Abstract
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 2019 has led to the development of a large number of vaccines, several of which are now approved for use in humans. Understanding vaccine-elicited antibody responses against emerging SARS-CoV-2 variants of concern (VOCs) in real time is key to inform public health policies. Serum neutralizing antibody titers are the current best correlate of protection from SARS-CoV-2 challenge in non-human primates and a key metric to understand immune evasion of VOCs. We report that vaccinated BALB/c mice do not recapitulate faithfully the breadth and potency of neutralizing antibody responses elicited by various vaccine platforms against VOCs, compared with non-human primates or humans, suggesting caution should be exercised when interpreting data obtained with this animal model.
Keywords: SARS-CoV-2, COVID-19, vaccines, mouse, non-human primates, humans, antibodies
Graphical abstract
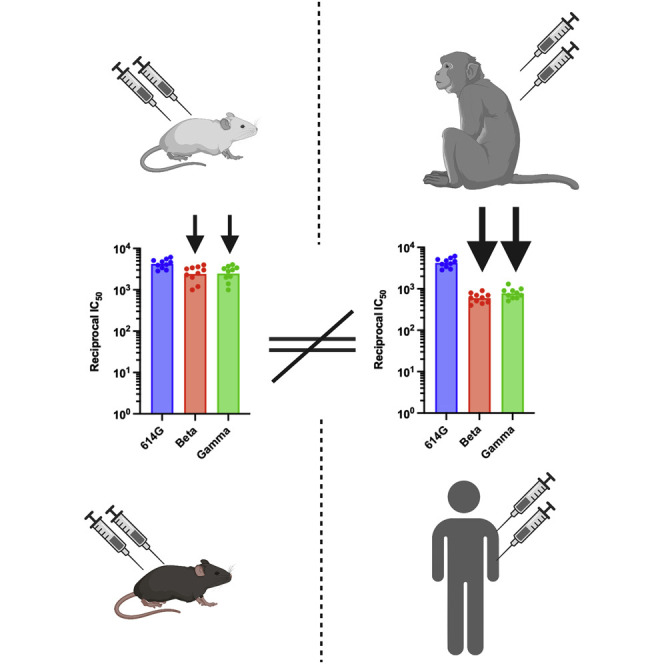
Understanding vaccine-elicited responses against SARS-CoV-2 variants of concern in real time is key to vaccine design. Walls et al. report that mice vaccination does not recapitulate the breadth and potency of neutralizing responses seen in non-human primates and humans.
Introduction
Vaccine candidates are typically evaluated in small mammals (e.g., mice) and non-human primates (NHPs) prior to cGMP manufacturing and human clinical trials. For SARS-CoV-2, serum neutralizing antibody titers represent the current best correlate of protection in NHP challenge studies (Arunachalam et al., 2021; Corbett et al., 2021) and in humans (Gilbert et al., 2021). Serum neutralizing antibody titers are also used as metrics in human clinical trials to benchmark new vaccine candidates (e.g., NCT05007951 comparing GBP510 to AZD1222) and the administration of neutralizing monoclonal antibodies has been shown to improve disease outcome for some patients with COVID-19 (Corti et al., 2021). As variants of SARS-CoV-2 emerge (Cameroni et al., 2021; Collier et al., 2021; Mlcochova et al., 2021; Tegally et al., 2020; Walls et al., 2022), it is necessary to evaluate their impact on serum neutralizing activity, as a proxy for vaccine efficacy, in order to inform public health policies worldwide and further vaccine development.
Results
We compared neutralizing antibody responses elicited following vaccination of BALB/c mice with three distinct AddaVax-adjuvanted protein subunit immunogens. Mice were immunized at weeks 0 and 3 with a clinical-stage, multivalent RBD-nanoparticle (RBD-NP) (Arunachalam et al., 2021; Walls et al., 2020a, 2021; Arunachalam et al., 2022; Song et al., 2022), an S “2P” prefusion-stabilized S (Pallesen et al., 2017; Walls et al., 2020a), or a next-generation prefusion-stabilized HexaPro S (Hsieh et al., 2020; Walls et al., 2021). Sera were collected 2, 5, and 8 weeks post-prime, and serum neutralizing activity (expressed as the dilution inhibiting 50% of entry: ID50) was evaluated using single-round vesicular stomatitis virus (VSV) pseudotyped with G614 S, Beta S (B.1.351: L18F, D80A, D215G, L242-L244 deletion, R246I, K417N, E484K, N501Y, D614G, A701V), or Gamma S (P.1: L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027, V1176F) in two distinct cell lines (VeroE6/TMPRSS2 [Lempp et al., 2021] and HEK293T/human ACE2 [Crawford et al., 2020]).
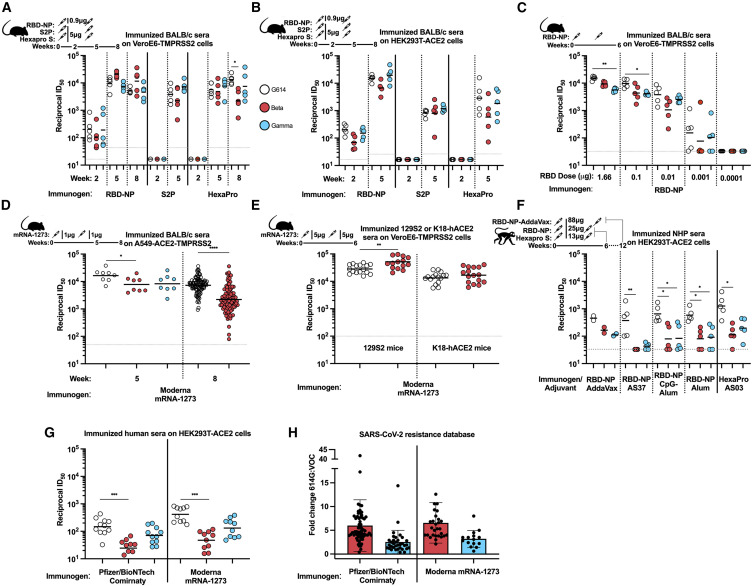
Of the three immunogens tested, only RBD-NP induced detectable serum neutralizing activity post-prime (2 weeks after immunization), in contrast to what we observed with AS03-adjuvanted HexaPro S in NHPs (Arunachalam et al., 2021; Walls et al., 2020a). With RBD-NP-elicited sera at week 2, we observed a 2.0-fold reduction and an equivalent neutralization potency against the Beta and Gamma variants of concern (VOCs) relative to G614 using VeroE6-TMPRSS2 cells, respectively (Figure 1A; Data S1). Using HEK293T-ACE2 cells, the corresponding decreases were 2.9- and 1.3-fold, respectively (Figure 1B; Data S1). All three protein immunogens induced robust neutralizing antibody responses 2 weeks after boost. Serum neutralizing activity relative to G614 at week 5 was increased 2.4-fold (Beta) and decreased 1.2-fold (Gamma) in mice immunized with RBD-NP, reduced 1.5-fold (Beta) and increased 2.0-fold (Gamma) for S2P, and reduced 1.2-fold (Beta) and increased 1.4-fold (Gamma) with HexaPro S-elicited sera using VeroE6-TMPRSS2 cells (Figure 1A; Data S1). These findings were overall consistent with the outcomes of the assays using HEK293T-ACE2 cells (Figure 1B; Data S1), with the exception of neutralizing antibody responses elicited by HexaPro S, which were reduced 4.9-fold against Beta and 2.8-fold against Gamma relative to G614, indicating that these are not antigen- or cell-type-specific observations. These results are consistent with a recent study showing a smaller than expected dampening of neutralizing activity against VOCs using protein subunit immunogens (Amanat et al., 2021).
Figure 1.
Vaccine-elicited serum neutralizing activity against VOCs in mice does not recapitulate observations in NHPs and humans
Results from mice in (A)–(E), NHPs in (F), and humans in (G)–(H).
(A) Neutralizing activity of sera from BALB/c mice immunized with RBD-NP, S2P, or HexaPro 2 weeks post-prime, 2 weeks after boost (5 weeks post-prime), or 5 weeks after boost (8 weeks post-prime with RBD-NP and Hexapro only) with VSV pseudotyped with SARS-CoV-2 G614 S, Beta S, or Gamma S on VeroE6-TMPRSS2 cells. Data from one out of at least two representative experiments shown; vaccination experiment performed once; n = 5 mice. Schematic above the figure shows animal model, antigen, dosing, and timeline of bleeds.
(B) Neutralizing activity of sera from BALB/c mice immunized with RBD-NP, S2P, or HexaPro 2 weeks post-prime or 2 weeks after boost (5 weeks post-prime) with VSV pseudotyped with SARS-CoV-2 G614 S, Beta S, or Gamma S on HEK-293T-ACE2 cells. Data from one out of at least two representative experiments shown; vaccination experiment performed once; n = 5 mice. Schematic above the figure shows organism, antigen, dosing, and timeline of bleeds.
(C) Neutralizing activity of sera from BALB/c mice immunized with different doses (ranging from 1.66–0.0001μg) RBD-NP analyzed 2 weeks after boost (6 weeks after prime) with VSV pseudotyped with SARS-CoV-2 G614 S, Beta S, or Gamma S on VeroE6-TMPRSS2 cells. Data from one of three representative experiments shown; vaccination experiment performed once; n = 5 mice/group. Schematic above the figure shows organism, antigen, dosing, and timeline of bleeds.
(D) Neutralizing activity of sera from BALB/c mice immunized with mRNA-1273 2 or 5 weeks after boost with VSV pseudotyped with SARS-CoV-2 G614 S, Beta S, or Gamma S on A549-ACE2-TMPRSS2 cells. Data from one out of at least two representative experiments shown; n = 8 or 104 mice.
(E) Neutralization of sera from 129S2 or K18-hACE2 mice immunized with mRNA-1273 3 weeks after boost with authentic SARS-CoV-2 G614 or Beta on VeroE6-TMPRSS2 cells. Data shown are from two independent experiments; vaccination experiment performed once; n = 16 mice.
(F) Neutralizing activity of sera from NHPs immunized with RBD-NP formulated in various adjuvants or HexaPro at peak titer (day 42 post-prime) with VSV pseudotyped with SARS-CoV-2 G614 S, Beta S, or Gamma S on HEK-293T-ACE2 cells. Data from one out of at least two representative experiments shown; vaccination experiment performed once; n = 2 pigtail macaques, n = 5 rhesus macaques.
(G) Neutralizing activity of sera from humans vaccinated either with Pfizer/BioNTech Comirnaty or Moderna mRNA-1273 on HEK-293T-ACE2 cells (Table S1). Data from one out of at least two representative experiments shown; vaccination experiment performed once; n = 10 Moderna mRNA-1273 vaccinees, n = 11 Pfizer/BioNTech Comirnaty vaccinees. Normalized curves and data fits for (A)–(G) are shown in Data S1.
(H) Fold change data from the SARS-CoV-2 resistance database (https://covdb.stanford.edu/page/susceptibility-data/) for sera obtained from humans vaccinated with either Pfizer/BioNTech Comirnaty or Moderna mRNA-1273 and assayed with G614 compared with either Beta (n = 62 Pfizer/BioNTech Comirnaty, n = 29 Moderna mRNA1273) or Gamma (n = 34 Pfizer/BioNTech Comirnaty, n = 15 Moderna mRNA-1273). Box plot shows mean with the standard deviation described by bars. Kruskal-Wallis with Dunn’s multiple comparisons or Mann Whitney statistical tests were used within a comparison group (i.e., comparing RBD-NP bleed 1 G614-Beta and G614-Gamma) and shown when significant (∗p = 0.033; ∗∗p = 0.002; ∗∗∗p < 0.0002).
Since immunogen dose has an effect on total serum neutralizing responses, we immunized BALB/c mice twice (at weeks 0 and 4) with RBD-NP at five different doses ranging from 1.66 to 0.0001 μg. Serum neutralizing activity was measured with G614, Beta, and Gamma VSV pseudotypes. Similar to above, the sera of mice vaccinated twice with 1.66 μg (RBD antigen dose) had 1.6- and 2.7-fold reductions in neutralization potency against Beta and Gamma, respectively (Figure 1C; Data S1). Sera elicited by two 0.1 μg vaccine doses had a 2.2- and 2.4-fold respective potency reductions against Beta and Gamma S VSV, whereas two 0.01 μg vaccine doses led to 3.6- and 1.5-fold respective potency reductions against Beta and Gamma S VSV. Mice immunized with two 0.001 μg doses experienced 2.0- and 1.5-fold reductions of serum neutralizing activity against Beta and Gamma S VSV pseudotypes, respectively (Figure 1C; Data S1). No serum neutralizing activity was observed for the lowest (0.0001 μg) dose, preventing further investigations of potential differences at this dose (Figure 1C; Data S1). These data show no correlation between vaccine dose and the ratio of neutralizing activity between G614 and VOCs.
To explore the generalizability of these observations, we immunized BALB/c mice twice (at weeks 0 and 3) with Moderna mRNA-1273 (Wu et al., 2021) and collected sera at week 5. Serum neutralizing activity 2 weeks after boost, measured in a different laboratory, was reduced ∼2-fold against both Beta and Gamma variant pseudoviruses compared with G614 S using A549/ACE2/TMPRSS2 cells (Figure 1D; Data S1) (Wu et al., 2021). To determine whether these findings were specific to BALB/c mice, we immunized K18-hACE2 transgenic and 129S2 mice with mRNA-1273 twice (3 weeks apart) and collected sera at week 6 (Ying et al., 2022). Serum neutralizing activity was evaluated using a focus-reduction neutralization test (FRNT) with authentic SARS-CoV-2 carrying the G614 S glycoprotein in a WA1/2020 background or a clinical isolate of the Beta VOC (Case et al., 2020a). We observed a 1.8- and 1.3-fold increased neutralization potency of the Beta variant compared with G614 for 129S2 and K18-hACE2 immunized mice, respectively (Figure 1E; Data S1). These data indicate that the unexpected apparent resilience of variants to polyclonal neutralizing antibody responses in mice appears to be independent of the mouse model, the vaccine platform, the nature of the antigen, the neutralization assay used, or the target cell type.
To understand whether these observations extend beyond mice, we analyzed sera obtained from pigtail macaques and rhesus macaques immunized with RBD-NP or HexaPro S protein subunit vaccines (Arunachalam et al., 2021; Walls et al., 2020a, 2021). Pigtail macaques (n = 2) were immunized with AddaVax-adjuvanted RBD-NP at weeks 0 and 4, and sera from week 12 were analyzed. Rhesus macaques were immunized at weeks 0 and 3 with RBD-NP adjuvanted with AS37 (n = 5), CpG-alum (n = 5), or alum (n = 5). Rhesus macaques (n = 5) immunized with HexaPro S adjuvanted with AS03 were used to compare RBD- and S-based vaccines. Rhesus macaque sera from week 6 were used for our analysis. All NHP serum neutralizing titers were lower than immunogen-matched mice titers. Serum neutralizing activity from pigtail macaques immunized with AddaVax-adjuvanted RBD-NP showed a 2.7-fold potency dampening against Beta and 4-fold against Gamma VOC compared with G614 S pseudovirus using HEK293T-ACE2 cells (Figure 1F; Data S1). Rhesus macaques immunized with AS37-adjuvanted RBD-NP had a reduction in neutralization potency of ≥11-fold (Beta) and 8.6-fold (Gamma), whereas a similar ∼6- to 8-fold decrease was detected with CpG-alum and alum for both Beta (8.2 for CpG-alum and 7.2 for alum) and Gamma (7.8 for CpG-alum and 6.4 for alum) using HEK293T-ACE2 cells (Figure 1F; Data S1). AS03-adjuvanted HexaPro S immunization led to 11.2- and 6.4-fold reductions in serum neutralizing activity against Beta and Gamma S VSV, respectively (Figure 1F; Data S1). These data recapitulate previous findings made with neutralization of authentic viruses (Arunachalam et al., 2021), suggesting that these outcomes are not specific to pseudovirus assays. Collectively, these data obtained with different clinical adjuvants indicate that vaccine-elicited neutralizing antibody responses in NHPs are significantly more affected by VOCs than those in BALB/c mice.
Analysis of serum neutralizing antibody titers in individuals vaccinated with either Pfizer/BioNTech Comirnaty or Moderna mRNA-1273 confirmed that the magnitudes of potency reduction against the Beta and Gamma variants relative to G614 are on par with our NHP data and that both are much greater than those observed in BALB/c mice (Figure 1G; Data S1; Table S1). Compared with G614 S, we observed 6.1-fold (Beta) and 2.1-fold (Gamma) reductions in neutralization potency with a cohort of humans immunized with Pfizer/BioNTech Comirnaty and 8.7-fold (Beta) and 3.2-fold (Gamma) reductions with a cohort of humans immunized with Moderna mRNA-1273 using a VSV pseudovirus assay with HEK293T/ACE2 target cells (Figure 1G; Data S1). These results are in line with the average attenuations of neutralizing antibody responses retrieved from the coronavirus antiviral and resistance database (https://covdb.stanford.edu/), which are 7.3 (Beta) and 4.1 (Gamma) for Pfizer/BioNTech Comirnaty and 7.8 (Beta) and 4.5 (Gamma) for Moderna mRNA-1273 (Figure 1H).
We next tested whether using mouse sera obtained 8 weeks post-prime would reveal larger differences between G614 and VOCs. At 8 weeks after RBD-NP prime (5 weeks after boost), mouse serum neutralizing activity remained mildly affected when comparing G614 to either Beta or Gamma S VSV pseudotyped viruses in VeroE6/TMPRSS2 cells (Figure 1A; Data S1). A mild attenuation (3.3-fold) of serum neutralizing activity was also observed 8 weeks post-prime against the Beta variant with a cohort of 104 BALB/c mice immunized with mRNA-1273 (Figure 1D; Data S1). In contrast, HexaPro S-immunized mice exhibited a 5.9-fold drop of serum neutralizing activity against Beta and 1.3-fold against Gamma S VSV pseudotyped viruses (Figure 1A; Data S1), more closely resembling the expected trends for NHP and human data (Figures 1F and 1G; Data S1). Vaccine-elicited BALB/c mice sera post-prime and after boost did not recapitulate observations made with NHPs and humans regarding the impact of SARS-CoV-2 variants on neutralizing activity regardless of overall titer, dose, time following vaccination, vaccine modality, target cell type, or antigen.
Discussion
Although BALB/c and other mice are widely used for evaluating vaccine candidates, our data suggest that they are not always an adequate animal model to evaluate the breadth and potency of neutralizing antibody responses against emerging SARS-CoV-2 VOCs, potentially due to their different immune repertoires compared with primates. For example, polyclonal neutralizing antibody responses resulting from infection or vaccination with ancestral SARS-CoV-2 isolates in humans and NHPs can be hyperfocused on position 484 in the RBD, which does not appear to be recapitulated in mice (Greaney et al., 2021; Walls et al., 2021). However, intranasal delivery of an S2P-based chimpanzee adenovirus-vectored vaccine (ChAd-SARS-CoV-2-S) in K18-hACE2 transgenic mice (Chen et al., 2021; Hassan et al., 2021) and intramuscular delivery of SARS-CoV-2 S mRNA-LNP to BALB/c mice (Martinez et al., 2021) show fold changes in line with the expected dampened human responses for Beta compared with G614, suggesting there could be exceptions to the generalizability of these findings. Further experiments will be necessary to understand the immunological basis of these findings, such as comparing vaccine-elicited responses in animals of both sexes, inbred versus outbred mouse strains (Gralinski et al., 2015), or exploring the microbial environment of the mice (Fiege et al., 2021). Another alternative may be to utilize humanized mice (Lee et al., 2014; Murphy et al., 2014; Nie et al., 2021; Walls et al., 2020a; Wang et al., 2020) to better match the antibody repertoire of primates as our data suggest that other animal models may be better suited to evaluate serum immune evasion of new variants as they emerge, even though mice are a cost- and time-effective option for evaluating overall immunogenicity.
Limitations of the study
This study is an observation of the dampening of neutralizing activity toward SARS-CoV-2 variants in mice compared with NHPs or humans. This study focused on characterizing the breadth and robust nature of these observations and not on the mechanisms behind these distinct differences in mice such as antibody repertoire, animal sex, inbred versus outbred mice, the microbial environment, etc., which will be important questions for future studies. Another limitation of this study is the low animal subject number, in some cases having an n of 2, and the study could be enhanced by larger numbers of animals within each vaccine type.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| SARS-CoV-2 G614 | Chen et al., 2021 | N/A |
| SARS-CoV-2 B.1.351 | Infected individual | N/A |
| Biological samples | ||
| VSV (G∗ΔG-luciferase) | Kaname et al., 2010 | N/A |
| SARS-CoV-2 vaccinated human sera | UWARN: COVID-19 (Table S1) | N/A |
| VSVΔG-firefly-luciferase | Whitt, 2010 | N/A |
| Experimental models: Cell lines | ||
| A549-hACE2-TMPRSS2 | N/A | N/A |
| Expi293F | Thermo Fisher | Cat #A14527 |
| HEK-ACE2 | BEI, Crawford et al., 2020 | NR-52511 |
| VeroE6-TMPRSS2 | Lempp et al., 2021 | N/A |
| BHK-21/WI-2 | Whitt, 2010 | N/A |
| HEK293T/17 Adherent | ATCC | Cat# CRL-11268 |
| Experimental models: Organisms/strains | ||
| Female BALB/c cByJ | Jackson Labs | Stock # 000,651 |
| Female BALB/c | Charles River | N/A |
| K18-hACE2 transgenic mice | Jackson Labs | #034860 |
| 129S2/SvPasCrl | Charles River | Cat # 287 |
| Male Pigtail macaques (Macaca nemestrina) | N/A | N/A |
| Male Rhesus macaques (Macaca mulatta) | N/A | N/A |
| Recombinant DNA | ||
| Hexapro S | (Hsieh et al., 2020) | N/A |
| SARS-CoV S-2P trimer | GeneArt (Walls et al., 2020b) | N/A |
| SARS-CoV-2 S full-length D614G (YP 009724390.1) del21 | Crawford et al., 2020 | Vector# BEI NR-52514 |
| SARS-CoV-2 full length S del21 Beta | GenScript (Walls et al., 2021) | N/A |
| SARS-CoV-2 full length S del21 Gamma | GenScript (Walls et al., 2021) | N/A |
| RBD-16GS-I53-50A | GenScript + subcloning (Walls et al., 2020a) | N/A |
| I53-50B.4.PT1 | Bale et al., 2016 | N/A |
| Software and algorithms | ||
| CoV Database | https://covdb.stanford.edu/page/susceptibility-data/ | N/A |
| Prism | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, David Veesler (dveesler@uw.edu).
Materials availability
Materials generated in this study will be made available on request after signing a materials transfer agreement with the University of Washington.
Experimental model and subject details
Cell lines
Expi293F (derived from 293 cells which are female) cells are derived from the HEK293F cell line (Life Technologies). Expi293F cells were grown in Expi293 Expression Medium (Life Technologies), cultured at 37°C with 8% CO2 and shaking at 130 rpm. HEK293T/17 is a female human embryonic kidney cell line (ATCC). The HEK-ACE2 (derived from HEK293T cells which are female) adherent cell line was obtained through BEI Resources, NIAID, NIH: Human Embryonic Kidney Cells (HEK293T) Expressing Human Angiotensin-Converting Enzyme 2, HEK293T-hACE2 Cell Line, NR-52511. The VeroE6-TMPRSS2 cell line is an African Green monkey Kidney cell line expressing TMPRSS2 (Lempp et al., 2021). The A549-hACE2-TMPRSS2 was produced from adenocarcinomic human alveolar basal epithelial cells.All adherent cells were cultured at 37°C with 5% CO2 in flasks with DMEM +10% FBS (Hyclone) + 1% penicillin-streptomycin. Cell lines were not tested for mycoplasma contamination nor authenticated.
Method details
Plasmid construction
The SARS-CoV-2 S ‘2P’ ectodomain trimer (BEI NR-52420) was synthesized by GenScript into pCMV with an N-terminal mu-phosphatase signal peptide and a C-terminal TEV cleavage site (GSGRENLYPQG), T4 fibritin foldon (GGGSGYIPEAPRDGQAYVRKDGEWVLLSTPL), and octa-histidine tag (GHHHHHHHH) (Walls et al., 2020b). The construct contains the 2P mutations (proline substitutions at residues 986 and 987; and an 682SGAG685 substitution at the furin cleavage site. The SARS-CoV-2 RBD (328RFPN331 and 528KKST531) was genetically fused to the N terminus of the trimeric I53-50A nanoparticle component using 16 glycine and serine residues (Walls et al., 2020a). The RBD-16GS-I53-50A fusion was cloned into pCMV/R using the Xba1 and AvrII restriction sites and Gibson assembly (Gibson et al., 2009). All RBD-bearing components contained an N-terminal mu-phosphatase signal peptide and a C-terminal octa-histidine tag. SARS-CoV-2 HexaPro construct is as previously described (Hsieh et al.) and placed into CMVR with an octa-his tag.
Transient transfection
Proteins were produced using endotoxin free DNA in Expi293F cells grown in suspension using Expi293F expression medium (Life Technologies) at 33°C, 70% humidity, 8% CO2 rotating at 150 rpm. The cultures were transfected using PEI-MAX (Polyscience) with cells grown to a density of 3.0 million cells per mL and cultivated for 3 days. Supernatants were clarified by centrifugation (5 min at 4000 rcf), addition of PDADMAC solution to a final concentration of 0.0375% (Sigma Aldrich, #409014), and a second spin (5 min at 4000 rcf).
Microbial protein expression and purification
The I53-50B.4.PT1 proteins (Bale et al., 2016) were expressed in Lemo21(DE3) (NEB) in LB (10 g Tryptone, 5 g Yeast Extract, 10 g NaCl) grown in 2 L baffled shake flasks or a 10 L BioFlo 320 Fermenter (Eppendorf). Cells were grown at 37°C to an OD600 ∼0.8, and then induced with 1 mM IPTG. Expression temperature was reduced to 18°C and the cells shaken for ∼16 h. The cells were harvested and lysed by microfluidization using a Microfluidics M110P at 18,000 psi in 50 mM Tris, 500 mM NaCl, 30 mM imidazole, 1 mM PMSF, 0.75% CHAPS. Lysates were clarified by centrifugation at 24,000 g for 30 min and applied to a 2.6 × 10 cm Ni Sepharose 6 FF column (Cytiva) for purification by IMAC on an AKTA Avant150 FPLC system (Cytiva). Protein of interest was eluted over a linear gradient of 30 mM to 500 mM imidazole in a background of 50 mM Tris pH 8, 500 mM NaCl, 0.75% CHAPS buffer. Peak fractions were pooled, concentrated in 10K MWCO centrifugal filters (Millipore), sterile filtered (0.22 μm) and applied to either a Superdex 200 Increase 10/300, or HiLoad S200 pg GL SEC column (Cytiva) using 50 mM Tris pH 8, 500 mM NaCl, 0.75% CHAPS buffer. I53-50A elutes at ∼0.6 column volume (CV). I53-50B.4PT1 elutes at ∼0.45 CV. After sizing, bacterial-derived components were tested to confirm low levels of endotoxin before using for nanoparticle assembly.
Protein purification
Proteins containing His tags were purified from clarified supernatants via a batch bind method where each clarified supernatant was supplemented with 1 M Tris-HCl pH 8.0 to a final concentration of 45 mM and 5 M NaCl to a final concentration of ∼310 mM. Talon cobalt affinity resin (Takara) was added to the treated supernatants and allowed to incubate for 15 min with gentle shaking. Resin was collected using vacuum filtration with a 0.2 μm filter and transferred to a gravity column. The resin was washed with 20 mM Tris pH 8.0, 300 mM NaCl, and the protein was eluted with 3 column volumes of 20 mM Tris pH 8.0, 300 mM NaCl, 300 mM imidazole. The batch bind process was then repeated and the first and second elutions combined. SDS-PAGE was used to assess purity. RBD-I53-50A fusion protein IMAC elutions were concentrated to >1 mg/mL and subjected to three rounds of dialysis into 50 mM Tris pH 7.4, 185 mM NaCl, 100 mM Arginine, 4.5% glycerol, and 0.75% w/v 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) in a hydrated 10K molecular weight cutoff dialysis cassette (Thermo Scientific). S ‘2P’ and HexaPro IMAC elution fractions were concentrated to ∼1 mg/mL and dialyzed three times into 50 mM Tris pH 8, 150 mM NaCl, 0.25% L-Histidine in a hydrated 10K molecular weight cutoff dialysis cassette (Thermo Scientific).
In vitro nanoparticle assembly and purification
Total protein concentration of purified individual nanoparticle components was determined by measuring absorbance at 280 nm using a UV/vis spectrophotometer (Agilent Cary 8454) and calculated extinction coefficients. The assembly steps were performed at room temperature with addition in the following order: RBD-I53-50A trimeric fusion protein, followed by additional buffer (50 mM Tris pH 7.4, 185 mM NaCl, 100 mM Arginine, 4.5% glycerol, and 0.75% w/v CHAPS) as needed to achieve desired final concentration, and finally I53-50B.4PT1 pentameric component (in 50 mM Tris pH 8, 500 mM NaCl, 0.75% w/v CHAPS), with a molar ratio of RBD-I53-50A:I53-50B.4PT1 of 1.1:1. All RBD-I53-50 in vitro assemblies were incubated at 2–8°C with gentle rocking for at least 30 min before subsequent purification by SEC in order to remove residual unassembled component. Different columns were utilized depending on purpose: Superose 6 Increase 10/300 GL column was used analytically for nanoparticle size estimation, a Superdex 200 Increase 10/300 GL column used for small-scale pilot assemblies, and a HiLoad 26/600 Superdex 200 pg column used for nanoparticle production. Assembled particles were purified in 50 mM Tris pH 7.4, 185 mM NaCl, 100 mM Arginine, 4.5% glycerol, and 0.75% w/v CHAPS, and elute at ∼11 mL on the Superose 6 column and in the void volume of Superdex 200 columns. Assembled nanoparticles were sterile filtered (0.22 μm) immediately prior to column application and following pooling of fractions.
Endotoxin measurements
Endotoxin levels in protein samples were measured using the EndoSafe Nexgen-MCS System (Charles River). Samples were diluted 1:50 or 1:100 in Endotoxin-free LAL reagent water, and applied into wells of an EndoSafe LAL reagent cartridge. Charles River EndoScan-V software was used to analyze endotoxin content, automatically back-calculating for the dilution factor. Endotoxin values were reported as EU/mL which were then converted to EU/mg based on UV/vis measurements. Our threshold for samples suitable for immunization was <50 EU/mg.
BALB/c mice for RBD-NP, S ‘2P’, and HexaPro protein immunizations
Female BALB/c mice (Stock # 000651, BALB/c cByJ mice) four weeks old were obtained from Jackson Laboratory, Bar Harbor, Maine, and maintained at the Comparative Medicine Facility at the University of Washington, Seattle, WA, accredited by the American Association for the Accreditation of Laboratory Animal Care International (AAALAC). Animal procedures were performed under the approvals of the Institutional Animal Care and Use Committee (IACUC) of University of Washington, Seattle, WA # 4470-01.
At six weeks of age, 8 female BALB/c mice per dosing group were vaccinated with a prime immunization, and three weeks later mice were boosted with a second vaccination (IACUC protocol 4470.01). Prior to inoculation, immunogen suspensions were gently mixed 1:1 vol/vol with AddaVax adjuvant (Invivogen, San Diego, CA) to reach a final concentration of 0.01 mg/mL antigen. Mice were injected intramuscularly into the quadriceps muscle of each hind leg using a 27-gauge needle (BD, San Diego, CA) with 50 μL per injection site (100 μL total) of immunogen under isoflurane anesthesia. To obtain sera all mice were bled two weeks after prime and boost immunizations. Blood was collected via submental venous puncture and rested in 1.5 mL plastic Eppendorf tubes at room temperature for 30 min to allow for coagulation. Serum was separated from red blood cells via centrifugation at 2,000 g for 10 min. Complement factors and pathogens in isolated serum were heat-inactivated via incubation at 56°C for 60 min. Serum was stored at 4°C or −80°C until use. The study was repeated twice.
For the dose dependency study, 5 6-week-old female mice were immunized with 1.66μg, 0.1μg, 0.01μg, 0.001μg or 0.0001μg RBD-NP. Prior to immunization immunogens at the indicated doses were gently mixed 1:1 (vol:vol) with AddaVax adjuvant (Invivogen, San Diego, CA). Mice were injected intramuscularly into the gastrocnemius muscle of each hind leg using a 27-gauge needle with 50 μL per injection site (100 μL total) of immunogen under isoflurane anesthesia. For sera collection, mice were bled via submental venous puncture 2 weeks following each inoculation. Serum was isolated from hematocrit via centrifugation at 2,000 g for 10 min, and stored at −80°C until use.
BALB/c mice for mRNA-1273 immunizations
Female BALB/c mice (6 to 8 weeks old) were obtained from Charles River Laboratories. Animal experiments were carried out in compliance with approval from the Animal Care and Use Committee of Moderna Inc. Mice were immunized with 1 μg of mRNA-1273 diluted in 50 μL of 1X phosphate-buffered saline (PBS), via intramuscular injection into the same hind leg for both prime and boost. Sera were obtained at 2 (week 5) and 5 (week 8) weeks post-boost for immune analysis. Data are from two independent experiments.All animal procedures done according to the approved SOPs.
K18-hACE2 mice for mRNA-1273 immunizations
7–9 week old female K18-hACE2 transgenic mice were purchased from Jackson Laboratories (#034860) and housed in a pathogen-free animal facility at Washington University in St. Louis. Animal studies were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Institutional Animal Care and Use Committee at the Washington University School of Medicine (Assurance number A3381-01). mRNA-1273 was produced as described previously (Ying et al., 2022). Mice were immunized with 5 μg of mRNA-1273 vaccine and boosted with the same dose three weeks later. Serum was obtained three weeks post-boost for immune analysis. Data are from two independent experiments.
1292S mice for mRNA-1273 immunizations
7–9 week old female 129S2 mice (strain: 129S2/SvPasCrl, Cat # 287) were obtained from Charles River Laboratories and housed in a pathogen-free animal facility at Washington University in St. Louis. Animal studies were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Institutional Animal Care and Use Committee at the Washington University School of Medicine (Assurance number A3381-01). Mice were immunized with 5 μg of mRNA-1273 vaccine and boosted with the same dose three weeks later. Serum was obtained three weeks post-boost for immune analysis. Data are from two independent experiments.
Pigtail macaques
Two adult 4.3–5.5 year old male Pigtail macaques (Macaca nemestrina) were immunized in this study. All animals were housed at the Washington National Primate Research Center (WaNPRC), an AAALAC International accredited institution. All experiments were approved by The University of Washington’s IACUC. Animals were singly housed in comfortable, clean, adequately-sized cages with ambient temperatures between 72–82°F. Animals received environmental enrichment for the duration of the study including grooming contact, perches, toys, foraging experiences and access to additional environment enrichment devices. Water was available through automatic watering devices and animals were fed a commercial monkey chow, supplemented daily with fruits and vegetables. Throughout the study, animals were checked twice daily by husbandry staff.
Two adult male Pigtail macaques were immunized with 250 μg of RBD-12GS-I53-50 nanoparticle (88 μg RBD antigen) with AddaVax at day 0 and day 28. Blood was collected every 14 days post-prime. Blood was collected in serum collection tubes and allowed to clot at room temperature. Serum was isolated after a 15 min spin at 1455 × g for 15 min and stored at −80°C until use. Prior to vaccination or blood collection, animals were sedated with an intramuscular injection (10 mg/kg) of ketamine (Ketaset®; Henry Schein). Prior to inoculation, immunogen suspensions were gently mixed 1:1 vol/vol with AddaVax adjuvant to reach a final concentration of 0.250 mg/mL antigen. The vaccine was delivered intramuscularly into both quadriceps muscles with 1 mL per injection site on days 0 and 28. All injection sites were shaved prior to injection. Animals were observed daily for general health (activity and appetite, urine/feces output) and for evidence of reactogenicity at the vaccine inoculation site (swelling, erythema, and pruritus) for up to 1 week following vaccination. They also received physical exams including temperature and weight measurements at each study time point. None of the animals became severely ill during the course of the study nor required euthanasia.
Rhesus macaques
Male rhesus macaques (Macaca mulatta) of Indian origin, aged 3–7 years were assigned to the study (Arunachalam et al., 2021). Animals were distributed between the groups such that the age and weight distribution were comparable across the groups. Animals were housed and maintained at the New Iberia Research Center (NIRC) of the University of Louisiana at Lafayette, an AAALAC International accredited institution, in accordance with the rules and regulations of the Guide for the Care and Use of Laboratory Animal Resources. The entire study (protocol 2020-8808-15) was reviewed and approved by the University of Louisiana at Lafayette IACUC. All animals were negative for SIV, simian T cell leukemia virus, and simian retrovirus.
Adapted from ref. (Arunachalam et al., 2021): All doses for RBD-NP were 25μg of RBD component. AS03 was kindly provided by GSK Vaccines. AS03 is an oil-in-water emulsion that contains 11.86 mg a-tocopherol, 10.69 mg squalene, and 4.86 mg polysorbate 80 (Tween-80) in PBS. AS37 is a TLR-7 agonist (200 μg mL−1) adsorbed to aluminium hydroxide (2 mg mL−1). For each dose, RBD–NP was diluted to 50 μg mL−1 (RBD component) in 250 μL of Tris-buffered saline (TBS) and mixed with an equal volume of AS03 or AS37. The dose of AS03 was 50% (v/v) (equivalent to one human dose), AS37 included 50 μg TLR-7 agonist and 0.5 mg aluminium hydroxide. CpG 1018 was kindly provided by Dynavax Technologies at a concentration of 12 mg mL−1. Alum (Alhydrogel 2%) was purchased from Croda Healthcare (batch 0001610348). For each dose of CpG-alum, 25 μg antigen (RBD component) in TBS was mixed with 0.75 mg alum and incubated on ice for 30 min. After 30 min of incubation, 1.5 mg of CpG 1018 was added and mixed rapidly. Each dose contained 1.5 mg CpG 1018 and 0.75 mg alum. For each dose of alum, 25 μg (RBD component) in TBS was mixed with 0.75 mg alum, matching the concentration of alum in the CpG-alum formulation, and incubated on ice for 30 min. Soluble HexaPro was diluted to 50 μg mL−1 in 250 μL TBS and mixed with an equal volume of AS03. All immunizations were administered via the intramuscular route in right forelimbs. The volume of each dose was 0.5 mL.
Pfizer and Moderna vaccinated human sera
Sera samples were collected from participants who had received both doses of the Pfizer or Moderna mRNA vaccine and were 7–30 days post second vaccine dose. Clinical data for these individuals are summarized in Table S1. Individuals were enrolled in the UWARN: COVID-19 in WA study at the University of Washington in Seattle, WA. This study was approved by the University of Washington Human Subjects Division Institutional Review Board (STUDY00010350).
Pseudovirus production
G614 SARS-CoV-2 S, B.1.351 S, and P.1 S pseudotyped VSV viruses were prepared as described previously (McCallum et al., 2021; Sauer et al., 2021; Walls et al., 2021). Briefly, HEK293T cells in DMEM supplemented with 10% FBS, 1% PenStrep seeded in 10-cm dishes were transfected with the plasmid encoding for the corresponding S glycoprotein using lipofectamine 2000 (Life Technologies) following manufacturer’s indications. One day post-transfection, cells were infected with VSV(G∗ΔG-luciferase) (Kaname et al., 2010) and after 2 h were washed five times with DMEM before adding medium supplemented with anti-VSV-G antibody (I1- mouse hybridoma supernatant, CRL- 2700, ATCC). Virus pseudotypes were harvested 18–24 h post-inoculation, clarified by centrifugation at 2,500 × g for 5 min, filtered through a 0.45 μm cut off membrane, concentrated 10 times with a 30 kDa cut off membrane, aliquoted and stored at −80°C.
Pseudovirus neutralization
HEK293-hACE2 cells (Crawford et al., 2020) or VeroE6-TMPRSS2 (Lempp et al., 2021) were cultured in DMEM with 10% FBS (Hyclone) and 1% PenStrep and 8ug/mL puromycin for TMPRSS2 maintenance with 5% CO2 in a 37°C incubator (ThermoFisher). One day prior to infection, 40 μL of poly-lysine (Sigma) was placed into 96-well plates and incubated with rotation for 5 min. Poly-lysine was removed, plates were dried for 5 min then washed 1 × with water prior to plating with 40,000 cells. The following day, cells were checked to be at 80% confluence. In an empty half-area 96-well plate a 1:3 serial dilution of sera was made in DMEM and diluted pseudovirus was then added to the serial dilution and incubated at room temperature for 30–60 min. After incubation, the sera-virus mixture was added to the cells at 37°C and 2 hours post-infection, 40 μL 20% FBS-2% PenStrep DMEM was added. After 17–20 hours VSV 40 μL/well of One-Glo-EX substrate (Promega) was added to the cells and incubated in the dark for 5–10 min prior reading on a BioTek plate reader. Measurements were done in at least duplicate. Non-vaccinated samples were used to detect background neutralization- there was none associated with these pseudoviruses. Relative luciferase units were plotted and normalized in Prism (GraphPad). Nonlinear regression of log(inhibitor) versus normalized response was used to determine IC50 values from curve fits. Kruskal Wallis tests were used to compare two groups to determine whether they were statistically different. Fold changes were determined by comparing individual animal IC50 and then averaging the individual fold changes for reporting.
Pseudovirus neutralization assay for BALB/c mRNA-1273 samples
Codon-optimized full-length spike genes (Wuhan-Hu-1 with G614, Beta, or Gamma) were cloned into pCAGGS vector. Spike genes of Beta and Gamma variants contain the following mutations: L18F-D80A-D215G-L242-244del-R246I-K417N-E484K-N501Y-D614G-A701V, and L18F-T20N-P26S-D138Y-R190S-K417T-E484K-N501Y-D614G-H655Y-T1027I-V1176F, respectively. To generate VSVΔG-based SARS-CoV-2 pseudovirus, BHK-21/WI-2 cells were transfected with the spike expression plasmid and infected by VSVΔG-firefly-luciferase as previously described (Whitt, 2010). A549-hACE2-TMPRSS2 cells were used as target cells for the neutralization assay. Lentivirus encoding hACE2-P2A-TMPRSS2 was made to generate A549-hACE2-TMPRSS2 cells which were maintained in DMEM supplemented with 10% fetal bovine serum and 1 μg/mL puromycin. To perform neutralization assay, mouse serum samples were heat-inactivated for 45 minutes at 56 Celsius and serial dilutions of the samples were made in DMEM supplemented with 10% fetal bovine serum. The diluted serum samples or culture medium (serving as virus only control) were mixed with VSVΔG-based SARS-CoV-2 pseudovirus and incubated at 37 Celsius for 45 minutes. The inoculum virus or virus-serum mix was subsequently used to infect A549-hACE2-TMPRSS2 cells for 18 hr at 37 Celsius. At 18 hr post infection, equal volume of One-Glo reagent (Promega; E6120) was added to culture medium for readout using BMG PHERastar-FS plate reader. The percentage of neutralization is calculated based on relative light units of the virus control, and subsequently analyzed using 4 parameter logistic curve (Prism 8).
Live virus focus-reduction neutralization test for K18-hACE2 and 1292S mRNA-1273 samples
SARS-CoV-2 G614 was produced by introducing the mutation into an infectious clone of WA1/2020 (Chen et al., 2021). B.1.351 was isolated from an infected individual. Both viruses were propagated on Vero-TMPRSS2 cells and subjected to deep sequencing to confirm the presence of expected substitutions. Focus-reduction neutralization tests (FRNTs) were performed as described (Case et al., 2020b). Briefly, serial dilutions of antibody were incubated with 102 FFU of SARS- CoV-2 for 1 h at 37°C. Immune complexes were added to VeroE6-TMPRSS2 cell monolayers and incubated for 1 h at 37°C prior to the addition of 1% (w/v) methylcellulose in MEM. Following incubation for 30 h at 37°C, cells were fixed with 4% paraformaldehyde (PFA), permeabilized and stained for infection foci with an oligoclonal mixture of ati-SARS-CoV-2 (SARS2-02, SARS2-11, SARS2-31, SARS2-38, SARS2-57, and SARS2-71 (VanBlargan et al., 2021); diluted to 1 mg/mL total mAb concentration). Antibody-dose response curves were analyzed using non-linear regression analysis (with a variable slope) (GraphPad Software). The antibody half-maximal inhibitory concentration (ID50) required to reduce infection was determined.
CoV database parameters
Data sheets were downloaded on December 1st, 2021 from https://covdb.stanford.edu/page/susceptibility-data/ selecting for either Pfizer BNT162b2 or Moderna mRNA1273 plasma Abs and either B.1.351 or P.1 variant and using the median fold change reported metric. Data sets where individuals were previously infected or had only 1 shot were excluded. Data sets where the fold-change comparator was not a Wuhan-Hu-1 derivative (WA1, G614) or datasets where the number of samples measured was below 3 were excluded.
Quantification and statistical analysis
2–104 samples were used per group and detailed in the Figure 1A figure legend. Experiments were all done in at least biologic and technical duplicates. Kruskal-Wallis with Dunn’s multiple comparisons or Mann Whitney statistical tests were used within a comparison group (i.e. comparing RBD-NP bleed 1 G614-Beta and G614-Gamma) and shown when significant (∗p = 0.033; ∗∗p = 0.002; ∗∗∗p < 0.0002).
Acknowledgments
We thank Hideki Tani (University of Toyama) for providing the reagents necessary for preparing VSV pseudotyped viruses and Wesley Van Voorhis for coordination of human sera samples. The authors thank Kathryn Guerriero, Brieann Brown, Solomon Wangari, Joel Ahrens, Naoto Iwayama, and William Garrison for their technical assistance with the pigtail macaque study This study was supported by the National Institute of Allergy and Infectious Diseases (DP1AI158186 and HHSN272201700059C to D.V., R01 AI157155 to M.S.D., and Influenza Research and Response [CEIRR] contract 75N93021C00014 to M.S.D.), National Institute of Health Office of Research Infrastructure Programs (U42OD011123 and P51-OD010425, Washington National Primate Research Center), a Pew Biomedical Scholars Award (D.V.), an Investigators in the Pathogenesis of Infectious Disease Awards from the Burroughs Wellcome Fund (D.V.), Fast Grants (D.V.), and the Bill & Melinda Gates Foundation (OPP1156262 to N.P.K. and D.V.). D.V. is an Investigator of the Howard Hughes Medical Institute.
Author contributions
A.C.W. and D.V. conceived the project and designed experiments. A.C.W., L.A.V., K.W., A.C., M.J.N., D.L., L.A., D.M.B., S.E., and K.S. performed experiments. M.N.P., J.C.K., M.C.M., E.K., M.J., A.B., B.F., M.A.O., N.B., P.S.A., L.S., K.R., L.C., D.H.F., F.V., B.P., and N.P.K. prepared immunogens and coordinated immunizations. A.C.W., D.H.F., M.S.D., D.K.E., N.P.K., and D.V. supervised the project and obtained funding. A.C.W. and D.V. analyzed the data and wrote the manuscript with input from all authors.
Declaration of interests
A.C.W., N.P.K., and D.V. are named as inventors on patent applications filed by the University of Washington based on the RBD-NP presented in this paper. N.P.K. is a co-founder, shareholder, paid consultant, and chair of the scientific advisory board of Icosavax, Inc., and the King lab has received an unrelated sponsored research agreement from Pfizer. M.S.D. is a consultant for Inbios, Vir Biotechnology, and Carnival Corporation, and on the Scientific Advisory Boards of Moderna and Immunome. The Diamond laboratory has received unrelated funding support in sponsored research agreements from Vir Biotechnology, Kaleido, and Emergent BioSolutions and past support from Moderna not related to these studies. K.W., A.C., and D.K.E are employees of Moderna and hold stock/stock options in the company. D.H.F. has equity interest in HDT Bio.
Published: August 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111299.
Supplemental information
Data and code availability
Data generated in this study are presented in Data S1 and data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Amanat F., Strohmeier S., Meade P.S., Dambrauskas N., Mühlemann B., Smith D.J., Vigdorovich V., Sather D.N., Coughlan L., Krammer F. Vaccination with SARS-CoV-2 variants of concern protects mice from challenge with wild-type virus. PLoS Biol. 2021;19:e3001384. doi: 10.1371/journal.pbio.3001384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunachalam P.S., Feng Y., Ashraf U., Hu M., Walls A.C., Edara V.V., Zarnitsyna V.I., Aye P.P., Golden N., Miranda M.C., et al. Durable protection against the SARS-CoV-2 Omicron variant is induced by an adjuvanted subunit vaccine. Sci Transl Med. 2022;14:eabq4130. doi: 10.1126/scitranslmed.abq4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunachalam P.S., Walls A.C., Golden N., Atyeo C., Fischinger S., Li C., Aye P., Navarro M.J., Lai L., Edara V.V., et al. Adjuvanting a subunit COVID-19 vaccine to induce protective immunity. Nature. 2021;594:253–258. doi: 10.1038/s41586-021-03530-2. [DOI] [PubMed] [Google Scholar]
- Bale J.B., Gonen S., Liu Y., Sheffler W., Ellis D., Thomas C., Cascio D., Yeates T.O., Gonen T., King N.P., et al. Accurate design of megadalton-scale two-component icosahedral protein complexes. Science. 2016;353:389–394. doi: 10.1126/science.aaf8818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Zepeda S.K., Culap K., Pinto D., VanBlargan L.A., De Marco A., di Iulio J., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2021;602:664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case J.B., Rothlauf P.W., Chen R.E., Liu Z., Zhao H., Kim A.S., Bloyet L.-M., Zeng Q., Tahan S., Droit L., et al. Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2. Cell Host Microbe. 2020;28:475–485.e5. doi: 10.1016/j.chom.2020.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case J.B., Bailey A.L., Kim A.S., Chen R.E., Diamond M.S. Growth, detection, quantification, and inactivation of SARS-CoV-2. Virology. 2020;548:39–48. doi: 10.1016/j.virol.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier D.A., The CITIID-NIHR BioResource COVID-19 Collaboration. De Marco A., Ferreira I.A.T., Meng B., Datir R.P., Walls A.C., Kemp S.A., Bassi J., Pinto D., et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature. 2021;593:136–141. doi: 10.1038/s41586-021-03412-7. [DOI] [PubMed] [Google Scholar]
- Corbett K.S., Nason M.C., Flach B., Gagne M., O’Connell S., Johnston T.S., Shah S.N., Edara V.V., Floyd K., Lai L., et al. Immune correlates of protection by mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. Science. 2021;373:eabj0299. doi: 10.1126/science.abj0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D., Purcell L.A., Snell G., Veesler D. Tackling COVID-19 with neutralizing monoclonal antibodies. Cell. 2021;184:4593–4595. doi: 10.1016/j.cell.2021.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford K.H.D., Eguia R., Dingens A.S., Loes A.N., Malone K.D., Wolf C.R., Chu H.Y., Tortorici M.A., Veesler D., Murphy M., et al. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses. 2020;12:513. doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiege J.K., Block K.E., Pierson M.J., Nanda H., Shepherd F.K., Mickelson C.K., Stolley J.M., Matchett W.E., Wijeyesinghe S., Meyerholz D.K., et al. Mice with diverse microbial exposure histories as a model for preclinical vaccine testing. Cell Host Microbe. 2021;29:1815–1827.e6. doi: 10.1016/j.chom.2021.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A., Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Gilbert P.B., Montefiori D.C., McDermott A.B., Fong Y., Benkeser D., Deng W., Zhou H., Houchens C.R., Martins K., Jayashankar L., et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science. 2021;375:43–50. doi: 10.1126/science.abm3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski L.E., Ferris M.T., Aylor D.L., Whitmore A.C., Green R., Frieman M.B., Deming D., Menachery V.D., Miller D.R., Buus R.J., et al. Genome wide identification of SARS-CoV susceptibility loci using the collaborative cross. PLoS Genet. 2015;11:e1005504. doi: 10.1371/journal.pgen.1005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29:463–476.e6. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A.O., Feldmann F., Zhao H., Curiel D.T., Okumura A., Tang-Huau T.-L., Case J.B., Meade-White K., Callison J., Chen R.E., et al. A single intranasal dose of chimpanzee adenovirus-vectored vaccine protects against SARS-CoV-2 infection in rhesus macaques. Cell Rep. Med. 2021;2:100230. doi: 10.1016/j.xcrm.2021.100230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.-L., Goldsmith J.A., Schaub J.M., DiVenere A.M., Kuo H.-C., Javanmardi K., Le K.C., Wrapp D., Lee A.G., Liu Y., et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science. 2020;369:1501–1505. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaname Y., Tani H., Kataoka C., Shiokawa M., Taguwa S., Abe T., Moriishi K., Kinoshita T., Matsuura Y. Acquisition of complement resistance through incorporation of CD55/decay-accelerating factor into viral particles bearing baculovirus GP64. J. Virol. 2010;84:3210–3219. doi: 10.1128/JVI.02519-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.-C., Liang Q., Ali H., Bayliss L., Beasley A., Bloomfield-Gerdes T., Bonoli L., Brown R., Campbell J., Carpenter A., et al. Complete humanization of the mouse immunoglobulin loci enables efficient therapeutic antibody discovery. Nat. Biotechnol. 2014;32:356–363. doi: 10.1038/nbt.2825. [DOI] [PubMed] [Google Scholar]
- Lempp F.A., Soriaga L.B., Montiel-Ruiz M., Benigni F., Noack J., Park Y.-J., Bianchi S., Walls A.C., Bowen J.E., Zhou J., et al. Lectins enhance SARS-CoV-2 infection and influence neutralizing antibodies. Nature. 2021;598:342–347. doi: 10.1038/s41586-021-03925-1. [DOI] [PubMed] [Google Scholar]
- Martinez D.R., Schäfer A., Leist S.R., De la Cruz G., West A., Atochina-Vasserman E.N., Lindesmith L.C., Pardi N., Parks R., Barr M., et al. Chimeric spike mRNA vaccines protect against Sarbecovirus challenge in mice. Science. 2021;373:991–998. doi: 10.1126/science.abi4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum M., Walls A.C., Sprouse K.R., Bowen J.E., Rosen L.E., Dang H.V., deMarco A., Franko N., Tilles S.W., Logue J., et al. Molecular basis of immune evasion by the delta and kappa SARS-CoV-2 variants. Science. 2021;374:1621–1626. doi: 10.1126/science.abl8506. [DOI] [PubMed] [Google Scholar]
- Mlcochova P., Kemp S., Dhar M.S., Papa G., Meng B., Mishra S., Whittaker C., Mellan T., Ferreira I., Datir R. SARS-CoV-2 B. 1.617. 2 Delta variant emergence, replication and sensitivity to neutralising antibodies. bioRxiv. 2021 doi: 10.1101/2021.05.08.443253. Preprint at. [DOI] [Google Scholar]
- Murphy A.J., Macdonald L.E., Stevens S., Karow M., Dore A.T., Pobursky K., Huang T.T., Poueymirou W.T., Esau L., Meola M., et al. Mice with megabase humanization of their immunoglobulin genes generate antibodies as efficiently as normal mice. Proc. Natl. Acad. Sci. USA. 2014;111:5153–5158. doi: 10.1073/pnas.1324022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie J., Xie J., Liu S., Wu J., Liu C., Li J., Liu Y., Wang M., Zhao H., Zhang Y., et al. Three epitope-distinct human antibodies from RenMab mice neutralize SARS-CoV-2 and cooperatively minimize the escape of mutants. Cell Discov. 2021;7:53. doi: 10.1038/s41421-021-00292-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallesen J., Wang N., Corbett K.S., Wrapp D., Kirchdoerfer R.N., Turner H.L., Cottrell C.A., Becker M.M., Wang L., Shi W., et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer M.M., Tortorici M.A., Park Y.-J., Walls A.C., Homad L., Acton O.J., Bowen J.E., Wang C., Xiong X., de van der Schueren W., et al. Structural basis for broad coronavirus neutralization. Nat. Struct. Mol. Biol. 2021;28:478–486. doi: 10.1038/s41594-021-00596-4. [DOI] [PubMed] [Google Scholar]
- Song J.Y., Choi W.S., Heo J.Y., Lee J.S., Jung D.S., Kim S.W., Park K.H., Eom J.S., Jeong S.J., Lee J., et al. Safety and immunogenicity of a SARS-CoV-2 recombinant protein nanoparticle vaccine (GBP510) adjuvanted with AS03: A randomised, placebo-controlled, observer-blinded phase 1/2 trial. EClinicalMedicine. 2022;51:101569. doi: 10.1016/j.eclinm.2022.101569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. 2020 doi: 10.1101/2020.12.21.20248640. Preprint at. [DOI] [Google Scholar]
- VanBlargan L.A., Adams L.J., Liu Z., Chen R.E., Gilchuk P., Raju S., Smith B.K., Zhao H., Case J.B., Winkler E.S., et al. A potently neutralizing SARS-CoV-2 antibody inhibits variants of concern by utilizing unique binding residues in a highly conserved epitope. Immunity. 2021;54:2399–2416.e6. doi: 10.1016/j.immuni.2021.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Fiala B., Schäfer A., Wrenn S., Pham M.N., Murphy M., Tse L.V., Shehata L., O’Connor M.A., Chen C., et al. Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2. Cell. 2020;183:1367–1382.e17. doi: 10.1016/j.cell.2020.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;183:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Miranda M.C., Schäfer A., Pham M.N., Greaney A., Arunachalam P.S., Navarro M.-J., Tortorici M.A., Rogers K., O’Connor M.A., et al. Elicitation of broadly protective sarbecovirus immunity by receptor-binding domain nanoparticle vaccines. Cell. 2021;184:5432–5447.e16. doi: 10.1016/j.cell.2021.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Sprouse K.R., Bowen J.E., Joshi A., Franko N., Navarro M.J., Stewart C., Cameroni E., McCallum M., Goecker E.A., et al. SARS-CoV-2 breakthrough infections elicit potent, broad, and durable neutralizing antibody responses. Cell. 2022;185:872–880.e3. doi: 10.1016/j.cell.2022.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A.D.M.E., van Kuppeveld F.J.M., Haagmans B.L., Grosveld F., Bosch B.-J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitt M.A. Generation of VSV pseudotypes using recombinant ΔG-VSV for studies on virus entry, identification of entry inhibitors, and immune responses to vaccines. J. Virol. Methods. 2010;169:365–374. doi: 10.1016/j.jviromet.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Choi A., Koch M., Elbashir S., Ma L., Lee D., Woods A., Henry C., Palandjian C., Hill A., et al. Variant SARS-CoV-2 mRNA vaccines confer broad neutralization as primary or booster series in mice. Vaccine. 2021;39:7394–7400. doi: 10.1016/j.vaccine.2021.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying B., Whitener B., VanBlargan L.A., Hassan A.O., Shrihari S., Liang C.-Y., Karl C.E., Mackin S., Chen R.E., Kafai N.M., et al. Protective activity of mRNA vaccines against ancestral and variant SARS-CoV-2 strains. Sci. Transl. Med. 2022;14:eabm3302. doi: 10.1126/scitranslmed.abm3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated in this study are presented in Data S1 and data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.