Abstract
During infectious disease outbreaks, inference of summary statistics characterizing transmission is essential for planning interventions. An important metric is the time-dependent reproduction number (Rt), which represents the expected number of secondary cases generated by each infected individual over the course of their infectious period. The value of Rt varies during an outbreak due to factors such as varying population immunity and changes to interventions, including those that affect individuals' contact networks. While it is possible to estimate a single population-wide Rt, this may belie differences in transmission between subgroups within the population. Here, we explore the effects of this heterogeneity on Rt estimates. Specifically, we consider two groups of infected hosts: those infected outside the local population (imported cases), and those infected locally (local cases). We use a Bayesian approach to estimate Rt, made available for others to use via an online tool, that accounts for differences in the onwards transmission risk from individuals in these groups. Using COVID-19 data from different regions worldwide, we show that different assumptions about the relative transmission risk between imported and local cases affect Rt estimates significantly, with implications for interventions. This highlights the need to collect data during outbreaks describing heterogeneities in transmission between different infected hosts, and to account for these heterogeneities in methods used to estimate Rt.
This article is part of the theme issue 'Technical challenges of modelling real-life epidemics and examples of overcoming these'.
Keywords: mathematical modelling, reproduction number, imported cases, branching processes, COVID-19, SARS-CoV-2
1. Introduction
Mathematical and computational models have been used during the COVID-19 pandemic to infer changes in transmissibility and to plan public health measures [1–7]. An important metric for assessing the effectiveness of current interventions during outbreaks is the time-dependent reproduction number (Rt—sometimes referred to informally as the ‘R number’), which represents the expected number of infections generated by someone infected at time t over the course of their infectious period [8–17]. This quantity varies during an outbreak in response to factors affecting transmission such as changes in public health measures, varying population immunity and pathogen evolution. If Rt remains below one, the number of cases each day will decrease; if instead Rt is persistently above one, the outbreak will grow. In the UK, the government has published estimates of Rt throughout the COVID-19 pandemic [18] alongside other values such as estimates of the epidemic growth rate and daily numbers of new reported cases, hospitalizations and deaths.
Different formal definitions of Rt have been proposed, most notably the instantaneous reproduction number and the case reproduction number [19]. The instantaneous reproduction number represents the expected number of infections generated (over the course of their infectious period) by someone who is infected at time t if transmission conditions do not change in the future (i.e. this quantity is a measure of instantaneous transmissibility). The case reproduction number, on the other hand, reflects the expected number of infections generated by someone who is infected at time t but accounts for changes in transmissibility that occur after time t (e.g. the subsequent introduction of public health measures). The instantaneous reproduction number has been proposed as the most appropriate definition to use for real-time inference, as this quantity reflects current transmissibility and does not require future changes in transmission conditions to be known [11]. For that reason, we use this definition of Rt for our analyses in this manuscript.
A range of methods has been developed for estimating Rt from outbreak data [11,12,20,21]. Two common approaches are the Cori method [8,9] and the Wallinga–Teunis method [22], which involve inferring the value of Rt from disease incidence time-series (i.e. time-series describing the number of new cases every day) and an estimate of the serial interval distribution (representing the time period between successive cases; specifically, the difference between the symptom onset times of infectors and infectees). Irrespective of the precise approach used to infer Rt, estimates can be updated and tracked as additional data become available during an outbreak.
Recent developments in the theory of Rt estimation include accounting for reporting delays [7] and considering the impacts of temporal changes in the serial interval [23]. Another consideration is the potential for heterogeneity in Rt between different subgroups in the population. The COVID-19 pandemic has highlighted that individuals in different settings (e.g. care homes as opposed to the wider population [24]) or with different characteristics (e.g. different ages [10,25–28] or vaccination statuses [29,30]) face different risks of both becoming infected and transmitting the virus. Shortly before the COVID-19 pandemic, the Cori method was extended to account for differences in the source locations of local and imported cases [9], but with an assumption that the expected numbers of onwards transmissions from each local case and each imported case are identical. With that assumption, that work illustrated that failing to differentiate between local and imported cases can lead to overestimation of the number of local infections and therefore overestimation of Rt [9].
Apart from their different origins, local and imported cases can differ in other ways. The risk of onwards transmission from an imported case may be different to the risk from a local case [31]. Imported cases may have visited regions with high case numbers and therefore respond more quickly to early signs of disease, isolating as soon as symptoms develop. This effect might be especially pronounced when a pathogen has first arrived in the local host population, when the infection risk may be higher outside the local population than within it. Imported cases may also be subject to increased testing for infection or pre-emptive home quarantine following travel, thereby lowering the risk of onwards transmission [32]. On the other hand, individuals who travel frequently may be likely to have more contacts with others than those who do not, potentially leading to a higher risk of onwards transmission for imported cases. For example, business travellers may participate in large numbers of meetings, thereby coming into contact with many other people. In either situation, an assumption that Rt is identical for both local cases and imported ones, as made previously [9], is not always appropriate.
In principle, the same disease incidence time-series can occur with different divisions of the transmission risk between local and imported infectors (figure 1). This has implications for pathogen control, since a scenario with substantial local transmission requires localized public health measures to disrupt chains of transmission and prevent spread. By contrast, a scenario with high transmission from imported cases may motivate travel restrictions to prevent importations. Here, we modify the Cori method to allow local and imported cases to have unequal risks of generating new infections. We analyse disease incidence time-series recorded during the COVID-19 pandemic in different locations. Our main goal is not to provide a novel methodological approach for estimating Rt, but rather to explore as simply as possible the potential consequences for estimates of Rt of failing to account for differences in the onwards transmission risk from local and imported cases. To allow other researchers to repeat our analyses for similar data, we provide an open-source Python software library including a user-friendly web interface (https://sabs-r3-epidemiology.github.io/branchpro). Our research demonstrates the importance of accounting for differences in the transmission risk between imported and local cases. More widely, it indicates that careful consideration of heterogeneity in the transmission risk between population subgroups may be necessary to make robust public health policy decisions.
Figure 1.
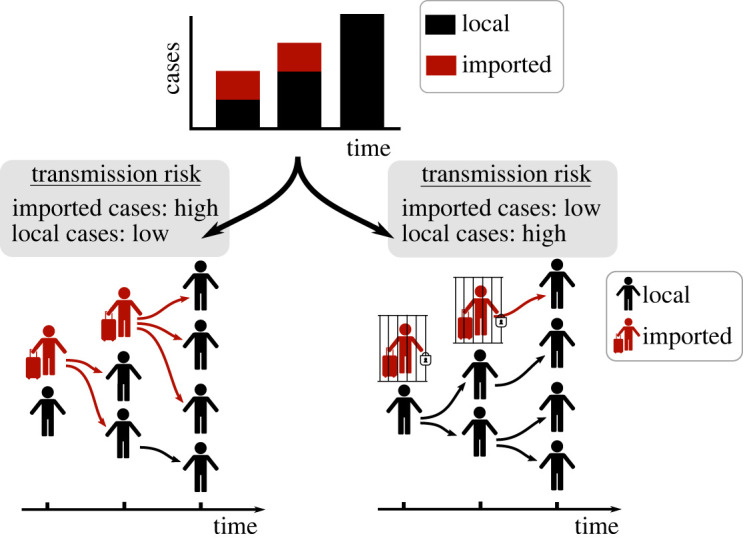
A disease incidence time-series dataset can be generated by different combinations of transmission risks from imported and local cases. In the first scenario (bottom left), observed cases are mostly due to infections by imported cases, whereas in the second scenario (bottom right), observed cases are mostly due to infections by local cases. In the bottom panels, red arrows represent infections generated by imported cases and black arrows represent infections generated by local cases. An individual who is infected by an imported case is classified as a local case, since they have themselves been infected locally. Despite the same overall incidence, the two scenarios shown correspond to different risks of sustained local transmission (the risk of sustained local transmission is higher in the second scenario—bottom right), with implications for public health measures. (Online version in colour.)
2. Methods
(a) . Inference of the time-dependent reproduction number
We modify the Cori method for estimating Rt [8,9] to account for differences in the onwards transmission risk between cases that arise locally compared with those originating elsewhere. In the underlying transmission model, new cases occur according to a time-varying branching process in which each local case is assumed to generate Rt new infections on average, and each imported case is expected to generate εRt new infections on average, where ε ≥ 0 indicates the relative transmission risk from an imported case compared with a local case. Here, we assume that Rt is the instantaneous reproduction number [8,9], representing the expected number of cases that an individual infected at time t is likely to generate over the course of their infection assuming that future pathogen transmissibility is fixed at the current level. Our focus is on estimating the extent of local transmission (i.e. the local time-dependent reproduction number [21]) characterized by Rt. As has been proposed previously [21], the value of Rt therefore reflects the potential for local transmission of the pathogen (rather than being an averaged quantity across both local and imported cases). Here, ε < 1 means that an imported case is responsible for fewer infections (on average) than a local case, whereas ε > 1 indicates that an imported case generates more infections.
The total number of new cases arising at time step t can be split according to the sources of infection, , where represents the number of new cases who were infected within the local population and represents the number of new cases who were infected elsewhere. The expected number of local cases at time step t (measured in days) is then given by
In this expression, the vector w is the (discrete) serial interval distribution with entries ws (which characterizes the times between successive cases in a chain of transmission; w1 is the probability that the serial interval is one day, w2 is the probability that the serial interval is two days, and so on).
We define the transmission potential at time step t to represent the expected number of local cases arising at time step t if Rt = 1. Thus, the transmission potential at time step t is given by . We assume that the number of local cases in time step t is drawn from a Poisson distribution with mean RtΛt(w, ε). Hence, the probability of observing the local incidence over a time window including τ + 1 days (assuming that Rt is constant during that time window), conditional each day on all previous incidence data, is given by
Data describing daily numbers of imported cases enter this expression through Λt(w, ε). The model therefore reflects how local cases arise using information about historical numbers of local and imported cases.
Assuming a gamma distributed prior for Rt, the posterior distribution for Rt over the time window [t − τ, t], conditional on w, ε and the observed incidence data (denoted p(Rt|w, ε, I≤t)—we represent this by p(·) rather than P(·) since the posterior is a continuous probability density function), is also a gamma distribution due to prior-likelihood conjugacy (see Cori et al. [8] and Thompson et al. [9] for further details). Specifically,
where for notational convenience, here and above we have combined the disease incidence data into the variable . In this expression, the parameters α > 0 and β > 0 are the shape and rate parameters of the gamma prior distribution for Rt. The function gamma(x, a, b) corresponds to the probability density function of a gamma distribution with shape parameter a and rate parameter b, so that
The inferred posterior, p(Rt|w, ε, I≤t), is based on local infectees appearing in the incidence data in the estimation window [t − τ, t], infected by local or imported infectors appearing in the incidence data at any time in [0, t − 1]. Estimates of Rt at successive time steps are generated by shifting the estimation window by one time step and repeating the inference procedure. The purpose of this estimation window (rather than estimating Rt based on infectees appearing in the incidence time-series on day t alone) is to increase the smoothness of successive Rt estimates, instead of inferring variations in Rt due to the inherent randomness in the epidemiological system (or any other factor affecting the numbers of cases observed each day; for example, daily fluctuations in the proportion of cases that are reported). This comes at the cost of missing changes in transmission occurring at a fine temporal resolution [8].
(b) . Accounting for uncertainty in the serial interval distribution
The approach described above involves estimating Rt using disease incidence time-series and an estimate of the serial interval distribution, accounting for differences in both the source location of infection and onwards transmission risk between local and imported cases. However, there is often significant uncertainty in the serial interval distribution. To account for this, we consider a scenario in which there is a set of equally plausible serial interval distributions, . For a single value of i, the entries of the vector w(i) correspond to the probability that the serial interval takes the value s days, conditional on w(i) being the true serial interval distribution.
In our analyses of COVID-19 data, we use a set of equally plausible serial intervals, , obtained from a previous study (see below). To account for this uncertainty in the serial interval distribution when estimating Rt, we first estimate Rt separately for each plausible serial interval distribution, w(i), giving the conditional posterior distribution p(Rt|w(i), ε, I≤t). We then combine these estimates to give a posterior distribution for Rt accounting for this uncertainty by calculating
(c) . Data and parameterization
In our main analyses, we consider five disease incidence time-series datasets collected in different locations during the COVID-19 pandemic. The key feature of these datasets is that information is available which allows locally originating cases to be differentiated from those infected elsewhere. The datasets are as follows:
-
(i)
Ontario, Canada (figure 2a(i)). Incidence data were obtained for the time period from 1 March to 20 April 2020 [33]. Cases were classified as imported if they reported travelling outside Ontario within 14 days prior to symptom onset. Cases with unknown recent travel status were assumed to have been infected locally.
-
(ii)
New South Wales, Australia (figure 2a(ii)). Incidence data were obtained for the time period from 1 March to 13 April 2020. Cases were classified as imported if they were reported as ‘overseas acquired’ in the Australian national COVID-19 database (see [32] for further details). Cases with unknown origin were assumed to have been infected locally.
-
(iii)
Victoria, Australia (figure 2a(iii)). Details as above for New South Wales.
-
(iv)
Hong Kong (figure 4a(i)). Incidence data were obtained for the time period from 23 January to 24 March 2020 [34]. Cases were classified as imported if they were listed as ‘imported case, confirmed’ in the Hong Kong Department of Health COVID-19 database (see [35] for further details). All other cases were classified as local cases.
-
(v)
Hainan Province, China (figure 4a(ii)). Incidence data were obtained for the time period from 22 January to 20 February 2020 [36]. Cases were classified as imported if they either reported travel outside Hainan Province in the 14 days prior to symptom onset or reported any recent travel to a known COVID-19 outbreak area. All other cases were classified as local cases.
Figure 2.
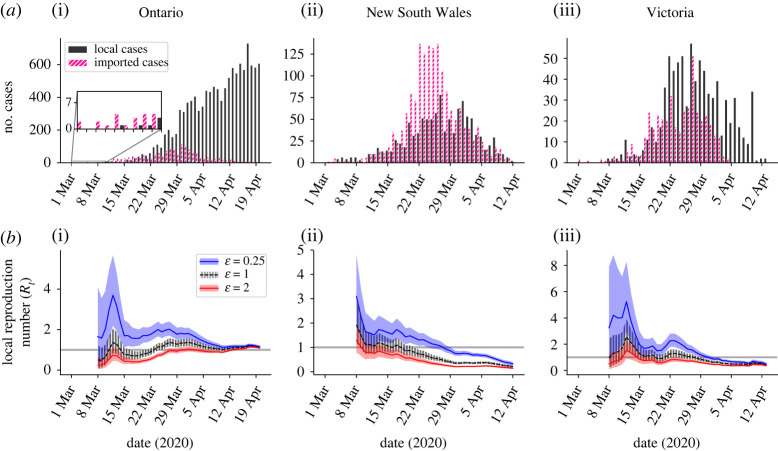
Inference of the local reproduction number (Rt) under different assumptions about the relative transmission risk from imported and local cases. (a) The COVID-19 incidence time-series datasets used in our main analyses, for Ontario (i), New South Wales (ii) and Victoria (iii). Black bars represent the daily numbers of local cases, and pink bars represent the daily numbers of imported cases. (b) Inferred Rt values for different assumed values of the relative transmission risk from an imported case compared with a local case (ε). The grey horizontal line represents the threshold Rt = 1, and shaded regions represent the 95% central credible interval of the Rt estimates. (Online version in colour.)
Figure 4.
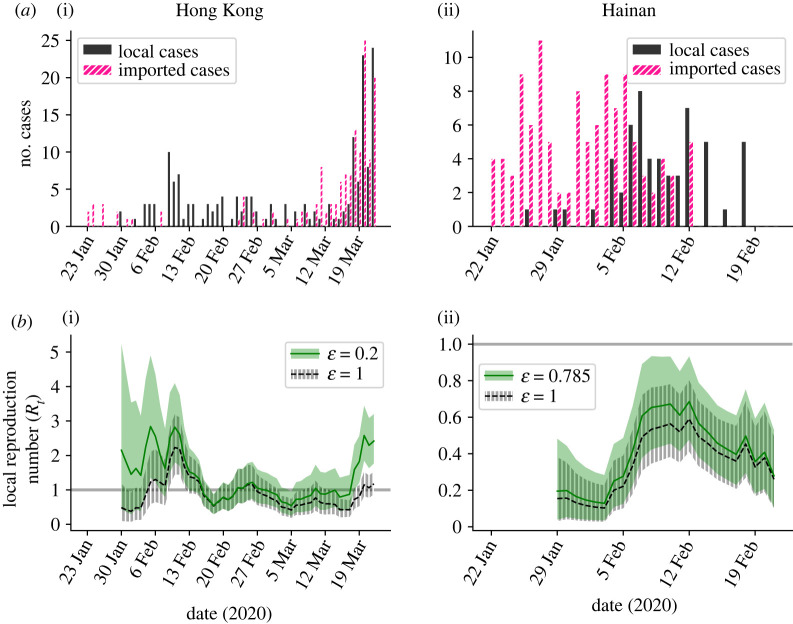
Inference of the local reproduction number (Rt) for estimated values of the relative transmission risk from imported and local cases. (a) The COVID-19 incidence time-series datasets used in our main analyses, for Hong Kong (i) and Hainan Province, China (ii). Black bars represent the daily numbers of local cases, and pink bars represent the daily numbers of imported cases. (b) Inferred Rt values for different assumed values of the relative transmission risk from an imported case compared with a local case (ε), for Hong Kong (i) and Hainan Province (ii). The grey horizontal line represents the threshold Rt = 1, and shaded regions represent the 95% central credible interval of the Rt estimates. The values ε = 0.2 for Hong Kong and ε = 0.785 for Hainan were estimated from alternative data sources, as described in the text. (Online version in colour.)
We chose to analyse the first three datasets in the main text due to their differing outbreak trajectories in the time periods considered. Specifically, the Ontario dataset represents a growing outbreak, the New South Wales dataset represents a full outbreak wave with a large number of imported cases compared with local cases and the Victoria dataset represents a full outbreak wave with more local cases than imported ones. We chose to analyse the fourth and fifth datasets because further information was available from those locations with which it was possible to approximate the value of ε. This allowed us to demonstrate inference of Rt in scenarios in which the relative transmission risk from imported and local cases is known.
In addition to our analyses in the main text, we considered datasets from other locations and display similar analyses in the electronic supplementary material, figures S1–S6; specifically, we considered COVID-19 disease incidence time-series datasets from five other Australian states, New Zealand and Hawaii, and we considered a disease incidence time-series dataset for MERS in Saudi Arabia in 2014–2015. The key feature of all these datasets is that information was available with which to classify cases as either local or imported. In the analysis of MERS in Saudi Arabia, imported cases were not those who had arrived from a geographically distinct location. Instead, in that analysis, imported cases were those who were likely to have been infected directly from the animal reservoir.
For the serial interval in all our analyses of COVID-19 incidence datasets, we considered an estimate for SARS-CoV-2 obtained by Nishiura et al. [37]. Specifically, those authors fitted a log-normal distribution to data from known infector–infectee transmission pairs using Markov chain Monte Carlo (MCMC), thereby obtaining a set of equally plausible possible serial interval distributions. We considered the set of serial interval distributions obtained by Nishiura et al. [37] using both certain and probable infector–infectee pairs while accounting for right-truncation (i.e. the possibility that a dataset detailing infector–infectee pairs observed when the outbreak is ongoing excludes some transmissions with longer serial intervals that have not yet occurred). For our inference procedure, we used n = 1000 randomly selected MCMC iterations from their analysis, where each iteration characterizes a continuous distribution. Since our approach considers the number of new cases each day, we require a discrete serial interval distribution. We therefore ‘discretized’ the continuous distributions into daily time steps using the method described by Cori et al. [8] (see web appendix 11 of that article). The set of n = 1000 serial interval distributions used in our analysis (i.e. ) is shown in the electronic supplementary material, figure S7.
We fixed the parameters of the gamma distributed prior for Rt so that both the mean and standard deviation were equal to five (to do this, we chose α = 1 and β = 0.2). The rationale for this choice is that a large standard deviation ensures that the prior is relatively uninformative, while a high mean ensures that the outbreak is unlikely to be determined as under control (Rt < 1) unless there is substantial evidence from the data supporting this conclusion. In all of our analyses of COVID-19 incidence data, Rt was estimated using a weekly sliding window, so that τ = 6 days. In the figures, the posterior distribution for Rt shown on day t is based on a sliding window that ends on day t (i.e. the sliding window [t − τ, t]).
(d) . Correctness and reproducibility of results
We followed a range of software development practices to guard against coding errors and to ensure code reusability: these included collaborative coding using Github to manage merging of code via pull requests, unit testing of functions and classes (with 100% test coverage) and continuous integration testing. To ensure reproducibility of results, all analyses for this paper can be rerun by cloning our Github repository (https://github.com/SABS-R3-Epidemiology/transmission-heterogeneity-results) and executed via a single command from the terminal.
3. Results
(a) . Effect of the relative transmission risk on estimates of Rt
To explore how different assumptions about the relative transmission risk from imported and local cases affect Rt estimates, we initially applied our method to data from the first three locations described in Methods (figure 2). We considered three different assumptions about the relative transmission risk. First, we assumed that imported cases were each expected to generate fewer infections than local cases (ε = 0.25; figure 2b—blue). Second, we assumed instead that imported cases were each expected to generate more infections than local cases (ε = 2; figure 2b—red). Third, we made the standard assumption [9] that the transmission risk from each local case was identical to the transmission risk from each imported case (ε = 1; figure 2b—black). These analyses highlight that different assumed values of ε lead to different inferred Rt values. As might be expected, assuming larger values of ε leads to smaller estimated values of Rt, since more transmission is then attributed to imported cases rather than local cases.
We then went on to consider the implications for public health policy of differences in the relative transmissibility of imported and local cases. For the dataset from Ontario (figure 2a(i)), the numbers of local cases broadly increased throughout the time period considered. A key question in that setting is ‘Is Rt > 1?’, since this determines whether sustained local transmission is likely to occur. If so, fast detection that Rt > 1 is crucial to allow interventions to be introduced quickly to prevent further exponential growth of the outbreak. In figure 3a(i), posterior mean estimates of Rt each day from 8 March to 20 April 2020 are shown for a range of values of ε. The first date on which the mean estimate of Rt is above one and remains above one thereafter is shown for different values of ε in figure 3b(i) (grey). This indicates that a smaller assumed value of ε leads to an earlier conclusion that Rt is greater than one for this dataset. The proportion of the period considered for which the mean Rt estimate is above one also depends on the assumed value of ε (figure 3c(i)).
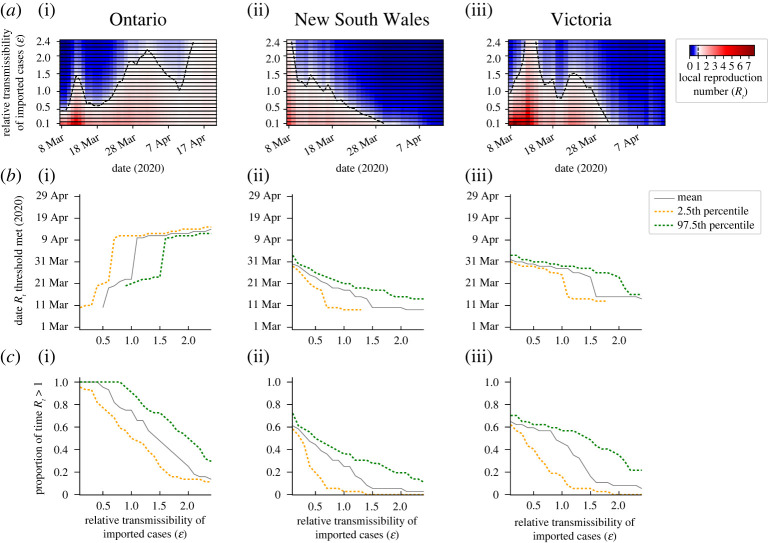
Figure 3.
Implications of differences in the assumed relative transmission risk from imported and local cases on policymaking. (a) Inferred mean Rt values for different values of the relative transmissibility of imported cases compared with local cases (ε). (b) Dates on which the estimated values of Rt cross policy-relevant thresholds (in scenarios where the thresholds are crossed at some stage in the outbreak; otherwise dates are not plotted). For Ontario (i), the date shown represents the first date when the estimated Rt value is above one and remains above one for the remainder of the time period considered (until 20 April 2020). This represents the first date when the outbreak is not inferred to be under control for the remainder of the time period. For New South Wales (ii) and Victoria (iii), the date shown represents the first date on which the estimated Rt value is below one and remains so for the remainder of the time period considered (until 13 April 2020). This represents the first date on which the outbreak could be concluded as being under control for the remainder of the time period. (c) The proportion of the time periods considered for which the inferred Rt values are above one (so the outbreak is not inferred to be under control). In (b,c), results are shown for the mean values of the posterior for Rt (grey), and well as for the 2.5th (yellow dotted) and 97.5th (green dotted) percentile values of the posterior for Rt (which span the 95% central credible interval). (Online version in colour.)
While a policymaker may choose to strengthen control measures when the mean estimate of Rt increases above one, a more risk-averse choice could be to conclude that the outbreak is not under control if an upper percentile of the posterior distribution of Rt exceeds one. For example, for the Ontario dataset, when ε = 1.2, the mean estimate of Rt is (and remains) above one from 11 April 2020 onwards (figure 3b(i), grey), whereas the 97.5th percentile estimate of Rt remains above one from the earlier date of 23 March 2020 onwards (figure 3b(i), green dashed). By using an approach like the one described here, policymakers can adjust their decision making according to their chosen level of risk aversion. This simply involves specifying the percentile value of Rt to track to guide decision making regarding strengthening and relaxing public health measures.
During the COVID-19 pandemic, public health measures have been relaxed in many regions and countries when the outbreak has been assessed as being under control. We therefore considered the incidence dataset from New South Wales and estimated when policymakers could conclude that Rt had fallen below one (figure 3b(ii)). In this scenario, a larger assumed value of ε led to an earlier date on which Rt was assessed to be below one (and remained below one thereafter). In order for policymakers to be more certain that Rt is below one when relaxing restrictions, one possibility is to conclude that Rt is below one when a high percentile value of the posterior for Rt has fallen below one. For example, for this dataset, if the mean estimate of Rt is considered and the value ε = 1.2 is assumed, then Rt is inferred to fall and remain below one on 15 March 2020 (figure 3b(ii), grey), whereas if instead the 97.5th percentile estimate of Rt is considered, then Rt is inferred to fall below one on the later date of 19 March 2020 (figure 3b(ii), green dashed).
As the final component of these analyses, we considered the disease incidence time-series dataset from Victoria and repeated the analysis that we conducted for the dataset from New South Wales. We found that, if a high value of ε is assumed, then the outbreak is inferred to be under control (Rt < 1) for the majority of the time period under consideration (figure 3c(iii)). However, if instead the value of ε is lower, then Rt may be estimated to be greater than one early in the outbreak. For small values of ε, so that initial estimated values of Rt are high, the most policy-relevant question may again be to determine when Rt has fallen below one (figure 3b(iii)).
(b) . Realistic values of the relative transmission risk
In the analyses presented in §3a, we demonstrated clearly that the assumed relative transmission risk between imported and local cases affects Rt estimates, impacting policy-relevant conclusions drawn from disease incidence time-series data. The relative transmission risk may differ between settings. In some scenarios, it may be possible to inform estimates of ε with real-world data. Here we provide two examples, in the context of SARS-CoV-2 transmission in Hong Kong and Hainan Province (the fourth and fifth disease incidence time-series datasets described in Methods). Additional possible approaches for estimating the value of ε are described in the Discussion.
First, we considered the dataset from Hong Kong (figure 4a(i)). A previous study [35] reconstructed the transmission network of cases in that region (between 23 January 2020 and 8 January 2021; although in principle a similar analysis could be conducted at a smaller spatial scale for shorter time periods, as would likely be most useful for early real-time estimation of Rt), inferring the ‘outdegree’ of imported and local cases. Based on the aggregated data shown in table 1 of that study, the mean outdegree was 0.74 for imported cases and 3.68 for local cases, which corresponds to a value of ε = 0.2. We therefore compared estimated values of Rt for ε = 0.2 (figure 4b(i), green) with analogous estimates under the standard assumption that ε = 1 (figure 4b(i), black). Since a value of ε = 0.2 leads to less transmission being attributed to imported infections than when ε = 1, estimated values of Rt are higher when ε = 0.2. In terms of decision making during an ongoing outbreak, time periods when the mean estimated value of Rt is greater than one for ε = 0.2 and less than one for ε = 1 may be particularly concerning. In these periods, the outbreak might erroneously be inferred as being under control if the incorrect assumption that ε = 1 is made. In the analysis shown in figure 4b(i), this is the case for 20.8% of the time period considered. Of course, similarly to the analyses presented in §3a, analogous analyses could be performed based on different percentile estimates of Rt rather than the mean estimated value.
Second, we considered the dataset from Hainan Province, China. A previous study [36] compared the epidemiological features of imported and local cases in that province and found that imported cases tended to belong to older age groups than local cases. We applied a contact matrix for China [38] to the age distributions of imported and local cases, and thus estimated the expected number of contacts per day for imported cases (10.5) and local cases (13.4). To approximate the value of ε, we divided the expected number of contacts per day for imported cases by the analogous value for local cases, giving ε = 0.785. We then compared estimates of Rt for that more realistic value of ε = 0.785 (figure 4b(ii), green) with estimates of Rt under the standard assumption that ε = 1 (figure 4b(ii), black). Since a value of ε = 1 is only slightly larger than ε = 0.785, and the data from Hainan Province suggest only limited local transmission, we found that incorrectly assuming that ε = 1 did not have a substantial effect on inferred Rt values for this dataset.
4. Discussion
Summary statistics for tracking pathogen transmissibility are increasingly used during infectious disease outbreaks to guide decision making. Throughout the COVID-19 pandemic, Rt has been estimated in regions and countries worldwide (see e.g. [7]). This metric is useful and straightforward to interpret, corresponding to the number of individuals that one infected host is expected (on average) to go on to infect. As well as providing information about whether an outbreak is growing or declining, the value of Rt can be used to determine the proportion of transmissions that must be prevented for a growing outbreak to decline.
In this article, we have presented a modified version of the commonly used Cori method for inferring Rt [8,9]. We have accounted for different transmission risks from local and imported cases, rather than assuming that the transmission risk is identical for individuals in these groups. We provide an accompanying online software tool for estimating Rt (https://sabs-r3-epidemiology.github.io/branchpro) where users can upload their own data (disease incidence time-series and an estimate of the serial interval distribution—or multiple equally plausible serial interval distributions as described in Methods). We have conducted a systematic analysis of the dependence of inferred Rt values on the assumed relative transmission risk from an imported case compared with a local case (ε; see figures 2 and 3). We also considered examples in which it was possible to approximate the value of ε from other data sources (figure 4). In general, larger assumed values of ε lead to smaller Rt estimates. This is important, since assuming an unrealistically high value of ε may lead to the outbreak being falsely determined as under control. When an outbreak is ongoing, we have shown that the speed at which local transmission can be inferred as being either under control or not depends on the assumed value of ε. This dependence on ε demonstrates clearly that whether or not an outbreak is under control cannot always be inferred accurately from summary statistics that do not account for differences in the transmission risk between imported and local cases (e.g. the growth rate of overall cases). We have also shown how different percentile estimates of Rt can be used to guide decision making, according to the policymaker's level of acceptable risk.
A previous approach for estimating Rt allows infectees to have been infected either within or outside the local population [9]. However, in that framework, an assumption is made that the transmission risk from a local case is identical to the analogous risk from an imported case. The potential for different transmission risks from imported and local cases has implications for optimizing interventions, since if the risk of transmission is predominantly from imported cases, then travel restrictions and interventions that prevent transmissions from imported cases (e.g. quarantine of incoming travellers) may be the optimal measures. If instead the transmission risk is highest from local cases, then interventions such as social distancing and face coverings that reduce transmission from all infected individuals in the population may be necessary. In scenarios in which a novel pathogen variant is being imported into a new location from somewhere it is already widespread, the composition of variants causing local and imported cases might affect the relative transmission risk [39]. However, we note that in our modelling framework it is only the imported cases themselves that are assumed to represent a different transmission risk (rather than all infected individuals in a chain of transmission starting with an imported case).
A recent, closely related study by Tsang et al. [31] involved estimating independent Rt values for local and imported cases throughout an outbreak. A benefit of that approach is that it does not require an assumption to be made about the relative transmission risk from each imported case than from each local case. However, there are substantial logistical challenges to estimating independent Rt values for local and imported cases: this requires local cases who were infected by other local cases to be distinguished from those who were infected by imported cases. This may be possible either on a small scale or in locations with extensive contact tracing [31,40], but, in many situations, it is infeasible. In the absence of data with which to estimate Rt for local and imported cases independently, and without known changes in the relative transmission risk from imported compared with local cases, then assuming a constant relative transmission risk between the two types of case as we have done seems reasonable. To obtain an idea about whether the relative transmission risk (i.e. the parameter ε in our model) is likely to be less than or greater than one, we considered examples in which we approximated ε using either a reconstructed transmission network or the age characteristics of local and imported cases (figure 4). In both examples that we considered, the estimated value of ε was less than one, suggesting a lower transmission risk from each imported case than from each local case in those settings. Other approaches for inferring ε are also possible. One way to estimate ε is to analyse data containing both local and imported cases in small-scale settings in which infector–infectee transmission pairs can be identified or estimated, such as household or contact tracing studies. Another option might be to perform forwards contact tracing on imported cases at a single stage of the outbreak. If the value of εRt can be estimated from the contact tracing data at that stage, ε could then be estimated from the population-level incidence data. The contribution of imported cases to transmission is likely to vary by the time in the outbreak and by location [32,41]. In principle, estimates of ε could be updated based on the latest available contact tracing data.
In our main analyses, we have considered scenarios in which imported cases are individuals who have been infected in other geographical locations. However, an imported case may be defined as any case with an infection source outside the local host population. In the electronic supplementary material, we consider an analysis of MERS cases in Saudi Arabia in 2014–2015 (electronic supplementary material, figures S5 and S6), where cases are likely to have arisen both via human-to-human transmission and from an animal reservoir (specifically, from dromedary camels [42]). In that analysis, imported cases are assumed to be those reporting regular contacts with camels. It is possible that those individuals typically live in less densely populated areas than individuals who do not have regular contacts with camels, meaning that the relative risk of an imported case transmitting the virus is lower on average than the analogous risk from a local case. Like our analyses of COVID-19 datasets, our analysis of the MERS incidence data illustrates that assumptions about the relative transmission risk between local and imported cases can affect estimates of Rt and conclusions about whether or not local human-to-human transmission is under control.
We also conducted an additional supplementary analysis in which we generated synthetic epidemic datasets and investigated further the conditions under which mischaracterizing the relative transmissibility of imported and local cases affects estimates of Rt substantially. Specifically, we generated synthetic data for different values of ε and different strengths of local transmission. We calculated the error in estimates of Rt if the standard assumption that ε = 1 is made (electronic supplementary material, figure S8). This suggests that the largest errors occur when the relative transmissibility of imported (compared with local) cases differs substantially from one, and when imported cases represent a high proportion of the overall cases observed in the population.
In the research that we have presented, we sought to explore the relationship between heterogeneities in the onwards transmission risk between different groups of infectious individuals and inferred values of Rt. Practical applications of this approach should consider incorporating additional features into the modelling framework. An important consideration when assessing pathogen transmissibility during outbreaks is that Rt represents the average number of onwards infections over multiple infected individuals and transmission events. However, different infected individuals may generate very different numbers of infections [10,43–45]. The potential for super-spreading events at which large numbers of infections occur could be built into the underlying transmission model and into the resulting Rt estimates, although it may then be impossible to generate an analytic expression for the posterior for Rt. We sought to demonstrate the general principle that population heterogeneity can affect estimates of Rt. To do this as simply as possible, we used a model with only two groups of infected hosts (i.e. local and imported cases) and assumed that individuals are classified accurately as either local or imported. However, the classification of hosts into distinct groups may be imperfect (as considered elsewhere in this theme issue [46]), and many different sources of heterogeneity exist within host populations. There may be substantial differences in the transmission risk between other subgroups of the population: for example, risk may vary by age [10,25,26] and vaccination status [29]. Geographically distinct populations could be linked in a transmission model, so that spatial heterogeneity in Rt can be explored. In principle, compartmental models can be developed in which a range of different sources of heterogeneity are included, and Rt may be estimated using those compartmental models. It might also be possible to include further sources of heterogeneity in a renewal equation framework as studied here. These possibilities represent interesting avenues for future research.
Here, we assumed that the data represent disease incidence time-series, and that the serial interval (the time between successive symptomatic cases in a transmission chain) is always positive. In reality, pre-symptomatic infections occur, and serial intervals may take negative values [47–49] with infectors developing symptoms after some of the individuals who they infect. While the assumption of a positive valued serial interval distribution has been made in many previous studies in which Rt has been estimated for different pathogens, this issue can be avoided by using the incidence of infections and the generation time distribution [47,49,50] rather than the incidence of cases and the serial interval distribution [11]. The subtle difference here is that incidence time-series of cases do not reflect the times at which individuals were first infected, but instead reflect the times at which individuals were recorded as infected (which occurs after infection, for example when individuals display symptoms). Use of the incidence of infections and the generation time distribution may require the incidence of infections to be inferred from the incidence of cases, for example using an assumed incubation period distribution and the Richardson–Lucy deconvolution technique [51]. We note that the serial interval distribution may be different to the generation time distribution (specifically, pre-symptomatic transmission can lead to shorter serial intervals than generation times [49]). Another potential extension to our research is incorporation of different serial interval (or generation time) distributions for local and imported cases [40], particularly given that part of an imported case's infectious period may occur before they enter the local population. Reconstructed transmission networks might provide insights into these distributions.
More broadly, we note that Rt is only one summary statistic for tracking changes in transmission during an infectious disease outbreak. This metric does not provide information about the speed of the outbreak, which is better measured by the growth rate of cases [52,53,54]. Furthermore, current incidence of reported cases, hospitalizations and deaths are also key inputs to policy decisions. For example, an outbreak with Rt close to one is likely to have more detrimental impacts if case numbers are high compared with if case numbers are low. Nonetheless, Rt has been useful for guiding interventions during the COVID-19 pandemic, in combination with these other statistics. We therefore contend that studies that improve understanding of the impacts of factors affecting Rt estimates, such as heterogeneity in the onwards transmission risk between different infectious hosts, are valuable and an important component of preparedness for future outbreaks.
Data accessibility
The user-friendly web interface for estimating Rt while accounting for different transmission risks from local and imported cases can be found at https://sabs-r3-epidemiology.github.io/branchpro. All data and computing scripts required to reproduce the results presented here are available at https://github.com/SABS-R3-Epidemiology/transmission-heterogeneity-results. The source code of the branchpro Python package, which we developed to perform the inference presented in this article, is available at https://github.com/SABS-R3-Epidemiology/branchpro. No restrictions exist on data availability.
The data are provided in the electronic supplementary material [55].
Authors' contributions
R.C.: formal analysis, investigation, software, validation, visualization and writing—review and editing; D.A.: formal analysis, investigation, software, validation, visualization and writing—review and editing; I.B.: formal analysis, investigation, software, validation, visualization and writing—review and editing; H.J.F.: formal analysis, investigation, software, validation, visualizationand writing—review and editing; S.M.: formal analysis, investigation, software, validation, visualization and writing—review and editing; A.A.: formal analysis, investigation, software, validation, visualization and writing—review and editing; M.R.: supervision; A.L.: supervision; D.G.: project administration, supervision and writing—review and editing; B.C.L.: project administration, supervision and writing—review and editing; R.N.T.: conceptualization, methodology, project administration, supervision, writing—original draft and writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
The authors declare that they have no competing interests.
Funding
This research was funded by the EPSRC via a doctoral training partnership studentship in the Department of Computer Science at the University of Oxford (R.C.), by the EPSRC via the CDT in Sustainable Approaches to Biomedical Science: Responsible and Reproducible Research – SABS:R3 grant no. EP/S024093/1 (I.B., H.J.F., S.M., A.A. and M.R.), by the Clarendon Fund (D.A.) and by the UKRI via grant no. EP/V053507/1 (R.N.T.). A.L.D. was funded as a Roche Pharmaceutical Research employee. The funders had no role in study design, data analysis, preparation of the manuscript or the decision to publish.
References
- 1.Brooks-Pollock E, Danon L, Jombart T, Pellis L. 2021. Modelling that shaped the early COVID-19 pandemic response in the UK. Phil. Trans. R. Soc. B 376, 20210001. ( 10.1098/rstb.2021.0001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson RN. 2020. Epidemiological models are important tools for guiding COVID-19 interventions. BMC Med. 18, 152. ( 10.1186/s12916-020-01628-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies NG et al., CMMID COVID-19 working group. 2020. The effect of non-pharmaceutical interventions on COVID-19 cases, deaths and demand for hospital services in the UK: a modelling study. Lancet Public Health 5, e375-e385. ( 10.1016/S2468-2667(20)30133-X) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moore S, Hill EM, Dyson L, Tildesley MJ, Keeling MJ. 2020. Modelling optimal vaccination strategy for SARS-CoV-2 in the UK. PLoS Comput. Biol. 17, e1008849. ( 10.1371/journal.pcbi.1008849) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dehning J, Zierenberg J, Spitzner EP, Wibral M, Neto JP, Wilczek M, Priesemann V. 2020. Inferring change points in the spread of COVID-19 reveals the effectiveness of interventions. Science 369, eabb9789. ( 10.1126/science.abb9789) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birrell P, Blake J, Van Leeuwen E, Gent N, De Angelis D. 2020. Real-time nowcasting and forecasting of COVID-19 dynamics in England: the first wave. Phil. Trans. R. Soc. B 376, 20200279. ( 10.1098/rstb.2020.0279) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abbott S et al. 2020. Estimating the time-varying reproduction number of SARS-CoV-2 using national and subnational case counts. Wellcome Open Res. 5, 112. ( 10.12688/wellcomeopenres.16006.1) [DOI] [Google Scholar]
- 8.Cori A, Ferguson NM, Fraser C, Cauchemez S. 2013. A new framework and software to estimate time-varying reproduction numbers during epidemics. Am. J. Epidemiol. 178, 1505-1512. ( 10.1093/aje/kwt133) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson RN et al. 2019. Improved inference of time-varying reproduction numbers during infectious disease outbreaks. Epidemics 29, 100356. ( 10.1016/j.epidem.2019.100356) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson RN et al. 2020. Key questions for modelling COVID-19 exit strategies. Proc. R. Soc. B 287, 20201405. ( 10.1098/rspb.2020.1405) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gostic KM et al. 2020. Practical considerations for measuring the effective reproductive number, Rt. PLoS Comput. Biol. 16, e1008409. ( 10.1371/journal.pcbi.1008409) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White LF, Moser CB, Thompson RN, Pagano M. 2020. Statistical estimation of the reproductive number from case notification data. Am. J. Epidemiol. 190, 611-620. ( 10.1093/aje/kwaa211) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishiura H, Chowell G. 2009. The effective reproduction number as a prelude to statistical estimation of time-dependent epidemic trends. In Mathematical and statistical estimation approaches in epidemiology, pp. 103-121. Dordrecht, The Netherlands: Springer. [Google Scholar]
- 14.Cowling BJ, Lau MSY, Ho LM, Chuang SK, Tsang T, Liu SH, Leung PY, Lo SV, Lau EH. 2010. The effective reproduction number of pandemic influenza: prospective estimation. Epidemiology 21, 842-846. ( 10.1097/EDE.0b013e3181f20977) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cauchemez S, Boëlle PY, Donnelly CA, Ferguson NM, Thomas G, Leung GM, Hedley AJ, Anderson RM, Valleron AJ. 2006. Real-time estimates in early detection of SARS. Emerg. Infect. Dis. 12, 110-113. ( 10.3201/eid1201.050593) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng Q, Liu Z, Cheng G, Huang J. 2020. Heterogeneity and effectiveness analysis of COVID-19 prevention and control in major cities in China through time-varying reproduction number estimation. Sci. Rep. 10, 21953. ( 10.1038/s41598-020-79063-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ackland GJ, Ackland JA, Antonioletti M, Wallace DJ. 2022. Fitting the reproduction number from UK coronavirus case data and why it is close to 1. Phil. Trans. R. Soc. A 380, 20210301. ( 10.1098/rstb.2021.0301) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.UK Government. 2021. The R value and growth rate. See https://www.gov.uk/guidance/the-r-value-and-growth-rate.
- 19.Fraser C. 2007. Estimating individual and household reproduction numbers in an emerging epidemic. PLoS ONE 2, e758. ( 10.1371/journal.pone.0000758) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Obadia T, Haneef R, Boëlle PY. 2012. The R0 package: a toolbox to estimate reproduction numbers for epidemic outbreaks. BMC Med. Inform. Decis. Mak. 12, 147. ( 10.1186/1472-6947-12-147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vegvari C et al. 2021. Commentary on the use of the reproduction number R during the COVID-19 pandemic. Stat. Methods Med. Res. 1, 09622802211037079. ( 10.1177/09622802211037079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wallinga J, Teunis P. 2004. Different epidemic curves for severe acute respiratory syndrome reveal similar impacts of control measures. Am. J. Epidemiol. 160, 509-516. ( 10.1093/aje/kwh255) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ali ST, Wang L, Lau EHY, Xu XK, Du Z, Wu Y, Leung GM, Cowling BJ. 2020. Serial interval of SARS-CoV-2 was shortened over time by nonpharmaceutical interventions. Science 369, 1106-1109. ( 10.1126/science.abc9004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ladhani SN et al. 2020. Increased risk of SARS-CoV-2 infection in staff working across different care homes: enhanced COVID-19 outbreak investigations in London care homes. J. Infect. 81, 621-624. ( 10.1016/j.jinf.2020.07.027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davies NG, Klepac P, Liu Y, Prem K, Jit M, Eggo RM, CMMID COVID-19 Working Group. 2020. Age-dependent effects in the transmission and control of COVID-19 epidemics. Nat. Med. 26, 1205-1211. ( 10.1038/s41591-020-0962-9) [DOI] [PubMed] [Google Scholar]
- 26.Keeling MJ et al. 2021. Predictions of COVID-19 dynamics in the UK: short-term forecasting and analysis of potential exit strategies. PLoS Comput. Biol. 17, e1008619. ( 10.1371/journal.pcbi.1008619) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lovell-Read FA, Shen S, Thompson RN. 2022. Estimating local outbreak risks and the effects of non-pharmaceutical interventions in age-structured populations: SARS-CoV-2 as a case study. J. Theor. Biol. 535, 110983. ( 10.1016/j.jtbi.2021.110983) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pooley CM, Doeschl-Wilson AB, Marion G. 2022. Estimation of age-stratified contact rates during the COVID-19 pandemic using a novel inference algorithm. Phil. Trans. R. Soc. A 380, 20210298. ( 10.1098/rsta.2021.0298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keeling MJ, Dyson L, Hill E, Moore S, Tildesley MJ. 2021. Road map scenarios and sensitivity: Step 4. [cited 4 Aug 2021]. See https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/993358/s1288_Warwick_RoadMap_Step_4.pdf.
- 30.Sachak-Patwa R, Byrne HM, Dyson L, Thompson RN. 2021. The risk of SARS-CoV-2 outbreaks in low prevalence settings following the removal of travel restrictions. Comms. Med. 1, 39. ( 10.1038/s43856-021-00038-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsang TK, Wu P, Lau EHY, Cowling BJ. 2021. Accounting for imported cases in estimating the time-varying reproductive number of COVID-19 in Hong Kong. J. Infect. Dis. 224, 783-787. ( 10.1093/infdis/jiab299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price DJ et al. 2020. Early analysis of the Australian COVID-19 epidemic. eLife 9, e58785. ( 10.7554/eLife.58785) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Government of Ontario. 2021. COVID-19 data: likely source of infection. See https://covid-19.ontario.ca/data/likely-source-infection.
- 34.Hong Kong Department of Health. 2021 Latest local situation of COVID-19. See https://data.gov.hk/en-data/dataset/hk-dh-chpsebcddr-novel-infectious-agent/resource/ec4b49af-83e0-4c71-a3ba-14120e453b9d.
- 35.Liu Y, Gu Z, Liu J. 2021. Uncovering transmission patterns of COVID-19 outbreaks: a region-wide comprehensive retrospective study in Hong Kong. EClinicalMedicine 36, 100929. ( 10.1016/j.eclinm.2021.100929) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu B et al. 2020. Compare the epidemiological and clinical features of imported and local COVID-19 cases in Hainan, China. Infect. Dis. Poverty 9, 143. ( 10.1186/s40249-020-00755-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishiura H, Linton NM, Akhmetzhanov AR. 2020. Serial interval of novel coronavirus (COVID-19) infections. Int. J. Infect. Dis. 93, 284-286. ( 10.1016/j.ijid.2020.02.060) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prem K, Cook AR, Jit M. 2017. Projecting social contact matrices in 152 countries using contact surveys and demographic data. PLoS Comp. Biol. 13, e1005697. ( 10.1371/journal.pcbi.1005697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Challen R et al. 2021. Early epidemiological signatures of novel SARS-CoV-2 variants: establishment of B.1.617.2 in England. medRxiv. ( 10.1101/2021.06.05.21258365) [DOI]
- 40.Li M, Liu K, Song Y, Wang M, Wu J. 2021. Serial interval and generation interval for imported and local infectors, respectively, estimated using reported contact-tracing data of COVID-19 in China. Front. Public Health 8, 577431. ( 10.3389/fpubh.2020.577431) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russell TW, Wu JT, Clifford S, Edmunds WJ, Kucharski AJ, Jit M, Centre for the Mathematical Modelling of Infectious Diseases COVID-19 working group. 2021. Effect of internationally imported cases on internal spread of COVID-19: a mathematical modelling study. Lancet Public Health 6, e12-e20. ( 10.1016/S2468-2667(20)30263-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haagmans BL et al. 2014. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect. Dis. 14, 140-145. ( 10.1016/S1473-3099(13)70690-X) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Endo A, Abbott S, Kucharski AJ, Funk S, CMMID COVID-19 working group. 2020. Estimating the overdispersion in COVID-19 transmission using outbreak sizes outside China. Wellcome Open Res. 5, 67. ( 10.12688/wellcomeopenres.15842.3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lloyd-Smith JO, Schreiber SJ, Kopp PE, Getz WM. 2005. Superspreading and the effect of individual variation on disease emergence. Nature 438, 355-359. ( 10.1038/nature04153) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Akhmetzhanov AR, Jung SM, Cheng HY, Thompson RN. 2021. A hospital-related outbreak of SARS-CoV-2 associated with variant Epsilon (B.1.429) in Taiwan: transmission potential and outbreak containment under intensified contact tracing, January–February 2021. Int. J. Infect. Dis. 110, 15-20. ( 10.1016/j.ijid.2021.06.028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li W, Bulekova K, Gregor B, White LF, Kolaczyk ED. 2022. Estimation of local time-varying reproduction numbers in noisy surveillance data. Phil. Trans. R. Soc. A 380, 20210303. ( 10.1098/rsta.2021.0303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hart WS, Maini PK, Thompson RN. 2021. High infectiousness immediately before COVID-19 symptom onset highlights the importance of continued contact tracing. eLife 10, e65534. ( 10.7554/eLife.65534) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Du Z, Xu X, Wu Y, Wang L, Cowling BJ, Meyers LA. 2020. Serial interval of COVID-19 among publicly reported confirmed cases. Emerg. Infect. Dis. 26, 1341-1343. ( 10.3201/eid2606.200357) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hart WS, Miller E, Andrews NJ, Waight P, Maini PK, Funk S, Thompson RN. 2022. Generation time of the alpha and delta SARS-CoV-2 variants: an epidemiological analysis. Lancet Infect. Dis. 22, 603-610. ( 10.1016/S1473-3099(22)00001-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hart WS, Abbott S, Endo A, Hellewell J, Miller E, Andrews N, Maini PK, Funk S, Thompson RN. 2022. Inference of the SARS-CoV-2 generation time using UK household data. eLife 11, e70767. ( 10.7554/eLife.70767) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goldstein E, Dushoff J, Ma J, Plotkin JB, Earn DJD, Lipsitch M. 2009. Reconstructing influenza incidence by deconvolution of daily mortality time series. Proc. Natl Acad. Sci. USA 106, 21829. ( 10.1073/pnas.0902958106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dushoff J, Park SW. 2021. Speed and strength of an epidemic intervention. Proc. R. Soc. B 288, 20201556. ( 10.1098/rspb.2020.1556) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pellis L et al. 2021. Challenges in control of Covid-19: short doubling time and long delay to effect of interventions. Phil. Trans. R. Soc. B 376, 20200264. ( 10.1098/rstb.2020.0264) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parag KV, Thompson RN, Donnelly CA. 2022. Are epidemic growth rates more informative than reproduction numbers? J. Roy. Stat Soc. 1, 1– 11. ( 10.1111/rssa.12867) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Creswell R et al. 2022. Heterogeneity in the onwards transmission risk between local and imported cases affects practical estimates of the time-dependent reproduction number. Figshare. ( 10.6084/m9.figshare.c.6070469) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Creswell R et al. 2022. Heterogeneity in the onwards transmission risk between local and imported cases affects practical estimates of the time-dependent reproduction number. Figshare. ( 10.6084/m9.figshare.c.6070469) [DOI] [PMC free article] [PubMed]
Data Availability Statement
The user-friendly web interface for estimating Rt while accounting for different transmission risks from local and imported cases can be found at https://sabs-r3-epidemiology.github.io/branchpro. All data and computing scripts required to reproduce the results presented here are available at https://github.com/SABS-R3-Epidemiology/transmission-heterogeneity-results. The source code of the branchpro Python package, which we developed to perform the inference presented in this article, is available at https://github.com/SABS-R3-Epidemiology/branchpro. No restrictions exist on data availability.
The data are provided in the electronic supplementary material [55].