Abstract
Background:
Hazardous alcohol consumption has significant adverse medical consequences. These effects may be mediated, in part, by alterations in DNA methylation. Thus, DNA methylation signatures in peripheral cells may provide biomarkers of the medical impact of alcohol use and the risk for future alcohol consumption.
Method:
Using a high density methylation array, we characterized epigenome-wide DNA methylation in saliva cells with respect to alcohol consumption in a large cohort of male European American veterans. In this study, DNA methylation of over 870,000 CpG DNA sites was profiled in 1,135 European American men. Alcohol consumption was assessed using the Alcohol Use Disorder Identification Test-Consumption (AUDIT-C). Linear regression was applied in an epigenome-wide association study (EWAS), adjusted for confounders. Gene set enrichment analysis was performed in the KEGG database with a correction for gene length.
Results:
We found that a total of 70 CpG sites reached EWAS-corrected significance (p<6E-08) with small effects on alcohol consumption for individual CpG sites, including 64 new CpG sites and six CpG sites that were previously reported as associated with alcohol use disorder, liver function, body mass index, and lipid metabolism. The most significant CpG site was located in SLC7A11(t=−11.34, p=2.66E-28), a gene involved specifically in cysteine and glutamate transportation. The 70 significant CpG sites were located on 44 genes, including genes involved in amino acid transport and metabolism systems. We identified 68 pathways with a false discovery rate <0.05.
Conclusion:
We identified novel DNA methylation sites associated with alcohol consumption. Results may shed light on peripheral mechanisms of alcohol consumption on adverse health outcomes among heavy drinkers.
Keywords: Alcohol consumption, DNA methylation, Epigenome-wide Association Study (EWAS)
Introduction
Alcohol consumption is a significant risk factor for psychiatric and medical diseases and is the seventh leading cause of death and disability worldwide (Burton and Sheron, 2018, Collaborators, 2018). Many of the mechanisms whereby alcohol consumption exerts adverse effects on health outcomes are unclear. Emerging evidence suggests that high levels of alcohol consumption alters epigenetic markers in both the central nervous system and peripheral tissues(Liu et al., 2018, Lussier et al., 2018, Berkel and Pandey, 2017), suggesting a role of epigenetic regulation consequent to alcohol exposure that may contribute to alcohol-related health outcomes.
DNA methylation (DNAm), the most commonly studied epigenetic mechanism, involves a process in which a methyl group is added to the cytosine pyrimidine without changing DNA sequence. DNAm is modulated by DNA methyltransferase which transfers a methyl group from S-adenosyl methionine (SAM) to the 5-position of cytosine in the context of cytosine-phosphate-guanine (CpG) dinucleotide. Hazardous alcohol consumption often causes folate and vitamin B deficiency; these are cofactors in methyl transfer reactions, and deficiency reduces the level of SAM(Williams et al., 1949). Thus, it has been hypothesized that DNAm alterations by hazardous alcohol consumption plays a critical role in regulating gene function(Berkel and Pandey, 2017, Palmisano and Pandey, 2017, Tulisiak et al., 2017).
In animals, decreased methylase activity and DNA hypomethylation of CpG sites were found in ethanol-treated developing brains(Garro et al., 1991). DNAm signatures at the nucleus accumbens core of male rhesus macaques reportedly differentiated light and heavy alcohol consumption(Cervera-Juanes et al., 2017). Alcohol exposure resulting in DNAm perturbation in developing brain can persist into adulthood in mice(Zhang et al., 2015). In humans, due to the limitation of access to living human brain, the majority of DNAm studies for the purpose of identifying biomarkers for alcohol consumption, as well as for guiding treatment have been conducted in peripheral cells (Lussier et al., 2018, Liu et al., 2018). DNAm of candidate genes that previously identified for a risk of Alcohol Use Disorder (AUD) through genetic association studies have been reported in relation to AUD and fetal alcohol spectrum disorder diagnoses, symptom severity, alcohol withdrawal, and treatment response (Zhang and Gelernter, 2017, Bruckmann et al., 2016, Heberlein et al., 2013, Hillemacher et al., 2015, van der Knaap et al., 2014, Laufer et al., 2015, Portales-Casamar et al., 2016). However, these studies were conducted in small samples and the findings were inconsistent. Recently, epigenome-wide association studies (EWAS) of DNAm in peripheral blood have identified widespread DNAm changes in the epigenome in individuals with hazardous alcohol use or AUD. The majority of the reported DNAm loci were found to be hypomethylated in individuals with AUD relative to those without AUD (Philibert et al., 2014, Bruckmann et al., 2017). The largest meta-EWAS to date in 13,317 individuals – a rather heterogenous sample -- identified hundreds of DNAm sites. A selected set of 144 CpGs enabled differentiation of alcohol drinking and non-alcohol drinking subjects, suggesting that DNAm sites could serve as biomarkers for alcohol consumption(Liu et al., 2018). A study conducted in isolated CD3+ T cells revealed 48 CpG sites associated with AUD treatment response(Bruckmann et al., 2017).
A notable limitation of DNAm candidate gene studies is that DNAm loci identified in such studies are rarely replicated in EWAS(Zhang and Gelernter, 2017). The findings of DNAm for AUD are also inconsistent among the reported EWAS to date, which may be due to either lack of power(Philibert et al., 2014, Hagerty et al., 2016), lack of correction of cell type effects on DNAm (Hagerty et al., 2016), using relatively low density of CpG array in epigenome(Zhang et al., 2013), and the sample heterogeneity expected to result from meta-analysis. Here, we report an EWAS for alcohol consumption in a comparatively large sample of U.S. veterans (N= 1,135) using a high density methylation array with 850,000 CpG sites in the epigenome. Because of the available source of DNA, the study aimed to understand peripheral mechanisms of alcohol consumption rather than exploring the neural mechanism of AUD, except to the extent that peripheral measures reflect central activity. We focused on European Americans to limit the confounding effects of ethnicity. Because the study population was predominantly male U.S. military veterans, we excluded women in our analysis to rule out sex effects on DNAm, lacking the power necessary to examine this important question.
Materials and methods
Study population
Our study sample included 1,135 European American (EA) male U.S. military veterans who participated in the National Health and Resilience in Veterans Study (NHRVS)(Stefanovics et al., 2018, Fuehrlein et al., 2018). The NHRVS is a nationally-representative study of U.S. veterans, where recruitment was conducted in October-December 2011. Participants were recruited from a research panel comprising more than 50,000 households developed and maintained by GfK Knowledge Networks, Inc. (Menlo Park, CA, USA). All participants provided informed consent. Demographic and substance use characteristics are presented in Table 1. This study was approved by the Human Subjects Subcommittee of the Veterans Affairs (VA) Connecticut Healthcare System and VA Office of Research & Development.
Table 1.
Demographic and clinical characteristics between non-hazardous andhazardous alcohol consumption
| None-hazardous Alcohol consumption (N = 807) | Hazardous alcohol consumption (N =328) | p-value | |
|---|---|---|---|
| Age (mean ± sd) | 64.70 ± 11.96 | 63.30 ± 12.64 | 8.55E-02 |
| Sex (male, %) | 100 | 100 | 1.0 |
| AUDIT_C (mean ± sd) | 1.12 ± 1.08 | 5.33 ± 1.94 | 3.10E-133 |
| Alcohol use disorder (%) | 42.5 | 62.5 | 1.49E-09 |
| Current smoking (%) | 9.91 | 17.38 | 6.77E-04 |
| Nicotine dependences (%) | 20.57 | 21.95 | 6.62E-01 |
| Drug use disorder (%) | 11.28 | 19.21 | 5.79E-04 |
| BMI (mean ± sd) | 29.15 ± 5.28 | 27.49 ± 4.47 | 1.09E-07 |
Continuous variables: two groups t-test
Binomial variables: two proportions z-Test
Significnace: P-value < 5.00E-02
AUDIT_C: Alcohol Use Disorder Identification Test; BMI: Body Mass Index
Alcohol consumption
Alcohol consumption was assessed via self-report using the Alcohol Use Disorder Identification Test-Consumption (AUDIT-C)(Bush et al., 1998). Scores on the AUDIT-C range from 0–12. The mean score in our samples was 2 ± 2.4 and 28.7% (N=326) screened positive for hazardous alcohol consumption with a cutoff score =>4. The distribution of AUDIT-C scores is shown in Supplemental Figure S1.
DNA methylation quantification and quality control
Genomic DNA (500 ng) was extracted from saliva samples using Oragene kits (DNA Genotek, Ottawa, Ontario, Canada) and treated with bisulfite reagents included in the EZ-96 DNAm kit (Zymo Research, Orange, CA, USA) following the manufacturer’s standard protocol. Bisulfite-converted DNA samples were used in the array-based DNAm assay, the Illumina Infinium Human MethylationEPIC BeadChip (Illumina, San Diego, CA, USA), which interrogates DNAm at >850,000 loci genome-wide. DNAm profiling was conducted at the Yale Center for Genome Analysis. The data processing and analytical framework is presented in Figure S2.
Quality control was conducted in R version 3.4.1 and performed based on a pipeline using the ‘minfi’ R package (Bioconductor 1.8.9)(Aryee et al., 2014). To ensure that only high-confidence probes are included, probes with detection p-value > 0.001 were removed. We also removed probes: 1) with annotated single nucleotide polymorphisms (SNPs) at SBE/CpG sites (via the Single Nucleotide Polymorphism database 137 [National Center for Biotechnology Information), 2) cross-hybridizing probes, and 3) sex chromosome probes. The Combat method in the ‘sva’ package(Leek et al., 2012) was applied to correct for batch effects to adjust for sample plate and cohort group. Data were normalized using the functional normalization method(Aryee et al., 2014) implemented in the ‘minfi’ R package, which uses internal control probes present on the array to control for between-array technical variation and known to outperform other existing approaches(Fortin et al., 2014). A total of 756,573 were left for subsequent analysis.
Cell type proportion in the peripheral saliva samples (e.g. CD34, CD14, and buccal cells) was estimated using a modified version of the Houseman and colleagues method(Houseman et al., 2012, Smith et al., 2015). To adjust for possible population stratification, a methylation-based principal component (PC) approach(Barfield et al., 2014) was conducted based on sets of CpG sites within 50 kb of SNPs using the 1000 Genomes Project variants with minor allele frequency (MAF) > 0.1(Barfield et al., 2014). The first 10 PCs were used in the analysis as covariates.
Statistical analysis
All statistical analyses were performed within R 3.4.0 (www.r-project.org). EWAS analysis was conducted using the ‘cpg.assoc’ function from the ‘minfi’ R package(Barfield et al., 2012), adjusted for age, current smoking status, and estimated cell proportions. We found no significant differences in estimated cell proportions between the non-hazardous and hazardous alcohol consumption groups (ps > 0.05). These covariates were nevertheless adjusted in the model as a “best practice” to correct for potential confounding effects. We also adjusted top the 10 PCs in the model to correct the effects of population admixture. The significance level was set at nominal p=6E-08, which is equivalent to Bonferroni correction for a significance level of p=0.05. Considering a possible close relationship between body weight and alcohol consumption, we did an additional analysis to test whether body mass index (BMI) confounded the effect of alcohol consumption on DNAm. (However note that in our sample, BMI was lower in the Hazardous alcohol consumption group than in the non-Hazardous alcohol consumption group – Table 1.) Further, we tested associations of EWAS-significant CpG sites with hazardous alcohol consumption that was defined with AUDIT-C score >=4. Methylation CpG sites associated with alcohol consumption at the p<5E-04 level (level chosen empirically to ensure a statistic power with a sufficient gene set) were selected for gene set enrichment analysis. Considering that gene length may lead to bias of gene-set enrichment analysis as a result of different numbers of CpG sites in different genes (Geeleher et al., 2013), we adjusted the gene length by using missMethyl program in BioConductor (http://bioconductor.org/packages/release/bioc/html/missMethyl.html). False discovery rate (FDR) < 0.05 was a cutoff for statistical significance.
Results
Epigenome-wide association study for alcohol consumption
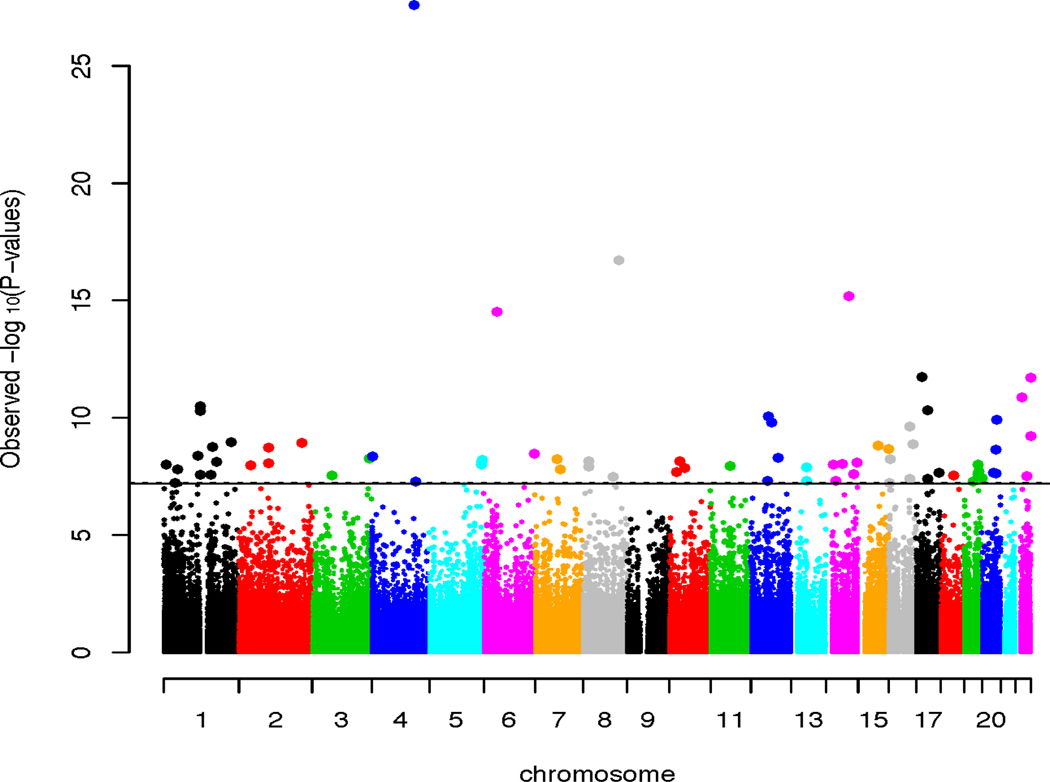
We identified 70 epigenome-wide significant (EWS) CpGs associated with AUDIT-C scores (Figure 1 and Table 2). Six of these 70 CpG sites were previously reported as associated with alcohol consumption or other alcohol-related phenotypes such as liver function and body weight, and 64 CpG sites were novel for alcohol consumption. Fifty-one out of 70 CpGs were located in a gene region: as expected, a majority of significant CpGs was in a gene body, while 16 CpGs were in promoter or UTR regions. Unlike previous reports showing that significantly more hypomethylation was associated with alcohol consumption(Liu et al., 2018, Hagerty et al., 2016), we found that methylation level was inversely associated with AUDIT-C scores at 37 CpG sits while methylation level was positively associated with AUDIT-C at 33 CpG sites.
Figure 1.
Manhattan plot of epigenome-wide association analysis with 756,573 CpG sites for alcohol consumption. X-axis shows chromosome locations of CpG sites. Y-axis shows -log p value for each CpG site. The line indicates threshold of epigenome-wide significance equivalent to Bonferroni correction (pnomial < 6E-08). The study was conducted for salivary DNA in 1,135 male veterans with European ancestry.
Table 2.
Significant CpG sites associated with alcohol consumption in 1,135 male European American veterans with and without adjusted for body mass index
| probe | CHR | Position | Gene | GROUP | CPG_ISLAND | t | p | t_adjBMI | p-adjBMI |
|---|---|---|---|---|---|---|---|---|---|
| cg06690548 | 4 | 139162808 | SLC7A11 | Body | −11.35 | 2.66E-28 | −11.70 | 7.43E-30 | |
| cg15837522 | 8 | 117892654 | −8.64 | 1.92E-17 | −8.96 | 1.42E-18 | |||
| cg06088069 | 14 | 75895604 | JDP2 | 5UTR | S_Shore | −8.20 | 6.5E-16 | −8.29 | 3.41E-16 |
| cg18120259 | 6 | 43894639 | LOC100132354 | Body | −8.00 | 3.05E-15 | −8.87 | 3E-18 | |
| cg15696816 | 17 | 17713886 | RAI1 | 3UTR | Island | 7.13 | 1.87E-12 | 7.30 | 5.34E-13 |
| cg12944146 | 22 | 50725609 | PLXNB2 | Body | Island | 7.12 | 1.97E-12 | 7.11 | 2.03E-12 |
| cg16834011 | 22 | 19931790 | COMT | 5UTR | S_Shelf | −6.84 | 1.33E-11 | −6.88 | 9.98E-12 |
| cg14476101 | 1 | 120255992 | PHGDH | Body | S_Shore | −6.71 | 3.17E-11 | −7.17 | 1.38E-12 |
| cg11637712 | 17 | 36665844 | ARHGAP23 | Body | N_Shore | 6.65 | 4.63E-11 | 6.74 | 2.62E-11 |
| cg16246545 | 1 | 120255941 | PHGDH | Body | S_Shore | −6.64 | 4.95E-11 | −7.13 | 1.88E-12 |
| cg08228578 | 12 | 57624193 | SHMT2 | Body | S_Shore | −6.55 | 8.91E-11 | −6.99 | 4.88E-12 |
| cg10589813 | 20 | 48809978 | S_Shore | −6.50 | 1.25E-10 | −6.68 | 3.74E-11 | ||
| cg21912872 | 12 | 68055270 | DYRK2 | 3UTR | −6.45 | 1.62E-10 | −6.03 | 2.3E-09 | |
| cg06469895 | 16 | 69418206 | TERF2 | Body | N_Shore | −6.39 | 2.39E-10 | −5.97 | 3.3E-09 |
| cg23041250 | 22 | 50725605 | PLXNB2 | Body | Island | 6.25 | 6.02E-10 | 6.16 | 1.02E-09 |
| cg05318849 | 1 | 223351252 | 6.15 | 1.1E-09 | 6.18 | 8.87E-10 | |||
| cg10440877 | 2 | 208378475 | −6.13 | 1.2E-09 | −5.71 | 1.41E-08 | |||
| cg19138349 | 16 | 80040260 | 6.12 | 1.33E-09 | 5.59 | 2.86E-08 | |||
| cg26170244 | 15 | 65176353 | 6.09 | 1.55E-09 | 5.67 | 1.85E-08 | |||
| cg16702725 | 1 | 161227516 | PCP4L1 | TSS1500 | N_Shore | 6.07 | 1.76E-09 | 6.16 | 9.99E-10 |
| cg05722266 | 2 | 96991791 | ITPRIPL1 | TSS200 | S_Shore | −6.06 | 1.9E-09 | −5.96 | 3.31E-09 |
| cg22411961 | 15 | 99318650 | IGF1R | Body | −6.04 | 2.13E-09 | −5.55 | 3.52E-08 | |
| cg01764256 | 20 | 45985832 | ZMYND8 | TSS200 | −6.03 | 2.21E-09 | −5.92 | 4.28E-09 | |
| cg03849851 | 6 | 166980629 | RPS6KA2 | Body | 5.96 | 3.32E-09 | 5.59 | 2.83E-08 | |
| cg00035969 | 1 | 112159097 | N_Shelf | 5.93 | 4.1E-09 | 5.79 | 8.99E-09 | ||
| cg01903185 | 4 | 1808586 | FGFR3 | Body | S_Shore | 5.92 | 4.37E-09 | 5.83 | 7.34E-09 |
| cg00508575 | 12 | 90050967 | ATP2B1 | TSS1500 | 5.89 | 5.02E-09 | 5.74 | 1.24E-08 | |
| cg09326269 | 3 | 189429439 | TP63 | Body | 5.88 | 5.29E-09 | 5.57 | 3.17E-08 | |
| cg04577162 * | 7 | 73667397 | RFC2 | Body | N_Shore | −5.87 | 5.79E-09 | −5.18 | 2.67E-07 |
| cg04725426 | 16 | 2131752 | TSC2 | Body | N_Shelf | 5.87 | 5.86E-09 | 5.90 | 4.75E-09 |
| cg16968128 | 5 | 176810555 | SLC34A1 | TSS1500 | 5.85 | 6.31E-09 | 5.74 | 1.22E-08 | |
| cg21998542 | 10 | 33605101 | NRP1 | Body | −5.84 | 6.93E-09 | −5.91 | 4.64E-09 | |
| cg07020366 | 8 | 19246614 | SH2D4A | Body | 5.83 | 7.19E-09 | 5.37 | 9.71E-08 | |
| cg06644515 | 1 | 173834831 | SNORD47/SNORD80 | TSS1500 | N_Shelf | −5.82 | 7.5E-09 | −5.44 | 6.49E-08 |
| cg17962756 | 5 | 172769199 | −5.82 | 7.91E-09 | −5.65 | 2.1E-08 | |||
| cg21050667 | 14 | 103624533 | 5.82 | 7.91E-09 | 5.59 | 2.8E-08 | |||
| cg06739059 | 2 | 96991826 | ITPRIPL1 | 5UTR | S_Shore | −5.80 | 8.65E-09 | −5.68 | 1.75E-08 |
| cg13409544 | 14 | 53159218 | ERO1A | Body | N_Shelf | 5.79 | 9.34E-09 | 5.63 | 2.32E-08 |
| cg26250989 | 1 | 6333502 | ACOT7 | Body | 5.78 | 9.55E-09 | 5.57 | 3.28E-08 | |
| cg13526915 | 14 | 24164078 | −5.78 | 9.75E-09 | −5.85 | 6.4E-09 | |||
| cg07463740 | 5 | 172737556 | −5.77 | 1.01E-08 | −5.55 | 3.63E-08 | |||
| cg10052860 | 19 | 45189438 | 5.77 | 1.01E-08 | 5.37 | 9.76E-08 | |||
| cg05190930 | 2 | 38701258 | LOC101929596 | Body | 5.77 | 1.04E-08 | 5.57 | 3.22E-08 | |
| cg13823144 | 11 | 66475721 | SPTBN2 | Body | 5.76 | 1.1E-08 | 5.68 | 1.71E-08 | |
| cg13825056 | 8 | 17186545 | MTMR7 | Body | −5.74 | 1.22E-08 | −5.53 | 4E-08 | |
| cg11408657 | 13 | 49147568 | 5.74 | 1.25E-08 | 5.76 | 1.09E-08 | |||
| cg27141751 | 10 | 50231850 | C10orf72 | Body | 5.72 | 1.39E-08 | 5.52 | 4.27E-08 | |
| cg03533141 | 1 | 45243518 | SNORD38A | TSS1500 | S_Shore | −5.70 | 1.54E-08 | −5.20 | 2.43E-07 |
| cg07446048 | 7 | 82353289 | −5.69 | 1.58E-08 | −5.20 | 2.41E-07 | |||
| cg07626482 | 19 | 47289503 | SLC1A5 | TSS1500 | N_Shore | −5.66 | 1.91E-08 | −6.02 | 2.34E-09 |
| cg00375132 | 10 | 22768370 | S_Shore | −5.65 | 2.02E-08 | −5.45 | 6.22E-08 | ||
| cg05476232 | 17 | 74361501 | 5.64 | 2.12E-08 | 5.43 | 6.8E-08 | |||
| cg13564967 | 20 | 36771794 | TGM2;TGM2 | Body | 5.64 | 2.17E-08 | 5.39 | 8.76E-08 | |
| cg01385679 * | 20 | 45033155 | ELMO2 | 5UTR | N_Shore | −5.63 | 2.27E-08 | −5.23 | 2.08E-07 |
| cg16940062 | 14 | 91763798 | CCDC88C | Body | 5.61 | 2.5E-08 | 5.76 | 1.11E-08 | |
| cg09109179 | 19 | 45268492 | 5.61 | 2.53E-08 | 5.49 | 5.06E-08 | |||
| cg26457483 | 1 | 120256112 | PHGDH | Body | S_Shore | −5.60 | 2.73E-08 | −5.99 | 2.79E-09 |
| cg19458269 * | 1 | 155949421 | ARHGEF2 | TSS1500 | S_Shore | −5.60 | 2.74E-08 | −5.31 | 1.32E-07 |
| cg24644830 * | 3 | 66533732 | LRIG1 | Body | −5.59 | 2.84E-08 | −5.043 | 5.36E-07 | |
| cg01589532 * | 22 | 38696599 | CSNK1E | Body | 5.57 | 3.13E-08 | 5.507 | 4.55E-08 | |
| cg06838966 | 8 | 98852306 | LAPTM4B | Body | 5.57 | 3.28E-08 | 5.51 | 4.55E-08 | |
| cg11701312 | 19 | 58897497 | RPS5 | TSS1500 | N_Shore | −5.54 | 3.75E-08 | −5.11 | 3.86E-07 |
| cg13545705 * | 16 | 67312573 | PLEKHG4 | TSS1500 | N_Shore | −5.53 | 4E-08 | −4.11 | 4.29E-05 |
| cg12192847 * | 17 | 36641717 | ARHGAP23 | Body | 5.53 | 4.04E-08 | 5.53 | 3.94E-08 | |
| cg12772028 | 14 | 33080373 | AKAP6 | Body | 5.50 | 4.7E-08 | 5.44 | 6.6E-08 | |
| cg02583484 * | 12 | 54677008 | HNRNPA1 | Body | S_Shelf | −5.50 | 4.83E-08 | −4.10 | 4.46E-05 |
| cg25989526 * | 13 | 49147573 | 5.49 | 4.87E-08 | 5.26 | 1.70E-07 | |||
| cg08151621 * | 19 | 28995456 | LOC100420587 | Body | −5.48 | 5.25E-08 | −5.22 | 2.16E-07 | |
| cg04425005 | 4 | 146783253 | ZNF827 | Body | −5.48 | 5.28E-08 | −5.52 | 4.32E-08 | |
| cg10254445 | 1 | 37197260 | −5.46 | 5.75E-08 | −5.40 | 7.98E-08 |
Note:
CpG sites with suggestive epigenom-wide significnace after adjusted for body mass index (BMI)
Multiple CpG sites in promoter regions reached epigenome-wide significance. For example, cg16834011 in the 5’UTR of COMT was inversely associated with alcohol consumption (t=−6.84; p=1.33E-11), while cg16702725 in TSS1500 region of PCP4L1 was positively associated with alcohol consumption (t=6.07; p=1.76E-09). Other CpGs in the promoter regions included cg01764256 in ZMYND8, cg00508575 in ATP2B1, cg06644515 in SNORD80, cg06739059 in ITPRIPL1, cg07626482 in SLC1A5, and cg11701312 in RPS5.
The most significant CpG, cg06690548, was located in the gene body of SLC7A11 (t=−11.35, p=2.66E-28), and was inversely associated with AUDIT-C score. This gene encodes a member of a heteromeric, sodium-independent, anionic amino acid transport system that is highly specific for cysteine and glutamate. Another CpG site in a sodium transporter gene, SLC1A5, was also inversely associated with alcohol consumption (cg07626482, t= −5.66; p=1.91E-08) while CpG, cg16968128, located at in a sodium channel gene, SLC34A1, showed positive association with AUDIT-C scores (t=5.85, p=6.31E-09). Unlike SLC7A11, SLC34A1 is involved in sodium-dependent phosphate transporter 2A (NaPi-IIa), which plays a role in the regulation of phosphate homeostasis.
Adjustment for BMI
Adding BMI into the analytic model, we found 58 out of 70 identified CpG sites remained EWS (Table 2). Twelve CpG sites were close to EWS with slightly reduced significance (p in a range of 2.67E-07~1.70E-07). Significant CpG sites on amino acid transporter genes (SLC7A11, SLC1A5, and SLC34A1) were still significantly associated with AUDIT-C. Taking into account the reduction in power consequent to adding an additional covariate, the results suggest that BMI is not a major confounding factor for EWAS on alcohol consumption in our cohort.
Differential DNAm between hazardous and non-hazardous alcohol consumption
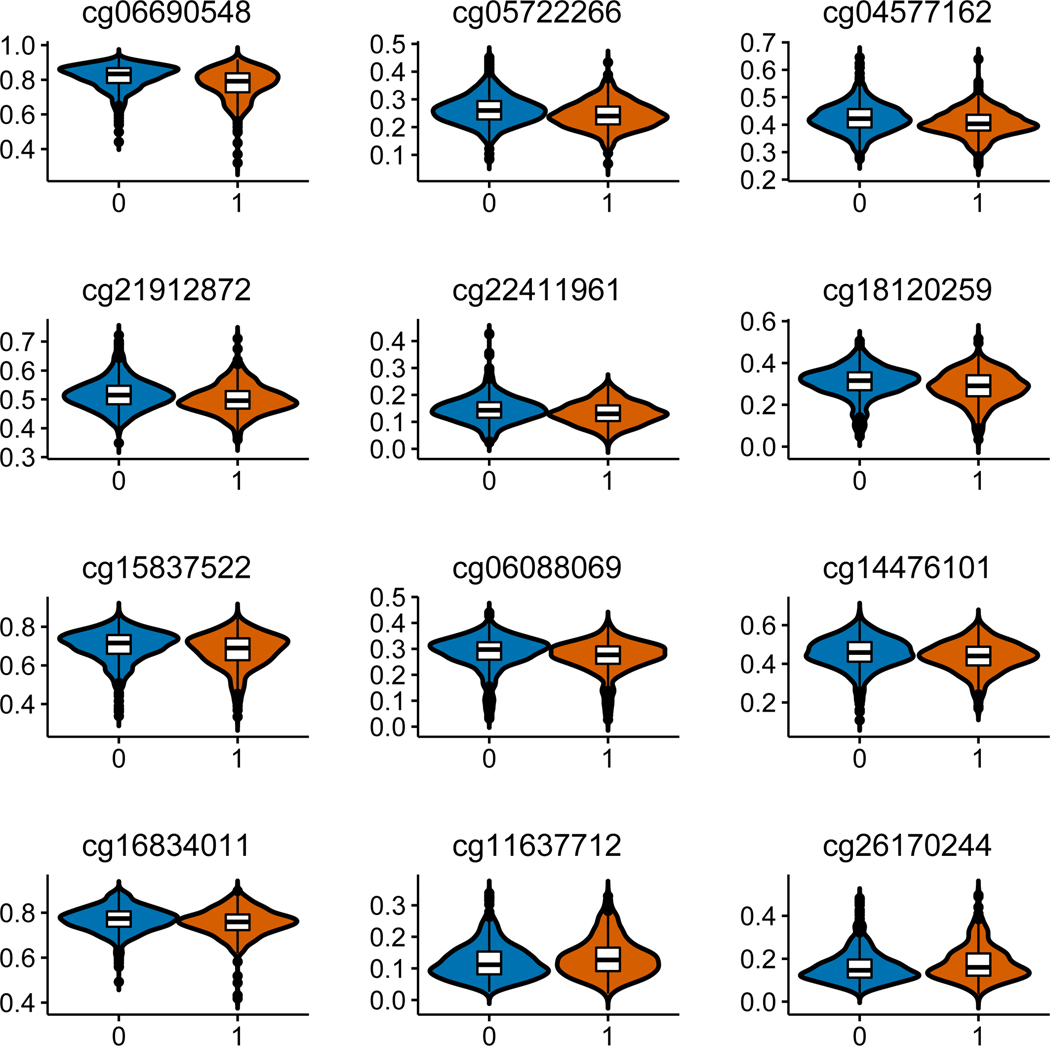
We conducted a secondary analysis to estimate the relationship of EWAS-significant CpG sites for alcohol consumption to hazardous alcohol drinking using an AUDIT-C score >=4 as a cutoff. We found 12 out of 70 CpG sites implicated in the primary analysis, also differentially methylated between hazardous and non-hazardous alcohol consumption with Bonferroni p < 0.05. The difference of DNAm at single CpG sites between hazardous and non-hazardous alcohol consumption was small across 12 significant DNAm loci. The difference of methylation β at each CpG site between hazardous and non-hazardous alcohol consumption was less than 5%. The largest differential DNAm CpG was 4% at cg06690548 in SLC7A11 (Supplemental Table S1). Figure 2 presents methylation level of 12 significant CpGs of DNAm between hazardous and non-hazardous alcohol consumption.
Figure 2.
Significant differential DNA methylation on 12 CpG sites between hazardous alcohol consumption and non-hazardous alcohol consumption. X-axis indicates hazardous alcohol consumption =1 and non-hazardous consumption =0; Y-axis indicates average beta-value of DNA methylation for each group. T-test was used to compare beta value between two groups (p < 0.0007).
Post-EWAS analysis and biological interpretation
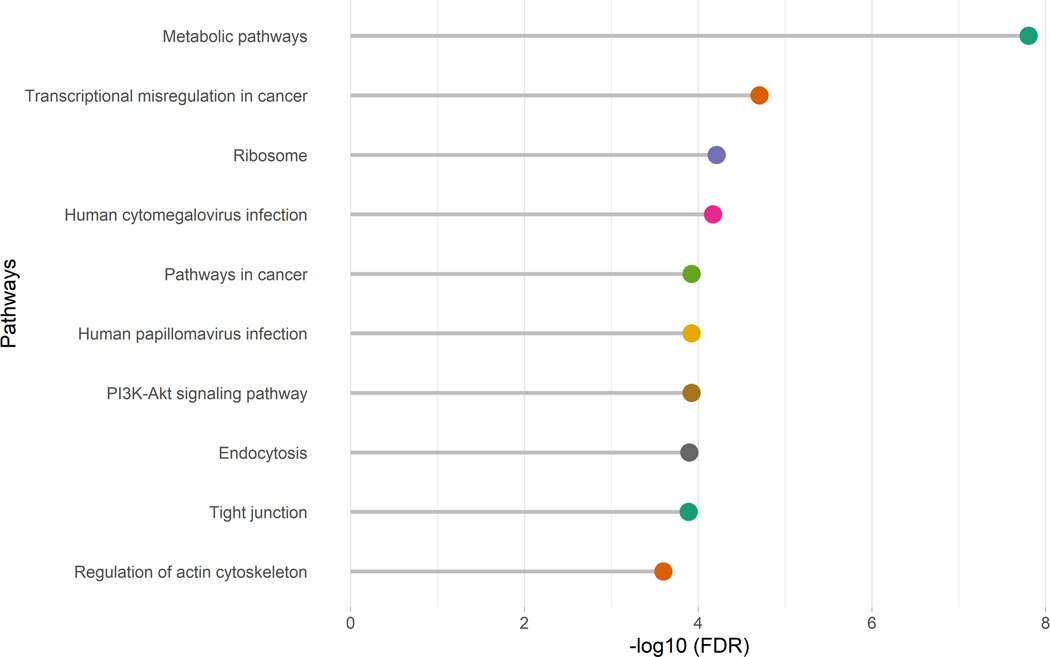
A total of 729 CpG sites with p <5E-04 were used for gene set enrichment analysis for alcohol consumption based on the sum of AUDIT-C score, not on hazaerdous use. These 729 CPG sites mapped to 527 genes. We found 68 significant pathways enriched in the KEGG database (Supplemental Table S2). Figure 3 presents the top 10 pathways associated with alcohol consumption. All significant pathways are presented in Table S2. Metabolic pathways was the most significant pathway associated with alcohol consumption (FDR=1.58E-08). Several pathways were relevant to cancer development, e.g. MicroRNAs in Cancer (FDR=5.96E-04), Proteoglycans in cancer (FDR=6.686E-04), Pathways in cancer (FDR=1.192E-04), Transcriptional misregulation in cancer (FDR=1.97E-05), and Pancreatic cancer (FDR=1.868E-02). Pathways involving viral or bacterial infection pathogenesis were also significant, e.g. Human cytomegalovirus infection (FDR=6.77E-05). Signaling pathways such as MAPK signaling pathway (FDR=5.96E-04), and Rap1 signaling,pathway(FDR=2.602E-04), were also statistically significant.
Figure 3.
Top 10 significant pathways in the KEGG database for alcohol consumption. CpG sites with p <= 5E-04 in epigenome-wide association analysis were selected for gene set enrichment analysis, which was adjusted with gene length by using missMethyl program in BioConductor. Significant level was set at false discovery rate <0.05.
Discussion
Using a high-density DNAm array, we completed an EWAS for alcohol consumption in a large EA male veteran population. We identified 64 novel differentially methylated loci in peripheral cells and replicated three previously reported loci associated with alcohol consumption (Liu et al., 2018) and three loci previously associated with alcohol-related medical conditions such as liver function (Nano et al., 2017) and body mass index (BMI) (Aslibekyan et al., 2015) (but not previously associated to alcohol use per se). Alcohol consumption-associated loci were located in genes involved in amino acid transport, synthesis, and metallization, and with important roles in cellular processes, development, and apoptosis. The alcohol-related genes identified from this EWAS were enriched in biological pathways related to cellular metabolism, infection, cancer, and signaling transduction. Because alcohol consumption is known to increase risks of cancer(Seitz et al., 2018, Bagnardi et al., 2013), infectious diseases(Trevejo-Nunez et al., 2015, Szabo, 1999), and metabolic syndrome (Freiberg et al., 2004), these findings suggest that the alteration of DNAm in peripheral cells associated with alcohol consumption may implicate DNAm in peripheral cells in alcohol-related medical outcomes.
It is important to consider the correlation of DNAm in saliva cells compared to other tissues. Emerging evidence suggests that a set of DNAm loci are highly correlated across different tissues including DNAm between blood and brain regions (Qi et al., 2018) and saliva and brain regions (Smith et al., 2015), suggesting that a proportion of DNAm sites in peripheral cells may be consistent with the methylation in neural cells. DNAm is correlated between saliva and intestinal mucosal cells within an individual; only approximately 11% of CpG sites are differentially methylated between saliva and intestinal mucosa (Hearn et al., 2019). A recent study reported 20 out of 26 CpG sites associated with smoking were consistent between blood and saliva cells (Barcelona et al., 2019). One interpretation of the highly correlated markers in different tissues is that pathophysiological processes resulting from environmental exposure and lifestyle (e.g. smoking, alcohol use) are involved in epigenetic alterations on a set of CpG sites across different tissues to regulate genes in response or counter response to such insults. Therefore, CpG sites identified in peripheral tissues such as blood and saliva could serve as surrogate markers for the phenotype of interest. This is especially useful for behavioral phenotypes considering the inaccessibility of live human brain. Study of peripheral DNAm has a very strong advantage of possible utility in identifying biomarkers, which, if identified in the brain, cannot be applied clinically.
We found that changes of DNAm associated with alcohol consumption were small: the DNAm differences between hazardous alcohol use and non-hazardous alcohol use were all <5%. These findings are consistent with expectation; unlike the effects of DNAm on oncogenes for cancers, emerging evidence has shown that intermediary DNAm level has become an epigenetic hallmark for many complex, non-malignant diseases and environmental stimuli (Leenen et al., 2016, Levenson, 2010). Differential DNAm between case and control groups of complex traits are typically <10% and often between 1–5% at single CpG sites. Some well-established methylation marks for hypertension, obesity, cardiovascular disease, major depression, and schizophrenia (Han et al., 2018, Ota et al., 2014) have shown small effects. Subtle DNAm changes associated with smoking and alcohol consumption have also been observed in previous studies (Gao et al., 2015, Harlaar and Hutchison, 2013, Liu et al., 2018). Although the underlying biology of mild DNAm alterations remains unaddressed, these methylation marks are often reliably detected across different phenotypic assessments, sample populations, and DNAm platforms. For example, several CpG sites in our study profiled by means of the Illumina EPIC platform were previously reported in studies using the Illumina 450K platform. Specifically, cg06690548 on SLC7A11 (p= 2.66E-28), cg16246545 on PHGDH (p=4.95E-11), cg06469895 on TERF2 (p=2.39E-10), and cg02583484 on HNRNPA1 (p=4.83E-08) were found to have significant associations alcohol consumption in our study and in Liu et al (Liu et al., 2018). Another four CpG sites, cg05993525 on TRERF1 (p=4.43E-07), cg2706351 on DENND2D (p=1.24E-06), cg0352740 on TXLNA (p=2.03E-06), and cg21180953 on SETBP1 (p=3.03E-6) were among the top significant genes associated with alcohol consumption in EAs(Liu et al., 2018) and, as noted, were of suggestive significance in our study. In a meta-analysis(Liu et al., 2018), p values for CpG sites on these four genes ranged from 3.2E-12 to 4.4E-15. Taken together, these data suggest that DNAm is fine-tuned in the epigenome for complex diseases and that pronounced regulation of gene transcription related to complex behaviors may generally not be expected. Instead, subtle changes of DNAm associated with alcohol consumption may be associated to a wide range of variability for alcohol consumption for this highly prevalent trait in the general population. This is rather similar to the case of stable genetic SNP markers detected by Genome-wide association studies which, for complex traits, generally have small-to-very small effect sizes.
Of note, several genes identified in our EWAS are involved in amino acid transport and metabolism. For example, 3 CpGs, cg14476101, cg16246545, and cg26457483 on PHGDH, were hypomethylated in hazardous alcohol users compared to non-hazardous alcohol users in our sample (p values ranging 2.73E-08 – 3.17E-11). This gene encodes the enzyme involved in the first step of serine biosynthesis. D-serine is a co-agonist for the N-Methyl-D-aspartic acid glutamate receptor and exogeneous D-serine has been showed to reduce alcohol consumption in animals (Seif et al., 2015). Serine was also demonstrated to ameliorate alcoholic fatty liver by accelerating serine-dependent homocysteine metabolism in mice and rats(Sim et al., 2015). Methylation of the same locus has been linked to BMI and waist circumference by EWAS (Wahl et al., 2017). These studies revealed that one of the three significant alcohol-related CpG sites on PHGDH, cg14476101, was also inversely associated with triglyceride levels (Truong et al., 2017) and liver enzymes (Nano et al., 2017).
The most significant CpG among these is cg06690548 on SLC7A11 (p=2.66E-028), which was inversely associated with alcohol consumption in our sample. As mentioned early, SLC7A11 is highly specific for cysteine and glutamate transportation. The cystine/glutamate antiporter brings cysteine into the cell in exchange for glutamate exported from the cell. The cysteine brought into the cell is often used as a substrate for glutathione, a “free radical” scavenger that protects the cell from injury. Thus, increased transcription of this antiporter gene could be a signal of cellular response to redox threat, which may implicate a mechanism for increased cancer risk from alcohol consumption (Habib et al., 2015, Linher-Melville et al., 2015). Similar to cg06690548 on SLC7A11, two other significant CpGs mapped on the SLC1A5 (cg07626482, t= −5.66; p=1.91E-08) and SLC34A1 (cg16968128, t=5.85, p=6.31E-09), genes that are involved in amino acid transportation and biosynthesis that may involve mechanisms of cancer. For example, SLC1A5 is a glutamate transporter reported to facilitate the uptake of glutamine in tumor cells, which in turn serves multiple metabolic functions within cells depending on the tumor oncogenotype and microenvironment (Altman et al., 2016). Methylation on these three genes, SLC7A11, SLC1A5 and SLC34A1, is also linked to liver function, lipid metabolism, and BMI (Nano et al., 2017, Wahl et al., 2017).
Alcohol has many peripheral effects also, raising our confidence in these findings; and in fact the best-replicated genetic variants influencing alcohol use disorder risk, map to genes encoding alcohol metabolizing enzymes that can act peripherally(Walters et al., 2018, Edenberg, 2007). Notwithstanding the noted limitations, the results of this study reveal possible peripheral mechanisms associated with alcohol consumption. They also provide further evidence of alcohol exposure’s impact on the epigenome and its role in alcohol-related health problems. The identified DNAm signatures may thus potentially serve as biomarkers of alcohol consumption and risk for alcohol-related health problems. Further research is needed to replicate these results in independent well-powered samples, as well as in more heterogeneous samples (e.g., women, different ethnic populations); and to evaluate the biological roles of the small effects of the identified CpG sites.
Supplementary Material
Acknowledgments and Disclosures
The National Health and Resilience in Veterans Study is supported by the U.S. Department of Veterans Affairs National Center for Posttraumatic Stress Disorder. This work was supported in part by National Institute on Drug Abuse (NIDA, R01 DA038632, Xu), grant R01-DA2690, and grant R01-AA11330.
The following competing interests for Dr. John H Krystal: 1) Consultant: Note: - The Individual Consultant Agreements listed below are less than $10,000 per year: AstraZeneca Pharmaceuticals; Biogen, Idec, MA; Biomedisyn Corporation; Bionomics, Limited (Australia); Concert Pharmaceuticals, Inc.; Heptares Therapeutics, Limited (UK); Janssen Research & Development; L.E.K. Consulting; Otsuka America Pharmaceutical, Inc.; Spring Care, Inc.; Sunovion Pharmaceuticals, Inc.; Takeda Industries; Taisho Pharmaceutical Co., Ltd; Scientific Advisory Board; Bioasis Technologies, Inc.; Biohaven Pharmaceuticals; Blackthorn Therapeutics, Inc.; Broad Institute of MIT and Harvard; Cadent Therapeutics; Lohocla Research Corporation; Pfizer Pharmaceuticals; Stanley Center for Psychiatric Research at the Broad Institute; 2) Stock: ArRETT Neuroscience, Inc.; Blackthorn Therapeutics, Inc.; Biohaven Pharmaceuticals Medical Sciences; Spring Care, Inc. Stock Options: Biohaven Pharmaceuticals Medical Sciences; 3) Income Greater than $10,000: Editorial Board
Editor - Biological Psychiatry; Patents and Inventions: Seibyl JP, Krystal JH, Charney DS. Dopamine and noradrenergic reuptake inhibitors in treatment of schizophrenia. US Patent #:5,447,948.September 5, 1995; Vladimir, Coric, Krystal, John H, Sanacora, Gerard - Glutamate Modulating Agents in the Treatment of Mental Disorders US Patent No. 8,778,979 B2 Patent Issue Date: July 15, 2014. US Patent Application No. 15/695,164:
Filing Date: 09/05/2017; Charney D, Krystal JH, Manji H, Matthew S, Zarate C., - Intranasal Administration of Ketamine to Treat Depression United States Application No. 14/197,767 filed on March 5, 2014; United States application or Patent Cooperation Treaty (PCT) International application No. 14/306,382 filed on June 17, 2014; Zarate, C, Charney, DS, Manji, HK, Mathew, Sanjay J, Krystal, JH, Department of Veterans Affairs “Methods for Treating Suicidal Ideation”, Patent Application No. 14/197.767 filed on March 5, 2014 by Yale University Office of Cooperative Research; Arias A, Petrakis I, Krystal JH. - Composition and methods to treat addiction; Provisional Use Patent Application no.61/973/961. April 2, 2014. Filed by Yale University Office of Cooperative Research; Chekroud, A., Gueorguieva, R., & Krystal, JH. “Treatment Selection for Major Depressive Disorder” [filing date 3rd June 2016, USPTO docket number Y0087.70116US00]. Provisional patent submission by Yale University; Yoon G, Petrakis I, Krystal JH - Compounds, Compositions and Methods for Treating or Preventing Depression and Other Diseases. U. S. Provisional Patent Application No. 62/444,552, filed on
January10, 2017 by Yale University Office of Cooperative Research OCR 7088 US01; Abdallah, C, Krystal, JH, Duman, R, Sanacora, G. Combination Therapy for Treating or Preventing Depression or Other Mood Diseases. U.S. Provisional Patent Application No. 047162-7177P1 (00754) filed on August 20, 2018 by Yale University Office of Cooperative Research OCR 7451 US01.
Footnotes
Other co-authors declare no completing interests.
References
- Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA (2014) Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30:1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslibekyan S, Demerath EW, Mendelson M, Zhi D, Guan W, Liang L, Sha J, Pankow JS, Liu C, Irvin MR, Fornage M, Hidalgo B, Lin LA, Thibeault KS, Bressler J, Tsai MY, Grove ML, Hopkins PN, Boerwinkle E, Borecki IB, Ordovas JM, Levy D, Tiwari HK, Absher DM, Arnett DK (2015) Epigenome-wide study identifies novel methylation loci associated with body mass index and waist circumference. Obesity (Silver Spring) 23:1493–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnardi V, Rota M, Botteri E, Tramacere I, Islami F, Fedirko V, Scotti L, Jenab M, Turati F, Pasquali E, Pelucchi C, Bellocco R, Negri E, Corrao G, Rehm J, Boffetta P, La Vecchia C (2013) Light alcohol drinking and cancer: a meta-analysis. Ann Oncol 24:301–308. [DOI] [PubMed] [Google Scholar]
- Barcelona V, Huang Y, Brown K, Liu J, Zhao W, Yu M, Kardia SLR, Smith JA, Taylor JY, Sun YV (2019) Novel DNA methylation sites associated with cigarette smoking among African Americans. Epigenetics 14:383–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barfield RT, Almli LM, Kilaru V, Smith AK, Mercer KB, Duncan R, Klengel T, Mehta D, Binder EB, Epstein MP, Ressler KJ, Conneely KN (2014) Accounting for population stratification in DNA methylation studies. Genet Epidemiol 38:231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barfield RT, Kilaru V, Smith AK, Conneely KN (2012) CpGassoc: an R function for analysis of DNA methylation microarray data. Bioinformatics 28:1280–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkel TD, Pandey SC (2017) Emerging Role of Epigenetic Mechanisms in Alcohol Addiction. Alcohol Clin Exp Res 41:666–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckmann C, Di Santo A, Karle KN, Batra A, Nieratschker V (2016) Validation of differential GDAP1 DNA methylation in alcohol dependence and its potential function as a biomarker for disease severity and therapy outcome. Epigenetics 11:456–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckmann C, Islam SA, MacIsaac JL, Morin AM, Karle KN, Di Santo A, Wust R, Lang I, Batra A, Kobor MS, Nieratschker V (2017) DNA methylation signatures of chronic alcohol dependence in purified CD3(+) T-cells of patients undergoing alcohol treatment. Sci Rep 7:6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton R, Sheron N (2018) No level of alcohol consumption improves health. Lancet. [DOI] [PubMed] [Google Scholar]
- Bush K, Kivlahan DR, McDonell MB, Fihn SD, Bradley KA (1998) The AUDIT alcohol consumption questions (AUDIT-C): an effective brief screening test for problem drinking. Ambulatory Care Quality Improvement Project (ACQUIP). Alcohol Use Disorders Identification Test. Arch Intern Med 158:1789–1795. [DOI] [PubMed] [Google Scholar]
- Cervera-Juanes R, Wilhelm LJ, Park B, Grant KA, Ferguson B (2017) Genome-wide analysis of the nucleus accumbens identifies DNA methylation signals differentiating low/binge from heavy alcohol drinking. Alcohol 60:103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborators GBDA (2018) Alcohol use and burden for 195 countries and territories, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ (2007) The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol research & health : the journal of the National Institute on Alcohol Abuse and Alcoholism 30:5–13. [PMC free article] [PubMed] [Google Scholar]
- Fortin JP, Labbe A, Lemire M, Zanke BW, Hudson TJ, Fertig EJ, Greenwood CM, Hansen KD (2014) Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome biology 15:503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freiberg MS, Cabral HJ, Heeren TC, Vasan RS, Curtis Ellison R, Third National H, Nutrition Examination S (2004) Alcohol consumption and the prevalence of the Metabolic Syndrome in the US.: a cross-sectional analysis of data from the Third National Health and Nutrition Examination Survey. Diabetes Care 27:2954–2959. [DOI] [PubMed] [Google Scholar]
- Fuehrlein BS, Kachadourian LK, DeVylder EK, Trevisan LA, Potenza MN, Krystal JH, Southwick SM, Pietrzak RH (2018) Trajectories of alcohol consumption in U.S. military veterans: Results from the National Health and Resilience in Veterans Study. The American journal on addictions. [DOI] [PubMed] [Google Scholar]
- Gao X, Jia M, Zhang Y, Breitling LP, Brenner H (2015) DNA methylation changes of whole blood cells in response to active smoking exposure in adults: a systematic review of DNA methylation studies. Clin Epigenetics 7:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garro AJ, McBeth DL, Lima V, Lieber CS (1991) Ethanol consumption inhibits fetal DNA methylation in mice: implications for the fetal alcohol syndrome. Alcohol Clin Exp Res 15:395–398. [DOI] [PubMed] [Google Scholar]
- Geeleher P, Hartnett L, Egan LJ, Golden A, Raja Ali RA, Seoighe C (2013) Gene-set analysis is severely biased when applied to genome-wide methylation data. Bioinformatics 29:1851–1857. [DOI] [PubMed] [Google Scholar]
- Hagerty SL, Bidwell LC, Harlaar N, Hutchison KE (2016) An Exploratory Association Study of Alcohol Use Disorder and DNA Methylation. Alcohol Clin Exp Res 40:1633–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han LKM, Aghajani M, Clark SL, Chan RF, Hattab MW, Shabalin AA, Zhao M, Kumar G, Xie LY, Jansen R, Milaneschi Y, Dean B, Aberg KA, van den Oord E, Penninx B (2018) Epigenetic Aging in Major Depressive Disorder. Am J Psychiatry 175:774–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlaar N, Hutchison KE (2013) Alcohol and the methylome: design and analysis considerations for research using human samples. Drug Alcohol Depend 133:305–316. [DOI] [PubMed] [Google Scholar]
- Hearn NL, Coleman AS, Ho V, Chiu CL, Lind JM (2019) Comparing DNA methylation profiles in saliva and intestinal mucosa. BMC Genomics 20:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberlein A, Muschler M, Frieling H, Behr M, Eberlein C, Wilhelm J, Groschl M, Kornhuber J, Bleich S, Hillemacher T (2013) Epigenetic down regulation of nerve growth factor during alcohol withdrawal. Addict Biol 18:508–510. [DOI] [PubMed] [Google Scholar]
- Hillemacher T, Weinland C, Lenz B, Kraus T, Heberlein A, Glahn A, Muschler MA, Bleich S, Kornhuber J, Frieling H (2015) DNA methylation of the LEP gene is associated with craving during alcohol withdrawal. Psychoneuroendocrinology 51:371–377. [DOI] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT (2012) DNA methylation arrays as surrogate measures of cell mixture distribution. BMC bioinformatics 13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufer BI, Kapalanga J, Castellani CA, Diehl EJ, Yan L, Singh SM (2015) Associative DNA methylation changes in children with prenatal alcohol exposure. Epigenomics 7:1259–1274. [DOI] [PubMed] [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD (2012) The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28:882–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leenen FA, Muller CP, Turner JD (2016) DNA methylation: conducting the orchestra from exposure to phenotype? Clin Epigenetics 8:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson VV (2010) DNA methylation as a universal biomarker. Expert Rev Mol Diagn 10:481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Marioni RE, Hedman AK, Pfeiffer L, Tsai PC, Reynolds LM, Just AC, Duan Q, Boer CG, Tanaka T, Elks CE, Aslibekyan S, Brody JA, Kuhnel B, Herder C, Almli LM, Zhi D, Wang Y, Huan T, Yao C, Mendelson MM, Joehanes R, Liang L, Love SA, Guan W, Shah S, McRae AF, Kretschmer A, Prokisch H, Strauch K, Peters A, Visscher PM, Wray NR, Guo X, Wiggins KL, Smith AK, Binder EB, Ressler KJ, Irvin MR, Absher DM, Hernandez D, Ferrucci L, Bandinelli S, Lohman K, Ding J, Trevisi L, Gustafsson S, Sandling JH, Stolk L, Uitterlinden AG, Yet I, Castillo-Fernandez JE, Spector TD, Schwartz JD, Vokonas P, Lind L, Li Y, Fornage M, Arnett DK, Wareham NJ, Sotoodehnia N, Ong KK, van Meurs JBJ, Conneely KN, Baccarelli AA, Deary IJ, Bell JT, North KE, Liu Y, Waldenberger M, London SJ, Ingelsson E, Levy D (2018) A DNA methylation biomarker of alcohol consumption. Mol Psychiatry 23:422–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussier AA, Morin AM, MacIsaac JL, Salmon J, Weinberg J, Reynolds JN, Pavlidis P, Chudley AE, Kobor MS (2018) DNA methylation as a predictor of fetal alcohol spectrum disorder. Clin Epigenetics 10:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nano J, Ghanbari M, Wang W, de Vries PS, Dhana K, Muka T, Uitterlinden AG, van Meurs JBJ, Hofman A, consortium B, Franco OH, Pan Q, Murad SD, Dehghan A (2017) Epigenome-Wide Association Study Identifies Methylation Sites Associated With Liver Enzymes and Hepatic Steatosis. Gastroenterology 153:1096–1106 e1092. [DOI] [PubMed] [Google Scholar]
- Ota VK, Noto C, Gadelha A, Santoro ML, Spindola LM, Gouvea ES, Stilhano RS, Ortiz BB, Silva PN, Sato JR, Han SW, Cordeiro Q, Bressan RA, Belangero SI (2014) Changes in gene expression and methylation in the blood of patients with first-episode psychosis. Schizophr Res 159:358–364. [DOI] [PubMed] [Google Scholar]
- Palmisano M, Pandey SC (2017) Epigenetic mechanisms of alcoholism and stress-related disorders. Alcohol 60:7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert RA, Penaluna B, White T, Shires S, Gunter T, Liesveld J, Erwin C, Hollenbeck N, Osborn T (2014) A pilot examination of the genome-wide DNA methylation signatures of subjects entering and exiting short-term alcohol dependence treatment programs. Epigenetics 9:1212–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portales-Casamar E, Lussier AA, Jones MJ, MacIsaac JL, Edgar RD, Mah SM, Barhdadi A, Provost S, Lemieux-Perreault LP, Cynader MS, Chudley AE, Dube MP, Reynolds JN, Pavlidis P, Kobor MS (2016) DNA methylation signature of human fetal alcohol spectrum disorder. Epigenetics Chromatin 9:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz HK, Bataller R, Cortez-Pinto H, Gao B, Gual A, Lackner C, Mathurin P, Mueller S, Szabo G, Tsukamoto H (2018) Alcoholic liver disease. Nat Rev Dis Primers 4:16. [DOI] [PubMed] [Google Scholar]
- Smith AK, Kilaru V, Klengel T, Mercer KB, Bradley B, Conneely KN, Ressler KJ, Binder EB (2015) DNA extracted from saliva for methylation studies of psychiatric traits: evidence tissue specificity and relatedness to brain. Am J Med Genet B Neuropsychiatr Genet 168B:36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovics EA, Potenza MN, Pietrzak RH (2018) The physical and mental health burden of obesity in U.S. veterans: Results from the National Health and Resilience in Veterans Study. J Psychiatr Res 103:112–119. [DOI] [PubMed] [Google Scholar]
- Szabo G (1999) CONSEQUENCES OF ALCOHOL CONSUMPTION ON HOST DEFENCE. Alcohol and Alcoholism 34:830–841. [DOI] [PubMed] [Google Scholar]
- Trevejo-Nunez G, Kolls JK, de Wit M (2015) Alcohol Use As a Risk Factor in Infections and Healing: A Clinician’s Perspective. Alcohol Res 37:177–184. [PMC free article] [PubMed] [Google Scholar]
- Tulisiak CT, Harris RA, Ponomarev I (2017) DNA modifications in models of alcohol use disorders. Alcohol 60:19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Knaap LJ, Schaefer JM, Franken IH, Verhulst FC, van Oort FV, Riese H (2014) Catechol-O-methyltransferase gene methylation and substance use in adolescents: the TRAILS study. Genes Brain Behav 13:618–625. [DOI] [PubMed] [Google Scholar]
- Walters RK, Adams MJ, Adkins AE, Aliev F, Bacanu S-A, Batzler A, Bertelsen S, Biernacka J, Bigdeli TB, Chen L-S, Clarke T-K, Chou Y-L, Degenhardt F, Docherty AR, Fontanillas P, Foo J, Fox L, Frank J, Giegling I, Gordon S, Hack L, Hartz SM, Heilmann-Heimbach S, Herms S, Hodgkinson C, Hoffmann P, Hottenga J-J, Kennedy MA, Alanne-Kinnunen M, Konte B, Lahti J, Lahti-Pulkkinen M, Ligthart L, Loukola A-M, Maher BS, Mbarek H, McIntosh AM, McQueen MB, Milaneschi Y, Palviainen T, Pearson JF, Peterson RE, Polimanti R, Ripatti S, Ryu E, Saccone NL, Salvatore JE, Sanchez-Roige S, Schwandt M, Sherva R, Streit F, Strohmaier J, Thomas N, Wang J-C, Webb BT, Wedow R, Wetherill L, Wills AG, Boardman JD, Chen D, Choi D-S, Copeland WE, Culverhouse RC, Dahmen N, Degenhardt L, Domingue BW, Elson SL, Frye M, Gäbel W, Ising M, Johnson EC, Keyes M, Kiefer F, Kramer J, Kuperman S, Lucae S, Lynskey MT, Maier W, Mann K, Männistö S, McClintick JN, Meyers JL, Müller-Myhsok B, Nurnberger JI, Palotie A, Preuss U, Räikkönen K, Reynolds MD, Ridinger M, Scherbaum N, Shuckit M, Soyka M, Treutlein J, Witt S, Wodarz N, Zill P, Adkins DE, Boden JM, Boomsma D, Bierut LJ, Brown SA, Bucholz KK, Cichon S, Costello EJ, de Wit H, Diazgranados N, Dick DM, Eriksson JG, Farrer LA, Foroud TM, Gillespie NA, Goate AA, Goldman D, Grucza RA, Hancock DB, Harris KM, Heath AC, Hesselbrock V, Hewitt JK, Hopfer C, Horwood J, Iacono W, Johnson EO, Kaprio JA, Karpyak V, Kendler KS, Kranzler HR, Krauter K, Lichtenstein P, Lind PA, McGue M, MacKillop J, Madden PAF, Maes H, Magnusson P, Martin NG, Medland SE, Montgomery GW, Nelson EC, Nöthen M, Palmer AA, Pedersen NL, Penninx BWJH, Porjesz B, Rice JP, Rietschel M, Riley BP, Rose R, Rujescu D, Shen P-H, Silberg J, Stallings MC, Tarter RE, Vanyukov MM, Vrieze S, Wall TL, Whitfield JB, Zhao H, Neale BM, Gelernter J, Edenberg HJ, Agrawal A (2018) Trans-ancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RJ, Berry LJ, Beerstecher E (1949) Individual Metabolic Patterns, Alcoholism, Genetotrophic Diseases. Proc Natl Acad Sci U S A 35:265–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CR, Ho MF, Vega MC, Burne TH, Chong S (2015) Prenatal ethanol exposure alters adult hippocampal VGLUT2 expression with concomitant changes in promoter DNA methylation, H3K4 trimethylation and miR-467b-5p levels. Epigenetics Chromatin 8:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Gelernter J (2017) Review: DNA methylation and alcohol use disorders: Progress and challenges. Am J Addict 26:502–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Herman AI, Kranzler HR, Anton RF, Zhao H, Zheng W, Gelernter J (2013) Array-based profiling of DNA methylation changes associated with alcohol dependence. Alcohol Clin Exp Res 37 Suppl 1:E108–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.