Abstract
Scorpion α-toxins bind at the pharmacologically-defined site-3 on the sodium channel and inhibit channel inactivation by preventing the outward movement of the voltage sensor in domain IV (IVS4), whereas scorpion β-toxins bind at site-4 on the sodium channel and enhance channel activation by trapping the voltage sensor of domain II (IIS4) in its outward position. However, limited information is available on the role of the voltage-sensing modules (VSM, comprising S1–S4) of domains I and III in toxin actions. We have previously shown that charge reversing substitutions of the innermost positively-charged residues in IIIS4 (R4E, R5E) increase the activity of an insect-selective site-4 scorpion toxin, Lqh-dprIT3-c, on BgNav1–1a, a cockroach sodium channel. Here we show that substitutions R4E and R5E in IIIS4 also increase the activity of two site-3 toxins, LqhαIT from Leiurusquinquestriatus hebraeus and insect-selective Av3 from Anemonia viridis. Furthermore, charge reversal of either of two conserved negatively-charged residues, D1K and E2K, in IIIS2 also increase the action of the site-3 and site-4 toxins. Homology modeling suggests that S2-D1 and S2-E2 interact with S4-R4 and S4-R5 in the VSM of domain III (III-VSM), respectively, in the activated state of the channel. However, charge swapping between S2-D1 and S4-R4 had no compensatory effects on gating or toxin actions, suggesting that charged residue interactions are complex. Collectively, our results highlight the involvement of III-VSM in the actions of both site 3 and site 4 toxins, suggesting that charge reversing substitutions in III-VSM allosterically facilitate IIS4 or IVS4 voltage sensor trapping by these toxins.
Keywords: Insect sodium channel, Scorpion α-toxin, Scorpion β-toxin, Homology modeling, Mutagenesis, Electrophysiology
1. Introduction
Voltage-gated sodium channels (Navs) initiate and propagate action potentials in most excitable cells. Navs activate rapidly upon membrane depolarization and then inactivate within milliseconds (Catterall, 2000; Hodgkin and Huxley, 1952). The pore-forming α-subunit of sodium channels consists of four homologous domains (I to IV), each containing six transmembrane segments (S1–S6). Segments S5, S6 and their interconnecting linker form the pore module (PM), while segments S1–S4 form the voltage-sensing module (VSM). The S4 segment in each domain contains repeated motifs of a positively charged Arg or Lys residue followed by two hydrophobic residues (Catterall, 2000). The S4 segments sense alterations in membrane potential and respond by outward movement during channel activation, initiating a conformational change that opens the channel pore (Armstrong, 1981; Catterall, 2010), which is then followed by fast inactivation.
Negative counter charges in S1–S3 of the VSM are highly conserved (Palovcak et al., 2014) and are proposed to interact with positively charged voltage sensor residues to coordinate the S4 translocation across a gating pore within the hydrophobic milieu of the membrane in response to membrane depolarization (reviewed by Groome and Bayless-Edwards, 2020). This ‘sliding helix’ model of ion channel gating has been supported by structural analysis coupled with disulfide cross-linking experiments of the VSM in prokaryotic sodium channels (DeCaen et al., 2011; DeCaen et al., 2009; DeCaen et al., 2008; Paldi and Gurevitz, 2010; Payandeh et al., 2012; Payandeh et al., 2011; Yarov-Yarovoy et al., 2012; Zhang et al., 2012a; reviewed by Groome, 2014) and with molecular dynamics simulations in eukaryotic channels (Delemotte et al., 2011; Gosselin-Badaroudine et al., 2012).
Due to their critical role in neuronal signaling and muscle fiber contractility, sodium channels are targeted by a variety of naturally-occurring neurotoxins, including venom toxins from scorpions and spiders. These toxins are thus valuable tools for exploring structure-function relations in sodium channels (Catterall, 1992; Cestele and Catterall, 2000; Dutertre and Lewis, 2010; Gordon, 1997; Nicholson, 2007; Terlau and Olivera, 2004). They are also potential leads for development of new selective insecticides because many of these venom toxins exhibit host specificity, being toxic to insects, but not to mammals (Krapcho et al., 1995; Strugatsky et al., 2005; Zlotkin et al., 2000; Zhu et al., 2020).
Scorpion α-toxins, several sea anemone toxins and some spider toxins inhibit channel fast inactivation upon binding to the pharmacologically defined site-3, which comprises residues in the VSM of domain IV and residues in the PM of domain I (Catterall et al., 2007; Gordon et al., 1992; Gur Barzilai et al., 2014; Gur et al., 2011; Gurevitz, 2012; Ma et al., 2013; Moran et al., 2007; Nicholson, 2007; Thomsen and Catterall, 1989; Wang et al., 2011). On the other hand, scorpion β-toxins enhance the activation of sodium channels upon binding to site-4, which is formed by residues in the VSM of domain II and the pore module of domain III (Gurevitz et al., 2007; Song et al., 2011; Zhang et al., 2011, 2012b).
The voltage sensor trapping model, in which toxins hinder the inward (deactivating) movement of the S4 segment from its outward activated state, illustrates the pivotal role of S4 in domains II and IV in the action of site-4 and site-3 toxins, respectively (Catterall et al., 2007; Cestele and Catterall, 2000; Zhang et al., 2011, b). However, the role of the VSMs of domains I and III in the action of these toxins is largely unclear. Song et al. (2011) previously reported that charge-reversing substitutions of the fourth and fifth gating charges in IIIS4, R4E and R5E, increased the sensitivity of the cockroach sodium channel, BgNav1–1a, to the insect-selective scorpion β-toxin, Lqh-dprIT3, suggesting that IIIS4 modulates the voltage-sensor trapping of IIS4. In this study, we examined the role of the VSM of domain III (III-VSM) in the action of two site-3 toxins, LqhαIT from the scorpion Leiurusquinquestriatus hebraeus and the insect-selective Av3 from the sea anemone Anemonia viridis, on the BgNav1–1a channel. We systematically conducted charge-reversal mutational analysis of the five positively-charged residues in IIIS4 and of four negatively-charged residues in IIIS1 and IIIS2 in BgNav1–1a, and examined the effects of the two site-3 toxins and the site-4 toxin Lqh-dprIT3 on these mutant channels. Our results show that alterations in III-VSM, which is not considered to constitute the binding site of site-3 or site-4 toxins, affect the action of both types of toxins, most likely by an allosteric mechanism.
2. Methods
2.1. Toxin production and functional analysis
Lqh-dprIT3-c (hereafter referred to as Lqh-dprIT3) and LqhαITof the scorpion Leiurusquinquestriatus hebraeus, and Av3 from the sea anemone Anemonia viridis, were produced in recombinant forms and analyzed as previously described (Moran et al., 2007; Strugatsky et al., 2005; Zilberberg et al., 1996).
2.2. Site-directed mutagenesis
Site-directed mutagenesis was performed via PCR using oligonucleotide primers and Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA, USA). The primers used in site-directed mutagenesis are summarized in Table S1. The mutations were verified by DNA sequencing.
2.3. Expression of BgNav1–1a channels in Xenopus oocytes
Xenopus oocytes were purchased from Xenopus 1 (Dexter, Michigan). The procedures for oocyte preparation and cRNA injection were identical to those described previously (Tan et al., 2002). For robust expression of BgNav1–1a channels, cRNA (1 ng) was co-injected into oocytes with D. melanogaster tipE cRNA (1:1 ratio), to enhance channel expression (Feng et al., 1995; Warmke et al., 1997).
2.4. Electrophysiological recording and analysis
Voltage dependent activation and fast inactivation were measured and analyzed using the two-electrode voltage clamp technique as previously described (Tan et al., 2005). Sodium currents were measured with a Warner OC-725C oocyte clamp and a Digidata 1440A interface. Data were sampled at 50 kHz and filtered at 2 kHz. Leak currents were corrected by p/4 subtraction. pCLAMP 10.2 was used for data acquisition and analysis. The maximal sodium current peak amplitude was limited to ˂ 2.0 μA to achieve optimal voltage control by adjusting the concentration of cRNA injected and timing of recording. The protocols to determine gating properties and toxin effects were identical to those described previously (Gao et al., 2014; Song et al., 2011). To examine the effects of Lqh-dprIT3, a 20 Hz train of 50, 5-ms depolarizing prepulses to 50 mV (conditioning pulses) were applied, followed by a 20 ms depolarizing test pulse between −80 and 65 mV from a holding potential of −120 mV. The voltage dependence of sodium channel conductance (G/V) was calculated by measuring peak current amplitude in response to test potentials ranging from −80 to 65 mV in 5 mV increments and divided by (V - Vrev), where V is the test potential and Vrev is the reversal potential for sodium ions. Peak conductance values were normalized to the maximal conductance (Gmax) and fitted with a two-state Boltzmann equation G/Gmax = [1 + exp (V–V1/2)/k]−1, where k is the slope factor, V is the command potential and V½ is the voltage for half-maximal activation.
The extent of channel modification by Lqh-dprIT3 was determined by the percentage of channels with the voltage dependence of activation shifted to negative membrane potentials, as derived from fractional amplitudes of double Boltzmann fits of the conductance-voltage relationship. The percentages of channel modification by LqhαIT and Av3 were measured by I20/Ipeak, where Ipeak is the peak current elicited by a 20 ms step depolarization to −10 mV and I20 is the non-inactivated current (due to toxin action) at the end of that depolarization. The ratio of I20/Ipeak reflects the potency of α-toxins affecting fast inactivation (Gao et al., 2014). Data are presented as means ± SEM. Statistical significance was determined by using one-way analysis of variance (ANOVA) with Scheffe’s post hoc analysis and significant value were set at p˂0.05.
2.5. Structural modeling
The secondary structure of Nav channels was predicted using JPred4 (Drozdetskiy et al., 2015) to confirm the preservation of helices. Multiple sequence alignment for BgNav domains I-IV, NavAb from the bacterium Arcobacter butzleri (Payandeh et al., 2011), and NaChBac from Bacillus halodurans was carried out using MUSCLE (Edgar, 2004). The structure of NavAb was downloaded from the Protein Data Bank (4EKW. pdb) and used as template. Homology modelling was performed using Modeller 2.0 (Payandeh et al., 2012; Sali and Blundell, 1993) and the best structure was selected based on the GA341 output. Energy minimization (1500 steps) to detect possible charge interactions was carried out using AmberTools. The convpdb.pl utility in the MMTSB tool set (Feig et al., 2004) was used to prepare the input for the AmberTools. The resulting protein structures were visualized using VMD (Humphrey et al., 1996). Multi-sequence alignment was conducted with Multalign Viewer in UCSF Chimera package (Pettersen et al., 2004).
3. Results
3.1. Substitutions R4E and R5E in IIIS4 increase the inhibitory effect of Av3 and LqhαIT on channel inactivation
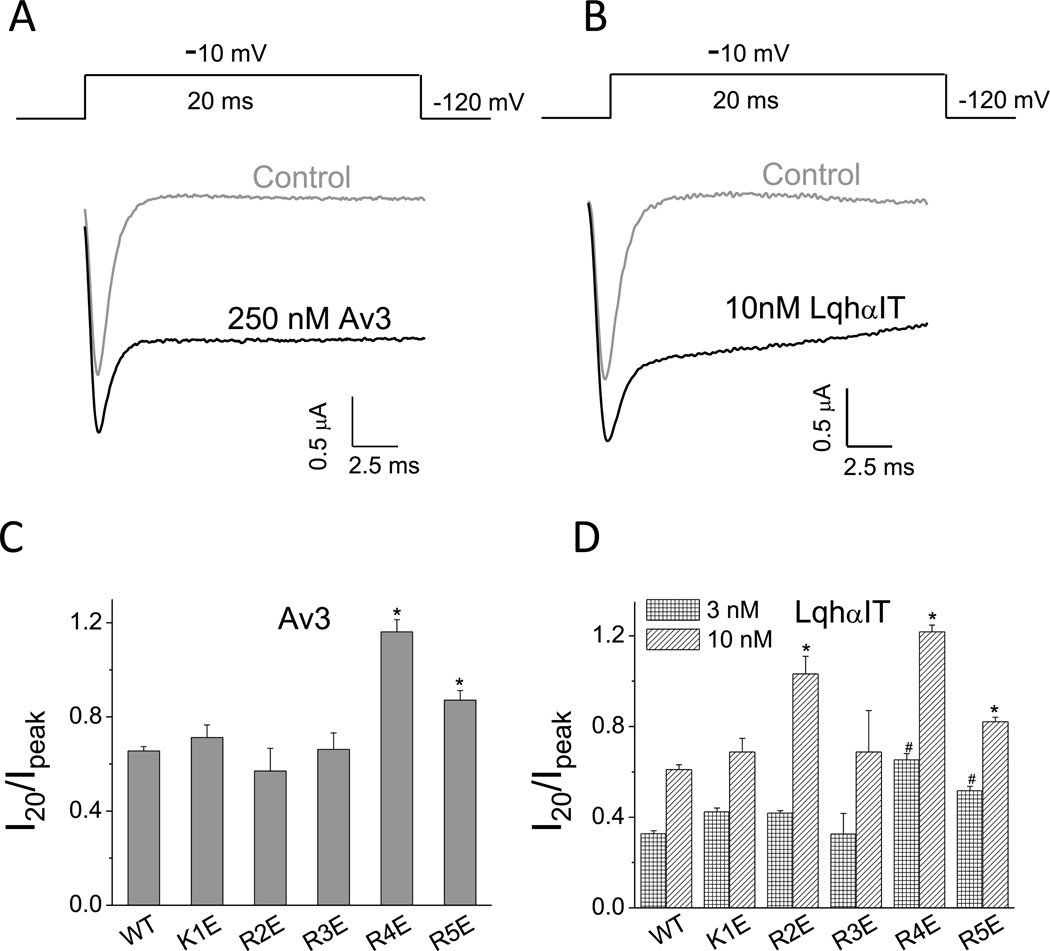
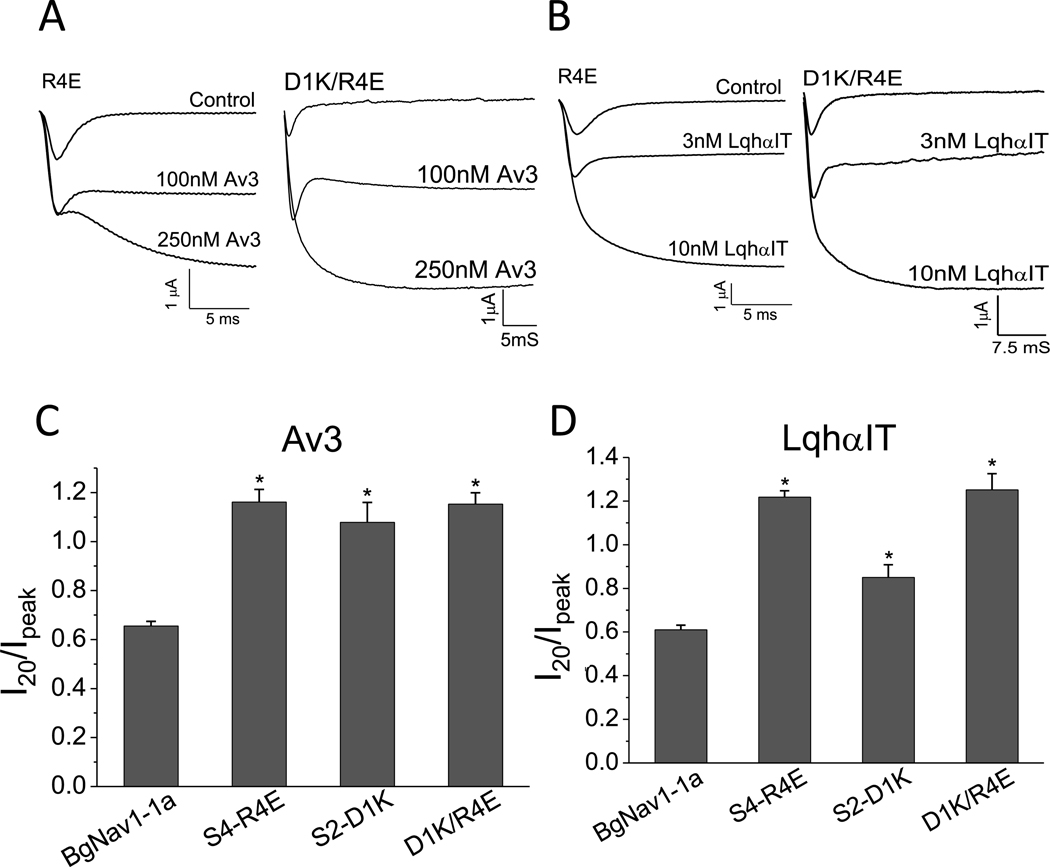
The inhibitory effect of Av3 and LqhαIT on fast inactivation of BgNav1–1a was quantified by determining the ratio of I20/Ipeak, as was previously described (Gao et al., 2014) (Fig. 1A and B). In addition, an increase in amplitude of the peak current was observed. The effect of charge reversal for each of the five positively-charged channel residues in IIIS4 was examined by measuring the activity of Av3 and LqhαIT on the channel mutants. Compared to WT BgNav1–1a, charge reversal of the 4th arginine (R4E) or 5th arginine (R5E) increased the inhibitory effect of either toxin (Fig. 1C and D; Fig. S1A), while charge reversals K1E or R3E had no effect on the action of either toxin at any dose tested. Substitution R2E revealed a significant effect on the action of LqhαIT at 10 nM, but not at 3 nM. Notably, in the absence of toxins, fast inactivation of the mutant channels was similar to that of the WT channel.
Fig. 1.
Substitutions R4E and R5E in IIIS4 increase the action of Av3 and LqhαIT. A-B, Representative sodium current traces in the absence or presence of Av3 (A) or LqhαIT (B). The protocols are presented above the current traces: the currents were elicited by a 20 ms test pulse to −10 mV, from a holding potential of −120 mV. C-D, Effects of charge reversal of the five positively-charged residues in IIIS4 on BgNav1–1a sensitivity to Av3 (C) or LqhαIT (D). The inhibitory effect on fast inactivation by the toxins was calculated by measuring the current that remained at 20 ms (I20) divided by the peak current (Ipeak). * denotes significant difference compared to WT BgNav1–1a channels (p˂ 0.05).
3.2. Charge reversal of two negative residues in IIIS2 facilitates the action of site-3 and site-4 toxins
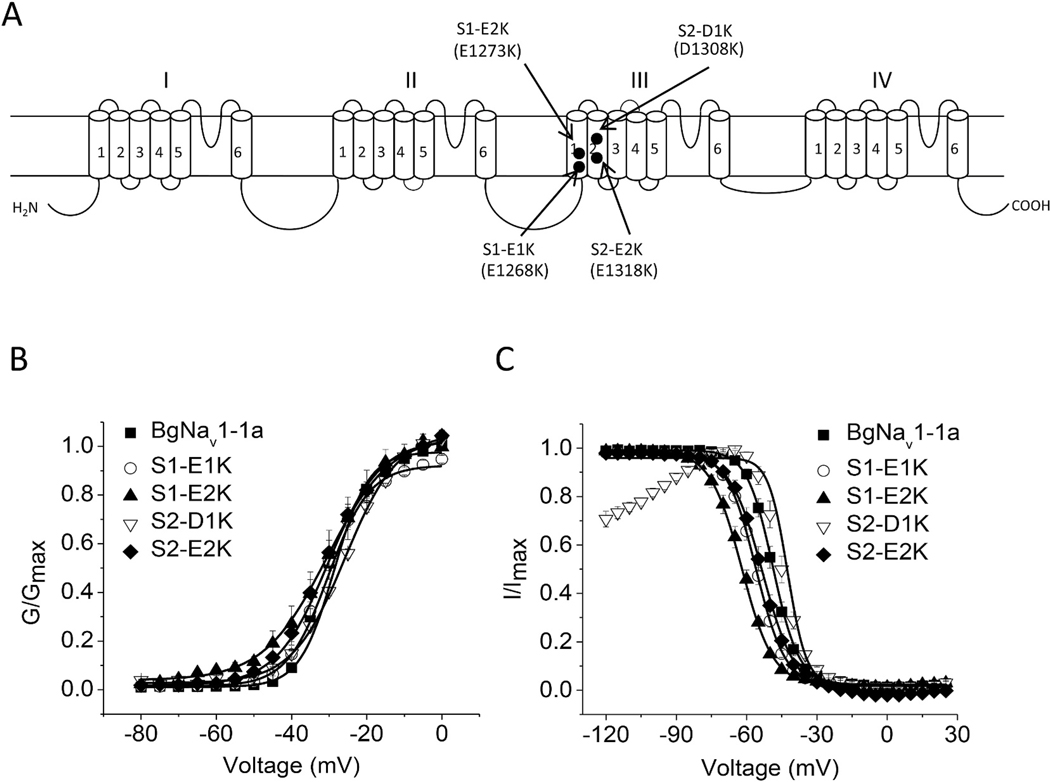
Negative countercharges in S1–S3 of the VSMs presumably interact with positively-charged voltage sensor residues (DeCaen et al., 2008, 2009, 2011; Chakrapani et al., 2010; Paldi and Gurevitz, 2010; Gosselin-Badaroudine et al., 2012; Yarov-Yarovoy et al., 2012; Groome and Winston, 2013) and so stabilize intermediate channel states during S4 translocation in response to alteration in membrane potential. These interactions may affect toxin binding, as it has been previously shown that substitution of the outermost negative charge E1290 in IIIS1 increases the sensitivity of a BgNaV1–1a splice variant to the site-4 toxin Lqh-dprIT3 (Song et al., 2011). To examine whether other counter-charges in S1 or S2 play a role in the action of site-3 and site-4 toxins, we examined the effect of toxins on mutations for each of the four highly conserved negatively-charged residues in the VSM of channel domain III. In S1, intracellular E1268 and E1273 were substituted with lysine (S1-E1K and S1-E2K, respectively). In S2, the extracellular D1308 and intracellular E1318 were also substituted with lysine, (S2-D1K and S2-E2K, respectively). All four channel mutants generated sufficient sodium currents in Xenopus oocytes enabling functional analyses.
The effects of these charge reversing mutations on channel gating are shown in Fig. 2. Compared to WT channels, substitutions S1-E1K, S1-E2K and S2-E2K rendered a shift in the voltage-dependence of channel fast inactivation to more hyperpolarizing voltages with no significant effect on channel activation (Fig. 2B and C; Table 1). In contrast, substitution S2-D1K caused a shift in the voltage-dependence of channel activation and fast inactivation to more depolarizing voltages (Fig. 2B and C; Table 1), and increased the extent of inactivation at potentials more hyperpolarized than −80 mV (Fig. 2C).
Fig. 2.
Voltage dependence of activation and inactivation of four charge reversal channel mutants at IIIS1 and IIIS2. A, Schematic diagram of a sodium channel indicating the positions of the charge reversing substitutions of four highly conserved positively-charged residues and negatively-charged residues in domain III. B–C, Voltage dependence of activation (B) and inactivation (C). Data are presented as mean ± SEM for 10–20 oocytes. * indicates a significant difference compared to that of BgNav1–1a channels only using a one-way ANOVA with Scheffe’s post hoc analysis (p < 0.05).
Table 1.
Voltage dependence of activation and inactivation of BgNav1–1a and mutants.
| Nav channel type | Activation |
Inactivation |
n | ||
|---|---|---|---|---|---|
| V1/2 (mV) | k (mV) | V1/2 (mV) | k (mV) | ||
|
| |||||
| BgNav1–1a | − 29.6 ± 1.3 | 4.6 ± 0.4 | − 49.2±1.0 | 4.7 ± 0.1 | 11 |
| IIIS1-E1K | − 31.3 ± 1.2 | 5.6 ± 0.2 | −56.1 ± 0.9* | 5.1 ± 0.1 | 26 |
| IIIS1-E2K | − 30.8 ± 1.0 | 7.4 ± 0.4 | − 61.9 ± 1.1* | 5.2 ± 0.1 | 13 |
| IIIS2-E2K | − 31.1 ± 1.6 | 6.6 ± 0.3 | − 53.7 ± 1.2* | 6.0 ± 0.2 | 13 |
| IIIS2-D1K | −26.0 ± 0.9* | 7.1 ± 0.2 | − 43.7 ± 1.2* | 3.7 ± 0.2 | 13 |
| IIIR4E | − 36.2 ± 1.8* | 4.6 ± 0.4 | − 49.2 ± 1.0 | 4.7 ± 0.1 | 11 |
| IIIS2-D1K/IIIR4E | − 41.2 ± 1.2* | 8.1 ± 0.2 | − 79.3 ± 1.5* | 8.8 ± 0.5 | 15 |
The voltage dependence values of conductance and inactivation were fitted with a two-state Boltzmann equation to determine V1/2, the voltage for half maximal conductance or inactivation, and k, the slope factor for conductance or inactivation. The values in the table represent the mean ± SEM, and n is the number of oocytes tested. The asterisks indicate significant differences as determined by one-way ANOVA (p < 0.05) with Scheffe’s post hoc analysis.
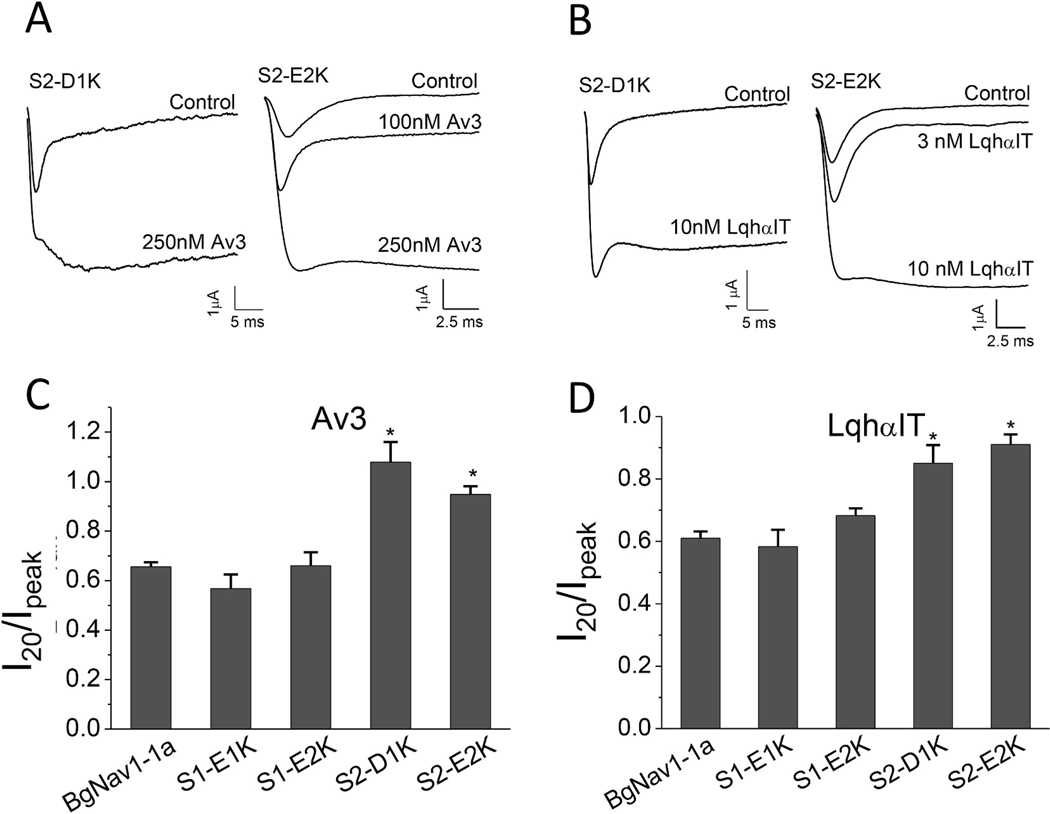
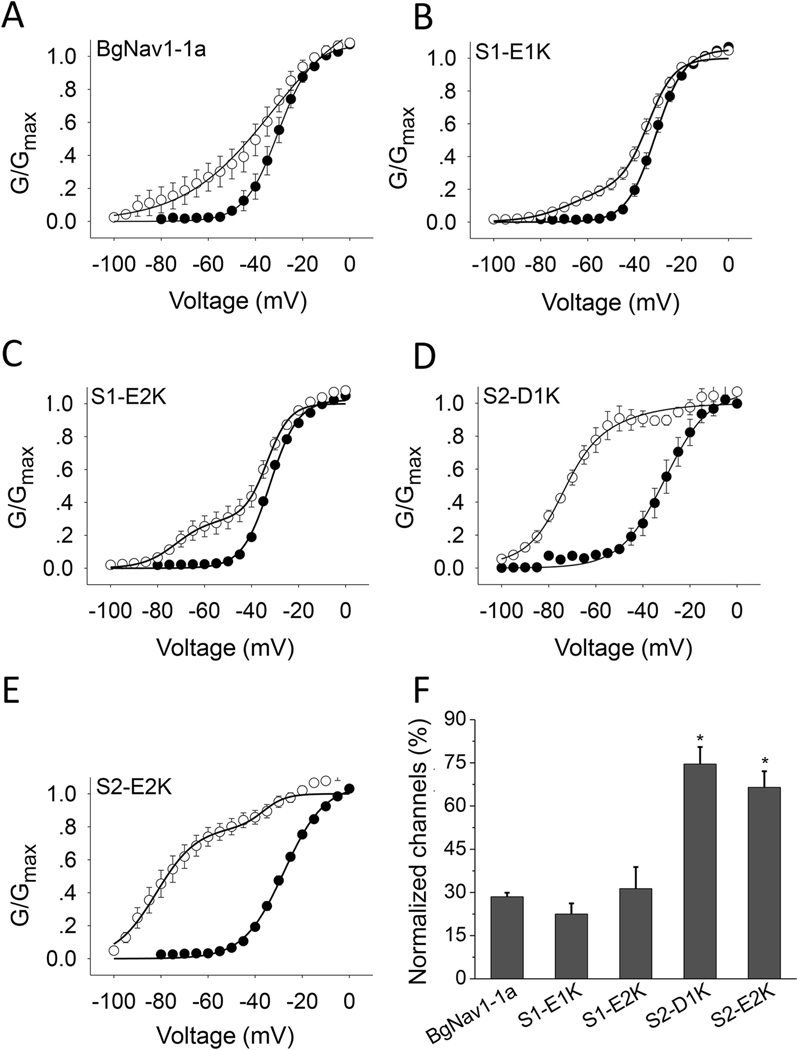
Charge reversing substitutions in S2, S2-D1K or S2-E2K, but not in S1, S1-E1K, S1-E2K, increased the effect of the site-3 toxins. Whereas both Av3 and LqhαIT induced greater non-inactivating currents in S2-D1K and S2-E2K channels compared to WT channels, their effect on S1-E1K and S1-E2K channel mutants was similar to that obtained on WT channels (Fig. 3; Fig. S1B). A similar profile was obtained when monitoring the effects of the charge reversing substitutions on the action of Lqh-dprIT3 site-4 toxin. At 300 nM, Lqh-dprIT3 enhanced BgNav1–1a activation, as indicated by a shift in the hyperpolarizing direction with 28% of the channels modified. This effect of toxin was increased in mutant IIIS2 channels (75% modified for S2-D1K channels and 67% for the S2-E2K channels (Fig. 4D–F; Table 2) but was unaltered by mutant IIIS1 channels (Fig. 4A–C, F; Table 2).
Fig. 3.
Charge reversal of two negatively-charged residues in IIIS2 increase the effect of site-3 toxins. A-B, Representative sodium current traces of channel mutants S2-D1K and S2-E2K in the absence or presence of Av3 (A) or LqhαIT (B). C-D, Effects of LqhαIT (C) and Av3 (D) on fast inactivation of the four charge reversal channel mutants. The toxin inhibitory effects on fast inactivation were calculated by measuring the current that remained after 20 ms (I20) divided by the peak current (Ipeak) for S1-E1K, S1-E2K and S2-E2K channel mutants. Since after 20 ms, the inactivation of S2-D1K was not complete, the remaining current of this channel mutant was determined only after 50 ms, and so the value measured was I50/Ipeak. *denotes significant difference compared to WT BgNav1–1a channels determined by one-way ANOVA with Scheffe’s post hoc analysis (p˂ 0.05).
Fig. 4.
Charge reversal of two negatively-charged residues in IIIS2 increase the effect of Lqh-dprIT3. A-E, Conductance-voltage relations in WT BgNav1–1a channels (A) or the four channel mutants substituted in domain III (B-E) in the absence (●) or presence (○) of 300 nMLqh-dprIT3. To measure the toxin effect, a 20 Hz train (50) of 5 ms depolarizing prepulses to 50 mV from a holding potential of −120 mV was followed by a series of 20 ms depolarizing test pulses between −80 and −65 mV. F, Percentage of modification of the WT and four mutant channels elicited by Lqh-dprIT3. * denotes a significant difference compared to WT BgNav1–1a channels, as determined by one-way ANOVA with Scheffe’s post hoc analysis (p˂ 0.05).
Table 2.
Voltage dependence of activation of BgNav1–1a and of the four negatively-charged channel mutants before and after the application of 300 nM Lqh-dprIT3.
| Toxin-free |
Lqh-dprIT3 (300 nM) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| V0.5k | V0.5 | k 1 | A1 (%) | V0.5 | k 2 | A2 (%) | ||||
|
| ||||||||||
| BgNav1–1a | −31.3 ± 2.2 | 4.9 ± 0.3 | −32.0 ± 1.9 | 3.9 ± 0.3 | 72 ± 5 | −68.0 ± 4.6 | 8.5 ± 1.0 | 28 ± 1 | ||
| IIIS1-E1K | −31.2 ± 1.1 | 5.2 ± 0.2 | −33.7 ± 1.2 | 4.4 ± 0.2 | 77 ± 4 | −70.7 ± 2.6 | 9.2 ± 0.5 | 23 ± 4 | ||
| IIIS1-E2K | −31.2 ± 0.5 | 5.7 ± 0.2 | −34.2 ± 1.0 | 4.7 ± 0.2 | 69 ± 7 | −75.0 ± 2.1 | 11.0 ± 0.8 | 31 ± 7 | ||
| IIIS2-D1K | −25.9 ± 0.8 | 7.6 ± 0.2 | −55.2 ± 5.8 | 4.3 ± 0.8 | 25 ± 6 | −75.0 ± 3.5 | 5.4 ± 0.5 | 75 ± 6a | ||
| IIIS2-E2K | −29.4 ± 1.2 | 7.5 ± 0.3 | −30.7 ± 1.8 | 4.8 ± 0.6 | 33 ± 6 | −69.9 ± 1.9 | 7.7 ± 0.9 | 67 ± 6a | ||
Each value represents the mean ± SEM for at least eight oocytes.
Significant differences from other mutants were determined by one-way ANOVA analysis of variance (p ˂ 0.05).
3.3. The D1K/R4E channel is equally sensitive to the site-3 toxins as D1K or R4E channels
To further examine whether electrostatic interactions between R4 in IIIS4 and negatively-charged residues in IIIS2 affect the gating of BgNaV1–1a and its interaction with site-3 and site-4 toxins, we constructed the double mutants S2-D1K/R4E and S2-E2K/R4E. Of these only the S2-D1K/R4E channels generated observable sodium currents in Xenopus oocytes. Compared to WT channels, S2-D1K/R4E channels exhibited a 12 mV hyperpolarizing shift in the voltage dependence of activation, and a 30 mV hyperpolarizing shift in the voltage dependence of inactivation (Table 1). The sensitivity of S2-D1K/R4E mutant channels to Av3 or LqhαIT was comparable, although not identical, to that of the single mutation channels, S2-D1K and R4E (Fig. 5). LqhαIT inhibited the fast inactivation of WT channels at 60%, S2-D1K channels at 85%, S4-R4E, and D1K/R4E channels at 100%. The sensitivity of the S2-D1K/ R4E mutant channels could not be evaluated in the presence of 300 nM Lqh-dprIT3 due to a marked decrease in the peak current amplitude along a significant increase in leak currents, which prevented measurements of channel activation at more negative potentials.
Fig. 5.
The sensitivity of S2-D1K/R4E double mutant channels to Av3 and LqhαIT compared to channel mutants S2-D1K and R4E. A-B, Representative sodium current traces of R4E and IIIS2-D1K/R4E channels in the absence or presence of Av3 (A) and LqhαIT (B). C-D, Effect of S2-D1K/R4E double substitution on channel sensitivity to Av3 (C) and LqhαIT (D) in comparison to the single channel substitution. * denotes significant differences compared to WT BgNav1–1a channels as determined by one-way ANOVA with Scheffe’s post hoc analysis (p˂ 0.05).
3.4. Putative electrostatic interactions at the voltage-sensing module of domain III
The results on gating for charge reversal substitutions at III-VSM followed by those for the analyses of toxin effects show that interference of electrostatic interactions in this channel region not only affect the coordinated movement of the voltage sensor, but also influence the actions of the various toxins. To illustrate the critical electrostatic interactions in III-VSM, we aligned the corresponding sequences of various sodium channels (Fig. S2A) and constructed a three-dimensional model of that region in BgNav1–1a (Fig. S2B) using NavAbas template in the activated state (4EKW.pdb). The model shows that during channel activation R3, R4 and R5 are in close proximity and may interact with E1290 (IIIS1) (Song et al., 2011), D1308 (S2-D1) and E1318 (S2-E2), respectively, of the negatively-charged residue cluster in domain III. These putative interactions resemble those suggested in disulfide-locking studies between the analogous S4 and S2 residues of the bacterial channel (NaChBac; Fig. S2A).
4. Discussion
Binding site-3 is recognized by a variety of structurally distinct toxins, such as scorpion α-toxins (Gordon et al., 2007), sea anemone toxins (Moran et al., 2006, 2007), and spider toxins (Nicholson, 2007; Rash and Hodgson, 2002). Inhibition of sodium channel fast inactivation by site-3 toxins involves channel residues of the VSM in domain IV and/or of the pore module in domain I (Gordon et al., 1992; Gur et al., 2011; Gurevitz, 2012; Ma et al., 2013; Thomsen and Catterall, 1989; Wang et al., 2011). In domain-swapped channels, these regions are in proximity, suggesting overlapping but non-identical toxin binding sites in these two regions. For example, the sea anemone toxin Av3 competes with LqhαIT for binding to insect sodium channels and similarly inhibits the inactivation process. However, Av3 does not bind at linker S3–S4 in domain IV of site-3 in the Drosophila sodium channel, but rather at a cleft in S6 of domain I (Gur Barzilai et al., 2014). In contrast, the D1701R substitution in S3–S4 at domain IV of the DmNav1 channel, which abolishes the activity of a scorpion α-toxin, does not affect Av3 action (Moran et al., 2007).
Here we show that substitution of VSM residues in domain III (R4, R5 in IIIS4 and D1, E2 in IIIS2) increases the action of LqhαIT and Av3 site-3 toxins. Together with findings from a previous study of a β-toxin (Song et al., 2011), our results indicate that substitutions of the innermost positively-charged residues R4 and R5 in IIIS4 enhance the action of both site-3 and site-4 toxins on BgNav1–1a channels. Our functional data and interaction model suggest that R4 and R5 provide the essential interactions with III-VSM negative charges during gating that enhance the actions of site-3 and site-4 toxins. These residues in III-VSM are unlikely to be directly involved in toxin binding to the channel, as was previously suggested using site-directed antibodies (Gordon et al., 1992; Thomsen and Catterall, 1989) and mutagenic dissection (Gur et al., 2011; Ma et al., 2013), thus raising questions as to the mechanism involved. Despite their differences in binding, both Av3 and LqhαIT prolong the fast-inactivation of S2-D1K (D1308K), S2-E2K (E1318K), R4E and R5E BgNav1–1a channel mutants (Figs. 1 and 3). Thus, differences in toxin interaction with site-3 seem irrelevant to the effects mediated by substitutions in III-VSM. The increased action of the toxins upon charge reversals in III-VSM may therefore reflect an allosteric effect on the coordinated movements of the VSMs in domains III and IV during fast-inactivation, a possibility consistent with findings that show that movements of S4 in sodium channels are not independent, but coupled (Chanda et al., 2004).
Binding site-4 (for scorpion β-toxins) involves channel residues in the VSM of domain II and in the pore module of domain III (Zhang et al., 2011, 2012b). We have previously shown that L1285P in IIIS1 and E1290 A/K at the extracellular end of IIIS1 enhance β-toxin action, possibly by facilitating VSM trapping in domain II (Song et al., 2011). Similar to the action of site-3 toxins, the increased action of Lqh-dprIT3 in the charge reversing channel mutants observed in this study could result from an allosteric effect between domains II and III.
In an attempt to visualize putative interactions among residues in III- VSM, we constructed a homology model of BgNav1–1a in the activated state, using as template the crystal structure of the analogous homotetrameric bacterial sodium channel NavAb (Payandeh et al., 2011). As shown in this model (Fig. S2B), the positive charge R3 likely interacts with E1290 during activation, consistent with results from a previous study on the role of E1290 in scorpion β-toxin action (Song et al., 2011). The model also suggests that R4 and R5 may interact with S2-D1 and S2-E2, respectively, consistent with our findings that substitutions D1K and E2K in IIIS2, but not E1K and E2K in IIIS1, markedly increased the activity of LqhαIT, Av3 and Lqh-dprIT3. D1and E2 in IIIS2 of BgNav1–1a are analogous in position to E283 and E293 in S2 of the voltage-dependent Shaker potassium channel, in which electrostatic interactions exist between E283 in S2 and R368 and R371 in S4, and between E293 in S2 and K374 in S4 (Papazian et al., 1995; Tiwari-Woodruff et al., 1997). Studies using NaChBac have suggested that specific interactions of S4 positively-charged residues with negative countercharges in S1 and S2 regulate S4 outward movement upon membrane depolarization (Chakrapani et al., 2010; DeCaen et al., 2008, 2009, 2011; Paldi and Gurevitz, 2010; Yarov-Yarovoy et al., 2012). Negative countercharges may stabilize the S4 sensors at rest, and, upon membrane depolarization, facilitate sequential ion pair interactions controlling the movement of the S4 voltage sensors during gating (DeCaen et al., 2008, 2009, 2011; Paldi and Gurevitz, 2010). These countercharges are also analogous in position to those in BgNav1–1a in our study. Clearly, the extracellular IIIS1 residue E1290, extracellular IIIS2 residue D1308, and intracellular IIIS2 residue E1318 play a critical role in BgNav1–1a gating, and disruption of the putative interactions of these residues with IIIS4 partners by charge reversing substitutions in III-VSM likely accounts for the facilitation of toxin effects at sites 3 and 4.
Interestingly, our data from the D1K/R4E double mutant showed that charge swapping between S2-D1 and S4-R4 could not compensate each other for the effects on either gating or toxin actions, suggesting complexity in the interaction of charged residues in III-VSM. Specific interactions of negatively and positively charged residues have been demonstrated functionally in prokaryotic sodium channels (DeCaen et al., 2008, 2009, 2011; Paldi and Gurevitz, 2010; Yarov-Yarovoy et al., 2012) by molecular dynamics simulations in eukaryotic sodium channels (Gosselin Badaroudine et al., 2012; Moran et al., 2014; Groome and Bayless-Edwards, 2020) and by cryo-EM determination of channel structure (Payandeh et al., 2011, 2012; Zhang et al., 2012; Pan et al., 2018; Shen et al., 2018). These studies support our premise that proximity of S1 and S2 negatively charged residues with specific S4 positively charged residues in III-VSM, but also point out the need for future investigation of the specific nature of these interactions during gating and channel response to toxin exposure.
Recent cryo-EM studies of structures of sodium channels in complex with scorpion and spider toxins predicted that toxin-induced conformational changes in the VSMs of domains II and IV are transduced allosterically to the extracellular linkers in the VSM of domain III (Clairfeuille et al., 2019; Shen et al., 2019).In the case of the scorpion α-toxin AaH2 (Androctonus australis Hectorα-toxin 2) on a chimeric channel, Nav1.7-NavPaS, which bears the pore module of a cockroach (Periplaneta americana) channel and the VSM of domain IV of the human channel Nav1.7 (Clairfeuille et al., 2019), not only that toxin binding at the VSM of domain IV rendered a considerable shift (over 13 Å) of the VSM in the intracellular direction, but it also induced conspicuous alterations at the extracellular loops S3–S4 and S1–S2 in the VSM of domain III (Fig. S3A). Similarly, noticeable alterations in the extracellular loops in the VSM of domain III have been shown in another cryo-EM study upon binding of a site-4 toxin Dc1a (from the venom of the desert spider Diguetiacanities) at the VSM of domain II of a cockroach NavPaS channel (Fig. S3B) (Shen et al., 2018). Based on these observations and our results, we suggest that charge reversal of ionizable residues in III-VSM may alter closed-state transitions in III-VSM that allosterically modulate extracellular linkers of the VSMs of domains II and IV. These alterations may prolong toxin binding to sites-3 and 4 and increase their effects. Together, this study not only extends the proposed role of III-VSM in sodium channel gating, but also demonstrates the allosteric effects among the VSMs, as reflected by alterations in the activities of site-3 and site-4 toxins.
Supplementary Material
Acknowledgement
We thank Prof. Boris Zhorov for composing Fig. S2. This study was partially supported by a grant (GM057440) from the National Institutes of Health to K.D., a grant (81273113) from the National Natural Science Foundation of China to R.G. and a grant (BK20170611) from the Natural Science Foundation of Jiangsu Province to Q.Z. Z.C. and G.W.W. were supported by NSF Grants DMS-1721024 and DMS-1761320, and NIH grant GM126189.
Footnotes
Declaration of competing interest
The authors declare that they have no conflicts of interest with the contents of this article.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ibmb.2021.103625.
References
- Armstrong CM, 1981. Sodium channels and gating currents. Physiol. Rev 61, 644–683. [DOI] [PubMed] [Google Scholar]
- Catterall WA, 1992. Cellular and molecular biology of voltage-gated sodium channels. Physiol. Rev 72, S15–S48. [DOI] [PubMed] [Google Scholar]
- Catterall WA, 2000. From ionic currents to molecular mechanisms: the structure and function of voltage-gated sodium channels. Neuron 26, 13–25. [DOI] [PubMed] [Google Scholar]
- Catterall WA, 2010. Ion channel voltage sensors: structure, function, and pathophysiology. Neuron 67, 915–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall WA, Cestele S, Yarov-Yarovoy V, Yu FH, Konoki K, Scheuer T, 2007. Voltage-gated ion channels and gating modifier toxins. Toxicon 49, 124–141. [DOI] [PubMed] [Google Scholar]
- Cestele S, Catterall WA, 2000. Molecular mechanisms of neurotoxin action on voltage- gated sodium channels. Biochimie 82, 883–892. [DOI] [PubMed] [Google Scholar]
- Chakrapani S, Sompornpisut P, Intharathep P, Roux B, Perozo E, 2010. The activated state of a sodium channel voltage sensor in a membrane environment. Proc. Natl. Acad. Sci. U. S. A 107, 5435–5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda B, Asamoah OK, Bezanilla F, 2004. Coupling interactions between voltage sensors of the sodium channel as revealed by site-specific measurements. J. Gen. Physiol 123, 217–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clairfeuille T, Cloake A, Infield DT, Llongueras JP, Arthur CP, Li ZR, Jian Y, Martin-Eauclaire M-F, Bougis PE, Ciferri C, Ahern CA, Bosmans F, Hackos DH, Rohou A, Payandeh J, 2019. Structural basis of α-scorpion toxin action on Nav channels. Science 363. [DOI] [PubMed] [Google Scholar]
- DeCaen PG, Yarov-Yarovoy V, Scheuer T, Catterall WA, 2011. Gating charge interactions with the S1 segment during activation of a Na+ channel voltage sensor. Proc. Natl. Acad. Sci. U. S. A 108, 18825–18830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaen PG, Yarov-Yarovoy V, Sharp EM, Scheuer T, Catterall WA, 2009. Sequential formation of ion pairs during activation of a sodium channel voltage sensor. Proc. Natl. Acad. Sci. U. S. A 106, 22498–22503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaen PG, Yarov-Yarovoy V, Zhao Y, Scheuer T, Catterall WA, 2008. Disulfide locking a sodium channel voltage sensor reveals ion pair formation during activation. Proc. Natl. Acad. Sci. U. S. A 105, 15142–15147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delemotte L, Tarek M, Klein ML, Amaral C, Treptow W, 2011. Intermediate states of the Kv1.2 voltage sensor from atomistic molecular dynamics simulations. Proc. Natl. Acad. Sci. U. S. A 108, 6109–6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdetskiy A, Cole C, Procter J, Barton GJ, 2015. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 43, W389–W394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutertre S, Lewis RJ, 2010. Use of venom peptides to probe ion channel structure and function. J. Biol. Chem 285, 13315–13320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC, 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig M, Karanicolas J, Brooks CL 3rd, 2004. MMTSB Tool Set: enhanced sampling and multiscale modeling methods for applications in structural biology. J. Mol. Graph. Model 22, 377–395. [DOI] [PubMed] [Google Scholar]
- Feng G, Deak P, Chopra M, Hall LM, 1995. Cloning and functional analysis of TipE, a novel membrane protein that enhances Drosophila para sodium channel function. Cell 82, 1001–1011. [DOI] [PubMed] [Google Scholar]
- Gao R, Du Y, Wang L, Nomura Y, Satar G, Gordon D, Gurevitz M, Goldin AL, Dong K, 2014. Sequence variations at I260 and A1731 contribute to persistent currents in Drosophila sodium channels. Neuroscience 268, 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D, 1997. A new approach to insect-pest control–combination of neurotoxins interacting with voltage sensitive sodium channels to increase selectivity and specificity. Invertebr. Neurosci 3, 103–116. [DOI] [PubMed] [Google Scholar]
- Gordon D, Karbat I, Ilan N, Cohen L, Kahn R, Gilles N, Dong K, Stuhmer W, Tytgat J, Gurevitz M, 2007. The differential preference of scorpion alpha-toxins for insect or mammalian sodium channels: implications for improved insect control. Toxicon 49, 452–472. [DOI] [PubMed] [Google Scholar]
- Gosselin-Badaroudine P, Delemotte L, Moreau A, Klein ML, Chahine M, 2012. Gating pore currents and the resting state of NaV1.3 voltage sensor domains. Proc. Natl. Acad. Sci. U.S.A 109, 19250–19255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D, Moskowitz H, Eitan M, Warner C, Catterall WA, Zlotkin E, 1992. Localization of receptor sites for insect-selective toxins on sodium channels by site- directed antibodies. Biochemistry 31, 7622–7628. [DOI] [PubMed] [Google Scholar]
- Groome JR, 2014. The voltage sensor module in sodium channels. Handb. Exp. Pharmacol 221, 7–31. [DOI] [PubMed] [Google Scholar]
- Groome JR, Bayless-Edwards L, 2020. Roles for countercharge in the voltage sensor domain of ion channels. Front. Pharmacol 11, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groome JR, Winston V, 2013. S1-S3 counter charges in the voltage sensor module of a mammalian sodium channel regulate fast inactivation. J. Gen. Physiol 141, 601–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur Barzilai M, Kahn R, Regev N, Gordon D, Moran Y, Gurevitz M, 2014. The specificity of Av3 sea anemone toxin for arthropods is determined at linker DI/SS2-S6 in the pore module of target sodium channels. Biochem. J 463, 271–277. [DOI] [PubMed] [Google Scholar]
- Gur M, Kahn R, Karbat I, Regev N, Wang J, Catterall WA, Gordon D, Gurevitz M, 2011. Elucidation of the molecular basis of selective recognition uncovers the interaction site for the core domain of scorpion alpha-toxins on sodium channels. J. Biol. Chem 286, 35209–35217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevitz M, 2012. Mapping of scorpion toxin receptor sites at voltage-gated sodium channels. Toxicon 60, 502–511. [DOI] [PubMed] [Google Scholar]
- Gurevitz M, Karbat I, Cohen L, Ilan N, Kahn R, Turkov M, Stankiewicz M, Stuhmer W, Dong K, Gordon D, 2007. The insecticidal potential of scorpion beta-toxins. Toxicon 49, 473–489. [DOI] [PubMed] [Google Scholar]
- Hodgkin AL, Huxley AF, 1952. A quantitative description of membrane current and its application to conduction and excitation in nerve. J. Physiol 117, 500–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K, 1996. VMD: visual molecular dynamics. J. Mol. Graph 14, 33–38. [DOI] [PubMed] [Google Scholar]
- Krapcho KJ, Kral RM Jr., Vanwagenen BC, Eppler KG, Morgan TK, 1995. Characterization and cloning of insecticidal peptides from the primitive weaving spider Diguetia canities. Insect Biochem. Mol. Biol 25, 991–1000. [DOI] [PubMed] [Google Scholar]
- Ma Z, Kong J, Gordon D, Gurevitz M, Kallen RG, 2013. Direct evidence that scorpion alpha-toxins (site-3) modulate sodium channel inactivation by hindrance of voltage-sensor movements. PloS One 8 e77758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran Y, Cohen L, Kahn R, Karbat I, Gordon D, Gurevitz M, 2006. Expression and mutagenesis of the sea anemone toxin Av2 reveals key amino acid residues important for activity on voltage-gated sodium channels. Biochemistry 45, 8864–8873. [DOI] [PubMed] [Google Scholar]
- Moran Y, Kahn R, Cohen L, Gur M, Karbat I, Gordon D, Gurevitz M, 2007. Molecular analysis of the sea anemone toxin Av3 reveals selectivity to insects and demonstrates the heterogeneity of receptor site-3 on voltage-gated Na+ channels. Biochem. J 406, 41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson GM, 2007. Insect-selective spider toxins targeting voltage-gated sodium channels. Toxicon 49, 490–512. [DOI] [PubMed] [Google Scholar]
- Paldi T, Gurevitz M, 2010. Coupling between residues on S4 and S1 defines the voltage-sensor resting conformation in NaChBac. Biophys. J 99, 456–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palovcak E, Delemotte L, Klein ML, Carnevale V, 2014. Evolutionary imprint of activation: the design principles of VSDs. J. Gen. Physiol 143, 145–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Li Z, Zhou Q, Shen H, Wu K, Huang X, Chen J, Zhang J, Zhu X, Lei J, Xiong W, Gong H, Xiao B, Yan N, 2018. Structure of the human voltage-gated sodium channel NaV1.4 in complex with beta1. Science 362. [DOI] [PubMed] [Google Scholar]
- Papazian DM, Shao XM, Seoh SA, Mock AF, Huang Y, Wainstock DH, 1995. Electrostatic interactions of S4 voltage sensor in Shaker K+ channel. Neuron 14, 1293–1301. [DOI] [PubMed] [Google Scholar]
- Payandeh J, Gamal El-Din TM, Scheuer T, Zheng N, Catterall WA, 2012. Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature 486, 135–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payandeh J, Scheuer T, Zheng N, Catterall WA, 2011. The crystal structure of a voltage-gated sodium channel. Nature 475, 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE, 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Rash LD, Hodgson WC, 2002. Pharmacology and biochemistry of spider venoms. Toxicon 40, 225–254. [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TL, 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol 234, 779–815. [DOI] [PubMed] [Google Scholar]
- Shen H, Li Z, Jiang Y, Pan X, Wu J, Cristofori-Armstrong B, Smith JJ, Chin YKY, Lei J, Zhou Q, King GF, Yan N, 2018. Structural basis for the modulation of voltage-gated sodium channels by animal toxins. Science 362. [DOI] [PubMed] [Google Scholar]
- Song W, Du Y, Liu Z, Luo N, Turkov M, Gordon D, Gurevitz M, Goldin AL, Dong K, 2011. Substitutions in the domain III voltage-sensing module enhance the sensitivity of an insect sodium channel to a scorpion beta-toxin. J. Biol. Chem 286, 15781–15788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strugatsky D, Zilberberg N, Stankiewicz M, Ilan N, Turkov M, Cohen L, Pelhate M, Gilles N, Gordon D, Gurevitz M, 2005. Genetic polymorphism and expression of a highly potent scorpion depressant toxin enable refinement of the effects on insect Na channels and illuminate the key role of Asn-58. Biochemistry 44, 9179–9187. [DOI] [PubMed] [Google Scholar]
- Tan J, Liu Z, Nomura Y, Goldin AL, Dong K, 2002. Alternative splicing of an insect sodium channel gene generates pharmacologically distinct sodium channels. J. Neurosci 22, 5300–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Liu Z, Wang R, Huang ZY, Chen AC, Gurevitz M, Dong K, 2005. Identification of amino acid residues in the insect sodium channel critical for pyrethroid binding. Mol. Pharmacol 67, 513–522. [DOI] [PubMed] [Google Scholar]
- Terlau H, Olivera BM, 2004. Conus venoms: a rich source of novel ion channel- targeted peptides. Physiol. Rev 84, 41–68. [DOI] [PubMed] [Google Scholar]
- Thomsen WJ, Catterall WA, 1989. Localization of the receptor site for alpha-scorpion toxins by antibody mapping: implications for sodium channel topology. Proc. Natl. Acad. Sci. U. S. A 86, 10161–10165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari-Woodruff SK, Schulteis CT, Mock AF, Papazian DM, 1997. Electrostatic interactions between transmembrane segments mediate folding of Shaker K+ channel subunits. Biophys. J 72, 1489–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Yarov-Yarovoy V, Kahn R, Gordon D, Gurevitz M, Scheuer T, Catterall WA, 2011. Mapping the receptor site for alpha-scorpion toxins on a Na+ channel voltage sensor. Proc. Natl. Acad. Sci. U. S. A 108, 15426–15431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmke JW, Reenan RA, Wang P, Qian S, Arena JP, Wang J, Wunderler D, Liu K, Kaczorowski GJ, Van der Ploeg LH, Ganetzky B, Cohen CJ, 1997. Functional expression of Drosophila para sodium channels. Modulation by the membrane protein TipE and toxin pharmacology. J. Gen. Physiol 110, 119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarov-Yarovoy V, DeCaen PG, Westenbroek RE, Pan CY, Scheuer T, Baker D, Catterall WA, 2012. Structural basis for gating charge movement in the voltage sensor of a sodium channel. Proc. Natl. Acad. Sci. U. S. A 109, E93–E102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JZ, Yarov-Yarovoy V, Scheuer T, Karbat I, Cohen L, Gordon D, Gurevitz M, Catterall WA, 2011. Structure-function map of the receptor site for beta-scorpion toxins in domain II of voltage-gated sodium channels. J. Biol. Chem 286, 33641–33651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ren W, DeCaen P, Yan C, Tao X, Tang L, Wang J, Hasegawa K, Kumasaka T, He J, Clapham DE, Yan N, 2012a. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature 486, 130–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JZ, Yarov-Yarovoy V, Scheuer T, Karbat I, Cohen L, Gordon D, Gurevitz M, Catterall WA, 2012b. Mapping the interaction site for a beta-scorpion toxin in the pore module of domain III of voltage-gated Na(+) channels. J. Biol. Chem 287, 30719–30728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Gao B, Peigneur S, Tytgat J, 2020. How a scorpion toxin selectively captures a prey sodium channel: the molecular and evolutionary basis uncovered. Mol. Biol. Evol 37 (11), 3149–3164. [DOI] [PubMed] [Google Scholar]
- Zilberberg N, Gordon D, Pelhate M, Adams ME, Norris TM, Zlotkin E, Gurevitz M, 1996. Functional expression and genetic alteration of an alpha scorpion neurotoxin. Biochemistry 35, 10215–10222. [DOI] [PubMed] [Google Scholar]
- Zlotkin E, Fishman Y, Elazar M, 2000. AaIT: from neurotoxin to insecticide. Biochimie 82, 869–881. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.