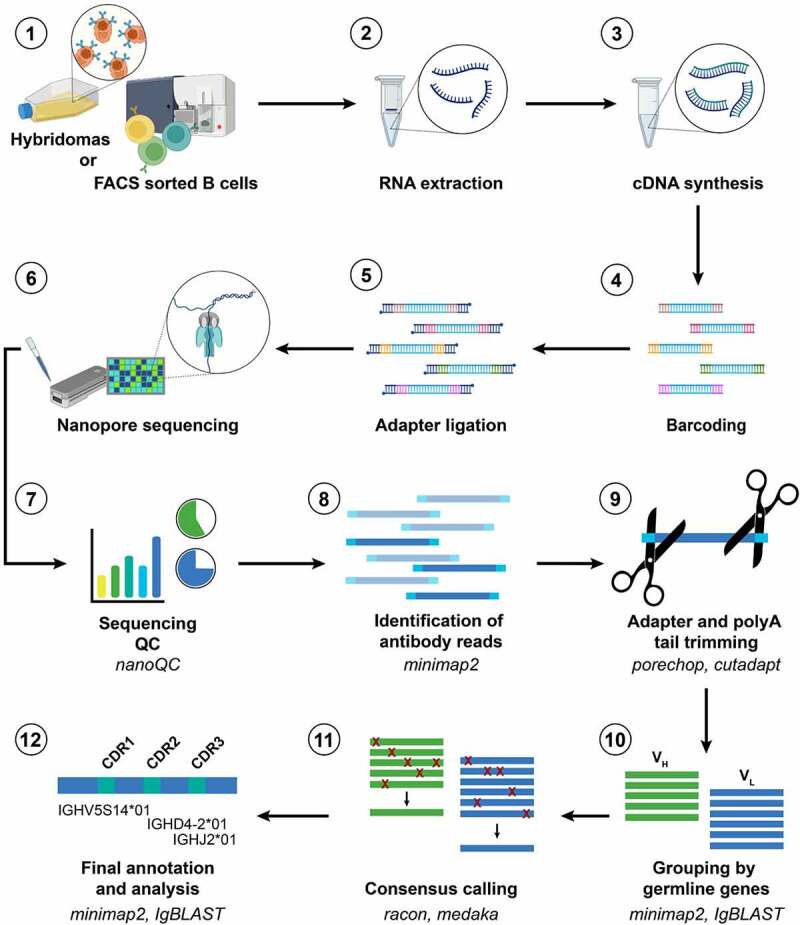
Figure 1.
NAb-seq workflow for parallel sequencing of full-length antibody heavy and light chain sequences from hybridoma cell lines and single B cells.
Long Description: A series of 12 steps are numbered 1 to 12. Step 1 depicts hybridomas or FACS sorted B cells (with images of hybridoma cells in culture and of B cells being sorted). The remaining steps are 2) RNA extraction (test tube plus RNA), 3) cDNA synthesis (test tube plus DNA), 4) barcoding (five DNA strands with colored ends), 5) adapter ligation (five DNA strands with blue added to each end), 6) Nanopore sequencing (images of a matchbox-shaped Flongle and of DNA strand passing through a protein nanopore), 7) sequencing quality control using nanoQC (graphs), 8) identification of antibody reads using minimap2 (colored DNA strands), 9) adapter and polyA tail trimming using porechop and cutadapt (scissors cutting a DNA strand), 10) grouping by germline genes using minimap2 and IgBLAST (parallel lines representing VH and VL sequences), 11) consensus calling using racon and medaka (VH and VL sequences with red crosses representing errors that are resolved to single sequence). Step 12 is final annotation and analysis using minimap2 and IgBLAST (green and blue line labeled with descriptors of an antibody gene sequence).
Figure 1 A series of images numbered 1 to 12 with word descriptions for each, and with arrows between images to indicate the workflow involved in Nanopore antibody sequencing.