Abstract
The first temperature-dependent proteins (expressed at 37°C, but not 26°C) to be identified in Yersinia pestis were antigens 3 (fraction 1), 4 (pH 6 antigen), and 5 (hereafter termed KatY). Antigens 3 and 4 are now established virulence factors, whereas little is known about KatY, except that it is encoded chromosomally, produced in abundance, possesses modest catalase activity, and is shared by Yersinia pseudotuberculosis, but not Yersinia enterocolitica. We report here an improved chromatographic method (DEAE-cellulose, calcium hydroxylapatite, and Sephadex G-150) that yields enzymatically active KatY (2,423 U/mg of protein). Corresponding mouse monoclonal antibody 1B70.1 detected plasminogen activator-mediated hydrolysis of KatY, and a polyclonal rabbit antiserum raised against outer membranes of Y. pestis was enriched for anti-KatY. A sequenced ∼16-kb Y. pestis DNA insert of a positive pLG338 clone indicated that katY encodes an 81.4-kDa protein (pI 6.98) containing a leader sequence of 2.6 kDa; the deduced molecular mass and pI of processed KatY were 78.8 kDa and 6.43, respectively. A minor truncated variant (predicted molecular mass of 53.6 kDa) was also expressed. KatY is similar (39 to 59% identity) to vegetative bacterial catalase-peroxidases (KatG in Escherichia coli) and is closely related to plasmid-encoded KatP of enterohemorrhagic E. coli O157:H7 (75% identity). katY encoded a putative Ca2+-binding site, and its promoter contained three homologues to the consensus recognition sequence of the pCD-encoded transcriptional activator LcrF. rbsA was located upstream of katY, and cybB, cybC, dmsABC, and araD were mapped downstream. These genes are not linked to katG or katP in E. coli.
The genome of Yersinia pestis, the causative agent of bubonic plague, consists of a ∼4,400-kb chromosome (35) plus three recently sequenced plasmids of ∼10 kb (pPCP) (24), ∼70 kb (pCD) (24, 50), and ∼100 kb (pMT) (24, 30). Y. pestis and closely related enteropathogenic yersiniae (Y. pseudotuberculosis and Y. enterocolitica) are noted for their ability to express temperature-dependent proteins at 37°C, but not 26°C. Many of these activities are encoded by pCD (pYV in the enteropathogenic yersiniae), where they function as regulators, modulators, or effectors of virulence, termed Yops (10, 11, 47). This form of control reflects upregulation of the pCD-encoded transcriptional activator LcrF (VirF in Y. enterocolitica) at host temperature but not room temperature (9, 23, 27). At least one such LcrF-mediated function promotes restriction in vitro (i.e., prevents the occurrence of vegetative growth at 37°C) unless the concentration of Na+ is reduced to ∼10 mM (16). However, addition of the amount of Ca2+ present in mammalian vascular fluid (≥2.5 mM) downregulates LcrF (11, 47), thereby permitting the occurrence of bacterial division regardless of the concentration of Na+ (16).
The classical study of Crumpton and Davies (12) defined three major additional temperature-dependent activities, termed antigens 3, 4, and 5. These proteins correspond to the pMT-encoded (51) capsular (fraction 1) antigen of Baker et al. (2), chromosomally encoded pH 6 antigen (29, 46), and chromosomally encoded antigen E (28) or p70 (53), respectively. Fraction 1 and pH 6 antigen are established virulence factors (47). In contrast, little is known about antigen 5 (hereafter termed KatY), except that it was one of two catalases present in Y. pestis (39) and that it exhibited comparably weak enzymatic activity (39). In addition, KatY was detected in Y. pseudotuberculosis, but not Y. enterocolitica (28, 40), identified in cytoplasm and periplasm (48, 53), and produced in great abundance even during restriction of vegetative growth in Ca2+-deficient medium (39, 40).
In this report, we provide an improved method capable of yielding essentially homogeneous KatY in an enzymatically active form and demonstrate that the resulting 78.8-kDa product possesses marked homology with known bacterial catalase-peroxidases, especially plasmid-encoded KatP of enterohemorrhagic Escherichia coli O157:H7 (6). In addition, we show that KatY undergoes degradation by pPCP-encoded plasminogen activator (47) in a manner similar to that previously described for hydrolysis of Yops (26, 39, 40, 53, 54, 56). Three LcrF-like binding sites within the promoter region of KatY were identified, as were sequences encoding a putative internal Ca2+-binding site and an evident false translational start site resulting in formation of a minor truncated derivative of 53.6 kDa. Genes neighboring katY were distinct from those known to be linked to either katG or katP in E. coli.
MATERIALS AND METHODS
Bacteria.
A nonpigmented (47) isolate of Y. pestis KIM10 carrying pPCP1 and pMT1 but not pCD1 (substrain D28) was used for production of KatY in fermentor vessels. Nonpigmented substrain D27 containing pPCP, pCD, and pMT was used to prepare rabbit polyclonal antiserum. Genomic DNA comprising the library (49) used to sequence katY was prepared from a pigmented Y. pestis KIM10 derivative carrying pMT, but not pCD or pPCP (substrain D46). A nonpigmented isolate of the latter (substrain D47) was used as a control in studies characterizing pPCP-mediated degradation of KatY. A pigmented isolate carrying pPCP and pMT, but not pCD (substrain D1), was used to raise rabbit polyclonal antisera against outer membranes. Nonpigmented Y. pestis EV76 and an isogenic derivative cured of pPCP were also used to compare pPCP-mediated degradation of KatY. E. coli HB101 transformed with pLG338 containing Y. pestis DNA (substrain D46) (49) provided the insert used to identify and characterize katY. Y. pseudotuberculosis PB1/0 and a derivative transformed with pPCP have been described previously (26).
Purification of KatY.
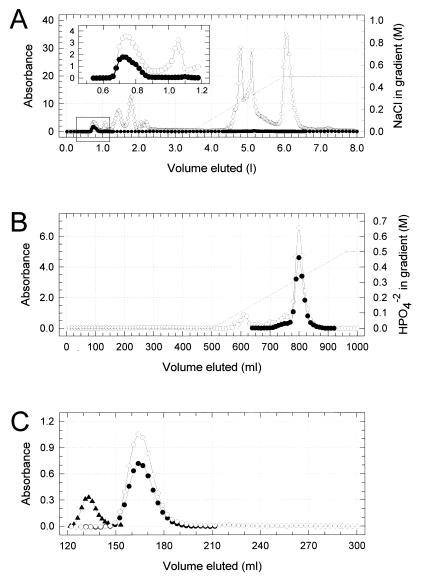
A modification of the method of Mehigh and Brubaker (39) capable of maintaining enzymatic activity was used for purification of KatY. Elution profiles and yields are shown in Fig. 1 and Table 1, respectively. Enzymatic activity at each step was determined by the method of Beers and Sizer (3) by monitoring the disappearance of peroxide at 240 nm. One unit of activity is defined as the amount of KatY needed to destroy 1 μmol of H2O2 in 1 min at 26°C (at pH 7.0). Yersiniae were cultivated in 2-liter Erlenmeyer flasks at 26°C for one transfer at 200 rpm on a New Brunswick Scientific model R-25 Gyrotory shaker (Edison, N.J.). The flasks contained a medium (250 ml each) consisting of 2.5% Sheffield NZ amine, type A (Quest International, Hoffman Estates, Ill.), the salt component (22) of Higuchi’s medium (final concentrations of 25 mM K2HPO4, 2.5 mM MgCl2, 10 mM citric acid, 0.1 mM FeSO4, and 0.01 mM MnCl2), 2.5 mM Na2S2O4, and 40 mM potassium l-gluconate, all adjusted to pH 7 with 10 M NaOH. These cultures were used to inoculate (10% [vol/vol]) 14-liter vessels containing 10 liters of the same medium at an optical density of 0.1 at 620 nm. The vessels were placed in a model MF-214 fermentor (New Brunswick Scientific), aerated at 37°C (12 liters/min at 500 rpm), and yersiniae were harvested by centrifugation (10,000 × g for 30 min at 4°C) after achieving late-logarithmic growth. After suspension in 0.033 M potassium phosphate buffer (pH 7.0 [phosphate buffer]), the bacteria were centrifuged again (10,000 × g for 30 min at 4°C) and then suspended at an optical density (620 nm) of ∼400 in 0.05 M Tris-Cl buffer (pH 7.5).
FIG. 1.
Elution profiles of pigmented material (A405) containing KatY from a cell extract of Y. pestis KIM10 (substrain D28) during stepwise chromatography on columns containing DEAE-cellulose (A [the insert illustrates details of the area of KatY elution shown in the box]), calcium hydroxylapatite (B), and Sephadex G-150 (C). A280 (○), A405 (●), and A615 for Blue dextran (▴) were used to determine the void volume for the column containing Sephadex G-150. See Materials and Methods for details.
TABLE 1.
Purification and yield of pigmented protein (A405) comprising KatY from Y. pestis KIM10 (substrain D28) cultivated at 37°C
| Preparation | Total vol (ml) | OD280 (U/ml) | Total OD280 (U) | OD405 (U/ml) | Total OD405 (U) | Protein concn (mg/ml) | Total protein concn (mg) | OD405/OD280 | OD405/mg of protein | % Yield | Fold purification |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Crude extract | 125 | 348 | 43,500 | 8.3 | 1,038 | 46.1 | 5,762.5 | 0.024 | 0.180 | 100 | 1 |
| DEAE-cellulose | 185 | 2.76 | 511 | 1.41 | 261 | 2.43 | 449.6 | 0.511 | 0.580 | 25.1 | 3.2 |
| Ca hydroxylapatite | 41 | 4.90 | 201 | 3.04 | 125 | 4.94 | 202.5 | 0.620 | 0.615 | 12.0 | 3.4 |
| Sephadex G-150 | 182 | 0.62 | 112 | 0.43 | 77 | 0.66 | 119.7 | 0.694 | 0.643 | 7.4 | 3.6 |
This suspension was passed through a French pressure cell (1,000 lb/in2), again adjusted to pH 7.5 by dropwise addition of 1 M NaOH, and centrifuged (10,000 × g for 30 min at 4°C), and the supernatant fluid was sterilized by addition of a few drops of CHCl3. After removal of residual CHCl3 by gentle aeration, the resulting crude extract was diluted in 0.05 M Tris-Cl buffer (pH 7.5) to achieve a concentration of ∼50 mg of protein/ml. A column (5 by 60 cm) of DEAE-cellulose (Whatman, Inc., Clifton, N.J.) received 125 ml of this preparation before application of the same buffer (2.7 ml/min). KatY eluted promptly, as judged by A405. Enriched samples were pooled and immediately applied to the surface of a column (2.5 by 60 cm) of calcium hydroxylapatite (Bio-Rad Laboratories, Hercules, Calif.). After application of 0.05 M Tris-Cl buffer (pH 7.5) (1 ml/min), KatY was eluted with a gradient of 0 to 0.5 M sodium phosphate in 0.05 M Tris-Cl buffer (pH 7.5). Material absorbing at 405 nm was pooled, concentrated by precipitation with 70% saturated (NH4)2SO4, centrifuged (10,000 × g for 30 min at 4°C), dialyzed for 6 h against 0.05 M Tris-Cl buffer (pH 7.5), and then applied to a column (1.5 by 200 cm) of Sephadex G-150 (Amersham Pharmacia Biotech, Inc., Piscataway, N.J.). The resulting preparation of KatY, eluted at 0.5 ml/min in the same buffer, was essentially homogeneous and stable for at least 6 months at 4°C. KatY prepared by this method was used to raise monoclonal antibodies (MAbs) in mice, and its N terminus was determined by direct sequencing as defined below.
Antisera.
The procedure used to raise MAbs entailed both intraperitoneal and subcutaneous injection of 100 μg of KatY in 50 μl of 1 part phosphate buffer plus 1 part TiterMax adjuvant (Vaxcel, Inc., Norcross, Ga.) into BALB/c mice on days 0 and 14. Upon detection of a positive reaction by immunoblotting, the animals were immunized once more (day 56) and euthanized 3 days later to extract splenocytes for fusion with the myeloma cell line Sp2/0. Fusion was performed with 50% (wt/vol) polyethylene glycol 1500 (Boehringer Mannheim Corp., Indianapolis, Ind.), and isolation of hybridoma clones was undertaken by established methods (20). Hybridoma cell lines were screened for the production of anti-KatY antibodies by enzyme-linked immunosorbent assay (ELISA). From 140 clones, 8 were selected on the basis of a strong immunological reaction and then cloned at least twice again by limiting dilution. These eight clones were further characterized by determination via ELISA and immunoblotting against 10 strains each of Y. pestis (all positive), Y. pseudotuberculosis (all positive), Y. enterocolitica (all negative), and E. coli (all negative). The MAb chosen for experimental work (termed 1B70.1) yielded a reaction in immunoblots that was indistinguishable from the remainder.
Rabbit polyclonal antiserum to outer membranes was obtained by isolating outer membranes from Y. pestis KIM10 substrain D1 by the method of Osborn et al. (45) as modified for Y. pestis (57); the organisms were grown at 37°C in the liquid medium described above. The preparation was adjusted to 2 mg of protein per ml of phosphate buffer and then emulsified with 1 volume of TiterMax adjuvant. Aliquots were used to immunize female New Zealand White rabbits by concomitant primary intramuscular (0.3 ml), intraperitoneal (0.4 ml), and subcutaneous (0.2 ml) injection; the animals also received 0.1 ml of the membrane preparation alone (without adjuvant) by intravenous injection (to achieve a total dose of 1 mg of protein). This process was repeated after 4 weeks and again after 3 weeks; test bleedings thereafter indicated occurrence of maximum titers after 2 additional weeks, at which time, the rabbits were anesthetized and all available blood was collected. The resulting sera were absorbed with disrupted and lyophilized cells of E. coli K-12 (20 mg/ml) by gentle aeration, first at 37°C for 30 min and then overnight at 4°C. Insoluble material was then removed by centrifugation (10,000 × g for 30 min at 4°C), and the absorption process was repeated twice; the remaining gamma globulin in the absorbed sera was purified by precipitation with (NH4)2SO4 and chromatography on DEAE-cellulose. Rabbit polyclonal antiserum raised against whole Y. pestis KIM10 was prepared by injection of female New Zealand White rabbits intravenously with 102, 104, and then 106 cells of substrain D27 at intervals of 4 weeks; blood was collected 3 weeks after the final injection and prepared as described above.
Amino acid sequencing.
N-terminal amino acid sequencing was performed by the Macromolecular Structure Facility (Department of Biochemistry, Michigan State University) on a model 494 Procise sequencer (PE Applied Biosystems, Foster City, Calif.). KatY was prepared for sequencing by the purification schema outlined above, and its ∼50-kDa truncated derivative was isolated after sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and transfer to a Sequi-Blot polyvinylidene difluoride membrane (Bio-Rad Laboratories) by the method of Matsudaira (38).
DNA sequencing.
Colony blotting identified a positive clone of the genomic library of Perry et al. (49), and its resident pLG338 (containing Y. pestis DNA) was purified by CsCl gradient ultracentrifugation. The approach used for sequencing this clone involved an in vitro transposon technique validated in collaboration with PE Applied Biosystems and previously tested during sequencing of human chromosome 19 clones (data not shown). Briefly, 1 μg of DNA corresponding to the entire plasmid was treated with a Primer Island transposition kit obtained from PE Applied Biosystems under the conditions recommended by the manufacturer. The transposon-treated DNA was electroporated into E. coli ElectroMax DH10B (Life Technologies, Gaithersburg, Md.), and trimethoprim-resistant clones were selected on appropriate antibiotic-containing plates. A total of 240 trimethoprim-resistant clones (those that had inherited a transposon-containing plasmid) were sequenced from both ends of the transposon by using dye terminator chemistry and primers SD118 and SD119 (13). The 480 sequencing reads (approximately 500 bases per read) represented a sequencing redundancy of 11. Base calling and assembly of sequences was performed by using PHRED/PHRAP and Consed (14, 19). A single contig of 13,641 nucleotides was obtained after assembly and subsequent removal of the vector sequences (which were easily identified by the presence of the two BamHI sites at the ends of the insert). Searching by BLAST for the N-terminal amino acid sequence of KatY obtained directly from the purified protein demonstrated the presence of katY within this contig. Sequence quality was determined by using a program termed Swedish, developed at the Human Genome Center, that automatically calculates error rates and ensures a cumulative error rate of less than 1 in 10,000 bases and a double-stranded coverage of <95% (24).
Annotation and analysis of DNA sequences.
Sequences were searched against current protein and nucleotide databases (including those from unfinished microbial sequencing projects) using BLAST (1).
Miscellaneous.
Colony immunoblotting was undertaken with MAb 1B70.1. The blocking solution used in these determinations was composed of 0.85% NaCl, 20 mM HEPES buffer (pH 7), 0.5% Triton X-100, 20% normal goat serum, and 0.01% thimerosal. SDS-PAGE and immunoblotting were performed as described previously (54). Protein was determined by the method of Lowry et al. (34).
Nucleotide sequence accession number.
The sequence of the Y. pestis insert has been submitted to the GenBank database under accession no. AF135170.
RESULTS
Purification of KatY.
Cells of Y. pestis KIM10 (substrain D28) were cultivated at 37°C, harvested, washed in phosphate buffer, and suspended in 0.05 M Tris-Cl buffer (pH 7.5). After disruption, KatY in the resulting extract was purified to near homogeneity by chromatography on columns containing DEAE-cellulose, calcium hydroxylapatite, and then Sephadex G-150 (Fig. 1). The native molecular mass of KatY was ∼300 kDa as judged by its pattern of elution on sizing resins (Fig. 1C and data not shown). Yields of protein and pigmented material absorbing at 405 nm and percentages of recovery are shown in Table 1. The crude extract and pooled 405-nm-absorbing material eluted from DEAE-cellulose, calcium hydroxylapatite, and Sephadex G-150 contained 39, 430, 1,635, and 2,423 U of catalase/mg of protein, respectively. KatY prepared by this method possessed the N-terminal amino acid sequence AEAPKTDS.
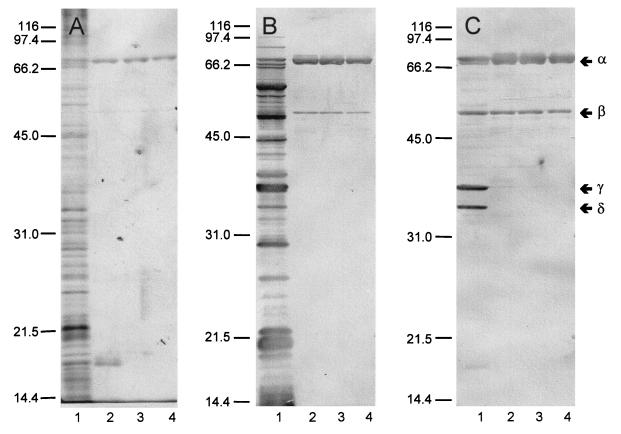
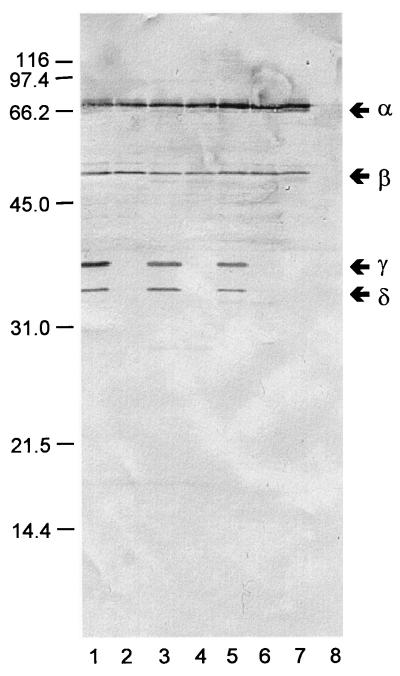
Immunoblots prepared with samples recovered during the four stages of purification (Fig. 2) revealed three unexpected findings. First, rabbit polyclonal antiserum raised against whole yersiniae (Fig. 2A) and MAb 1B70.1 directed against KatY (Fig. 2C) reacted as anticipated. However, an equally strong reaction was provided by a polyclonal rabbit antiserum obtained by immunization with a highly purified preparation of outer membranes of Y. pestis KIM10 (substrain D28) (Fig. 2B). This observation is consistent with the possibility that KatY exists, at least in part, in association with the outer membrane. Second, all samples of ∼70-kDa KatY (α-KatY [as estimated by SDS-PAGE]) contained a minor ∼50-kDa component (β-KatY) that reacted with the three antisera. None of the steps used for purification, including sizing on Sephadex G-150, eliminated β-KatY. Third, the crude cell extract contained two smaller antigens of ∼36 (γ-KatY) and ∼34 (δ-KatY) kDa that reacted with the MAb. These derivatives were entirely removed during initial chromatography on DEAE-cellulose, were not regenerated thereafter (Fig. 2C), and were absent in comparable crude cell extracts of yersiniae lacking pPCP (Fig. 3). These findings suggest that γ-KatY and δ-KatY arose as Pla-mediated degradation products of the larger α and possibly β forms of KatY.
FIG. 2.
Immunoblots prepared with rabbit polyclonal antiserum raised against whole cells of Yersinia pestis KIM10 substrain D27 (A), rabbit polyclonal antiserum raised against purified outer membranes of Y. pestis KIM10 substrain D28 (B), or mouse MAb 1B70.1 raised against KatY. Lanes: 1, crude cell extract of Y. pestis KIM10 (substrain D28); 2, eluate obtained upon chromatography on DEAE-cellulose; 3, eluate obtained upon chromatography on calcium hydroxylapatite; 4, eluate obtained upon chromatography on Sephadex G-150. The positions of the α, β, γ, and δ forms of KatY are shown in the right margin.
FIG. 3.
Immunoblot prepared with mouse MAb 1B70.1 raised against KatY versus whole cells of Y. pestis KIM10 substrain D28 carrying pPCP (lane 1), Y. pestis KIM10 substrain D47 cured of pPCP (lane 2), Y. pestis EV76 carrying pPCP (lane 3), Y. pestis EV76 cured of pPCP (lane 4), Y. pseudotuberculosis PB1 transformed with pPCP (lane 5), Y. pseudotuberculosis PB1 lacking pPCP (lane 6), E. coli HB101 transformed with pLG338 containing katY (lane 7), and E. coli HB101 alone (lane 8). The positions of the α, β, γ, and δ forms of KatY are shown in the right margin.
Cloning and sequence analyses.
The genomic library of Perry et al. (49) comprising ∼2,000 clones of E. coli HB101 transformed with pLG338 containing genomic Y. pestis DNA was screened by colony immunoblotting with MAb 1B70.1. The presence of KatY in cell extracts of a positive clone was verified by immunoblotting (Fig. 3, lane 7). The nucleotide sequence of the insert of the recombinant plasmid isolated from this clone was determined following transposon bombing (Fig. 4); the region encoding AEAPKTDS (previously identified as the N-terminal amino acid sequence of KatY) was located within the assembled contig (13,641 bp). Beginning with ATG, the coding region of the open reading frame (ORF) containing this sequence predicted a putative protein of 737 amino acid residues with a deduced molecular mass of 81.4 kDa and a pI of 6.98. This product possessed a 2.6-kDa prokaryotic leader signal sequence of 23 amino acids, which, upon processing, leaves a protein of 726 amino acid residues with a deduced molecular mass of 78.8 kDa (possessing an N terminus identical to that determined by direct sequencing) and a pI of 6.43. This molecular mass is larger than, but nevertheless consistent with, the value of ∼70 kDa previously estimated by SDS-PAGE for α-KatY.
FIG. 4.
Sequence of a 3,460-bp fragment from the Y. pestis KIM10 (substrain D46) genome that contains the ORF (amino acids 82 to 2292) encoding catalase-peroxidase katY and the ORFs (amino acids 2372 to 2752 and 2903 to 3430) that are homologous with the cybC and cybB genes encoding cytochromes b562 (61) and b561 (44) of E. coli, respectively. A potential ribosome-binding site (RBS) and putative −10 and −35 promoter areas of katY are underlined. An inverted repeat sequence 12 bp downstream of the termination codon and potential binding sites 1, 2, and 3 for the LcrF-like transcriptional regulator upstream of katY are shown by dashed arrows under and over the nucleotide sequence, respectively. N-terminal amino acid sequences of the native (amino acids 24 to 31) and truncated (amino acids 250 to 254) KatY that were determined by protein sequencing are designated by italic letters. Peroxidase motifs (amino acids 96 to 109 and 255 to 268) are in boldface and are underlined.
β-KatY.
An SDS-PAGE gel of purified KatY was prepared as for immunoblotting, but the protein within the region containing β-KatY was removed and subjected to N-terminal amino acid sequencing. The resulting N terminus was AMNDEE, a sequence occurring immediately after a methionine residue located prior to the second peroxidase motif (Fig. 4). If the C terminus remains identical to that of α-KatY, then β-KatY would exist as a peptide of 488 amino acid residues with a predicted molecular mass of 53.6 kDa and a pI of 5.78. This value is in close agreement with that determined by SDS-PAGE (∼50 kDa).
Homologues.
A BLAST comparison of the ORF encoding KatY with entries in the GenBank database showed significant similarities with several bifunctional bacterial catalase-peroxidases (Table 2). The closest homology (75%) occurred with KatP encoded by the large plasmid of enterohemorrhagic E. coli O157:H7 (6); the relatedness to the vegetative catalase-peroxidase of E. coli K-12 (KatG) and that of other bacteria was also extensive.
TABLE 2.
Amino acid sequence identities and similarities of katY from Y. pestis KIM10 (substrain D28) and other bacterial catalase-peroxidasesa
| Organism | Enzyme | % Identity | % Similarity | Length (no. of amino acids) | Accession no. | Reference |
|---|---|---|---|---|---|---|
| Escherichia coli O157:H7 | KatP | 75 | 86 | 736 | X89017 | 6 |
| Bacillus stearothermophilus | PerA | 60 | 72 | 735 | M29876 | 33 |
| Mycobacterium tuberculosis | KatG | 59 | 71 | 739 | X68081 | 64 |
| Mycobacterium bovis | KatG | 59 | 71 | 751 | X83277 | 21 |
| Mycobacterium intracellulare | MI85 | 58 | 71 | 757 | M86741 | 42 |
| Mycobacterium fortuitum | KatGI | 57 | 70 | 752 | Y07865 | 41 |
| Escherichia coli K-12 | KatG (HPI)b | 55 | 67 | 726 | M21516 | 60 |
| Salmonella typhimurium | KatG (HPI) | 55 | 67 | 727 | X53001 | 32 |
| Synechococcus sp. strain PPCC 7942 | HPI | 52 | 64 | 754 | D61378 | 43 |
| Rhodobacter capsulatum | CpeA | 43 | 54 | 576 | X71420 | 15 |
Alignment and values were calculated with the Web-based BLAST 2 Sequences tool with the program BLASP, version 2.0.5, and the matrix O BLOSUM62 (36).
HPI, hydroperoxidase I.
Features of katY.
Amino acids 96 to 109 and 255 to 268 (Fig. 4) are active-site motifs (encoding proximal and distal histidine imidazoles facilitating binding to heme) typical of other bacterial catalase-peroxidases (63). In addition, katY contained a sequence encoding a putative calcium-binding site. The likely function of this site is to maintain the structural integrity of the enzyme as described for the diheme cytochrome c peroxidase of Pseudomonas aeruginosa (17). A comparative alignment of this domain among other catalase-peroxidases showed diversity in amino acids 1 to 6 and conservation in amino acids 7 to 13 (Table 3). Analysis of the promoter immediately upstream (∼500 bp) from the initiation codon of katY revealed a potential ribosome-binding site as well as three putative transcriptional activator binding sequences characterized by direct or inverted repeats (Fig. 4); no ORFs were identified within this region. A potential rho-independent termination sequence containing a 12-bp dyad was found downstream from the termination codon (Fig. 4); sequences known to encode membrane-associated helixes were not detected.
TABLE 3.
Alignment of different catalase-peroxidases within the area of the putative calcium-binding site of KatY from Y. pestis KIM10
| Sequence | Enzyme | Amino acid at position:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | ||
| Consensusa | D | X | DNS | ILV or FYW | DENS or TG | DNQ or GHR | GP | LIV or MC | DENQ or STAG | X | X | DE | LIVM or FYW | |
| Y. pestis | KatY | D | R | S | D | G | K | K | V | S | L | S | D | L |
| E. coli O157:H7 | KatP | K | Q | S | D | G | K | K | V | S | L | A | D | L |
| B. stearothermophilus | PerA | R | E | L | P | K | K | V | S | I | A | D | L | |
| M. tuberculosis | KatG | A | A | P | G | N | I | K | V | S | F | A | D | L |
| M. bovis | KatG | A | A | P | G | N | I | K | V | S | F | A | D | L |
| M. intracellulare | MI85 | S | A | S | G | G | K | K | I | S | L | A | D | L |
| M. fortuitum | KatGI | S | A | S | D | G | K | K | I | S | L | A | D | L |
| E. coli K-12 | KatG | I | Q | K | E | S | G | K | A | S | L | A | D | I |
| S. typhimurium | KatG | K | A | S | L | A | D | I | ||||||
| Synechococcus spp. | HPI | S | A | A | T | G | A | T | V | A | D | V | ||
| R. capsulatum | CpeA | D | R | G | G | A | G | A | S | V | A | D | V | |
Consensus motif of EF-hand calcium-binding domain (Prosite, PDOC00018).
KatY is synthesized by yersiniae during expression of the low-calcium response (40). It was therefore of interest to determine if the three putative transcriptional activator binding sequences in the katY promoter were potentially recognizable by LcrF, the transcriptional activator of the Yersinia virulence regulon. We therefore aligned these sequences with the consensus site recognized by LcrF (62). Two of these repeats (no. 1 and 3) are almost identical, located on the complementary chain, and overlap the −10 box of the promoter and initiation codon (Fig. 4). In contrast, sequence 2 overlaps the ribosome-binding site. As shown in Table 4, all three sequences possess significant homology with the consensus sequence for LcrF.
TABLE 4.
Alignment of putative binding sites for LcrF
| Binding site | Sequence |
|---|---|
| Consensus for LcrF | TTTTaGYcTtTata |
| No. 1 | TTTTAACATGATTb |
| No. 2 | TTTTAGTTAAAGG |
| No. 3 | TTTTAACATATATb |
Nucleotides conserved in ≥60% of the sequences are in uppercase letters; others are in lowercase letters. Y is C or T (62).
Sequence from a complement DNA chain is shown.
Linked genes and sequences.
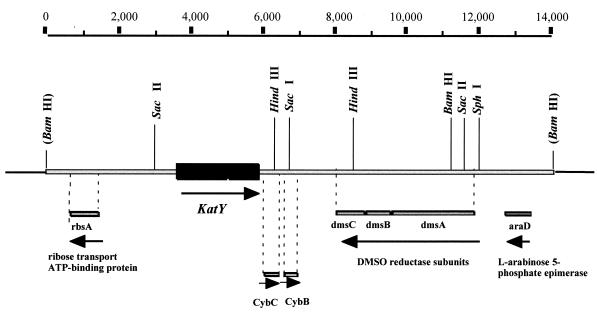
A homologue to E. coli ribose transport ATP-binding protein (rbsA) (38% identity, 59% similarity) was detected upstream of katY (Fig. 5). Two ORFs encoding proteins homologous to cytochrome b562 (CybC) (48% identity, 67% similarity) (61) and cytochrome b561 (CybB) (61% identity, 77% similarity) (44) of E. coli were identified immediately downstream (Fig. 4 and 5). These genes were followed by an evident operon encoding the dimethyl sulfoxide reductase subunits DmsA (82% identity, 90% similarity), DmsB (85% identity, 93% similarity), and DmsC (60% identity, 69% similarity) and then l-arabinose 5-phosphate epimerase (araD) (67% identity, 75% similarity) (Fig. 5). These genes are not linked to katG or with each other on the E. coli K-12 chromosome, where katG was mapped between metB and ppc at 89.2 min (4).
FIG. 5.
Physical map of the Yersinia pestis KIM10 (substrain D46) katY region. katY is represented by the hatched box, the rest of the chromosomal region is represented by a gray bar, and selected restriction sites present in the segment are indicated. The homologue genes identified in the region sequenced upstream and downstream of KatY are represented by gray bars below the chromosome. Arrows indicate the direction of transcription. Base pair coordinates are depicted on the top part of the figure. DMSO, dimethyl sulfoxide.
DISCUSSION
KatY is produced in great abundance, as evidenced by the marked reddish-brown color of concentrated cell extracts prepared after growth at 37°C in enriched media. The existence of this pigmented protein may account for the extremely high level of catalase previously reported for Y. pestis (7). Nevertheless, the activity of KatY following sizing was modest in comparison to that of a second enzyme (39); thus, we were not fully convinced that KatY was a typical catalase until its primary sequence was established in this study. Further work will be required to completely characterize the second catalase (now assumed to be vegetative KatG) and to determine if the previously observed low specific activity of KatY reflects denaturation or some other uncontrolled variable. In either event, it is now clear that katY is related to other chromosomally encoded bacterial catalase-peroxidase genes (43 to 60% identity) and that the plasmid-encoded KatP of enterohemorrhagic E. coli O157:H7 (6) is a close homologue (75% identity).
As often noted for other proteins, the molecular mass of native KatY (termed α-KatY in this context) as calculated from its DNA sequence (78.8 kDa) was larger than that approximated by SDS-PAGE (∼70 kDa). Since related catalase-peroxidases exist as tetramers (31, 37), KatY may also be composed of four identical subunits. In this case, its native molecular mass would be 315.2 kDa, in close agreement with the value of ∼300 kDa determined by molecular sieving. The smaller derivative, termed β-KatY (∼50 kDa by SDS-PAGE) was detected by MAb 1B70.1 in all preparations (including those obtained from E. coli transformed with pLG338 containing katY). β-KatY may therefore exist as a rare spontaneously truncated derivative of α-KatY capable of forming a mixed oligomer with the full-length subunits. In this context, it may be significant that a methionine residue in KatY immediately precedes the N terminus determined for β-KatY. This observation is consistent with the possibility that the sequence encoding MAMNDEE occasionally functions as a false translational start signal.
Of interest was the occurrence of two smaller forms of KatY, termed γ-KatY (∼36 kDa) and δ-KatY (∼34 kDa), as estimated by SDS-PAGE. These derivatives were only expressed in organisms harboring pPCP and, unlike β-KatY, were removed during the first step used to purify α-KatY. The most likely explanation for the existence of γ-KatY and δ-KatY is that they are cleaved from α-KatY (and possibly β-KatY) by plasminogen activator via the same process that, as noted above, serves to catalyze the posttranslational degradation of Yops.
Degradation of KatY by plasminogen activator, a known outer membrane protein (26, 55, 58), is consistent with the earlier finding that significant KatY was associated with the periplasm (48), an anatomical niche also shared by other bacterial catalases (63). Further study will be required to determine if the observed hydrolysis of KatY occurred exclusively within periplasm or after leakage or translocation to the outer membrane. The observation that rabbit antiserum raised against purified outer membranes of Y. pestis was rich in antibodies directed against KatY suggests its possible release from the periplasm. Sufficient homogeneous soluble plasminogen activator can now be prepared (26) to undertake this hydrolytic reaction in vitro with the prospect of defining the amino acid sequence or sequences of KatY which serve as hydrolytic substrates.
Analysis of katY provided useful information regarding its structure, including identification of active-site motifs encoding histidine residues involved in heme binding and a putative calcium-binding site. The absence of internal sequences known to facilitate binding to membranes is consistent with a traditional periplasmic location for KatY. The three potential LcrF-binding sequences discovered in the katY promoter may account, in part, for the observed thermoregulation of KatY, especially during restriction. However, since mutants lacking pCD (and thus LcrF) also exhibit temperature-dependent synthesis of KatY, it now seems probable that a second temperature-activated regulator is encoded elsewhere in the genome. Further evidence favoring this notion is the finding that transcription of katY occurs almost immediately after a shift from 26°C to 37°C, whereas that of known LcrF-mediated functions requires significantly longer incubation (data not shown).
H2O2 generated by professional phagocytes fulfills key roles in mediating oxygen-dependent processes of bacterial killing (5). Accordingly, a logical function for KatY would be to destroy H2O2 generated during phagocytosis. As judged by this potential ability as well as its thermoregulation and abundance, we assume that KatY will emerge as an important virulence factor of Y. pestis and Y. pseudotuberculosis. Although certain Yops have been implicated in providing resistance to killing by professional phagocytes (10, 11), this phenomenon is known to be dependent upon many variables, especially the multiplicity of infection. When the latter is minimized to avoid cytotoxicity (8, 18), there are little or no differences in intracellular survival between the wild-type cells of these two yersiniae and mutants now known to lack pCD or pYV (25, 52, 59). Attempts to establish a role for KatY in mediating resistance to killing by professional phagocytes are currently in progress.
ACKNOWLEDGMENTS
The work carried out at Lawrence Livermore National Laboratory was performed under the auspices of the U.S. Department of Energy, contract no. W-7405-Eng-48; that undertaken at Michigan State University was supported by Department of Defense contract DAAA15-93-K-0012 from the U.S. Army Research, Development, and Engineering Command.
We are grateful to Robert D. Perry for providing access to his genetic library. The excellent technical assistance of Janet M. Fowler is gratefully acknowledged.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Meyers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Baker E E, Somer H, Foster L W, Meyer E, Meyer K F. Studies on immunization against plague. I. The isolation and characterization of the soluble antigen of Pasteurella pestis. J Immunol. 1952;68:131–145. [PubMed] [Google Scholar]
- 3.Beers R F, Jr, Sizer I W. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J Biol Chem. 1952;195:133–140. [PubMed] [Google Scholar]
- 4.Blattner F R, Plunkett G, III, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose J, Mau B, Shau Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 5.Brubaker R R. Mechanisms of bacterial virulence. Annu Rev Microbiol. 1985;39:21–50. doi: 10.1146/annurev.mi.39.100185.000321. [DOI] [PubMed] [Google Scholar]
- 6.Brunder W, Schmidt H, Karch H. KatP, a novel catalase-peroxidase encoded by the large plasmid of enterohaemorrhagic Escherichia coli O157:H7. Microbiology. 1996;142:3305–3315. doi: 10.1099/13500872-142-11-3305. [DOI] [PubMed] [Google Scholar]
- 7.Burrows T W, Farrell J M, Gilette W A. The catalase activity of Pasteurella pestis and other bacteria. Br J Exp Pathol. 1964;45:579–588. [PMC free article] [PubMed] [Google Scholar]
- 8.Charnetzky W T, Shuford W W. Survival and growth of Yersinia pestis within macrophages and an effect of the loss of the 47-megadalton plasmid on growth in macrophages. Infect Immun. 1985;47:234–241. doi: 10.1128/iai.47.1.234-241.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornelis G, Sluiters C, Lambert de Rouvroit C, Michiels T. Homology between VirF, the transcriptional activator of the Yersinia virulence regulon, and AraC, the Escherichia coli arabinose operon regulator. J Bacteriol. 1989;171:254–262. doi: 10.1128/jb.171.1.254-262.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornelis G R. The Yersinia deadly kiss. J Bacteriol. 1998;180:5495–5504. doi: 10.1128/jb.180.21.5495-5504.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cornelis G R, Boland A, Boyd A P, Geuijen C, Iriarte M, Neyt C, Sory M-P, Stainier I. The virulence plasmid of Yersinia, an antihost genome. Microbiol Mol Biol Rev. 1998;62:1315–1352. doi: 10.1128/mmbr.62.4.1315-1352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crumpton M Y, Davies D A L. An antigenic analysis of Pasteurella pestis by diffusion of antigens and antibodies in agar. Proc R Soc Lond Ser B. 1956;145:109–134. doi: 10.1098/rspb.1956.0021. [DOI] [PubMed] [Google Scholar]
- 13.Devine S E, Boeke J D. Efficient integration of artificial transposons into plasmid targets in vivo: a useful tool for DNA mapping, sequencing and genetic analysis. Nucleic Acids Res. 1994;22:3765–3772. doi: 10.1093/nar/22.18.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ewing B, Hillier L, Wendl M C, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 15.Forkl H, Vandekerckhove J, Drews G, Tadros M H. Molecular cloning, sequence analysis and expression of the gene for catalase-peroxidase (cpeA) from the photosynthetic bacterium Rhodobacter capsulatus B10. Eur J Biochem. 1993;214:251–258. doi: 10.1111/j.1432-1033.1993.tb17918.x. [DOI] [PubMed] [Google Scholar]
- 16.Fowler J M, Brubaker R R. Physiological basis of the low calcium response in Yersinia pestis. Infect Immun. 1994;62:5234–5241. doi: 10.1128/iai.62.12.5234-5241.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fulop V, Ridout C J, Greenwood C, Hajdu J. Crystal structure of the dihaem cytochrome c peroxidase from Pseudomonas aeruginosa. Structure. 1995;11:1225–1233. doi: 10.1016/s0969-2126(01)00258-1. [DOI] [PubMed] [Google Scholar]
- 18.Goguen J D, Walker W S, Hatch T P, Yother J. Plasmid-determined cytotoxicity in Yersinia pestis and Yersinia pseudotuberculosis. Infect Immun. 1986;51:788–794. doi: 10.1128/iai.51.3.788-794.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 20.Harlow E, Lane D. Antibodies. A laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 21.Heym B, Alzari P M, Honore N, Cole S T. Missense mutations in the catalase-peroxidase gene, katG, are associated with isoniazid resistance in Mycobacterium tuberculosis. Mol Microbiol. 1995;15:235–245. doi: 10.1111/j.1365-2958.1995.tb02238.x. [DOI] [PubMed] [Google Scholar]
- 22.Higuchi K, Kupferberg L L, Smith J L. Studies on the nutrition and physiology of Pasteurella pestis. III. Effects of calcium ions on the growth of virulent and avirulent strains of Pasteurella pestis. J Bacteriol. 1959;77:317–321. doi: 10.1128/jb.77.3.317-321.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoe N P, Goguen J D. Temperature sensing in Yersinia pestis: translation of the LcrF activator protein is thermally regulated. J Bacteriol. 1993;175:7901–7909. doi: 10.1128/jb.175.24.7901-7909.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu P, Elliott J, McCready P, Skowronski E, Garnes J, Kobayashi A, Brubaker R R, Garcia E. Structural organization of virulence-associated plasmids of Yersinia pestis. J Bacteriol. 1998;180:5192–5202. doi: 10.1128/jb.180.19.5192-5202.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janssen W A, Surgalla M J. Plague bacillus: survival within host phagocytes. Science. 1969;163:950–952. doi: 10.1126/science.163.3870.950. [DOI] [PubMed] [Google Scholar]
- 26.Kutyrev V, Mehigh R J, Motin V L, Pokrovskaya M S, Smirnov G B, Brubaker R R. Expression of the plague plasminogen activator in Yersinia pseudotuberculosis and Escherichia coli. Infect Immun. 1999;67:1359–1367. doi: 10.1128/iai.67.3.1359-1367.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lambert de Rouvroit C, Sluiters C, Cornelis G R. Role of the transcriptional activator, VirF, and temperature in the expression of the pYV plasmid genes of Yersinia enterocolitica. Mol Microbiol. 1992;6:395–409. [PubMed] [Google Scholar]
- 28.Lawton W D, Fukui G M, Surgalla M J. Studies on the antigens of Pasteurella pestis and Pasteurella pseudotuberculosis. J Immunol. 1960;84:475–479. [PubMed] [Google Scholar]
- 29.Lindler L E, Klempner M S, Straley S C. Yersinia pestis pH 6 antigen: genetic, biochemical, and virulence characterization of a protein involved in the pathogenesis of bubonic plague. Infect Immun. 1990;58:2569–2577. doi: 10.1128/iai.58.8.2569-2577.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindler L E, Plano G V, Burland V, Mayhew G F, Blattner F R. Complete DNA sequence and detailed analysis of the Yersinia pestis KIM5 plasmid encoding murine toxin and capsular antigen. Infect Immun. 1998;66:5731–5742. doi: 10.1128/iai.66.12.5731-5742.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loewen P. Probing the structure of catalase HPH of Escherichia coli—a review. Gene. 1996;179:39–44. doi: 10.1016/s0378-1119(96)00321-6. [DOI] [PubMed] [Google Scholar]
- 32.Loewen P C, Triggs B L, George C S, Hrabarchuk B E. Genetic mapping of katG, a locus that affects synthesis of the bifunctional catalase-peroxidase hydroperoxidase I in Escherichia coli. J Bacteriol. 1985;162:661–667. doi: 10.1128/jb.162.2.661-667.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loprasert C, Negoro S, Okada H. Cloning, nucleotide sequence, and expression in Escherichia coli of the Bacillus stearothermophilus peroxidase gene (perA) J Bacteriol. 1989;171:4871–4875. doi: 10.1128/jb.171.9.4871-4875.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 35.Lucier T S, Brubaker R R. Determination of genome size, macrorestriction pattern polymorphism, and nonpigmentation-specific deletion in Yersinia pestis by pulsed-field gel electrophoresis. J Bacteriol. 1992;174:2078–2086. doi: 10.1128/jb.174.7.2078-2086.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madden T L, Tatvsov R L, Zhang J. Applications of network BLAST server. Methods Enzymol. 1996;266:131–141. doi: 10.1016/s0076-6879(96)66011-x. [DOI] [PubMed] [Google Scholar]
- 37.Marcinkeviciene J A, Magliozzo R S, Blanchard J S. Purification and characterization of the Mycobacterium smegmatis catalase-peroxidase involved in isoniazid activation. J Biol Chem. 1995;270:22290–22295. doi: 10.1074/jbc.270.38.22290. [DOI] [PubMed] [Google Scholar]
- 38.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 39.Mehigh R J, Brubaker R R. Major stable peptides of Yersinia pestis synthesized during the low-calcium response. Infect Immun. 1993;61:13–22. doi: 10.1128/iai.61.1.13-22.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mehigh R J, Sample A K, Brubaker R R. Expression of the low-calcium response in Yersinia pestis. Microb Pathog. 1989;6:203–217. doi: 10.1016/0882-4010(89)90070-3. [DOI] [PubMed] [Google Scholar]
- 41.Menéndez M C, Ainsa J A, Martin C, Garcia M J. katGI and katGII encode two different catalases-peroxidases in Mycobacterium fortuitum. J Bacteriol. 1997;179:6880–6886. doi: 10.1128/jb.179.22.6880-6886.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morris S L, Nair J, Rouse D A. The catalase-peroxidase of Mycobacterium intracellulare: nucleotide sequence analysis and expression in Escherichia coli. J Gen Microbiol. 1992;138:2362–2370. doi: 10.1099/00221287-138-11-2363. [DOI] [PubMed] [Google Scholar]
- 43.Mutsuda M, Ishikawa T, Takeda T, Shigeoka S. The catalase-peroxidase of Synechococcus PCC 7942: purification, nucleotide sequence analysis and expression in Escherichia coli. Biochem J. 1996;316:251–257. doi: 10.1042/bj3160251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakamura H, Murakami H, Yamato I, Anraku Y. Nucleotide sequence of the cybB gene encoding cytochrome b561 in Escherichia coli K12. Mol Gen Genet. 1988;212:1–5. doi: 10.1007/BF00322437. [DOI] [PubMed] [Google Scholar]
- 45.Osborn M J, Gander J E, Parsi E, Carson J. Mechanism of assembly of the outer membrane of Salmonella typhimurium. J Biol Chem. 1972;247:3962–3972. [PubMed] [Google Scholar]
- 46.Payne D, Tatham D, Williamson E D, Titball R W. The pH 6 antigen of Yersinia pestis binds to β1-linked galactosyl residues in glycosphingolipids. Infect Immun. 1998;66:4545–4548. doi: 10.1128/iai.66.9.4545-4548.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perry R D, Fetherston J D. Yersinia pestis—etiologic agent of plague. Clin Microbiol Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perry R D, Lucier T S, Sikkema D J, Brubaker R R. Storage reservoirs of hemin and inorganic iron in Yersinia pestis. Infect Immun. 1993;61:32–39. doi: 10.1128/iai.61.1.32-39.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perry R D, Pendrak M L, Schuetze P. Identification and cloning of a hemin storage locus involved in the pigmentation phenotype of Yersinia pestis. J Bacteriol. 1990;172:5929–5937. doi: 10.1128/jb.172.10.5929-5937.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perry R D, Straley S C, Fetherston J D, Rose D J, Gregor J, Blattner F R. DNA sequencing and analysis of the low-Ca2+-response plasmid pCD1 of Yersinia pestis KIM5. Infect Immun. 1998;66:4611–4623. doi: 10.1128/iai.66.10.4611-4623.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Protsenko O A, Anisimov P I, Mosarov O T, Donnov N P, Popov Y A, Kokushkin A M. Detection and characterization of Yersinia pestis plasmids determining pesticin I, fraction 1 antigen and mouse toxin synthesis. Genetika. 1983;19:1081–1090. [PubMed] [Google Scholar]
- 52.Richardson M, Harkness T K. Intracellular Pasteurella pseudotuberculosis: multiplication in cultured spleen and kidney cells. Infect Immun. 1970;2:631–639. doi: 10.1128/iai.2.5.631-639.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sample A K, Brubaker R R. Post-translational regulation of Lcr plasmid-mediated peptides in pesticinogenic Yersinia pestis. Microb Pathog. 1987;3:239–248. doi: 10.1016/0882-4010(87)90057-x. [DOI] [PubMed] [Google Scholar]
- 54.Sample A K, Fowler J M, Brubaker R R. Modulation of the low calcium response in Yersinia pestis by plasmid-plasmid interaction. Microb Pathog. 1987;2:443–453. doi: 10.1016/0882-4010(87)90051-9. [DOI] [PubMed] [Google Scholar]
- 55.Sodeinde O A, Goguen J D. Nucleotide sequence of the plasminogen activator gene of Yersinia pestis: relationship to ompT of Escherichia coli and gene E of Salmonella typhimurium. Infect Immun. 1989;57:1517–1523. doi: 10.1128/iai.57.5.1517-1523.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sodeinde O A, Sample A K, Brubaker R R, Goguen J D. Plasminogen activator/coagulase gene of Yersinia pestis is responsible for degradation of plasmid-encoded outer membrane proteins. Infect Immun. 1988;56:2749–2752. doi: 10.1128/iai.56.10.2749-2752.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Straley S C, Brubaker R R. Cytoplasmic and membrane proteins of yersiniae cultivated under conditions simulating mammalian intracellular environment. Proc Natl Acad Sci USA. 1981;78:1224–1228. doi: 10.1073/pnas.78.2.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Straley S C, Brubaker R R. Localization in Yersinia pestis of peptides associated with virulence. Infect Immun. 1982;36:129–135. doi: 10.1128/iai.36.1.129-135.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Straley S C, Harmon P A. Growth in mouse peritoneal macrophages of Yersinia pestis lacking established virulence determinants. Infect Immun. 1984;45:649–654. doi: 10.1128/iai.45.3.649-654.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Triggs-Raine B L, Doble B W, Mulvey M R, Sorby P A, Loewen P C. Nucleotide sequence of KatG, encoding HPI of Escherichia coli. J Bacteriol. 1988;170:4415–4419. doi: 10.1128/jb.170.9.4415-4419.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trower N K. PCR cloning, sequence analysis and expression of the cybC genes encoding soluble cytochrome b-562 from Escherichia coli B strain OP7 and K strain NM522. Biochim Biophys Acta. 1993;1143:109–111. doi: 10.1016/0005-2728(93)90223-3. [DOI] [PubMed] [Google Scholar]
- 62.Wattiau P, Cornelis G R. Identification of DNA sequences recognized by VirF, the transcriptional activator of the Yersinia Yop regulon. J Bacteriol. 1994;176:3878–3884. doi: 10.1128/jb.176.13.3878-3884.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Welinder K G. Bacterial catalase-peroxidases are gene duplicated members of the plant peroxidase superfamily. Biochim Biophys Acta. 1991;1080:215–220. doi: 10.1016/0167-4838(91)90004-j. [DOI] [PubMed] [Google Scholar]
- 64.Zhang Y, Heym B, Allen B, Young D, Cole S. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992;358:591–593. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]