Abstract
The general secretion pathway of gram-negative bacteria is responsible for extracellular secretion of a number of different proteins, including proteases and toxins. This pathway supports secretion of proteins across the cell envelope in two distinct steps, in which the second step, involving translocation through the outer membrane, is assisted by at least 13 different gene products. Two of these components, the cytoplasmic membrane proteins EpsL and EpsM of Vibrio cholerae, have been purified and characterized. Based on gel filtration analysis, both purified EpsM(His)6 and wild-type EpsL present in an Escherichia coli Triton X-100 extract are dimeric proteins. EpsL and EpsM were also found to interact directly and form a Triton X-100 stable complex that could be precipitated with either anti-EpsL or anti-EpsM antibodies. In addition, when the L and M proteins were coexpressed in E. coli, they formed a stable complex and protected each other from proteolytic degradation, indicating that these two proteins interact in vivo and that no other Eps protein is required for their association. Since EpsL is predicted to contain a large cytoplasmic domain, while EpsM is predominantly exposed on the periplasmic side, we speculate that these components might be part of a structure that is involved in bridging the inner and outer membranes. Furthermore, since EpsL has previously been shown to interact with the autophosphorylating cytoplasmic membrane protein EpsE, we hypothesize that this trimolecular complex might be involved in regulating the opening and closing of the secretion pore and/or transducing energy to the site of outer membrane translocation.
In gram-negative bacteria, a number of different pathways for extracellular secretion have been identified (4, 9, 43). One of these pathways is the general secretion, or type II, pathway, which involves two steps (43). Proteins containing an N-terminal signal peptide are first translocated across the cytoplasmic membrane via the sec machinery. Then, after folding in the periplasm and, in some cases, assembly into multimeric protein complexes, they cross the outer membrane in a separate step. The second step requires between 12 and 16 genes, depending on species, and the homologous genes and gene products have been designated by letters A through O and S (for review, see reference 40).
Cholera toxin and several other putative virulence factors, including protease(s), neuraminidase, lipase, and chitinase, are translocated via the type II pathway in Vibrio cholerae (reference 46 and unpublished results). This process is assisted by the products of the epsC-epsN genes and vcpD (32, 37, 44, 46), which exhibit a high degree of similarity to genes required for type II secretion in other bacterial species, including Klebsiella oxytoca, Pseudomonas aeruginosa, Aeromonas hydrophila, Erwinia carotovora, Erwinia chrysanthemi, and Xanthomonas campestris (1, 3, 12, 13, 21, 23, 28, 41). In addition, sequences homologous to the eps genes are present in Escherichia coli; however, they do not appear to support secretion of cholera toxin or the related heat-labile enterotoxin. It is possible that these genes are not expressed under normal laboratory growth conditions, as was suggested by Francetic and Pugsley (14). Alternatively, their products may assist in secretion of other, as yet unidentified, extracellular proteins. Some of the eps components are also similar to genes required for assembly and export of pili in P. aeruginosa, Neisseria gonorrhoeae, and V. cholerae; for transfer of T-DNA from Agrobacterium tumifaciens to plants; and for DNA uptake in Bacillus subtilis (2, 8, 19, 22, 25, 27, 35).
While evidence for the function of some of the individual gene products of the secretion apparatus has been obtained, the mechanism for outer membrane translocation is still poorly understood. The D protein is present in the outer membrane and forms a large oligomeric ring of 12 to 18 subunits, which has been visualized by electron microscopy (5). Protein D is thought to be the actual pore through which secreted proteins are translocated (5, 26, 29). The E protein is located on the cytoplasmic side of the cytoplasmic membrane and might act as a kinase that regulates the secretion process or as an ATPase that supplies energy required for outer membrane translocation or biogenesis of the secretion apparatus (45). The O protein is located in the cytoplasmic membrane and is responsible for N-terminal processing and methylation of four other secretion-mediating proteins, G, H, I, and J (36). Finally, the S protein, which is a lipoprotein and might be specific to Klebsiella and Erwinia, appears to stabilize and assist protein D in its outer membrane localization (10, 16, 17). Although no function has been described for the other components, it is believed that several of them might have structural roles. Since the proteins G, H, I, and J are similar to pilin subunits with respect to their size and sequence identity at the N-terminal domain, it has been suggested that they might form a pseudopilus-like structure, through or along which secretion occurs (38). However, convincing evidence for such a structure has yet to be provided (39).
It is generally believed that the proteins required for secretion act in concert to form a functional secretion apparatus within the cell envelope. However, only limited evidence for specific intermolecular interactions, characteristic of a large multiprotein complex, exists. Our previous work indicated that expression of the cytoplasmic membrane protein EpsL results in stabilization and cytoplasmic membrane association of EpsE (45). Genetic suppressor studies suggest that protein E (Pseudomonas XcpR) also interacts with protein G (XcpT) (24). In addition, genetic analysis with the λ CI repressor as a reporter for dimerization has demonstrated that protein E may be a dimer in vivo (49), although purified EpsE(His)6 is monomeric (45). Cross-linking analysis of P. aeruginosa whole cells suggests that protein G, in turn, can form heterodimers with proteins H, I, and J (30). No protein-protein interactions have thus far been demonstrated for proteins C, F, K, M, and N, although we have observed that K and/or M appear to stabilize the EpsE-EpsL interaction (45).
In this study, we extend the analysis of the secretion apparatus of V. cholerae by characterizing two of its components, the cytoplasmic membrane proteins EpsL and EpsM. Very little is known about these proteins, with the exception that both of them may play a role in the membrane association of EpsE. In addition, membrane topology analysis of the EpsL and EpsM homologues in P. aeruginosa and E. carotovora suggests that they are cytoplasmic membrane proteins with a single membrane-spanning domain (6, 42). We show here that, in addition to forming homodimers, EpsL and EpsM also form a stable complex with each other. This interaction occurs in the absence of other Eps proteins and appears to stabilize EpsL and prevent it from proteolytic degradation. Given the subcellular location and the membrane topology of these components, their role in extracellular secretion is discussed.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Characteristics of the strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| MC1061 | F−lac | 7 |
| M15 | F lac | Qiagen |
| TG1 | lacIq | 31 |
| V. cholerae | ||
| TRH7000 | El Tor, Δ(ctxAB), wild type with respect to extracellular protein secretion | 20 |
| Mut8 | TRH7000 with a Kmr insertion in the epsL gene | 45 |
| PU3 | TRH7000 with a Tn5-transposon in the epsM gene | 37 |
| Plasmids | ||
| pMMB67 | Broad-host-range plasmid, Apr | 15 |
| pMMB587 | epsM(His)6 in pQE70 | This study |
| pMMB606 | epsM(His)6 in pMMB67EH replicon | This study |
| pMS44 | epsL in pMMB67HE | 46 |
| pMS45 | (His)6epsL in pQE30 | This study |
| pMS46 | (His)6epsL in pMMB67EH | This study |
| pMS47 | epsL(His)6 in pQE60 | This study |
| pMS48 | epsL(His)6 in pMMB67EH | This study |
| pMS58 | epsL epsM in pMMB67HE | This study |
| pREP4 | lacIq, Kmr | Qiagen |
| pQE30 | Codons for six N-terminal histidine residues, Apr | Qiagen |
| pQE60 | Codons for six C-terminal histidine residues, Apr | Qiagen |
| pQE70 | Codons for six C-terminal histidine residues, Apr | Qiagen |
| pWD615 | etxB (E. coli heat-labile enterotoxin B subunit gene), Tcr | 11 |
Purification of EpsL.
E. coli M15 containing plasmids pREP4 and pMS45 [encoding (His)6epsL] or pMS47 [encoding epsL(His)6] was grown in 500 ml of Luria broth (LB) (34) containing 200 μg of ampicillin and 50 μg kanamycin per ml at 30°C to an optical density at 600 nm (OD600) of 0.6 to 0.7. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 2 mM, and after growth for 3 h, the cells were harvested by centrifugation at 4°C. The cells were resuspended in 2.5 ml of 0.2 M Tris (pH 8.0)–5 mM β-mercaptoethanol. To the suspended cells, 5 ml of 0.2 M Tris (pH 8.0)–1 M sucrose–5 mM β-mercaptoethanol, 0.5 ml of 10 mg of lysozyme per ml, 0.5 ml of 10 mM EDTA, 0.5 ml of 10 mg of DNase per ml, and 16 ml of H2O containing 5 mM β-mercaptoethanol were added. The samples were incubated at room temperature for 15 min, and then 25 ml of 2% Triton X-100–20 mM MgCl2–5 mM β-mercaptoethanol was added. Samples were incubated for an additional 15 min and then centrifuged at 25,000 × g for 30 min at 4°C. The pellets containing His-tagged EpsL were resuspended in 10 ml of urea buffer (8 M urea–0.1 M NaPO4-0.01 M Tris (pH 8.0), 5 mM β-mercaptoethanol). Samples were incubated for 1 h at room temperature and then centrifuged at 25,000 × g for 30 min. Imidazole was added to the supernatants to a final concentration of 10 mM, and the supernatants were applied to a 3-ml Ni2+-nitrilotriacetic acid-agarose column (Qiagen). The column was extensively washed with urea buffer (pH 6.3) containing 40 mM imidazole, and His-tagged EpsL was eluted with a 50 to 200 mM imidazole step gradient in urea buffer (pH 6.3). Fractions of 3 ml were collected and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining. Both (His)6EpsL and EpsL(His)6 eluted with 120 mM imidazole.
Purification of EpsM.
E. coli TG1 carrying plasmid pMMB587 [epsM(His)6] was grown in 2 liters of LB containing 100 μg of ampicillin per ml at 37°C to an OD650 of 0.3. IPTG was added to a final concentration of 0.05 mM, and the culture was grown overnight. Cells were harvested by centrifugation, suspended in 44.0 ml of buffer I (50 mM NaPO4 buffer [pH 8.0], 0.3 M NaCl), and lysed by sonication in the presence of 1.0 mg of lysozyme per ml. DNase I was added to a final concentration of 10 U/ml in the presence of 10 mM MgCl2 and incubated at room temperature for 10 min. The lysate was centrifuged at 70,000 × g for 45 min at 4°C, and the pellet was extracted with 50 ml of buffer II (50 mM NaPO4, buffer [pH 8.0], 0.3 M NaCl, 30 mM n-octyl-β-d-glucopyranoside, 60 mM imidazole). After a second centrifugation at 70,000 × g the supernatant was applied to a 25.0-ml metal chelate column, POROS 20MC (PerSeptive Biosystems, Framingham, Mass.), preloaded with NiSO4 and equilibrated with buffer II. After being washed with buffer II, proteins were eluted with a linear gradient of 60 to 700 mM imidazole in buffer II. EpsM(His)6 eluted at approximately 300 mM imidazole.
Purified EpsM(His)6 was blotted onto Immobilon membranes, and the N-terminal amino acid sequence was determined by an automated Edman degradation procedure. Fifteen residues matched exactly the sequence predicted by the nucleotide sequence of the gene, determined previously (46). However, only one of the two predicted N-terminal Met residues was present in the protein produced from the synthetic gene, since this was the way PCR primers were designed for the construction of the EpsM(His)6 protein. It is still unknown, therefore, whether the wild-type EpsM protein carries one or two N-terminal methionines. As shown below, the recombinant EpsM containing only one N-terminal Met residue and six C-terminal His residues was functional in V. cholerae.
Gel filtration.
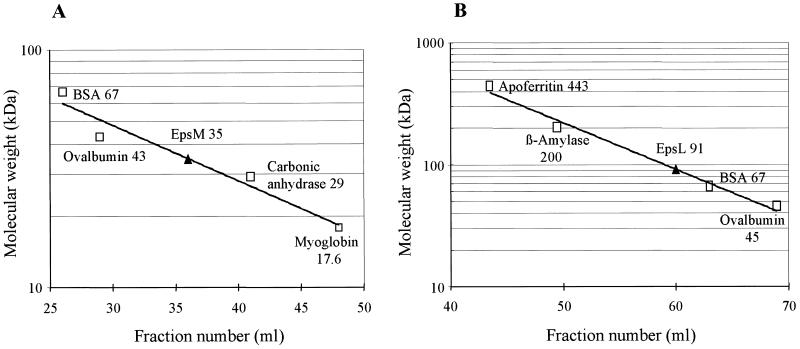
Approximately 1.5 mg of EpsM(His)6 in 0.5 ml of buffer III (50 mM NaPO4 [pH 8.0], 150 mM NaCl, 30 mM n-octyl-β-d-glucopyranoside) was loaded onto a 10- by 300-mm Superdex 200 HR column (Pharmacia Biotech) at a flow rate of 0.25 ml/min. Fractions of 1 ml were collected. E. coli Triton X-100 extract corresponding to a 2.5-ml overnight culture of MC1061/pMS44 expressing EpsL without IPTG induction was applied to a 16- by 600-mm Sephacryl S-300 column (Pharmacia Biotech) at a flow rate of 0.5 ml/min. Fractions of 3 ml were collected. Elution was monitored by measurement of OD280, SDS-PAGE, and Coomassie blue staining. For the analysis of EpsL, the fractions were also subjected to immunoblot analysis with anti-EpsL antibodies. Reference proteins apoferritin (443 kDa), β-amylase (200 kDa), bovine serum albumin (BSA) (67 kDa), ovalbumin (43 kDa), carbonic anhydrase (29 kDa), and myoglobin (17.6 kDa) were analyzed under the same conditions and used to estimate the molecular masses of EpsM(His)6 and EpsL.
Antibody production.
Antibodies against EpsL and EpsM were raised in New Zealand White rabbits by using purified EpsL(His)6 (in 1 M urea) and EpsM(His)6 and Freund’s adjuvant according to the method of Harlow and Lane (18).
Purification and biotinylation of anti-EpsL and anti-EpsM IgG.
Anti-EpsL(His)6 or anti-EpsM(His)6 antisera (1.5 ml) were applied to a 1-ml protein G Sepharose 4 Fast flow column (Pharmacia Biotech) equilibrated with 20 mM NaPO4 (pH 7.0). Following extensive washing with the same buffer, immunoglobulin G (IgG) was eluted with 0.1 M glycine (pH 2.9) into tubes containing 0.1 volume of 1 M Tris-HCl (pH 8.0). Six milligrams of IgG was then dialyzed against 0.1 M Na-carbonate buffer (pH 8.8), and 0.6 mg of d-biotinoyl-ɛ-aminocaproic acid N-hydroxysuccinimide ester was added. Following incubation at room temperature for 2 h, the samples were dialyzed against Tris-buffered saline (TBS).
Cell fractionation.
Wild-type, epsL, and epsM mutant strains of V. cholerae TRH7000 were grown in M9 medium supplemented with 0.4% glucose, 0.4% Casamino Acids, and 0.1 mg of thymine per ml to late log phase. The growth medium was separated from the cells by centrifugation, and the cells were treated with 5,000 U of polymyxin B per ml in phosphate-buffered saline (PBS) for 15 min on ice to release periplasmic content. The samples were centrifuged, and the supernatants containing periplasmic proteins were transferred to new tubes. The pellets were resuspended in PBS and sonicated. Following centrifugation to separate soluble cytoplasmic proteins from total membranes, cytoplasmic membrane proteins were extracted from the membrane pellets with 1% Triton X-100 and 10 mM MgCl2. Cytoplasmic and outer membrane proteins were separated by centrifugation.
Preparation of Triton X-100 soluble cell extracts.
V. cholerae or E. coli strains expressing different eps genes were grown in M9 growth medium supplemented with glucose (0.4%), Casamino Acids (0.4%), and thymine (0.1 mg/ml, for V. cholerae only) at 37°C to late log phase. Cells from 1-ml cultures were centrifuged and resuspended in 50 μl of 0.2 M Tris (pH 8.0). To the suspended cells, 100 μl of 0.2 M Tris (pH 8.0)–1.0 M sucrose, 10 μl of 10 mM EDTA, 10 μl of 10 mg of lysozyme per ml, and 320 μl of H2O were added. DNase was added to a final concentration of 20 μg/ml. Samples were incubated for 10 min at room temperature, and 0.5 ml of 2% Triton X-100–20 mM MgCl2 was added. Samples were incubated at 4°C for 30 min, followed by centrifugation at 14,000 × g for 30 min. The supernatants were subjected to SDS-PAGE and immunoblotting with anti-EpsL or anti-EpsM antiserum and horseradish peroxidase-conjugated goat anti-rabbit IgG. The blots were developed with Supersignal chemiluminescent substrate (Pierce). Densitometric analysis of images of scanned immunoblots was performed by using the Scion Image program (Scion Corp.).
Immunoprecipitation.
Anti-EpsL and anti-EpsM antisera were incubated with equal volumes of preswollen protein G-Sepharose in TBS overnight at 4°C, after which the Sepharose was extensively washed with TBS. In order to precipitate EpsL or EpsM from 100 μl of bacterial Triton X-100 cell extract, 10 μl of protein G-Sepharose containing either anti-EpsL or anti-EpsM IgG was added. This was done in a total volume of 1 ml of TBS containing 1% Triton X-100 and 1 mM EDTA. Following incubation on a rocker for 2 h at 4°C, the samples were centrifuged, and the supernatants were removed. The Sepharose beads were washed two times with 1 ml of TBS–1% Triton X-100–1 mM EDTA and once with 1 ml of TBS. Twenty microliters of SDS sample buffer was added, and the samples were incubated at 68°C for 10 min and centrifuged. The supernatants (10 μl) were analyzed by SDS-PAGE and immunoblotting with biotinylated anti-EpsL or anti-EpsM IgG and horseradish peroxidase-conjugated streptavidin. Peroxidase activity was visualized with Supersignal chemiluminescent substrate (Pierce).
Determination of toxin B subunit secretion.
Wild-type and epsL and epsM mutant strains of V. cholerae TRH7000 containing plasmid pWD615 were grown in M9 medium supplemented with 0.4% glucose, 0.4% Casamino Acids, and 0.1 mg of thymine per ml to late log phase. The growth medium was separated from the cells by centrifugation, and the cells were treated with 5,000 U of polymyxin B per ml to release periplasmic content. The amount of B subunit pentamers present in the growth media and periplasmic extract was determined by GM1 enzyme-linked immunosorbent assay as described previously (37, 47).
RESULTS
Construction and purification of histidine-tagged Eps proteins.
In order to purify EpsL and EpsM, they were produced as histidine-tagged proteins with six histidine residues at either the N or C terminus. Neither the N- nor C-terminal histidine constructs of EpsL displayed full complementation activity when produced in a V. cholerae epsL mutant. In both cases, secretion was increased four- to fivefold compared to the epsL mutant, but was reduced two- to threefold compared to wild-type V. cholerae (Table 2). In comparison, plasmid-encoded wild-type epsL could restore secretion of toxin to wild-type levels in the epsL mutant strain (46). In contrast, EpsM(His)6 protein appeared to be produced as a fully functional protein (Table 2), as determined by its ability to restore toxin secretion in the epsM mutant strain to a level comparable with that obtained in the presence of plasmid-encoded wild-type epsM (37).
TABLE 2.
Complementation of secretion defects by His-tagged EpsL and EpsM
| Strain | Plasmid-encoded protein | Fraction of toxin B subunit in:
|
|
|---|---|---|---|
| Medium | Cells | ||
| TRH7000(wt)a/pWD615 | 0.83 | 0.17 | |
| Mut8 (epsL)/pWD615 | 0.07 | 0.93 | |
| Mut8/pWD615/pMS46 | (His)6EpsL | 0.31 | 0.69 |
| Mut8/pWD615/pMS48 | EpsL(His)6 | 0.38 | 0.62 |
| PU3 (epsM)/pWD615 | 0.22 | 0.78 | |
| PU3/pWD615/pMMB606 | EpsM(His)6 | 0.77 | 0.23 |
wt, wild type with regard to Eps function.
When N- or C-terminal histidine-tagged EpsL proteins were overproduced in E. coli, they were found in aggregates, and, therefore, they were purified under denaturing conditions (8 M urea) by affinity chromatography on Ni2+-nitrilotriacetic acid-agarose (not shown). EpsM(His)6, on the other hand, could be purified under native conditions from either octylglucoside or the Triton X-100 extract of E. coli membranes (not shown).
EpsM and EpsL form dimers.
The size of purified EpsM(His)6 was determined by gel filtration on a Superdex 200 HR column. EpsM(His)6 eluted in a single peak, and its elution was compared with that of known molecular mass markers. The results indicated that EpsM(His)6 was present in solution as a 35-kDa protein (Fig. 1A). This is in good agreement with the expected size for a dimer of the 18.5 kDa molecule predicted from the translation of the epsM gene sequence (46). The molecular size of EpsL in solution could not be determined by analysis of purified material, since EpsL was purified under denaturing conditions. Instead, gel filtration analysis was performed with a Triton X-100 extract from E. coli expressing wild type epsL. Figure 1B shows the result from a gel filtration experiment in which the mass of EpsL was 91 kDa. The average molecular mass for Triton X-100-soluble EpsL from independent experiments was 101 kDa, which is 2.2 times the size of the predicted molecular mass for EpsL (46), suggesting that EpsL also forms dimers when produced in E. coli in the absence of other Eps proteins.
FIG. 1.
Size determination of EpsM(His)6 and EpsL. Fractionation of purified EpsM(His)6 and EpsL present in an E. coli Triton X-100 extract was performed on a Superdex 200 HR column (A) and Sephacryl S-300 column (B), respectively. Fractions of 1.0 ml (A) or 3.0 ml (B) were analyzed by OD280 as well as SDS-PAGE and Coomassie brilliant blue staining or immunoblotting with anti-EpsL antibodies. Elution of EpsM(His)6 and EpsL was compared with the elution of standard proteins of known molecular mass. The volume of eluent at which the individual standard proteins emerged from the column was plotted against the logarithm of their molecular mass and fit by linear regression. The elution peak of EpsM(His)6 and EpsL is indicated by a triangle, and the apparent molecular mass for these proteins was calculated from these data.
EpsL and EpsM are associated with the cytoplasmic membrane.
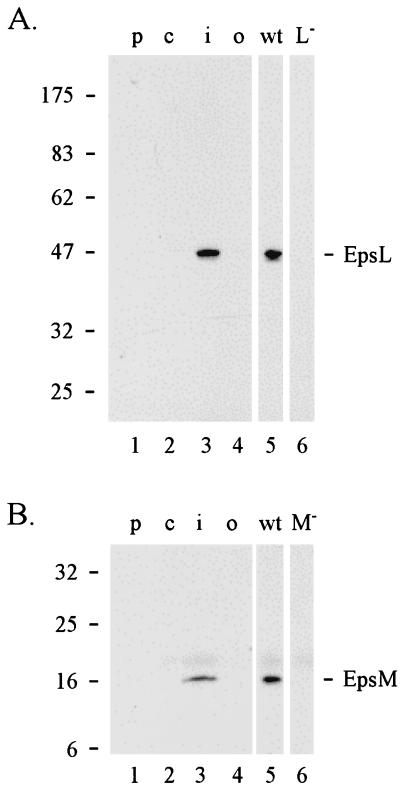
Although both EpsL and EpsM proteins are required for outer membrane translocation of cholera toxin, they are predicted to be cytoplasmic membrane proteins based on structural analysis of their primary sequence (not shown). In addition, analysis of fusion proteins composed of the EpsL and EpsM homologues XcpY and XcpZ in P. aeruginosa and either alkaline phosphatase or β-lactamase suggests that the L and M proteins are cytoplasmic membrane proteins (6). In order to determine the subcellular location of EpsL and EpsM proteins, wild-type V. cholerae cells were separated into periplasmic, cytoplasmic, and membrane fractions by polymyxin B treatment, sonication, and centrifugation. Cytoplasmic membrane proteins were then extracted with Triton X-100 and separated from outer membrane proteins by centrifugation. All fractions were analyzed by SDS-PAGE and immunoblotting with anti-EpsL(His)6 and anti-EpsM(His)6 antibodies (Fig. 2). The majority of both EpsL and EpsM were found in the Triton X-100-soluble extract (Fig. 2A and B, lanes 3), suggesting that they are most likely located in the cytoplasmic membrane.
FIG. 2.
Subcellular localization of EpsL (A) and EpsM (B). Wild-type V. cholerae was fractionated into periplasmic (lane 1), cytoplasmic (lane 2), Triton X-100-soluble inner membrane (lane 3), and Triton X-100-insoluble outer membrane fractions (lane 4) as described in Materials and Methods. The samples were subjected to SDS-PAGE and immunoblot analysis by using anti-EpsL (A) or anti-EpsM (B) antibodies. For comparison and specificity of the antibodies, total cell extracts of the wild type (lane 5) and epsL mutant (L−; lane 6 in panel A) or epsM mutant (M−; lane 6 in panel B) were also analyzed.
Interaction between EpsL and EpsM.
We have previously demonstrated that EpsL is responsible for stabilization and partial membrane association of EpsE. We also found that coexpression of proteins K and M, with proteins E and L, resulted in a further increase of membrane-associated EpsE, suggesting that proteins K and/or M can stabilize the EpsE-EpsL interaction (45). In order to test for a direct physical interaction between these proteins, coimmunoprecipitation experiments were performed.
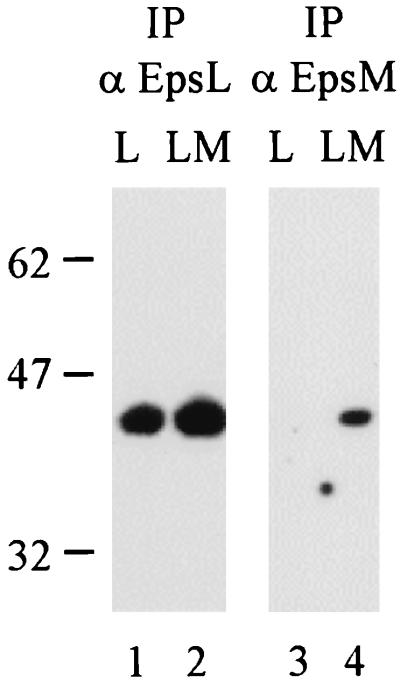
Triton X-100 extracts of wild-type V. cholerae TRH7000, Mut8 (epsL mutant), and PU3 (epsM mutant) were subjected to immunoprecipitation with anti-EpsL or anti-EpsM antibodies coupled to protein G-Sepharose. Precipitated samples were then subjected to SDS-PAGE and immunoblot analysis with either biotinylated anti-EpsL (Fig. 3A) or anti-EpsM (Fig. 3B) IgG. The results demonstrated, first of all, that native EpsL could be immunoprecipitated from the wild-type cell extract and then recognized on a blot with biotinylated anti-EpsL IgG (Fig. 3A, lane 1), suggesting that although the EpsL(His)6 protein used for the development of the anti-EpsL antibodies was denatured, the antibodies can recognize and precipitate the presumably native form of EpsL present in the Triton extract. Second, the results indicated that EpsL can be coimmunoprecipitated with EpsM by using anti-EpsM antibodies (Fig. 3A, lane 4) and, likewise, that EpsM can be precipitated with EpsL by using anti-EpsL antibodies from wild-type extract (Fig. 3B, lane 1). Control experiments showed that EpsL could not be precipitated with anti-EpsM antibodies from an epsM mutant strain (Fig. 3A, lane 5), indicating that in order to precipitate with anti-EpsM, EpsL needs to be part of a complex with EpsM and that the coprecipitation is not due to cross-reactivity between the anti-EpsM antibodies and EpsL. The observation was also true for EpsM. EpsM could not be precipitated with anti-EpsL antibodies from an epsL mutant (Fig. 3B, lane 3), again demonstrating the specificity of the antibodies. These results demonstrate that EpsL and EpsM form a complex that can be extracted with the nonionic detergent Triton X-100.
FIG. 3.
Coimmunoprecipitation of EpsL and EpsM. Triton X-100 extracts from wild-type (wt) and epsM (M−) and epsL (L−) mutant V. cholerae strains were immunoprecipitated (IP) with either anti-EpsL or anti-EpsM antibodies (α) prior to SDS-PAGE and immunoblot analysis with biotinylated anti-EpsL IgG (A) or anti-EpsM IgG (B) and horseradish peroxidase-conjugated streptavidin.
No other Eps proteins are required for the EpsL-EpsM interaction.
In order to test whether additional Eps proteins are required for the EpsL-EpsM interaction, the epsL and epsM genes were expressed in E. coli in the absence or presence of other eps genes. Triton X-100-soluble E. coli material was subjected to immunoprecipitation with either anti-EpsL or EpsM antibodies, and then analyzed by SDS-PAGE and immunoblotting with biotinylated anti-EpsL IgG (Fig. 4). EpsL could be precipitated with anti-EpsL antibodies from E. coli extracts containing either EpsL only or EpsL and EpsM (Fig. 4, lanes 1 and 2). In contrast, EpsL could not be precipitated with the anti-EpsM antibodies in the absence of the EpsM protein (Fig. 4, lane 3), again demonstrating the specificity of the antibodies and also suggesting that E. coli does not produce an EpsM homologue that can interact with EpsL. In addition, EpsL was precipitated with the anti-EpsM antibodies from extracts containing EpsL and EpsM only (Fig. 4, lane 4), demonstrating that no other Eps protein is needed for the EpsL-EpsM interaction. Thus, expression of epsL and epsM is sufficient to obtain stable complex formation between these two components.
FIG. 4.
EpsL and EpsM can form a stable complex in E. coli in the absence of other Eps proteins. Triton X-100-soluble extracts of E. coli producing EpsL (L) or EpsL and EpsM (LM) were immunoprecipitated (IP) with either anti-EpsL (lanes 1 and 2) or anti-EpsM (lanes 3 and 4) antibodies (α). Samples were subjected to SDS-PAGE and immunoblot analysis with biotinylated anti-EpsL IgG and horseradish peroxidase-coupled streptavidin.
EpsL is stabilized by EpsM when expressed in E. coli.
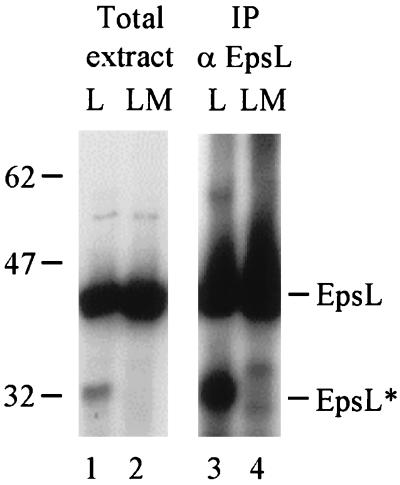
When total E. coli extracts were analyzed by immunoblotting, it was observed that a fraction of EpsL was being degraded when produced in the absence of EpsM (Fig. 5, lane 1). That the degradation product of 32 kDa was indeed of EpsL origin was also demonstrated by its ability to be immunoprecipitated with anti-EpsL antibodies (Fig. 5, lane 3). In the presence of EpsM, this degradation product was absent, suggesting that EpsM not only interacts with EpsL, but also stabilizes EpsL and prevents its degradation (Fig. 5, lanes 2 and 4). Consistent with this, densitometric analysis of the immunoblot indicated that the amount of full-length EpsL increased approximately twofold in the presence of EpsM (data not shown).
FIG. 5.
EpsL is stabilized by EpsM. Total cell extracts of E. coli expressing either epsL (L) alone or epsL and epsM (LM) were analyzed by SDS-PAGE and immunoblotting with biotinylated anti-EpsL antibodies (lanes 1 and 2) or immunoprecipitated (IP) with anti-EpsL antibodies (αEpsL) prior to SDS-PAGE and immunoblotting with biotinylated anti-EpsL IgG (lanes 3 and 4). The immunoprecipitated blot was exposed for a longer time than those in Fig. 4 in order to clearly show the EpsL degradation product (EpsL*).
DISCUSSION
EpsL and EpsM are components of the general secretion apparatus in V. cholerae. This multicomponent system is required for extracellular secretion of cholera toxin and other putative virulence factors. Although both the L and M proteins are specifically required for the outer membrane translocation step, they appear to be localized to the cytoplasmic membrane based on subcellular fractionation and detergent extraction analysis (Fig. 2). Somewhat surprisingly, cytoplasmic membrane location has also been observed for several other components of the type II secretion apparatus in addition to EpsL and EpsM, either by similar cell fractionation studies or by topological analysis using phoA and blaM probes (6, 42, 48). In addition, computer analysis (50) of the primary structure of most of these factors predicts that they are inner membrane proteins. The majority of these proteins are thought to span the cytoplasmic membrane only once, and some of them, including EpsL, are predicted to contain large cytoplasmic domains. Others, such as EpsM, may have large periplasmic domains, and therefore may be part of a structure that bridges the inner and outer membranes.
In this study, we have shown that in addition to forming homodimers, EpsL and EpsM are also involved in protein-protein interactions with each other. We demonstrated that EpsL and EpsM could be coimmunoprecipitated with both anti-EpsL and anti-EpsM antibodies, suggesting that EpsL and EpsM form a stable complex in vivo. Although there is a possibility that the EpsL-EpsM complex was formed during or after the extraction of the two proteins from the membrane, the fact that EpsL was stabilized against proteolytic degradation by EpsM when produced in E. coli suggests that these proteins are likely to interact in vivo, and the observed complex is not an artifact of the extraction procedure. The notion that EpsL also protected EpsM from proteolytic degradation in E. coli supports this conclusion (data not shown), as do our observations with the V. cholerae epsL and epsM mutants, in which removal of either Eps protein resulted in the destabilization and degradation of the other Eps protein (not shown). Finally, during the review of the manuscript, additional support for the interaction between proteins L and M was presented by Michel et al. (33), who showed that the absence of the EpsM homologue XcpZ in P. aeruginosa resulted in a reduced amount of the EpsL homologue XcpY. Similarly, mutations in the xcpY gene resulted in a lower level of XcpZ.
Previously we found that EpsE is associated with the cytoplasmic membrane via EpsL and that this interaction appeared to be stabilized by EpsM and/or EpsK (45). The observation that EpsL and EpsM form a stable complex in vivo suggests that, at least for EpsM, its effect on the EpsL-EpsE interaction may be through this direct interaction with EpsL. EpsM may exert its effect by interacting directly with and stabilizing the L protein, resulting in a conformational change within EpsL that allows for increased affinity for EpsE. Since EpsM is largely exposed on the periplasmic side, it is very likely to also interact with other components of the secretion apparatus that contain a periplasmic domain and, possibly, with outer membrane proteins. If this is the case, EpsM would be part of a structure that bridges the two membranes. This would then connect the EpsE protein in the cytoplasm with the outer membrane via its interaction with EpsL. Since we have previously demonstrated that EpsE has autophosphorylation properties, it was suggested that EpsE is a protein kinase that might regulate the extracellular secretion process (45). It might do so through its interaction with EpsL and EpsM and, through these, a protein in the outer membrane. This could result in transmembrane signaling that opens the secretory channel or pore, permitting extracellular proteins to cross the outer membrane. If the demonstrated autophosphorylation activity of EpsE is only an intermediate step and EpsE is in fact an ATPase, proteins L and M could possibly transduce energy from EpsE and the cytoplasmic membrane to the site of outer membrane translocation. Finally, there is also a possibility that EpsM, due to its predicted exposure to the periplasm, might be one of the components within the apparatus that recognize and/or target secreted proteins to the putative secretion pore.
ACKNOWLEDGMENTS
We thank D. A. Lawrence for critically reading the manuscript.
This work was supported by the American Red Cross and by the U.S. Department of Agriculture (92-37204-7839) and Research Excellence Fund from the State of Michigan to M.B.
REFERENCES
- 1.Akrim M, Bally M, Ball G, Tommassen J, Teerink H, Filloux A, Lazdunski A. Xcp-mediated protein secretion in Pseudomonas aeruginosa: identification of two additional genes and evidence for regulation of xcp gene expression. Mol Microbiol. 1993;10:431–443. doi: 10.1111/j.1365-2958.1993.tb02674.x. [DOI] [PubMed] [Google Scholar]
- 2.Albano M, Dubnau D A. Cloning and characterization of a cluster of linked Bacillus subtilis late competence mutations. J Bacteriol. 1989;171:5376–5385. doi: 10.1128/jb.171.10.5376-5385.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bally M, Filloux A, Akrim M, Ball G, Lazdunski A, Tommassen J. Protein secretion in Pseudomonas aeruginosa: characterization of seven xcp genes and processing of secretory apparatus components by prepilin peptidase. Mol Microbiol. 1992;6:1121–1131. doi: 10.1111/j.1365-2958.1992.tb01550.x. [DOI] [PubMed] [Google Scholar]
- 4.Binet R, Letoffe S, Ghigo J M, Delepelaire P, Wandersman C. Protein secretion by Gram-negative bacterial ABC exporters—a review. Gene. 1997;192:7–11. doi: 10.1016/s0378-1119(96)00829-3. [DOI] [PubMed] [Google Scholar]
- 5.Bitter W, Koster M, Latijnhouwers M, de Cock H, Tommassen J. Formation of oligomeric rings by XcpQ and PilQ, which are involved in protein transport across the outer membrane of Pseudomonas aeruginosa. Mol Microbiol. 1998;27:209–219. doi: 10.1046/j.1365-2958.1998.00677.x. [DOI] [PubMed] [Google Scholar]
- 6.Bleves S, Lazdunski A, Filloux A. Membrane topology of three Xcp proteins involved in exoprotein transport by Pseudomonas aeruginosa. J Bacteriol. 1996;178:4297–4300. doi: 10.1128/jb.178.14.4297-4300.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casadaban M C, Cohen S N. Analysis of gene control signal by DNA fusion in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 8.Christie P J, Ward J J, Jr, Gordon M P, Nester E W. A gene required for transfer of T-DNA to plants encodes an ATPase with autophosphorylating activity. Proc Natl Acad Sci USA. 1989;86:9677–9681. doi: 10.1073/pnas.86.24.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collazo C M, Galan J E. The invasion-associated type-III protein secretion system in Salmonella—a review. Gene. 1997;192:51–59. doi: 10.1016/s0378-1119(96)00825-6. [DOI] [PubMed] [Google Scholar]
- 10.Daefler S, Guilvout I, Hardie K R, Pugsley A P, Russel M. The C-terminal domain of the secretin PulD contains the binding site for its cognate chaperone, PulS, and confers PulS dependence on pIVf1 function. Mol Microbiol. 1997;24:465–475. doi: 10.1046/j.1365-2958.1997.3531727.x. [DOI] [PubMed] [Google Scholar]
- 11.Dallas W S. Conformity between heat-labile toxin genes from human and porcine enterotoxigenic Escherichia coli. Infect Immun. 1983;140:647–652. doi: 10.1128/iai.40.2.647-652.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.d’Enfert C, Ryter A, Pugsley A P. Cloning and expression in Escherichia coli of the Klebsiella pneumoniae genes for production, surface localization and secretion of the lipoprotein pullulanase. EMBO J. 1987;6:3531–3538. doi: 10.1002/j.1460-2075.1987.tb02679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dums F, Dow J M, Daniels M J. Structural characterization of protein export genes of the bacterial phytopathogen Xanthomonas campestris pathovar campestris: relatedness to export systems of other Gram-negative bacteria. Mol Gen Genet. 1991;229:357–364. doi: 10.1007/BF00267456. [DOI] [PubMed] [Google Scholar]
- 14.Francetic O, Pugsley A P. The cryptic general secretory pathway (gsp) operon of Escherichia coli K-12 encodes functional proteins. J Bacteriol. 1996;178:3544–3549. doi: 10.1128/jb.178.12.3544-3549.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fürste J P, Pansegrau W, Frank R, Blocker H, Scholz P, Bagdasarian M, Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 16.Hardie K R, Lory S, Pugsley A P. Insertion of an outer membrane protein in Escherichia coli requires a chaperone-like protein. EMBO J. 1996;15:978–988. [PMC free article] [PubMed] [Google Scholar]
- 17.Hardie K R, Seydel A, Guilvout I, Pugsley A P. The secretin-specific, chaperone-like protein of the general secretory pathway: separation of proteolytic protection and piloting functions. Mol Microbiol. 1996b;22:967–976. doi: 10.1046/j.1365-2958.1996.01539.x. [DOI] [PubMed] [Google Scholar]
- 18.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 19.Häse C C, Bauer M E, Finkelstein R A. Genetic characterization of mannose-sensitive hemagglutinin (MSHA)-negative mutants of Vibrio cholerae derived by Tn5 mutagenesis. Gene. 1994;150:17–25. doi: 10.1016/0378-1119(94)90852-4. [DOI] [PubMed] [Google Scholar]
- 20.Hirst T R, Sanchez J, Kaper J B, Hardy S J S, Holmgren J. Mechanism of toxin secretion by Vibrio cholerae investigated in strains harboring plasmids that encode heat-labile enterotoxins of Escherichia coli. Proc Natl Acad Sci USA. 1984;81:7752–7756. doi: 10.1073/pnas.81.24.7752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howard S P, Critch J, Bedi A. Isolation and analysis of eight exe genes and their involvement in extracellular protein secretion and outer membrane assembly in Aeromonas hydrophila. J Bacteriol. 1993;175:6695–6703. doi: 10.1128/jb.175.20.6695-6703.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iredell J R, Manning P A. The toxin-co-regulated pilus of Vibrio cholerae O1: a model for type 4 pilus biogenesis? Trends Microbiol. 1994;2:187–192. doi: 10.1016/0966-842x(94)90109-i. [DOI] [PubMed] [Google Scholar]
- 23.Jiang B, Howard S P. The Aeromonas hydrophila exeE gene, required both for protein secretion and normal outer membrane biogenesis, is a member of a general secretion pathway. Mol Microbiol. 1992;6:1351–1361. doi: 10.1111/j.1365-2958.1992.tb00856.x. [DOI] [PubMed] [Google Scholar]
- 24.Kagami Y, Ratliff M, Surber M, Martinez A, Nunn D N. Type II protein secretion by Pseudomonas aeruginosa: genetic suppression of a conditional mutation in the pilin-like component XcpT by the cytoplasmic component XcpR. Mol Microbiol. 1998;27:221–233. doi: 10.1046/j.1365-2958.1998.00679.x. [DOI] [PubMed] [Google Scholar]
- 25.Kaufman M R, Shaw C E, Jones I D, Taylor R K. Biogenesis and regulation of the Vibrio cholerae toxin-coregulated pilus: analogies to other virulence factor secretory systems. Gene. 1993;126:43–49. doi: 10.1016/0378-1119(93)90588-t. [DOI] [PubMed] [Google Scholar]
- 26.Kazmierczak B I, Mielke D L, Russel M, Model P. pIV, a filamentous phage protein that mediates phage export across the bacterial cell envelope, forms a multimer. J Mol Biol. 1994;238:187–198. doi: 10.1006/jmbi.1994.1280. [DOI] [PubMed] [Google Scholar]
- 27.Lauer P, Albertson N H, Koomey M. Conservation of genes encoding components of a type IV pilus assembly/two-step protein export pathway in Neisseria gonorrhoeae. Mol Microbiol. 1993;8:357–368. doi: 10.1111/j.1365-2958.1993.tb01579.x. [DOI] [PubMed] [Google Scholar]
- 28.Lindeberg M, Collmer A. Analysis of eight out genes in a cluster required for pectic enzyme secretion by Erwinia chrysanthemi: sequence comparison with secretion genes from other gram-negative bacteria. J Bacteriol. 1992;174:7385–7397. doi: 10.1128/jb.174.22.7385-7397.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linderoth N A, Model P, Russel M. Essential role of a sodium dodecyl sulfate-resistant protein IV multimer in assembly-export of filamentous phage. J Bacteriol. 1996;178:1962–1970. doi: 10.1128/jb.178.7.1962-1970.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu H M, Motley S T, Lory S. Interactions of the components of the general secretion pathway: role of Pseudomonas aeruginosa type IV pilin subunits in complex formation and extracellular protein secretion. Mol Microbiol. 1997;25:247–259. doi: 10.1046/j.1365-2958.1997.4561818.x. [DOI] [PubMed] [Google Scholar]
- 31.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 32.Marsh J W, Taylor R K. Identification of the Vibrio cholerae type 4 prepilin peptidase required for cholera toxin secretion and pilus formation. Mol Microbiol. 1998;6:1481–1492. doi: 10.1046/j.1365-2958.1998.01031.x. [DOI] [PubMed] [Google Scholar]
- 33.Michel G, Bleves S, Ball G, Lazdunski A, Filloux A. Mutual stabilization of the XcpZ and XcpY components of the secretory apparatus in Pseudomonas aeruginosa. Microbiology. 1998;144:3379–3386. doi: 10.1099/00221287-144-12-3379. [DOI] [PubMed] [Google Scholar]
- 34.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 35.Nunn D, Bergman S, Lory S. Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. J Bacteriol. 1990;172:2911–2919. doi: 10.1128/jb.172.6.2911-2919.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nunn D N, Lory S. Cleavage, methylation, and localization of the Pseudomonas aeruginosa export proteins XcpT, -U, -V, and -W. J Bacteriol. 1993;175:4375–4382. doi: 10.1128/jb.175.14.4375-4382.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Overbye L J, Sandkvist M, Bagdasarian M. Genes required for extracellular secretion of toxin are clustered in Vibrio cholerae. Gene. 1993;132:101–106. doi: 10.1016/0378-1119(93)90520-d. [DOI] [PubMed] [Google Scholar]
- 38.Pugsley A P. Processing and methylation of PulG, a pilin-like component of the general secretory pathway of Klebsiella oxytoca. Mol Microbiol. 1993;9:295–308. doi: 10.1111/j.1365-2958.1993.tb01691.x. [DOI] [PubMed] [Google Scholar]
- 39.Pugsley A P, Possot O. The general secretory pathway of Klebsiella oxytoca: no evidence for relocalization or assembly of pilin-like PulG protein into a multiprotein complex. Mol Microbiol. 1993;10:665–674. doi: 10.1111/j.1365-2958.1993.tb00938.x. [DOI] [PubMed] [Google Scholar]
- 40.Pugsley A P, Francetic O, Possot O M, Sauvonnet N, Hardie K R. Recent progress and future directions in studies of the main terminal branch of the general secretory pathway in Gram-negative bacteria—a review. Gene. 1997;192:13–19. doi: 10.1016/s0378-1119(96)00803-7. [DOI] [PubMed] [Google Scholar]
- 41.Reeves P J, Whitcombe D, Wharam S, Gibson M, Allison G, Bunce N, Barallon R, Douglas P, Mulholland V, Stevens S, Walker D, Salmond G P C. Molecular cloning and characterization of 13 out genes from Erwinia carotovora subspecies carotovora: genes encoding members of a general secretion pathway (GSP) widespread in Gram-negative bacteria. Mol Microbiol. 1993;8:443–456. doi: 10.1111/j.1365-2958.1993.tb01589.x. [DOI] [PubMed] [Google Scholar]
- 42.Reeves P J, Douglas P, Salmond G P C. Beta-lactamase topology probe analysis of the OutO NMePhe peptidase, and six other Out protein components of the Erwinia carotovora general secretion pathway apparatus. Mol Microbiol. 1994;12:445–457. doi: 10.1111/j.1365-2958.1994.tb01033.x. [DOI] [PubMed] [Google Scholar]
- 43.Russel M. Macromolecular assembly and secretion across the bacterial cell envelope: type II protein secretion systems. Review J Mol Biol. 1998;279:485–499. doi: 10.1006/jmbi.1998.1791. [DOI] [PubMed] [Google Scholar]
- 44.Sandkvist M, Morales V, Bagdasarian M. A protein required for secretion of cholera toxin through the outer membrane of Vibrio cholerae. Gene. 1993;123:81–86. doi: 10.1016/0378-1119(93)90543-c. [DOI] [PubMed] [Google Scholar]
- 45.Sandkvist M, Bagdasarian M, Howard S P, DiRita V J. Interaction between the autokinase EpsE and EpsL in the cytoplasmic membrane is required for extracellular secretion in Vibrio cholerae. EMBO J. 1995;14:1664–1673. doi: 10.1002/j.1460-2075.1995.tb07155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sandkvist M, Overbye-Michel L, Hough L P, Morales V M, Bagdasarian M, Koomey M, DiRita V J, Bagdasarian M. General secretion pathway (eps) genes required for toxin secretion and outer membrane biogenesis in Vibrio cholerae. J Bacteriol. 1997;179:6994–7003. doi: 10.1128/jb.179.22.6994-7003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Svennerholm A-M, Holmgren J. Identification of Escherichia coli heat-labile enterotoxin by means of a ganglioside immunosorbent assay (GM1-ELISA) procedure. Curr Microbiol. 1978;1:19–23. [Google Scholar]
- 48.Thomas J D, Reeves P J, Salmond G P C. The general secretion pathway of Erwinia carotovora subsp. carotovora: analysis of the membrane topology of OutC and OutF. Microbiology. 1997;143:713–720. doi: 10.1099/00221287-143-3-713. [DOI] [PubMed] [Google Scholar]
- 49.Turner L R, Olson J W, Lory S. The XcpR protein of Pseudomonas aeruginosa dimerizes via its N-terminus. Mol Microbiol. 1997;26:877–887. doi: 10.1046/j.1365-2958.1997.6201986.x. [DOI] [PubMed] [Google Scholar]
- 50.von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]