partners lack the Ccw12p protein. The different phenotype characteristics of cell wall protein mutants are summarized in Table 3.
FIG. 9.
Mating ability of ccw12 mutant. A total of 106 cells of the MATα strain SEY6210 (wild type [WT]) or MEY12A (ccw12) were mixed with the same number of cells of the MATa strain SEY6211 (WT) or MEY12B (ccw12) and incubated for 6 h in 150 μl of YPD medium at 30°C. Serial 1:10 dilutions were made, and 5 μl of each dilution was plated on a minimal medium lacking adenine and lysine, allowing the growth of diploids only (starting from a 10−1 dilution).
FIG. 10.
Agglutination of the ccw12 mutant. The formation of cell agglutinates was monitored by mixing 2 × 107 cells of either SEY6210 (wild type [WT]) or MEY12A (ccw12) with either SEY6211 (WT) or MEY12B (ccw12), as described in Materials and Methods.
TABLE 3.
Phenotypes of different cell wall protein mutantsa
| Strain | Growth | Sensitivity to:
|
Mating | Agglutination | |
|---|---|---|---|---|---|
| Calcofluor white | Congo red | ||||
| WT | Normal | − | − | + | + |
| ccw12 | Slow | ++ | ++ | − | − |
| ccw13 | Normal | − | − | + | n.d. |
| ccw14 | Normal | ++ | ++ | + | n.d. |
Different phenotypes of wild-type SEY6211 (WT) and the mutant strains MEY12A (ccw12), MEY 13 (ccw13), and MEY14 (ccw14) are summarized. n.d., not determined.
DISCUSSION
Various components of the yeast cell wall are covalently linked to one another, as recently demonstrated and summarized by Kollár et al. (19). Chitin can be β-1,4 linked to the nonreducing end of β-1,3-glucan (20) as well as to the nonreducing end of β-1,6-glucan (19). In the latter case, the reducing end of this β-1,6-glucan is bound to β-1,3-glucan, thus forming a bridge between chitin and β-1,3-glucan. To the nonreducing end of β-1,6-glucan, a specific group of mannoproteins, containing a GPI-derived glycan part, is covalently attached (16, 23, 25).
All these different covalent linkages between wall components certainly contribute to the rigidity of the yeast cell wall. Another major protein modification of almost all cell wall mannoproteins, the addition of short O-linked sugar chains, has not been expected to play a similar role. However, two observations have been reported: (i) it has long been known that mild alkaline conditions, typically used for β-elimination reactions, cause drastic size changes in cell wall mannoprotein fractions (28); (ii) certain knockout mutants of PMT genes, which are responsible for protein O-mannosylation, lead to an osmolabile phenotype, although a significant decrease in the total mannose content of the cell wall was not observed (9). Sugar chains O-linked to proteins could affect the secretory process of these cell wall proteins, or the sugar chains, although short, could contribute directly to cell wall rigidity. Either way these observations indicate that not only polysaccharide components, but cell wall proteins as well, should be considered important for cell wall rigidity. This could either be caused by O-mannosylated cell wall proteins as structural elements, or it could be related to the function of these proteins as extracellular enzymes; they could contribute to rigidity through the reaction they catalyze, for example, a transglycosylation.
Considering that protein-bound short O-linked sugar chains could be directly involved in building a rigid cell wall, the question of course arises of how this may be achieved. One possibility is the existence of a relatively acid-stable phosphate link between these chains and other cell wall polymers, a type of linkage existing in bacterial teichoic acids or in the arabino-protein of the Volvox extracellular matrix (11, 44). To test this hypothesis, it was investigated whether 33P gets incorporated into the insoluble yeast cell wall material remaining after the removal of soluble wall components by SDS. Indeed a significant amount of radioactivity was found in the insoluble fraction. About 90% of this radioactivity could be solubilized by laminarinase. Since its mobility on SDS gels was affected by PNGase F, it seemed obvious that 33P has been incorporated into mannoproteins covalently linked to cell walls. The postulated acid-stable phosphodiester, however, could not be detected. The structures finally determined, P-6-Man-1,2-Man-1,6-Man and P-6-Man-1,6-Man, correspond to linkages which have previously been identified as part of N-linked glycan chains on several soluble yeast glycoproteins, for example, on mannoproteins extractable with a citrate buffer and on carboxypeptidase (1, 15). Biosynthetically these phosphomonoesters arise via a Man-1-P transfer from GDP-Man, yielding Man-1-P-6-Man-R and a subsequent release of the terminal mannose (2, 31). The Man-1-P link is extremely acid labile (31) and would not have been preserved in the wall material studied here.
Since the first part of the paper gave a negative result—that the postulated stable phosphodiester does not exist—we tried in the second part to at least characterize the wall-bound phosphate. Surprisingly, phosphate incorporation into covalently bound cell wall proteins seems to be restricted to a few proteins or complex cell wall components containing mannoproteins. The possibility of a high-molecular-weight phosphorylated complex is suggested by the observation that all the 33P radioactivity solubilized by β-glucanase is retained in the stacking portion of an SDS gel (Fig. 7B). When biotinylated walls were prepared as described previously (27) it was indeed possible to demonstrate quite a significant amount of covalently bound protein material in the stacking gel after SDS electrophoresis (Fig. 7A). The corresponding proteins were purified and after partial N-terminal sequencing identified as Ccw12p, Ccw13p, and Ccw14p (Table 2). The corresponding genes had partly been identified previously in a different context (34, 35, 46); only for CCW14 has the protein been studied and shown to represent a cell wall component called Icwp, inner cell wall protein (26). The protein material in the stacking gel disappeared when CCW14 together with the other two genes was disrupted (Fig. 7A). Similarly, the amount of 33P incorporated into cell wall mannoproteins of this mutant was also slightly reduced, and the radioactivity was shifted into the separating gel. The fact that the 33P radioactivity was still associated with cell wall proteins of the triple mutant indicates, however, that further proteins besides Ccw14p get phosphorylated and that the running behavior of these proteins differs in the triple mutant from that of the wild type. This indicates some interaction or covalent connection of these various cell wall proteins. The question, however, of which cell wall protein(s) the phosphate is linked to has to remain unanswered, unfortunately.
It has been suggested that the covalently bound cell wall proteins are linked to the complex described by Kollár et al. (19) via a transmannosidase reaction, transferring the protein moiety together with part of its GPI anchor to the β-1,6-glucan (16, 23, 25). Since this transfer has been postulated to occur within the oligomannose portion of the GPI anchor, the newly attached mannoprotein should contain a fairly acid-stable phosphodiester link between ethanolamine and mannose (19). So far only indirect evidence for such a link has been presented (16). Although 90% of the 33PO43− incorporated into cell wall material (and which was not extractable by SDS under reducing conditions) could be solubilized by laminarinase, radioactivity was not associated with any of the protein bands previously characterized as Ccw1p to Ccw5p (27) (Fig. 7A). Although the cell wall proteins Ccw1p to Ccw5p are not completely identified so far, they do react with a β-1,6-glucan antibody (16a), and according to the biosynthetic scheme postulated (19, 23), they should incorporate 33P. Finally in this context it should be pointed out that the cell wall proteins associated with the stacking gel (see above) did also react with the anti-β-1,6-glucan antibody (16a). Since the genes of these proteins (CCW12, CCW13, and CCW14) show a putative GPI-anchoring sequence, a part of their 33P content may be due to the phosphodiester postulated by Kapteyn et al. (16).
The disruption of the three genes coding for the cell wall proteins described herein revealed some apparent phenotypes of the corresponding mutants. The ccw12 mutant was particularly interesting, since a marked decrease in the mating ability of this strain was detected. Although it seemed unlikely that the mating defect was caused by a decrease in agglutination (24), the agglutination ability of the mutant was tested and found to be decreased (Fig. 10). However, when the presence of agglutinins in the ccw12 mutant was investigated, no change in the amount of a- or α-agglutinin in the cell walls of the corresponding mating types could be seen, either on immunoblots of laminarinase cell wall extracts or by fluorescence microscopy of cells labeled with fluorescein isothiocyanate conjugates of a- or α-agglutinin antibodies (data not shown). Thus, it has to be speculated that the inability of the mutant to agglutinate is due to an inaccessibility of agglutinins at the cell surface or to some other change in the structure of the cell wall. Analysis of other biotin-labeled cell wall proteins released by laminarinase showed that the lack of Ccw12p, as well as the lack of the other two proteins studied in this work, did not influence the presence and amount of any other protein in the cell wall.
Besides the described mating defect, the ccw12 mutant showed a significantly weakened cell wall, as indicated by the increased sensitivity to calcofluor white and Congo red. A similar effect has been reported previously (26) and is confirmed here for the Ccw14p (also called Icw1p) knockout.
The mutation of the third cell wall protein identified here, Ccw13p, did not yield a phenotype for the properties tested. It has to be mentioned that the gene coding for this protein, called DAN1, is repressed under aerobic growth conditions (35), a property reported also for another cell wall protein, Tir1p (40). Therefore, it can be assumed that Ccw13p plays a particular role restricted to anaerobic growth and that only a basal, constitutive level of Ccw13p was found in the cells studied here. Although it is presently not understood which cell wall functions may be specific in anaerobically grown yeasts, one may speculate that a whole set of cell wall proteins may be specifically produced during fermentative growth.
Thus, a number of open questions remain, including the central one of this paper, about the role of protein O-linked manno-oligosaccharides for cell wall stability. Cell wall structure and biosynthesis have to be explored further, which will yield interesting and most likely even surprising results.
ACKNOWLEDGMENTS
We are especially grateful to R. Deutzmann for protein sequencing. Thanks are also due to J. C. Kapteyn for analyzing some of our gels with a β-1,6-glucan-specific antibody.
This work has been supported by the Deutsche Forschungsgemeinschaft (SFB 521) and by Fonds der Chemischen Industrie.
REFERENCES
- 1.Ballou L, Hernandez L M, Alvarado E, Ballou C E. Revision of the oligosaccharide structure of yeast carboxypeptidase Y. Proc Natl Acad Sci USA. 1990;87:3368–3372. doi: 10.1073/pnas.87.9.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bretthauer R K, Kozak L P, Irwin W E. Phosphate and mannose transfer from guanosine diphosphate mannose to yeast mannan acceptors. Biochem Biophys Res Commun. 1969;37:820–827. doi: 10.1016/0006-291x(69)90965-6. [DOI] [PubMed] [Google Scholar]
- 3.Cappellaro C, Hauser K, Mrsa V, Watzele M, Watzele G, Gruber C, Tanner W. Saccharomyces cerevisiae a- and alpha-agglutinin: characterization of their molecular interaction. EMBO J. 1991;10:4081–4088. doi: 10.1002/j.1460-2075.1991.tb04984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cappellaro C, Mrsa V, Tanner W. New potential cell wall glucanases of Saccharomyces cerevisiae and their involvement in mating. J Bacteriol. 1998;180:5030–5037. doi: 10.1128/jb.180.19.5030-5037.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubois M, Gilles K A, Hamilton J K, Rebers P A, Smith F. Colorimetric method for the determination of sugars and related substances. Anal Chem. 1956;28:350–356. doi: 10.1038/168167a0. [DOI] [PubMed] [Google Scholar]
- 6.Eamus D G, Jennings D H. Water, turgor and osmotic potentials of fungi. In: Ayres P G, Body L, editors. Water, fungi and plants. Cambridge, England: Cambridge University Press; 1986. pp. 27–47. [Google Scholar]
- 7.Fleet G H. Cell walls. In: Rose A H, Harrison G H, editors. The yeasts. 2nd ed. Vol. 4. London, England: Academic Press; 1991. pp. 199–277. [Google Scholar]
- 8.Frevert J, Ballou C E. Saccharomyces cerevisiae structural cell wall mannoprotein. Biochemistry. 1985;24:753–759. doi: 10.1021/bi00324a033. [DOI] [PubMed] [Google Scholar]
- 9.Gentzsch M, Tanner W. The PMT gene family: protein O-glycosylation in Saccharomyces cerevisiae is vital. EMBO J. 1996;15:5752–5759. [PMC free article] [PubMed] [Google Scholar]
- 10.Gietz D, Jean A S, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Godl K, Hallmann A, Wenzl S, Sumper M. Differential targeting of closely related ECM glycoproteins: the pherophorin family from Volvox. EMBO J. 1997;16:25–34. doi: 10.1093/emboj/16.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hakomori S. A rapid permethylation of glycolipid and polysaccharide catalyzed by methylsulfinyl carbanion in dimethyl sulfoxide. J Biochem. 1964;55:205–207. [PubMed] [Google Scholar]
- 13.Hardwick K G, Boothroyd J C, Rudner A D, Pelham H R B. Genes that allow yeast cells to grow in the absence of the HDEL receptor. EMBO J. 1992;11:4187–4194. doi: 10.1002/j.1460-2075.1992.tb05512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartley J L, Donelson J E. Nucleotide sequence of the yeast plasmid. Nature. 1980;286:860–865. doi: 10.1038/286860a0. [DOI] [PubMed] [Google Scholar]
- 15.Hernandez L M, Ballou L, Alvarado E, Tsai P K, Ballou C E. Structure of the phosphorylated N-linked oligosaccharides from the mnn9 and mnn10 mutants of Saccharomyces cerevisiae. J Biol Chem. 1989;264:13648–13659. [PubMed] [Google Scholar]
- 16.Kapteyn J C, Montijn R C, Vink R, de la Cruz J, Llobell A, Douwes J E, Shimoi H, Lipke P N, Klis F M. Retention of Saccharomyces cerevisiae cell wall proteins through a phosphodiester-linked beta-1,3-/beta-1,6-glucan heteropolymer. Glycobiology. 1996;6:337–345. doi: 10.1093/glycob/6.3.337. [DOI] [PubMed] [Google Scholar]
- 16a.Kapteyn, J. C. Personal communication.
- 17.Klebl F, Tanner W. Molecular cloning of a cell wall exo-β-1,3-glucanase from Saccharomyces cerevisiae. J Bacteriol. 1989;171:6259–6264. doi: 10.1128/jb.171.11.6259-6264.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klis F M. Cell wall assembly in yeast. Yeast. 1994;10:851–869. doi: 10.1002/yea.320100702. [DOI] [PubMed] [Google Scholar]
- 19.Kollár R, Reinhold B B, Petrakova E, Yeh H J C, Ashwell G, Drgonova J, Kapteyn J C, Klis F M, Cabib E. Architecture of the yeast cell wall. β(1→6)Glucan interconnects mannoproteins, β(1→3)glucan, and chitin. J Biol Chem. 1997;272:17762–17775. doi: 10.1074/jbc.272.28.17762. [DOI] [PubMed] [Google Scholar]
- 20.Kollár R, Petrakova E, Ashwell G, Robbins P W, Cabib E. Architecture of the yeast cell wall: the linkage between chitin and beta(1→3)-glucan. J Biol Chem. 1995;270:1170–1178. doi: 10.1074/jbc.270.3.1170. [DOI] [PubMed] [Google Scholar]
- 21.Kuranda M J, Robbins P W. Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J Biol Chem. 1991;266:19758–19767. [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Lipke P N, Ovalle R. Cell wall architecture in yeast: new structure and new challenges. J Bacteriol. 1998;180:3735–3740. doi: 10.1128/jb.180.15.3735-3740.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lipke P N, Wojciechowicz D, Kurjan J. AGα1 is the structural gene for the Saccharomyces cerevisiae α-agglutinin, a cell surface glycoprotein involved in cell-cell interactions during mating. Mol Cell Biol. 1989;9:3155–3165. doi: 10.1128/mcb.9.8.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montijn R, Von Rinsum J, Van Schagen F A, Klis F. Glucomannoproteins in the cell walls of Saccharomyces cerevisiae contain a novel type of carbohydrate side chain. J Biol Chem. 1994;269:19338–19342. [PubMed] [Google Scholar]
- 26.Moukadiri I, Armero J, Abad A, Sentandreu R, Zueco J. Identification of a mannoprotein present in the inner layer of the cell wall of Saccharomyces cerevisiae. J Bacteriol. 1997;179:2154–2162. doi: 10.1128/jb.179.7.2154-2162.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mrsa V, Seidl T, Gentzsch M, Tanner W. Specific labeling of cell wall proteins by biotinylation. Identification of four covalently linked O-mannosylated proteins of Saccharomyces cerevisiae. Yeast. 1997;13:1145–1154. doi: 10.1002/(SICI)1097-0061(19970930)13:12<1145::AID-YEA163>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 28.Nakajima T, Ballou C E. Characterization of the carbohydrate fragments obtained from Saccharomyces cerevisiae mannan by alkaline degradation. J Biol Chem. 1974;249:7679–7684. [PubMed] [Google Scholar]
- 29.Nimtz M, Noll G, Pâques E-P, Conradt H S. Carbohydrate structures of a human tissue plasminogen activator variant expressed in recombinant Chinese hamster ovary cells. FEBS Lett. 1990;271:14–18. doi: 10.1016/0014-5793(90)80361-l. [DOI] [PubMed] [Google Scholar]
- 30.Robinson J S, Klionsky D J, Banta L M, Emr S D. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenfeld L, Ballou C E. Genetic control of yeast mannan structure. Biochemical basis for the transformation of Saccharomyces cerevisiae somatic antigen. J Biol Chem. 1974;249:2319–2321. [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.San Segundo P, Correa J, Vazquez de Aldana C R, del Rey F. SSG1, a gene encoding a sporulation-specific 1,3-β-glucanase in Saccharomyces cerevisiae. J Bacteriol. 1993;175:3823–3837. doi: 10.1128/jb.175.12.3823-3837.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seidl J, Tanner W. Characterization of two new genes down-regulated by α-factor. Yeast. 1997;13:809–817. doi: 10.1002/(SICI)1097-0061(199707)13:9<809::AID-YEA141>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 35.Sertil O, Cohen B D, Davies K J A, Lowry C V. The DNA1 gene of S. cerevisiae is regulated in parallel with the hypoxic genes, but by a different mechanism. Gene. 1997;192:199–205. doi: 10.1016/s0378-1119(97)00028-0. [DOI] [PubMed] [Google Scholar]
- 36.Shimoi H, Kitagaki H, Ohmori H, Iimura Y, Ito K. Sed1p is a major cell wall protein of Saccharomyces cerevisiae in the stationary phase and is involved in lytic enzyme resistance. J Bacteriol. 1998;180:3381–3387. doi: 10.1128/jb.180.13.3381-3387.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimoi H, Limura Y, Obata T. Molecular cloning of CWP1: a gene encoding a Saccharomyces cerevisiae cell wall protein solubilized with Rarobacter faecitabidus protease I. J Biochem. 1995;118:302–311. doi: 10.1093/oxfordjournals.jbchem.a124907. [DOI] [PubMed] [Google Scholar]
- 38.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in S. cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silve S, Volland C, Garnier C, Jund R, Chevallier M R, Haguenauer-Tsapis R. Membrane insertion of uracil permease, a polytopic yeast plasma membrane protein. Mol Cell Biol. 1991;11:1114–1124. doi: 10.1128/mcb.11.2.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toh-e A, Yasunaga S, Nirogi H, Tanaka K, Oguchi T, Matsui Y. Three yeast genes, PIR1, PIR2 and PIR3, containing internal tandem repeats, are related to each other, and PIR1 and PIR2 are required for tolerance to heat shock. Yeast. 1993;9:481–494. doi: 10.1002/yea.320090504. [DOI] [PubMed] [Google Scholar]
- 41.Tsai P-K, Frevert J, Ballou C E. Carbohydrate structure of Saccharomyces cerevisiae mnn9 mannoprotein. J Biol Chem. 1984;259:3805–3811. [PubMed] [Google Scholar]
- 42.Valentin E, Herrero W, Pastor J F I, Sentandreu R. Solubilization and analysis of mannoprotein molecules from the cell wall of S. cerevisiae. J Gen Microbiol. 1984;130:1419–1428. [Google Scholar]
- 43.Van Der Vaart J M, Caro L H P, Chapman J W, Klis F M, Verrips C T. Identification of three mannoproteins in the cell wall of Saccharomyces cerevisiae. J Bacteriol. 1995;177:3104–3110. doi: 10.1128/jb.177.11.3104-3110.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Holst O, Christoffel V, Fründ R, Moll H, Sumper M. A phosphodiester bridge between two arabinose residues as a structural element of an extracellular glycoprotein of Volvox carteri. Eur J Biochem. 1989;181:345–350. doi: 10.1111/j.1432-1033.1989.tb14730.x. [DOI] [PubMed] [Google Scholar]
- 45.Varki A, Kornfeld S. Structural studies of phosphorylated high mannose-type oligosaccharides. J Biol Chem. 1980;255:10847–10858. [PubMed] [Google Scholar]
- 46.Viswanathan M, Muthukumar G, Cong Y S, Lenard J. Seripauperins of Saccharomyces cerevisiae: a new multigene family encoding serine-poor relatives of serine-rich proteins. Gene. 1994;148:149–153. doi: 10.1016/0378-1119(94)90249-6. [DOI] [PubMed] [Google Scholar]
- 47.Watari J, Takata Y, Ogawa M, Sahara H, Koshino S, Onnela M L, Airaksinen U, Jaatinen R, Penttila M, Keranen S. Molecular cloning and analysis of the yeast flocculation gene FLO1. Yeast. 1994;10:211–225. doi: 10.1002/yea.320100208. [DOI] [PubMed] [Google Scholar]
- 48.Watzele M, Klis F, Tanner W. Purification and characterization of the inducible a agglutinin of Saccharomyces cerevisiae. EMBO J. 1988;7:1483–1488. doi: 10.1002/j.1460-2075.1988.tb02966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]