Abstract
Genes encoding ribosomal proteins and other components of the translational apparatus are coregulated to efficiently adjust the protein synthetic capacity of the cell. Ssb, a Saccharomyces cerevisiae Hsp70 cytosolic molecular chaperone, is associated with the ribosome-nascent chain complex. To determine whether this chaperone is coregulated with ribosomal proteins, we studied the mRNA regulation of SSB under several environmental conditions. Ssb and the ribosomal protein rpL5 mRNAs were up-regulated upon carbon upshift and down-regulated upon amino acid limitation, unlike the mRNA of another cytosolic Hsp70, Ssa. Ribosomal protein and Ssb mRNAs, like many mRNAs, are down-regulated upon a rapid temperature upshift. The mRNA reduction of several ribosomal protein genes and Ssb was delayed by the presence of an allele, EXA3-1, of the gene encoding the heat shock factor (HSF). However, upon a heat shock the EXA3-1 mutation did not significantly alter the reduction in the mRNA levels of two genes encoding proteins unrelated to the translational apparatus. Analysis of gene fusions indicated that the transcribed region, but not the promoter of SSB, is sufficient for this HSF-dependent regulation. Our studies suggest that Ssb is regulated like a core component of the ribosome and that HSF is required for proper regulation of SSB and ribosomal mRNA after a temperature upshift.
Molecular chaperones of the heat shock protein 70-kDa (Hsp70) class are highly conserved proteins that bind unfolded polypeptides, preventing nonproductive interactions that can lead to misfolding or protein aggregation. Hsp70s are composed of three domains: a conserved 44-kDa ATPase segment, an 18-kDa domain which is the binding site for unfolded polypeptides, and a 10-kDa variable region at the C terminus. A variety of cellular processes such as protein synthesis, protein folding, and polypeptide translocation across organellar membranes are assisted by these molecular chaperones (5).
The budding yeast Saccharomyces cerevisiae contains two major classes of cytosolic Hsp70 chaperones, Ssa and Ssb (3). This report focuses on the ribosome-associated chaperone, Ssb. The SSB Hsp70 family is composed of two genes, SSB1 and SSB2. In contrast to SSA genes, SSB genes are not heat inducible; in fact, their expression is reduced after a heat shock. The SSB-encoded proteins have greater than 99% identity. In this paper, both SSB genes will be collectively referred to as SSB unless specified otherwise. Strains containing gene disruptions for both SSB genes are hypersensitive to certain translation inhibitors and are cold sensitive (24). These two phenotypes are completely suppressed by one copy of either of the SSB genes but not by a constitutive overexpression of the heat-inducible SSA genes (4). Ssb associates with translating ribosomes and can be cross-linked to the nascent polypeptide chain (24, 28). Its association with translating ribosomes is resistant to treatment with high concentrations of salt, implying that Ssb associates with the ribosome like a core component of this apparatus.
Genes encoding many components of the translational machinery have a coordinated regulation in response to environmental changes even though they are dispersed throughout the genome (9). Cells increase or decrease their ribosomal protein (RP) mRNA pools based on growth conditions to accommodate their needs for protein synthetic capacity. For example, upon a carbon upshift (i.e., when glucose is added to a culture growing on a poor carbon source such as ethanol or glycerol) the mRNA levels for RP genes increase. Upon amino acid limitation, cells elicit a response known as stringent control which induces transcription of amino acid biosynthetic genes and reduces the mRNA levels of RP genes (37). A promoter sequence found in some RP genes, known as the RPG box, is required to regulate transcription of RP genes upon a carbon upshift and amino acid limitation. This promoter sequence is the binding site for the transcriptional regulator Rap1 (6, 13, 22, 25).
While expression of RP genes, SSB, and many other genes is decreased, a set of genes called heat shock genes is induced upon temperature upshift (21). It has been reported that both transcription and mRNA stability of RP genes are reduced after a heat shock (12, 16), but sequences required for this regulation have not been identified. In contrast, the induction of heat shock genes has been well characterized. The transcriptional activator of many heat-inducible genes, the heat shock factor (HSF), is a homotrimer that binds sequences known as heat shock elements (HSEs) in the promoters of heat-inducible genes (23). Upon an increase in temperature, HSF is activated, resulting in augmentation of transcription from HSE-containing promoters. The yeast HSF monomer is composed of four domains: an N-terminal activation domain, a DNA binding domain, an oligomerization domain, and a C-terminal activation domain. One HSF allele, EXA3-1, has a single base substitution in the DNA binding domain that changes the proline at position 214 to glutamine (10). This mutation reduces the ability of HSF to bind to the HSE, resulting in a delay in the induction of heat shock genes upon a temperature upshift.
Unlike heat-inducible proteins, RPs are coordinately expressed to allow the cell to vary ribosome abundance depending on its needs for protein synthesis. Ssb, while a member of a heat shock family of proteins, is a component of translating ribosomes. To better understand Ssb’s regulation, we analyzed SSB mRNA levels under three different growth conditions (carbon upshift, amino acid starvation, and heat shock) known to affect RP gene expression. We report that SSB and RP genes are regulated in a coordinated manner. In addition, to elucidate components of the heat shock regulation of SSB and RP mRNA, we analyzed mRNA levels in cells containing a defective heat shock response due to the presence of the EXA3-1 mutation. Our results indicate that the negative regulation of RP and SSB mRNAs after a heat shock is HSF dependent.
MATERIALS AND METHODS
S. cerevisiae strains.
With the exception of F113 (21), yeast strains used in this study have the following genotype: ura3-52 lys1 lys2 trp1-Δ1 his3-11,15 leu2-3,112. The F113 genotype is MATa can1 ino1-13 ura3-52 (22). The wild-type strains were DS10 (MATa) and JH27A (MATα). The following strains carry the additional alleles in parentheses: NL113 (ssb1:LEU2 and ssb2:HIS3), MH297 (EXA3-1) (10), and NL95 (ssb1:LEU2 ssb2:HIS3 EXA3-1 URA3). The EXA3-1 allele was introduced into cells containing ssb1 ssb2 disrupted by mating an ssb1:LEU2 ssb2:HIS3 strain (JN208) (24) with an EXA3-1 strain (MH297) in which the EXA3-1 allele was genetically marked by the URA3 gene. The resulting haploids (NL95 and NL113) were confirmed by marker segregation and by using Northern blot analysis to measure the transcript levels of an internal control RPL11 (previously RPL16A) in a wild-type strain and the EXA3-1 strain after a heat shock.
Bacterial strains, transformations, plasmids, and gene fusion constructions.
DH5α was the preferred Escherichia coli strain for general cloning procedures [genotype: φ80dlacZΔM15 endA1 recA1 hsdR17 (rK− mK+) supE44 thi-1 λ− gyrA relA1 F− ΔlacZYA-argF]. E. coli cells were transformed by CaCl2 procedures (20), and yeast strains were transformed by the lithium acetate procedure described elsewhere (8). Plasmids used in this study include pRS313-U2 (generously provided by Warren Heideman [27]), YEpCUP1-HSE-M-lacZ (a gift from Dennis Thiele [35]), pSSB-URA3, pEC302, pJHSSB1P, and pCUP-SSB1. To construct pSSB-URA3, the 5′ untranslated region and promoter of the SSB1 gene were isolated from the plasmid pEC302 as a 617-bp EcoRI-XbaI fragment. This fragment includes DNA sequences from the polylinker which encode the restriction sites for XbaI and SalI. The fragment was cloned directionally into pUC18 digested with EcoRI and XbaI to create pJHSSB1P. The 962-bp PstI-HindIII fragment from YEp24 containing the URA3 coding region and 19 nucleotides from the 5′-end untranslated region was inserted into pJHSSB1P digested with PstI and HindIII to create pJHSSB1P-URA3. This construct contained the SSB1 promoter fused to the coding region of the URA3 gene. The SSB-URA fusion gene was cloned into the yeast centromeric pRS314 by removing the insert from pJHSSB1P-URA3 by HindIII and EcoRI digestion. This fragment was inserted directionally into pRS314 digested with EcoRI and BamHI (filled in) to create pSSB-URA3.
To construct pCUP1-SSB, a modified version of the CUP1 promoter designated CUP1 hse-m and the first half of the SSB1 gene were amplified by two separate PCRs. The CUP1 hse-m promoter was amplified from plasmid YEpCUP1-HSE-M-lacZ with two oligonucleotides: CupSSBa (TTT TCT CGA GCG AGA TGA AAT GAA TAG C) and CupSSBb (CTG TAA TGA TCC TAT ATG ATA TTG CAC TAA C). The amplified first half of the SSB1 gene started from the first guanine nucleotide in the transcript and ended at the BglII site in the middle of the gene. This SSB1 fragment was amplified with the following oligonucleotides: CupSSBc (GCA ATA TCA TAT AGG ATC ATT ACA GTA TTT TAA TTG) and CupSSBd (CTT CGT CGA TTT GAG AC). The two PCR products were fused to each other by PCR-mediated overlapping extension (14), and the resulting 2.6-kb PCR fragment placed the first guanine nucleotide of the SSB1 transcript immediately after the first start of transcription found in the CUP1 hse-m promoter. The 2.6-kb PCR fragment has two restriction sites at the ends (XhoI and BglII) that were used to digest and subclone it into a 6.5-kb XhoI-BglII-opened pRS314-SSB1 vector. This 6.5-kb XhoI-BglII pRS314-SSB1 fragment provided the other half of the SSB1 gene to the Cup1-SSB1 fusion after the BglII site. This plasmid was used to transform NL113 (ssb1 ssb2) and NL95 (ssb1 ssb2 EXA3-1) strains. This gene fusion was functional, as it completely suppressed the ssb1 ssb2 mutant phenotypes of cold sensitivity and hypersensitivity to translation inhibitors. In addition, transcript levels from this gene fusion increased upon addition of copper (data not shown). The analysis of this gene fusion did not require addition of copper to the medium due to appropriate basal expression under our growth conditions.
Chemicals.
Yeast extract, peptone, and yeast nitrogen base without amino acids were from Difco Laboratories (Detroit, Mich.); dextrose and 3-amino-1,2,4-triazole (3AT) were from Sigma Chemical Co. (St. Louis, Mo.); SeaKem agarose was from FMC Corp. (Rockland, Maine).
Growth conditions.
The carbon upshift was performed by growing cells in glycerol-based medium (YPG [1% yeast extract, 2% peptone, 5% glycerol]) to exponential growth (A600 between 0.4 and 0.8) and then adding glucose to the culture to a final concentration of 2%. For amino acid starvation, 10 mM 3AT was added to exponentially growing cells cultured in SD minimal medium (29). The heat shock response was elicited by growing cells on YPD (1% yeast extract, 2% peptone, 2% dextrose) rich medium at 23°C and then shifting the medium to 39°C. The temperature upshift for stable messages (i.e., 50% reduction of basal levels takes longer than 5 min) was done by moving a 25-ml aliquot of the cell suspension to a 39°C prewarmed 250-ml flask. For short-half-life mRNAs such as that for URA3, the temperature upshift was done by mixing approximately 10 ml of prewarmed YPD medium at 68°C into a 15-ml culture growing at 23°C and placing it in a 39°C water bath. In each condition, aliquots of the cell suspension were taken at times indicated and total RNA was prepared from pelleted cells by the heat-freeze method (31).
Northern (RNA) blot analysis and primer extension analysis.
Northern blot analysis was done by separating 4 to 10 μg of total RNA in a 1% agarose-formaldehyde gel, transferring the gel to a nylon membrane, and hybridizing it with radiolabeled probes (specific activity ≈ 107 cpm/μg) made by random priming (2). Probes were generated with [α-32P]dCTP (3,000 Ci/mmol) (DuPont NEN). After stringent washes, filters were exposed to a detection screen with a PhosphorImager, and the signal was quantified with the ImageQuant software package (Molecular Dynamics). Differences in RNA loading on Northern blots were normalized with indicated loading controls on each experiment. rRNA was used to normalize loaded amounts of RNA in heat shock experiments since even the levels of relatively stable transcripts decrease significantly during the time course of the experiment, making quantitative analysis difficult. The validity of this method has been confirmed elsewhere (10).
For primer extension analysis, a 20-mer oligonucleotide (5′-GAT AGC ACC TTG GAA AAC AC-3′) complementary to the 5′ ends of the SSB1 and SSB2 coding regions was used to prime cDNA synthesis. To normalize amounts of RNA used per reaction, a primer with sequence complementary to the small nuclear RNA (snRNA) U4 (CGG ACG AAT CCT CAC TGA TAT GC) was included on each reaction. Gel-purified oligonucleotides were radiolabeled at the 5′ end with T4 polynucleotide kinase and [γ-32P]ATP (3,000 Ci/mmol) (DuPont NEN) (2). Radiolabeled primers were hybridized to 20 to 30 μg of total RNA at 90°C for 3 min and then quickly chilled on ice. Annealed primers were extended with avian myeloblastosis virus reverse transcriptase (Promega, Madison, Wis.), and primer extension products were separated with 6% polyacrylamide gels. Signals were visualized with a PhosphorImager and quantified with the ImageQuant software package.
RP nomenclature.
RPs of S. cerevisiae have been named in this work according to the new nomenclature described in the work of Mager et al. (18).
RESULTS
SSB mRNA levels increase upon a carbon upshift.
Most RP genes possess a transcriptional regulatory sequence designated an RPG box (CCC ATA CAT CT) (39, 40) and a T-rich region important for constitutive expression (30). Analysis of the SSB promoter sequence indicates that both genes possess a T-rich region; the SSB1 promoter also has a putative RPG box centered at 261 bases upstream of the adenine of the start codon with the sequence CCC ATA CAC CG. These common structural features and the fact that both are components of ribosomes suggested to us that SSB and RP gene regulation may be coordinated.
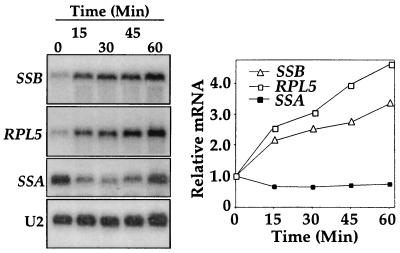
It has been observed that mRNA levels of RPs rapidly increase by more than twofold upon a carbon upshift (13, 15). We analyzed SSB mRNA levels by Northern blot analysis after supplying glucose to wild-type cells growing in glycerol-based medium. As shown in Fig. 1, SSB mRNA levels increased by more than twofold within 30 min of carbon upshift, similar to the mRNA increase of the RP gene RPL5 (previously called L1a). In contrast, the mRNA levels of genes encoding another cytosolic chaperone, Ssa, did not increase but rather transiently decreased upon glucose addition.
FIG. 1.
mRNA levels of SSB genes upon a carbon upshift. At time zero, glucose was added to wild-type cells growing in glycerol-based medium. At left, signals from Northern blotting of several transcripts upon a carbon upshift are shown. The graph at right shows quantification of the signals after normalization to snRNA U2 levels.
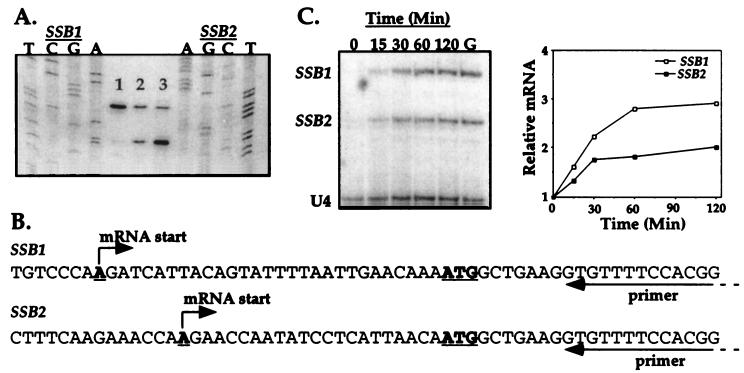
The SSB chaperone family is composed of two genes, SSB1 and SSB2, which are more than 90% identical. To determine whether both SSB transcripts increase upon a carbon upshift, we analyzed the mRNA levels of both genes in wild-type cells. Primer extension analysis was used to distinguish between the two transcripts. We designed an oligonucleotide complementary to the 5′ end of the SSB coding sequence. Due to the complete sequence identity of SSB1 and SSB2 in this region, both SSB transcripts are able to efficiently serve as templates during a primer extension reaction. As shown in Fig. 2A, the analysis of the SSB mRNA species in a wild-type strain resulted in two predominant bands (lane 2) representing products extending 29 and 22 bases beyond the initiation codon ATG. It seemed likely that each band represented the transcript from a single SSB gene, but it was unclear which band was the extension product of which mRNA. We reasoned that increasing the gene dosage of either SSB1 or SSB2 would result in higher amounts of the respective mRNA than that expressed from a single chromosomal copy. Consequently, we overexpressed SSB1 or SSB2 by supplying wild-type cells with a high-copy-number plasmid containing either SSB1 or SSB2. Overexpression of SSB1 resulted in enhanced levels of the upper band, while overexpression of SSB2 caused an increase in the signal of the lower band (Fig. 2A, lanes 1 and 3, respectively). Interestingly, overexpression of the SSB1 gene appears to decrease SSB2 mRNA levels, although the reverse does not appear to be true. We conclude that the upper band is an extension of the SSB1 gene and that the lower band is an extension of the SSB2 gene. This experiment localized the 5′ end of the SSB1 and SSB2 genes to 29 and 22 nucleotides, respectively, upstream from the A of the initiating ATG codon (Fig. 2B).
FIG. 2.
Localization of the 5′ ends of the SSB1 and SSB2 genes and analysis of their mRNA levels upon a carbon upshift. (A) Primer extension reactions are shown in the center: lane 1, strain overexpressing the SSB1 gene; lane 2, wild-type cells; lane 3, strain overexpressing the SSB2 gene. Sequencing reactions are shown on the left for the SSB1 gene and on the right for the SSB2 gene. (B) Sequences from the SSB1 and SSB2 genes showing the start site of transcription relative to the initiation ATG codon. Sequences complementary to the primer are located above the arrow labeled as primer. (C) mRNA levels of SSB1 and SSB2 genes after a carbon upshift. Samples of the culture were collected at the indicated times after glucose addition. At left is shown a sequencing gel which separates the primer extension products. Lane G shows results from cells grown on glucose-based medium. The graph shows the quantification of the signal obtained after normalization to snRNA U4.
The individual increase in mRNA levels of SSB1 and SSB2 after a carbon upshift was studied by primer extension analysis with the same oligonucleotide that was used to localize the start site of transcription of these RNAs. We also included in the reactions a primer that complements sequences of the U4 snRNA to normalize amounts of RNA used in reactions. Both SSB1 and SSB2 mRNA species increased upon a carbon upshift, although SSB1 mRNA increased to a greater extent than did SSB2 mRNA (Fig. 2C).
The stringent control response results in a coordinated reduction in the mRNA levels of SSB and RP genes.
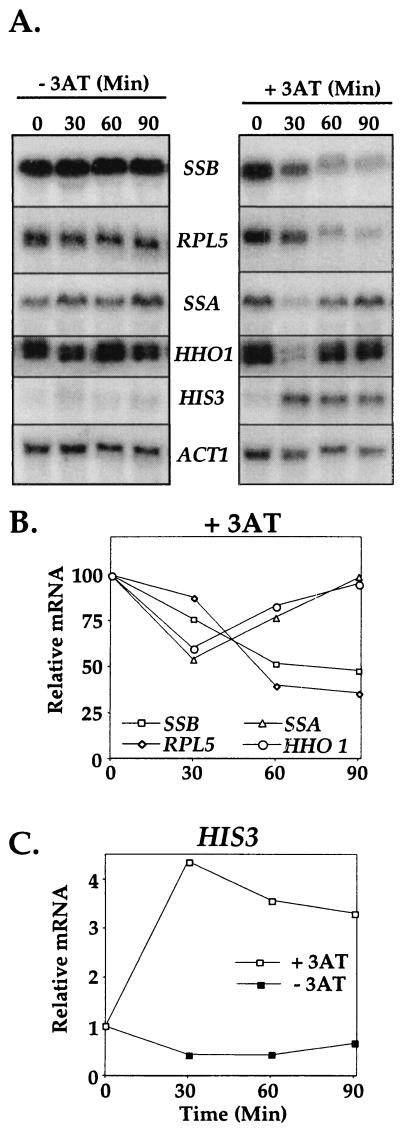
Amino acid starvation elicits a cellular response known as stringent control resulting in the transcriptional activation of amino acid biosynthetic genes and reduction in the expression of RP genes (37). We hypothesized that if Ssb is a component of the ribosome it would have a pattern of regulation similar to that of RP genes not only under carbon upshift but under other nutritional conditions as well. We expanded the analysis of SSB mRNA regulation by studying its mRNA levels after amino acid starvation. The stringent response was induced by adding 3AT, a competitive inhibitor of an enzyme required for histidine biosynthesis (22), to cells growing in minimal medium. Total RNA was prepared from aliquots of the culture taken at various times after 3AT addition. Northern blot analysis was used to quantify mRNA levels of SSB and the RP gene RPL5. As negative controls, we analyzed mRNAs of two genes whose products do not function in translation: SSA, which encodes another cytosolic Hsp70 chaperone, and HHO1, the histone H1 gene. RPL5 and SSB mRNA levels gradually decreased, dropping below 50% of the control levels by 90 min after addition of 3AT. We observed a rapid and transient decrease of SSA and histone H1 mRNAs 30 min after 3AT addition with a return to basal levels by 90 min (Fig. 3). To examine the effectiveness of the 3AT treatment, we measured the mRNA levels of HIS3, a histidine biosynthetic gene. As we expected, after amino acid limitation the mRNA levels of HIS3 increased up to fourfold. Thus, we conclude that SSB is regulated in coordination with RP genes during amino acid starvation, while the gene encoding the other cytosolic Hsp70, Ssa, is not.
FIG. 3.
mRNA levels of various transcripts after amino acid limitation. 3AT was added to cells (strain F113) growing in SD minimal medium at time zero. RNA samples were analyzed by Northern blotting. (A) (Left) Control from cells that had no 3AT (−3AT) treatment. (Right) Signals obtained from cells treated with 3AT. Genes analyzed were SSB, RPL5 (RP), SSA (a yeast cytosolic Hsp70), and HHO1 (histone H1). HIS3 was used as a control to detect the efficiency of the treatment. (B and C) Quantification of the mRNA levels in the presence of 3AT (+3AT) and of HIS3 mRNA with (+3AT) and without (−3AT) 3AT, as shown in panel A, after normalization to actin transcript levels.
EXA3-1, an allele of HSF1 which encodes the yeast HSF, causes a delay in mRNA reduction of SSB and RP genes upon a heat shock.
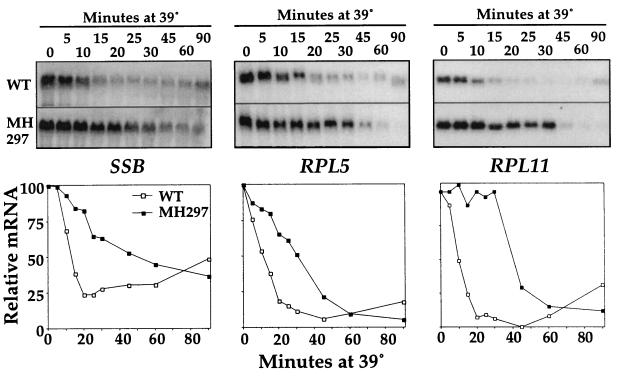
As shown previously, the mRNA levels of RP genes and SSB decrease drastically upon a heat shock in a wild-type strain (16, 38). However, the cause of the negative regulation is not known. Since induction of many heat shock genes is dependent on the transcriptional activator HSF, we asked whether HSF activity has an effect on the negative regulation of SSB and RP genes. Therefore, we analyzed the heat shock regulation of several mRNAs in a mutant strain (MH297) containing the EXA3-1 HSF allele. Cells were grown in rich medium to mid-log phase at 23°C and then shifted to 39°C. Aliquots of the culture were taken at the indicated times, and mRNA levels were quantified by Northern blot analysis. The SSB mRNA reduction was much less rapid in MH297 than in a wild-type strain (Fig. 4). For example, after 20 min of a heat shock the SSB mRNA levels in wild type were reduced by greater than 75% while the levels in the MH297 strain were reduced by less than 20%. A similar delay in the reduction of mRNA levels was seen for the RP mRNAs of RPL5, RPL11, RPS14 (previously known as CRY1 and CRY2), and RPS17 (formerly known as RP51) (Fig. 4 and data not shown).
FIG. 4.
mRNA levels of ribosomal components upon a heat shock in the wild-type (WT) strain and in cells containing the EXA3-1 mutation. JH27A (wild-type) and MH297 (EXA3-1) cells growing in YPD medium at 23°C were rapidly shifted to 39°C. Aliquots of the culture were collected at the times indicated after the temperature upshift, and extracted RNA was subjected to Northern blot analysis. (Top) Northern blot images. (Bottom) Quantification after normalization to rRNA (see Materials and Methods).
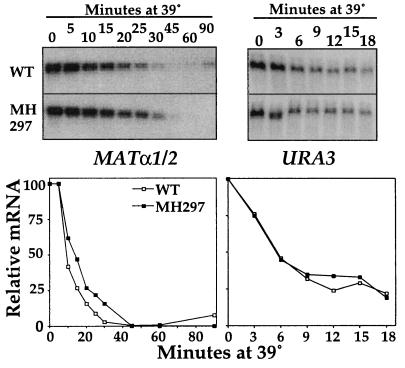
It is known that most mRNA species are reduced in abundance upon a heat shock. To determine whether EXA3-1 has a global effect on the negative regulation of genes after a heat shock, we tested the effect of the mutation on the mRNA levels of two genes whose products are unrelated to the protein synthesis process. URA3 encodes a biosynthetic enzyme required for uracil synthesis; MATα encodes the mating pheromone α-factor. Little or no difference was observed in the rate of decrease of either of these two mRNAs upon a heat shock (Fig. 5), indicating that EXA3-1 does not have a global effect on the regulation of all mRNAs upon temperature upshift.
FIG. 5.
Levels of MATα1/2 and URA3 transcripts upon a heat shock in the wild-type (WT) (JH27A) strain and in cells containing the EXA3-1 mutation (MH297). Experiments were performed as described for Fig. 4. In the case of the short-half-life message URA3, samples were taken at 0 to 18 min after a temperature upshift (see Materials and Methods). (Top) Northern blot images. (bottom) Graphs showing the quantification of signal after normalization to rRNA.
The SSB promoter is not sufficient to decrease its mRNA levels in an HSF-dependent manner.
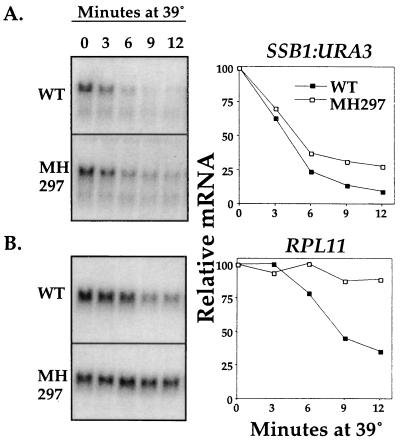
To aid in understanding the regulation of mRNA levels by HSF, we wanted to determine the sequences in the SSB gene essential for this regulation. To test the sufficiency of the SSB1 promoter, we utilized an SSB1:URA3 engineered fusion to analyze the SSB1 promoter contribution to the URA3 mRNA regulation in the wild type and the MH297 mutant strain. This gene fusion expressed a functional protein that allowed growth of cells lacking a functional genomic URA3 gene in medium lacking uracil (data not shown). Since the ura3-52 genomic allele in this strain background does not encode detectable transcript (data not shown), we were assured that URA3 mRNA detected in our experiments was expressed from the SSB1 promoter. URA3 was appropriate for this analysis because the decrease of its mRNA after heat shock is unaffected by EXA3-1 mutation (Fig. 5B).
Wild-type and MH297 strains were transformed with pSSB-URA. As shown in Fig. 6, the EXA3-1 mutation in MH297 did not have an obvious effect on the reduction of URA3 mRNA produced from the SSB1:URA3 gene fusion after a temperature upshift. In contrast, the internal control of the RP transcript encoded by the chromosomal copy of RPL11 showed a significant delay in its rapid reduction in the MH297 strain. These results indicate that the SSB promoter is not sufficient to regulate its mRNA after heat shock in an HSF-dependent manner.
FIG. 6.
mRNA levels of an SSB1:URA3 fusion in wild-type (WT) and mutant MH297 cells after a heat shock. The experiment was performed as described for URA3 in Materials and Methods. Northern blot images of each transcript are shown at left. (A) SSB1:URA3 fusion; (B) RPL11. Quantification of the signals detected in panels A and B is shown on the right. Signals were normalized to rRNA levels.
Sequences within the transcribed region of SSB are sufficient to regulate SSB transcript levels after a heat shock in an HSF-dependent manner.
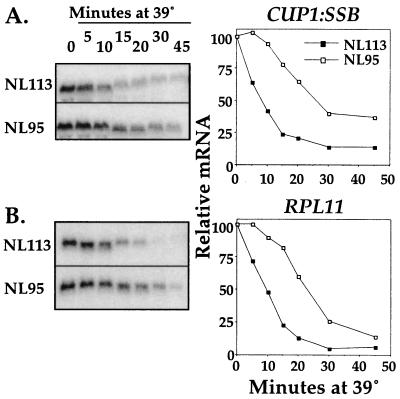
Since the promoter of SSB1 was not sufficient for the HSF-dependent SSB mRNA regulation upon a heat shock, we tested whether the transcribed region of the SSB1 gene possesses sequences sufficient for such regulation. We designed a gene fusion that contained the entire SSB1 transcribed region downstream of the CUP1 promoter (Fig. 7). Since the native CUP1 promoter is induced by a heat shock through the action of HSF, we used a CUP1 hse-m promoter which has no functional interaction with HSF (Cup1 hse-m) (35). Therefore, transcription from this promoter is not heat induced (data not shown). The fusion was created in such a way that the start of transcription of CUP1 hse-m is the first nucleotide in the SSB1 transcript. This CUP1:SSB1 fusion suppressed both phenotypes (cold sensitivity and hypersensitivity to translation inhibitors) of a mutant strain containing deletions of both SSB genes. In addition, primer extension analysis showed that the 5′ end of the CUP1:SSB1 transcript is similar in size to that of the native SSB1 transcript (data not shown).
FIG. 7.
Influence of the EXA3-1 mutation on the transcript levels of a CUP1:SSB1 fusion. The experiment was done as described in the legend to Fig. 5. At left, Northern blot images for CUP1:SSB1 fusion (A) and RPL11 (B) are shown. At right is shown quantification of the signals after a heat shock with rRNA as a loading control.
This CUP1:SSB1 fusion was used to transform two strains that have the following genotypes: ssb1 ssb2 EXA3-1 (NL95) and ssb1 ssb2 (NL113). Analysis of the CUP1:SSB1 fusion showed rapid reduction of its mRNA levels in NL113. This reduction was retarded in the presence of the EXA3-1 allele. The delay in the heat shock regulation of the CUP1:SSB1 transcript in NL95 is comparable to what was observed for the internal control, the genomic RP gene RPL11. After 20 min of the temperature upshift, the mRNA levels of both RPL11 and CUP1:SSB1 were about threefold higher in NL95 than in NL113. Therefore, the sequences included in the SSB1 transcribed region are sufficient to provide a heat shock mRNA regulation dependent on HSF.
DISCUSSION
It has been established that cells regulate components of the ribosome according to their growth rates and stress conditions that compromise protein synthesis (19, 39). Here we have demonstrated that mRNA levels of the Hsp70 molecular chaperone Ssb are coregulated with RP mRNA levels under three different growth conditions: carbon upshift, amino acid starvation, and heat shock. Other data indicate a relationship of Ssb with ribosomes. Ssb, which is present within the cell in a two- to three-times molar excess over ribosomes (28), is associated with the ribosome-nascent chain complex in a salt-resistant manner (28). In addition, strains lacking Ssb are more sensitive to certain translation inhibitors (24). Hence, the regulation reported here coupled with previous results supports the hypothesis that Ssb should be considered an important component of the translational apparatus.
The regulation of SSB and SSA, which encode two major classes of cytosolic Hsp70s, is strikingly different. These two Hsp70s have evolved with similar protein structures as evidenced by the fact that they are more than 60% identical in sequence, but their regulation is different, presumably because of their different functional niches within the cell. Ssa has been implicated in the refolding of proteins partially denatured upon exposure to increased temperatures and in regulation of the heat shock response. Therefore, it is not surprising that SSA expression is induced by a heat shock to cope with the protein damage generated at high temperatures. In contrast, expression of Ssb, as well as of RPs, is repressed after a temperature upshift. Apparently, it is advantageous for cells not to expend energy in translation under conditions in which proteins will be jeopardized by denaturation conditions and subjected to aggregation. Also, both SSB and RP gene mRNA levels are regulated according to the growth rates of the cell. For example, we observed that mRNA levels of SSB and RP genes decline upon starvation for amino acids when the level of protein synthesis is dropping. Moreover, SSB mRNA levels increase rapidly upon addition of a rich carbon source when the rates of protein synthesis in a cell are rising. Therefore, the synthesis of Ssb and structural RPs is carefully regulated by the cell, presumably to minimize the energy used in forming the translational apparatus, which comprises about 16 to 18% of cellular protein.
The mechanism of down-regulation of RP mRNA levels upon a heat shock is not well understood. Regulation at the level of transcription and mRNA degradation have been suggested (12, 16). We found the negative heat shock regulation of both SSB and four RP genes analyzed to be dependent on HSF. This conclusion is based on our observation that a strain containing the HSF allele EXA3-1 has a delay in the reduction of mRNA levels of SSB and RP genes upon a heat shock. Moreover, the EXA3-1 mutation did not affect the negative heat shock mRNA regulation of two proteins whose function is not related to the translation machinery, the mating α-factor gene MATα1/2 and the URA3 gene transcripts. The rate of reduction for these two mRNAs after a temperature upshift was similar in wild type and in cells containing the EXA3-1 mutation. These data suggest that two mechanisms are responsible for reducing mRNA species upon a heat shock: a global mechanism that is HSF independent and a specific mechanism for RP mRNAs that requires HSF.
Considering the results reported here, it is somewhat surprising that another HSF allele (hsf1-m3/mas3) was reported to have no effect on the mRNA regulation of RP genes upon a heat shock (7). This HSF allele was thought to abolish its heat activation, reducing drastically the induction of heat-inducible genes (34). Recently, it was found that a mutant strain containing the hsf1-m3 allele did not have an obvious decrease in the induction of several heat shock proteins (17, 36), including Hsp104. However, we have observed that the EXA3-1 mutation appears to retard the heat induction of Hsp104 as well as that of other Hsp proteins (10). Consistent with this result, the EXA3-1 mutation also delays the reduction of SSB and RP transcripts. It is possible that the hsf1-m3 mutation does not significantly affect the activity important in the up-regulation of Hsp104 as well as the down-regulation of genes encoding ribosomal components.
We can envision two general ways in which HSF may be acting in the regulation of SSB and RP genes: (i) HSF may act as a transcriptional repressor of SSB and RP genes or (ii) HSF is a transcriptional activator of a heat-inducible regulatory factor needed for the down-regulation of RP and SSB genes. We favor the second idea, that the EXA3-1 mutation causes a delay in the expression of a heat-inducible factor, for several reasons. If HSF is a repressor, it must be binding to cryptic sequences since neither SSB gene contains a canonical HSE. There is a precedent for the binding of a transcriptional activator or repressor to variant sequences in different modes of regulation (32). However, alignment of SSB and the RP genes studied here did not reveal a conserved sequence present in these genes which might act as a novel binding site for HSF acting as a repressor. In addition, our results show that the transcribed region of SSB is sufficient to regulate its mRNA levels after a heat shock in an HSF-dependent manner, while the promoter is not. This result does not preclude HSF acting directly as a repressor since binding of repressors to the coding region has been found previously (11, 33). However, such cases appear to be rare, particularly in yeast. Finally, previous experiments suggest that a factor needs to be synthesized de novo for the appropriate mRNA regulation of RP genes upon a heat shock (12, 26). Together, these observations suggest that HSF acts as a transcriptional activator of a heat-inducible factor, not as a transcriptional repressor of SSB and RP genes.
What might the function of such a heat-inducible factor be? Such a factor could be or could activate a heat-inducible transcriptional repressor or an RNase activity specific for these genes or mRNAs. Unfortunately, it is very difficult to determine directly whether the differences in SSB regulation in wild-type and HSF mutant cells are at the transcriptional or the posttranscriptional level due to unique problems in studying RNA regulation during the heat shock response. The methods currently available to measure RNA stability are inadequate under these conditions. The addition of drugs such as 1,10-phenanthroline and thiolutin, which inhibit the vast majority of mRNA synthesis, induces the synthesis of heat shock mRNAs (1). The temperature-sensitive mutation in the RNA polymerase subunit RPB1 commonly used (26) requires a “heat shock” to inhibit RNA synthesis, but the temperature inactivation of the RPB1 subunit is too slow to eliminate induction of a heat-inducible factor (data not shown). In the absence of additional experimental data, we favor the simplest idea, that HSF is required for induction of an RNase or of a factor required for activation of an RNase. This RNase or factor may recognize an RNA secondary structure common to both SSB and RP mRNAs which is not recognizable at the primary sequence level.
The experiments reported here emphasize an HSF-dependent mode of regulation of mRNA levels upon a temperature upshift. This mode of regulation does not affect all genes, as the rates of decrease of MATα1/2 and URA3 mRNAs were not significantly affected by the EXA3-1 mutation. There is certainly a second mechanism of regulation, as both these mRNAs, as well as many others, decrease in levels upon a heat shock. The regulatory mechanism unaffected by the EXA3-1 mutation which controls the expression of many genes, including SSB and RP genes, may be a general cessation of transcription of non-heat-inducible genes, as pulse-labeling experiments indicate that transcription of RP genes in S. cerevisiae is reduced after a heat shock (16). However, this regulatory pathway is overlaid by an HSF-dependent mechanism that down-regulates SSB and RP mRNAs, perhaps by hastening the degradation of existing mRNA molecules. This HSF-dependent mechanism is an extremely efficient way for the cell to couple the activation of heat-inducible genes with the negative regulation of the translational apparatus, by linking both modes of regulation to the same regulatory molecules. This coupling implies that the down-regulation of the translational apparatus is an important component of the regulatory pathways that the cell has evolved to cope with environmental stress.
ACKNOWLEDGMENTS
We thank Warren Heideman, Alan Hinnebusch, Dennis Thiele, and John Woolford for providing plasmids and strains and Bonnie K. Baxter, Philip James, and Christine Pfund for thoughtful comments on the manuscript.
This work was supported by NIH grant 5RO1 GM31107 (E.A.C.) and NIH Predoctoral Fellowship GM18507-02 (N.L.).
REFERENCES
- 1.Adams C, Gross D. The yeast heat shock response is induced by conversion of cells to spheroplasts and by potent transcriptional inhibitors. J Bacteriol. 1991;173:7429–7435. doi: 10.1128/jb.173.23.7429-7435.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F, Brent R, Kingston R, Moore D, Seidman J G, Smith J, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1997. [Google Scholar]
- 3.Boorstein W R, Ziegelhoffer T, Craig E A. Molecular evolution of the HSP70 multigene family. J Mol Evol. 1994;38:1–17. doi: 10.1007/BF00175490. [DOI] [PubMed] [Google Scholar]
- 4.Craig E A, Jacobsen K. Mutations in cognate gene of Saccharomyces cerevisiae HSP70 result in reduced growth rates at low temperatures. J Biol Chem. 1985;5:3517–3524. doi: 10.1128/mcb.5.12.3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craig E A, Ziegelhoffer T, Nelson J, Laloraya S, Halladay J. Complex multigene family of functionally distinct Hsp70s of yeast. Cold Spring Harbor Symp Quant Biol. 1995;XV:441–449. doi: 10.1101/sqb.1995.060.01.049. [DOI] [PubMed] [Google Scholar]
- 6.Donovan D M, Pearson N J. Transcriptional regulation of ribosomal proteins during a nutritional upshift in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:2429–2435. doi: 10.1128/mcb.6.7.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galego L, Barahona I, Alves A-P, Vreken P, Raué H A, Planta R J, Rodrigues-Pousada C. Known heat-shock proteins are not responsible for stress-induced rapid degradation of ribosomal protein mRNAs in yeast. Yeast. 1994;9:583–588. doi: 10.1002/yea.320090604. [DOI] [PubMed] [Google Scholar]
- 8.Gietz R D, Schiestl R H, Willems A R, Woods R A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 9.Gorenstein C, Warner J R. Coordinate regulation of the synthesis of eukaryotic ribosomal proteins. Proc Natl Acad Sci USA. 1976;73:1547–1551. doi: 10.1073/pnas.73.5.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halladay J, Craig E. A heat shock transcription factor with reduced activity suppresses a yeast HSP70 mutant. Mol Cell Biol. 1995;15:4890–4897. doi: 10.1128/mcb.15.9.4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herrero P, Ramirez M, Martinez-Campa C, Moreno F. Identification and characterization of two transcriptional repressor elements within the coding sequences of the Saccharomyces cerevisiae HXK2 gene. Nucleic Acids Res. 1996;24:1822–1828. doi: 10.1093/nar/24.10.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herruer M H, Mager W H, Raué H A, Vreken P, Wilms E, Planta R J. Mild temperature shock affects transcription of the yeast ribosomal protein genes as well as the stability of their mRNAs. Nucleic Acids Res. 1988;16:7917–7929. doi: 10.1093/nar/16.16.7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herruer M H, Mager W H, Woudt L P, Nieuwint R T M, Wassenaar G M, Groeneveld P, Planta R J. Transcriptional control of yeast ribosomal protein synthesis during carbon-source upshift. Nucleic Acids Res. 1987;15:10133–10144. doi: 10.1093/nar/15.24.10133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 15.Kief D R, Warner J R. Coordinate control of syntheses of ribosomal ribonucleic acid and ribosomal proteins during nutritional shift-up in Saccharomyces cerevisiae. Mol Cell Biol. 1981;1:1007–1015. doi: 10.1128/mcb.1.11.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim C H, Warner J R. Mild temperature shock alters the transcription of a discrete class of Saccharomyces cerevisiae genes. Mol Cell Biol. 1983;3:457–465. doi: 10.1128/mcb.3.3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindquist S, Kim G. Heat-shock protein 104 expression is sufficient for thermotolerance in yeast. Proc Natl Acad Sci USA. 1996;93:5301–5306. doi: 10.1073/pnas.93.11.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mager W H, Planta R J, Ballesta J-P G, Lee J C, Mizuta K, Suzuki K, Warner J R, Woolford J. A new nomenclature for the cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Nucleic Acids Res. 1997;25:4872–4875. doi: 10.1093/nar/25.24.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mager W H, Planta R J. Coordinate expression of ribosomal protein genes in yeast as a function of cellular growth rate. Mol Cell Biochem. 1991;104:181–187. doi: 10.1007/BF00229818. [DOI] [PubMed] [Google Scholar]
- 20.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 21.Miller M J, Xuong N-H, Geiduschek E P. Quantitative analysis of the heat shock response of Saccharomyces cerevisiae. J Bacteriol. 1982;151:311–327. doi: 10.1128/jb.151.1.311-327.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moehle C M, Hinnebusch A G. Association of RAP1 binding sites with stringent control of ribosomal protein gene transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:2723–2735. doi: 10.1128/mcb.11.5.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morimoto R I, Tissieres A, Georgopoulos C. The biology of heat shock proteins and molecular chaperones. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 24.Nelson R J, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig E A. The translation machinery and seventy kilodalton heat shock protein cooperate in protein synthesis. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- 25.Neuman-Silberberg F D, Bhattacharya S, Broach J R. Nutrient availability and the Ras/cyclic AMP pathway both induce expression of ribosomal protein genes in Saccharomyces cerevisiae but by different mechanisms. Mol Cell Biol. 1995;15:3187–3196. doi: 10.1128/mcb.15.6.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nonet M, Scafe C, Sexton J, Young R. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol. 1987;7:1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parviz F, Hall D D, Markwardt D D, Heideman W. Transcriptional regulation of CLN3 expression by glucose in Saccharomyces cerevisiae. J Bacteriol. 1998;180:4508–4515. doi: 10.1128/jb.180.17.4508-4515.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfund C, Lopez-Hoyo N, Ziegelhoffer T, Schilke B A, Lopez-Buesa P, Walter W A, Wiedmann M, Craig E A. The molecular chaperone SSB from S. cerevisiae is a component of the ribosome-nascent chain complex. EMBO J. 1998;17:3981–3989. doi: 10.1093/emboj/17.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rose M D, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 30.Rotenberg M O, Woolford J L., Jr Tripartite upstream promoter element essential for expression of Saccharomyces cerevisiae ribosomal protein genes. Mol Cell Biol. 1986;6:674–680. doi: 10.1128/mcb.6.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitt M E, Brown T A, Trumpower B L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shore D, Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencers and activator elements. Cell. 1987;51:721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- 33.Sinclair D A, Kornfeld G D, Dawes I W. Yeast intragenic transcriptional control: activation and repression sites within the coding region of the Saccharomyces cerevisiae LPD1 gene. Mol Cell Biol. 1994;14:214–225. doi: 10.1128/mcb.14.1.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith B J, Yaffe M P. Uncoupling thermotolerance from the induction of heat shock proteins. Proc Natl Acad Sci USA. 1991;88:11091–11094. doi: 10.1073/pnas.88.24.11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamai K T, Liu X, Silar P, Sosinowski T, Thiele D J. Heat shock transcription factor activates yeast methallothionein gene expression in response to heat and glucose starvation via distinct signalling pathways. Mol Cell Biol. 1994;14:8155–8165. doi: 10.1128/mcb.14.12.8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Treger J M, Schmitt A P, Simon J R, McEntee K. Transcriptional factor mutations reveal regulatory complexities of heat shock and newly identified stress genes in Saccharomyces cerevisiae. J Biol Chem. 1998;273:26875–26879. doi: 10.1074/jbc.273.41.26875. [DOI] [PubMed] [Google Scholar]
- 37.Warner J R, Gorenstein C. Yeast has a true stringent response. Nature. 1978;275:338–339. doi: 10.1038/275338a0. [DOI] [PubMed] [Google Scholar]
- 38.Werner-Washburne M, Becker J, Kosics-Smithers J, Craig E A. Yeast Hsp70 RNA levels vary in response to the physiological status of the cell. J Bacteriol. 1989;171:2680–2688. doi: 10.1128/jb.171.5.2680-2688.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woolford J L, Jr, Warner J R. The ribosome and its synthesis. In: Broach J R, Pringle J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces: genome dynamics, protein synthesis, and energetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 587–626. [Google Scholar]
- 40.Woudt L P, Smit A B, Mager W H, Planta R J. Conserved sequence elements upstream of the gene encoding yeast ribosomal protein L25 are involved in transcription activation. EMBO J. 1986;5:1037–1040. doi: 10.1002/j.1460-2075.1986.tb04319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]