Abstract

While ruthenium arene complexes have been widely investigated for their medicinal potential, studies on homologous compounds containing a tridentate tris(1-pyrazolyl)methane ligand are almost absent in the literature. Ruthenium(II) complex 1 was obtained by a modified reported procedure; then, the reactions with a series of organic molecules (L) in boiling alcohol afforded novel complexes 2–9 in 77–99% yields. Products 2–9 were fully structurally characterized. They are appreciably soluble in water, where they undergo partial chloride/water exchange. The antiproliferative activity was determined using a panel of human cancer cell lines and a noncancerous one, evidencing promising potency of 1, 7, and 8 and significant selectivity toward cancer cells. The tested compounds effectively accumulate in cancer cells, and mitochondria represent a significant target of biological action. Most notably, data provide convincing evidence that the mechanism of biological action is mediated by the inhibiting of mitochondrial calcium intake.
Introduction
Complexes of d-block metals possess unique properties otherwise not available to organic compounds and thus offer significant medicinal potential.1−3 In particular, platinum compounds have been used in clinical treatments against various types of cancer;4−6 however, despite their undisputed efficacy, they exhibit serious drawbacks, such as negative side effects, phenomena of intrinsic or acquired resistance, a limited number of treatable tumors, and the necessity of hospitalization for the intravenous administration.7−10 These facts have stimulated research to develop new drugs based on other transition-metal elements.11−13 Specifically, a variety of ruthenium complexes have shown great promise.14,15 Besides the prototypal NAMI-A, KP1019, and related ruthenium(III) salts that entered clinical trials,14,16,17 half-sandwich organometallic complexes based on the RuII–arene scaffold have attracted considerable attention. In particular, RAPTA compounds, featured by the amphiphilic 1,3,5-triaza-7-phosphaadamantane ligand (PTA), have emerged as prominent and are currently pointing to clinical trials.18,19 The popularity of RAPTAs and the easy accessibility of related structures have steered the way to the exploration of a considerable number of derivatives with a diversity of arenes and coligands (Figure 1A,B).20−22 However, a suitable combination of electronic factors should be formulated to avoid the removal of the arene moiety and the consequent disaggregation of the complexes in aqueous media, which is a disliking characteristic for a drug candidate.23−27
Figure 1.
(A) structure of RAPTA-C, leading compound of the RAPTA family; (B) general structure of ruthenium(II)–arene complexes investigated as anticancer drugs (R = alkyl/aryl; X = neutral or ionic ligand; L,L′ = pair of neutral ligands of bidentate neutral/ionic ligand); (C, D) structures of ruthenium–tpm compounds assessed for their cytotoxicity (L = MeCN, DMSO, PMePh2; n = 2–4).
Tris(pyrazolyl)methane (tpm) and ring-substituted derivatives are homologous to arenes in that both types of compounds are neutral and may behave as six-electron ligands towards transition-metal centers. However, while the arene–metal bond possesses a π-backbonding component, tpm is essentially a strong donor and provides substantial stability to the resulting complexes.28,29 Moreover, tripodal coordination with three nitrogen atoms (κ3) is usually observed, although alternative modes are possible,30,31 and the interchange between tri- and bidentate coordination might play some key role in metal-catalyzed organic transformations.32−34 In sharp contrast with the related arene systems and the fact that NAMI-A and KP1019 contain a mono-pyrazolyl ring, ruthenium(II)–tpm complexes remain almost unexplored for their medicinal potential heretofore. Indeed, to the best of our knowledge, studies are limited to sparse DNA binding experiments35,36 and the assessment of the in vitro cytotoxicity of complexes [RuCl(κ3-tpm)(L)2]PF6 (L = MeCN, DMSO, PMePh2) and [RuCl(κ3-tpm)(LL)]PF6 (LL= diphosphine) against breast MCF-7 and cervical HeLa cancer cell lines (Figure 1C,D).37 In the latter case, while the MeCN and DMSO adducts revealed inactive, the introduction of phosphine ligands resulted in IC50 values falling in the low micromolar range. Presumably, the lack of biological studies is also a consequence of the paucity of straightforward routes to add diversity to the {RuII–tpm} scaffold.
Here, we report a general synthetic strategy to access a new family of robust ruthenium(II)–tpm complexes and an extensive investigation of their anticancer activity.
Results and Discussion
Synthesis and Structural Characterization of Ruthenium Complexes
The ruthenium–tpm complex 1, containing two triphenylphosphine ligands, was obtained from commercial RuCl3·3H2O via a straightforward two-step procedure that was optimized with respect to the literature (Scheme 1).38,39 In particular, toluene was found to be the optimal solvent for the reaction of tpm with [RuCl2(PPh3)3], allowing to collect the desired product in 95% yield (gram scale). Following a previous work reporting the thermal substitution of one PPh3 with PTA (1,3,5-triaza-7-phosphaadamantane),40 we investigated the possibility of modifying the ruthenium coordination set in 1 by introducing different types of ligands (i.e., different N-heterocyclic ligands; a phosphite that should provide more hydrophilicity than PPh3; two isocyanides with an aryl and alkyl substituent, respectively). Quantitative PPh3/MeCN replacement was achieved by heating a solution of 1 in acetonitrile at reflux, affording complex 2 in 95% yield (Scheme 1). Note that nitrile ligands usually behave as labile ones and are easily replaced by phosphines when coordinated to group 841−44 or other transition metals.45,46 In the present case, the reversed reaction is probably favored by steric factors arising from two bulky triphenylphosphines occupying adjacent coordination sites in 1.38
Scheme 1. Synthesis of Ruthenium(II) Tris(pyrazolyl)methane Complexes Investigated in This Work (1–9); Cy = C6H11.

To enable other substitution reactions, we found ethanol to be the best solvent; the reactions of 1 with a slight excess of various N-heterocyclic donors, trimethylphosphite and isocyanides, were carried out in ethanol at reflux and proceeded straightforwardly to afford novel complexes 3–9, which were isolated in good to nearly quantitative yields (Scheme 1).
The IR spectra of 4 and 5 (solid state, Figures S1–S9 in the Supporting Information) display the absorption attributed to the NH moiety at ca. 3450 cm–1, while the OH group belonging to 6 was detected at 3668 cm–1. The intense band due to the carbon–nitrogen triple bond in the isocyanide complexes occurs at 2141 (CNCy, 8) and 2147 cm–1 (CNCH2P(O)(OEt)2, 9). These values are almost coincident with those of the respective free isocyanides, indicating a scarce metal to isocyanide backdonation.47−51
The NMR spectra of 2–9 (in CDCl3, see Figures S10–S35) contain single sets of resonances, and the signals related to tpm are not significantly affected by the nature of the varying ligand (L). More precisely, the ring carbons resonate in the ranges 149.2–144.0 ppm (Cα), 109.0–107.3 ppm (Cβ), and 136.0–133.4 ppm (Cγ), whereas the methylidyne group gives rise to resonances at about 12 ppm (1H) and 74 ppm (13C). Inequivalence of the NMR resonances of the three pyrazolyl rings is in accordance with the chirality of the metal atom. The 31P NMR spectra display the resonance related to PPh3 falling at 44.8 (7)–52.5 (3) ppm; the resonance associated with the additional phosphorus ligand in 9 and 7 occurs at 15.7 and 138.0 ppm, respectively.
The structures of 3, 4, 5, 6, 7, and 8 were confirmed by single-crystal X-ray diffraction analyses; views of these structures are shown in Figure 2 with relevant bond lengths and angles reported in Table 1. Complexes 3–8 display a distorted octahedral geometry, as found in precursor 1 and related [RuCl(κ3–tpm)(PPh3)(L)]+ complexes.37,38,52 The Ru(1)–N(3) and Ru(1)–N(5) distances (Table 1) are comparable in 3–8 since they are trans to Cl(1) and P(1), respectively, in all complexes. In contrast, the Ru(1)–N(1) bond displays similar values in 3–6 [2.056(4), 2.045(5), 2.070(4), and 2.077(3) Å], being in the trans position to an aromatic N(7) ligand, whereas it is slightly elongated in 7 [2.180(2) Å] and 8 [2.1428(19) Å], where better π-acceptors P(OMe)3 and CNCy are present in the trans position. The nature of the varying ligand, L, slightly affects the Ru(1)–Cl(1) distance, reaching the lowest values in complexes 7 and 8 [2.3950(10) and 2.3942(6) Å, respectively]; the strength of the ruthenium–chloride bond is likely to be correlated with the cytotoxic activity of the complexes (vide infra). The Ru(1)–C(21) bond length in 8 [1.918(2) Å] is the shortest reported for an octahedral RuII center bonded to CNXyl, for which typical Ru–C values fall in the range 1.95–2.04 Å.53−56 This is likely to be since CNXyl is trans to N(1) in 8.
Figure 2.
View of the molecular structures (ORTEP drawing) of the cations of 3, 4, 5, 6, 7, and 8. Displacement ellipsoids are at the 30% probability level. CCDC reference numbers are 2167597 (3), 2167598 (4), 2167599 (5), 2167600 (6), 2167601 (7), and 2167602 (8).
Table 1. Selected Bond Lengths (Å) and Angles (°) for 3, 4, 5, 6, 7, and 8.
| 3 | 4 | 5 | 6 | 7 | 8 | |
|---|---|---|---|---|---|---|
| Ru(1)–N(1) | 2.056(4) | 2.045(5) | 2.070(4) | 2.077(3) | 2.180(2) | 2.1428(19) |
| Ru(1)–N(3) | 2.060(4) | 2.055(5) | 2.069(4) | 2.061(3) | 2.085(2) | 2.0798(17) |
| Ru(1)–N(5) | 2.103(4) | 2.106(5) | 2.104(4) | 2.100(3) | 2.131(2) | 2.106(2) |
| Ru(1)–P(1) | 2.3063(13) | 2.3021(16) | 2.3252(13) | 2.3184(8) | 2.3209(9) | 2.3125(6) |
| Ru(1)–Cl(1) | 2.3980(13) | 2.4045(15) | 2.4252(12) | 2.4021(8) | 2.3950(10) | 2.3942(6) |
| Ru(1)–Xa | 2.076(4) | 2.103(5) | 2.082(4) | 2.094(3) | 2.2181(10) | 1.918(2) |
| N(1)–Ru(1)–N(3) | 87.66(17) | 87.71(18) | 87.68(16) | 87.29(11) | 87.81(8) | 87.79(7) |
| N(1)–Ru(1)–N(5) | 84.36(17) | 84.39(19) | 85.02(17) | 84.69(10) | 81.02(8) | 82.67(7) |
| N(3)–Ru(1)–N(5) | 84.23(17) | 84.1(2) | 83.50(17) | 84.15(11) | 82.88(9) | 82.06(7) |
| P(1)–Ru(1)–Cl(1) | 95.74(5) | 98.16(6) | 96.68(4) | 97.75(3) | 91.04(3) | 92.98(2) |
| P(1)–Ru(1)–Xa | 94.14(12) | 95.35(14) | 94.84(12) | 95.51(8) | 93.35(3) | 92.44(7) |
| Cl(1)–Ru(1)–Xa | 88.62(12) | 87.92(14) | 87.37(12) | 88.03(8) | 96.29(3) | 91.20(6) |
X = N(7), 3–6; P(2), 7; C(21), 8.
The ligands of complexes 4, 5, and 6 contain NH (4 and 5) or OH (6) groups involved in hydrogen bonds. In particular, the N(8)H(8) benzimidazole group of 4 forms a hydrogen bond with the chloride counterion Cl(3) [N(8)–H(8) 0.88 Å, H(8)···Cl(3) 2.15 Å, N(8)···Cl(3) 3.027(6) Å, N(8)–H(8)–Cl(3) 173.7°]. Similarly, the O(1)H(1a) group of 6 is involved in a similar H-bond with the counterion [O(1)–H(1a) 0.848 Å, H(1a)···Cl(2) 2.19 Å, O(1)···Cl(2) 3.027(3) Å, O(1)–H(1a)–Cl(2) 175.7°]. In contrast, the N(8)H(8) group of the pyrazole ligand of 5 forms an intramolecular H-bond with the Cl(1) ligand [N(8)–H(8) 0.88 Å, H(8)···Cl(1) 2.525 Å, N(8)···Cl(1) 3.039(5) Å, N(8)–H(8)–Cl(1) 118.2°].
Solubility, Partition Coefficient, and Stability in Aqueous Media
A detailed study on the behavior of complexes 1–9 in aqueous media was performed: experimental procedures are provided in Experimental Section, and the results are presented in Table 2.
Table 2. Solubility in Water (D2O), Octanol/Water Partition Coefficient (log Pow), and Residual Ruthenium Complex in D2O (after 48 h) and DMSO-d6/DMEM-d (1:4 v/v, Except 1:3 v/v in the Case of 4; after 24 h) Solutions Maintained at 37 °C.
| compound | solubility/10–3 mol·L–1 (D2O, 21°C)a | log Pow | residual complex % in D2Oa,b | residual complex % in DMEM-d/ DMSO-d6a,b |
|---|---|---|---|---|
| 1 | 1.18 ± 0.05 | 69c | 88 | |
| 2 | 4.5 | –0.33 ± 0.07 | 89 | 51 |
| 3 | 3.4 | –0.15 ± 0.03 | 100 | 100 |
| 4 | 2.4 | 0.58 ± 0.06 | 100 | 93 |
| 5 | 3.4 | –0.05 ± 0.05 | 100 | 98 |
| 6 | 1.9 | 1.11 ± 0.07 | 100 | 100 |
| 7 | 3.1 | –0.02 ± 0.05 | 100 | 95 |
| 8 | 1.1 | 0.34 ± 0.01 | 92 | 98 |
| 9 | 4.6 | –0.20 ± 0.05 | 93 | 97 |
Calculated by 1H NMR (Me2SO2 as the internal standard).
Sum of chloro (2–9) and aquo (2W–9W) complexes.
DMSO-d6/D2O (4:1 v/v) mixture.
NMR experiments (1H and 31P) were used to study the speciation of 2–9 in D2O; preparation of the samples required 2 h (t0) stirring of a suspension of each complex in D2O; then, NMR analyses of the solutions pointed out the occurrence of a chloride–water exchange process (Scheme 2).
Scheme 2. Chloride Dissociation Reaction of Ruthenium(II) Tris(pyrazolyl)methane Complexes in Water.
NMR data of the resulting dicationic aquo species 2W–9W are reported in the Supporting Information (Figures S36–S43); the representative spectra of 3 dissolved in D2O are shown in Figures S44–S45. The aquo complexes were detected in a variable relative amount according to the ligand L (0–90%) after 2 h, as sharp NMR signals; 2W–9W became the largely prevalent species in solution after 48 h in all cases (see below) when the system had probably reached equilibrium. A significant increase of the molar conductivity of a 10–3 M solution of 6 in H2O was recognized over 24 h, in alinement with the increase by one unit of the net cationic charge of the ruthenium species (see Experimental Section). The pH of a 10–3 M H2O solution of 3, this compound undergoing almost complete aquation over 48 h (Table S1), was monitored over time, revealing a substantially constant and close to neutrality value [pH(t0) = 6.78, pH(24h) = 6.66, pH(48h) = 6.36]. This evidence reveals that the chloride/water exchange process is not followed by deprotonation of the H2O ligand, differently from what is documented for RAPTA compounds.57 The chloride/water substitution is fully reversible, in that a mixture of 3 and 3W (isolated from the D2O solution) fully reverted to 3 in CD3OD during 48 h (see page S32 and Figures S48–S49 in the SI).
Note that leading ruthenium(II)–arene complexes, such as RAPTA compounds, are considered prodrugs, and it has been proposed that the dissociation of chloride ligand(s) in the physiological environment is key to activation, enabling the binding of the metal center with biosubstrates.18,58−60 However, the kinetics of chloride/water exchange typically occurs in a much shorter time scale (≤30 min) for the arene complexes.58−60
Compound 1 is insoluble in water, and its behavior was evaluated in a DMSO-d6/D2O mixture. Under these conditions, chloride displacement was not recognized, whereas partial substitution of PPh3 with one solvent molecule was suggested by the appearance of a new signal at −7.2 ppm in the 31P NMR spectrum.
The D2O solubility of 2–9 was determined by 1H NMR using dimethyl sulfone (Me2SO2) as the internal standard, ranging between 1.1 and 4.6 mM, with reference to the sum of the chloro and aquo complex observed after 2 h (see above).
The octanol–water partition coefficients (log Pow) of 1–9 were measured by a UV–vis method; log Pow values of 2–9 were obtained approx. 20 min after the dissolution and thus are representative of the monocationic chloro complexes 2–9, as suggested by 1H and 31P NMR spectra recorded on a D2O solution of 3 after the same time (Figures S46–S47). The log Pow values are compiled in Table 2 and reflect an overall amphiphilic or moderate lipophilic character. More in detail, 1, which contains two triphenylphosphine ligands, and 6, featured by a hydroxy-pyridine ligand, revealed to be the most lipophilic compounds of the series. Conversely, the incorporation of acetonitrile, 1-methylimidazole, pyrazole, trimethylphosphite, and diethyl isocyanomethyl phosphonate as ligands leads to slightly negative log Pow values.
The D2O solutions of RuII–tpm complexes were then maintained at 37 °C and monitored for 48 h. Apart from the chloride dissociation process, no other changes were observed, except for a minor degradation in the cases of 2, 8, and 9, ascribable to the release of L (see Experimental Section for details). In fact, signals of dissociated PPh3 and tpm were not found in the NMR spectra.
The behavior of 1–9 was also analyzed in deuterated cell culture medium. The solutions were diluted with a variable amount of DMSO-d6 to obtain an appreciable solubility and then kept at 37 °C for 24 h. The compounds were found to be stable even in these conditions, and only minor degradation of the starting materials was detected by 1H and 31P NMR (1–12%, 49% for 2). The 31P NMR spectra of 1 and 3–9 resembled those recorded in D2O–DMSO-d6 and D2O solutions, respectively, while four different signals were detected in the final 31P NMR spectrum of 2 (Figure S50). In the case of 2, in addition to chloride/water exchange, it is possible that replacement of the acetonitrile ligand by solvent molecules occurs. On the other hand, the NMR spectra of 3–9 in DMSO–DMEM after 24 h only contained the resonances related to the starting complexes and the respective aquo species; therefore, DMSO coordination must be ruled out. The presence of ca. 0.1 M chloride ion in the medium slowed down the chloride/water substitution from 1–9, being almost negligible in the initial solution, and decreased the relative amount of aquo species after 48 h with respect to the analogous experiments in pure D2O (see Table S1 in the Supporting Information). The lower percentage of the aquo complex is exhibited by 7, 8, and 9, in accordance with the electron-withdrawing property of trimethylphosphite and isocyanide ligands, presumably strengthening the ruthenium–chloride bond.61,62
Antiproliferative Activity
Primary screening of antiproliferative activity of the family of ruthenium–tpm complexes 1–9 was performed by the commonly used MTT assay on five cancer cell lines of various origins: MCF-7 (breast), HeLa (cervical), 518A2 (melanoma), HCT116 (colon), and RD (rhabdomyosarcoma). In addition, normal human fibroblasts MRC5pd30 were used to assess the toxicity of the complexes on a noncancerous cell line. The results are summarized in Table 3. The IC50 values (concentration that causes 50% inhibition of cell proliferation) obtained for 1–9 are compared to those obtained for clinically used cisplatin under the same experimental conditions.
Table 3. IC50 Values (μM) Determined by the MTT Test after 72 h of Treatmenta.
| MCF-7 | HeLa | 518A2 | HCT116 | RD | MRC5pd30 | SIb | |
|---|---|---|---|---|---|---|---|
| 1 | 2.4 ± 0.6 | 4.0 ± 0.4 | 2.6 ± 0.4 | 1.5 ± 0.1 | 2.2 ± 0.2 | 5.8 ± 0.7 | 2.3 |
| 2 | 32 ± 4 | 53 ± 4 | 26 ± 4 | 25 ± 2 | 26 ± 5 | 76 ± 1 | 2.4 |
| 3 | 38 ± 8 | 91 ± 3 | 33 ± 5 | 25 ± 1 | 27 ± 4 | 83.6 ± 0.5 | 2.0 |
| 4 | 37 ± 7 | 36 ± 1 | 35 ± 4 | 25 ± 2 | 25 ± 3 | 56.6 ± 0.7 | 1.8 |
| 5 | 32 ± 5 | 54 ± 6 | 31 ± 3 | 30 ± 2 | 23 ± 2 | 63 ± 4 | 1.9 |
| 6 | 46 ± 7 | 54 ± 14 | 38 ± 8 | 31 ± 2 | 38 ± 6 | 82 ± 4 | 2.0 |
| 7 | 6 ± 1 | 10 ± 2 | 6.8 ± 0.8 | 6.7 ± 0.4 | 6 ± 1 | 24 ± 1 | 3.4 |
| 8 | 10 ± 2 | 15 ± 1 | 10 ± 2 | 8 ± 2 | 6.6 ± 0.7 | 19.7 ± 0.4 | 2.0 |
| 9 | 43 ± 7 | 36 ± 2 | 38 ± 7 | 35 ± 6 | 24 ± 3 | 50.8 ± 0.4 | 1.5 |
| cisplatin | 13 ± 3c | 14 ± 3c | 2.6 ± 0.7d | 8 ± 1c | 4.6 ± 0.3 | 11.7 ± 0.8c | 1.1 |
Most of the investigated compounds possess a moderate activity with IC50 values in tens of micromolar. The antiproliferative activity appears significantly influenced by the nature of the ligand (L) and only partially correlated with the log Pow values. Thus, among the tested compounds, three complexes stand out. Namely, compound 1 featured the highest degree of lipophilicity within the series (log Pow = 1.18) and 7 showed the best antiproliferative activity with IC50 values in a single-digit micromolar range in almost all of the investigated cancer cell lines. Furthermore, the potency of these two complexes is comparable to or even better than that of clinically used cisplatin, depending on the specific cell line. On the other hand, the substantially lower activity of 6 (log Pow = 1.11), compared to that of 1, is associated with the presence of the benzimidazole ligand in place of triphenylphosphine.
In addition to 1 and 7, complex 8 (log Pow = 0.34), bearing a cyclohexyl isocyanide ligand, also showed very good activity, still roughly comparable to cisplatin. The beneficial effect of incorporating the cyclohexyl moiety (Cy) within anticancer metal compounds was previously recognized and attributed to its compact and hydrophobic structure.65−67
Notably, the cytotoxic effects of 1–9 on noncancerous human skin fibroblasts MRC5pd30 were significantly lower, demonstrating selectivity toward cancer over noncancerous cells.
It should be mentioned that complexes of formula [RuCl(κN-Py)(PPh3)(η6-p-cymene)]+ (Py = substituted pyridine) were previously assessed for their cytotoxicity toward the HL60 leukemia tumor cell line, displaying IC50 values in the 5–15 μM range.68
The MTT assay, employed in the previously described experiment, is based on the mitochondrial reduction of MTT dye to formazan in living cells. However, several ruthenium complexes have been shown to affect mitochondrial metabolism.69,70 Moreover, the ruthenium–tris-pyrazolylmethane complexes studied in this work contain triphenylphosphine, which is known to impart specific features and activate additional modes of action to the related metal complexes.68,71,72 The data obtained by the MTT assay could then be overestimated due to the possible impact of the investigated compounds on mitochondrial metabolism. Therefore, the experiments focused on antiproliferative activity were repeated using an assay based on a mechanism other than mitochondrial metabolism, namely, Sulphorhodamine B (SRB) assay. This method relies on the stoichiometric binding of SRB dye to proteins in cells. The amount of dye is a proxy for cell mass and thus the number of cells in a sample. For SRB testing (and all further experiments as well), the HCT116 cell line was chosen in which the most active compound 1 exhibited the most promising anticancer activity (the lowest IC50 value).
As shown in Table S2 in the Supporting Information, the values obtained by the SRB assay were similar to (in the range of experimental error) or slightly lower than those from the MTT assay. This indicates that mitochondrial succinate dehydrogenase is likely not inhibited by the ruthenium complexes. Notably, the SRB assay confirmed the same trend in the biological activity of all tested complexes as found by MTT, with 1 being the most effective drug, followed by 7 and 8.
Intracellular Accumulation
To reveal a possible relationship between the cellular uptake and the in vitro activity of the investigated complexes, the ruthenium content of HCT116 cells after 24 h of incubation with complexes 1–9 was determined by inductively coupled plasma mass spectrometry (ICP-MS). The viabilities of the cells after the treatment ranged from 93 to 97%, as verified by the trypan blue exclusion assay so that the results were not affected by elevated permeability of compromised cell membranes of dying/dead cells. The results are summarized in Table 4.
Table 4. Accumulation of Ruthenium in HCT116 Cells after Treatment with Ruthenium Complexes (15 μM, 24 h)a,b.
| ng Ru/106 cells | log Pow | |
|---|---|---|
| 1 | 145.1 ± 5.9 | 1.18 ± 0.05 |
| 2 | 29.6 ± 2.9 | –0.33 ± 0.07 |
| 3 | 66.1 ± 8.0 | –0.15 ± 0.03 |
| 4 | 54.9 ± 1.3 | 0.58 ± 0.06 |
| 5 | 58.4 ± 3.0 | –0.05 ± 0.05 |
| 6 | 27.2 ± 1.1 | 1.11 ± 0.07 |
| 7 | 63.3 ± 6.0 | –0.02 ± 0.05 |
| 8 | 59.4 ± 0.6 | 0.34 ± 0.01 |
| 9 | 16.2 ± 1.4 | –0.20 ± 0.05 |
log Pow values are also reported for comparison.
Data for intracellular Ru concentration represent the mean ± standard error of the mean (SEM) from two independent experiments.
The inspection of data in Tables 3 and 4 reveals a correlation between antiproliferative activity and intracellular accumulation of the tested ruthenium complexes (Pearson’s correlation coefficient calculated for IC50 and accumulated Ru r = −0.76, see also Figure S51). This indicates that the ability of individual complexes to cross the cell membrane and accumulate in cells significantly contributes to their respective biological activity. In agreement with this view, the most active complex 1 accumulates in cells much more effectively than the other compounds.
The overall collection of data (speciation in aqueous solutions, log Pow, average IC50, cellular uptake) suggests that the antiproliferative activity of the complexes might be related to adequate lipophilicity of the active species. This condition can be achieved with coligand L (see Scheme 1) providing hydrophobic character and/or disfavoring the conversion into more hydrophilic biscationic complexes via chloride/water exchange (Scheme 2). More precisely, 7 and 8 display log Pow values of −0.02 and 0.34, respectively; however the electron-withdrawing character of L limits the elimination of the chloride and the consequent formation of 7W–8W (16% in DMEM-d/DMSO-d6 after 24 h, see Table S1). On the other hand, for instance, 4 and 6 are considerably more lipophilic than 7 and 8, but the former complexes generate a higher fraction of biscations (4W and 6W, respectively, 59 and 70% in DMEM-d/DMSO-d6), which are expected to be less prone than the parent monocationic complexes to pass through the cell membrane.73 Structural factors might also disfavor the penetration of the membrane for [{Ru}–OH2]2+ species compared to the corresponding [{Ru}–Cl]+.
The three most potent compounds, i.e., 1, 7, and 8, were selected for further studies to elucidate their mechanism of action.
Mechanism of Cell Death Induced by Ru Complexes
The previous test revealed that complexes 1, 7, and 8 possess an interesting antiproliferative activity. However, these tests cannot distinguish between the cytostatic (growth arrest and inhibition of division) and cytotoxic (loss of viability) effects.74,75 Therefore, we were interested in whether such complexes could induce cell death and, if so, by what mechanism. For this purpose, a commonly used Annexin-V/Propidium iodide (PI) assay was used, and the results were evaluated by flow cytometry (FACS). The typical densitograms obtained from FACS are shown in Figure S52, and Figure 3 shows a quantitative evaluation of the results. All tested complexes effectively induced apoptosis in HCT116 cells (Annexin-V-positive/PI-negative cells), whereas the percentage of cells undergoing necrosis was negligible (0.2–0.3%). This result was confirmed by measuring apoptosis/necrosis in real time immediately after the treatment (Figure S53).
Figure 3.
Bar graph of early apoptotic cells (% of the total population) in HCT116 cells treated with Ru complexes 1, 7, and 8 for 24 h at their equitoxic concentration (4xIC50,72h) quantified by FACS. Positive controls staurosporine (10 μM) and EtOH (5% v/v) were included as well-known apoptosis and necrosis inducers, respectively. Data represent mean ± SEM from three independent measurements; * indicates a significant (p < 0.05) difference from control, untreated cells.
Intracellular Distribution of Ru
To assess the subcellular distribution of selected complexes (1, 7, and 8) in HCT116 cells, cell fractionation was carried out following the treatment for 5 and 24 h. A FractionPREP Cell Fractionation kit (BioVision) was employed for the assay. According to the manufacturer’s information, the isolated fractions contain the nucleus (total nucleus soluble proteins, including the nuclear membrane proteins), cytosol (total cellular soluble proteins from cytoplasm), membrane/particulate (total cellular membrane proteins including cellular organelles and organelles membrane proteins), and cytoskeleton + DNA (total cellular insoluble proteins, genomic DNA). In addition, the ruthenium content in each fraction was determined by ICP-MS.
As shown in Figure 4, ruthenium from Ru–tpm complexes was detected primarily on the membrane/particulate fraction, so approximately 92–98% of total intracellular Ru was associated with this fraction. This means that the tested complexes preferentially localize in the membrane of organelles like mitochondria or endoplasmic reticulum. However, a small but not insignificant portion of Ru was also associated with nuclear and genomic DNA-containing fractions (2–8%), and the amount of Ru in these fractions increased with incubation time. It has been shown that in the case of cisplatin, whose anticancer mode of action is accepted as mediated by its interaction with DNA,76 only ca. 2–3% of intracellular platinum reaches the nucleus and binds to DNA.77,78 Thus, although the amount of ruthenium in fractions containing nuclear components (DNA and proteins) is minor, the mechanism of antiproliferative activity via interaction and chromatin damage cannot be excluded.
Figure 4.
Relative distribution of selected Ru complexes in subcellular fractions after 5 and 24 h treatment of HCT116 cells with equimolar concentrations (10 μM) of 1, 7, and 8. The sum of Ru concentration in all sample compartments was taken as 100%. The experiment was performed in duplicate. Data represents mean ± SEM from two measurements.
Additionally, many Ru complexes from the literature have been shown to act via DNA-damaging mechanisms.79 The complexes studied in this work contain one chlorido ligand, which is prone to substitution by water in a low-chloride environment such as the intracellular one, thus possibly favoring the DNA binding. Despite not collecting relevant evidence from the stability studies (see Section 2), the additional potential role of κ3 to κ2 switching of tpm coordination should not be ruled out in principle (see the Introduction). Next, experiments were performed to investigate the possibility of DNA-damaging potency in the overall biological activity of the three leading complexes.
One of the experimental criteria applied to prove DNA binding to be responsible for the cytotoxicity of metal-based drugs is based on the observation that the drug exhibits higher toxicity in the cells deficient in DNA repair.80,81 The reason lies in the fact that the ability of DNA lesions to induce cell death is inversely dependent on the capacity of the cells to repair the damage. Therefore, an experiment focused on the effect of the studied complexes in wild-type Chinese hamster ovary cell line CHO-K1 and its mutant NER-deficient counterpart MMC-2 was performed to clarify the involvement of nuclear DNA damage in mediating cellular sensitivity to 1, 7, and 8.
As shown in Table S3, the MMC-2 cells were significantly more sensitive to the treatment with cisplatin (IC50 values being almost ten times lower than the IC50 values found for parental cells CHO-K1). This result confirms that unrepaired DNA damage caused by cisplatin contributes markedly to its antiproliferative activity, in agreement with the DNA-damage-mediated mechanism of action of cisplatin. However, the IC50 values found for selected ruthenium complexes were nearly the same for both MMC-2 and parental CHO-K1 cells, indicating that NER-reparable DNA lesions do not play a significant role in the mechanism of activity of Ru–tpm complexes. Notwithstanding the above results, it should also be noted that cytoskeletal proteins are also present in the DNA-containing fraction. Therefore, to evaluate the possible effect of Ru–tpm complexes on the cytoskeleton, the morphology of cytoskeletal polymers, such as actin and tubulin filaments that participate in many vital cell functions, including division, morphogenesis, phagocytosis, and motility, was monitored by confocal microscopy. As shown in Figures S54 and S55, incubation with complexes 1, 7, or 8 did not significantly affect the structure, shape, and layout of either tubulin (Figure S54) or actin (Figure S55) networks, even at concentrations causing a significant antiproliferative effect (IC50). Thus, the antiproliferative activity of the investigated complexes seems to be unlikely related to the damage of the two major components of the cellular cytoskeleton, i.e., actin or tubulin filaments.
Real-Time Cell Growth Monitoring
The results described above reveal DNA damage as an unlikely cause of the biological action of the studied Ru complexes. Therefore, real-time impedance monitoring of cellular responses was used further to elucidate their mechanism of action. It has been shown that bioactive compounds produce specific time-dependent cell response profiles (TCRPs), predictive of the mechanism of action of the investigated molecules.82−84 Furthermore, a comparison of the shape characteristics of the TCRPs obtained for 1, 7, and 8 with those published for classes of compounds acting through various mechanisms82 revealed that the TRCPs of these three complexes (Figure 5) significantly differ from those characteristic for DNA-damaging agents. This result further supports the view that DNA is not a major target of the Ru–tpm complexes tested in this work.
Figure 5.

Interaction of HCT116 cells with 1, 7, and 8 at their 40, 160, and 160 μM concentrations, respectively, monitored by a real-time cell analyzer (RTCA). The vertical dashed lines indicate the start of the treatment after allowing the cells to adhere to microelectrodes and grow for 24 h. Cell indices were normalized to account for differences in cell counts across the wells prior to the treatment.
Interestingly, TRCPs obtained for 1, 7, and 8 resemble the profile of deoxycycline,82 a known inhibitor of mitochondria in cancer cells. This inhibitor has been shown to impair mitochondrial function by reducing mitochondrial membrane potential and mitochondrial respiration.85 Thus, the result of this experiment suggested that the mechanism of action of Ru–tpm complexes could be related to the impairment of mitochondria. Moreover, the majority of the intracellular Ru from 1, 7, and 8 was localized in the fraction comprising cellular organelles, including mitochondria (Figure 4). In isolated mitochondria, 467 ± 66, 127 ± 24, and 70 ± 15 pg Ru/106 cells were found when the cells were treated with 1, 7, and 8 respectively, indicating that Ru–tpm complexes accumulate in these organelles, although not exclusively. These facts support the hypothesis that the mitochondria may represent one of the significant targets of biological action of the tested complexes. Therefore, several functional assays were used to confirm whether and how the Ru–tpm complexes affect mitochondria in HCT116 cells.
Effect on Mitochondrial Membrane Potential
To provide evidence for the hypothesis that Ru–tris-pyrazolylmethane complexes tested in this work promote tumor cell death by a mitochondria-dependent mechanism, changes in the mitochondrial membrane potential of HCT116 cells after treatment with 1, 7, and 8 were determined by the TMRE assay. TMRE (ethyl ester of tetramethylrhodamine) is a cell-permeant fluorescent dye that accumulates in negatively charged mitochondria in a charge-dependent manner and is therefore used as a marker of mitochondrial membrane potential. If mitochondria depolarize or lose their integrity, the TMRE fluorescence intensity decreases accordingly.
After 5 h treatment, the HCT116 cells were stained with TMRE, and fluorescence changes reflecting mitochondrial membrane depolarization in response to the ruthenium complexes were observed by flow cytometry. Quantitative analysis (Figures 6 and S56) revealed a significant decrease of TMRE fluorescence in treated cells compared with control (untreated) cells, which indicated a marked mitochondrial depolarization in HCT116 cells due to the action of ruthenium complexes. The effect was concentration-dependent and more pronounced for 1 and 8. Instead, 7 was less potent in reducing mitochondrial membrane potential, but its effect was still significant.
Figure 6.
Bar graph showing median of TMRE intensity normalized to the intensity of the control. Cells were treated with equimolar and equitoxic concentrations of Ru complexes. Data represent mean ± SEM from three independent experiments.
Effect on Oxidative Phosphorylation
Mitochondria are key players in cellular bioenergetics, producing the majority of ATP by oxidative phosphorylation (OXPHOS). Therefore, the next experiment was performed to reveal whether the activity of the Ru complexes toward mitochondria also comprises respiratory chain uncoupling and inhibition of ATP syntheses. Mitochondrial Tox-Glo assay quantifies fluorescence signal originating from cell membrane permeabilization (nonspecific cell death) and luminescence signal generated by luciferase in the presence of ATP. Thus, it can distinguish primary mitochondrial disfunction from secondary cytotoxic events. Cells are treated and grown in a glucose-containing or glucose-free (galactose-supplemented) medium. In the presence of glucose, the cells may preferentially rely on glycolysis to meet bioenergetics needs and are relatively unresponsive to mitochondrial toxins. However, under glucose-free conditions (e.g., in the presence of galactose), the cells necessarily use OXPHOS to generate ATP and are more responsive to mitochondrial perturbation. If the drug disrupts oxidative phosphorylation, then a decrease in the ATP signal becomes observable.
As demonstrated in Figure 7, well-known OXPHOS inhibitor Antimycin A (used as a positive control) significantly decreased ATP production by cells growing in galactose-supplemented media. Simultaneously, the viability of cells was unaffected, as demonstrated by no changes in fluorescence signals (Figure 7). This indicates that a decrease in ATP synthesis results from OXPHOS inhibition rather than the overall cytotoxic effect. In contrast, no effect on ATP synthesis was observed for the investigated Ru complexes, neither in glucose-containing nor glucose-free medium. A decrease in ATP production was noticed only at the highest concentrations of complexes 1 and 8; however, it was accompanied by a decrease in viability. Thus, reducing ATP levels under these conditions likely resulted from the cytotoxic effect instead of OXPHOS inhibition. In summary, the primary mechanism of action of Ru–tris-pyrazolylmethane complexes does not consist in inhibition of OXPHOS; in other words, these compounds do not behave as specific mitotoxicants from the point of view of OXPHOS.
Figure 7.
Graphs showing the fluorescence signal intensity (in black, cell viability) and luminescence (in red, ATP) normalized to the untreated control. Cells grown in a glucose-containing medium are marked with a rectangle, while cells grown in a glucose-free medium are marked with a circle. Antimycin A was used as a positive control of OXPHOS inhibition.
Effect on Calcium Homeostasis
The previous results revealed that the investigated ruthenium complexes, although affecting mitochondria, do not uncouple mitochondrial energy metabolism. Thus, another mechanism underlying the effect of the investigated Ru complexes on mitochondria must be in play. Until now, multiple mechanisms of mitochondrial toxicity have been reported besides OXPHOS inhibition.86,87 Mitochondria are important in intracellular signal transduction and tuning of calcium (Ca2+) homeostasis. In stress conditions, intracellular Ca2+ is often elevated, and functional mitochondria serve as a potent Ca2+-buffer system.88 Much of the function of the mitochondria relies on Ca2+ homeostasis and effective Ca2+ signaling. Calcium regulates mitochondrial dynamics and functionality, thus affecting various cellular processes, including the mitochondrial pathway, enzyme activity, etc. Then, the fine modulation of mitochondrial calcium homeostasis plays a fundamental role in many processes involving these organelles.89 Cancer cells have therefore evolved mechanisms to modulate mitochondrial Ca2+ transport in order to sustain their metabolic demand and ensure their survival.90 The complex role of mitochondrial calcium in cancer has been thoroughly summarized in several recent reviews.90−92 Therefore, we decided to assess the effect of selected complexes on Ca2+ homeostasis using cytoplasmic and mitochondrial Ca2+ fluorescent sensors. As demonstrated in Figure 8A, complex 1 reduced Ca2+ flux into the mitochondria induced by ionomycin, a potent ionophore commonly used for this assay. Similarly, a decrease in Ca2+ flux into the mitochondria was also observed for complexes 7, 8, and the less active 9 (Figure 8C), although their effects were less pronounced (7 and 8) or insignificant (9), in agreement with their noticeably lower activity (Table 3). This effect was accompanied by an increase in the concentration of Ca2+ in the cytoplasm (Figure 8B,D) and was most prominent for complex 1, consistently with its greatest antiproliferative activity. The elevation of cytoplasmic calcium can be related to the inhibition of mitochondrial Ca2+ intake, as cytoplasmic calcium cannot be transferred to the mitochondria. However, it cannot be ruled out that this increase could also be related to other concurrent factors, such as a release of Ca2+ from the endoplasmic reticulum or an effect on Ca2+ channels in the cytoplasmic membrane. Thus, the tested complexes behave similarly to other well-known mitochondrial Ca2+ uptake inhibitors.93−97 The enhanced cytoplasmic concentration of Ca2+ ions can then lead to apoptosis via calcineurin-mediated proapoptotic protein activation,98 calpain proteases activation,99 or autophagy by mTOR inhibition.100 These data imply that the mechanism of biological action of the new complexes may also be mediated by disruption of Ca2+ homeostasis. On the other hand, in addition to disrupting Ca2+ homeostasis, other mechanisms may be involved in the biological effects of the here investigated compounds.
Figure 8.
(A, C) Effect on the mitochondrial influx of calcium. Cells pretreated for 2.5 h with 2.5, 5, or 10 μM Ru complex 1 (panel A) or with 10 μM 1, 7, 8, or 9 (panel C) were stained with 5 μM Rhod-2 (mitochondrial Ca2+ indicator) and treated with 5 μM ionomycin. (B, D) Effect on the cytosolic concentration of Ca2+. Cells were treated for 2.5 h with 2.5, 5, or 10 μM Ru complex 1 (panel B) or with 10 μM 1, 7, 8, or 9 (panel D) and subsequently stained with 5 μM Fluo-4 (cytoplasmic Ca2+ indicator). In both experiments, the fluorescence intensity was recorded in PBS with 2 mM CaCl2.
Cytotoxicity in Three-Dimensional (3D) Spheroids
To emphasize the possibility that the tested complexes might be promising candidates for further preclinical testing and to improve the relevance of our in vitro results, we used 3D cell cultures (spheroids), which are much better at replicating in vivo environment than traditional two-dimensional (2D) cultures. Cells grow in complex 3D cultures in an environment closely reflecting the tumor microenvironment, such as nutrient and oxygen gradients, intercellular and cell-extracellular matrix interactions, and heterogeneity.101 Hence, 3D growth of immortalized established cell lines or primary cell cultures is regarded as a more stringent and representative model for performing in vitro drug screening.101−103 Moreover, 3D cell culture models using human cells can circumvent the drawbacks of animal models that, aside from the high cost and ethical considerations, cannot always recapitulate human diseases or capture the side effect of drugs accurately. Also, the tumor spheroids have exhibited several features of the in vivo solid tumors. The similarities in the drug responsiveness among the tumor spheroids and the animal models might largely be due to their similarities in enhanced cellular interactions via adhesion and secretion of soluble factors of tumors that lead to low pH and hypoxia.104
Therefore, we assessed the ability of 1, 7, and 8 to inhibit spheroid formation and growth. HCT116 cells were cultured under 3D cell culture conditions for 96 h to grow up to a tissue mass of around 100 μm in diameter, as described in Experimental Section. The spheroids were then treated with various concentrations of complexes for an additional 72 h. The Cell Titer-Glo 3D cell viability assay was used to determine IC50 values. The results (Table 5) confirm that Ru–tpm complexes tested in this experiment, which showed very good activity in conventional 2D cell cultures, also exhibit substantial activity in the 3D spheroids formed from HCT116 cells being significantly more effective than clinically used cisplatin. According to what is observed on 2D cultures, 1 excels in its activity, being approximately 18-fold more effective than cisplatin.
Table 5. IC50 Values (μM) for the HCT116 Cell Line Determined by the Cell Titer-Glo Test after 72 h of Treatment.
| compound | HCT116 (3D) |
|---|---|
| 1 | 2.5 ± 0.6 |
| 7 | 12.2 ± 2.5 |
| 8 | 12.1 ± 2.7 |
| cisplatin | 44.6 ± 1.8 |
The effect on the morphology of 3D spheroids of HCT116 cells is shown in Figure S57. The spheroids formed by the control, untreated cells displayed round-shape morphology with a well-defined surrounding edge (Figure S57, panel A). However, after treatment with 7, 8, or cisplatin (Figure S57, panels C–E), spheroids displayed heterogeneous morphology with several dissociated cell clumps. Furthermore, these dissociated cell clumps were found to a much greater extent in the samples treated with 1; under the condition of the experiment, the spheroids treated with this complex were almost completely disintegrated (Figure S57, panel B). Thus, also this morphologic study showed superior activity of Ru–tris-pyrazolylmethane complexes in the 3D spheroids formed from colon cancer HCT116 cells.
Conclusions
The search for metal drugs alternative to platinum compounds, which are currently administered in clinical treatment against several types of tumors, is an ultimate demand of research. Ruthenium complexes have aroused a great interest in this regard due to their versatile anticancer activity and the scarce toxicity of the metal element, and some ruthenium(II)–arene compounds (RAPTA complexes) are pointing to clinical trials with great promise. Tris-pyrazolylmethane (tpm) is a versatile ligand behaving as a neutral six-electron donor like the arene moiety, but parallel studies on the anticancer potential of Ru–tpm species are lacking in the literature, apparently due to some synthetic drawbacks. Here, we report a straightforward route to access a family of novel, robust cationic ruthenium(II)–tpm complexes differing from each other in one key ligand, modulating both the amphiphilic character and the strength of the ruthenium–chloride bond, which may be implicated in the activation mechanism.
Three complexes of the series, showing a favorable combination of these two factors, displayed micromolar potency against a panel of human cancer cells (comparable to that of conventional cisplatin). Notably, the promising potency of these leading Ru–tpm complexes was bolstered by the results obtained with the 3D spheroids formed from cancer cells, which are much better at replicating in vivo environments than traditional 2D cultures. Moreover, these complexes demonstrated selectivity toward cancer over noncancerous cells. Our further data prove that Ru–tpm complexes effectively induced apoptosis in cancer cells, whereas the percentage of cells undergoing necrosis was negligible. The complexes are taken up in large amounts by cancer cells, and a correlation was observed between antiproliferative activity and intracellular accumulation. In vitro growth inhibition studies were completed by investigating the mechanism by which the studied complexes inhibit the growth of cancer cells. The results of these experiments showed that the mitochondria represent a significant target. Although mitochondria are key players in cellular bioenergetics, producing the majority of ATP by oxidative phosphorylation (OXPHOS), the primary mechanism of action of Ru–tpm complexes does not consist of inhibition of OXPHOS. In contrast, we conclude, based on the present findings, that the biological action is mediated by disruption of calcium homeostasis due to the inhibiting of mitochondrial calcium intake. To the best of our knowledge, this is the first study demonstrating a mechanism of antiproliferative activity of ruthenium complexes in cancer cells that involves the regulation of mitochondrial calcium homeostasis. However, the regulation of mitochondrial calcium homeostasis may not be the only mechanism by which the present complexes act. Ruthenium complexes are generally considered to be multifactorial agents, so the biological activity of Ru–tpm complexes may include other factors whose identification was beyond the scope of this work. Nevertheless, disruption of calcium homeostasis undoubtedly contributes to the overall activity of these complexes. Work is currently underway to evaluate other possible mechanistic contributions to the overall biological activity and the in vivo tumor efficacy of this new class of antitumor metal complexes.
Experimental Section
General Remarks
Reactants and solvents were purchased from Alfa Aesar, Merck, Strem, or TCI Chemicals and were of the highest purity available. Tris(1-pyrazolyl)methane (tpm) was prepared according to the published procedure.105 Reactions were conducted under a N2 atmosphere using standard Schlenk techniques, and all products were stored in air once isolated. All compounds are >95% pure by elemental analysis. Solvents were used as received unless otherwise stated. Toluene and diethyl ether were dried with the solvent purification system mBraun MB SPS5, while methanol was distilled from calcium hydride and isopropanol from magnesium. IR spectra of solid samples were recorded on an Agilent Cary630 FTIR spectrometer. IR spectra were processed with Spectragryph software.106 NMR spectra were recorded at 298 K on a JEOL JNM-ECZ500R instrument equipped with a Royal HFX Broadband probe. Chemical shifts (expressed in parts per million) are referenced to the residual solvent peaks (1H, 13C)107 or external standard (31P to H3PO4). 1H and 13C{1H} NMR spectra were assigned with the assistance of 1H–13C (gs-HSQC and gs-HMBC) correlation experiments.108 Elemental analyses were performed on a Vario MICRO cube instrument (Elementar). Conductivity measurements were performed at 25 °C using an XS COND 8 instrument (cell constant = 1.0 cm–1).109,110 pH measurements were performed with an Orion pH meter equipped with a Hamilton glass pH electrode. ESI-Q/ToF flow injection analysis was carried out using a 1200 Infinity HPLC (Agilent Technologies), coupled to a Jet Stream ESI interface (Agilent) with a Quadrupole-Time-of-Flight tandem mass spectrometer 6530 Infinity Q-TOF (Agilent Technologies).
Synthesis and Characterization of Complexes
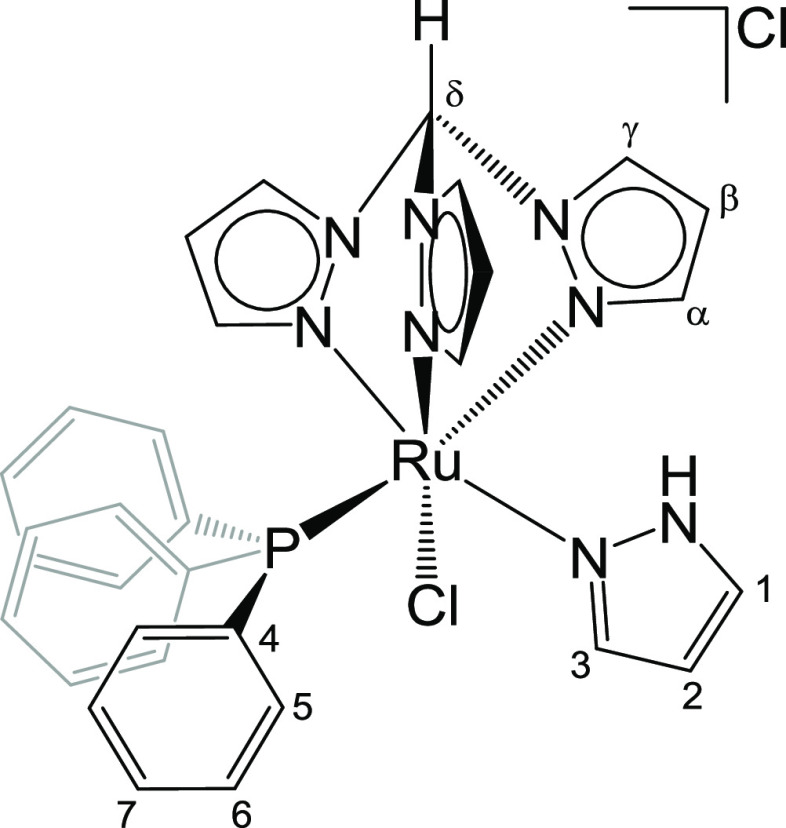
[RuCl(κ3-tpm)(PPh3)2]Cl, 1 (Chart 1)
Chart 1. Synthesis and Structure of 1 (Labeling Refers to Carbon Atoms).
The title compound was prepared using a slightly modified literature procedure (Chart 1).38,39
Step (a): Synthesis of [RuCl2(PPh3)3]
A solution of RuCl3·3H2O (350 mg, 1.34 mmol) and triphenylphosphine (1.75 g, 6.70 mmol) in distilled methanol (20 mL) was heated at reflux for 4 h. Thus, complex [RuCl2(PPh3)3] precipitated as a brown-purple solid. This solid was separated by filtration under a N2 atmosphere, washed with methanol and three times with diethyl ether, and finally dried under vacuum for several hours. The isolated material contained minor amounts of ineliminable PPh3 and O=PPh3, according to 31P NMR (resonances at −5.5 and 29.2 ppm, respectively), which did not affect step (b).
Step (b): Synthesis of [RuCl2(κ3-tpm)(PPh3)2]
Complex [RuCl2(PPh3)3], as obtained from step (a), and an equimolar amount of tpm (286 mg, 1.34 mmol) were taken in 30 mL of anhydrous toluene under vigorous agitation. A yellow solid rapidly precipitated, and the mixture was left to stir overnight. The solid was filtered, washed with toluene and three times with diethyl ether, and finally dried under vacuum. Yellow solid, yield 1.13 g (95%). Anal. calcd for C46H40Cl2N6P2Ru: C, 60.66; H, 4.43; N, 9.23; Cl, 7.79. Found: C, 60.35; H, 4.47; N, 9.13; Cl, 7.90. IR (solid state): ṽ/cm–1 = 3138w, 3121w, 3101w, 3061w, 3054w, 1515w, 1480m, 1467m, 1454m, 1440m, 1433m, 1411m, 1404m, 1383w, 1303m, 1291m, 1259m, 1254m, 1222m, 1187m, 1090s, 1075m, 1048m, 1024w, 998w, 988w, 852m, 799m, 776m, 768m, 754s, 742s, 696s, 691s, 681m. 1H NMR (CDCl3): δ/ppm = 12.21 (s, 1H, CδH); 8.86 (d, 1H, 3JHH = 2.7 Hz, CγH trans to P); 8.64 (d, 2H, 3JHH = 2.8 Hz, CγH trans to Cl); 7.29–7.26 (m, 3H, C4H); 7.15–7.07 (m, 12H, C3H + C2H); 6.83 (d, 2H, 3JHH = 2.2 Hz, CαH trans to P); 5.98 (t, 2H, 3JHH = 2.6 Hz, CβH trans to P); 5.50 (t, 1H, 3JHH = 2.7 Hz, CβH trans to Cl); 5.12 (d, 1H, 3JHH = 2.4 Hz, CαH trans to Cl). 31P{1H} NMR (CDCl3): δ/ppm = 40.138
Preparation of [RuCl(κ3-tpm)(PPh3)(NCMe)]Cl, 2 (Chart 2)
Chart 2. Structure of 2 (Labeling Refers to Carbon Atoms).
A solution of 1 (200 mg, 0.22 mmol) in 25 mL of acetonitrile (MeCN) was heated at reflux for 3 h. The solvent was evaporated under reduced pressure, and the obtained solid was washed with diethyl ether and dried under vacuum. Yellow solid, yield 144 mg (95%). Anal. calcd for C30H28Cl2N7PRu: C, 52.26; H, 4.09; N, 14.22; Cl, 10.28. Found: C, 52.08; H, 3.98; N, 14.26; Cl, 10.40. IR (solid state): ṽ/cm–1 = 3109w, 3055w, 2958w, 2919w, 2278w (ṽN=C), 1620w-br, 1507w, 1483w, 1450w, 1433m, 1408m, 1375w, 1289m, 1277w, 1252w, 1223w, 1187w, 1090s, 1053w, 1048w, 997w, 987w, 857w, 971s, 779s, 767s, 750s, 756s, 695s. 1H NMR (CDCl3): δ/ppm = 12.29 (s, 1H, CδH); 8.90, 8.75, 8.71 (d, 3H, 3JHH = 2.9 Hz, CγH); 8.14 (d-br, 1H, CαH); 6.91, 6.55 (d, 2H, 3JHH = 2.2 Hz, CαH); 7.41–7.27 (m, 15H, C3H + C4H + C6H); 6.43 (s-br, 1H CβH); 6.07, 5.96 (t, 2H, 3JHH = 2.6 Hz, CβH); 2.16 (s, 3H, C2H). 13C{1H} NMR (CDCl3): δ/ppm = 148.0, 147.2, 144.1 (Cα); 135.6, 135.4, 133.6 (Cγ); 134.1 (C4, 3JPC = 9.4 Hz); 132.3 (C3, 1JPC = 42.3 Hz); 130.1 (C6); 128.3 (C5, 4JPC = 9.3 Hz); 127.9 (C1); 108.3, 108.2, 108.0 (Cβ); 74.4 (Cδ); 4.7 (C2). 31P{1H} NMR (CDCl3): δ/ppm = 48.1 (Chart 2).
General Procedure for the Synthesis of Complexes [RuCl(κ3-tpm)(PPh3)(L)]Cl
A solution of 1 and the proper ligand L in ethanol or anhydrous isopropanol was heated at reflux for a variable time. After cooling to room temperature, the volatiles were evaporated under reduced pressure. The crude product was redissolved in the minimum volume of dichloromethane, precipitated with diethyl ether, filtered, and dried under vacuum.
[RuCl(κ3-tpm)(PPh3){NCHN(Me)(CH)2}]Cl, 3 (Chart 3)
Chart 3. Structure of 3 (Labeling Refers to Carbon Atoms).
From 1 (150 mg, 0.165 mmol) and N-methylimidazole (14.5 μL, 0.182 mmol) in ethanol (10 mL). Reaction time: 16 h. Yellow solid, yield 114 mg (94%). Anal. calcd for C32H31Cl2N8PRu: C, 52.61; H, 4.28; N, 15.34; Cl, 9.71. Found: C, 52.42; H, 4.17; N, 15.47; Cl, 9.56. IR (solid state): ṽ/cm–1 = 3107w-br, 2982w-br, 1481w, 1449w-br, 1434m, 1407m, 1289m, 1250m, 1222w, 1087s, 1053m, 858m, 792s, 755s, 748s-br, 695s. 1H NMR (CDCl3): δ/ppm = 12.22 (s, 1H, CδH); 8.99, 8.77, 8.73 (d, 3H, 3JHH = 2.8 Hz, CγH); 7.40, 7.13, 6.72 (d, 3H, 3JHH = 2.2 Hz, CαH); 7.34 (t, 3H, 3JHH = 7.3 Hz, C7H); 7.22–7.18 (m, 7H, C5H + CHImid); 7.06 (m, 6H, C6H); 6.69 (t-br, 1H, CHImid); 6.30 (t-br, 1H, CβH); 6.15–6.14 (m, 2H, CβH + CHImid); 5.96 (t, 1H, 3JHH = 2.6 Hz, CβH); 3.40 (s, 3H, C4H). 13C{1H} NMR (CDCl3): δ/ppm = 148.5, 148.2, 144.1 (Cα); 143.3 (Imid); 135.6, 135.1, 133.4 (Cγ); 133.9 (d, 3JCP = 9.5 Hz, C7); 132.9 (d, 1JCP = 40.1 Hz, C5); 130.4 (Imid); 129.7 (C8); 128.1 (d, 2JCP = 9.1 Hz, C6); 120.2 (Imid); 108.0, 108.0, 107.9 (Cβ); 74.5 (Cδ); 34.6 (C4). 31P{1H} NMR (CDCl3): δ/ppm = 52.5. Crystals suitable for X-ray diffraction were collected by slow diffusion of diethyl ether into a solution of 3 in methanol/dichloromethane (1:2 v/v) (Chart 3).
[RuCl(κ3-tpm)(PPh3){NCHNHC(CH)4C}]Cl, 4 (Chart 4)
Chart 4. Structure of 4 (Labeling Refers to Carbon Atoms).
From 1 (200 mg, 0.22 mmol) and benzimidazole (40 mg, 0.33 mmol) in ethanol (15 mL). Reaction time: 16 h. Yellow solid, yield 129 mg (77%). Anal. calcd for C35H31Cl2N8PRu: C, 54.83; H, 4.08; N, 14.62; Cl, 9.25. Found: C, 54.92; H, 3.98; N, 14.76; Cl, 9.11. IR (solid state): ṽ/cm–1 = 3138w, 3104w, 3059w, 2933w-br, 1669w, 1619w, 1587w, 1517w, 1480w, 1449w, 1433m, 1409m, 1304w, 1289m, 1273m, 1245m, 1222m, 1186w, 1152w, 1091m, 1053m, 856m, 794m, 774m, 746s, 740s, 696s. 1H NMR (CDCl3): δ/ppm = 12.06 (s-broad, 1H, NH); 11.98 (s, 1H, CδH); 8.97, 8.73, 8.66 (d-br, 3H, CγH); 7.76 (s-br, 1H, C1H); 7.56 (d, 1H, 3JHH = 8.1 Hz, C3H or C6H); 7.34 (t, 3H, 3JHH = 7.7 Hz, C11H); 7.16–6.96 (m, 15H, C4H or C5H + C9H + C10H + 2CαH); 6.67 (d-br, 1H, CαH); 6.60 (t-br, 1H, C4H or C5H); 6.15, 6.10, 5.92 (t-br, 3H, CβH); 5.09 (d-br, 1H, C3H or C6H). 13C{1H} NMR (CDCl3): δ/ppm = 150.7, 148.7, 145.0 (Cα); 148.1, 140.6 (C2 + C7); 135.4, 134.6, 133.2 (Cγ); 134 (d-br, C10); 132.9 (C1); 132.3 (d, 1JCP = 39.2 Hz, C8); 130.1 (C11); 128.4; (d, 2JCP = 9.0 Hz, C9); 123.5 (C4); 122.2 (C5); 117.5 (C6); 113.1 (C3); 108.7, 108.2, 107.8 (Cβ); 74.8 (Cδ). 31P{1H} NMR (CDCl3): δ/ppm = 51.1. Crystals suitable for X-ray diffraction were collected by slow diffusion of diethyl ether into a dichloromethane solution of 4 (Chart 4).
[RuCl(κ3-tpm)(PPh3){NNH(CH)3}]Cl, 5 (Chart 5)
Chart 5. Structure of 5 (Labeling Refers to Carbon Atoms).
From 1 (150 mg, 0.165 mmol) and pyrazole (12 mg, 0.181 mmol) in ethanol (8 mL). Reaction time: 8 h. Yellow solid, yield 106 mg (90%). Anal. calcd For C31H29Cl2N8PRu: C, 51.96; H, 4.08; N, 15.64; Cl, 9.90. Found: C, 51.86; H, 3.99; N, 15.55; Cl, 10.01. IR (solid state): ṽ/cm–1 = 3452w-br (ṽN-H), 3140w-br, 3109w-br, 2976w, 2924w, 2871w, 1518w, 1483w, 1450w, 1434w, 1407m, 1378w, 1350w, 1290m, 1283m, 1251m, 1223w, 1185w, 1156w, 1126w, 1114w, 1090s, 1050m, 1043m, 999w, 987w, 859m, 790m, 785–775s-br, 696s, 686m. 1H NMR (CDCl3): δ/ppm = 12.33 (s-br, 1H, NH); 11.79 (s, 1H, CδH); 9.05, 8.81, 8.75 (d, 3H, 3JHH = 2.8 Hz, CγH); 7.41, 7.09, 6.64 (d-br, 3H, CαH); 7.36 (t, 3H, 3JHH = 7.5 Hz, C7H); 7.24 (d-br, 1H, C3H); 7.19 (t, 6H, 3JHH = 7.7 Hz, C5H); 6.94 (m, 7H, C6H + C1H); 6.31 (m, 2H, CβH + C2H); 6.13, 6.05 (t, 2H, 3JHH = 2.6 Hz, CβH). 13C{1H} NMR (CDCl3): δ/ppm = 148.7, 148.2, 144.0 (Cα); 141.3 (C1); 136.0, 135.4, 133.6 (Cγ); 133.7 (d, 3JCP = 9.5 Hz, C6); 132.2 (d, 1JCP = 41.0 Hz, C4); 130.3 (C3); 130.0 (C7); 128.4 (d, 2JCP = 9.2 Hz, C5); 108.5, 107.7 (Cβ); 108.2 (Cβ + C2); 74.5 (Cδ). 31P{1H} NMR (CDCl3): δ/ppm = 49.9. Crystals suitable for X-ray diffraction were collected by slow diffusion of diethyl ether into a dichloromethane solution of 5 (Chart 5).
[RuCl(κ3-tpm)(PPh3){N(CH)3C(OH)CH}]Cl, 6 (Chart 6)
Chart 6. Structure of 6 (labeling refers to carbon atoms).
From 1 (150 mg, 0.165 mmol) and 3-pyridinol (25 mg, 0.26 mmol) in ethanol (10 mL). Reaction time: 16 h. Yellow solid, yield 108 mg (88%). Anal. calcd for C33H30Cl2N7OPRu: C, 53.30; H, 4.07; N, 13.19; Cl, 9.54. Found: C, 53.22; H, 4.02; N, 13.20; Cl, 9.65. IR (solid state): ṽ/cm–1 = 3668w-br (ṽO-H), 3110w-br, 3052w, 2980w, 2797w, 2773w, 2989w, 2622w, 1589w, 1481w, 1451w, 1436m, 1408m, 1382w, 1288m, 1273m, 1257m, 1222w, 1189w, 1158w, 1087m, 1054w, 1029w, 996w, 988w, 892w, 857m, 792m, 777m, 752s, 699s. 1H NMR (CDCl3): δ/ppm = 11.53 (s, 1H, CδH); 9.47 (s-br, 1H, OH); 8.72, 8.62, 8.50 (d-br, 3H, CγH); 7.83 (s-br, 1H, C5H); 7.60 (s-br, 1H, C1H); 7.40 (d, 1H, 3JHP = 8.4 Hz, C3); 7.34 (t, 3H, 3JHP = 7.4 Hz, C9); 7.23 (d-br, 1H, CαH); 7.14 (t, 6H, 3JHP = 7.6 Hz, C7); 7.02 (d, 1H, 3JHH = 2.2 Hz, CαH); 6.90 (t, 6H, 3JHP = 8.8 Hz, C8); 6.71 (dd, 1H, 3JHH = 8.3 Hz, 3JHH = 5.6 Hz, C4); 6.63 (d, 1H, 3JHH = 2.2 Hz, CαH); 6.19 (t-br, 1H, CβH); 6.09 (t, 1H, 3JHH = 2.5 Hz, CβH); 6.01 (t, 1H, 3JHH = 2.4 Hz, CβH). 13C{1H} NMR (CDCl3): δ/ppm = 153.8 (C2); 148.9, 148.8, 144.5 (Cα); 135.4 (Cγ); 134.6, 133.4 (Cγ); 133.9 (d, 3JCP = 9.3 Hz, C8); 132.0 (d, 1JCP = 40.7 Hz, C6); 130.1 (C9); 128.4 (d, 2JCP = 9.0 Hz, C7); 124.0 (C4); 123.4 (C3); 108.8, 108.6, 108.3 (Cβ); 74.8 (Cδ). C1and C5not observed. 31P{1H} NMR (CDCl3): δ/ppm = 50.6. Λm (H2O, c = 1.0 × 10–3 M, t0) = 118 S·cm2·mol–1; Λm (H2O, c = 1.0 × 10–3 M, after 24 h) = 240 S·cm2·mol–1. Crystals suitable for X-ray diffraction were collected by slow diffusion of diethyl ether into a dichloromethane solution of 6 (Chart 6).
[RuCl(κ3-tpm)(PPh3){P(OMe)3}]Cl, 7 (Chart 7)
Chart 7. Structure of 7 (Labeling Refers to Carbon Atoms).
From 1 (100 mg, 0.11 mmol) and trimethylphosphite {P(OMe)3} (16.9 μL, 0.14 mmol) in anhydrous isopropanol (6 mL). Reaction time: 7 h. Yellow solid, yield 78 mg (94%). Anal. calcd for C31H34Cl2N6O3P2Ru: C, 48.19; H, 4.44; N, 10.88; Cl, 9.18. Found: C, 48.28; H, 4.39; N, 10.67; Cl, 9.27. IR (solid state): ṽ/cm–1 = 3118w, 3086w, 3055w, 2952w, 2952–2846w, 1484w, 1461w, 1454w, 1435w, 1405w, 1304w, 1292m, 1257w, 1224w, 1184w, 1093m, 1054–1021s (ṽP-O), 990w, 983w, 856m, 793s, 769s, 762s, 749m, 740m, 728m, 695s, 684m. 1H NMR (CDCl3): δ/ppm = 12.19 (s, 1H, CδH); 8.92, 8.69 (d, 2H, 3JHH = 2.8 Hz, CγH); 8.65 (d-br, 1H, CγH); 8.15, 6.83, 6.37 (d, 3H, 3JHH = 2.2 Hz, CαH); 7.4 (t, 6H, 3JHH = 7.6 Hz, C3H); 7.34 (t, 3H, 3JHH = 7.3 Hz, C5H); 7.24 (t, 6H, 3JHH = 7.5 Hz, C4H); 6.34, 5.97 (t-br, 2H, CβH); 5.90 (t, 1H, 3JHH = 2.6 Hz, CβH); 3.46 (d, 3H, 2JHP = 10.2 Hz, C1H). 13C{1H} NMR (CDCl3): δ/ppm = 149.2, 146.6, 146.1 (Cα); 135.8, 133.9, 133.8 (Cγ); 134.4 (d, JCP = 9.21 Hz, C3); 133.4 (d, 1JCP = 43.5 Hz, C2); 129.9 (d, 4JCP = 2.3 Hz, C5); 128.0 (d, JCP = 9.4 Hz, C4); 109.0, 108.0, 107.3 (Cβ); 74.0 (Cδ); 53.2 (d, 2JCP = 8.2 Hz, C1). 31P{1H} NMR (CDCl3): δ/ppm = 138.0 (d, 2JPP = 57.8 Hz, P(OMe)3); 44.8 (d, 2JPP = 57.8 Hz, PPh3). Crystals suitable for X-ray diffraction were collected by slow diffusion into hexane from an acetone solution of 7 (Chart 7).
[RuCl(κ3-tpm)(PPh3)(CNCy)]Cl, 8 (Chart 8)
Chart 8. Structure of 8 (Labeling Refers to Carbon Atoms).
From 1 (200 mg, 0.22 mmol) and cyclohexyl isocyanide (30.3 μL, 0.242 mmol) in ethanol (15 mL). Reaction time: 3 h. Yellow solid, yield 157 mg (99%). Anal. calcd for C35H36Cl2N7PRu: C, 55.48; H, 4.79; N, 12.94; Cl, 9.36. Found: C, 55.25; H, 4.67; N, 12.88; Cl, 9.26. IR (solid state): ṽ/cm–1 = 3103w-br, 3058w, 2937w, 2926w, 2863w, 2854w, 2141s (ṽC=N), 1483w, 1448m, 1437m, 1434m, 1410m, 1403m, 1288s, 1255m, 1250m, 1222w, 1093s, 1084m, 1053m, 856m, 792s, 774s, 758s, 745s, 695s, 688s. 1H NMR (CDCl3): δ/ppm = 12.28 (s, 1H, CδH); 8.91, 8.72, 8.67 (d, 3H, 3JHH = 2.8 Hz, CγH); 8.06, 6.22, 6.03 (d, 3H, 3JHH = 2.2 Hz, CαH); 7.49 (t, 6H, 3JHH = 9.0 Hz, C7H); 7.40 (t, 3H, 3JHH = 7.3 Hz, C9H); 7.31 (t, 6H, 3JHH = 8.6 Hz, 3JHH = 4.3 Hz, C8H); 6.38 (t-br, 1H, CβH); 6.07, 5.91 (t, 2H, 3JHH = 2.5 Hz, CβH); 3.90 (m, 1H, C2-H); 1.90 (m, 2H, C4H); 1.66 (m, 2H, C3H); 1.59 (m, 2H, C4H); 1.49 (m, 1H, C5H); 1.30 (m, 3H, C3H + C5H). 13C{1H} NMR (CDCl3): δ/ppm = 155.9 (C1); 147.3, 145.9, 144.7 (Cα); 135.9, 134.3, 133.4 (Cγ); 134.2 (C7, 3JPC = 9.6 Hz); 132.8 (C6, 1JPC = 44.8 Hz); 130.2 (C6, 4JPC = 2.21 Hz); 128.4 (C8, 3JPC = 9.6 Hz); 108.2, 108.0, 107.8 (Cβ); 74.2 (Cδ); 55.4 (C2); 33.7 (C4); 24.9 (C5); 23.3 (C3). 31P{1H} NMR (CDCl3): δ/ppm = 49.6. Crystals suitable for X-ray diffraction were collected by slow diffusion of diethyl ether into a CH2Cl2 solution of 8 (Chart 8).
[RuCl(κ3-tpm)(PPh3){CNCH2P(O)(OEt2)2}]Cl, 9 (Chart 9)
Chart 9. Structure of 9 (Labeling Refers to Carbon Atoms).
From 1 (200 mg, 0.22 mmol) and diethyl isocyanomethyl phosphonate (38.8 μL, 0.242 mmol) in ethanol (15 mL). Reaction time: 3 h. Yellow solid, yield 159 mg (92%). Anal. calcd for C34H37Cl2N7O3P2Ru: C, 49.46; H, 4.52; N, 11.88; Cl, 8.59. Found: C, 49.33; H, 4.57; N, 11.96; Cl, 8.68. HR-ESI-MS: [M]+m/z = 790.1171 (theoretical for [C34H37ClN7O3P2Ru]+: m/z = 790.1165). IR (solid state): ṽ/cm–1 = 3149w, 3123w, 3109w, 3061w, 3001w, 2981w, 2933w, 2905w, 2147s (ṽC=N), 1515w, 1483w, 1479w, 1455w, 1445w, 1437m, 1433m, 1410m, 1397w, 1339w, 1293m, 1258m, 1245s (ṽP=O), 1227m, 1191w, 1164w, 1159w, 1090s, 1055m, 1042s, 1011s, 990m, 982m, 971s, 938m, 918w, 8895w, 857m, 836w, 815m, 793m, 768m, 763s, 750s, 745s, 699s, 695s, 682m. 1H NMR (CDCl3): δ/ppm = 12.24 (s, 1H, CδH); 8.86, 8.77 (d, 2H, 3JHH = 2.8 Hz, CγH); 8.64 (d-br, 1H, CγH); 8.12, 6.85, 6.44 (d, 3H, 3JHH = 2.2 Hz, CαH); 7.47–7.39 (m, 9H, C6H + C8H); 7.31 (m, 6H, C7H); 6.38 (t-br, 1H, CβH); 6.13, 5.90 (t, 2H, 3JHH = 2.5 Hz, CβH); 4.11–3.86 (m, 6H, C2H + C3H); 1.20 (t, 3H, 3JHH = 7.2 Hz, C4H); 0.99 (t, 3H, 3JHH = 7.1 Hz, C4H). 13C{1H} NMR (CDCl3): δ/ppm = 163.5 (C1); 148.3, 146.0, 145.1 (Cα); 135.7, 134.6, 133.4 (Cγ); 134.1 (d, 2JCP = 9.7 Hz, C6); 132.2 (d, 1JCP = 45.6 Hz, C5); 130.4 (C8); 128.5 (d, 3JCP = 9.7 Hz, C7); 108.1, 108.1, 107.9 (Cβ); 74.2 (Cδ); 64.0 (d, 2JCP = 6.8 Hz, C3); 63.7 (d, 2JCP = 6.6 Hz, C3); 41.1 (d, 1JCP = 156.3 Hz, C2); 16.4, 16.1 (d, 3JCP = 5.6 Hz, C4). 31P{1H} NMR (CDCl3): δ/ppm = 47.2 (s, PPh3); 15.7 (s, PCH2) (Chart 9).
X-ray Crystallography
Crystal data and collection details for 3·2CH3OH, 4·0.5Et2O·solv, 5·CH2Cl2·2H2O, 6·solv, 7·CH3COCH3·H2O, and 8·2H2O are reported in Table 6. Data were recorded on a Bruker APEX II diffractometer equipped with a PHOTON2 detector using Mo Kα radiation. The structures were solved by direct methods and refined by full-matrix least-squares based on all data using F2.111 Hydrogen atoms were fixed at calculated positions and refined using a riding model.112 The crystals of 4·0.5Et2O·solv and 6·solv contain some highly disordered solvent molecules that have been treated using the SQUEEZE routine of PLATON.113,114 The refined crystal structures of 4·0.5Et2O·solv and 5·CH2Cl2·2H2O contain large difference peaks; these are located in the proximity of disordered chloride ions or CH2Cl2 molecules.
Table 6. Crystal Data and Measurement Details for 3·2CH3OH, 4·0.5Et2O·solv, 5·CH2Cl2·2H2O, 6·solv, 7·CH3COCH3·H2O, and 8·2H2O.
| 3·2CH3OH | 4·0.5Et2O·solv | 5·CH2Cl2·2H2O | 6·solv | 7·CH3COCH3·H2O | 8·2H2O | |
|---|---|---|---|---|---|---|
| formula | C34H39Cl2N8O2PRu | C37H36Cl2N8O0.5PRu | C32H35Cl4N8O2PRu | C33H30Cl2N7OPRu | C34H42Cl2N6O5P2Ru | C35H40Cl2N7O2PRu |
| FW | 794.67 | 803.68 | 837.52 | 743.58 | 848.64 | 793.68 |
| T, K | 100(2) | 100(2) | 100(2) | 100(2) | 100(2) | 100(2) |
| λ, Å | 0.71073 | 0.71073 | 0.71073 | 0.71073 | 0.71073 | 0.71073 |
| crystal system | triclinic | monoclinic | triclinic | triclinic | monoclinic | monoclinic |
| space group | P1̅ | P21/c | P1̅ | P1̅1̅ | C2/c | P21/c |
| a, Å | 10.1428(8) | 20.5145(12) | 9.7016(8) | 11.1525(4) | 41.370(8) | 17.110(2) |
| b, Å | 13.2743(11) | 20.6593(12) | 10.0588(8) | 12.4862(5) | 9.756(2) | 10.1025(13) |
| c, Å | 13.9235(11) | 19.0486(11) | 19.2103(16) | 14.7685(6) | 19.152(4) | 21.524(3) |
| α, deg | 72.776(2) | 90 | 97.728(2) | 87.2220(10) | 90 | 90 |
| β, deg | 77.299(2) | 100.126(2) | 103.181(3) | 73.5550(10) | 91.34(3) | 106.679(8) |
| γ, deg | 85.255(3) | 90 | 97.790(2) | 89.3910(10) | 90 | 90 |
| cell volume, Å3 | 1746.4(2) | 7947.3(8) | 1781.7(3) | 1970.09(13) | 7728(3) | 3564.1(8) |
| Z | 2 | 8 | 2 | 2 | 8 | 4 |
| Dc, g·cm–3 | 1.511 | 1.343 | 1.561 | 1.253 | 1.459 | 1.479 |
| μ, mm–1 | 0.692 | 0.607 | 0.828 | 0.606 | 0.674 | 0.677 |
| F(000) | 816 | 3288 | 852 | 756 | 2488 | 1632 |
| crystal size, mm | 0.16 × 0.13 × 0.10 | 0.22 × 0.16 × 0.14 | 0.22 × 0.18 × 0.15 | 0.18 × 0.15 × 0.12 | 0.24 × 0.21 × 0.18 | 0.18 × 0.13 × 0.10 |
| θ limits, deg | 1.5649–26.998 | 1.467–25.998 | 2.074–25.997 | 1.633–27.227 | 1.970–26.000 | 1.9759–26.998 |
| reflections collected | 32 233 | 142 792 | 29 925 | 27 297 | 54 659 | 72 769 |
| independent reflections | 75 857 [Rint = 0.0850] | 15 547 [Rint = 0.1422] | 6987 [Rint = 0.0724] | 8722 [Rint = 0.0465] | 7594 [Rint = 0.0553] | 7780 [Rint = 0.0673] |
| data/restraints/parameters | 7585/17/428 | 15 547/8/871 | 6987/39/445 | 8722/0/407 | 7594/130/473 | 7780/6/445 |
| goodness on fit on F2 a | 1.193 | 1.075 | 1.036 | 1.097 | 1.038 | 1.064 |
| R1 (I > 2σ(I))b | 0.0628 | 0.0799 | 0.0610 | 0.0486 | 0.0336 | 0.0308 |
| wR2 (all data)c | 0.1366 | 0.1881 | 0.1604 | 0.1053 | 0.0772 | 0.0767 |
| largest diff. peak and hole, e Å–3 | 1.972/–1.232 | 3.653/–1.407 | 3.443/–2.006 | 1.132/–0.668 | 1.028/–0.793 | 0.651/–0.568 |
Goodness on fit on F2 = [Σw(FO2 – FC2)2/(Nref – Nparam)]1/2, where w = 1/[σ2(FO2) + (aP)2 + bP], where P = (FO2 + 2FC2)/3; Nref = number of reflections used in the refinement; Nparam = number of refined parameters.
R1 = Σ||FO| – |FC||/Σ|FO|.
wR2 = [Σw(FO2 – FC2)2/Σw(FO2)2]1/2, where w = 1/[σ2(FO2) + (aP)2 + bP], where P = (FO2 + 2FC2)/3.
Behavior in Aqueous Media
(a) Solubility in Water
A suspension of the selected ruthenium complex (3–5 mg) in a D2O solution (0.7 mL) containing Me2SO2 as the internal standard115 (3.36 × 10–3 M) was vigorously stirred at 21 °C for 2 h. The resulting saturated solution was filtered over celite, transferred into an NMR tube, and analyzed by 1H NMR and 31P NMR spectroscopy (delay time = 3 s; number of scans = 20). The concentration (solubility) was calculated by the relative integral (starting complex + aquo complex, CβH signal in the 1H spectrum) with respect to Me2SO2 (δ/ppm = 3.14). The results are compiled in Table 2. NMR data are given in the Supporting Information (Figures S36–S47).
(b) Stability in D2O and DMSO-d6/D2O
The same samples prepared at point (a) were used in this experiment, apart from complex 1 that was dissolved in DMSO-d6/D2O 4:1 v/v solution (0.7 mL; [Ru] = approx. 3 mg) containing Me2SO2 as the standard.115 After 1H and 31P{1H} NMR analyses described above at point (a) (time = t0), samples were heated at 37 °C for 48 h. After cooling to room temperature, 1H and 31P NMR spectra were recorded. The percentage of starting complex + related aquo complex was calculated by the relative integral (CβH signal in the 1H spectrum) with respect to Me2SO2 (c = 3.3 × 10–3 mol·L–1; δ/ppm = 3.14 in D2O; δ/ppm = 2.95 in DMSO-d6/D2O 4:1 v/v), see Table 2 and S1.
(c) Octanol/Water Partition Coefficients (log Pow)
Partition coefficients (Pow); IUPAC: KD partition constant,116 defined as Pow = corg/caq, where corg and caq are the molar concentrations of the selected compound in the organic and aqueous phase, respectively, were determined by the shake-flask method and UV–vis measurements.62,117,118 Deionized water and 1-octanol were vigorously stirred for 24 h to enable saturation of both phases and then separated by centrifugation. A stock solution of the selected ruthenium compound (ca. 2 mg) was prepared by first adding MeOH (50 μL, to help solubilization), followed by octanol-saturated water (2.5 mL). The solution was diluted with octanol-saturated water (ca. 1:3 v/v ratio, cRu ≈ 10–4 M, so that 1.5 ≤ A ≤ 2.0 at λmax), and its UV–vis spectrum was recorded (Aaq0). An aliquot of the solution (Vaq = 1.2 mL) was transferred into a test tube, and water-saturated octanol (Vorg = Vaq = 1.2 mL) was added. The mixture was vigorously stirred for 20 min at 21 °C then centrifuged (5000 rpm, 5 min). The UV–vis spectrum of the aqueous phase was recorded (Aaq), and the partition coefficient was calculated as Pow = (Aaq0 – Aaq)/Aaqf, where Aaq and Aaqf are the absorbances in the aqueous phase before and after partition with the organic phase, respectively.118 For 1, an inverse procedure was followed, starting from a solution of the compound in water-saturated octanol. The partition coefficient was calculated as Pow = Aorg/(Aorg0 – Aorg), where Aorg0 and Aorg are the absorbances in the organic phase before and after partition with the aqueous phase, respectively. The wavelength of the maximum absorption of each compound (280–380 nm range) was used for UV–vis quantitation. The procedure was repeated three times for each sample (from the same stock solution); the results are given as mean ± standard deviation (Table 2). Naphthoquinone was used as a reference compound (log P = 1.8 ± 0.2; literature:119 1.71).
(d) Stability in Cell Culture Medium
Powdered DMEM cell culture medium (1000 mg/L glucose and l-glutamine, without sodium bicarbonate and phenol red; D2902; Sigma-Aldrich) was dissolved in D2O (10 mg/mL), according to the manufacturer’s instructions. The solution of deuterated cell culture medium (“DMEM-d”) was treated with Me2SO2 (6.6 × 10–3 M) and NaH2PO4/Na2HPO4 (0.10 M, pD = 7.5)120−122 and then stored at 4 °C under N2. The selected ruthenium compound (2–3 mg) was dissolved in DMSO-d6 (0.14 mL; 0.18 mL for 4) and then diluted with DMEM-d up to 0.75 mL total volume (cRu ca. 4 × 10–3 M). The mixture was stirred for 30 min, then filtered over celite, and transferred into an NMR tube. The resulting yellow solution was analyzed by 1H and 31P NMR (delay time = 3 s; number of scans = 20) and then heated at 37 °C for 24 h. After cooling to room temperature, NMR analyses were repeated. Compound 1 was instead dissolved in a DMSO-d6/DMEM-d 4:1 v/v solution (0.7 mL; [Ru] = approx. 3 × 10–3 M) containing Me2SO2 as the internal standard.115 The percentage of the starting complex + related aquo complex was calculated by the relative integral with respect to Me2SO2 (δ/ppm = 3.16 in DMSO-d6/DMEM-d 1:4 and 1:3 v/v; δ/ppm = 2.95 in DMSO-d6/DMEM-d 4:1 v/v).
Biological Studies
Cell Lines, Culture Conditions, and Stock Solutions of Ru Complexes
The human cervical carcinoma HeLa cells and human colorectal carcinoma cells HCT116 were kindly supplied by Professor B. Keppler, University of Vienna (Austria). Human rhabdomyosarcoma RD cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA). Human breast cancer MCF-7 cells, human skin melanoma 518A2 cells, and human MRC5pd30 cells derived from normal lung tissue were purchased from the European Collection of Authenticated Cell Cultures (ECACC) (Salisbury, U.K.). Chinese hamster ovary CHO-K1 cell line (wild type) and its derivative MMC-2 carrying the ERCC3/XPB mutation (NER-deficient) cell line were kindly supplied by Dr. M. Pirsel, Cancer Research Institute, Slovak Academy of Sciences, Bratislava (Slovakia).
All of the cell lines were cultivated in DMEM medium (high glucose 4.5 g L–1, PAA, Pasching, Austria) supplemented with gentamycin (50 μg mL–1, Serva, Heidelberg, Germany) and 10% heat-inactivated fetal bovine serum (PAA). The medium for MRC5pd30 cells was further supplemented by 1% nonessential amino acids (Sigma-Aldrich, Prague, Czech Republic). All cells were cultured as adherent monolayers in a humidified incubator at 37 °C in a 5% CO2 atmosphere and subcultured twice a week with an appropriate plating density.
For biological studies, stock solutions of Ru complexes were prepared by dissolving the compounds in DMSO to a final concentration of 10 mM and subsequently diluted to the media to the required concentration. The concentration of Ru in media used in the experiment was verified by flameless atomic absorption spectrometry (FAAS). The final DMSO concentration in the cell culture medium did not exceed 1% (v/v) to avoid DMSO toxicity.
Antiproliferative Activity
In vitro antiproliferative activity of Ru complexes was determined by the MTT or, alternatively, SRB assay as already described,84 after 72 h of incubation of cells with various concentrations of the Ru complex. The reported IC50 values are an average of three independent experiments, each consisting of three replicates per concentration.
Cellular Uptake
In these experiments, 1 × 106 HCT116 cells were seeded on 100 mm Petri dishes. After overnight preincubation in a drug-free medium, the cells were treated with the Ru complexes (15 μM) for 24 h. Afterward, the cells were extensively washed with PBS (37 °C), detached using 0.25% trypsin, washed twice with ice-cold PBS, and counted by an automatic cell counter. The cell pellets were digested using a microwave acid (HCl) digestion system (CEM Mars). The quantity of Ru taken up by the cells was determined by inductively coupled plasma mass spectrometry (ICP-MS). All experiments were carried out in triplicate.
Annexin-V/PI Staining
The type of cell death (apoptosis/necrosis) caused by studied Ru complexes was determined by flow cytometry using Annexin-V and propidium iodide staining after 24 h of treatment. HCT116 cells were seeded in a six-well plate at a density of 150000 cells/well. After overnight cultivation, the cells were treated with studied complexes and incubated for 24 h. Afterward, the cells from individual wells were collected. Pellets were resuspended in the Annexin-V/PI staining solution (BD Pharmingen), and the samples were analyzed using a BD FACSverse flow cytometer.
Real-Time Apoptosis/Necrosis
Type of cell death and its kinetics were measured using the Real Time-Glo Annexin-V Apoptosis and Necrosis Assay (Promega). HCT116 cells were seeded at a density of 8 × 103 cells/50 μL in a 96-well black plate and incubated overnight. The cells were then treated with the Ru complexes, and immediately afterward, kinetic analysis began. Staurosporine (10 μM) and ethanol (5%) were used as positive controls of apoptosis and necrosis, respectively. Luminescence (integration of 1000 ms) and fluorescence (λex: 485 nm; λem: 535 nm) were detected by a SPARK reader (Tecan, Manedorf, Switzerland).
Real-Time Cell Growth Monitoring
We employed an xCELLigence RTCA SP Instrument (ROCHE) for monitoring cell growth in real time. First, the background of the 96-well E-Plate was read (100 μL of cultivation media). Then, the cells were added to the E-Plate (2000 cells/well in 50 μL of media), and the measurement was started. After 21 h, Ru complexes were added at various concentrations, and impedance was monitored for 72 h. An arbitrary unit CI (cell index) is a quantitative measure in which the status of the cells (number and morphology of attached cells) is reflected.
Cell Fractionation
The HCT116 cells were seeded at a density of 1.5 × 106 cells/10 mL Petri dish and incubated overnight. Then, the cells were treated with 10 μM Ru complexes, incubated for 5 or 24 h, harvested, and exhaustively washed with PBS. The cell pellets were processed by the FractionPREP Cell Fractionation Kit (BioVision) according to the manufacturer’s instructions, yielding four subcellular fractions: cytosol, membrane/particulate, nuclear, and cytoskeletal. The Ru content in each fraction was evaluated by ICP-MS. The measurement was performed in triplicate.
Isolation of Mitochondria
The HCT116 cells were seeded at a density of 3 × 106 cells/10 mL Petri dish and incubated overnight. The cells were treated with Ru complexes (10 μM) for 24 h, harvested, and washed with ice-cold PBS, and mitochondrial fractions were extracted by the Mitochondrial Isolation Kit (MITOISO2, Sigma-Aldrich) according to the manufacturer’s instructions. The Ru content in each sample was evaluated by ICP-MS. The measurement was performed in triplicate.
Effect on Mitochondrial Membrane Potential
Effects of Ru complexes on mitochondrial membrane potential were assessed by TMRE staining of HCT116 cells after treatment. First, HCT116 cells were seeded on a six-well plate at a 1.5 × 105 cells/well density. The next day, the cells were treated with equimolar (10 μM) and equitoxic (twofold or fourfold IC50,72h) concentrations of Ru complexes for 5 h. Then, the cells were collected and stained with 100 nM TMRE in a complete DMEM medium for 30 min at 37 °C in the dark. The TMRE-containing medium was then replaced with PBS, and cells were analyzed by a BD FACSverse flow cytometer (λex = 488 nm, λem = 586 nm).
Effect on Oxidative Phosphorylation
The Mitochondrial Tox-Glo assay was used to determine whether studied Ru complexes influence oxidative phosphorylation in HCT116 cells. Two culture media were prepared: glucose-containing, serum-free RPMI medium (RPMI 1640, + l-glutamine, 10 mM D-glucose (Gibco, Thermo Fisher Scientific)) supplemented with sodium bicarbonate and glucose-free, serum-free RPMI medium (RPMI 1640, + l-glutamine, no glucose (Gibco, Thermo Fisher Scientific)) supplemented with 10 mM galactose (Sigma) and sodium bicarbonate. Cells were seeded at a density of 1.5 × 103 cells/well in 50 μL in a 96-well black plate in media containing either glucose or galactose (vide supra) and incubated overnight. Then, the cells were treated with 50 μL of Ru complexes in various concentrations as indicated. Cells were cultivated for 90 min at 37 °C. Cell membrane permeability and ATP quantity were determined by the Mitochondrial Tox-Glo assay according to the manufacturer’s protocol. The first step was to add a fluorogenic peptide substrate (bis-AAF-R110) to measure the dead cell protease activity. Bis-AAF-R110 substrate cannot cross the intact membrane of live cells and therefore gives an insignificant signal with viable cells relative to nonviable cells. The second step of the procedure is adding an ATP detection reagent, resulting in cell lysis and generating a luminescent signal proportional to the amount of ATP present. The final fluorescent and luminescent signals were detected on multimode reader SPARK (Tecan, Manedorf, Switzerland).
Calcium Flux
The distribution of calcium ions in cytosol and mitochondria was studied using calcium-sensitive fluorescent probes. HCT116 cells were seeded at a density of 2 × 105 cells/well on six-well plates. After overnight cultivation, the cells were treated with Ru complexes at indicated concentrations for 2.5 h. The culture medium was then replaced with either 5 μM Rhod-2 or Fluo-4 in PBS supplemented with 2 mM CaCl2 and incubated for 30 min at 37 °C. At last, cells were harvested and suspended in PBS (with 2 mM CaCl2) or in PBS (with 2 mM CaCl2) supplemented with ionomycin (5 μM) for 30 min at 37 °C and analyzed using a BD FACSverse flow cytometer.
Cytotoxicity in Colonospheres
A spontaneous spheroid formation was used to generate colonospheres derived from the HCT116 cell line. The HCT116 cells were seeded on 96-well ultralow attachment plates (1400 cells/well) and cultivated in a tumor sphere-forming medium (DMEM/F12, supplemented with B27 (Invitrogen), BSA, bFGF (10 ng mL–1), and EGF (20 ng mL–1)) for 4 days. After the incubation period, colonospheres were treated with Ru complexes for another 3 days. The viability of colonospheres was determined by the Cell Titer-Glo 3D cell viability assay (Promega) according to the manufacturer’s instructions. The reported IC50 values are the average of three independent experiments, each consisting of three replicates per concentration level. Bright-field images of the spheroids were taken as well to determine the effect on the morphology of the spheroids.
Confocal Microscopy Imaging of Actin and Tubulin
HCT116 cells were seeded on coverslips precoated with 0.1% gelatin in six-well culture plates at a density of 1 × 105 cells/well. The following day, the cells were treated with the tested compounds at concentrations corresponding to IC50 for 24 h. Following the treatment, the cells were washed with PBS, fixed with 4% p-formaldehyde, washed, permeabilized with 0.1% Triton X-100, and blocked. Samples for actin staining were blocked with 1.5% BSA for 1 h and then stained with Alexa Fluor 488-conjugated Phalloidin (Thermo Fisher Scientific, 1:50 dilution, 20 min). Samples for tubulin staining were blocked with 5% goat serum for 1 h and incubated with primary antibody (anti-α-tubulin, Abcam, 1:200 dilution, 1 h) and Alexa Fluor-conjugated secondary antibody (goat antirabbit, Abcam, 1:500 dilution, 1 h). Both groups of samples were mounted with ProLong Diamond Antifade Mountant with DAPI (Invitrogen). Cells were visualized on a confocal microscope Leica TCS SP8 SMD.
Acknowledgments
The authors thank the University of Pisa (PRA_2020_39) and the Czech Science Foundation (Grant no. 21-27514S) for financial support. The authors thank Dr. Hana Kostrhunová from the Institute of Biophysics of the ASCR in Brno for performing experiments focused on the evaluation of the morphology of actin and tubulin filaments by confocal microscopy.
Glossary
Abbreviations Used
- δ
chemical shift in parts per million downfield from tetramethylsilane
- μ
micro
- Å
angstrom(s)
- °C
degrees celsius
- 3D
three-dimensional
- 518A2
human skin melanoma cells
- aq
aqueous
- ATP
adenosine triphosphate
- BSA
bovine serum albumin
- CHO-K1
Chinese Hamster ovary cell line
- CI
cell index
- Cy
cyclohexyl
- DMSO
dimethyl sulfoxide
- DMEM
Dulbecco’s modified Eagle’s medium
- Et
ethyl
- FAAS
flameless atomic absorption spectrometry
- FACS
fluorescence-activated cell sorting
- FIA
flow injection analysis
- HCT116
human colon tumor cell line 116
- HeLa
cervical cancer cell line
- HR-ESI-MS
high-resolution electron spray ionization mass spectroscopy
- IC50
concentration that causes 50% inhibition of cell proliferation
- ICP-MS
inductively coupled plasma mass spectrometry
- KP1019
indazolium trans-[tetrachlorobis(1H-indazole)ruthenate(III)]
- log Pow
octanol–water partition coefficient
- MCF-7
human breast cancer cells (Michigan Cancer Foundation-7)
- Me
methyl
- mTOR
mammalian target of rapamycin
- MTT
(3-(4,5-dimethylthiazol-2-yl)2,5-diphenyl-tetrazolium bromide)
- NAMI-A
imidazolium trans-[tetrachloro(dimethylsulfoxide)imidazoleruthenate(III)]
- NER
nucleotide excision repair
- NMR
nuclear magnetic resonance
- ORTEP
Oak Ridge thermal ellipsoid plot
- OXPHOS
oxidative phosphorylation
- PBS
phosphate-buffered saline
- Ph
phenyl
- PI
propidium iodide
- ppm
part(s) per million
- PTA
1,3,5-triaza-7-phosphaadamantane
- Q-TOF
quadrupole-time-of-flight
- RAPTA
ruthenium arene 1,3,5-triaza-7-phosphaadamantane complexes
- RD
rhabdomyosarcoma
- RT
room temperature
- RTCA
real-time cell analyzer
- SI
selectivity index
- SRB
sulphorhodamine B
- SEM
standard error of the mean
- SRB
sulphorhodamine B
- TCRPs
time-dependent cell response profiles
- TMRE
tetramethylrhodamine ethyl ester
- tpm
tris(pyrazolyl)methane
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jmedchem.2c00722.
Accession Codes
CCDC reference numbers 2167597 (3), 2167598 (4), 2167599 (5), 2167600 (6), 2167601 (7) and 2167602 (8) contain the supplementary crystallographic data for the X-ray studies reported in this work. Authors will release the atomic coordinates upon article publication.
Author Contributions
# J.C. and A.G. contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Anthony E. J.; Bolitho E. M.; Bridgewater H. E.; Carter O. W. L.; Donnelly J. M.; Imberti C.; Lant E. C.; Lermyte F.; Needham R. J.; Palau M.; Sadler P. J.; Shi H.; Wang F.-X.; Zhang W.-Y.; Zhang Z. Metallodrugs are unique: opportunities and challenges of discovery and development. Chem. Sci. 2020, 11, 12888–12917. 10.1039/D0SC04082G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marloye M.; Berger G.; Gelbcke M.; Dufrasne F. A survey of the mechanisms of action of anticancer transition metal complexes. Future Med. Chem. 2016, 8, 2263–2286. 10.4155/fmc-2016-0153. [DOI] [PubMed] [Google Scholar]
- Boros E.; Dyson P. J.; Gasser G. Classification of metal-based drugs according to their mechanisms of action. Chem 2020, 6, 41–60. 10.1016/j.chempr.2019.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.-C.; Miller J. J.. Platinum-Based Anticancer Drugs. In Encyclopedia of Inorganic and Bioinorganic Chemistry; Wiley, 2011. [Google Scholar]
- Ghosh S. Cisplatin: The first metal based anticancer drug. Bioorg. Chem. 2019, 88, 102925 10.1016/j.bioorg.2019.102925. [DOI] [PubMed] [Google Scholar]
- Riddell I. A.; Lippard S. J. Cisplatin and Oxaliplatin: Our current understanding of their actions. Met. Ions Life Sci. 2018, 18, 1–42. [DOI] [PubMed] [Google Scholar]
- Oun R.; Moussa Y. E.; Wheate N. J. The side effects of platinum-based chemotherapy drugs: a review for chemists. Dalton Trans. 2018, 47, 6645–6653. 10.1039/C8DT00838H. [DOI] [PubMed] [Google Scholar]
- Siddik Z. H. Cisplatin mode of cytotoxic action and molecular basis of resistance. Oncogene 2003, 22, 7265–7279. 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- Peng K.; Liang B.-B.; Liu W.; Mao Z.-W. What blocks more anticancer platinum complexes from experiment to clinic: major problems and potential strategies from drug design perspectives. Coord. Chem. Rev. 2021, 449, 214210 10.1016/j.ccr.2021.214210. [DOI] [Google Scholar]
- Qi L.; Luo Q.; Zhang Y.; Jia F.; Zhao Y.; Wang F. Advances in toxicological research of the anticancer drug cisplatin. Chem. Res. Toxicol. 2019, 32, 1469–1486. 10.1021/acs.chemrestox.9b00204. [DOI] [PubMed] [Google Scholar]
- Murray B. S.; Dyson P. J. Recent progress in the development of organometallics for the treatment of cancer. Current Opinion. Chem. Biol. 2020, 56, 28–34. [DOI] [PubMed] [Google Scholar]
- Hanif M.; Hartinger C. G. Anticancer metallodrugs: where is the next cisplatin?. Future Med. Chem. 2018, 10, 615–617. 10.4155/fmc-2017-0317. [DOI] [PubMed] [Google Scholar]
- Mjos K. D.; Orvig C. Metallodrugs in medicinal inorganic chemistry. Chem. Rev. 2014, 114, 4540–4563. 10.1021/cr400460s. [DOI] [PubMed] [Google Scholar]
- Thota S.; Rodrigues D. A.; Crans D. C.; Barreiro E. J. Ru(II) compounds: next-generation anticancer metallotherapeutics?. J. Med. Chem. 2018, 61, 5805–5821. 10.1021/acs.jmedchem.7b01689. [DOI] [PubMed] [Google Scholar]
- Zeng L.; Gupta P.; Chen Y.; Wang E.; Ji L.; Chao H.; Chen Z.-S. The development of anticancer ruthenium(II) complexes: from single molecule compounds to nanomaterials. Chem. Soc. Rev. 2017, 46, 5771–5804. 10.1039/C7CS00195A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alessio E. Thirty years of the drug candidate NAMI-A and the myths in the field of ruthenium anticancer compounds: a personal perspective. Eur. J. Inorg. Chem. 2017, 2017, 1549–1560. 10.1002/ejic.201600986. [DOI] [Google Scholar]
- Trondl R.; Heffeter P.; Kowol C. R.; Jakupec M. A.; Berger W.; Keppler B. K. NKP-1339, the first ruthenium-based anticancer drug on the edge to clinical application. Chem. Sci. 2014, 5, 2925–2932. 10.1039/C3SC53243G. [DOI] [Google Scholar]
- Murray B. S.; Babak M. V.; Hartinger C. G.; Dyson P. J. The development of RAPTA compounds for the treatment of tumors. Coord. Chem. Rev. 2016, 306, 86–114. 10.1016/j.ccr.2015.06.014. [DOI] [Google Scholar]
- Rausch M.; Dyson P. J.; Nowak-Sliwinska P. Recent Considerations in the application of RAPTA-C for cancer treatment and perspectives for its combination with immunotherapies. Adv. Therap. 2019, 2, 1900042 10.1002/adtp.201900042. [DOI] [Google Scholar]
- Steel T. R.; Walsh F.; Wieczorek-Błauz A.; Hanif M.; Hartinger C. G. Monodentately-coordinated bioactive moieties in multimodal half-sandwich organoruthenium anticancer agents. Coord. Chem. Rev. 2021, 439, 213890 10.1016/j.ccr.2021.213890. [DOI] [Google Scholar]
- Singh A. K.; Pandey D. S.; Xu Q.; Braunstein P. Recent advances in supramolecular and biological aspects of arene ruthenium(II) complexes. Coord. Chem. Rev. 2014, 270–271, 31–56. 10.1016/j.ccr.2013.09.009. [DOI] [Google Scholar]
- Swaminathan S.; Haribabu J.; Balakrishnan N.; Vasanthakumar P.; Karvembu R. Piano stool Ru(II)-arene complexes having three monodentate legs: A comprehensive review on their development as anticancer therapeutics over the past decade. Coord. Chem. Rev. 2022, 459, 214403 10.1016/j.ccr.2021.214403. [DOI] [Google Scholar]
- Chotard F.; Malacea-Kabbara R.; Balan C.; Bodio E.; Picquet M.; Richard P.; Ponce-Vargas M.; Fleurat-Lessard P.; Le Gendre P. Atom transfer radical addition catalyzed by ruthenium-arene complexes bearing a hybrid phosphine-diene ligand. Organometallics 2018, 37, 812–820. 10.1021/acs.organomet.7b00851. [DOI] [Google Scholar]
- Biancalana L.; Pampaloni G.; Zacchini S.; Marchetti F. Synthesis, characterization and behavior in water/DMSO solution of Ru(II) arene complexes with bioactive carboxylates. J. Organomet. Chem. 2018, 869, 201–211. 10.1016/j.jorganchem.2018.05.020. [DOI] [Google Scholar]
- Egger A. E.; Hartinger C. G.; Renfrew A. K.; Dyson P. J. Metabolization of [Ru(η6-C6H5CF3)(pta)Cl2]: a cytotoxic RAPTA-type complex with a strongly electron withdrawing arene ligand. J. Biol. Inorg. Chem. 2010, 15, 919–927. 10.1007/s00775-010-0654-x. [DOI] [PubMed] [Google Scholar]
- Bugarcic T.; Habtemariam A.; Stepankova J.; Heringova P.; Kasparkova J.; Deeth R. J.; Johnstone R. D. L.; Prescimone A.; Parkin A.; Parsons S.; Brabec V.; Sadler P. J. The contrasting chemistry and cancer cell cytotoxicity of bipyridine and bipyridinediol ruthenium(II) arene complexes. Inorg. Chem. 2008, 47, 11470–11486. 10.1021/ic801361m. [DOI] [PubMed] [Google Scholar]
- Biancalana L.; Batchelor L. K.; Ciancaleoni G.; Zacchini S.; Pampaloni G.; Dyson P. J.; Marchetti F. Versatile coordination of acetazolamide to ruthenium(II) p-cymene complexes and preliminarycytotoxicity studies. Dalton Trans. 2018, 47, 9367–9384. 10.1039/C8DT01555D. [DOI] [PubMed] [Google Scholar]
- Bigmore H. R.; Lawrence S. C.; Mountford P.; Tredget C. S. Coordination, organometallic and related chemistry of tris(pyrazolyl)methane ligands. Dalton Trans. 2005, 635–651. 10.1039/b413121e. [DOI] [PubMed] [Google Scholar]
- Reger D. L. Tris(pyrazolyl) methane ligands: the neutral analogs of tris(pyrazolyl) borate ligands. Comments Inorg. Chem. 1999, 21, 1–28. 10.1080/02603599908020413. [DOI] [Google Scholar]
- Semeniuc R. F.; Reger D. L. Metal complexes of multitopic, third generation poly(pyrazolyl)-methane ligands: multiple coordination arrangements. Eur. J. Inorg. Chem. 2016, 2016, 2253–2271. 10.1002/ejic.201600116. [DOI] [Google Scholar]
- Bhambri S.; Tocher D. A. Synthesis and characterisation of ruthenium(II) arene complexes containing κ3- and κ2-poly(pyrazolyl)borates and methanes. J. Chem. Soc. Dalton Trans. 1997, 18, 3367–3372. 10.1039/a702674i. [DOI] [Google Scholar]
- Muñoz-Molina J. M.; Belderrain T. R.; Perez P. J. Group 11 tris(pyrazolyl) methane complexes: structural features and catalytic applications. Dalton Trans. 2019, 48, 10772–10781. 10.1039/C9DT01661A. [DOI] [PubMed] [Google Scholar]
- Martins L. M. D. R. S.; Pombeiro A. J. L. Tris(pyrazol-1-yl)methane metal complexes for catalytic mild oxidative functionalizations of alkanes, alkenes and ketones. Coord. Chem. Rev. 2014, 265, 74–88. 10.1016/j.ccr.2014.01.013. [DOI] [Google Scholar]
- Martins L. M. D. R. S. C-scorpionate complexes: Ever young catalytic tools. Coord. Chem. Rev. 2019, 396, 89–102. 10.1016/j.ccr.2019.06.009. [DOI] [Google Scholar]
- Waywell P.; Gonzalez V.; Gill M. R.; Adams H.; Meijer A. J. H. M.; Williamson M. P.; Thomas J. A. Structure of the complex of [Ru(tpm)(dppz)py]2+ with a B-DNA oligonucleotide-a single-substituent binding switch for a metallo-Intercalator. Chem. - Eur. J. 2010, 16, 2407–2417. 10.1002/chem.200901758. [DOI] [PubMed] [Google Scholar]
- Foxon S. P.; Metcalfe C.; Adams H.; Webb M.; Thomas J. A. Electrochemical and photophysical properties of DNA metallo-intercalators containing the ruthenium(II) tris(1-pyrazolyl)methane unit. Inorg. Chem. 2007, 46, 409–416. 10.1021/ic0607134. [DOI] [PubMed] [Google Scholar]
- Walker J. M.; McEwan A.; Pycko R.; Tassotto M. L.; Gottardo C.; Th’ng J.; Wang R.; Spivak G. J. [Tris(pyrazolyl)methane]ruthenium complexes capable of inhibiting cancer cell growth. Eur. J. Inorg. Chem. 2009, 2009, 4629–4633. 10.1002/ejic.200900766. [DOI] [Google Scholar]
- Field L. D.; Messerle B. A.; Soler L.; Buys I. E.; Hambley T. W. Polypyrazolylmethane complexes of ruthenium. J. Chem. Soc., Dalton Trans. 2001, 1959–1965. 10.1039/b103939n. [DOI] [Google Scholar]
- Shaw A. P.; Ryland B. L.; Norton J. R.; Buccella D.; Moscatelli A. Electron exchange involving a sulfur-stabilized ruthenium radical cation. Inorg. Chem. 2007, 46, 5805–5812. 10.1021/ic700580u. [DOI] [PubMed] [Google Scholar]
- Miguel S.; Diez J.; Gamasa M. P.; Lastra M. E. Synthesis and structural features of new ruthenium(II) complexes containing the scorpionate ligands tris(pyrazol-1-yl)methanesulfonate (Tpms) and tris(pyrazol-1-yl)methane (Tpm). Eur. J. Inorg. Chem. 2011, 2011, 4745–4755. 10.1002/ejic.201100621. [DOI] [Google Scholar]
- Belli R. G.; Wu Y.; Ji H.; Joshi A.; Yunker L. P. E.; McIndoe J. S.; Rosenberg L. Competitive ligand exchange and dissociation in Ru indenyl complexes. Inorg. Chem. 2019, 58, 747–755. 10.1021/acs.inorgchem.8b02915. [DOI] [PubMed] [Google Scholar]
- Lentz D.; Michael-Schulz H. Syntheses, structure determination and reactions of phosphine substituted derivatives of (μ3-FC)2Fe3(CO)9. Z. Anorg. Allg. Chem. 1992, 618, 111–120. 10.1002/zaac.19926180120. [DOI] [Google Scholar]
- Albano V. G.; Busetto L.; Marchetti F.; Monari M.; Zacchini S.; Zanotti V. Synthesis and characterization of new diiron and diruthenium μ-aminocarbyne complexes containing terminal S-, P- and C-ligands. Z. Naturforsch. B 2007, 62, 427–438. 10.1515/znb-2007-0317. [DOI] [Google Scholar]
- Bresciani G.; Biancalana L.; Pampaloni G.; Zacchini S.; Ciancaleoni G.; Marchetti F. A comprehensive analysis of the metal–nitrile bonding in an organo-diiron system. Molecules 2021, 26, 7088. 10.3390/molecules26237088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker P. K.; Harman M. E.; Hursthouse M. B.; Karaulov A. I.; Lavery A. J.; Malik K. M. A.; Muldoon D. J.; Shawcross A. Nitrile exchange reactions of MI2(CO)3(NCMe)2. X-ray crystal structures of mixed-ligand seven-coordinate complexes. J. Organomet. Chem. 1995, 494, 205–221. 10.1016/0022-328X(95)05405-E. [DOI] [Google Scholar]
- Su B.-K.; Liu Y.-H.; Peng S.-M.; Liu S.-T. An anthyridine-based pentanitrogen donor switches from mono- to tetradentate with Pd(II) Ions. Organometallics 2021, 40, 4110–4119. 10.1021/acs.organomet.1c00569. [DOI] [Google Scholar]
- Biancalana L.; Ciancaleoni G.; Zacchini S.; Pampaloni G.; Marchetti F. Carbonyl-isocyanide mono-substitution in [Fe2Cp2(CO)4]: A re-visitation. Inorg. Chim. Acta 2020, 517, 120181. [Google Scholar]
- Compare νCN for cyclohexyl isocyanide in 8 (2147 cm-1) with [RuCl2(CO)2(PPh3)(CyNC)] (2160 cm-1) and [RuCpCl(PPh3)(CyNC)] (2130 cm-1)49,50
- Faraone F.; Piraino P.; Marsala V.; Sergi S. Tricarbonyldichloro(thiocarbonyl)ruthenium(II) and related complexes: synthesis and reactions to give aminomercaptocarbene and isocyanide complexes. J. Chem. Soc., Dalton Trans. 1977, 859–861. 10.1039/DT9770000859. [DOI] [Google Scholar]
- Bruce M. I.; Wallis R. C. Cyclopentadienyl-ruthenium and -osmium chemistry. XIII* Some complexes containing isonitrile ligands. Aust. J. Chem. 1981, 34, 209–213. 10.1071/CH9810209. [DOI] [Google Scholar]
- Bartalucci N.; Belpassi L.; Marchetti F.; Pampaloni G.; Zacchini S.; Ciancaleoni G. Ubiquity of cis-halide to isocyanide direct interligand interaction in organometallic complexes. Inorg. Chem. 2018, 57, 14554–14563. 10.1021/acs.inorgchem.8b02088. [DOI] [PubMed] [Google Scholar]
- Wilson D. C.; Nelson J. H. Reactions of ruthenium(II) tris(pyrazolyl)borate and tris(pyrazolyl)methane complexes with diphenylvinylphosphine and 3,4-dimethyl-1-phenylphosphole. J. Organomet. Chem. 2003, 682, 272–289. 10.1016/j.jorganchem.2003.07.010. [DOI] [Google Scholar]
- Niu Y.; Han F.; Zhang Q.; Xie T.; Lu L.; Li S.; Xia H. Off/On Fluorescent chemosensors for organotin halides based on binuclear ruthenium complexes. Angew. Chem., Int. Ed. 2013, 52, 5599–5603. 10.1002/anie.201209549. [DOI] [PubMed] [Google Scholar]
- Cuervo D.; Menéndez-Pedregal E.; Díez J.; Gamasa M. P. Mononuclear ruthenium(II) complexes bewring the (S,S)-iPr-pybox ligand. J. Organomet. Chem. 2011, 696, 1861–1867. 10.1016/j.jorganchem.2011.02.025. [DOI] [Google Scholar]
- Tubaro C.; Bertinazzo D.; Monticelli M.; Saoncella O.; Volpe A.; Basato M.; Badocco D.; pastore P.; Graiff C.; Venzo A. Synthesis and reactivity of cationic Bis(N-heterocyclic dicarbene) ruthenium(II) complexes. Eur. J. Inorg. Chem. 2014, 2014, 1524–1532. 10.1002/ejic.201301583. [DOI] [Google Scholar]
- Cotton F. A.; Yokochi A. Three reactions of Ru25+ compounds of the passlewheel type that lead to cleavage of the Ru-Ru bond. Inorg. Chim. Acta 1998, 275-276, 557–561. 10.1016/S0020-1693(98)00136-4. [DOI] [Google Scholar]
- Scolaro C.; Hartinger C. G.; Allardyce C. S.; Keppler B. K.; Dyson P. J. Hydrolysis study of the bifunctional antitumour compound RAPTA-C, [Ru(η6-p-cymene)Cl2(pta)]. J. Inorg. Biochem. 2008, 102, 1743–1748. 10.1016/j.jinorgbio.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Wang F.; Habtemariam A.; van der Geer E. P. L.; Fernández R.; Melchart M.; Deeth R. J.; Aird R.; Guichard S.; Fabbiani F. P. A.; Lozano-Casal P.; Oswald I. D. H.; Jodrell D. I.; Parsons S.; Sadler P. J. Controlling ligand substitution reactions of organometallic complexes: tuning cancer cell cytotoxicity. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 18269–18274. 10.1073/pnas.0505798102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scolaro C.; Bergamo A.; Brescacin Delfino L.; Delfino R.; Cocchietto M.; Laurenczy G.; Geldbach T. J.; Sava G.; Dyson P. J. In vitro and in vivo evaluation of ruthenium(II)-arene PTA complexes. J. Med. Chem. 2005, 48, 4161–4171. 10.1021/jm050015d. [DOI] [PubMed] [Google Scholar]
- Meier-Menches S. M.; Gerner C.; Berger W.; Hartinger C. G.; Keppler B. K. Structure–activity relationships for ruthenium and osmium anticancer agents – towards clinical development. Chem. Soc. Rev. 2018, 47, 909–928. 10.1039/C7CS00332C. [DOI] [PubMed] [Google Scholar]
- Biancalana L.; Batchelor L. K.; Dyson P. J.; Zacchini S.; Schoch S.; Pampaloni G.; Marchetti F. α-Diimine homologues of cisplatin: synthesis, speciation in DMSO/water and cytotoxicity. New J. Chem. 2018, 42, 17453–17463. 10.1039/C8NJ04195D. [DOI] [Google Scholar]
- Biancalana L.; Batchelor L. K.; Funaioli T.; Zacchini S.; Bortoluzzi M.; Pampaloni G.; Dyson P. J.; Marchetti F. α-Diimines as versatile, derivatizable ligands in ruthenium(II) p-cymene anticancer complexes. Inorg. Chem. 2018, 57, 6669–6685. 10.1021/acs.inorgchem.8b00882. [DOI] [PubMed] [Google Scholar]
- Kostrhunova H.; Petruzzella E.; Gibson D.; Kasparkova J.; Brabec V. A new anticancer Pt(IV) prodrug that acts by mechanisms involving DNA damage and different epigenetic effects. Chem.–Eur. J. 2019, 25, 5235–5245. 10.1002/chem.201805626. [DOI] [PubMed] [Google Scholar]
- Rothemund M.; Bär S. I.; Rehm T.; Kostrhunova H.; Brabec V.; Schobert R. Antitumoral effects of mitochondria-targeting neutral and cationic cis-[bis(1,3-dibenzylimidazol-2-ylidene)Cl(L)]Pt(II) complexes. Dalton Trans. 2020, 49, 8901–8910. 10.1039/D0DT01664K. [DOI] [PubMed] [Google Scholar]
- Marchetti F.; Di Nicola C.; Pettinari R.; Pettinari C.; Aiello I.; La Deda M.; Candreva A.; Morelli S.; De Bartolo L.; Crispini A. Zinc(II) complexes of acylpyrazolones decorated with a cyclohexyl group display antiproliferative activity against human breast cancer cells. Eur. J. Inorg. Chem. 2020, 2020, 1027–1039. 10.1002/ejic.201900775. [DOI] [Google Scholar]
- Akhmetova V. R.; Akhmadiev N. S.; Abdullin M. F.; Dzhemileva L. U.; Dyakonov V. A. Synthesis of new N,N-Pd(Pt) complexes based on sulfanyl pyrazoles, and investigation of their in vitro anticancer activity. RSC Adv. 2020, 10, 15116–15123. 10.1039/C9RA09783J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanda E.; Busto N.; Biancalana L.; Zacchini S.; Biver T.; Garcia B.; Marchetti F. Anticancer and antibacterial potential of robust Ruthenium(II) arene complexes regulated by choice of α-diimine and halide ligands. Chem. Biol. Interact. 2021, 344, 109522 10.1016/j.cbi.2021.109522. [DOI] [PubMed] [Google Scholar]
- Sáez R.; Lorenzo J.; Prieto M. J.; Font-Bardia M.; Calvet T.; Omeñaca N.; Vilaseca M.; Moreno V. Influence of PPh3 moiety in the anticancer activity of new organometallic ruthenium complexes. J. Inorg. Biochem. 2014, 136, 1–12. 10.1016/j.jinorgbio.2014.03.002. [DOI] [PubMed] [Google Scholar]
- Zeng L.; Gupta P.; Chen Y.; Wang E.; Ji L.; Chao H.; Chen Z.-S. The development of anticancer ruthenium(ii) complexes: from single molecule compounds to nanomaterials. Chem. Soc. Rev. 2017, 46, 5771–5804. 10.1039/C7CS00195A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K.; Zhao Z.-Z.; Bo H.-B.; Hao X.-J.; Wang J.-Q. Applications of ruthenium complex in tumor diagnosis and therapy. Front. Pharmacol. 2018, 9, 1323 10.3389/fphar.2018.01323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parveen S.; Hanif M.; Movassaghi S.; Sullivan M. P.; Kubanik M.; Shaheen M. A.; Söhnel T.; Jamieson S. M. F.; Hartinger C. G. Cationic Ru(η6-p-cymene) complexes of 3-hydroxy-4-pyr(id)ones – lipophilic triphenylphosphine as co-ligand is key to highly stable and cytotoxic anticancer agents. Eur. J. Inorg. Chem. 2017, 2017, 1721–1727. 10.1002/ejic.201601163. [DOI] [Google Scholar]
- Biancalana L.; Zacchini S.; Ferri N.; Lupo M. G.; Pampaloni G.; Marchetti F. Tuning the cytotoxicity of ruthenium(II) para cymene complexes by mono-substitution at a triphenylphosphine/phenoxydiphenylphosphine ligand. Dalton Trans. 2017, 46, 16589–16604. 10.1039/C7DT03385K. [DOI] [PubMed] [Google Scholar]
- Schoch S.; Batchelor L. K.; Funaioli T.; Ciancaleoni G.; Zacchini S.; Braccini S.; Chiellini F.; Biver T.; Pampaloni G.; Dyson P. J.; Marchetti F. Diiron complexes with a bridging functionalized allylidene ligand: synthesis, structural aspects, and cytotoxicity. Organometallics 2020, 39, 361–373. 10.1021/acs.organomet.9b00813. [DOI] [Google Scholar]; LogPow values for monocationic diiron complex [Fe2Cp2(CO)(μ-CO){μ-η1:η3-C(3-C6H4OH)CHCNMe2}]CF3SO3, [FeFe]+[CF3SO3]–, and the corresponding neutral complex derived from cyanide addition, [FeFe(CN)], are −0.34 and 1.00, respectively (UV-Vis measurements)
- Eastman A. Improving anticancer drug development begins with cell culture: misinformation perpetrated by the misuse of cytotoxicity assays. Oncotarget 2017, 8, 8854–8866. 10.18632/oncotarget.12673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzayans R.; Andrais B.; Murray D. Do multiwell plate high throughput assays measure loss of cell viability following exposure to genotoxic agents?. Int. J. Mol. Sci. 2017, 18, 1679. 10.3390/ijms18081679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddell I. A.; Lippard S. J.. Metallo-Drugs: Development and Action of Anticancer Agents; Sigel A.; Sigel H.; Freisinger E.; Sigel R. K. O., Eds.; De Gruyter: Berlin/Munich/Boston, 2018; pp 1–42. [Google Scholar]
- Novohradsky V.; Zerzankova L.; Stepankova J.; Vrana O.; Raveendran R.; Gibson D.; Kasparkova J.; Brabec V. New insights into the molecular and epigenetic effects of antitumor Pt(IV)-valproic acid conjugates in human ovarian cancer cells. Biochem. Pharmacol. 2015, 95, 133–144. 10.1016/j.bcp.2015.04.003. [DOI] [PubMed] [Google Scholar]
- Novohradsky V.; Zanellato I.; Marzano C.; Pracharova J.; Kasparkova J.; Gibson D.; Gandin V.; Osella D.; Brabec V. Epigenetic and antitumor effects of platinum(IV)-octanoato conjugates. Sci. Rep. 2017, 7, 3751 10.1038/s41598-017-03864-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabec V.; Kasparkova J. Ruthenium coordination compounds of biological and biomedical significance. DNA binding agents. Coord. Chem. Rev. 2018, 376, 75–94. 10.1016/j.ccr.2018.07.012. [DOI] [Google Scholar]
- Johnson N. P.; Butour J.-L.; Villani G.; Wimmer F. L.; Defais M.; Pierson V.; Brabec V.. Metal Antitumor Compounds: The Mechanism of Action of Platinum Complexes. In Ruthenium and Other Non-Platinum Metal Complexes in Cancer Chemotherapy, Progress in Clinical Biochemistry and Medicine; Springer, 1989; Vol. 10, pp 1–24. [Google Scholar]
- Brabec V.; Hrabina O.; Kasparkova J. Cytotoxic platinum coordination compounds. DNA binding agents. Coord. Chem. Rev. 2017, 351, 2–31. 10.1016/j.ccr.2017.04.013. [DOI] [Google Scholar]
- Abassi Y. A.; Xi B.; Zhang W. F.; Ye P. F.; Kirstein S. L.; Gaylord M. R.; Feinstein S. C.; Wang X. B.; Xu X. Kinetic cell-based morphological screening: prediction of mechanism of compound action and off-target effects. Chem. Biol. 2009, 16, 712–723. 10.1016/j.chembiol.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novohradsky V.; Yellol J.; Stuchlikova O.; Santana M. D.; Kostrhunova H.; Yellol G.; Kasparkova J.; Bautista D.; Ruiz J.; Brabec V. Organoruthenium complexes with C∧N ligands are highly potent cytotoxic agents that act by a new mechanism of action. Chem. - Eur. J. 2017, 23, 15294–15299. 10.1002/chem.201703581. [DOI] [PubMed] [Google Scholar]
- Novohradsky V.; Zerzankova L.; Stepankova J.; Kisova A.; Kostrhunova H.; Liu Z.; Sadler P. J.; Kasparkova J.; Brabec V. A dual-targeting, apoptosis-inducing organometallic half-sandwich iridium anticancer complex. Metallomics 2014, 6, 1491–1501. 10.1039/C4MT00112E. [DOI] [PubMed] [Google Scholar]
- Tan Q.; Yan X.; Song L.; Yi H.; Li P.; Sun G.; Yu D.; Li L.; Zeng Z.; Guo Z. Induction of mitochondrial dysfunction and oxidative damage by antibiotic drug doxycycline enhances the responsiveness of glioblastoma to chemotherapy. Med. Sci. Monit. 2017, 23, 4117–4125. 10.12659/MSM.903245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L.; Neuzil J. Targeting mitochondria as an anticancer strategy. Cancer Commun. 2019, 39, 63. 10.1186/s40880-019-0412-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.-T.; Lin K.-H.; Huang C.-J.; Wei A.-C. MitoTox: a comprehensive mitochondrial toxicity database. BMC Bioinf. 2021, 22, 369 10.1186/s12859-021-04285-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L.; Wen N.; Xia M.; Zhang Y. U.; Liu W.; Xu Y. E.; Sun L. Calcium efflux from the endoplasmic reticulum regulates cisplatin-induced apoptosis in human cervical cancer HeLa cells. Oncol. Lett. 2016, 11, 2411–2419. 10.3892/ol.2016.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgi C.; Agnoletto C.; Bononi A.; Bonora M.; De Marchi E.; Marchi S.; Missiroli S.; Patergnani S.; Poletti F.; Rimessi A.; Suski J. M.; Wieckowski M. R.; Pinton P. Mitochondrial calcium homeostasis as potential target for mitochondrial medicine. Mitochondrion 2012, 12, 77–85. 10.1016/j.mito.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterea A. M.; El Hiani Y.. The Role of Mitochondrial Calcium Signaling in the Pathophysiology of Cancer Cells. In Calcium Signaling: Advances in Experimental Medicine and Biology; Islam M., Ed.; Springer, 2020; Vol. 1131. [DOI] [PubMed] [Google Scholar]
- Graier W. F.; Malli R. Mitochondrial calcium: a crucial hub for cancer cell metabolism?. Transl. Cancer Res. 2017, 6, S1124–S1127. 10.21037/tcr.2017.08.28. [DOI] [Google Scholar]
- Dejos C.; Gkika D.; Cantelmo A. R. The two-way relationship between calcium and metabolism in cancer. Front. Cell Dev. Biol. 2020, 8, 573747 10.3389/fcell.2020.573747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Garcia S.; Prado-Garcia H. Mitochondrial calcium: transport and modulation of cellular processes in homeostasis and cancer. Int. J. Oncol. 2019, 54, 1155–1167. 10.3892/ijo.2019.4696. [DOI] [PubMed] [Google Scholar]
- Nathan S. R.; Wilson J. J. Synthesis and evaluation of a ruthenium-based mitochondrial calcium uptake inhibitor. J. Vis. Exp. 2017, 128, 56527 10.3791/56527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de J García-Rivas G.; Carvajal K.; Correa F.; Zazueta C. Ru360, a specific mitochondrial calcium uptake inhibitor, improves cardiac post-ischaemic functional recovery in rats in vivo. Br. J. Pharmacol. 2006, 149, 829–837. 10.1038/sj.bjp.0706932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods J. J.; Lovett J.; Lai B.; Harris H. H.; Wilson J. J. Redox stability controls the cellular uptake and activity of ruthenium-based inhibitors of the mitochondrial calcium uniporter. Angew. Chem., Int. Ed. 2020, 59, 1433–785. 10.1002/anie.202000247. [DOI] [PubMed] [Google Scholar]
- Woods J. J.; Nemani N.; Shanmughapriya S.; Kumar A.; Zhang M. Q.; Nathan S. R.; Thomas M.; Carvalho E.; Ramachandran K.; Srikantan S.; Stathopulos P. B.; Wilson J. J.; Madesh M. A selective and cell-permeable mitochondrial calcium uniporter (MCU) inhibitor preserves mitochondrial bioenergetics after hypoxia/reoxygenation injury. ACS Cent. Sci. 2019, 5, 153–166. 10.1021/acscentsci.8b00773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. G.; Pathan N.; Ethell I. M.; Krajewski S.; Yamaguchi Y.; Shibasaki F.; McKeon F.; Bobo T.; Franke T. F.; Reed J. C. Ca2+-induced apoptosis through calcineurin dephosphorylation of BAD. Science 1999, 284, 339–343. 10.1126/science.284.5412.339. [DOI] [PubMed] [Google Scholar]
- Carafoli E.; Molinari M. Calpain: a protease in search of a function?. Biochem. Biophys. Res. Commun. 1998, 247, 193–203. 10.1006/bbrc.1998.8378. [DOI] [PubMed] [Google Scholar]
- Høyer-Hansen M.; Bastholm L.; Szyniarowski P.; Campanella M.; Szabadkai G.; Farkas T.; Bianchi K.; Fehrenbacher N.; Elling F.; Rizzuto R.; Stenfeldt Mathiasen I.; Jäättelä M. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-beta, and Bcl-2. Mol. Cell 2007, 25, 193–205. 10.1016/j.molcel.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Zanoni M.; Piccinini F.; Arienti C.; Zamagni A.; Santi S.; Polico R.; Bevilacqua A.; Tesei A. 3D tumor spheroid models for in vitro therapeutic screening: a systematic approach to enhance the biological relevance of data obtained. Sci. Rep. 2016, 6, 19103 10.1038/srep19103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma C. R.; Zimmermann M.; Agarkova I.; Kelm J. M.; Krek W. 3D cell culture systems modeling tumor growth determinants in cancer target discovery. Adv. Drug Delivery Rev. 2014, 69–70, 29–41. 10.1016/j.addr.2014.03.001. [DOI] [PubMed] [Google Scholar]
- Wernitznig D.; Kiakos K.; Del Favero G.; Harrer N.; Machat H.; Osswald A.; Jakupec M. A.; Wernitznig A.; Sommergruber W.; Keppler B. K. First-in-class ruthenium anticancer drug (KP1339/IT-139) induces an immunogenic cell death signature in colorectal spheroids in vitro. Metallomics 2019, 11, 1044–1048. 10.1039/c9mt00051h. [DOI] [PubMed] [Google Scholar]
- Ravi M.; Paramesh V.; Kaviya S. R.; Anuradha E.; Solomon F.D.P. 3D cell culture systems: advantages and applications. J. Cell. Physiol. 2015, 230, 16–26. 10.1002/jcp.24683. [DOI] [PubMed] [Google Scholar]
- Reger D. L.; Grattan T.; Brown K. J.; et al. Syntheses of tris(pyrazolyl)methane ligands and {[tris(pyrazolyl)methane]Mn(CO)3}SO3CF3 complexes: comparison of ligand donor properties. J. Organomet. Chem. 2000, 607, 120–128. 10.1016/S0022-328X(00)00290-4. [DOI] [Google Scholar]
- Menges F.″Spectragryph - optical spectroscopy software″, Version 1.2.5, @ 2016-2017, http://www.effemm2.de/spectragryph.
- Fulmer G. R.; Miller A. J. M.; Sherden N. H.; Gottlieb H. E.; Nudelman A.; Stoltz B. M.; Bercaw J. E.; Goldberg K. I. NMR chemical shifts of trace impurities: common laboratory solvents, organics, and gases in deuterated solvents relevant to the organometallic chemist. Organometallics 2010, 29, 2176–2179. 10.1021/om100106e. [DOI] [Google Scholar]
- Willker W.; Leibfritz D.; Kerssebaum R.; Bermel W. Gradient selection in inverse heteronuclear correlation spectroscopy. Magn. Reson. Chem. 1993, 31, 287–292. 10.1002/mrc.1260310315. [DOI] [Google Scholar]
- Jutand A. The use of conductivity measurements for the characterization of cationic palladium(II) complexes and for the determination of kinetic and thermodynamic data in palladium-catalyzed reactions. Eur. J. Inorg. Chem. 2003, 2003, 2017–2040. 10.1002/ejic.200300069. [DOI] [Google Scholar]
- Geary W. J. The use of conductivity measurements in organic solvents for the characterisation of coordination compounds. Coord. Chem. Rev. 1971, 7, 81–122. 10.1016/S0010-8545(00)80009-0. [DOI] [Google Scholar]
- Sheldrick G. M. Crystal structure refinement with SHELXL. Acta Crystallogr., Sect. C: Struct. Chem. 2015, 71, 3–8. 10.1107/S2053229614024218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flack H. D. On enantiomorph-polarity estimation. Acta Crystallogr., Sect. A: Found. Crystallogr. 1983, 39, 876–881. 10.1107/S0108767383001762. [DOI] [Google Scholar]
- Spek A. L. Single-crystal structure validation with the program PLATON. J. Appl. Crystallogr. 2003, 36, 7–13. 10.1107/S0021889802022112. [DOI] [Google Scholar]
- Spek A. L. Structure validation in chemical crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2009, 65, 148–155. 10.1107/S090744490804362X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundlöf T.; Mathiasson M.; Bekiroglu S.; Hakkarainen B.; Bowden T.; Arvidsson T. Survey and qualification of internal standards for quantification by 1H NMR spectroscopy. J. Pharm. Biomed. Anal. 2010, 52, 645–651. 10.1016/j.jpba.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Rice N. M.; Irving H. M. N. H.; Leonard M. A. Nomenclature for liquid-liquid distribution (solvent extraction) (IUPAC Recommendations 1993). Pure Appl. Chem. 1993, 65, 2373–2396. 10.1351/pac199365112373. [DOI] [Google Scholar]
- OECD Guidelines for testing of chemicals, in OECD, Paris: 1995; Vol. 107
- Dearden J. C.; Bresnen G. M. The measurement of partition coefficients. Quant. Struct.-Act. Relat. 1988, 7, 133–144. 10.1002/qsar.19880070304. [DOI] [Google Scholar]
- Currie D. J.; Lough C. E.; Silver R. F.; Holmes H. L. Partition coefficients of some conjugated heteroenoid compounds and 1,4-naphthoquinones. Can. J. Chem. 1966, 44, 1035–1043. 10.1139/v66-154. [DOI] [Google Scholar]
- Calculated by the formula pD = pH* + 0.4, where pH* is the value measured for H2O-calibrated pH-meter121,122
- Westcott C. C.pH Measurements; Academic Press: New York, 1978. [Google Scholar]
- Covington A. K.; Paabo M.; Robinson R. A.; Bates R. G. Use of the glass electrode in deuterium oxide and the relation between the standardized pD (paD) scale and the operational pH in heavy water. Anal. Chem. 1968, 40, 700–706. 10.1021/ac60260a013. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.