Abstract
Cattle at high-risk for bovine respiratory disease on entry to western Canadian feedlots are often treated metaphylactically with antimicrobials from the macrolide class. High levels of resistance to macrolides have been reported in Mannheimia haemolytica isolates from clinical samples, but it is less clear whether this trend extends to the broader feedlot population. The objective was to describe near-term [< 40 days on feed (DOF)] changes in the recovery and susceptibility of M. haemolytica isolates from healthy feedlot calves after metaphylactic exposure to tulathromycin. Eight cohorts of 100 calves (n = 800) were sampled via deep nasopharyngeal swab at entry processing (i.e., before metaphylaxis, at 1 DOF) and again at 13 DOF. Ten calves from each cohort (n = 80) were randomly sampled a third time at 36 DOF. Recovery of M. haemolytica isolates across all cohorts increased over the study period, from 33% (95% CI: 26.5 to 40.2%) at 1 DOF to 75% (95% CI: 71.4 to 78.3%) at 36 DOF. A significant shift in the minimum inhibitory concentration (MIC) distribution of tulathromycin from 1 DOF (MIC90 ≤ 8 μg/mL) to 13 DOF (MIC90 > 64 μg/mL) was observed. A subset of 36 isolates from 13 DOF screened for macrolide resistance genes via multiplex polymerase chain reaction all harbored the msrE and mphE genes. Recovery of M. haemolytica at 13 and 36 DOF did not decline in response to metaphylactic use of tulathromycin; conversely, we inferred the potential for rapid inter-pen spread of a macrolide-resistant clone by 13 DOF in 6 of 8 pens under selective pressure from antimicrobial use.
Résumé
Changements dans la sensibilité phénotypique des isolats de Mannheimia haemolytica aux antibiotiques de la classe des macrolides au début de la période d’alimentation après l’utilisation métaphylactique de tulathromycine chez les veaux des parcs d’engraissement de l’Ouest canadien. Les bovins à risque élevé de maladies respiratoires bovines à leur entrée dans les parcs d’engraissement de l’Ouest canadien sont souvent traités métaphylactiquement avec des antimicrobiens de la classe des macrolides. Des taux élevés de résistance aux macrolides ont été signalés chez les isolats de Mannheimia haemolytica provenant d’échantillons cliniques, mais il est moins clair si cette tendance s’étend à la population plus large des parcs d’engraissement. L’objectif était de décrire les changements à court terme [< 40 jours d’alimentation (DOF)] dans la récupération et la sensibilité des isolats de M. haemolytica provenant de veaux sains en parc d’engraissement après une exposition métaphylactique à la tulathromycine. Huit cohortes de 100 veaux (n = 800) ont été échantillonnées via un prélèvement nasopharyngé profond lors du traitement d’entrée (i.e., avant la métaphylaxie, à 1 DOF) et à nouveau à 13 DOF. Dix veaux de chaque cohorte (n = 80) ont été échantillonnés au hasard une troisième fois à 36 DOF. La récupération des isolats de M. haemolytica dans toutes les cohortes a augmenté au cours de la période d’étude, passant de 33 % (IC 95 % : 26,5 à 40,2 %) à 1 DOF à 75 % (IC 95 % : 71,4 à 78,3 %) à 36 DOF. Un changement significatif dans la distribution de la concentration minimale inhibitrice (MIC) de la tulathromycine de 1 DOF (MIC90 ≤ 8 μg/mL) à 13 DOF (MIC90 > 64 μg/mL) a été observé. Un sous-ensemble de 36 isolats de 13 DOF criblés pour les gènes de résistance aux macrolides via une réaction d’amplification en chaîne par polymérase multiplex hébergeaient tous les gènes msrE et mphE. L’isolement de M. haemolytica à 13 et 36 DOF n’a pas diminué en réponse à l’utilisation métaphylactique de la tulathromycine; à l’inverse, nous avons suggéré le potentiel de propagation rapide entre les enclos d’un clone résistant aux macrolides par 13 DOF dans 6 des 8 enclos sous la pression sélective de l’utilisation d’antimicrobiens.
(Traduit par Dr Serge Messier)
Introduction
Bovine respiratory disease (BRD) is a multifactorial disease characterized by complex interactions between bacterial and viral pathogens, a variable host immune response, and ecological conditions arising from a multiphasic beef production system (1). Consistent prevention of BRD is described as difficult and costly (2), owing to the many known risk factors that combine to predispose feedlot cattle to acute respiratory disease (3). In the absence of sensitive and rapid diagnostic tools to support more targeted treatment decisions, the beef finishing industry depends on antimicrobial metaphylaxis to control the incidence of acute BRD in high-risk cattle (4). In a recent study of 36 western Canadian feedlots, most individually dosed antimicrobial use (AMU) was administered as metaphylaxis to address the risk of BRD; furthermore, cattle categorized as being high risk for developing BRD were more likely than low-risk cattle to receive macrolides for BRD metaphylaxis (5). Results from a network meta-analysis of injectable antibiotic options at feedlot arrival suggested that macrolide antimicrobials (e.g., tulathromycin) were the most effective approach for reducing BRD incidence in feedlot settings (6).
Mannheimia haemolytica is one of several bacterial pathogens implicated in the clinical presentation of BRD. Most M. haemolytica isolates (88%) recovered from a large Canadian survey (2007 to 2010) of mixed-risk feedlot cattle sampled at arrival and > 30 d on feed (DOF) were pan-susceptible to 21 tested antimicrobials, and distributions of minimum inhibitory concentrations (MICs) were representative of a highly susceptible bacterial population (7). In the same 3-year period after tulathromycin, a 15-membered ring macrolide, was approved for use in Canadian feedlot cattle, more than 99% of M. haemolytica isolates recovered at arrival and > 60 DOF were susceptible to a concentration of tulathromycin several orders of magnitude lower than the resistance breakpoint (≥ 64 μg/mL) (8). A more recent review concerning the prevalence of antimicrobial resistance (AMR) in M. haemolytica described a general decrease in the susceptibility of isolates to antimicrobial drugs, including macrolides, in the last decade (9); in particular, high levels of resistance were reported for M. haemolytica isolates recovered from American cattle exposed to metaphylactic macrolides in stocker facilities at < 30 DOF (10). There was relatively limited resistance to tulathromycin (< 5%) in M. haemolytica isolates from beef cattle in a recent survey (2017 to 2019) of mixed-origin animals on entry to Alberta feedlots (11), but samples in that study were exclusively collected before administration of antimicrobials.
This report highlights preliminary results of interest from a larger-scale effort to develop strategies that actualize the potential of genomic diagnostics to support prudent antimicrobial drug selection in beef production (12). The objective was to describe changes in the recovery and susceptibility to macrolides of M. haemolytica isolates from healthy feedlot calves after metaphylactic exposure to tulathromycin. A related objective concerns identification of resistance genes associated with reduced susceptibility to macrolides in select isolates in which follow-up analyses might offer important insights. Throughout the text, susceptibility to macrolides is discussed with reference to the relevant subclass defined by the size of the lactone ring (i.e., 15- or 16-membered) and the hypothesized similarity in prevalence of resistance. This work fills a knowledge gap as it pertains to AMR in BRD on Canadian feedlots, given that longitudinal data from multiple points in the early feeding period (< 60 DOF) is underrepresented in the published literature.
Materials and methods
The study protocol and procedures described here were approved by the Animal Care Committee at the University of Saskatchewan (AUP 20190069). Recently weaned steers of various beef breeds were sourced from a local auction market and shipped to a research feedlot at the Livestock and Forage Centre of Excellence (LFCE) in Clavet, Saskatchewan. One cohort of 100 calves was purchased each week for 8 wk, from October 6th through December 1st, 2020. The sampled animals originated from a combined total of 292 unique farming operations, ensuring a heterogeneous mix of calves were present in each cohort. Calves were kept in a holding pen overnight after arriving at the LFCE and processed the following morning; their weight ranged from 211 to 291 kg (mean: 253 kg). All calves (N = 800) were processed following an industry standard protocol that included placement of an identification ear tag, verification of castration, and subcutaneous administration of the following: a Mannheimia haemolytica and modified-live viral vaccine (Pyramid 5 + Presponse; Boehringer Ingelheim Animal Health, Burlington, Ontario), a clostridial vaccine (Ultrachoice 7; Zoetis, Kirkland, Quebec), and metaphylactic tulathromycin at a dose of 2.5 mg/kg based on the average weight of the cohort (Draxxin; Zoetis). These calves also received a growth implant (Ralgro; Merck Animal Health, Kirkland, Quebec), and a topical anthelmintic (Solmectin; Solvet, Calgary, Alberta).
After processing, the cohort of 100 calves was moved to its home pen where it remained intact (i.e., no re-sorting) for the duration of the 45-day study. The outdoor, dirt-floor study pens were filled consecutively and adjacent pen pairs shared fence-line water troughs. The presence of a satellite building created a “buffer zone” that separated the first 4 pens from the last 4 pens. Calves were fed a balanced, high-forage starter feed ration and monensin (33 mg/kg of dry matter); no other in-feed antimicrobials were administered to prevent or manage disease. Experienced feedlot personnel monitored the animals daily for signs of illness. A post-metaphylactic interval of 7 d was observed, after which sick calves were treated in accordance with established feedlot protocols. Relatively few animals became sick with BRD in this study (n = 28), and no more than 7 clinical cases were identified in any single pen-cohort across the feeding period. The overall rates of BRD morbidity and mortality were 3.5 and 0.25%, respectively.
The same 800 calves were sampled at time of processing (Day 1 on feed) and again 12 d later (Day 13 on feed). A random subset of 10 calves from each pen-cohort were sampled a third time (n = 80), at 36 DOF. At each time point, calves were restrained in a hydraulic chute and sampled via 3 deep nasopharyngeal (DNP) swabs; a neck extender was used to stabilize the calves’ heads during sampling. A single-use paper towel was used to wipe clean the external nares, and a double-guarded culture swab (Continental Plastic, Delevan, Wisconsin, USA) was directed into the ventral meatus of the nostril. The polyester-tipped swab was advanced through the inner sheath and vigorously rotated against the pharyngeal mucosa for 5 to 6 rotations. The swab was withdrawn into the inner sheath and outer guard before removal from the nostril, and approximately 3 cm of swab tip was cut and placed in a vial containing 3 mL of liquid Amies transport medium. Two additional samples were obtained from alternating nostrils using the same procedure, and all 3 DNP swabs per calf were pooled in the same vial. Sample collectors used clean gloves for each calf.
Samples were transported to the University of Saskatchewan for same-day processing. Pooled DNP samples were vortexed for 1 min and a 300 μL aliquot was submitted to Prairie Diagnostic Services (PDS) for bacterial culture and susceptibility testing. One each of sheep blood and chocolate agar plates were inoculated with 10 μL of sample and incubated at 35°C in 5% CO2 for 48 h. Bacterial colonies were examined at 18 and 42 h of incubation; colonies suspected of being Mannheimia haemolytica or other respiratory isolates of interest were confirmed using a MALDI-TOF MS Microflex LT instrument and MALDI Biotyper software (Bruker Corporation, Billerica, Massachusetts, USA). A single M. haemolytica colony from each sample was tested for susceptibility to antimicrobials via serial broth microdilution using the Sensititre platform and commercially available Bovine BOPO7F AST Plate (ThermoFisher Scientific, Waltham, Massachusetts, USA). The recovery and susceptibility data were exported from a central repository to Stata/IC 14.2 for descriptive and statistical analyses. Null binomial response models were fit using generalized estimating equations (xtgee) to account for clustering of observations by pen-cohort. The MICs were interpreted with reference to the breakpoints established by the Clinical Laboratory Standards Institute (CLSI). Isolates with intermediate MIC values were categorized as “susceptible” for all analyses.
A subset of macrolide-resistant M. haemolytica isolates from the second time point (13 DOF) were purposively selected for follow-up polymerase chain reaction (PCR) analyses. Representative gamithromycin and tulathromycin-resistant isolates from calves with disparate farms of origin were identified from 6 of 8 pen-cohorts (6 isolates per cohort, n = 36 isolates). Isolates were revived from frozen stocks on sheep blood agar, and broth cultures were grown overnight in brain heart infusion media containing 1% (w/v) glucose at 37°C with shaking. Genomic DNA was extracted using a modified salting out procedure (13), and ethanol-precipitated DNA was resuspended in 100 μL of TE buffer [10 mM Tris-HCl (pH 8.0), 1 mM EDTA] and stored at 4°C overnight. The concentration of genomic DNA was determined the next day by measuring absorbance at 260 nm.
Selected isolates were screened for macrolide resistance genes erm42, mphE, and msrE using a multiplex PCR assay, as described (14). Expected product sizes were 173 bp (erm42), 271 bp (mphE), and 395 bp (msrE). Primers that amplified a region of the 23S rRNA gene of gammaproteobacteria (720 bp in M. haemolytica) were likewise present in the assay and served as an internal control. Positive controls for the erm42, mphE, and msrE primer sets in the multiplex assay were synthesized DNA fragments containing a sequence unrelated to the actual target [partial cpn60 sequences (15)] flanked by annealing sites for the forward and reverse primers. Products generated from the positive control templates differed in size from the actual targets (282, 334, and 509 bp, respectively) to permit detection of cross-contaminated samples. Following synthesis (Integrated DNA Technologies), DNA segments were ligated into vector pGem T Easy (Promega) and the resultant plasmid constructs were used to transform Escherichia coli DH5α cells. Inserts were confirmed by sequencing, and purified plasmids were used as positive amplification controls. No-template controls were also included in PCR experiments. Following PCR, products were resolved by agarose gel electrophoresis for visual interpretation of results.
Results
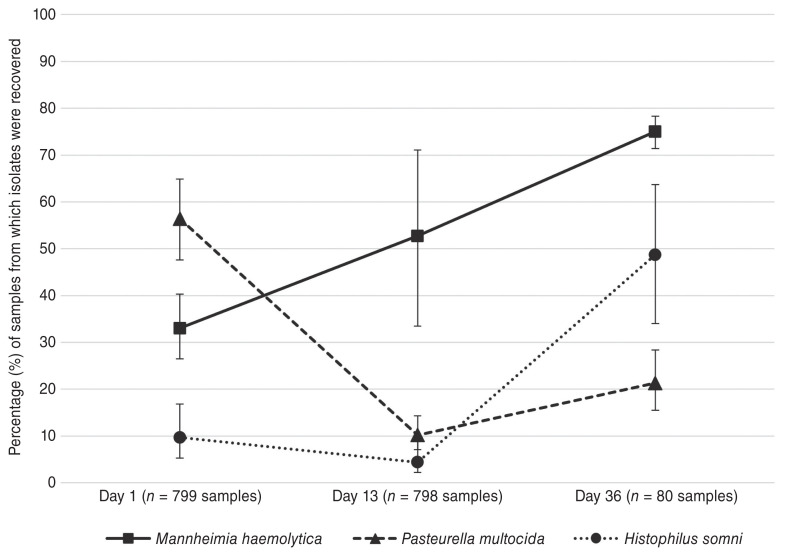
Percentages of DNP samples from which M. haemolytica and other BRD-associated pathogens of interest were recovered are displayed in Figure 1. Population-averaged estimates are shown with 95% confidence intervals (CI) adjusted for the clustering of observations by pen-cohort. The recovery of M. haemolytica isolates across all pen-cohorts increased over the study period, from 33% (95% CI: 26.5 to 40.2%) at 1 DOF, to 52.7% (33.5 to 71.1%) at 13 DOF, and 75% (71.4 to 78.3%) at 36 DOF. Although inter-cohort variability in M. haemolytica detection was most pronounced at 13 DOF, substantial increases in recovery at the second time point were nevertheless observed in 6 of 8 pen-cohorts. In contrast, recovery of the other BRD pathogens decreased from 1 to 13 DOF; the decline was especially marked for P. multocida [56.4% (47.5 to 64.8%) at 1 DOF to 10.2% (7.1 to 14.3%) at 13 DOF]. The recovery of both pathogens rebounded at 36 DOF and was particularly marked for H. somni [4.4% (2.2 to 8.8%) at 13 DOF to 48.7% (34.0 to 63.7%) at 36 DOF].
Figure 1.
Percentage (%) of samples from which M. haemolytica, P. multocida, and H. somni isolates were recovered at 1, 13, and 36 DOF. The number of samples submitted was n = 799 at 1 DOF, n = 798 at 13 DOF, and n = 80 at 36 DOF. Proportions are shown with 95% confidence intervals, adjusted for clustering by pen-cohort.
The number and raw, unadjusted percentage of recovered and macrolide-resistant M. haemolytica isolates at each time point are reported in Table 1. Population-averaged estimates of AMR prevalence were calculated for M. haemolytica isolates from all pen-cohorts at 1 DOF (n = 264), 13 DOF (n = 421), and 36 DOF (n = 60). One M. haemolytica isolate [0.4%, (95% CI: 0.8 to 2.1%)] at the first time point was resistant to both gamithromycin and tulathromycin (i.e., 15-membered ring macrolides). At 13 DOF, resistance to gamithromycin and tulathromycin increased significantly to 67% (range: 44.6 to 83.6%) and 65.6% (range: 45.4 to 81.3%), respectively. An increase in the prevalence of resistance to 16-membered ring macrolide tilmicosin [5.2% (range: 2.0 to 12.5%)] was less marked, and only 3 isolates [0.7% (range: 0.1 to 4.0%)] were resistant to tildipirosin. Resistance at 36 DOF was 73.6% (range: 57.8 to 85.1%), 67.6% (range: 43.8 to 84.8%), 11.6% (range: 4.4 to 27.2%), and 1.6% (range: 0.3 to 10.0%) for gamithromycin, tulathromycin, tilmicosin, and tildipirosin, respectively. The considerably smaller number of isolates at this time point is reflected in the relative uncertainty around these prevalence estimates. Two or fewer M. haemolytica isolates were resistant to any of ampicillin, florfenicol, or tetracycline at any time point; resistance to Category 1 antimicrobials, including third-generation cephalosporins and fluoroquinolones, was not observed. The percentage of M. haemolytica isolates with inhibited growth at the only tested sulphadimethoxine concentration (256 μg/mL) decreased from 91.7% at 1 DOF to 12.4% at 13 DOF.
Table 1.
Number of DNP samples, recovered M. haemolytica (Mh) isolates, and macrolide-resistant Mh isolates across all pen-cohorts at each of 1, 13, and 36 DOF. The unadjusted percentages of total and resistant isolates are reported in parentheses.
| Sampling time point | |||
|---|---|---|---|
|
|
|||
| 1 DOF | 13 DOF | 36 DOF | |
| Number of animals sampled | 799 | 798 | 80 |
| Number of Mh isolates recovered (% of total DNP samples) | 264 (33.0) | 421 (52.8) | 60 (75.0) |
| Number of resistant Mh isolates (% of total Mh isolates) | |||
| 15-membered ring macrolides | |||
| Gamithromycin | 1 (0.4) | 351 (83.4) | 44 (73.3) |
| Tulathromycin | 1 (0.4) | 341 (81.0) | 40 (66.7) |
| 16-membered ring macrolides | |||
| Tildipirosin | 0 (0) | 3 (0.7) | 1 (1.7) |
| Tilmicosin | 0 (0) | 25 (5.9) | 7 (11.7) |
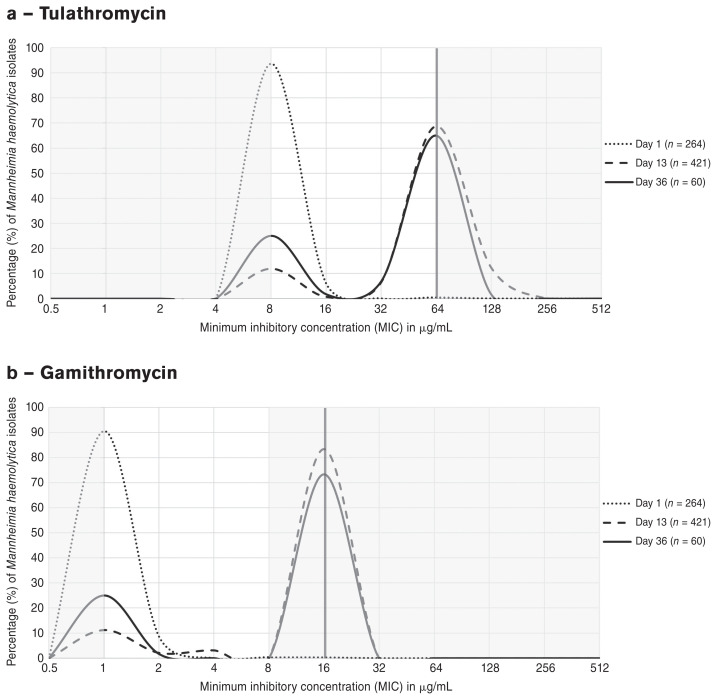
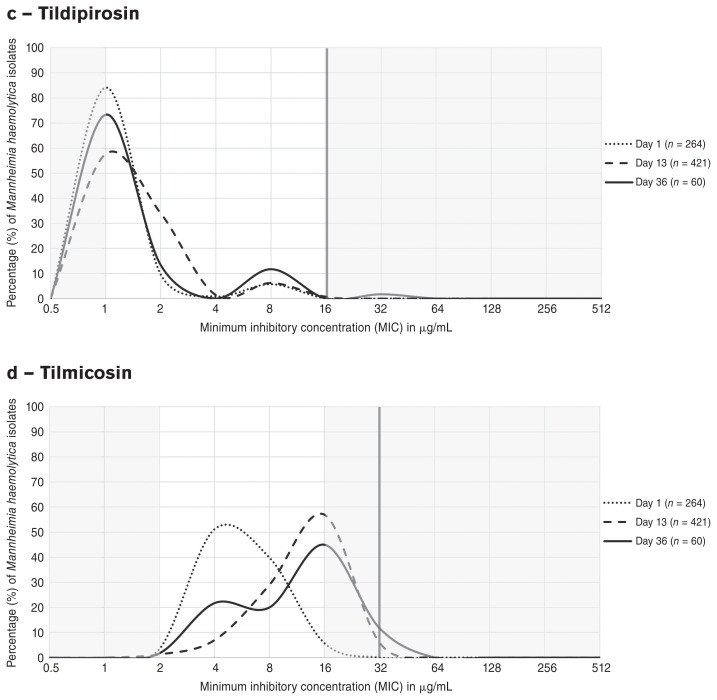
Distributions of minimum concentrations of macrolide antimicrobials that inhibited growth of M. haemolytica isolates at 1, 13, and 36 DOF are depicted as line graphs in Figure 2 a–d. A dramatic shift in the MIC distribution of tulathromycin (a) from 1 DOF (MIC90 ≤ 8 μg/mL) to 13 DOF (MIC90 > 64 μg/mL) was observed; a rightward shift of similar magnitude was noted for gamithromycin (b), for which the MIC90 increased from ≤ 1 to > 8 μg/mL between the same time points. This trend was also present to a lesser extent for tilmicosin (d) and was captured by the unimodal shift from 1 DOF (MIC50 = 4 μg/mL) to 13 DOF (MIC50 = 16 μg/mL). A marked decrease from 1 DOF (> 80%) to 13 DOF (< 60%) in the proportion of M. haemolytica isolates with inhibited growth at the lowest concentration of tildipirosin (c) was likewise apparent. At 36 DOF, a modest “reverse” shift toward increased susceptibility was observed to varying degrees for all 4 macrolides; at this time point, the lowest concentration of antimicrobial on the BOPO7F plate inhibited growth of a larger proportion of M. haemolytica isolates than at 13 DOF.
Figure 2.
Shifts in the MIC distributions of macrolide antimicrobials (a) tulathromycin, (b) gamithromycin, (c) tildipirosin, and (d) tilmicosin for Mannheimia haemolytica isolates from all pen-cohorts at 1 DOF (n = 264, dotted lines), 13 DOF (n = 421, dashed lines) and 36 DOF (n = 60, solid lines). The vertical red lines and shaded grey areas represent the resistance breakpoints established by CLSI and the concentrations outside the range tested for the macrolides on the BOPO7F plate, respectively.
The MIC of gamithromycin was > 8 μg/mL for all M. haemolytica isolates selected for PCR analyses; the MIC of tulathromycin was 64 μg/mL and > 64 μg/mL for n = 31 and n = 5 selected isolates, respectively. Results from positive and negative control PCRs and the internal amplification control were as expected. All tested isolates from 13 DOF were PCR positive for the gammaproteobacterial 23S rRNA target and macrolide-resistance genes msrE and mphE. The erm42 gene was not detected in any selected isolate.
Discussion
Much of the available literature evaluating AMR in BRD-associated pathogens draws on clinical or post-mortem samples from diseased cattle exposed to multiple courses and/or classes of antimicrobials (9). However, results presented here contribute to a more fulsome understanding of the recovery and susceptibility of M. haemolytica isolates in the general feedlot population following metaphylaxis with tulathromycin, particularly as it pertains to the early feeding period (< 40 DOF). A nationally representative surveillance effort initiated in 2019 and led by the Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) reported on AMR in M. haemolytica isolates detected on feedlot arrival and at animal rehandling (16); there was significant variation in the timing of rehandling, but it occurred later in the feeding period on average (i.e., 60 DOF, often at re-implantation of cattle) [Sheryl Gow, written communication]. The second sampling time point in this study (13 DOF) was selected to coincide with the maximum estimated duration of action for injectable tulathromycin (14 d) (17) and is reflective of the immediate-term impacts on the nasopharyngeal microbiota in metaphylactically treated cattle. The third sampling time point (36 DOF) was chosen to investigate medium-term trends in recovery and susceptibility within the larger study’s 45-day timeline.
Mannheimia haemolytica and other BRD-associated pathogens were isolated in greater proportions in this study than in other recent investigations of healthy Canadian feedlot cattle from mixed origins (11,18). The unadjusted prevalence of M. haemolytica in beef-type cattle was 13.8% in a cross-sectional survey of over 2800 Canadian animals at feedlot entry (11), consistent with published Canadian estimates (7,8). This team isolated M. haemolytica from 33% of cattle from all pen-cohorts at arrival processing, an estimate more comparable to those reported for auction-sourced calves entering Saskatchewan feedlots (28 to 40%) (19,20). Injectable tulathromycin appeared to limit colonization of commercial feedlot cattle with Mannheimia spp. through ≥ 60 DOF in (21); indeed, reductions in M. haemolytica recovery at 76 DOF (20%) and ≥ 90 DOF (23%) were described for tulathromycin-treated cattle in references (19) and (20), respectively. In contrast, the prevalence of M. haemolytica increased from feedlot arrival (10%) to 40 DOF (55%) in the experimental group of ranch-direct calves receiving metaphylactic gamithromycin, a 15-membered ring macrolide (18). Recovery of M. haemolytica at 13 DOF did not decline in response to the metaphylaxis in this study, and an increase in prevalence from arrival to 36 DOF (75%) was observed across all 8 pen-cohorts of tulathromycin-treated cattle. Given the very few calves that became sick in this study, there was no evidence that this increase was associated with calf morbidity or post-metaphylactic treatments for BRD.
Several Canadian studies reported high levels of resistance to macrolides (> 70%) in M. haemolytica isolates from BRD cases (22) and from the lower respiratory tract of tulathromycin-treated cattle (23). It is less clear whether these trends extend to the broader feedlot population and the healthy animals included therein; for example, metaphylactic treatment of 2 experimental groups (18) did not appear to select for macrolide resistance through 40 DOF, as evidenced by the generally low MICs for all BRD-associated pathogens. Conversely, significant increases in resistance to tulathromycin (20 to 36%) and tilmicosin (28%) were reported at follow-up sampling of commercial feedlot calves exposed to a metaphylactic macrolide (19,20). In this study, significant increases in resistance to 15-membered ring macrolides (65 to 67%) at 13 DOF were akin to those described by Snyder et al (24) in their investigation of AMR in M. haemolytica isolates from American stocker cattle that received metaphylactic tulathromycin. When high-risk calves were sampled via DNP 10 to 14 d after entry processing at the time of revaccination, upwards of 98% of the tested isolates from 123 culture-positive calves were resistant to both gamithromycin and tulathromycin (24). Similarly substantial increases in the prevalence of resistance to tilmicosin (99.2%), enrofloxacin (99.2%), and florfenicol (68.3%) (24) were not present in the M. haemolytica isolates at 13 or 36 DOF in the study herein.
The timing of second and/or subsequent sample collection might explain the apparent contrast between the recovery and AMR prevalence data reported here and the generally lower estimates from previous investigations of healthy feedlot cattle in Canada (7,8,18–20). Specifically, the concurrence of this second sample with both peak BRD occurrence and the upper limit of metaphylactic drug activity (4,17) distinguished this from other longitudinal studies of AMR prevalence later in the feeding period (40 to 180 DOF). Noyes et al (7) speculated that the likelihood of M. haemolytica recovery increased when metaphylactic AMU in high-risk cattle selected for resistant bacterial populations that spread among pen-mates. Although the prevalence of phenotypic AMR was negligible at 1 DOF in this study, perhaps relevant resistant clones were not identified on arrival given that: i) representativeness of only 1 sample of the respiratory tract flora cannot be guaranteed (25); and ii) only a single M. haemolytica colony from each culture-positive sample was selected for susceptibility testing. Differential expression of AMR genes owing to stress, subclinical illness, and/or exposure to antimicrobials might likewise impacted the dynamics reported here (26,27). Planned analyses to investigate clonal relatedness of M. haemolytica isolates across time will help this team to better understand the role of contagious spread in metaphylactically treated cattle.
An examination of the MIC distributions by antimicrobial drug can illuminate complex trends in susceptibility beyond the susceptible/resistant dichotomy established by clinical breakpoints; these distributions are more often included as tables (7,22) than as plots in Figures 2 a–d. Depicted this way, it is possible to visualize the dramatic reductions in susceptibility to macrolides across the population of M. haemolytica isolates under selective pressure from metaphylactic tulathromycin (13 DOF). The partial return to phenotypic susceptibility in the absence of AMU exposure at 36 DOF is likewise more easily appreciated with this format. The MIC distribution for tylosin, a 16-membered ring macrolide, revealed little in the way of susceptibility trends. Across all time points, the MIC90 for tylosin was > 32 μg/mL and therefore outside of the upper limit of concentrations tested for this drug. There were no significant shifts across time in the distributions of MICs for all other tested antimicrobial drugs, with the notable exception of sulphadimethoxine. The proportion of M. haemolytica isolates which did not grow at the only tested sulphadimethoxine concentration shifted in tandem with gamithromycin and tulathromycin at 13 and 36 DOF; this was suggestive of a co-selection mechanism resulting in decreased susceptibility to both 15-membered ring macrolides and sulfonamides in these isolates.
The near total absence of resistance to tetracycline in the M. haemolytica isolates from this study was unexpected. Resistance to oxytetracycline (MIC ≥ 8 μg/mL) was the most frequently observed phenotype (10.0%) in 281 M. haemolytica isolates from mixed-origin beef cattle at arrival to feedlot in (11); in contrast, there were no oxytetracycline-resistant isolates at 1 DOF in this study. Furthermore, only a single oxytetracycline-resistant isolate was detected at either 13 or 36 DOF. This deviated markedly from the comparatively higher prevalence of oxytetracycline resistance in isolates from the rehandling time points in both the 2020 CIPARS feedlot surveillance data (22.9%) (Sheryl Gow, written communication) and a recent study of auction-sourced feedlot cattle in Saskatchewan (52.3%) (19). Antimicrobials in the tetracycline class comprise most of both parenteral and in-feed AMU on western Canadian feedlots (5,22), and cattle in the comparison studies had likely or confirmed exposures to metaphylactic or in-feed (19) tetracyclines as part of their management protocols. It is notable that the cattle in this study did not receive in-feed chlortetracycline for prevention of histophilosis, an observation that might suggest a potential role for continuous, subtherapeutic AMU in the maintenance of tetracycline resistance in M. haemolytica isolates (21,27).
Genes conferring resistance to macrolides (erm42, mphE, msrE) have been identified on integrative conjugative elements (ICEs) in M. haemolytica isolates from North American cattle (28). Various combinations of these antimicrobial resistance genes (ARGs) were detected in clinical Pasteurellaceae isolates via multiplex PCR and linked to distinct resistance phenotypes to the 15- and 16-membered ring macrolides in (14,29). Screening with the assay described in (14) revealed that all representative M. haemolytica isolates from 13 DOF harbored both msrE and mphE, genes which are organized in an operon-like structure and sufficient to confer greatly elevated MICs to both gamithromycin (> 8 μg/mL) and tulathromycin (≥ 64 μg/mL) in this study as elsewhere (14,29,30). Elevated MICs to tildipirosin were not detected in M. haemolytica isolates lacking erm42 in (14,31), a finding that is mirrored here and emphasized the potential for distinct pathways to resistance by macrolide subclass. These genetic data were suggestive of the rapid inter-pen spread of a macrolide-resistant M. haemolytica clone by 13 DOF. In their study of isolates from BRD-affected cattle treated with gamithromycin, DeDonder et al (30) noted that a single genetic subtype was associated with most observed macrolide resistance; however, the erm42 gene was not detected, and the msrE-mphE operon was part of an ICE in all cases.
Ongoing genetic characterization of these isolates will help to clarify whether the msrE-mphE genes were associated with other ARGs, including possibly sul2, a gene linked to reduced susceptibility to sulfonamides and frequently located on mobile genetic elements (MGEs) (21,32). Furthermore, and because in-feed exposure to antimicrobials may have broader implications for the selection of particular multi-drug resistant phenotypes, planned analyses might reveal differences in either size or composition of ICE/MGE in these calves compared to those fed chlortetracycline and/or tylosin. Mutations in the 23S rRNA conferring high-level resistance to macrolides in Pasteurellaceae species have been identified (28,33) and might also have an as-of-yet unrecognized role in the resistance patterns described here. Conjugal transfer of ICE-linked ARGs between species of the Pasteurellaceae family has been reported (34), but phenotypic evidence suggests it is unlikely to have featured prominently through 36 DOF in this group of calves. Resistance to macrolides was not detected at any time point for either P. multocida or H. somni (data not shown) and decreases in recovered proportions of these isolates following metaphylaxis were consistent with highly susceptible bacterial populations.
Findings reported here offered unique insights into dynamics of AMR in BRD-associated pathogens immediately after metaphylactic drug exposure. This work was distinct from the existing literature in this field in several respects, including: i) its focus on the general feedlot population in the early feeding period (< 40 DOF) and therefore near-term impacts of metaphylaxis on nasopharyngeal microbiota; and ii) its longitudinal design, whereby the same animals from 8 large pen-cohorts were systematically sampled across multiple time points. These results advanced our understanding of the population-level impact on AMR of exposure to metaphylactic macrolides in calves at high risk for BRD.
Acknowledgments
This research was conducted with funding from the Saskatchewan Agriculture Development Fund, and with graduate student support from the Western College of Veterinary Medicine. The authors extend their gratitude to the many colleagues who contributed their expertise to the collection, handling, and processing of these samples. CVJ
Footnotes
Use of this article is limited to a single copy for personal study. Anyone interested in obtaining reprints should contact the CVMA office (hbroughton@cvma-acmv.org) for additional copies or permission to use this material elsewhere.
References
- 1.Hilton WM. BRD in 2014: Where have we been, where are we now, and where do we want to go? Anim Health Res Rev. 2014;15:120–122. doi: 10.1017/S1466252314000115. [DOI] [PubMed] [Google Scholar]
- 2.Griffin D. Antibiotic metaphylaxis to control respiratory disease. Cattle Production Library CL-606. 2006. [Last accessed June 7, 2022]. Available from: http://www.4cattlemen.com/ncba2007/newsroom/PR102GriffinAntibiotic.pdf.
- 3.Taylor JD, Fulton RW, Lehenbauer TW, Step DL, Confer AW. The epidemiology of bovine respiratory disease: What is the evidence for predisposing factors? Can Vet J. 2010;51:1095–1102. [PMC free article] [PubMed] [Google Scholar]
- 4.Ives SE, Richeson JT. Use of antimicrobial metaphylaxis for the control of bovine respiratory disease in high-risk cattle. Vet Clin North Am Food Anim Pract. 2015;31:341–350. doi: 10.1016/j.cvfa.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 5.Brault SA, Hannon SJ, Gow SP, et al. Antimicrobial use on 36 beef feedlots in western Canada: 2008–2012. Front Vet Sci. 2019;6:329. doi: 10.3389/fvets.2019.00329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Connor AM, Hu D, Totton SC, et al. A systematic review and network meta-analysis of injectable antibiotic options for the control of bovine respiratory disease in the first 45 days post arrival at the feedlot. Anim Health Res Rev. 2019;20:163–181. doi: 10.1017/S1466252320000031. [DOI] [PubMed] [Google Scholar]
- 7.Noyes NR, Benedict KM, Gow SP, et al. Mannheimia haemolyticain feedlot cattle: Prevalence of recovery and associations with antimicrobial use, resistance, and health outcomes. J Vet Intern Med. 2015;29:705–713. doi: 10.1111/jvim.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alexander TW, Cook S, Klima CL, Topp E, McAllister TA. Susceptibility to tulathromycin in Mannheimia haemolytica isolated from feedlot cattle over a 3-year period. Front Microbiol. 2013;4:297. doi: 10.3389/fmicb.2013.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Credille B. Antimicrobial resistance in Mannheimia haemolytica: Prevalence and impact. Anim Health Res Rev. 2020;21:196–199. doi: 10.1017/S1466252320000109. [DOI] [PubMed] [Google Scholar]
- 10.Woolums AR, Karisch BB, Frye JG, et al. Multidrug resistant Mannheimia haemolytica isolated from high-risk beef stocker cattle after antimicrobial metaphylaxis and treatment for bovine respiratory disease. Vet Microbiol. 2018;221:143–152. doi: 10.1016/j.vetmic.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 11.Andrés-Lasheras S, Ha R, Zaheer R, et al. Prevalence and risk factors associated with antimicrobial resistance in bacteria related to bovine respiratory disease — A broad cross-sectional study of beef cattle at entry into Canadian feedlots. Front Vet Sci. 2021;8:710. doi: 10.3389/fvets.2021.692646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.University of Saskatchewan. Genomic ASSETS for Livestock. [Last accessed June 7, 2022]. Available from: https://research-groups.usask.ca/genomic-assets/index.php.
- 13.Martin-Platero AM, Valdivia E, Maqueda M, Martinez-Bueno M. Fast, convenient, and economical method for isolating genomic DNA from lactic acid bacteria using a modification of the protein “salting-out” procedure. Anal Biochem. 2007;366:102–104. doi: 10.1016/j.ab.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 14.Rose S, Desmolaize B, Jaju P, Wilhelm C, Warrass R, Douthwaite S. Multiplex PCR to identify macrolide resistance determinants in Mannheimia haemolytica and Pasteurella multocida. Antimicrob Agents Ch. 2012;56:3664–3669. doi: 10.1128/AAC.00266-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vancuren SJ, Hill JE. Update on cpnDB: A reference database of chaperonin sequences. Database (Oxford) 2019 doi: 10.1093/database/baz033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beef Cattle Research Council. Antibiotic resistance in bovine respiratory disease bacteria. [Last accessed June 7, 2022]. Available from: https://www.beefresearch.ca/factsheet.cfm/antibiotic-resistance-in-bovine-respiratory-disease-bacteria-315.
- 17.Apley M. Bovine respiratory disease: Pathogenesis, clinical signs, and treatment in lightweight calves. Vet Clin North Am Food Anim Pract. 2006;22:399–411. doi: 10.1016/j.cvfa.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 18.Guo Y, McMullen C, Timsit E, et al. Genetic relatedness and antimicrobial resistance in respiratory bacteria from beef calves sampled from spring processing to 40 days after feedlot entry. Vet Micro. 2020;240:108478. doi: 10.1016/j.vetmic.2019.108478. [DOI] [PubMed] [Google Scholar]
- 19.Wennekamp TR. Biosecurity and bovine respiratory disease on beef operations in western Canada [Master’s thesis] Saskatoon, Saskatchewan: University of Saskatchewan; 2020. [Google Scholar]
- 20.Erickson NEN, Ngeleka M, Lubbers BV, Trokhymchuk A. Changes in the rates of field isolation and antimicrobial susceptibility of bacterial pathogens collected from fall-placed feedlot steers between arrival at the feedlot and 90 to 120 days on feed. Bovine Pract. 2017;51:165–173. [Google Scholar]
- 21.Holman DB, Timsit E, Booker CW, Alexander TW. Injectable antimicrobials in commercial feedlot cattle and their effect on the nasopharyngeal microbiota and antimicrobial resistance. Vet Micro. 2018;214:140–147. doi: 10.1016/j.vetmic.2017.12.015. [DOI] [PubMed] [Google Scholar]
- 22.Anholt RM, Klima C, Allan N, et al. Antimicrobial susceptibility of bacteria that cause bovine respiratory disease complex in Alberta, Canada. Front Vet Sci. 2017;4:207. doi: 10.3389/fvets.2017.00207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Timsit E, Hallewell J, Booker C, Tison N, Amat S, Alexander TW. Prevalence and antimicrobial susceptibility of Mannheimia haemolytica, Pasteurella multocida, and Histophilus somni isolated from the lower respiratory tract of healthy feedlot cattle and those diagnosed with bovine respiratory disease. Vet Micro. 2017;208:118–125. doi: 10.1016/j.vetmic.2017.07.013. [DOI] [PubMed] [Google Scholar]
- 24.Snyder E, Credille B, Berghaus R, Giguère S. Prevalence of multi drug antimicrobial resistance in Mannheimia haemolytica isolated from high-risk stocker cattle at arrival and two weeks after processing. J Anim Sci. 2017;95:1124–1131. doi: 10.2527/jas.2016.1110. [DOI] [PubMed] [Google Scholar]
- 25.Lubbers BV, Turnidge J. Antimicrobial susceptibility testing for bovine respiratory disease: Getting more from diagnostic results. Vet J. 2015;203:149–154. doi: 10.1016/j.tvjl.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 26.Poole K. Bacterial stress responses as determinants of antimicrobial resistance. J Antimicrob Chemother. 2012;67:2069–2089. doi: 10.1093/jac/dks196. [DOI] [PubMed] [Google Scholar]
- 27.Food and Agriculture Organization of the United Nations Drivers, dynamics and epidemiology of antimicrobial resistance in animal production. 2016. [Last accessed June 7, 2022]. Available from: https://www.fao.org/3/i6209e/i6209e.pdf.
- 28.Klima CL, Holman DB, Cook SR, et al. Multidrug resistance in Pasteurellaceae associated with bovine respiratory disease mortalities in North America from 2011 to 2016. Front Microbiol. 2020;11:2817. doi: 10.3389/fmicb.2020.606438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beker M, Rose S, Lykkebo CA, Douthwaite S. Integrative and conjugative elements (ICEs) in Pasteurellaceae species and their detection by multiplex PCR. Front Microbiol. 2018;9:1329. doi: 10.3389/fmicb.2018.01329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeDonder KD, Harhay DM, Apley MD, et al. Observations on macrolide resistance and susceptibility testing performance in field isolates collected from clinical bovine respiratory disease cases. Vet Microbiol. 2016;192:186–193. doi: 10.1016/j.vetmic.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 31.Michael GB, Eidam C, Kadlec K, et al. Increased MICs of gamithromycin and tildipirosin in the presence of the genes erm(42) and msr(E)-mph(E) for bovine Pasteurella multocida and Mannheimia haemolytica. J Antimicrob Chemother. 2012;67:1555–1557. doi: 10.1093/jac/dks076. [DOI] [PubMed] [Google Scholar]
- 32.Clawson ML, Murray RW, Sweeney MT, et al. Genomic signatures of Mannheimia haemolytica that associate with the lungs of cattle with respiratory disease, an integrative conjugative element, and antibiotic resistance genes. BMC Genomics. 2016;17:1–14. doi: 10.1186/s12864-016-3316-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olsen AS, Warrass R, Douthwaite S. Macrolide resistance conferred by rRNA mutations in field isolates of Mannheimia haemolytica and Pasteurella multocida. J Antimicrob Chemother. 2015;70:420–423. doi: 10.1093/jac/dku385. [DOI] [PubMed] [Google Scholar]
- 34.Klima CL, Zaheer R, Cook SR, et al. Pathogens of bovine respiratory disease in North American feedlots conferring multidrug resistance via integrative conjugative elements. J Clin Microbiol. 2014;52:438–448. doi: 10.1128/JCM.02485-13. [DOI] [PMC free article] [PubMed] [Google Scholar]