Abstract
Delivery of nucleic acids, such as mRNA, to immune cells has become a major focus in the past decade with ionizable lipid nanoparticles (LNPs) emerging as a clinically-validated delivery platform. LNPs—typically composed of ionizable lipids, cholesterol, phospholipids, and polyethylene glycol lipids —have been designed and optimized for a variety of applications including cancer therapies, vaccines, and gene editing. However, LNPs have only recently been investigated for delivery to T cells, which has various therapeutic applications including the engineering of T cell immunotherapies. While several LNP formulations have been evaluated for mRNA delivery, recent work has demonstrated that the utilization of cholesterol analogs may enhance mRNA delivery. Other studies have shown that cholesterols modified with hydroxyl groups can alter endocytic recycling mechanisms. Here, we engineered a library of LNPs incorporating hydroxycholesterols to evaluate their impact on mRNA delivery to T cells by leveraging endosomal trafficking mechanisms. Substitution of 25% and 50% 7α-hydroxycholesterol for cholesterol in LNPs enhanced mRNA delivery to primary human T cells ex vivo by 1.8-fold and 2.0-fold, respectively. Investigation of endosomal trafficking revealed that these modifications also increase late endosome production and reduce the presence of recycling endosomes. These results suggest that hydroxyl modification of cholesterol molecules incorporated into LNP formulations provides a mechanism for improving delivery of nucleic acid cargo to T cells for a range of immunotherapy applications.
Keywords: Lipid nanoparticles, mRNA delivery, Cholesterol, Endosomal trafficking
1. Introduction
Over the past decade, immunotherapy has become a critical tool in the treatment of a variety of conditions from autoimmune disorders such as psoriasis to blood cancers and solid tumors [1-3]. These immunotherapies span a wide range of modalities from antibody-based inhibitors to genetically-engineered immune cells [4,5]. Messenger RNA (mRNA)-based immunotherapies have piqued significant interest due to their transient nature [6,7]. Because mRNA is translated in the cytosol, it avoids many risks associated with genomic integration such as insertional mutagenesis and allows for temporal control over the immunotherapy [8,9]. Further, optimization of in vitro transcribed mRNA design and manufacturing techniques has enabled the scaled-up production of highly potent mRNA [10-12]. These benefits have led to the utilization of mRNA immunotherapies in clinical applications such as vaccines for influenza, Zika virus, and COVID-19 [13-16], as well as for cancer therapies such as chimeric antigen receptor (CAR) T cell therapy and tumor-infiltrating T cell therapy [17-19].
Currently, the primary technology utilized in clinical settings for mRNA delivery to immune cells is electroporation–a method in which electric pulses permeabilize cell membranes to allow for mRNA delivery into the cytosol [20-22]. However, electroporation of immune cells is primarily limited to ex vivo applications, which greatly increases the cost and labor burden of manufacturing these therapies [23-25]. Moreover, electroporation ex vivo can be highly toxic to target cells and may alter genomic expression [20,26,27]. Thus, there exists a technological gap and need for a platform that delivers mRNA cargo efficiently and with low toxicity to immune cells in a manner that enables future translation to in vivo clinical applications.
An alternative strategy for mRNA delivery to immune cells is the use of ionizable lipid nanoparticles (LNPs). We have recently shown that, compared to electroporation, LNPs are a particularly promising delivery platform as they can effectively deliver mRNA to T cells while inducing lower toxicity in T cells during ex vivo transfection [34,35]. Furthermore, LNP platforms are translatable to in vivo mRNA delivery applications [2,14,15,36-39]. LNPs have been utilized in several therapeutic applications to deliver a variety of nucleic acid cargos—including DNA, siRNA, miRNA, and mRNA—to a variety of cell types including immune cells [28-32]. The clinical success of these LNP platforms has led to the FDA approval of Alnylam's Onpattro [33] as well as emergency use authorization of LNPs for the Moderna and Pfizer/BioNTech COVID-19 mRNA vaccines [14,16]. However, limitations of LNPs include liver accumulation which can prevent the optimal biodistribution required to achieve delivery to extrahepatic tissues [40], accelerated blood clearance due to PEGylation [41], and endosomal recycling – a process in which cells exocytose lipid-containing exogenous materials to maintain homeostasis [42]. Thus, while LNPs have enabled the delivery of nucleic acids for vaccines and therapeutics, significant delivery challenges remain.
To overcome these challenges, investigations have explored altering LNP formulation ratios, modifying excipients, and incorporating novel LNP components. LNPs are typically composed of four main components in addition to their nucleic acid cargo: (i) ionizable lipids, (ii) phospholipids, (iii) cholesterol, and (iv) lipid-anchored polyethylene glycol [32]. The relative ratio between these components has been extensively explored and has been a focus of novel LNP development due to the demonstrated impact of these excipient ratios on LNP performance, biodistribution, and stability [35,43-46]. Other studies have investigated the addition of novel components to LNP formulations which may alter the physiochemical properties that impact LNP potency and biodistribution. For example, DOTAP is a permanently charged cationic lipid that induces internal charge changes to the LNP and may be linked to target cell delivery [43]. Other examples of novel additions include targeting moieties that enable cell-specific delivery [32,47]. Additional work has been conducted to optimize LNP formulations for the coencapsulation of multiple nucleic acid cargos [48,49]. These previous investigations that focus on the substitution of individual components have primarily explored the impact of ionizable lipids on target cell specificity and functional delivery of nucleic acid cargos, or the impact of phospholipids (e.g., DOPE, DSPC, DOTAP) on cargo encapsulation, biodistribution, and protein corona development [48,50-52]. Recently, the role of cholesterol in LNP formulations and its impact on endosomal trafficking of LNPs in cells has also been investigated. Specifically, it has been shown that LNP formulations in which cholesterol is replaced with naturally occurring phytosterols demonstrate improved mRNA delivery in several cell lines [42,53]. Such improvements have been linked to endosomal recycling mechanisms involving the Niemann Pick C1 (NPC1) enzyme [53]. Previous enzyme-ligand binding studies conducted on NPC1 and various cholesterol analogs revealed that certain modifications can reduce recognition of cholesterol by the recycling enzyme NPC1 [54,55]. In the present study, a new class of cholesterol analogs, hydroxycholesterols, are evaluated for their impact on LNP-mediated mRNA delivery to T cells. The addition of a hydroxyl group to various locations along the cholesterol molecule can alter the binding kinetics between the modified cholesterol and NPC1 [56]. The goal of this alteration is to ultimately reduce NPC1 recognition of cholesterol during the endosomal trafficking of LNPs. However, cholesterol recognition by membrane proteins remains a critical step for LNP uptake/endocytosis [57]. Therefore, this study evaluated the substitution of six hydroxycholesterol candidates at four different substitution percentages to determine if these substitutions improve the delivery of mRNA to T cells. The resulting library of LNP candidates was screened in vitro and ex vivo, and a subset of top performers were investigated for dose response behavior and endosomal trafficking. Ultimately, it was shown that certain hydroxycholesterol substitutions into LNPs enhance mRNA delivery to T cells.
2. Materials and methods
2.1. Ionizable lipid synthesis
The ionizable lipid (C14–4) was synthesized (Supplemental Fig. 1) using Michael addition chemistry [34]. The polyamine core (Enamine Inc., Monmouth Junction, NJ) was reacted with an excess of epoxideterminated C14 alkyl chains (epoxytetradecane, Sigma Aldrich, St. Louis, MO). This 48 h reaction was conducted at 80 °C under gentle stirring. A Rotovap R-3000 (Buchi, New Castle, DE) was used to dry the product which was subsequently resuspended in ethanol. This resulting solution was the ionizable lipid used in all LNP formulations.
2.2. mRNA synthesis
The luciferase (Fluc) gene sequence was codon-optimized, synthesized, and cloned into a mRNA production plasmid. mRNA was transcribed from the plasmid using MegaScript T7 RNA polymerase (Invitrogen, Waltham, MA), precipitated using LiCl, and purified by cellulose chromatography. The m1Ψ UTP nucleoside-modified mRNA, containing a 101 nucleotide-long poly(A) tail, produced by this plasmid was capped with a trinucleotide cap1 analogue (TriLink, San Diego, CA) during transcription. Agarose gel electrophoresis, mRNA sequencing, standard J2 dot plots, and assays assessing INF induction in human monocyte-derived dendritic cells were used to analyze the mRNA. mRNA was stored at −80 °C.
2.3. Lipid nanoparticle formulation
Lipid nanoparticles (LNPs) were formulated by preparing an ethanol phase containing C14–4 ionizable lipid, 1,2-Dioleoyl-sn-glycero-3-phos-phoethanolamine (DOPE) (Avanti Polar Lipids, Alabaster, AL), 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] (PEG) (Avanti Polar Lipids), cholesterol (Avanti Polar Lipids), and X-hydroxycholesterol. An aqueous phase was prepared containing 25 μg luciferase mRNA in 300 μL of 10 mM citric acid. LNPs were formulated via chaotic mixing of the ethanol and aqueous phases in a microfluidic device at a 1:3 volume ratio using pump33DS syringe pumps (Harvard Apparatus, Holliston, MA) [58]. LNPs were subsequently dialyzed against 1× PBS in 20 kDa molecular weight cutoff dialysis cassettes for 2 h and sterile filtered through 0.22 μm filters.
2.4. LNP encapsulation efficiency
Encapsulation efficiency was determined by the Quant-iT™ RiboGreen™ RNA Assay Kit (Thermo Fisher Scientific). LNPs were diluted to mRNA concentrations of 2 μg/mL into either 1× TE buffer or 0.1% Triton X-100 in 1× TE buffer (Sigma Aldrich). LNPs were incubated on a shaker for 20 min at 300 rpm. 100 μL of each solution were then pipetted into an assay plate, and RiboGreen™ reagent was pipetted into each well, as per manufacturer instructions. A plate reader, Infinite M Plex plate reader (Tecan, Morissville, NC), was then used to read fluorescence at an excitation of 490 nm and an emission of 520 nm. Encapsulation efficiency was then reported as a calculated value, , where RX is the RNA content in the Triton X-100 and RTE is the RNA content in the TE buffer.
2.5. Dynamic light scattering
10 μL of LNPs were diluted 100× in 1× PBS in 4 mL disposable cuvettes for dynamic light scattering (DLS) measurements on the Zetasizer Nano (Malvern Instruments, Malvern, UK). LNP size (Z-average diameter) and polydispersity index (PDI) are reported as the mean ± standard deviation (n=3 measurements).
2.6. Surface zeta potential
For zeta potential measurements, 20 μL of each LNP solution was diluted 50× in water in DTA1070 zeta potential cuvettes (Malvern Panalytical, Malvern, UK) and measured on the Zetasizer Nano instrument. LNP zeta potential is reported as the mean ± standard deviation (n=3 measurements).
2.7. LNP pKa measurements
The pKa of each LNP formulation was measured by 6-(p-Toluidino)-2-naphthalenesulfonic Acid (TNS) assays. Briefly, buffered solutions of 150 mM sodium chloride, 20 mM sodium phosphate, 25 mM ammonium citrate, and 20 mM ammonium acetate were adjusted to pH values of 2 to 12 in 0.5 increments. LNPs were added to each pH-adjusted solution in a 96-well plate, and TNS was then added to each well for a final TNS concentration of 6 μM. The resulting fluorescence was read on the Infinite M Plex plate reader. The resulting data was fit with a sigmoidal regression, and pKa was calculated as the pH at which the fluorescence intensity was 50% of its maximum value.
2.8. LNP library design
The library screen involved the evaluation of 6 hydroxycholesterols (cholesterols with hydroxyl groups added to various locations along the cholesterol molecule): 7α-hydroxycholesterol (Abcam, Cambridge, MA), 7β-hydroxycholesterol (Sigma Aldrich), 19-hydroxycholesterol (Cayman Chemicals, Ann Arbor, MI), 20(S)-hydroxycholesterol (Abcam, Cambridge, MA), 24(S)-hydroxycholesterol (Cayman Chemicals, Ann Arbor, MI), 25-hydroxycholesterol (Abcam, Cambridge, MA). The library's base formulation excipient molar percentages were 35% C14–4, 16% DOPE, 46.5% Cholesterol, and 2.5% PEG. The six hydroxycholesterol candidates were incorporated into these formulations by substituting cholesterol with hydroxycholesterol at various molar substitution percentages (12.5%, 25%, 50%, 100%). The molar percentages of the excipients for these candidate formulations were maintained at 35% C14–4, 16% DOPE, 46.5% total cholesterol, and 2.5% PEG where total cholesterol constituted cholesterol and the hydroxycholesterol substitute.
2.9. Cell culture and mRNA LNP treatment
Jurkats, an immortalized T cell line, (ATCC no. TIB-152) were cultured in RPMI-1640 with l-glutamine (Thermo Fisher Scientific, Waltham, MA) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (P/S). Primary human T cells (CD3+) were collected from healthy donors and procured from the Human Immunology Core at Penn Medicine. Primary human T cells were subsequently combined in RPMI-1640 media with l-glutamine, 10% FBS, and 1% P/S at a 1:1 ratio of CD4+ to CD8+ T cells. Primary human T cells were subsequently activated with Human T-activator CD3/CD28 Dynabeads (Thermo Fisher Scientific) at a 1:1 bead to cell ratio, as this is necessary for LNP-mediated mRNA delivery to primary human T cells (Supplemental Fig. 2). For all screening, cells were plated in 96-well plates at 60,000 cells per well in 60 μL of media. LNPs were then added to the wells at the desired mRNA dosage concentration (e.g., 60 ng per 60,000 cells). The mRNA concentration of LNPs was determined using A260 absorbance on an Infinite M Plex plate reader (Tecan, Morissville, NC) using Tecan's NanoQuant plate. Cells were incubated for 24 h before functional readout assays were performed.
2.10. Luciferase expression and cell viability assays
For luciferase expression assays, 96-well plates were spun at 300 g for 7 min to pellet cells. Supernatant media was removed, and cells were resuspended in 50 μL 1 × lysis buffer (Promega, Madison, WI) and 100 μL of luciferase assay substrate (Promega). Following 10 min of incubation, a plate reader was used to read the luminescent signal from each well. Luminescence was normalized within each plate to the S2 LNP formulation. Results shown are the mean ± the standard deviation of 3 biological replicates with at least 3 technical replicates per plate.
For cell viability assays, 60 μL of CellTiter-Glo™ (Promega) was added to each well. Following 10 min of incubation, a plate reader was used to read luminescent signal from each well. Luminescence was normalized within each plate to untreated cells to calculate percent cell viability.
2.11. Colocalization of LNPs with acidic organelles using confocal microscopy
LNPs were diluted in 1× PBS to 10 ng of luciferase mRNA per μL. Vybrant™ DiO Cell-Labeling Solution (Thermo Fisher Scientific) was added to LNP solutions at a 1:75 ratio by volume. Jurkat cells were treated at 60 ng of luciferase mRNA per 60,000 cells in 60 μL of media for 3 h. Cells were collected, centrifuged at 300 g for 5 min, resuspended in RPMI media containing LysoTracker™ Deep Red (Thermo Fisher Scientific) (1:3000), and incubated for an additional 1 h. Cells were collected, centrifuged at 300 g for 5 min, and resuspended in 1× PBS. Cell-containing PBS solution was then allowed to sediment for 15 min onto a chambered microscope slide, Nunc™ Lab-Tek™ II 4-well Chamber Slide with removable wells (Thermo Fisher Scientific). PBS was aspirated off, and slides were incubated with 4% formaldehyde (Sigma Aldrich) for 10 min to fix cells [59]. Slides were then washed two times with 1 × PBS for 5 min each while rocking gently. Finally, chamber walls were removed, and cover slips were placed on the slides to prepare the samples for confocal microscopy imaging. Images were taken using a Zeiss LSM 710 confocal microscope (Zeiss, Oberkochen, Germany) with a x63 oil lens. Excitation and emission for DiO were 484 nm and 501 nm, respectively. Excitation and emission for Lysotracker were 647 nm and 668 nm, respectively.
2.12. Endosomal trafficking assay
Jurkats were treated at either 60 or 150 ng of luciferase mRNA per 60,000 cells for 4 h. Cells were collected, centrifuged at 300 g for 5 min, and resuspended in 1× PBS. Cell-containing PBS solution was then allowed to sediment for 15 min onto a chambered microscope slide. PBS was aspirated off, and slides were incubated with 4% formaldehyde for 10 min to fix cells. All subsequent washes and incubations were done while gently rocking the slides. Slides were then washed with 1 × PBS for 5 min and incubated with 0.3% Tween-20 in 1× PBS for 15 min [60]. Next, slides were washed with 1× PBS and subsequently blocked with 2% bovine serum albumin (BSA) in 1× PBS for 30 min. Following the blocking step, slides were incubated for 1 h with 1:100 dilutions of either Rab5A #46449, Rab7 #9367, or Rab11 #5589 XP® Rabbit mAb (Cell Signaling Technology, Danvers, MA). Following 2 washes in 1 × PBS for 5 min each, cells were covered and incubated with 1:1000 dilutions of Anti-rabbit IgG (H + L), F(ab’)2 Fragment (Alexa Fluor 647® Conjugate) #4144 (Cell Signaling Technology) for 1 h. Following 2 additional 1× PBS washes, slides were prepared using cover slips and ProLong™ Gold Antifade mountant (Thermo Fisher Scientific). Images were taken using a Zeiss LSM 710 confocal microscope (Zeiss, Oberkochen, Germany) with a x63 oil lens. Excitation and emission for the secondary antibody were 650 nm and 671 nm, respectively.
2.13. Imaging software and analysis
For colocalization calculations, channels were overlayed with one another and analyzed via coloc2, Fiji's built-in colocalization package [61]. Spearman's rank correlation coefficient was recorded for a total of 5 image views, with a combined total of at least 75 cells. The Spearman’s rank correlation coefficient was used as it is more resistant than other correlation metrics (e.g., Pearson's) against relevant outliers which would include cells that produce oversaturated signals [62].
For total quantification of Rab5, Rab7, and Rab11 expression in endosomal trafficking assays, regions of interest were selected around cells using Fiji. Integrated density for each cell was then recorded with at least 50 total cells in each treatment group. The per cell Rab expression was then averaged and reported.
3. Results and discussion
3.1. LNP library design and synthesis
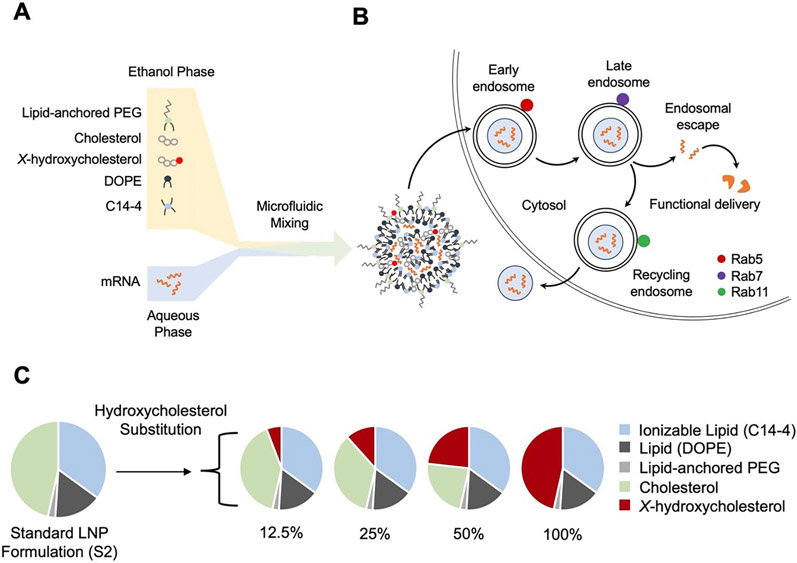
In this study, a library of 24 LNPs was synthesized and evaluated for mRNA delivery to T cells. The LNPs were synthesized using microfluidic mixing of an ethanol phase containing C14–4 ionizable lipid, cholesterol, X-hydroxycholesterol, DOPE, and lipid-anchored PEG with a citric acid phase containing luciferase mRNA (Fig. 1A). The base formulation of the library (S2) was a previously optimized formulation with the following excipients and molar ratio percentages: 35% C14–4 ionizable lipid, 46.5% cholesterol, 16% DOPE, and 2.5% PEG [45,48,63]. This formulation was previously demonstrated to have potent T cell transfection comparable to electroporation [35]. Notably, a significant molar percentage of the LNP formulation is made up of cholesterol, which is modulated in cells in a homeostatic manner by endocytic recycling [54,55,64]. Previous work has demonstrated that endocytic recycling results in endocytosed LNPs being exocytosed from target cells [65]. These exocytic pathways, specifically Niemann Pick type C1 (NPC1) mediated recycling, have been identified as core contributors to reduced functional delivery of nucleic acid cargos [66]. In the recycling process, endosomal trafficking enzymes, such as NPC1, recognize lipids, especially cholesterol, and migrate these lipid components to the cell membrane [67], releasing any cargo in the endosome into the extracellular environment. However, enzyme-ligand binding studies on cholesterol have demonstrated that the addition of hydroxyl groups to cholesterol molecules alter their binding kinetics with NPC1, providing an opportunity to mitigate endocytic recycling [56]. To investigate endosomal trafficking, we characterized the progression of candidate LNPs through different stages of the endosome. Endosomal trafficking through the cell can be tracked by the Ras-associated binding (Rab) family of proteins [68] including Rab5, Rab7, and Rab11, which associate with the early, late, and recycling endosomes, respectively (Fig. 1B) [69]. Given that LNPs typically release mRNA cargo into the cytoplasm after reaching and escaping the late endosome, it is apparent that LNPs that can reach the late endosome without being subsequently recycled have the greatest propensity for functional delivery [70]. Given these observations, we hypothesize that hydroxycholesterol substitution into LNP formulations may reduce recycling of endocytosed LNPs out of the cell. Thus, we explored the impact of hydroxycholesterol substitution into LNP formulations on functional delivery, toxicity, and endosomal trafficking in T cells.
Fig. 1.
Engineering lipid nanoparticles (LNP) with hydroxycholesterol substitution: motivation, design, and synthesis. (A) Schematic of LNP components, formulation, and expected structure. An ethanol phase containing lipid-anchored PEG (polyethylene glycol), cholesterol, X-hydroxycholesterol, DOPE (1,2-Dioleoyl-snglyeero-3-phosphoethanolamine), and C14–4 ionizable lipid and an aqueous phase containing mRNA are mixed in a microfluidic device to produce LNPs. (B) Diagram of LNP delivery into a T cell and the endosomal trafficking mechanisms involving the Rab family of proteins. Rab5, Rab7, and Rab11 associate with the early, late, and recycling endosomes, respectively. (C) Design of an LNP library incorporating the substitution of various hydroxycholesterols for unmodified cholesterol.
First, an LNP library was designed by substituting hydroxycholesterols into the S2 base formulation at various substitution percentages of unmodified cholesterol (12.5%, 25%, 50%, or 100%) (Fig. 1C). Each of the hydroxycholesterol substitutes is abbreviated A1, A2, A3, B1, B2, and B3, respectively. “A” substitutes (i.e., A1, A2, A3) refer to analogs that have hydroxyl group additions to the ring structure, or body, of the cholesterol molecule. “B” substitutes (i.e., B1, B2, B3) refer to analogs that have hydroxyl group additions at the hydrophobic pole, or tail, of the cholesterol molecule. In total, six hydroxycholesterol analogs were evaluated: 7α-hydroxycholesterol (A1), 7β-hydroxycholesterol (A2), 19-hydroxycholesterol (A3), 20(S)-hydroxycholesterol (B1), 24(S)-hydroxycholesterol (B2), and 25-hydroxycholesterol (B3) (Fig. 2A). Further, LNP formulations were named by both the hydroxycholesterol substitute and percentage substitution. For example, the A2–50 formulation is a 50% substitution of A2 into the S2 formulation.
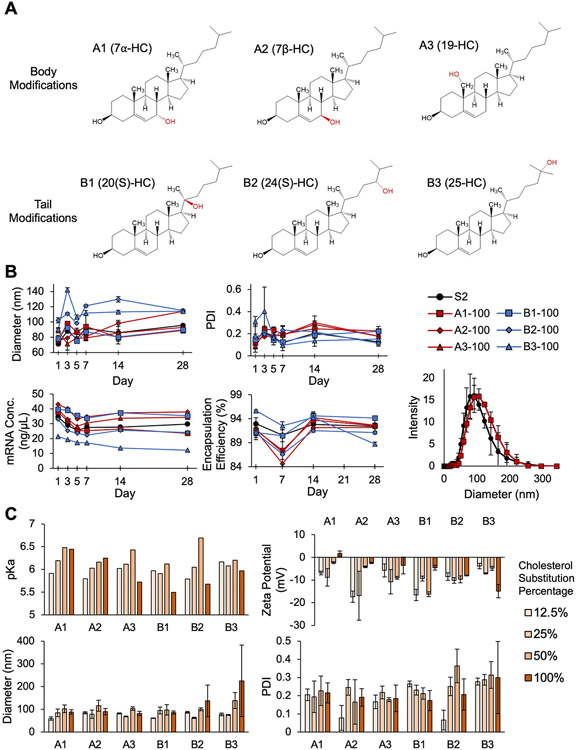
Fig. 2.
Characterization and stability of LNP formulations containing cholesterol analogs. (A) Structures of the six hydroxycholesterols (7α-HC, 7β-HC, 19-HC, 20(S)-HC, 24(S)-HC, and 25-HC) grouped by the location of the hydroxyl modification on the cholesterol molecule. LNPs containing cholesterol modifications to the body of the molecule are denoted by “A” (i.e., A1, A2, A3), and LNPs containing cholesterol modifications to the tail of the molecule are denoted by “B” (i.e., B1, B2, B3). (B) Measurements of z-average diameter, PDI, mRNA concentration, and encapsulation efficiency for S2 LNPs and LNP formulations with 100% cholesterol substitution taken over 28 days to assess LNP stability. The sample DLS curves show representative size distributions of LNP formulations S2 and A1-100 at day 3. n = 3. Error bars denote standard deviation. (C) The pKa, zeta potential, z-average diameter, and PDI measurements for the entire LNP library. n = 3. Error bars denote standard deviation.
These particular hydroxycholesterol substitutes were selected based on NPC1-binding profiles, location of hydroxyl group additions along the cholesterol molecule, and commercial availability [56]. Interestingly, many of these cholesterol analogs are found naturally in the body and result from the processing of cholesterol by reactive oxygen species or enzymes [71]. For example, 7α-hydroxycholesterol is a bile acid precursor [72], and 20(S)-hydroxycholesterol participates in the Smoothened oncoprotein signaling pathway [73]. Therefore, several of these hydroxycholesterols have natural bioactivity compared to other, previously investigated, cholesterol analogs (e.g., phytosterols) [74].
3.2. Effects of hydroxycholesterol substitution on LNP stability
Because chemical interactions between lipid components enable the formation of energetically stable membranes, LNPs with hydroxycholesterol substitutions were first evaluated for their stability [75]. In particular, the presence of cholesterol in lipid membranes contributes to membrane stability and the intrinsic curvature of lipid bilayers by producing an ordering effect [57,76]. As the cholesterol molecule contains a hydroxyl group at its head—which serves as a hydrophilic pole—it is amphipathic, enabling the molecule to align with neighboring lipids perpendicular to the lipid membrane [57]. Further, this integration of the cholesterol molecule is energetically favorable due to nonpolar interactions between cholesterol and lipids [77]. Thus, though the introduction of an additional hydroxyl group to the cholesterol molecule in LNPs is intended to interfere with NPC1 binding, such modifications may also alter the nonpolar and electrostatic interactions that cholesterol has with other LNP excipients, potentially leading to LNP instability. Therefore, we explored the effect of hydroxycholesterol substitution into LNP formulations on particle stability.
To observe the impact of these hydroxycholesterols, LNP formulations containing 100% substitutions were evaluated and compared to the standard S2 formulation. Stability was observed via measurements of z-average diameter, polydispersity index (PDI), mRNA concentration, and encapsulation efficiency over a 28-day period [78]. In terms of LNP diameter and PDI, most LNPs with a 100% substitution of hydroxycholesterols maintained sizes between 60 and 100 nm and PDIs below 0.25 over the 28-day period with minimal fluctuations in these measurements after initial stabilization. Temporal trends in mRNA concentration and encapsulation efficiency were also similar between most LNP candidates and S2 (Fig. 2B). Thus, we conclude that these LNPs remained stable throughout the 28-day observation period. Notably, however, B2–100 and B3–100 exhibited some unstable characteristics. B2–100 and B3–100 both had average diameters above 100 nm and trended lower in mRNA concentration. Furthermore, B3–100 had significant variation in PDI over the 28-day period. B2–100 and B3–100 both incorporate tail modifications of the cholesterol molecule, and we expect that the addition of a hydroxyl group at the tail end of these cholesterol molecules altered their amphipathic nature, potentially preventing alignment within the lipid membrane and resulting in this observed instability. Given this instability, it is possible that PDI and z-average diameter variance observed in days 1–5 for the B-series analogues may be partially explained by degradation products and lipid aggregation [79]. Alternatively, body modifications to cholesterol (A1, A2, A3) would not affect the polarity of cholesterol as significantly, given that the hydroxyl group on unmodified cholesterol is located at the head of the molecule, making it close in proximity to any such body modifications. Generally, the body-modified cholesterols produced stable LNPs, and tail-modified cholesterols produced LNPs with minimal changes in stability.
To further investigate an equilibrium between cholesterol and hydroxycholesterol in LNP formulations, we characterized LNPs that had 12.5%, 25%, 50%, and 100% substitutions of their respective hydroxycholesterol candidate for cholesterol using pKa, zeta potential, z-average diameter, and PDI (Fig. 2C). The pKa of an LNP is the pH at which it is 50% protonated, which indicates how efficiently the LNP changes charge from positive to negative in acidic environments to facilitate endosomal escape. Trends indicate that for LNPs featuring substitutions of body-modified hydroxycholesterols, increasing substitution percentage is associated with increasing pKa, suggesting that such substitutions may play a role in endosomal escape as the LNPs may be protonated at earlier stages of endosomal trafficking where pH is higher. Zeta potential measurements ranged from −17.3 mV to 1.67 mV but did not demonstrate any obvious trends with respect to the hydroxycholesterol candidate or substitution percentage. While A1, A2, A3 (body modifications), and B1 remained within expected ranges for diameter and PDI, B2 and B3 had increased particle diameters and were more polydisperse. This further confirms the previous conclusion that tail modifications at the 24 or 25 terminus of the cholesterol molecule may disrupt normal LNP formation, possibly by reducing cholesterol's ordering effect. Ultimately, these characterization parameters revealed that most of the hydroxycholesterol substitutes in consideration do not significantly affect LNP formation and stability over a range of substitution percentages.
3.3. In vitro screen of LNP library
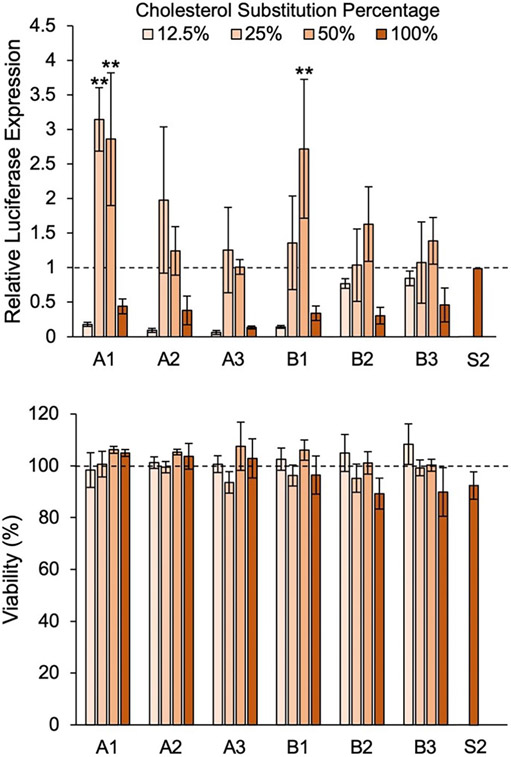
After observing the stability and physiochemical properties of the LNP library, we conducted assays to determine functional delivery of mRNA by each LNP formulation. As cholesterol in the LNP is critical to membrane fusion [57], removing it completely from our formulations would likely result in reduced cellular uptake. Thus, we explored 4 substitution percentages (12.5%, 25%, 50%, and 100%) of the 6 hydroxycholesterols, generating a library of 24 LNP formulations. This library was evaluated for functional mRNA delivery and cytotoxicity in Jurkats, an immortalized T cell line, as compared to the standard S2 formulation. Specifically, a reporter assay in which translated luciferase-encoding mRNA was measured was used as a readout for functional delivery [34,79]. This screen showed that for each hydroxycholesterol substitute, relative luciferase expression across substitution percentages tended to be unimodal. The lowest delivery was observed in LNPs with 12.5% and 100% substitutions, while LNPs with moderate substitutions of 25% and 50% resulted in higher delivery (Fig. 3). The screen further showed that A1–25, A1–50, and B1–50 produced statistically significant improvements in mRNA delivery to Jurkats as compared to S2 with 2.1-fold, 1.9-fold, and 1.7-fold increases in luminescence, respectively. Further, none of the LNP formulations in the library produced significant changes in cell viability, suggesting that the incorporation of hydroxycholesterol substitutes into LNPs does not induce cytotoxicity. Ultimately, A1–25, A1–50, and B1–50 demonstrated increased delivery of mRNA with no significant changes in LNP toxicity in vitro, marking them as potentially promising candidates for ex vivo applications.
Fig. 3.
Screen of LNP library for luciferase mRNA delivery and viability in Jurkats to identify top formulations. Jurkat cells were treated with LNP formulations at 60 ng mRNA / 60,000 cells for 24 h. Luciferase expression was normalized to cells treated with the standard LNP formulation (S2), and background luminescence was subtracted. Percent viability of cells treated with LNPs was determined by normalization to untreated cells. Legend denotes percent substitution of each hydroxycholesterol substitute into the S2 formulation. n = 3 biological replicates. Error bars denote standard deviation. An ANOVA was used to determine if treatment group means differed significantly. **: p < 0.01 in Tukey's honest significance test between LNP candidate and S2.
3.4. Ex vivo screen of LNPs containing top-performing hydroxycholesterol candidates in primary human T cells
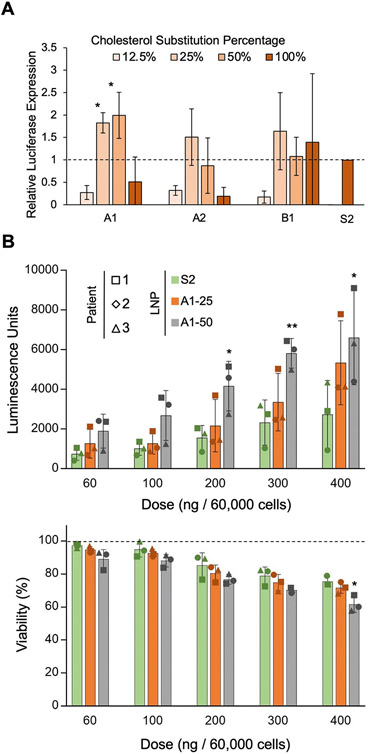
To further explore the translatability of these modified LNPs to ex vivo applications, we screened 12 LNPs from the first library—those containing the A1, A2, and B1 substitutes—in primary human T cells. A1 and B1 were selected because A1–25, A1–50, and B1–50 performed better than S2 in the in vitro screen. Although none of the A2 candidates significantly improved delivery of mRNA in vitro, A2-containing LNPs were included in this ex vivo screen as A1 and A2 are stereoisomers with similar chemical profiles. To deliver mRNA to primary T cells, cells were activated via CD3/CD28 dependent pathways; these expansion triggers may alter the membrane homeostasis of the cells. As such, all substitution percentages for the 3 selected hydroxycholesterol substitutes (A1, A2, B2) were evaluated in this ex vivo screen to re-optimize substitution percentages for ex vivo applications. Despite patient-to-patient variability, the screen revealed that A1–25 and A1–50 significantly improved mRNA delivery to primary T cells by 1.8-fold and 2.0-fold, respectively, as compared to S2 (Fig. 4). Interestingly, A1–25 outperformed A1–50 in terms of mRNA delivery in vitro while A1–50 outperformed A1–25 in mRNA delivery ex vivo. This result may be due to the inherent differences in sterol homeostasis exhibited by activated primary T cells and immortalized T cells [80]. Ultimately, A1–25 and A1–50 were the top performers from the ex vivo screen.
Fig. 4.
Screen of LNPs formulated with top-performing hydroxycholesterol substitutes in primary human T cells. (A) Luciferase expression in primary human T cells treated with LNP formulations containing A1, A2, or B1 hydroxycholesterols or S2 at a dose of 300 ng mRNA / 60,000 cells for 24 h. n = 3 biological replicates. Error bars denote standard deviation. An ANOVA was performed to determine if treatment group means differed significantly. *: p < 0.05 in student t-test between LNP candidate and S2. (B) Luciferase expression and relative viability of primary human T cells treated with S2, A1–25, and A1–50 at various doses. Luciferase expression was normalized to cells treated with the standard LNP formulation (S2), and background luminescence was subtracted. Percent viability was determined by normalizing to untreated cells. Legend denotes percent substitution of each hydroxycholesterol substitute into the S2 formulation. Each patient is represented by a different marker, n = 3 biological replicates. Error bars denote standard deviation. *: <0.05, **: p < 0.01 in student t-test between from S2 and either A1–25 or A1–50 LNPs.
To further validate these findings, a dose response was conducted in primary human T cells, and it showed that A1–25 and A1–50 sustain improvements in mRNA delivery to T cells at dosages ranging from 60 to 400 ng of mRNA per 60,000 cells with little to no significant decrease in cell viability. This suggests that A1–25 and A1–50 can potentially be utilized in ex vivo applications, such as CAR T cell therapy, to increase mRNA delivery efficiency without increasing toxicity towards target cells.
3.5. Endosomal colocalization and trafficking of hydroxycholesterol-substituted LNP formulations
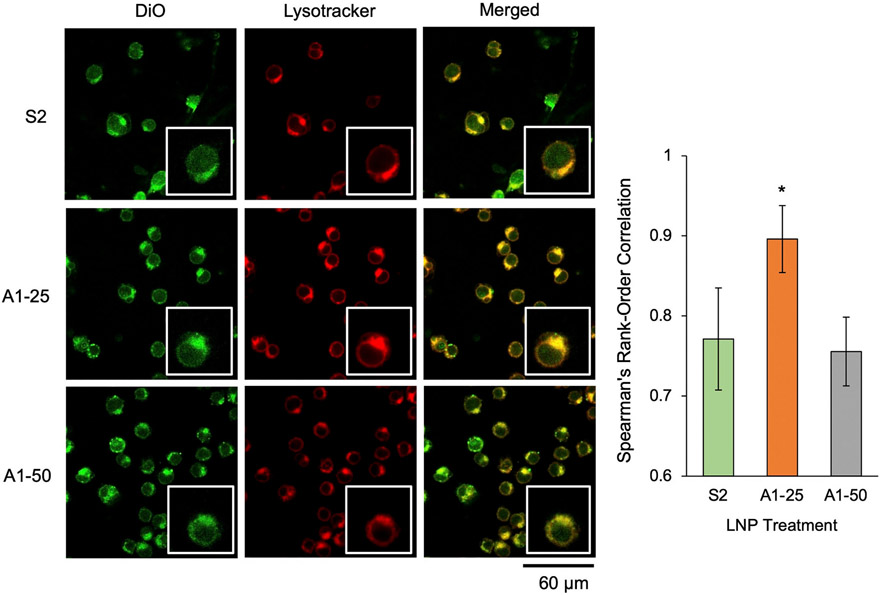
To better understand the impact of these hydroxycholesterol substitutions on endosomal uptake, retention, and recycling, a colocalization assay was utilized to assess the accumulation of top-performing LNPs (A1–25 and A1–50) within acidic organelles in T cells. Specifically, Jurkat cells were utilized, as opposed to human primary T cells, to avoid the effects of patient-to-patient variability seen in the ex vivo screen. Lysotracker was used to mark spherical, acidic organelles, the majority of which are endosomes and lysosomes, while LNPs were labeled with DiO, a lipophilic dye [81,82]. Confocal microscopy imaging was conducted after 4 h of LNP incubation with Jurkats, and images were subsequently analyzed for colocalization of LNPs and endosomes/lysosomes. A1–25 demonstrated increased colocalization with these acidic organelles, suggesting that the A1–25 particle either enters cells at higher rates or remains in endosomes for longer periods of time (Fig. 5). It has previously been observed that for LNPs to release cargo and enable mRNA translation, LNPs must reach the late endosome [42,70]. As such, either increases in particle uptake or a greater frequency of LNPs reaching the late endosome could explain this observed colocalization trend. This is further validated in the context of improved functional delivery of luciferase mRNA by A1–25 in Jurkats.
Fig. 5.
Endosomal uptake and colocalization of mRNA LNPs with varying 7α-hydroxycholesterol substitutions with endosomes in Jurkats. Confocal microscopy images of Jurkat cells treated with DiO-labeled LNPs at 60 ng mRNA/ 60,000 cells and stained with Lysotracker. Images were merged, background was subtracted, and Spearman's rank-order correlation was used to quantify association between LNPs and acidic organelles in cells. Single cell insets have been included for each image to provide visual reference, and insets are 2× magnification of the original image. Colocalization statistics (i.e., Spearman's rank-order correlation) were obtained from 5 fields of view (at least 90 cells in total) of each treatment group. Error bars denote standard deviation. An ANOVA was performed to determine if group means differed significantly. *: p < 0.05 in student t-test with Bonferroni p-value correction between colocalization statistics from S2 and either A1–25 or A1–50 LNPs.
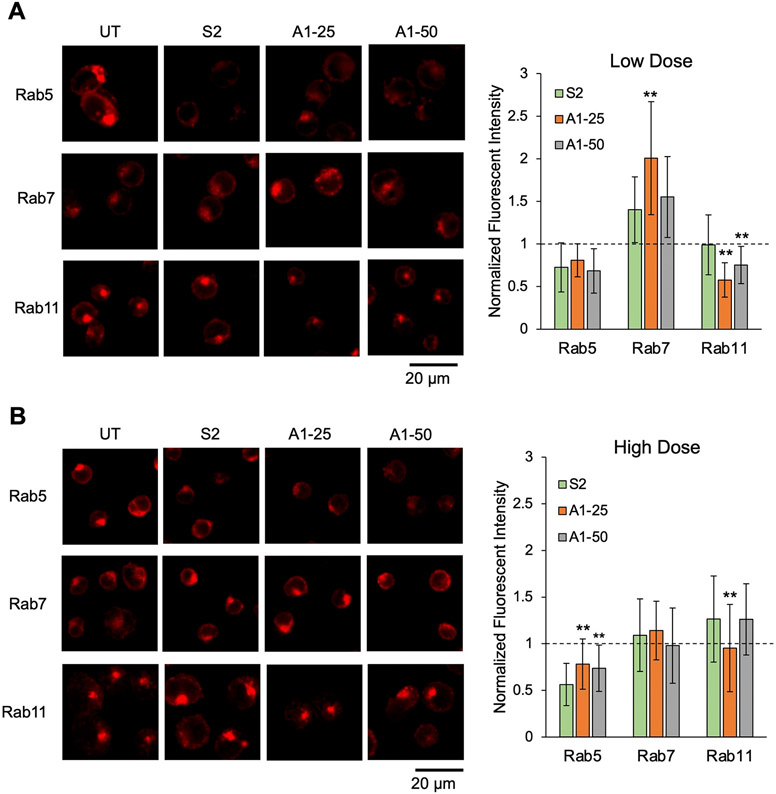
We then investigated the effect of hydroxycholesterol substitution on endosomal trafficking behavior using S2, A1–25, and A1–50. Specifically, three proteins that associate with various stages of the endosome were assessed: Rab5, Rab7, and Rab11. Rab5 tends to associate with early endosomes which provides insight on cell uptake of LNPs [83]. Rab7 associates with the late endosome and has been previously shown to be the direct precursor stage to endosomal escape and functional delivery [42]. Rab11 associates with the recycling endosome, which includes the exocytosis of endocytosed LNPs [64,84]. To assess trafficking using these Rab proteins as markers for endosomal progression, Jurkat cells were treated for 4 h with S2, A1–25, and A1–50 and imaged via confocal microscopy. Subsequent image analysis showed differing trends at low and high mRNA LNP doses (Fig. 6). At the low dose (Fig. 6A), Rab5 expression was slightly reduced compared to untreated cells. This is most likely explained by the transition of Rab5-associated early endosomes to Rab7-associated late endosomes induced by the introduction of exogenous materials (i.e., LNPs), leading to less early endosomes. This is further supported by increased Rab7 expression observed in all 3 LNP formulation treatment groups as compared to untreated cells, indicating the presence of additional late endosomes. Notably, A1–25 demonstrates significantly higher Rab7 expression as compared to S2, which is expected given that late endosomes are most closely associated with endosomal escape, and A1–25 significantly enhanced mRNA delivery in Jurkats compared to S2 in previous screens [42]. As the colocalization experiments were also performed at this low dose, these findings suggest that the high colocalization of A1–25 and endosomes observed in Fig. 5 was likely between A1–25 and late endosomes. Regarding the Rab11-associated recycling pathway, A1–25 and A1–50 induced significantly lower Rab11 expression than S2, with no LNP treatments showing increased expression compared to untreated cells. In conjunction with improved functional delivery results, decreased Rab11 expression in cells treated A1–25 and A1–50 suggests that these two LNPs did not induce as much of a recycling response as S2. In all, these results demonstrate that a low-dose delivery of A1–25 induced an increase of late endosomes. Furthermore, a low-dose delivery of A1–25 and A1–50 both induced a decrease of recycling endosomes.
Fig. 6.
Characterization of LNP endosomal trafficking. Confocal microscopy images of Jurkats stained with antibodies for Rab5, Rab7, or Rab11. Cells were either untreated (UT) or treated with S2, A1–25, or A1–50 LNPs at (A) 60 ng mRNA / 60,000 cells or (B) 150 ng mRNA / 60,000 cells. Rab5, Rab7, and Rab11 expression was quantified by averaging the fluorescent signal from at least 50 cells in each treatment group. Fluorescent intensity signal was normalized to untreated cells. Error bars denote standard deviation. An ANOVA was used within each Rab protein group to determine if group means differed significantly. *: <0.05, **: p < 0.01 in student t-test with Bonferroni p-value correction between from S2 and either A1–25 or A1–50 LNPs.
When observing these endosomal stages in Jurkat cells treated with higher LNP doses, there were a few notable differences (Fig. 6B). Though Rab5 expression remained lower than untreated cells in all groups, high-dose treatment with A1–25 and A1–50 LNPs significantly increased Rab5 expression compared to S2. Given that A1–25 and A1–50 both showed increased functional delivery of mRNA to Jurkats in our initial screens, we suspect that this increase in early endosome generation is the result of increased cellular uptake of these LNPs compared to S2. Additionally, it is possible that the hydroxycholesterol molecules in A1–25 and A1–50 may be causing morphological changes to the LNP that impact cellular uptake, but further characterization needs to be conducted to evaluate this hypothesis [53]. Though A1–25 and A1–50 increased Rab5 expression, there was no significant difference observed between LNP treatments in terms of Rab7 expression. This could be due to several factors including saturation of the endocytic pathway as seen in other delivery technologies [85]. However, despite the similar Rab7 levels observed across LNP treatment groups, A1–25 maintains a reduction in recycling endosomes, further defining a relationship between the presence of hydroxycholesterols and endosomal recycling. Thus, these results demonstrate that the high-dose delivery of A1–25 features increased early endosomes with a decrease in recycling endosomes relative to S2, while A1–50 achieves only an increase in early endosomes relative to S2.
Though the expression of these endosomal markers varies for these LNPs when administered at low and high doses, A1–25 and A1–50 have demonstrated improved mRNA delivery in vitro and ex vivo at various concentrations across this range. This may suggest that different mechanisms could be responsible for the improved delivery of A1-substituted LNPs depending on dose, with increased late endosomes leading to their enhanced endosomal escape at low doses and increased early endosomes indicating their improved cellular uptake in high doses. However, at both doses, the top-performing A1–25 LNP demonstrated decreased recycling endosomes, confirming the importance of this mechanism for potent mRNA delivery to T cells. Thus, the expression of these various endosomal markers suggests that 25% and 50% substitutions of A1 for unmodified cholesterol in the S2 formulation markedly improves functional delivery of mRNA to T cells through a combination of mechanisms that may vary with dose—including improved cellular uptake, increased generation of late endosomes, and reduced endosomal recycling.
4. Conclusion
In conclusion, we demonstrated that hydroxycholesterol substitution into LNP formulations does not significantly impact LNP stability, increases functional delivery of mRNA in vitro and ex vivo, and maintains similar cytotoxicity profiles as unmodified formulations. Furthermore, A1–25 and A1–50 emerged as top performers in our LNP screens. In particular, A1–25 demonstrated higher colocalization with the endosome, increased generation of late endosomes, and reduced endosomal recycling. More broadly, the results of the present study suggest that incorporation of 7α-hydroxycholesterol into LNP formulations at 25% and 50% substitution percentages for unmodified cholesterol in our S2 formulation improved mRNA LNP delivery to T cells. However, further characterization on the morphology of these modified LNPs is needed as well as further studies on the impact of incorporating these hydroxycholesterols into other LNP formulations. With these results, hydroxycholesterol substitution for cholesterol in LNPs presents itself as a potentially promising modification to increase the efficiency of various mRNA-based T cell therapeutic applications including cancer immunotherapy and vaccine development.
Supplementary Material
Acknowledgements
M.J.M. acknowledges support from a Burroughs Wellcome Fund Career Award at the Scientific Interface (CASI), a US National Institutes of Health (NIH) Director's New Innovator Award (DP2 TR002776), a grant from the American Cancer Society (129784-IRG-16-188-38-IRG), the National Institutes of Health (NCI R01 CA241661, NCI R37 CA244911, and NIDDK R01 DK123049), and a US National Science Foundation CAREER Award (CBET-2145491). M.M.B. was supported by a NIH F31 Ruth L. Kirschstein Predoctoral Individual National Research Service Award (F31CA260922-01). K.L.S. was supported by a US National Science Foundation Graduate Research Fellowship (Award 1845298).
Footnotes
Declaration of competing interest
The authors declare the following competing financial interest(s): M. J.M., M.M.B., and S.K.P. are inventors on a patent related to this work filed by the Trustees of the University of Pennsylvania (63251255).
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jconrel.2022.05.020.
References
- [1].Gall JS, Kalb RE, Infliximab for the treatment of plaque psoriasis, Biol. Targets Ther 2 (2008) 115–124, 10.2147/btt.s2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].June CH, O’Connor RS, Kawalekar OU, Ghassemi S, Milone MC, CAR T cell immunotherapy for human cancer, Science. 359 (2018) 1361–1365, 10.1126/science.aar6711. [DOI] [PubMed] [Google Scholar]
- [3].Chai LF, Prince E, Pillarisetty VG, Katz SC, Challenges in assessing solid tumor responses to immunotherapy, Cancer Gene Ther. 27 (2020) 528–538, 10.1038/s41417-019-0155-1. [DOI] [PubMed] [Google Scholar]
- [4].Chhabra N, Kennedy J, A review of Cancer immunotherapy toxicity II: adoptive cellular therapies, kinase inhibitors, monoclonal antibodies, and oncolytic viruses, J. Med. Toxicol (2021), 10.1007/s13181-021-00835-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Li D, Li X, Zhou WL, Huang Y, Liang X, Jiang L, Yang X, Sun J, Li Z, Han WD, Wang W, Genetically engineered T cells for cancer immunotherapy, Signal Transduct. Target. Ther 4 (2019) 35, 10.1038/s41392-019-0070-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gong N, Sheppard NC, Billingsley MM, June CH, Mitchell MJ, Nanomaterials for T-cell cancer immunotherapy, Nat. Nanotechnol 16 (2021) 25–36, 10.1038/s41565-020-00822-y. [DOI] [PubMed] [Google Scholar]
- [7].Mitchell MJ, Billingsley MM, Haley RM, Wechsler ME, Peppas NA, Langer R, Engineering precision nanoparticles for drug delivery, Nat. Rev. Drug Discov 20 (2021) 101–124, 10.1038/s41573-020-0090-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Guevara ML, Persano F, Persano S, Advances in lipid nanoparticles for mRNA-based Cancer immunotherapy, Front. Chem 8 (2020) 963, 10.3389/fchem.2020.589959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Miao L, Zhang Y, Huang L, mRNA vaccine for cancer immunotherapy, Mol. Cancer 20 (2021) 41, 10.1186/s12943-021-01335-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Pardi N, Hogan MJ, Porter FW, Weissman D, mRNA vaccines — a new era in vaccinology, Nat. Rev. Drug Discov 17 (2018) 261–279, 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Foster JB, Barrett DM, Karikó K, The emerging role of in vitro-transcribed mRNA in adoptive T cell immunotherapy, Mol. Ther 27 (2019) 747–756, 10.1016/j.ymthe.2019.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Soundara Rajan T, Gugliandolo A, Bramanti P, Mazzon E, In vitro-transcribed mRNA chimeric antigen receptor T cell (IVT mRNA CAR T) therapy in hematologic and solid tumor management: a preclinical update, Int. J. Mol. Sci 21 (2020) 6514, 10.3390/ijms21186514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wong SS, Webby RJ, An mRNA vaccine for influenza, Nat. Biotechnol 30 (2012) 1202–1204, 10.1038/nbt.2439. [DOI] [PubMed] [Google Scholar]
- [14].Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Pérez Marc G, Moreira ED, Zerbini C, Bailey R, Swanson KA, Roychoudhury S, Koury K, Li P, Kalina WV, Cooper D, Frenck RW, Hammitt LL, Türeci Ö, Nell H, Schaefer A, Ünal S, Tresnan DB, Mather S, Dormitzer PR, Şahin U, Jansen KU, Gruber WC, Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine, N. Engl. J. Med 383 (2020) 2603–2615, 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Richner JM, Himansu S, Dowd KA, Butler SL, Salazar V, Fox JM, Julander JG, Tang WW, Shresta S, Pierson TC, Ciaramella G, Diamond MS, Modified mRNA vaccines protect against Zika virus infection, Cell. 168 (2017) 1114–1125.e10, 10.1016/j.cell.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, Diemert D, Spector SA, Rouphael N, Creech CB, McGettigan J, Khetan S, Segall N, Solis J, Brosz A, Fierro C, Schwartz H, Neuzil K, Corey L, Gilbert P, Janes H, Follmann D, Marovich M, Mascola J, Polakowski L, Ledgerwood J, Graham BS,Bennett H, Pajon R, Knightly C, Leav B, Deng W, Zhou H, Han S, Ivarsson M, Miller J, Zaks T, Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine, N. Engl. J. Med 384 (2021) 403–416, 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Schultz L, Mackall C, Immunotherapy with genetically modified T cells, in: Cooper LJN, Mittendorf EA, Moyes J, Prabhakaran S (Eds.), Immunother. Transl. Cancer Res, John Wiley & Sons, Inc., Hoboken, NJ, USA, 2018, pp. 101–114, 10.1002/9781118684535.ch8. [DOI] [Google Scholar]
- [18].Kalos M, Levine BL, Porter DL, Katz S, Grupp SA, Bagg A, June CH, T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia, Sci. Transl. Med 3 (2011), 95ra73–95ra73, 10.1126/scitranslmed.3002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Idorn M, thor Straten P, Svane IM, Met Ö, Transfection of tumor-infiltrating T cells with mRNA encoding CXCR2, in: Rhoads RE (Ed.), Synth. MRNA, Springer, New York, New York, NY, 2016, pp. 261–276, 10.1007/978-1-4939-3625-0_17. [DOI] [PubMed] [Google Scholar]
- [20].Stewart MP, Sharei A, Ding X, Sahay G, Langer R, Jensen KF, In vitro and ex vivo strategies for intracellular delivery, Nature. 538 (2016) 183–192, 10.1038/nature19764. [DOI] [PubMed] [Google Scholar]
- [21].Smits E, Ponsaerts P, Lenjou M, Nijs G, Van Bockstaele DR, Berneman ZN, Van Tendeloo VFI, RNA-based gene transfer for adult stem cells and T cells, Leukemia. 18 (2004) 1898–1902, 10.1038/sj.leu.2403463. [DOI] [PubMed] [Google Scholar]
- [22].Barrett DM, Zhao Y, Liu X, Jiang S, Carpenito C, Kalos M, Carroll RG, June CH, Grupp SA, Treatment of advanced leukemia in mice with mRNA engineered T cells, Hum. Gene Ther 22 (2011) 1575–1586, 10.1089/hum.2011.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wang X, Rivière I, Clinical manufacturing of CAR T cells: foundation of a promising therapy, Mol. Ther. - Oncolytics 3 (2016) 16015, 10.1038/mto.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Vormittag P, Gunn R, Ghorashian S, Veraitch FS, A guide to manufacturing CAR T cell therapies, Curr. Opin. Biotechnol 53 (2018) 164–181, 10.1016/j.copbio.2018.01.025. [DOI] [PubMed] [Google Scholar]
- [25].Atsavapranee ES, Billingsley MM, Mitchell MJ, Delivery technologies for T cell gene editing: applications in cancer immunotherapy, EBioMedicine. 67 (2021), 103354, 10.1016/j.ebiom.2021.103354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Nickoloff JA (Ed.), Animal Cell Electroporation and Electrofusion Protocols, Humana Press, Totowa, N.J, 1995. [Google Scholar]
- [27].Dullaers M, Breckpot K, Van Meirvenne S, Bonehill A, Tuyaerts S, Michiels A, Straetman L, Heirman C, De Greef C, Van Der Bruggen P, Thielemans K, Side-by-side comparison of Lentivirally transduced and mRNA-Electroporated dendritic cells: implications for Cancer immunotherapy protocols, Mol. Ther 10 (2004) 768–779, 10.1016/j.ymthe.2004.07.017. [DOI] [PubMed] [Google Scholar]
- [28].Kulkarni JA, Myhre JL, Chen S, Tam YYC, Danescu A, Richman JM, Cullis PR, Design of lipid nanoparticles for in vitro and in vivo delivery of plasmid DNA, nanomedicine Nanotechnol, Biol. Med 13 (2017) 1377–1387, 10.1016/j.nano.2016.12.014. [DOI] [PubMed] [Google Scholar]
- [29].Kotelianski V, Zatsepin T, Kotelevtsev Y, Lipid nanoparticles for targeted siRNA delivery – going from bench to bedside, Int. J. Nanomedicine 11 (2016) 3077–3086, 10.2147/IJN.S106625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lee SWL, Paoletti C, Campisi M, Osaki T, Adriani G, Karnm RD, Mattu C, Chiono V, MicroRNA delivery through nanoparticles, J. Control. Release 313 (2019) 80–95, 10.1016/j.jconrel.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Swingle KL, Hamilton AG, Mitchell MJ, Lipid nanoparticle-mediated delivery of mRNA therapeutics and vaccines, Trends Mol. Med 27 (2021) 616–617, 10.1016/j.molmed.2021.03.003. [DOI] [PubMed] [Google Scholar]
- [32].Kim M, Jeong M, Hur S, Cho Y, Park J, Jung H, Seo Y, Woo HA, Nam KT, Lee K, Lee H, Engineered ionizable lipid nanoparticles for targeted delivery of RNA therapeutics into different types of cells in the liver, Sci. Adv 7 (2021), 10.1126/sciadv.abf4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Garber K, Alnylam launches era of RNAi drugs, Nat. Biotechnol 36 (2018) 777–778, 10.1038/nbt0918-777. [DOI] [PubMed] [Google Scholar]
- [34].Billingsley MM, Singh N, Ravikumar P, Zhang R, June CH, Mitchell MJ, Ionizable lipid nanoparticle-mediated mRNA delivery for human CAR T cell engineering, Nano Lett. 20 (2020) 1578–1589, 10.1021/acs.nanolett.9b04246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Billingsley MM, Hamilton AG, Mai D, Patel SK, Swingle KL, Sheppard NC, June CH, Mitchell MJ, Orthogonal Design of Experiments for optimization of lipid nanoparticles for mRNA engineering of CAR T cells, Nano Lett. (2021), acs. nanolett. 1 cO2503, 10.1021/acs.nanolett.1c02503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Akinc A, Maier MA, Manoharan M, Fitzgerald K, Jayaraman M, Barros S, Ansell S, Du X, Hope MJ, Madden TD, Mui BL, Semple SC, Tam YK, Ciufolini M, Witzigmann D, Kulkarni JA, van der Meel R, Cullis PR, The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs, Nat. Nanotechnol 14 (2019) 1084–1087, 10.1038/S41565-019-0591-y. [DOI] [PubMed] [Google Scholar]
- [37].Kulkarni JA, Cullis PR, van der Meel R, Lipid nanoparticles enabling gene therapies: from concepts to clinical utility, Nucleic Acid Ther. 28 (2018) 146–157, 10.1089/nat.2018.0721. [DOI] [PubMed] [Google Scholar]
- [38].Metzloff AE, Billingsley MM, Mitchell MJ, Lighting the way to personalized mRNA immune cell therapies, Sci. Adv 8 (2022) eabo2423, 10.1126/sciadv.abo2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Su FY, Zhao QH, Dahotre SN, Gamboa L, Bawage SS, Silva Trenkle AD, Zamat A, Phuengkham H, Ahmed R, Santangelo PJ, Kwong GA, In vivo mRNA delivery to virus-specific T cells by light-induced ligand exchange of MHC class I antigen-presenting nanoparticles, Sci. Adv 8 (2022) eabm7950, 10.1126/sciadv.abm7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lorenzer C, Dirin M, Winkler AM, Baumann V, Winkler J, Going beyond the liver: Progress and challenges of targeted delivery of siRNA therapeutics, J. Control. Release 203 (2015) 1–15, 10.1016/j.jconrel.2015.02.003. [DOI] [PubMed] [Google Scholar]
- [41].Besin G, Milton J, Sabnis S, Howell R, Mihai C, Burke K, Benenato KE, Stanton M, Smith P, Senn J, Hoge S, Accelerated blood clearance of lipid nanoparticles entails a biphasic humoral response of B-1 followed by B-2 lymphocytes to distinct antigenic moieties, ImmunoHorizons. 3 (2019) 282–293, 10.4049/immunohorizons.1900029. [DOI] [PubMed] [Google Scholar]
- [42].Herrera M, Kim J, Eygeris Y, Jozic A, Sahay G, Illuminating endosomal escape of polymorphic lipid nanoparticles that boost mRNA delivery, Biomater. Sci 9 (2021) 4289–4300, 10.1039/D0BM01947J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Cheng Q, Wei T, Farbiak L, Johnson LT, Dilliard SA, Siegwart DJ, Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas gene editing, Nat. Nanotechnol 15 (2020) 313–320, 10.1038/S41565-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Chen D, Love KT, Chen Y, Eltoukhy AA, Kastrup C, Sahay G, Jeon A, Dong Y, Whitehead KA, Anderson DG, Rapid discovery of potent siRNA-containing lipid nano particles enabled by controlled microfluidic formulation, J. Am. Chem. Soc 134 (2012) 6948–6951, 10.1021/ja301621z. [DOI] [PubMed] [Google Scholar]
- [45].Kauffman KJ, Dorkin JR, Yang JH, Heartlein MW, DeRosa F, Mir FF, Fenton OS, Anderson DG, Optimization of lipid nanoparticle formulations for mRNA delivery in vivo with fractional factorial and definitive screening designs, Nano Lett. 15 (2015) 7300–7306, 10.1021/acs.nanolett.5b02497. [DOI] [PubMed] [Google Scholar]
- [46].Swingle KL, Billingsley MM, Bose SK, White B, Palanki R, Dave A, Patel SK, Gong N, Hamilton AG, Alameh MG, Weissman D, Peranteau WH, Mitchell MJ, Amniotic fluid stabilized lipid nanoparticles for in utero intra-amniotic mRNA delivery, J. Control. Release 341 (2022) 616–633, 10.1016/j.jconrel.2021.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Yu MK, Park J, Jon S, Targeting strategies for multifunctional nanoparticles in Cancer imaging and therapy, Theranostics. 2 (2012) 3–44, 10.7150/thno.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Ball RL, Hajj KA, Vizelman J, Bajaj P, Whitehead KA, Lipid nanoparticle formulations for enhanced co-delivery of siRNA and mRNA, Nano Lett. 18 (2018) 3814–3822, 10.1021/acs.nanolett.8b01101. [DOI] [PubMed] [Google Scholar]
- [49].Farbiak L, Cheng Q, Wei T, Álvarez-Benedicto E, Johnson LT, Lee S, Siegwart DJ, All-in-one dendrimer-based lipid nanoparticles enable precise HDR-mediated gene editing in vivo, Adv. Mater 33 (2021) 2006619, 10.1002/adma.202006619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ermilova I, Swenson J, DOPC versus DOPE as a helper lipid for gene-therapies: molecular dynamics simulations with DLin-MC3-DMA, Phys. Chem. Chem. Phys 22 (2020) 28256–28268, 10.1039/D0CP05111J. [DOI] [PubMed] [Google Scholar]
- [51].Zhang R, El-Mayta R, Murdoch TJ, Warzecha CC, Billingsley MM, Shepherd SJ, Gong N, Wang L, Wilson JM, Lee D, Mitchell MJ, Helper lipid structure influences protein adsorption and delivery of lipid nanoparticles to spleen and liver, Biomater. Sci 9 (2021) 1449–1463, 10.1039/D0BM01609H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Gan Z, Lokugamage MP, Hatit MZC, Loughrey D, Paunovska K, Sato M, Cristian A, Dahlman JE, Nanoparticles containing constrained phospholipids deliver mRNA to liver immune cells in vivo without targeting ligands, Bioeng. Transl. Med 5 (2020), 10.1002/btm2.10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Patel S, Ashwanikumar N, Robinson E, Xia Y, Mihai C, Griffith JP, Hou S, Esposito AA, Ketova T, Welsher K, Joyal JL, Almarsson Ö, Sahay G, Naturally-occurring cholesterol analogues in lipid nanoparticles induce polymorphic shape and enhance intracellular delivery of mRNA, Nat. Cornmun 11 (2020) 983, 10.1038/s41467-020-14527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Pfeffer SR, NPC intracellular cholesterol transporter 1 (NPC1)-mediated cholesterol export from lysosomes, J. Biol. Chem 294 (2019) 1706–1709, 10.1074/jbc.TM118.004165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Infante RE, Wang ML, Radhakrishnan A, Kwon HJ, Brown MS, Goldstein JL, NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes, Proc. Natl. Acad. Sci 105 (2008) 15287–15292, 10.1073/pnas.0807328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Infante RE, Abi-Mosleh L, Radhakrishnan A, Dale JD, Brown MS, Goldstein JL, Purified NPC1 Protein, J. Biol. Chem 283 (2008) 1052–1063, 10.1074/jbc.M707943200. [DOI] [PubMed] [Google Scholar]
- [57].Yang ST, Kreutzberger AJB, Lee J, Kiessling V, Tamm LK, The role of cholesterol in membrane fusion, Chem. Phys. Lipids 199 (2016) 136–143, 10.1016/j.chemphyslip.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Shepherd SJ, Warzecha CC, Yadavali S, El-Mayta R, Alameh M-G, Wang L,Weissman D, Wilson JM, Issadore D, Mitchell MJ, Scalable mRNA and siRNA lipid nanoparticle production using a parallelized microfluidic device, Nano Lett. 21 (2021) 5671–5680, 10.1021/acs.nanolett.1c01353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Tsang M, Gantchev J, Ghazawi FM, Litvinov IV, Protocol for adhesion and immunostaining of lymphocytes and other non-adherent cells in culture, BioTechniques. 63 (2017) 230–233, 10.2144/000114610. [DOI] [PubMed] [Google Scholar]
- [60].Lukas JR, Aigner M, Denk M, Heinzl H, Buri an M, Mayr R, Carbocyanine postmortem neuronal tracing: influence of different parameters on tracing distance and combination with immunocytochemistry, J. Histochem. Cytochem 46 (1998) 901–910, 10.1177/002215549804600805. [DOI] [PubMed] [Google Scholar]
- [61].Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, Fiji: an open-source platform for biological-image analysis, Nat. Methods 9 (2012) 676–682, 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Schober P, Boer C, Schwarte LA, Correlation coefficients: appropriate use and interpretation, Anesth. Analg 126 (2018) 1763–1768, 10.1213/ANE.0000000000002864. [DOI] [PubMed] [Google Scholar]
- [63].Cheng Q, Wei T, Jia Y, Farbiak L, Zhou K, Zhang S, Wei Y, Zhu H, Siegwart DJ, Dendrimer-based lipid nanoparticles deliver therapeutic FAH mRNA to normalize liver function and extend survival in a mouse model of Hepatorenal Tyrosinemia type I, Adv. Mater 30 (2018) 1805308, 10.1002/adma.201805308. [DOI] [PubMed] [Google Scholar]
- [64].Hölttä-Vuori M, Tanhuanpää K, Möbius W, Somerharju P, Ikonen E, Modulation of cellular cholesterol transport and homeostasis by Rab11, Mol. Biol. Cell 13 (2002) 3107–3122, 10.1091/mbc.e02-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Sahay G, Querbes W, Alabi C, Eltoukhy A, Sarkar S, Zurenko C, Karagiannis E, Love K, Chen D, Zoncu R, Buganim Y, Schroeder A, Langer R, Anderson DG, Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling, Nat. Biotechnol 31 (2013) 653–658, 10.1038/nbt.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Wang H, Tam YYC, Chen S, Zaifman J, van der Meel R, Ciufolini MA, Cullis PR, The Niemann-pick Cl inhibitor NP3.47 enhances gene silencing potency of lipid nanoparticles containing siRNA, Mol. Ther 24 (2016) 2100–2108, 10.1038/mt.2016.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Lusa S, Blom TS, Eskelinen EL, Kuismanen E, Månsson JE, Simons K,Ikonen E, Depletion of rafts in late endocytic membranes is controlled by NPC1-dependent recycling of cholesterol to the plasma membrane, J. Cell Sci 114 (2001) 1893–1900. [DOI] [PubMed] [Google Scholar]
- [68].Zerial M, McBride H, Rab proteins as membrane organizers, Nat. Rev. Mol. Cell Biol 2 (2001) 107–117, 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- [69].Beztsinna N, de Matos MBC, Walther J, Heyder C, Hildebrandt E, Leneweit G, Mastrobattista E, Kok RJ, Quantitative analysis of receptor-mediated uptake and pro-apoptotic activity of mistletoe lectin-1 by high content imaging, Sci. Rep 8 (2018) 2768, 10.1038/s41598-018-20915-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Patel S, Ashwanikumar N, Robinson E, DuRoss A, Sun C, Murphy-Benenato KE, Mihai C, Almarsson Ö, Sahay G, Boosting intracellular delivery of lipid nano particle-encapsulated mRNA, Nano Lett. 17 (2017) 5711–5718, 10.1021/acs.nanolett.7b02664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Litwack G, Metabolism of fat, carbohydrate, and nucleic acids, in, Hum. Biochem. Elsevier; (2018) 395–426, 10.1016/B978-0-12-383864-3.00014-4. [DOI] [Google Scholar]
- [72].Anderson KE, Kok E, Javitt NB, Bile acid synthesis in man: metabolism of 7α-hydroxycholesterol-14C and 26-hydroxycholesterol-3H, J. Clin. Invest 51 (1972) 112–117, 10.1172/JCI106780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Nachtergaele S, Mydock LK, Krishnan K, Rammohan J, Schlesinger PH, Covey DF, Rohatgi R, Oxysterols are allosteric activators of the oncoprotein smoothened, Nat. Chem. Biol 8 (2012) 211–220, 10.1038/nchembio.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Kim J, Jozic A, Sahay G, Naturally derived membrane lipids impact nanoparticle-based messenger RNA delivery, Cell. Mol. Bioeng 13 (2020) 463–474, 10.1007/s12195-020-00619-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Maeki M, Fujishima Y, Sato Y, Yasui T, Kaji N, Ishida A, Tani H, Baba Y, Harashima H, Tokeshi M, Understanding the formation mechanism of lipid nanoparticles in microfluidic devices with chaotic micromixers, PLoS One 12 (2017), eO187962, 10.1371/journal.pone.0187962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Chen Z, Rand RP, The influence of cholesterol on phospholipid membrane curvature and bending elasticity, Biophys. J 73 (1997) 267–276, 10.1016/S0006-3495(97)78067-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Kessel A, Ben-Tal N, May S, Interactions of cholesterol with lipid bilayers: the preferred configuration and fluctuations, Biophys. J 81 (2001) 643–658, 10.1016/S0006-3495(01)75729-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Ball R, Bajaj P, Whitehead K, Achieving long-term stability of lipid nanoparticles: examining the effect of pH, temperature, and lyophilization, Int. J. Nanomedicine 12 (2016) 305–315, 10.2147/IJN.S123062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Ryals RC, Patel S, Acosta C, McKinney M, Pennesi ME, Sahay G, The effects of PEGylation on LNP based mRNA delivery to the eye, PLoS One 15 (2020), e0241006, 10.1371/journal.pone.0241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Kidani Y, Bensinger SJ, Modulating cholesterol homeostasis to build a better T cell, Cell Metab. 23 (2016) 963–964, 10.1016/j.cmet.2016.05.015. [DOI] [PubMed] [Google Scholar]
- [81].Yapici NB, Bi Y, Li P, Chen X, Yan X, Mandalapu SR, Faucett M, Jockusch S, Ju J, Gibson KM, Pavan WJ, Bi L, Highly stable and sensitive fluorescent probes (LysoProbes) for lysosomal labeling and tracking, Sci. Rep 5 (2015) 8576, 10.1038/srep08576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Kulkarni JA, Witzigmann D, Leung J, van der Meel R, Zaifman J, Darjuan MM, Grisch-Chan HM, Thöny B, Tam YYC, Cullis PR, Fusion-dependent formation of lipid nanoparticles containing macromolecular payloads, Nanoscale. 11 (2019) 9023–9031, 10.1039/C9NR02004G. [DOI] [PubMed] [Google Scholar]
- [83].The early endosome: a busy sorting station for proteins at the crossroads, Histol. Histopathol (2009) 99–112, 10.14670/HH-25.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Takahashi S, Kubo K, Waguri S, Yabashi A, Shin H-W, Katoh Y, Nakayama K, Rab11 regulates exocytosis of recycling vesicles at the plasma membrane, J. Cell Sci (2012) jcs.102913, 10.1242/jcs.102913. [DOI] [PubMed] [Google Scholar]
- [85].Teo SLY, Rennick JJ, Yuen D, Al-Wassiti H, Johnston APR, Pouton CW, Unravelling cytosolic delivery of cell penetrating peptides with a quantitative endosomal escape assay, Nat. Commun 12 (2021) 3721, 10.1038/s41467-021-23997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.