Abstract
Synthetic mRNA technology is a promising avenue for treating and preventing disease. Key to the technology is the incorporation of modified nucleotides such as N1-methylpseudouridine (m1Ψ) to decrease immunogenicity of the RNA. However, relatively few studies have addressed the effects of modified nucleotides on the decoding process. Here, we investigate the effect of m1Ψ and the related modification pseudouridine (Ψ) on translation. In a reconstituted system, we find that m1Ψ does not significantly alter decoding accuracy. More importantly, we do not detect an increase in miscoded peptides when mRNA containing m1Ψ is translated in cell culture, compared with unmodified mRNA. We also find that m1Ψ does not stabilize mismatched RNA-duplex formation and only marginally promotes errors during reverse transcription. Overall, our results suggest that m1Ψ does not significantly impact translational fidelity, a welcome sign for future RNA therapeutics.
Keywords: N1-methylpseudouridine, mRNA vaccine, ribosome, fidelity, translation, decoding
Graphical abstract
The COVID-19 mRNA vaccines contain the modified nucleoside N1-methylpseudouridine to bypass innate immune responses and increase translation in vivo. Kim et al. show that the modification has minimal impact on the yield and accuracy of translation.
Introduction
The remarkable effectiveness of mRNA vaccines against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and their record-setting approval (Baden et al., 2021; Polack et al., 2020) have generated considerable interest in synthetic mRNA therapeutics. This technology promises many advantages, including simplicity of production, ease of customization, and relatively low cost, as RNA can be readily made in vitro with high yields (Baronti et al., 2018). Importantly, RNAs produced in this manner can be synthesized with a cap structure just like eukaryotic mRNAs. These synthetic mRNAs, when successfully delivered to the cytoplasm of eukaryotic cells, are well recognized as templates for protein synthesis by the ribosomes. Unlike direct delivery of proteins—which are typically limited to the extracellular space—proteins translated from synthetic mRNAs can be targeted to remain within the cell, directed for export, or even deposited in membranes. Furthermore, these mRNAs are non-integrating and readily degraded by ubiquitous cellular RNases, presumably making them safer than comparable DNA therapeutics (Sahin et al., 2014).
The potential of mRNA therapeutics has been documented in the literature for several decades. In one of the first reports, Wolff and colleagues demonstrated that injection of mRNAs for several reporter genes into the muscles of mice could produce protein products with yields similar to comparable injections of DNA plasmids (Wolff et al., 1990). Two years later, Jirikowski and colleagues showed that injection of vasopressin mRNA into the hypothalamus of Brattleboro rats resulted in expression of the protein and temporary reversal of their diabetes insipidus (Jirikowski et al., 1992). However, various obstacles hindered development of the technology until present times. Key challenges were the lack of an efficient delivery method and the instability and immunogenicity of in vitro-transcribed mRNAs (Damase et al., 2021; Van Hoecke and Roose, 2019; Sahin et al., 2014).
Over the past decade, advances in lipid nanotechnology and materials sciences have provided efficient and safe delivery systems for synthetic mRNAs (Damase et al., 2021; Guan and Rosenecker, 2017; Kaczmarek et al., 2017). At the same time, increased understanding of mRNA biochemistry and cellular mechanisms have addressed stability and immunogenicity concerns. In particular, the immunogenicity of in-vitro-transcribed mRNA has been attributed to the activation of cell surface, endosomal, and cytoplasmic RNA sensors (Akira et al., 2006; Freund et al., 2019; Karikó et al., 2004, 2005; Weissman et al., 2000) that monitor for the presence of viral RNAs. Incorporation of modified nucleotide monophosphates into mRNA during its synthesis, along with careful purification of the modified mRNA, was found to suppress the activation of these sensors (Freund et al., 2019; Karikó et al., 2005, 2008, 2011). Substitution of uridine by pseudouridine (Ψ), a modification abundant in tRNAs and rRNAs, was one of the first modifications found to exhibit such effects on the innate immune response (Anderson et al., 2010; Karikó et al., 2008). Later studies documented similar effects for 5-methylcytidine and 2-thiouridine and observed that modified mRNAs produced 10- to 100-fold more protein compared with unmodified mRNAs (Kormann et al., 2011). Recently, N1-methylpseudouridine (m1Ψ), the modification used in the current mRNA SARS-CoV-2 vaccines, was found to possess superior characteristics to Ψ; m1Ψ elicited less immunogenicity and increased protein production by more than an order of magnitude relative to Ψ (Andries et al., 2015; Parr et al., 2020; Svitkin et al., 2017).
While much effort has been devoted to understanding the mechanisms by which nucleotides modifications suppress the innate immune response and increase protein yield, relatively few studies have investigated their effects on the fidelity of protein synthesis. The accuracy of tRNA selection by the ribosome when encountering modified mRNAs is an important consideration, as the fidelity of the protein product might be a critical factor in RNA-based treatments. In the case of Ψ, studies have reported conflicting results as that to the effect of the modification on ribosome accuracy. Studies by the Yu group found that introduction of Ψ to stop codons induced readthrough in vitro and in vivo (Adachi and Yu, 2020; Fernández et al., 2013; Karijolich and Yu, 2011)—an effect rationalized as unusual base pairing between the codon and anticodon that is only allowed when modified bases are used (Fernández et al., 2013; Parisien et al., 2012). Studies from other groups examining the effects of Ψ on amino acid misincorporation provided conflicting results and suggested that the modification has little effect on the fidelity of protein synthesis (Eyler et al., 2019; Hoernes et al., 2016, 2019; Nir et al., 2022; Svidritskiy et al., 2016). To the best of our knowledge, however, there have been no such studies on the effects of m1Ψ on tRNA selection by the ribosome and subsequent translation, even though m1Ψ is the current choice for mRNA vaccines.
To address this knowledge gap, we explored the effect of m1Ψ and Ψ on decoding using various in vitro systems. In a reconstituted system, the substitution of a single uridine with m1Ψ in a model mRNA was found to slightly decrease the observed rate of peptide-bond formation. However, m1Ψ did not significantly alter the overall accuracy of tRNA selection by the ribosome, whereas Ψ marginally increased the incorporation of near- and non-cognate amino acids. Liquid chromatography with tandem mass spectrometry (LC-MS/MS) analysis of SARS-CoV-2 spike protein products produced in HEK 293 cells via electroporation of mRNAs containing only uridine, Ψ, or m1Ψ showed that translation of either modified mRNA did not lead to a detectable increase in miscoded proteins. High-resolution melt analysis found that Ψ, but not m1Ψ, increased the stability of mismatched duplexes. Primer extension assays and quantification of reverse transcriptase errors across the modifications via deep sequencing revealed that the modifications are read differently by reverse transcriptases. Taken together, our findings provide important insights into the effects of m1Ψ on translation and its usage in future mRNA therapeutics.
Results
Validation of Ψ and m1Ψ modifications in synthetic model mRNAs
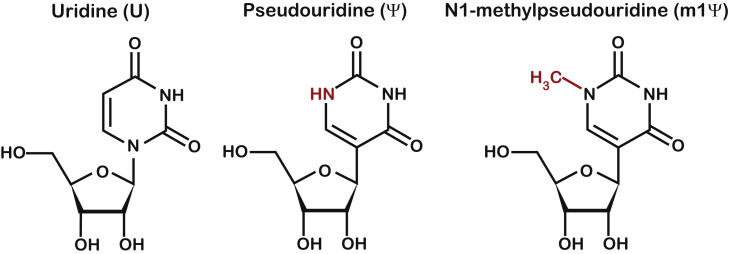
Our initial goal was to determine how mRNA modifications in synthetic mRNAs modify their base-pairing properties and impact the decoding process. In particular, we sought to systemically examine how the modifications alter the incorporation of all near- and non-cognate amino acids. To do so, we resorted to our reconstituted bacterial system, which allowed us to carry out well-defined reactions with each of the 20 tRNA isoacceptors as well as release factors (Keedy et al., 2018). To program ribosomes with modified mRNAs, we obtained three chemically synthesized model mRNAs—one with no modification, and the other two having a single substitution of uridine for Ψ or m1Ψ (Figure 1 ). Before proceeding, we verified that each model mRNA contained the correct modification using LC-MS/MS analysis. To generate standards for Ψ and m1Ψ, we treated their respective nucleotide triphosphate with calf intestinal phosphatase (CIP). Standards for the four canonical nucleotides, along with Ψ and m1Ψ, were analyzed with an Agilent 6490 QQQ triple-quadrupole LC mass spectrometer to validate mass transitions and retention times. We then subjected our mRNAs to P1 nuclease digestion and CIP treatment to generate single nucleosides. Using the parameters determined from analysis of the standards, analysis of the digested RNAs yielded peaks with distinct retention times and/or mass transitions for U, Ψ, and m1Ψ. Furthermore, Ψ and m1Ψ peaks were only observed in the respective RNA (Figure S1). Analysis of the A, C, G, and U peaks confirmed that the base composition of each mRNA was correct (Figure S1D).
Figure 1.
Chemical structures of uridine and its modifications
Chemical structure of uridine, pseudouridine, and N1-methylpseudouridine. The N1 of both modified nucleosides are marked in red.
m1Ψ slightly decreases the rate of peptide-bond formation
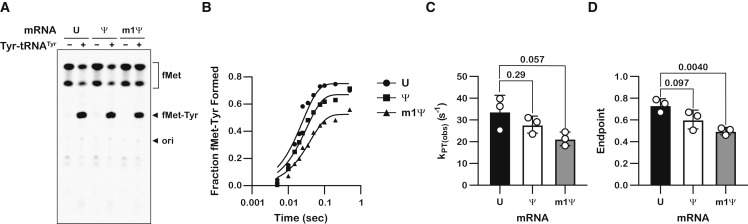
Using our reconstituted bacterial translation system, we measured the observed rates of peptide-bond formation (k pep) (Zaher and Green, 2010) in the presence of our model mRNAs. Briefly, programmed ribosomes displayed either UAC, ΨAC, or m1ΨAC in the A site, which codes for the incorporation of tyrosine (Tyr). The initiation complexes also carried f-[35S]-Met-tRNAfMet in the P site, which allowed us to follow dipeptide formation by electrophoretic thin-layer chromatography (TLC) (Youngman et al., 2004). As expected, in the presence of a Tyr-tRNATyr•EFTu•GTP ternary complex, we observe robust accumulation of dipeptide products for all three initiation complexes after 10 s of incubation (Figure 2A). Next, we used a pre-steady-state quench approach to measure the observed k pep in the presence of 1 μM ribosome complexes and a 2.5 μM tRNATyr•EFTu•GTP ternary complex. At this sub-saturating concentration of reactants, changes to the observed k pep reflect alterations to k cat and K 1/2 (Zhang et al., 2016), allowing us to monitor contributions from both parameters. Ψ and, to a greater extent, m1Ψ were found to reduce k pep in the presence of the cognate ternary complex, with measured k pep values of 40, 31, and 25 s−1 for the UAC-, ΨAC- and m1ΨAC-programmed complexes, respectively (Figures 2B and 2C). This is consistent with reports from multiple groups showing that Ψ slows translation (Eyler et al., 2019; Hoernes et al., 2016; Karikó et al., 2008). Similarly, we observe modest, but significant, reductions in the endpoint of the reactions, from ∼0.7 for the unmodified mRNA to ∼0.5 for the mRNA containing m1Ψ (Figure 2D). However, given that other steps during protein synthesis, including translocation, have been estimated to be much slower that peptide-bond formation, it is highly likely that the reduction in k pep seen here is inconsequential to the overall protein yield from the modified mRNAs. Indeed, in eukaryotic extracts, the modifications appear not to affect overall protein synthesis yield (Hoernes et al., 2019).
Figure 2.
Pseudouridine and N1-methylpseudouridine slow down peptidyl transfer by the ribosome
(A) A representative electrophoretic TLC of triplicates showing the translation products of three different initiation complexes—UAC mRNA, ΨAC mRNA, or m1ΨAG mRNA—in the absence and presence of cognate tRNA (Tyr-tRNATyr tRNA) ternary complex.
(B) A representative time course plot of triplicates showing the kinetics of f-Met-Tyr peptide formation in the presence of unmodified (U), pseudouridine (Ψ)-containing, and N1-methylpseudouridine (m1Ψ)-containing A-site UAC codon.
(C and D) Bar graph showing the measured observed rates of peptide-bond formation and reaction endpoints, respectively, in the presence of 1 μM initiation complex and 2.5 μM tRNATyr•EFTu•GTP ternary complex.
Plotted are the average values determined from three independent time courses, with error bars representing the standard deviation around the mean. p values are denoted above the plots.
m1Ψ preserves the fidelity of tRNA selection by the ribosome
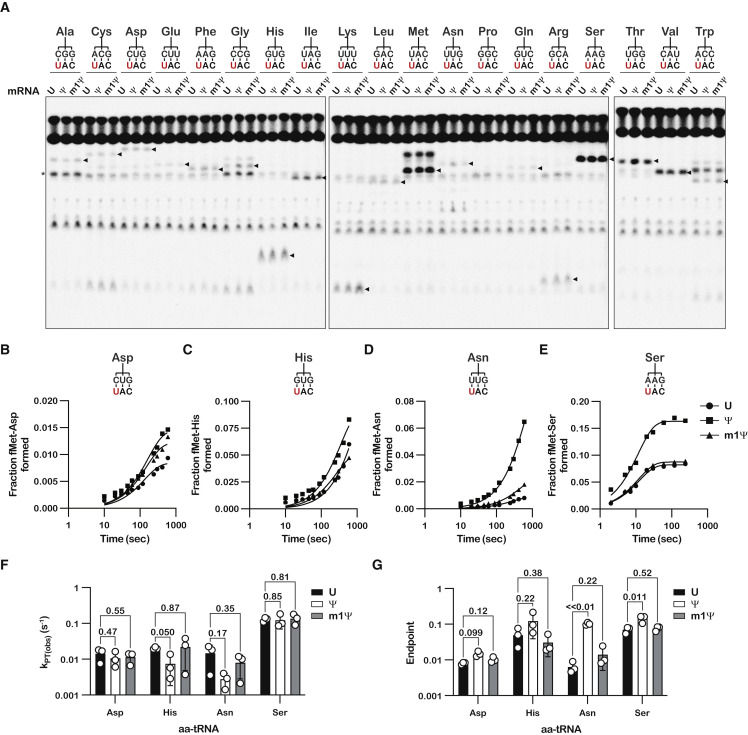
Having established that Ψ and m1Ψ only modestly affect the cognate codon-anticodon interaction, we next sought to assess whether the modifications alter the incorporation frequency of near- and non-cognate aminoacyl-tRNAs (aa-tRNAs). As mentioned earlier, conflicting data exist as to whether or not Ψ promotes miscoding (Adachi and Yu, 2020; Eyler et al., 2019; Fernández et al., 2013; Hoernes et al., 2016, 2019; Karijolich and Yu, 2011; Nir et al., 2022; Svidritskiy et al., 2016). This raises the possibility that m1Ψ may also stabilize mismatched codon-anticodon interactions and allow ribosomes to accept near- or non-cognate tRNAs. To address this, we conducted a peptidyl-transfer (PT) reactivity survey between the initiation complexes and all 19 near/non-cognate aa-tRNA isoacceptors (Keedy et al., 2018). The survey revealed that, with few exceptions, the overall reactivity profile was similar among the different initiation complexes after 10 s of incubation with ternary complexes (Figure 3A). We note that Tyr-tRNATyr is present at low concentrations in the tRNA mix—even after many attempts to deacylate it prior to aminoacylation—as evidenced by detection of a contaminating fMet-Tyr dipeptide in all reactions. However, this contaminant was readily distinguished by its distinct electrophoretic TLC migration (Figure 3A). Furthermore, we have previously shown that each dipeptide has a unique retention factor (Rf) on our electrophoretic TLC (eTLC) assay, thus allowing identification (Keedy et al., 2018; Pierson et al., 2016; Simms et al., 2014). Altogether, the reactivity survey suggested that the modifications have little impact on overall accuracy of translation.
Figure 3.
Unlike Ψ, m1Ψ has little to no effect on the accuracy of tRNA selection
(A) Representative electrophoretic TLCs of triplicates showing dipeptide formation reactions (10 s time point) of near/non-cognate tRNAs with UAC, ΨAC, or m1ΨAG initiation complexes. Arrowheads denote the translation product of each respective aa-tRNA.
(B–E) Representative time courses of triplicates of near-cognate tRNA ternary complexes (Asp, His, Asn, and Ser) and UAC, ΨAC, or Met-m1ΨmAG initiation complexes. The codon (UAC; modification in red) and near-cognate tRNA are indicated.
(F and G) Bar graph showing the measured observed rates of peptide-bond formation and reaction endpoints, respectively, in the presence of 1 μM initiation complex and 2.5 μM denoted ternary complex.
Plotted are the average values determined from three independent time courses, with error bars representing the standard deviation around the mean. p values are denoted above the plots.
To better understand the effect of the modifications on miscoding, we focused on near-cognate aa-tRNAs and other aa-tRNAs that exhibited reactivity with the initiation complexes. We initially measured the observed k pep values for the near-cognate Asp-tRNAAsp, His-tRNAHis, Asn-tRNAAsn, and Ser-tRNASer substrates (Figures 3B–3F). Of note, the first three aa-tRNAs correspond to mismatches involving the modification itself (U•C, U•G, and U•U, respectively), whereas the latter corresponds to an A•A mismatch 3′ to the modification. For all four reactions, Ψ and m1Ψ did not alter the observed k pep values significantly (Figure 3F). Interestingly, Ψ was found to increase the endpoint for the near-cognate interactions, especially in the presence of the Asn-tRNAAsn ternary complex, which involves a U:U mismatch, whereas m1Ψ did not (Figure 3G). These observations suggest that Ψ modestly promotes miscoding by altering the proofreading phase of tRNA selection. Miscoding was not limited to these near-cognate aa-tRNAs, as similar observations were made for the remaining near-cognate aa-tRNAs: Cys-tRNACys and Phe-tRNAPhe (Figures S2JA and S2K). Interestingly, Ψ had the most dramatic impact on a mismatch involving its neighbor, i.e., reactions with Ser-tRNASer, having an A•A mismatch to its 3′ (Figure 3E), suggesting that Ψ alters the base-pairing properties of the entire codon-anticodon interaction. Supporting this proposal is the observation that Ψ increased endpoint values for many non-cognate aa-tRNAs, which harbor more than one mismatch (Figure S2). By contrast, m1Ψ did not increase k pep or endpoint values significantly for all tested near- and non-cognate aa-tRNAs (Figures 3 and S2). Ultimately, our data suggest that although Ψ modestly increases mispairing during tRNA selection, m1Ψ behaves similarly to uridine.
m1Ψ suppresses near-stop codon recognition by release factors
In addition to RNA-RNA interactions that occur within the decoding center between the codon of mRNAs and anticodon of tRNAs, RNA-protein interactions also occur during stop-codon recognition by release factors (RFs). In fact, decoding of the stop codons is completely distinct than that of sense codons (Youngman et al., 2004). Fortuitously, our choice of the UAC A-site codon above, which is a near-stop codon for UAA and UAG, allowed us to evaluate how Ψ and m1Ψ impact misreading of sense codons by RFs (Figure S3A). Bacteria have two RFs (Youngman et al., 2004), with overlapping specificities: RF1, which recognizes UAA and UAG, and RF2, which recognizes UAA and UGA. RF1 was observed to react much faster with the UAC complex than RF2 (Figures S3B–S3D), with rates of hydrolysis of 0.2 and 0.03 s−1, respectively. This was expected since RF1 exhibits promiscuity toward the third base of the stop codon—the same position where mismatches occur as the factor decodes the UAC near-stop codon. We also observed that m1Ψ significantly inhibited near-stop codon recognition by both factors, as we measured an endpoint of 0.3–0.4 compared with ∼0.7 observed for uridine (Figure S3E). Ψ, by contrast, only slightly decreased the rate of hydrolysis by RF1 (0.08 s−1) and not by RF2 (0.04 s−1) and had no effect on the endpoint (Figures S3D and S3E). These observations not only reinforce the distinctions between the interactions of Ψ and m1Ψ with the decoding center but also suggest that m1Ψ suppresses premature termination during protein synthesis.
m1Ψ-modified SARS-CoV-2 spike protein mRNA is translated faithfully by eukaryotic ribosomes
So far, our analysis of the decoding process in the presence of modified mRNA focused on short model mRNAs added to a reconstituted bacterial system. These chemically synthesized mRNAs harbor a single modification, which allows us to systemically characterize their impact on the overall accuracy of tRNA selection. However, they are not ideal when evaluating how therapeutic mRNAs, which can harbor thousands of modified nucleotides, are translated. For example, all the constituent uridines in the mRNA vaccines are substituted with m1Ψ. To probe whether such pervasive modification alters the accuracy of the ribosome in conditions more closely resembling those found in vivo, we generated SARS-CoV-2 spike protein mRNAs in which all constituent uridines remained unmodified or were completely replaced with Ψ or m1Ψ (Figure 4A). The template sequence is identical to that used in the BNT162b2 (Pfizer) mRNA coronavirus 2019 (COVID-19) vaccine (Jeong et al., 2021; Polack et al., 2020), except for the addition of sequences encoding His and FLAG tags at the 5′ and 3′ ends (Table S1), respectively, and the mRNAs were similarly capped.
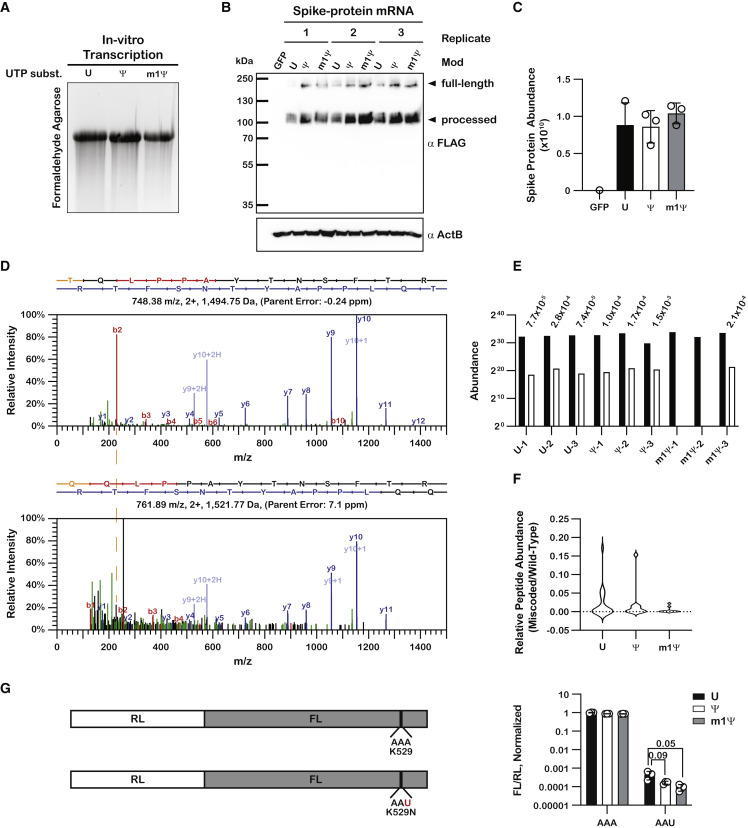
Figure 4.
The presence of m1Ψ in mRNA does not increase amino acid misincorporation frequency during translation in human cells
(A) UV-transillumination image of an ethidium bromide-stained formaldehyde agarose gel used to visualize the mRNA constructs transfected into HEK 293 cells.
(B) Immunoblot analysis of total cell lysate from transfected HEK 293 cells. The SARS-CoV-2 spike protein is known to undergo post-translational processing, and products corresponding to their respective sizes can be seen, as denoted by the arrows.
(C) Bar graphs showing the abundance of the wild-type SARS-CoV-2 spike protein isolated from on-bead digestion, as determined by label-free quantitation from LC-MS/MS analysis.
(D) Fragmentation spectra of the wild-type peptide TQLPPAYTNSFTR and its substituent miscoded product, QQLPPAYTNSFTR. b and y ions are denoted in red and blue, respectively, while the substituted amino acid is denoted in orange. The difference in the m/z for the b2–b4 peaks between the wild-type peptide (top) and miscoded product (bottom) corresponds to a threonine to glutamine substitution. The dashed orange line indicates the shift in the mass of the b2 ion. The nominal mass difference between threonine and glutamine is 27 Da.
(E) Bar graphs showing the abundance of TQLPPAYTNSFTR and its miscoded substituent, QQLPPAYTNSFTR. The miscoded peptide was not detected in samples m1Ψ-1 and m1Ψ-2. The relative peptide abundance is denoted above each pair.
(F) Violin plots showing the distribution of relative peptide abundances for miscoded spike protein peptides translated from U, Ψ-containing, or m1Ψ-containing mRNA. Only those peptides where both the wild-type and the miscoded species were detected were included.
(G) On the left is a schematic of the dual-luciferase reporter system used to assess miscoding frequency in HEK 293 cells. On the right are bar graphs showing normalized luminescence values for U, Ψ-containing, and m1Ψ-containing mRNA. Plotted are the average values of three biological replicates with error bars representing the standard deviation around the mean. Unpaired t tests did not show statistically significant differences in miscoding frequency between the U, Ψ-containing, and m1Ψ-containing mRNA.
To assess the integrity of spike protein products produced in human cells, we electroporated the unmodified and modified mRNAs into HEK 293 cells. Electroporation of a GFP plasmid control measured electroporation efficiency, which was estimated at 90% (Figure S4A). More importantly, immunoblot analysis showed that the yields of spike protein products were similar regardless of modification status of the electroporated mRNA (Figures 4B and S4B). This was expected, as HEK 293 cells do not express TLR3, the receptor responsible for activating the innate immune response to unmodified mRNAs (Andries et al., 2015). Moreover, previous studies investigating translation of Ψ-containing mRNAs in HEK 293 cells reported similar results (Eyler et al., 2019; Hoernes et al., 2019). In addition to comparable yields, we also noted that regardless of the modification status of the mRNA, the resultant spike protein appeared to be processed into smaller products in the HEK 293 cells (Figure 4B), as has been previously noted (Ou et al., 2020). Additionally, the processed protein products migrated as heterogeneous bands (Figure 4B), suggesting that protein glycosylation also occurred as expected (Watanabe et al., 2020). Having established that incorporation of Ψ and m1Ψ into the mRNAs have little to no effect on protein yield under our experimental conditions, we next sought to assess the impact of the modifications on miscoding using MS approaches. Spike protein products were purified with anti-FLAG magnetic beads, with a GFP-electroporated sample serving as a negative control. Following extensive washing, the samples were subjected to on-bead trypsin digestion and analyzed by LC-MS/MS.
After searching the datasets against the spike protein in the presence of the human proteome, we successfully identified 39% of the spike protein sequence (Figure S4D). Reassuringly, label-free quantification of spike protein abundance did not detect significant levels of spike protein in the GFP control sample (<100-fold relative to the spike protein mRNA samples) (Figure 4C). Additionally, spike protein abundance was largely similar across all U, Ψ, and m1Ψ samples (Figure 4C), consistent with the immunoblot analysis (Figures 4B and S4B), which suggested that the modifications did not significantly alter protein yield.
To identify miscoded peptides, we conducted an error-tolerant search to identify candidates for further analysis. Results were checked in Scaffold to select the highest confidence candidates. To further ensure that we did not miss any miscoded peptides, we also conducted a second search against an in silico-generated spike protein library, comprising all single substitution protein products arising from miscoding events at every uridine in the spike protein sequence, to identify additional candidates. The sequences of all chosen candidates were then combined and added to the search library for a final confirmation search of the datasets.
Miscoded peptides identified in the final search were quantified from the MS1 scans using Proteome Discoverer and were further validated using Skyline. Miscoded peptides that were not assigned quantitation values by Proteome Discoverer were manually quantified using Skyline if good peaks could be found. An example set of predicted fragmentation spectra for a wild-type peptide and its miscoded substituent is shown in Figure 4D. We then estimated the error frequency by dividing the abundance of the miscoded peptide by that of the parent faithful peptide (Figure 4E; Table S2). Contrary to previous reports, which documented an increase in miscoding frequency when Ψ-containing luciferase mRNA was translated in HEK 293 cells (Eyler et al., 2019), we did not observe an increase in miscoding frequency for our Ψ-containing spike protein mRNA (Figure 4F). These distinctions could be due to the different proteins being analyzed. It is also feasible that our analysis missed rare miscoding events due to low coverage of any single miscoded peptide species. As such, the frequency of such events may be higher in the presence of Ψ-containing mRNA than we observed. However, our data suggest that for the most abundant miscoding events, Ψ does not significantly alter their frequency. Given the lack of observed effect for Ψ, the presence of the modification in the mRNA is not likely to have a biological impact on the fidelity of the protein products. More importantly, we do not observe an increase for our m1Ψ-containing mRNA (Figure 4F).
To add further confidence in our observations that the modifications do not significantly affect translational fidelity, we conducted a more sensitive assessment of miscoding in the presence of Ψ- and m1Ψ-containing mRNAs using a dual-luciferase reporter assay system (Kramer and Farabaugh, 2007). For this reporter, we mutated a critical lysine residue, encoded by AAA, in the active site of firefly luciferase, to an asparagine encoded by AAU (Figure 4G). Any observed firefly luciferase signal is then the result of miscoding events where the asparagine is miscoded as a lysine. By normalizing the signal to a wild-type control, we were able to assess how the presence of Ψ or m1Ψ affected miscoding frequency. The reporter construct RNA (Table S1) was transcribed in vitro and electroporated into HEK 293 cells using the same methods as for the spike protein mRNA. We found that neither the presence of Ψ nor the presence of m1Ψ in the RNA induced a detectable and/or significant increase in miscoding (Figure 4G), consistent with our LC-MS/MS analysis.
We also tested the mRNAs in a wheat germ system to broadly investigate how the modifications affect other eukaryotic systems. The protein yields were assessed by measuring 35S-methionine incorporation into full-length polypeptides. Consistent with our results in the HEK 293 system, we did not observe significant differences in spike protein yield in the presence of either Ψ- or m1Ψ-containing mRNAs relative to unmodified mRNA (Figure S5A). We then subjected affinity-purified spike protein to LC-MS/MS analysis to assess miscoding events. Here, we searched the datasets against the computationally generated library used to search the HEK 293 datasets. An example set of predicted fragmentation spectra for a wild-type peptide and its miscoded substituent is shown in Figure S5C. As before, we estimated the error frequency by dividing the abundance of the miscoded peptide by that of the parent faithful peptide (Table S3). In wheat germ, translation of Ψ-containing mRNA induced a modest increase in miscoding frequency for some peptides (∼1.5-fold) (Figure S5D). However, and similar to what we observe in HEK 293 cells (Figure 4F), the relative abundance of most miscoded peptides did not change significantly in the presence of Ψ-containing mRNA. More importantly, as seen with the HEK 293 dataset, translation of the m1Ψ-containing mRNA in wheat germ extracts did not increase miscoding frequency (Figure S5D). A second MS analysis found similar trends in a different set of miscoded peptides, with only marginal increases in miscoded peptides over wild-type peptides (Figure S5D). While the relative abundance values were much higher in the second test, this could be due to inaccurate quantification, as the abundance of overall protein was very low. Collectively, our findings indicate that m1Ψ does not significantly increase miscoding during translation.
Unlike Ψ, m1Ψ does not stabilize mismatches during RNA-duplex formation
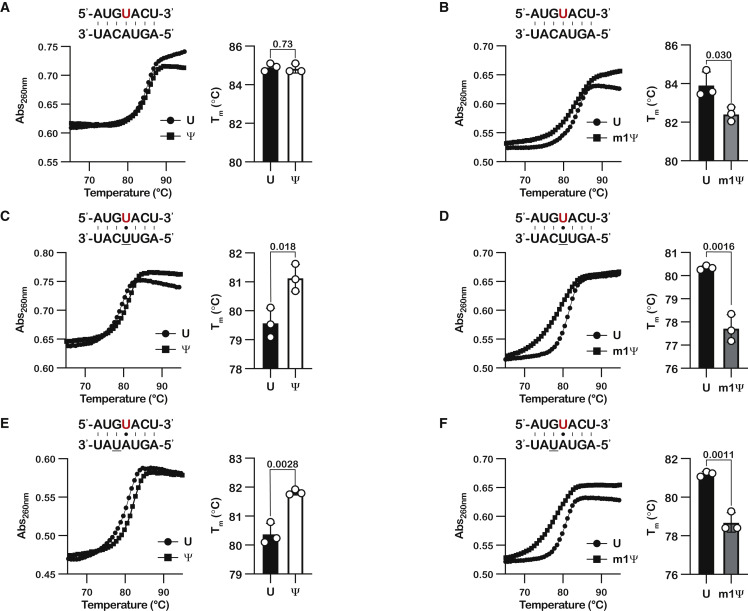
Our data on the error frequency by the ribosome during the decoding of Ψ implied that this modification stabilizes mismatches between the codon and anticodon while m1Ψ does not. This distinction between the two modified bases might be due to their interactions with the ribosome decoding center, in which Ψ is allowed to sample different conformations, while m1Ψ is sterically restricted to fewer conformations. Alternatively, the distinctions could arise from inherent differences between the two modifications, irrespective of the environment. Indeed, previous studies on Ψ suggest that its introduction increased that stability of UA, UG, and UU base pairs by 0.3–0.8 kcal/mol (Kierzek et al., 2014). Interestingly, Ψ was reported to stabilize mRNA structure regardless of the sequence context, while the impact of m1Ψ on mRNA stability was dependent on the identity of its neighbors (Mauger et al., 2019), in agreement with the idea that the modifications differently alter RNA-duplex formation. As the effect of m1Ψ on mismatch stability remained unclear, we examined the effect of Ψ and m1Ψ on the stability of mismatched duplexes by measuring the melting-point temperatures (Tm) of duplex RNAs created by pairing our three model mRNAs with variants of their reverse complement. We initially generated 4 reverse complement RNAs, containing A, U, C, and G opposite to the nucleotide of interest. Consistent with earlier studies, Ψ was found to have no effect on the stability of the Watson-Crick UA base pair, as we measured a Tm of 84.9°C for the UA duplex and 84.8°C for the ΨA duplex (Figure 5A). By contrast, m1Ψ was found to decrease the Tm by more than 1°C (Figure 5B). More importantly, whereas Ψ increased that stability of all tested mismatches—especially for the UU pairing (Tm of ∼79.5°C for UU compared with >81°C for ΨU)—m1Ψ significantly decreased their Tm by as much as by 3°C (Figures 5A–5D and S6A–S6D). These observations hint that stabilization of mismatched codon-anticodon pairings by Ψ may be the reason for the slight increase in near- and non-cognate tRNA binding.
Figure 5.
Ψ stabilizes formation of mismatched RNA duplexes, including neighboring mismatches, whereas N1-methylpseudourdine does not
(A–F) Scatterplots showing the change in absorbance at 254 nm as a function of temperature for the indicated duplexes, with an accompanying bar graph showing the determined melting temperature for the same duplexes. Plotted are the means of three replicates, with the error bars representing the standard deviation around the mean. p values are denoted above the bar graphs. (A), (C), and (E) correspond to duplexes containing Ψ, while (B), (D), and (F) correspond to duplexes containing m1Ψ. The modified base is denoted in red.
We also noticed that in addition to miscoding events that correspond to mismatches between Ψ and the aa-tRNA anticodon, we observed errors caused by mismatches that did not involve the modification itself but its neighbors (Figure 3). As a result, we speculated that Ψ stabilizes mismatches involving its neighbor bases. To test this hypothesis, we generated a reverse complement that, when paired with our model mRNA, harbored a GU mismatch 5′ immediate to the modification. We found that Ψ increased the Tm of this neighbor-mismatched duplex by more than 1°C, whereas m1Ψ significantly decreased the Tm by more than 2°C (Figures 5E and 5F).
Ψ, but not m1Ψ, increases errors by reverse transcriptases
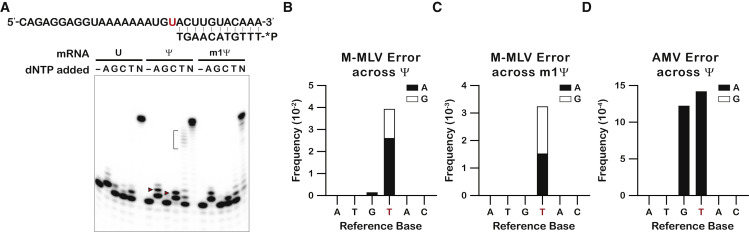
Our data on the relative stability of mismatched duplexes containing Ψ and m1Ψ suggested that these modifications are likely to alter the accuracy of other processes that utilize RNA templates or nucleotide triphosphates (NTPs). Previous reports have documented increased error frequency during reverse transcription (RT) of RNAs containing Ψ (Potapov et al., 2018). Similarly, RNA polymerases were found to increase the frequency of misincorporation events when incorporating ΨTP instead of UTP (Potapov et al., 2018). It is worth noting, however, that these studies used enzymatic reactions to generate the modified mRNA. Since the modification is likely to affect the accuracy of neighboring nucleotide incorporation during transcription, the source of error during RT could be ambiguous. Our model mRNAs bypass these issues since they only harbor a single modification that was synthetically incorporated. As a result, we could isolate the effects of the modifications on the error rate of RT. To this end, we first investigated the effects of the modification on the accuracy of Moloney murine leukemia virus (M-MLV) reverse transcriptase using a primer-extension assay. A radiolabeled primer complementary to all bases upstream of the modified base was annealed to the three model mRNAs. RT was initiated by adding either only one of the four canonical deoxynucleotide triphosphates (dNTPs), no dNTPs, or all four dNTPs. PAGE analysis of the resultant cDNA products revealed that uridine and its two substituents allowed for efficient incorporation of dAMP, as expected (Figure 6A). By contrast, and consistent with our analysis of duplex stability for Ψ-neighbor mismatches, a second event of dAMP incorporation was observed to occur more robustly for the Ψ-containing mRNA. Hence, even when correctly base paired with A, Ψ increases the misincorporation frequency downstream. Testing the incorporation of the other three nucleotides also allowed for a direct assessment of how the modifications alter mispairing frequency in the active site of the reverse transcriptase. As expected, and consistent with earlier reports (Potapov et al., 2018), Ψ was found to increase the levels of misincorporation products, particularly those corresponding to U•C and U•T mismatches.
Figure 6.
The fidelity of reverse transcriptase enzymes is altered in the presence of Ψ
(A) Phosphorimage of a denaturing urea gel used to follow primer extension by M-MLV in the presence of the indicated modified template and dNTP substrates. Shown is a representative gel of duplicates.
(B–D) Bar graphs showing the frequency of variant bases in the codon containing the modified base, as well as the previous codon, as detected by deep sequencing. Bars represent the sum of all variant frequencies, with the proportion for each variant denoted by color. Reference bases are shown below, with the modified base denoted in red. No variants in the two-codon window were detected as statistically significant for the AMV-reverse transcribed m1Ψ RNA.
For RT of the mRNA containing m1Ψ, we also detected an increase in misincorporated products, albeit to a much lesser level compared with the mRNA containing Ψ. Interestingly, for RT reactions containing dTMP, we observed significant accumulation of high-order cDNA products corresponding to 10–11 incorporation events only in the presence of the Ψ-containing mRNA, suggesting that the enzyme successfully crossed the stretch of adenosines in the mRNA template. In turn, this suggests that the enzyme was able to carry out three misincorporations of dTMP across Ψ, G, and U before successfully crossing the stretch of As. These observations add support to our proposal that Ψ dramatically affects the base-pairing properties of its neighbors.
While the primer-extension assay above enabled the assessment of increased misincorporation events that occur on Ψ and m1Ψ, it did not enable quantification of the error rate of RT in the presence of all nucleotides. To quantify this error rate for each modification, we conducted deep sequencing of RT cDNA products for each mRNA. Briefly, an oligonucleotide linker was ligated to each of our model mRNAs, and cDNA synthesis was carried out using a primer complementary to the ligated adapter. We used both avian myeloblastosis virus (AMV) and M-MLV reverse transcriptases for first-strand synthesis, as each RT is known to have distinct error profiles (Potapov et al., 2018). We attempted to amplify our cDNAs with primers containing unique molecular identifiers (UMIs) but were unable to do so. Instead, we proceeded with conventional deep sequencing and acquired between 10 and 15 million reads per sample. Reads were then processed to remove adapters and the linker from the 5′ and 3′ ends. Since we were unable to utilize UMIs to account for library amplification and sequencing artifacts, we processed our data as if searching for sub-clonal variants to better discriminate variation from sequencing error. To go about this, we mapped our processed reads to the reference sequence and searched for variants using deepSNV (Gerstung et al., 2014). While this method could not calculate a true error rate, it did provide clues as to how the modifications influence transcriptase error during library construction. The Ψ and m1Ψ reads were compared with their corresponding U reads for each reaction. Interestingly, similar to the ribosome, reactions with the mRNA containing Ψ increased error frequency, while reactions with the mRNA containing m1Ψ had a much lesser effect. For M-MLV, Ψ induced a greater incidence of T → A and T → G substitutions as well as increasing the error rate of the upstream base from G → A (Figure 6B). m1Ψ induced a similar error profile but with frequencies that were an order of magnitude smaller (Figure 6C). Ψ also had similar effects on AMV reactions, albeit with lower frequencies than with M-MLV (Figure 6D). However, the same error profile was not observed in analysis of the AMV m1Ψ dataset, with none of the mentioned substitutions detected. Thus, our deep-sequencing data corroborate our primer extension results, showing an increased rate of mutation from T to A and T to G, particularly for Ψ. This also explains why we observe higher molecular weight products when conducting primer extension assays with only TTP or CTP. The increased rate of G to A mutation in the preceding base, which is downstream to the modified base during RT, also supports the presence of the polyT products observed after gel electrophoresis. Likewise, the difference in error rates between Ψ and m1Ψ we saw in our sequencing data is consistent with differences in the frequency of misincorporated products that we observed in our primer extension assays.
Discussion
Here, we systemically characterized the effects of m1Ψ and Ψ on protein synthesis in well-defined in vitro systems and assessed whether these effects are relevant to the application of these modification in real-life examples, such as mRNA vaccines. We first investigated the effect of nucleotide modifications on the rate of peptide-bond formation in the context of our model mRNAs. We found m1Ψ to reduce the overall rate of peptide-bond formation in a bacterial reconstituted system (Figure 2). However, this decrease in k pep did not seem to affect the overall yield of protein synthesis in eukaryotic cells or extracts (Figures 4B, S4B, and S5A). These results highlight an interesting pattern also seen with Ψ, where studies utilizing in vitro bacterial systems report a significant reduction for the rate of peptide-bond formation in the presence of the modification (Hoernes et al., 2016), whereas studies employing eukaryotic extracts document no decrease in the overall protein yield when modified RNA is used (Eyler et al., 2019; Hoernes et al., 2019). There are at least two non-mutually exclusive models that could explain this discrepancy between the bacterial system and eukaryotic cell-free extracts. Firstly, it is highly possible that even with the significant decrease in k pep, translation remains rate limited by another step during initiation or elongation. Secondly, the overall elongation rate (including peptide-bond formation and translocation) of eukaryotic ribosomes has been estimated to be at least 4-fold slower than that of bacterial ribosomes (Dennis and Bremer, 1974; Riba et al., 2019; Vogel and Jensen, 1994; Young and Bremer, 1976), suggesting that eukaryotic ribosomes may be less affected by changes to k pep.
Regardless of whether or not the modifications slow translation, it has been shown that mRNAs harboring m1Ψ can produce as much as 10-fold more protein compared with unmodified mRNAs (Andries et al., 2015; Parr et al., 2020; Svitkin et al., 2017). At least two mechanisms have been proposed for this increased protein production: (1) unlike unmodified mRNAs, mRNAs with m1Ψ are more effective at evading Toll-like receptors (TLRs) (Andries et al., 2015), and (2) modified mRNAs mitigate PKR-mediated activation of the integrated stress response, which in turn prevents repression of translation initiation (Anderson et al., 2010; Svitkin et al., 2017).
We next assessed the effect of m1Ψ on the accuracy of tRNA selection by the ribosome by systematically examining all possible amino acid substitutions on our model mRNA templates (Figure 3). Unlike Ψ, which was found to increase the error frequency by several near- and non-cognate aa-tRNAs, m1Ψ did not increase the observed rates of amino acid misincorporation. The increased miscoding by the ribosome on a Ψ-modified UAC codon is similar to that recently reported by Eyler et al. with a Ψ-modified UUU codon (Eyler et al., 2019), suggesting that this effect is not sequence-context specific. However, we note that the increase in the error frequencies that we observed is relatively modest and not likely to contribute to aberrant-protein production. Indeed, LC-MS/MS analysis of SARS-CoV-2 spike protein produced from mRNAs containing Ψ and m1Ψ exhibited no detectable increase in miscoded protein in human cell culture and only a modest increase in a wheat germ system (Figures 4B, S4B, and S5A). Together, our observations indicate that m1Ψ maintains the fidelity of protein synthesis, while Ψ can marginally decrease the accuracy of the ribosome depending on the conditions.
Interestingly, this distinction between Ψ and m1Ψ appears to be due to inherent differences between the two nucleotides and their base-pairing properties during duplex-RNA formation. Similar to other reports (Kierzek et al., 2014), Ψ was found to stabilize all tested mismatches, especially those involving uridine:pyrimidine base pairs, as assessed by UV-melt curve analysis (Figures 5 and S6). Furthermore, the modification even stabilized duplexes involving mismatches with its neighbor. By contrast, m1Ψ destabilized all tested mismatched duplexes.
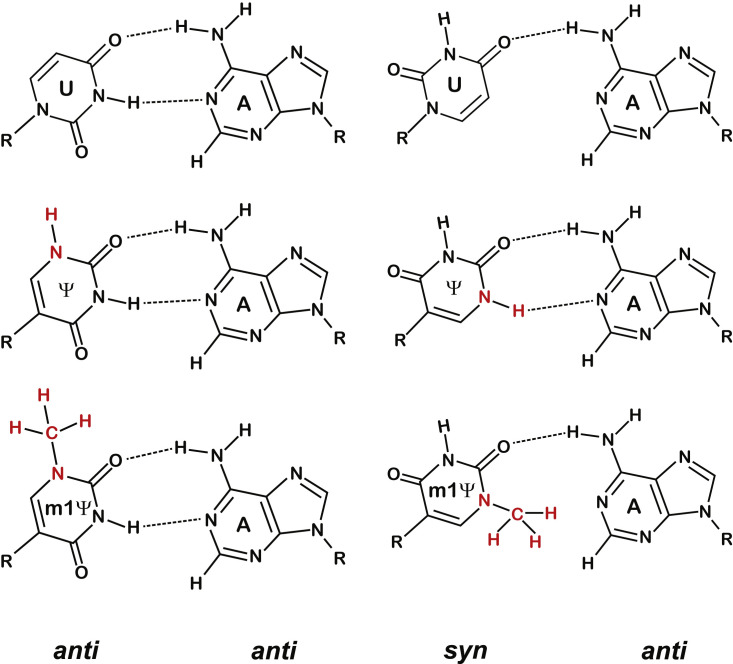
This dissimilarity between the two modified bases in their ability to mispair during duplex-RNA formation has ramifications beyond codon-anticodon interactions during protein synthesis. For example, it appears that only Ψ increases dNMP misincorporation by the reverse transcriptases M-MLV and AMV (Figure 6A). What is the structural basis for these differences between Ψ and m1Ψ? While our data do not directly address this question, others have proposed models rationalizing the ability of Ψ to stabilize duplex RNA (Davis, 1995; Deb et al., 2019; Hudson et al., 2013; Kierzek et al., 2014). A new imino proton becomes available when uridine is isomerized to Ψ—C5 of uridine is replaced with N1 of Ψ—which gives Ψ two hydrogen-bond donors at N1 and N3. This means that Ψ is capable of forming equivalent hydrogen-bond interactions with adenosine, for example, regardless of whether it adopts a typical anti conformation or the atypical syn conformation (Figure 7 ). Indeed, a Ψ:U mismatch can form two distinct structures with different hydrogen-bonding interactions (Kierzek et al., 2014). On the other hand, the introduction of a methyl group to N1 of m1Ψ disrupts the ability of N1 to donate a hydrogen bond, limiting the types of pairings that m1Ψ can form with other bases (Figure 7). Furthermore, better base stacking by Ψ has been suggested to play a role in its ability to stabilize RNA duplexes (Davis, 1995; Deb et al., 2019; Hudson et al., 2013; Kierzek et al., 2014), and as such, it is likely that m1Ψ is incapable of equivalent stacking interactions. Even so, the lack of a noticeable effect by m1Ψ on decoding and the superior in vivo characteristics of m1Ψ-modified mRNAs support usage of this modification in mRNA-based therapeutics.
Figure 7.
Ψ can form additional base-pairing interactions with adenosine while m1Ψ cannot
U-A, Ψ-A, and m1Ψ-A base pairs shown in either the anti-anti (left) or syn-anti (right) conformation.
Limitations of the study
Here, we utilized a multidisciplinary approach to study the effects of the mRNA modification m1Ψ on the decoding process by the ribosome, as well as on cDNA synthesis by reverse transcriptases. However, our approach does have some limitations. For instance, although the use of the bacterial reconstituted system allowed us to systematically examine how m1Ψ alters tRNA selection by the ribosome, our observations may not be completely reflective of occurrences during translation in eukaryotes. In addition, the task of detecting miscoded peptides by MS is a difficult one. We focused our analysis to those miscoded peptides detected with high confidence to investigate how the modification affects translational fidelity in a clinically relevant manner. As such, we may have missed other miscoded peptides due to complexities with peak assignment for low-frequency events. Although such rare events would have minimal impact on application of the modification in therapeutics, more extended characterization of these events may shed insights on how the modification impacts local RNA structure on the ribosome.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti-mouse IgG-HRP | Thermo Fisher Scientific | Cat#31430; RRID:AB_228307 |
| Anti-FLAG M2 magnetic beads | Millipore Sigma | Cat#M8823; RRID:AB_2637089 |
| Anti-Renilla Luciferase antibody | Millipore Sigma | Cat#MAB4400; RRID:AB_95116 |
| Anti-β actin mouse monoclonal antibody | Cell Signaling Technology | Cat#12262; RRID:AB_2566811 |
| Anti-FLAG M2 mouse monoclonal antibody | Millipore Sigma | Cat#F1804; RRID:AB_262044 |
| Bacterial and virus strains | ||
| Escherichia coli (Migula) Castellani and Chalmers | ATCC | Cat#29417 |
| Chemicals, peptides, and recombinant proteins | ||
| Pseudo-UTP | APExBio | Cat#B7972 |
| N1-Methylpseudo-UTP | APExBio | Cat#B8049 |
| tRNA from E. coli MRE 600 | Roche | Cat#10109541001 |
| Passive Lysis Buffer | Promega | Cat#E1941 |
| M-MLV Reverse Transcriptase | Promega | Cat#M1701 |
| AMV Reverse Transcriptase | Promega | Cat#M5101 |
| EasyTag™ L-[35S]-Methionine | PerkinElmer | Cat#NEG709A500UC |
| ATP, [γ-32P] | PerkinElmer | Cat#NEG002A100UC |
| 3′-O-Me-m7G(5’)ppp(5′)G RNA Cap Structure Analog | NEB | Cat#S1411S |
| MEGAscript T7 Transcription Kit | Invitrogen | Cat#AM1334 |
| T4 RNA Ligase 2, truncated | NEB | Cat#M0242S |
| T4 Polynucleotide Kinase | NEB | Cat#M0201S |
| CircLigase ssDNA Ligase | Lucigen | Cat#CL4111K |
| Nuclease P1 from Penicillium citrinum | Millipore Sigma | Cat#N8630 |
| Calf Intestinal Phosphatase | NEB | Cat#M0290 |
| Critical commercial assays | ||
| Dual-Luciferase Reporter Assay System | Promega | Cat#E1910 |
| Wheat Germ Extract | Promega | Cat#L4380 |
| Deposited data | ||
| Mass Spectrometry Data | This Study | ProteomeXchange: PXD029291 |
| NGS Data for reverse transcriptase error | This Study | GEO: GSE186464 |
| Mendeley Data | This Study | https://doi.org/10.17632/m5sjksybcn.1 |
| Experimental models: Cell lines | ||
| Flp-In-T-REx-293 | Thermo Fisher Scientific | Cat#R78007; RRID:CVCL_U427 |
| Oligonucleotides | ||
| /5Phos/AGATCGGAAGAGCGTCGTGTAG GGAAAGAGT GTAGATCTCGGTGGTCG C/iSp18/CACTCA/iSp18/TTCAGACGTG TGCTCTTCCGATCTATTGATGGTG CCTACAG |
IDT | N/A |
| 5′rAppCTGTAGGCACCATCAAT/3ddC/3′ | Millipore Sigma | N/A |
| 5′-CAGAGGAGGUAAAA AAAUGUACUUGUACAAA-3′ |
Millipore Sigma | N/A |
| 5′-CAGAGGAGGUAAAA AAAUGYACUUGUACAAA-3′ |
Dharmacon | N/A |
| 5′ CAGAGGAGGUAAAAAA AUG m1YAC UUG UACAAA 3′ | Genelink | N/A |
| Recombinant DNA | ||
| Pfizer-Spike_pUC57 | GenScript; See Table S1 for sequence and primers | N/A |
| Dual Luciferase Reporter | This Work; See Table S1 for sequence and primers | N/A |
| Software and algorithms | ||
| Proteome Discoverer | Thermo Fisher Scientific | Cat#OPTON-31040; RRID:SCR_014477 |
| GraphPad Prism | GraphPad | RRID:SCR_002798 |
| deepSNV | Bioconductor | RRID:SCR_006214 |
| ImageQuant TL | Cytiva | RRID:SCR_018374 |
| Scaffold | Proteome Software | RRID:SCR_014345 |
| MSnbase | Bioconductor | RRID:SCR_019317 |
| FastQC | N/A | RRID:SCR_014583 |
| Samtools | N/A | RRID:SCR_002105 |
| NGmerge | N/A | |
| Cutadapt | N/A | RRID:SCR_011841 |
| BBMap | N/A | RRID:SCR_016965 |
| Image Lab | Bio-Rad | RRID:SCR_014210 |
| SkyLine | University of Washington | RRID:SCR_014080 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Hani Zaher (hzaher@wustl.edu).
Materials availability
Plasmids generated in this study are available from the lead contact.
Experimental model and subject details
Cell culture conditions
Flp-In T-REx 293 cells (Thermo Fischer) were cultured using standard protocols in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco) and supplemented with 10% heat-inactivated FBS (Gibco), 1 × Penicillin-Streptomycin-Glutamine (Gibco) and 1 × MEM Non-Essential Amino Acids (Gibco). Cells were tested for mycoplasma contamination using the Universal Mycoplasma Detection Kit (ATCC) every 6 months.
Method details
Purification of E. Coli ribosomes and translation factors
70S tight-couple ribosomes were purified from Escherichia coli MRE600 (ATCC29417) as described (Moazed et al., 1986). Translation factors were purified as previously described (Zaher and Green, 2009).
mRNAs used for assays
Unmodified control mRNA (5′-CAGAGGAGGUAAAAAAAUGUACUUGUACAAA-3′; coding sequence underlined) was purchased from Millipore Sigma. Modified mRNAs containing Ψ (5′ CAGAGGAGGUAAAAAA AUG ΨAC UUG UACAAA 3′) and m1Ψ (5′ CAGAGGAGGUAAAAAA AUG m1ΨAC UUG UACAAA 3′) were purchased from Dharmacon and GeneLink, respectively.
Validation of RNA modifications by liquid chromatography-mass spectrometry/mass spectrometry
Free adenosine, guanosine, and cytosine standards were purchased from Acros Organics and uridine was purchased from Tokyo Chemical Industry. To generate Ψ and m1Ψ standards, 50 nmol of UTP (NEB), ΨTP (ApexBio), and m1ΨTP (ApexBio) were diluted to a final volume of 25 μL in 100 mM Tris pH 7.5 buffer and incubated with 10 U of calf intestinal alkaline phosphatase (CIP; NEB) at 37°C for 4 h. For analysis of the synthetic RNA, 1 nmol of synthetic RNA was digested by nuclease P1 (Millipore Sigma, 10 Units) at 50°C for 150 min. Afterward, Tris pH 7.5 was added to a final concentration of 100 mM to adjust the pH and 10 U of CIP was added. The reaction was incubated at 37°C for an additional 90 min to dephosphorylate the nucleotide 5′-monophosophates. For all reactions treated with CIP, the resulting nucleosides were filtered using a 0.22 μm pore size syringe filter. To run each sample, 10 μL was loaded onto a Zorbax Eclipse Plus C18 column (2.1 × 50 mm, 1.8 micron) paired with an Agilent 6490 QQQ triple-quadrupole LC mass spectrometer. Runs were analyzed using multiple-reaction monitoring in positive-ion mode. The transitions used were: 268.1→136 (A), 244.1→112 (C), 284.2→152 (G), 245.1→113 (U), 245.1→125 (Ψ), 259.1→139 (m1Ψ).
Charging of tRNAs
f-[35S]-Met-tRNAfMet was prepared as previously described (Walker and Fredrick, 2008). The remaining tRNAs were aminoacylated by incubating 150 μM total RNA mix (Roche) with 0.4 mM of the appropriate amino acid, ∼5 μM tRNA synthetase, and 2 mM ATP in charging buffer (100 mM K-HEPES pH 7.6, 20 mM MgCl2, 10 mM KCl, and 1 mM DTT) at 37°C for 30 min. Aminoacylated tRNAs were then purified by phenol/chloroform extraction, ethanol precipitated, and resuspended in aa-tRNA buffer (20 mM KOAc pH 5.2 and 1 mM DTT).
Formation of ribosome initiation complexes
Initiation complexes (IC) were prepared as previously described (Pierson et al., 2016). Briefly, 2 μM 70S ribosomes, 3 μM of each IF1, IF2, IF3, [35S]-fMet-tRNAfMet, 2 mM GTP, and 6 μM of mRNA was incubated in 1 × polymix buffer (Jelenc and Kurland, 1979) (95 mM KCl, 5 mM NH4Cl, 5 mM Mg(OAc)2, 0.5 mM CaCl2, 8 mM putrescine, 1 mM spermidine, 10 mM K2HPO4 pH 7.5, 1 mM DTT) at 37°C for 30 min. Initiation complexes were then isolated via centrifugation over sucrose cushion (composed of 1.1 M sucrose, 20 mM Tris-HCl pH 7.5, 500 mM NH4Cl, 0.5 mM EDTA, and 10 mM MgCl2) for 2 h at 287,000 × g at 4°C. Resultant pellets were resuspended in 1 × polymix buffer and stored at −80°C. The radioactivity of the resuspended pellet was compared to input radioactivity in order to determine the concentration of the prepared IC.
Kinetics of peptidyl transfer
To prepare ternary complexes, EF-Tu, and GTP were incubated for 15 min at 37°C. aa-tRNA was added to the mixture, and the resulting ternary complex was incubated for an additional 15 min. The final concentration of EF-Tu, GTP, and aa-tRNA was 30 μM, 2 mM, and ∼5 μM, respectively. Kinetics of peptidyl transfer was performed by mixing equivalent volume of IC (1 μM) and TC at 37°C; either manually or using an RQF-3 quench flow instrument, depending on how fast the reaction was. The reaction was stopped at different time points using KOH to a final concentration of 500 mM. Dipeptide products were separated on cellulose TLC plates by electrophoresis in pyridine acetate at pH 2.8 (Youngman et al., 2004). The TLC plates were exposed to a phosphor screen overnight and the screens were imaged using a GE Typhoon scanner. All reactions were done in triplicate.
fMet release assay
Peptide release assays were performed in polymix buffer at 37°C. Equal volumes of initiation complexes and methylated release factor (either RF1 or RF2) (Pierson et al., 2016) were mixed to a final concentration of 0.5 μM and 10 μM, respectively. The reaction was stopped by adding twice the volume of the reaction of 50 mM sodium acetate pH 5.0 containing 40 mM EDTA at varying time points. Released f-[35S]-Met was separated from f-[35S]-Met-tRNAfMet by electrophoretic TLC and imaged via phosphorimaging as above. All reactions were done in triplicate.
In vitro transcription of Pfizer spike protein mRNA
The sequence corresponding to the complete BNT162b2 mRNA Covid-19 vaccine (Pfizer) was modified to encode a SARS-CoV-2 spike protein with an N-terminal His tag and a C-terminal FLAG tag. The rest of the CDS sequence, the 5′-UTR, 3′-UTR, and the polyA sequence remained unchanged (Table S1). A plasmid containing the entire sequence under a T7 promoter was synthesized by GenScript and the sequence was verified using Sanger sequencing. The DNA template for in vitro transcription was generated by PCR amplification using primers described in Table S1. Capped mRNAs were generated using the MEGAscript T7 Kit (Thermo Fisher Scientific) according to manufacturer’s instructions, except that GTP concentration was reduced from 7.5 mM to 1.5 mM and cap analogue 3′-O-Me-m7G(5’)ppp(5′)G (NEB) was added to a final concertation of 6 mM. To generate Ψ and m1Ψ-modified mRNAs, UTP was substituted with ΨTP or m1ΨTP, respectively. Following transcription, mRNAs were purified using the LiAC method per manufacturer’s instructions, and their integrity assessed using denaturing formaldehyde agarose electrophoresis (Sambrook and Russell, 2006).
Expression and purification of spike protein in HEK 293 cells
90% confluent cells were washed with 1 × PBS, harvested, and resuspended in 1 × PBS at a density of ∼2 × 107 cells/mL. 5 μg of RNA or 4 μg of pCDNA5-EGFP was mixed with 100 μL of cells. Electroporation was carried out using a Neon Transfection System (Thermo Fisher Scientific) as per manufacturer’s instructions. Following electroporation, cells were allowed to recover in 1 mL DMEM media with 10% FBS without antibiotics for 30 min at 37°C, before plating them on a 10 cm dish and incubating an additional 24 h at 37°C. Media was then removed and cells were collected in 10 mL conical tubes, washed with PBS, and lysed in 1 mL of FLAG-IP buffer (50 mM HEPES pH 7.5, 0.3 M KCl, 10% glycerol, 0.5% Triton, 1 mM EDTA, 0.5 mM DTT and supplemented with protease inhibitor from Gold Biotechnology). 20 μL was used for immunoblot analysis and the rest was flash frozen in liquid nitrogen. For purification, the lysate was thawed on ice and applied to 100 μL of anti-FLAG magnetic beads (Millipore Sigma). The beads were washed 5 times with FLAG-IP buffer without detergent and stored at −80°C prior to on-bead trypsin digest. All reactions were done in triplicate.
On-bead digestion and LC-MS/MS analysis of spike protein produced in HEK 293 cells
Beads were reduced with 10 mM TCEP and alkylated with 25 mM Iodoacetamide, followed by digestion with trypsin at 37°C overnight. The digest was separated from the beads using a magnetic stand and acidified with 1% TFA before being cleaned up with C18 tip. The extracted peptides were dried down and each sample was resuspended in 10 μL 5% ACN/0.1% FA. 5 μL was analyzed by LC-MS using a Dionex RSLCnano HPLC coupled to a Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific) using a 2-h gradient. Peptides were resolved using a 75 μm × 50 cm PepMap C18 column (Thermo Fisher Scientific).
Peptides were eluted at 300 nL/min from a 75 μm × 50 cm PepMap C18 column (Thermo Fisher Scientific) using the following gradient: time = 0–4 min, 2% B isocratic; 4–8 min, 2–10% B; 8–83 min, 10–25% B; 83–97 min, 25–50% B; 97–105 min, 50–98% B. Mobile phase A consisted of 0.1% formic acid and mobile phase B consisted of 0.1% formic acid in acetonitrile. The instrument was operated in data-dependent acquisition mode in which each MS1 scan was followed by higher-energy collisional dissociation (HCD) of as many precursor ions in a 2 s cycle (top speed method). The mass range for the MS1 done using the FTMS was 365–1800 m/z with resolving power set to 60,000 @ 400 m/z and the automatic gain control (AGC) target set to 1,000,000 ions with a maximum fill time of 100 ms. The selected precursors were fragmented in the ion trap using an isolation window of 1.5 m/z, an AGC target value of 10,000 ions, a maximum fill time of 100 ms, a normalized collision energy of 35, and activation time of 30 ms. Dynamic exclusion was performed with a repeat count of 1, exclusion duration of 30 s, and a minimum MS ion count for triggering MS/MS set to 5000 counts.
Identification of amino acid substitutions in the HEK 293 dataset
Two strategies were utilized to identify possible miscoded peptides. In the first approach, raw MS files were searched using the MASCOT search engine (version 2.5) against the Pfizer spike protein sequence using an error-tolerant search allowing for all amino acid substitutions. Mascot search parameters included Trypsin/P specificity, up to 2 missed cleavages, a fragment ion mass tolerance of 0.60 Da and a parent ion tolerance of 10 ppm, and variable modifications of carbamidomethyl cysteine and oxidized methionine. In the second approach, raw MS files were searched against an in silico generated SARS-CoV-2 spike protein library consisting of every possible single substitution protein product arising from a miscoding event at each uridine in the sequence. Searches against the computationally generated library with the Mascot search engine were launched in Proteome Discoverer (version 2.4) using the same parameters as the first approach. The sequences of candidate miscoded peptides identified from both methods, as determined by greater than 90% peptide probability in Scaffold (version 4.8.2) with Mascot Ion Score >15, were concatenated to a mock sequence for a final confirmation search. The final search was performed using the MASCOT search engine launched in Proteome Discoverer against the Human reference proteome (Uniprot.org, downloaded 05/2019) modified to include the mock sequence and the wild-type Pfizer spike protein sequence. Mascot results of the final search were loaded into Skyline-daily (University of Washington, version 20) to check proper peak picking for each peptide of interest. Each peak was manually inspected for good fragment ion coverage and elution times in line with the MS/MS identification time of the peptide. The sum of the top 3 isotopes were then exported for each wild-type and miscoded peptide for further analysis.
Dual-luciferase reporter assays in HEK 293 cells
Dual-luciferase reporter mRNAs were transcribed in vitro using the same method as for the spike protein mRNA. The AAA codon coding for K529 was mutated to AAU using site-directed mutagenesis. RNAs were electroporated into HEK 293 cells using a similar method as for the spike protein mRNA, except cells were cultured in 6-well plates instead of dishes and RNA amounts were scaled down accordingly. After recovery, cells were lysed using passive lysis buffer (Promega) and luminescence was measured using the Dual-Luciferase Reporter Assay System (Promega) on a Tecan plate reader equipped with an automated injection system. All reactions were done in triplicate.
In vitro translation of spike protein using wheat germ extracts
Wheat-germ extracts were purchased from Promega and used per manufacturer’s instructions. For 35S-incorporation assays, translation reactions (10 μL volume) were initiated in the presence of 6 μCi of EasyTag L-[35S]-Methionine (PerkinElmer). Reactions were stopped by adding an equivalent volume of HU buffer (8 M Urea, 5% SDS, 200 mM Tris pH 6.8, 100 mM DTT). Protein products were resolved using 8% SDS PAGE and transferred to a PVDF membrane using a semi-dry transfer apparatus (Bio-Rad). Protein products were visualized using phosphorimaging as described earlier. The PVDF membrane was then subjected to immunoblotting using anti-FLAG (Millipore Sigma) and anti-Renilla luciferase (Millipore Sigma) antibodies.
For spike protein purification from wheat-germ extracts, the reaction volume was increased to 400 μL and unlabeled methionine was used. Following incubation, the reaction was diluted to 5 mL in FLAG-IP buffer and incubated with 50 μL of prewashed anti-FLAG Agarose beads (Millipore Sigma) for 3 h at 4°C. Beads were washed 4 times with FLAG-IP buffer in spin columns. Bound proteins were eluted using HU buffer and resolved on 8% SDS PAGE. The bands corresponding to the size of spike protein were cut and subjected to in-gel trypsin digestion. All reactions were done in duplicates.
In-gel digestion and LC-MS/MS analysis of spike protein produced in wheat germ extracts
Coomassie-stained gel bands containing the spike protein were excised and cut into smaller pieces. Gel slices were destained twice with 25 mM ammonium bicarbonate in 50% acetonitrile for 30 min at 37°C. Gel slices were then reduced with 50 mM DTT in 50 mM ammonium bicarbonate at 60°C for 10 min, followed by alkylation with 200 mM chloroacetamide in 50 mM ammonium bicarbonate in the dark at room temperature for 1 h. Gel slices were washed twice with 25 mM ammonium bicarbonate in 50% acetonitrile for 30 min at 37°C and then dehydrated with 100% acetonitrile for 10 min at room temperature. Excess solvent was removed by speed-vacuum. After dehydration, gel slices were rehydrated in a solution of 0.01 mg/mL trypsin in 100 mM ammonium bicarbonate and digested for 16 h at 37°C. Digested peptides were extracted using a 50% acetonitrile, 0.1% trifluoracetic acid solution by incubation at 37°C for 10 min. The extraction process was repeated two additional times and the three extractions were pooled with the digest solution. Samples were dried completely in a speed-vacuum and resuspended in ∼0.1–1% trifluoroacetic acid to achieve a pH of ∼3. Samples were then concentrated and desalted using OMIX C18 pipette tips (Agilent) and eluted with a 0.1% acetic acid, 75% acetonitrile solution.
Prepared peptides were analyzed with a Q-Exactive Plus mass spectrometer (Thermo Fisher Scientific) after reversed-phase nano-HPLC separation with a 25-cm analytical C18 resin column (Acclaim PepMap RSLC; Thermo Fisher Scientific) and a 5 to 95% acetonitrile step gradient in 0.1% formic acid (FA) at a flow rate of 250 nL/min for 70 min. The mass spectrometer was operated in the data-dependent mode to automatically switch between full-scan MS and MS/MS acquisition. Data-dependent acquisitions were obtained using Xcalibur 4.0 software in positive-ion mode. MS1 spectra were measured at a resolution of 70,000 with an automatic gain control of 1 × 106, a maximum ion time of 40 msec, and a mass range of 300-1,800 m/z. Up to 12 MS2 scans, with a charge state of 2–4, were triggered at a resolution of 17,500, an automatic gain control of 5 × 105 with a maximum ion time of 120 msec, a 1.6-m/z isolation window, and a normalized collision energy of 28. MS1 scans that triggered MS2 scans were dynamically excluded for 30 s.
Identification of amino acid substitutions in the wheat germ extract dataset
The resulting MS datasets were searched against an artificially generated SARS-COV-2 spike protein database, consisting of every possible single substitution product arising from a miscoding event at each uridine in the sequence, using Proteome Discoverer (version 2.5.0.400; Thermo Fisher Scientific) and a list of common protein contaminants. Peptides were assigned by SEQUEST HT, allowing a maximum of two missed tryptic cleavages, a minimum peptide length of 6, a precursor mass tolerance of 10 ppm, and fragment mass tolerances of 0.02 Da. Carbamidomethylation of Cys and oxidation of Met were specified as static and dynamic modifications, respectively. Only peptides where both the wild-type peptide and the substitution product were detected were further analyzed.
Duplex-RNA melting analysis
A Varian Cary-100 spectrophotometer with a Peltier-controlled cuvette holder was used to monitor the change in absorbance at a wavelength of 260 nm. RNAs were first annealed in a reaction containing equal molarities of each RNA, calculated from optical density measurements and absorbance values at 260 nm, with the goal of having a final absorbance value for the annealed RNA of about 0.5–0.6 at room temperature. RNAs were heated in water to 80°C for 5 min to denature. They were then cooled to room temperature and buffer was added to a final concentration of 50 mM Tris-HCl, 100 mM NaCl, 10 mM MgCl2 pH 7.9. Annealed RNAs were then incubated for 3 min at the starting temperature of 40°C in the Varian Cary-100 spectrophotometer. Following incubation, the temperature ramp was set to 1°C/min and absorbance values were measured in 0.2°C increments until the final temperature of 96°C was reached. All assays were done in triplicate.
Primer-extension assays
50 nmol of primer was 5′-end labeled using ∼70 mCi [γ-32P] ATP (6000 Ci/mmol) and T4 polynucleotide kinase (PNK) in a total reaction volume of 10 μL. The reaction was incubated at 37°C for 30 min, and then at 65°C for 15 min to inactivate PNK before diluting to a final volume of 50 μL. Radiolabeled primer and synthetic RNA, at ∼70 nM and ∼4 uM, respectively, were reverse transcribed with Promega M-MLV according to manufacturer instructions, using the radiolabeled primer and only providing a single deoxy nucleotide triphosphate (either adenosine, guanosine, cytosine, or thymidine), no dNTPs, or all four dNTPs. Samples were separated by urea denaturing polyacrylamide gel electrophoresis and imaged on a GE Typhoon scanner. All reactions were done in duplicate.
cDNA library generation for high-throughput sequencing
We used a short-RNA cloning protocol similar to the one used for ribosome profiling (Ingolia et al., 2009). Briefly, ∼100 pmole of the synthetic mRNAs used in our reconstituted translation reactions (unmodified, pseudouridine-containing, and N1-methylpseudouridine-containing) were ligated to 150 pmole of short adenylated DNA oligonucleotide, 5′rAppCTGTAGGCACCATCAAT/3ddC/3′, at their 3′ end using truncated T4 RNA ligase 2 (NEB) in a total volume of 10 μL. The ligated products were purified using denaturing urea PAGE and subjected to reverse transcription using M-MLV (Promega) or AMV (Promega) enzymes and RS-1 primer (/5Phos/AGATCGGAAGAGCGTCGTGTAGGGAAAGAGT GTAGATCTCGGTGGTCGC/iSp18/CACTCA/iSp18/TTCAGACGTGTGCTCTTCCGATCTATTGATGGTGCCTACAG). Following PAGE purification, cDNA products were circularized using CircLigase kit (Epicentre). Pilot PCR was then used to determine the optimal numbers of cycles required to amplify the cDNA. Following PCR amplification with unique barcoded primers, the DNA libraries were purified using native PAGE and then analyzed for length and purity using Agilent Bioanalyzer.
Sequencing and bioinformatic analysis
Prepared cDNAs were sequenced as paired 75-nt NextSeq runs at the Genome Technology Access Center (GTAC) of Washington University in St. Louis. Samples were demultiplexed based on their 6-nt barcode and checked for initial quality using FastQC (Andrews et al., 2010). Overhangs from each paired end set were removed, such that only consensus sequence between each mate pair remained, and then merged to form a single consensus read using NGmerge (Gaspar, 2018). Stitched reads were processed with Cutadapt (Martin, 2011) to remove the 17-nt linker sequence and any reads not containing the linker sequence were discarded. Reads were mapped to the unmodified control RNA sequence using BBMap (Bushnell and Work, 2014) in local mode with “very slow” settings. Mapped sam files were sorted, converted to the bam format, and indexed using Samtools. Reads were analyzed using deepSNV (Gerstung et al., 2014), with only mapped reads having high mapping quality (MAPQ ≥ 20 out of 41) and high-quality bases (Q ≥ 30) counted. Error rates for the nucleotides surrounding the modification (A T G X A C, where X is the modified nucleotide) were analyzed. Deletions were ignored as they appeared to be an artifact of sequencing; deletions were completely absent when mapping quality filter was set to ≥25.
Quantification and statistical analysis
Quantification of RNA modifications
The presence of modified nucleosides in the synthetic RNAs was checked by comparing retention times and nucleoside to base ion mass transitions to those of the standards. The nucleoside composition of each synthetic RNA was calculated using the diode array detector (DAD) signal intensities as follows: the integrated peak area of each nucleoside in the synthetic RNA (A, U, C, and G) was compared to the integrated peak area of one of the other nucleosides in the synthetic RNA, and then normalized to the corresponding ratio of areas in the standards (eg. ). The normalized value was then multiplied by the reference count (A: 15, U: 6, C: 3, G: 7) to generate a count value. The process was repeated for the other two nucleoside comparisons and the average of the three counts was used as the final count for each nucleoside.
Analysis of peptidyl transfer kinetics and fMet release assays
Imaged phosphor screens were quantified using Image Lab (Bio-Rad). Fractional radioactivity corresponding to the dipeptides were plotted against time, and observed rates and endpoints were determined using a one-phase association equation in Prism (GraphPad). Differences in observed rates and endpoints were tested for statistical significance using an unpaired t test in Prism.
Quantification of spike protein in HEK 293 cells
Spike protein detected on the immunoblot was quantified by normalizing the FLAG signal to actin signal using ImageQuant (Cytiva). Differences in normalized signal were tested for statistical significance by one-way ANOVA in Prism (GraphPad).
Quantification of amino acid substitutions in the HEK 293 dataset
Relative abundance was calculated by comparing the label-free quantitation value of each miscoded peptide to the quantitation value of the parent, faithful peptide, as determined by manual peak picking in Skyline.
Analysis of dual-luciferase reporter assays in HEK 293 cells
Firefly luminescence values were normalized to corresponding in-frame, unaltered Renilla luminescence values. Differences between normalized values were tested for statistical significance using an unpaired t test in Prism (GraphPad).
Quantification of amino acid substitutions in the wheat germ extract dataset
Relative abundance was calculated by comparing the label-free quantitation value of each miscoded peptide to the value of the parent, faithful peptide. In the event only one of the technical replicate values for a peptide was missing, quantitation values were log2 transformed and the missing values imputed using a maximum likelihood-based method from the MSnbase R package (Gatto and Lilley, 2012; Gatto et al., 2021).
Duplex-RNA melting point determination
To determine the melting point, absorbance values were first normalized to the initial absorbance value. The first derivative of the data was calculated and fit to a Gaussian function to find the peak, which corresponds to the melting point of the duplex, using Prism (GraphPad). Differences in melting point were tested for statistical significance using an unpaired t test in Prism (GraphPad).
Acknowledgments
We thank the lab of Dr. Roberto Galletto for their assistance in obtaining the UV-melt data. We thank the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital in St. Louis, MO, USA, and the Institute of Clinical and Translational Sciences (ICTS) at Washington University in St. Louis for the use of the Genome Technology Access Center, which provided sequencing services. The Siteman Cancer Center is supported in part by an NCI Cancer Center Support Grant #P30 CA091842, and the ICTS is funded by the National Institutes of Health’s NCATS Clinical and Translational Science Award (CTSA) program grant #UL1 TR002345. This work was supported by a grant from the National Institutes of Health to H.S.Z. (R01GM141474).
Author contributions
K.Q.K., B.D.B., S.-C.T., C. Jing, C. Jungers, J.Z., L.L.Y., S.D., and H.S.Z. carried out the experimental work. K.Q.K. and H.S.Z. conceived the work. K.Q.K., B.D.B., R.D.V., and H.S.Z. wrote the manuscript. H.S.Z. supervised the work.
Declaration of interests
The authors declare no competing interests.
Published: August 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111300.
Supplemental information
Data and code availability
-
•
The mass spectrometry data are available in the ProteomeXchange database under accession number PXD029291 within the PRIDE repository. The reverse transcriptase sequencing data is available at GEO under accession number GSE186464. Both are publicly available as of the date of publication and accession numbers are listed in the key resources table. Source data and original immunoblots and phosphorimages have been deposited at Mendeley at https://doi.org/10.17632/m5sjksybcn.1 and are publicly available as of the date of publication. The DOI is also listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Adachi H., Yu Y.T. Pseudouridine-mediated stop codon readthrough in s. cerevisiae is sequence context–independent. RNA. 2020;26:1247–1256. doi: 10.1261/rna.076042.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S., Uematsu S., Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Anderson B.R., Muramatsu H., Nallagatla S.R., Bevilacqua P.C., Sansing L.H., Weissman D., Karikó K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010;38:5884–5892. doi: 10.1093/nar/gkq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S., Krueger F., Segonds-Pichon A., Biggins L., Krueger C., Wingett S. 2010. FastQC. [Google Scholar]
- Andries O., Mc Cafferty S., De Smedt S.C., Weiss R., Sanders N.N., Kitada T. N1-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control Release. 2015;217:337–344. doi: 10.1016/j.jconrel.2015.08.051. [DOI] [PubMed] [Google Scholar]
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baronti L., Karlsson H., Marušič M., Petzold K. A guide to large-scale RNA sample preparation. Anal. Bioanal. Chem. 2018;410:3239–3252. doi: 10.1007/s00216-018-0943-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell B., Work R. 2014. BBMap: A Fast, Accurate, Splice-Aware Aligner; pp. 3–5. [Google Scholar]
- Damase T.R., Sukhovershin R., Boada C., Taraballi F., Pettigrew R.I., Cooke J.P. The limitless future of RNA therapeutics. Front. Bioeng. Biotechnol. 2021;9:628137–628224. doi: 10.3389/fbioe.2021.628137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis D.R. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 1995;23:5020–5026. doi: 10.1093/nar/23.24.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb I., Popenda Ł., Sarzyńska J., Małgowska M., Lahiri A., Gdaniec Z., Kierzek R. Computational and NMR studies of RNA duplexes with an internal pseudouridine-adenosine base pair. Sci. Rep. 2019;9:16278–16313. doi: 10.1038/s41598-019-52637-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis P.P., Bremer H. Differential rate of ribosomal protein synthesis in Escherichia coli B/r. J. Mol. Biol. 1974;84:407–422. doi: 10.1016/0022-2836(74)90449-5. [DOI] [PubMed] [Google Scholar]
- Eyler D.E., Franco M.K., Batool Z., Wu M.Z., Dubuke M.L., Dobosz-Bartoszek M., Jones J.D., Polikanov Y.S., Roy B., Koutmou K.S. Pseudouridinylation of mRNA coding sequences alters translation. Proc. Natl. Acad. Sci. USA. 2019;116:23068–23074. doi: 10.1073/pnas.1821754116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández I.S., Ng C.L., Kelley A.C., Wu G., Yu Y.T., Ramakrishnan V. Unusual base pairing during the decoding of a stop codon by the ribosome. Nature. 2013;500:107–110. doi: 10.1038/nature12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund I., Eigenbrod T., Helm M., Dalpke A.H. RNA modifications modulate activation of innate toll-like receptors. Genes. 2019;10:E92. doi: 10.3390/genes10020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar J.M. NGmerge: merging paired-end reads via novel empirically-derived models of sequencing errors. BMC Bioinf. 2018;19:536. doi: 10.1186/s12859-018-2579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto L., Lilley K.S. MSnbase-an R/Bioconductor package for isobaric tagged mass spectrometry data visualization, processing and quantitation. Bioinformatics. 2012;28:288–289. doi: 10.1093/bioinformatics/btr645. [DOI] [PubMed] [Google Scholar]
- Gatto L., Gibb S., Rainer J. MSnbase, efficient and elegant R-based processing and visualization of raw mass spectrometry data. J. Proteome Res. 2021;20:1063–1069. doi: 10.1021/acs.jproteome.0c00313. [DOI] [PubMed] [Google Scholar]
- Gerstung M., Papaemmanuil E., Campbell P.J. Subclonal variant calling with multiple samples and prior knowledge. Bioinformatics. 2014;30:1198–1204. doi: 10.1093/bioinformatics/btt750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan S., Rosenecker J. Nanotechnologies in delivery of mRNA therapeutics using nonviral vector-based delivery systems. Gene Ther. 2017;24:133–143. doi: 10.1038/gt.2017.5. [DOI] [PubMed] [Google Scholar]
- Van Hoecke L., Roose K. How mRNA therapeutics are entering the monoclonal antibody field. J. Transl. Med. 2019;17:54. doi: 10.1186/s12967-019-1804-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoernes T.P., Clementi N., Faserl K., Glasner H., Breuker K., Lindner H., Hüttenhofer A., Erlacher M.D. Nucleotide modifications within bacterial messenger RNAs regulate their translation and are able to rewire the genetic code. Nucleic Acids Res. 2016;44:852–862. doi: 10.1093/nar/gkv1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoernes T.P., Heimdörfer D., Köstner D., Faserl K., Nußbaumer F., Plangger R., Kreutz C., Lindner H., Erlacher M.D. Eukaryotic translation elongation is modulated by single natural nucleotide derivatives in the coding sequences of mRNAs. Genes. 2019;10:E84. doi: 10.3390/genes10020084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson G.A., Bloomingdale R.J., Znosko B.M. Thermodynamic contribution and nearest-neighbor parameters of pseudouridine-adenosine base pairs in oligoribonucleotides. RNA. 2013;19:1474–1482. doi: 10.1261/rna.039610.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia N.T., Ghaemmaghami S., Newman J.R.S., Weissman J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelenc P.C., Kurland C.G. Nucleoside triphosphate regeneration decreases the frequency of translation errors. Proc. Natl. Acad. Sci. USA. 1979;76:3174–3178. doi: 10.1073/pnas.76.7.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong D.-E., McCoy M., Artiles K., Ilbay O., Fire A., Nadeau K., Park H., Betts B., Boyd S., Hoh R., et al. 2021. Assemblies-of-putative-SARS-CoV2-spike-encoding-mRNA-sequences-for-vaccines-BNT-162b2-and-mRNA-1273. [Google Scholar]
- Jirikowski G.F., Sanna P.P., Maciejewski-Lenoir D., Bloom F.E., Paolo S.P., Dominique M.-L. Reversal of diabetes insipidus in Brattleboro rats: intrahypothalamic injection of vasopressin mRNA. Science. 1992;255:996–998. doi: 10.1126/science.1546298. [DOI] [PubMed] [Google Scholar]
- Kaczmarek J.C., Kowalski P.S., Anderson D.G. Advances in the delivery of RNA therapeutics: from concept to clinical reality. Genome Med. 2017;9:60. doi: 10.1186/s13073-017-0450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karijolich J., Yu Y.T. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature. 2011;474:395–398. doi: 10.1038/nature10165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karikó K., Ni H., Capodici J., Lamphier M., Weissman D. mRNA is an endogenous ligand for toll-like receptor 3. J. Biol. Chem. 2004;279:12542–12550. doi: 10.1074/jbc.M310175200. [DOI] [PubMed] [Google Scholar]
- Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Karikó K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S., Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karikó K., Muramatsu H., Ludwig J., Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011;39:e142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keedy H.E., Thomas E.N., Zaher H.S. Decoding on the ribosome depends on the structure of the mRNA phosphodiester backbone. Proc. Natl. Acad. Sci. USA. 2018;115:E6731–E6740. doi: 10.1073/pnas.1721431115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierzek E., Malgowska M., Lisowiec J., Turner D.H., Gdaniec Z., Kierzek R. The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic Acids Res. 2014;42:3492–3501. doi: 10.1093/nar/gkt1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kormann M.S.D., Hasenpusch G., Aneja M.K., Nica G., Flemmer A.W., Herber-Jonat S., Huppmann M., Mays L.E., Illenyi M., Schams A., et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 2011;29:154–157. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- Kramer E.B., Farabaugh P.J. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA. 2007;13:87–96. doi: 10.1261/rna.294907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10. [Google Scholar]
- Mauger D.M., Cabral B.J., Presnyak V., Su S.V., Reid D.W., Goodman B., Link K., Khatwani N., Reynders J., Moore M.J., McFadyen I.J. mRNA structure regulates protein expression through changes in functional half-life. Proc. Natl. Acad. Sci. USA. 2019;116:24075–24083. doi: 10.1073/pnas.1908052116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D., Stern S., Noller H.F. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. Journal of Molecular Biology. 1986;187:399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- Nir R., Hoernes T.P., Muramatsu H., Faserl K., Karikó K., Erlacher M.D., Sas-Chen A., Schwartz S. A systematic dissection of determinants and consequences of snoRNA-guided pseudouridylation of human mRNA. Nucleic Acids Res. 2022;50:4900–4916. doi: 10.1093/nar/gkac347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisien Marc, Yi Chengqi, Pan Tao. Rationalization and prediction of selective decoding of pseudouridine-modified nonsense and sense codons. RNA. 2012;18:355–367. doi: 10.1261/rna.031351.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr C.J.C., Wada S., Kotake K., Kameda S., Matsuura S., Sakashita S., Park S., Sugiyama H., Kuang Y., Saito H. N 1-Methylpseudouridine substitution enhances the performance of synthetic mRNA switches in cells. Nucleic Acids Res. 2020;48:E35. doi: 10.1093/nar/gkaa070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson W.E., Hoffer E.D., Keedy H.E., Simms C.L., Dunham C.M., Zaher H.S. Uniformity of peptide release is maintained by methylation of release factors. Cell Rep. 2016;17:11–18. doi: 10.1016/j.celrep.2016.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potapov V., Fu X., Dai N., Corrêa I.R., Jr., Tanner N.A., Ong J.L. Base modifications affecting RNA polymerase and reverse transcriptase fidelity. Nucleic Acids Res. 2018;46:5753–5763. doi: 10.1093/nar/gky341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riba A., Di Nanni N., Zavolan M., Mittal N., Arhné E., Schmidt A., Zavolan M. Protein synthesis rates and ribosome occupancies reveal determinants of translation elongation rates. Proc. Natl. Acad. Sci. USA. 2019;116:15023–15032. doi: 10.1073/pnas.1817299116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U., Karikó K., Türeci Ö. MRNA-based therapeutics-developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- Green M.R., Sambrook J. Separation of RNA according to size: electrophoresis of RNA through agarose gels containing formaldehyde. Cold Spring Harb. Protoc. 2022;2022 doi: 10.1101/pdb.prot101758. pdb.prot4050. [DOI] [PubMed] [Google Scholar]
- Simms C.L., Hudson B.H., Mosior J.W., Rangwala A.S., Zaher H.S. An active role for the ribosome in determining the fate of oxidized mRNA. Cell Rep. 2014;9:1256–1264. doi: 10.1016/j.celrep.2014.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svidritskiy E., Madireddy R., Korostelev A.A. Structural basis for translation termination on a pseudouridylated stop codon. J. Mol. Biol. 2016;428:2228–2236. doi: 10.1016/j.jmb.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin Y.V., Cheng Y.M., Chakraborty T., Presnyak V., John M., Sonenberg N. N1-methyl-pseudouridine in mRNA enhances translation through eIF2α-dependent and independent mechanisms by increasing ribosome density. Nucleic Acids Res. 2017;45:6023–6036. doi: 10.1093/nar/gkx135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel U., Jensen K.F. The RNA chain elongation rate in Escherichia coli depends on the growth rate. J. Bacteriol. 1994;176:2807–2813. doi: 10.1128/jb.176.10.2807-2813.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker S.E., Fredrick K. Preparation and evaluation of acylated tRNAs. Methods. 2008;44:81–86. doi: 10.1016/j.ymeth.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman D., Ni H., Scales D., Dude A., Capodici J., McGibney K., Abdool A., Isaacs S.N., Cannon G., Karikó K. HIV gag mRNA transfection of dendritic cells (DC) delivers encoded antigen to MHC class I and II molecules, causes DC maturation, and induces a potent human in vitro primary immune response. J. Immunol. 2000;165:4710–4717. doi: 10.4049/jimmunol.165.8.4710. [DOI] [PubMed] [Google Scholar]
- Wolff J.A., Malone R.W., Williams P., Chong W., Acsadi G., Jani A., Felgner P.L. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- Young R., Bremer H. Polypeptide-chain-elongation rate in Escherichia coli B/r as a function of growth rate. Biochem. J. 1976;160:185–194. doi: 10.1042/bj1600185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman E.M., Brunelle J.L., Kochaniak A.B., Green R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell. 2004;117:589–599. doi: 10.1016/s0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]
- Zaher H.S., Green R. Quality control by the ribosome following peptide bond formation. Nature. 2009;457:161–166. doi: 10.1038/nature07582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher H.S., Green R. Hyperaccurate and error-prone ribosomes exploit distinct mechanisms during tRNA selection. Mol. Cell. 2010;39:110–120. doi: 10.1016/j.molcel.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Ieong K.W., Mellenius H., Ehrenberg M. Proofreading neutralizes potential error hotspots in genetic code translation by transfer RNAs. RNA. 2016;22:896–904. doi: 10.1261/rna.055632.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The mass spectrometry data are available in the ProteomeXchange database under accession number PXD029291 within the PRIDE repository. The reverse transcriptase sequencing data is available at GEO under accession number GSE186464. Both are publicly available as of the date of publication and accession numbers are listed in the key resources table. Source data and original immunoblots and phosphorimages have been deposited at Mendeley at https://doi.org/10.17632/m5sjksybcn.1 and are publicly available as of the date of publication. The DOI is also listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.