Abstract
A strain of Bacillus designated TA2.A1, isolated from a thermal spring in Te Aroha, New Zealand, grew optimally at pH 9.2 and 70°C. Bacillus strain TA2.A1 utilized glutamate as a sole carbon and energy source for growth, and sodium chloride (>5 mM) was an obligate requirement for growth. Growth on glutamate was inhibited by monensin and amiloride, both inhibitors that collapse the sodium gradient (ΔpNa) across the cell membrane. N,N-Dicyclohexylcarbodiimide inhibited the growth of Bacillus strain TA2.A1, suggesting that an F1F0-ATPase (H type) was being used to generate cellular ATP needed for anabolic reactions. Vanadate, an inhibitor of V-type ATPases, did not affect the growth of Bacillus strain TA2.A1. Glutamate transport by Bacillus strain TA2.A1 could be driven by an artificial membrane potential (ΔΨ), but only when sodium was present. In the absence of sodium, the rate of ΔΨ-driven glutamate uptake was fourfold lower. No glutamate transport was observed in the presence of ΔpNa alone (i.e., no ΔΨ). Glutamate uptake was specifically inhibited by monensin, and the Km for sodium was 5.6 mM. The Hill plot had a slope of approximately 1, suggesting that sodium binding was noncooperative and that the glutamate transporter had a single binding site for sodium. Glutamate transport was not affected by the protonophore carbonyl cyanide m-chlorophenylhydrazone, suggesting that the transmembrane pH gradient was not required for glutamate transport. The rate of glutamate transport increased with increasing glutamate concentration; the Km for glutamate was 2.90 μM, and the Vmax was 0.7 nmol · min−1 mg of protein. Glutamate transport was specifically inhibited by glutamate analogues.
Bacteria display a remarkable capacity to survive and grow in extremely hostile environments. Entire groups of organisms (e.g., thermophiles, halophiles, and acidophiles) have adapted their lifestyles to these extreme environments. Even within a given group, a very wide range of environmental limits may be tolerated. In general terms, microorganisms are able to grow over a wide range of pH values, from 0 to 11.0 (1). For example, alkaliphiles grow between pH 9.0 and 11.5 and maintain their intracellular pH between 8.4 and 9.0 (2, 10, 13, 17). Most aerobic alkaliphiles belong to the genus Bacillus (10, 13, 17). Bacillus species are gram-positive sporeformers which have been isolated from a diverse range of environments, including those of neutral, acidic, and alkaline pH. Few bacteria growing at extremes of both pH and temperature have been described (18, 19, 22). For example, a novel group of anaerobic alkaliphilic bacteria capable of optimal growth at 55°C and pH 9.3 (upper limit of pH 10.3) was described by Li et al. (18, 19). These bacteria extended the combined range of temperature and pH for bacterial groups, and one of these organisms, Clostridium paradoxum, was subsequently shown to maintain a pH gradient of 1.3 pH units at an extracellular pH of 9.0 (2). Bacteria that grow aerobically at alkaline pH and extreme temperature (optimum growth temperature above 65°C) have not previously been described.
Bacteria that grow at alkaline pH are faced with bioenergetic problems in terms of chemiosmotic energy generation and solute transport driven by the proton motive force (Δp) (14). The alkaliphilic bacteria studied maintain an intracellular pH lower than the extracellular pH (inverted pH gradient). It is generally accepted that alkaliphiles use sodium/proton antiporters to acidify the cytosol and generate an inwardly directed sodium motive force (ΔpNa) (16). The use of sodium as a coupling ion circumvents the problem of a low Δp. Growth at extremes of temperature also poses the additional problem of a leaky cytoplasmic membrane to protons (28). Konings and coworkers (3, 4, 8, 25–27) and Holtom et al. (9) have demonstrated high proton permeability of membranes at high temperature, and some thermophilic bacteria overcome this problem by using sodium as a coupling ion for solute transport.
In this study, we describe the growth and bioenergetic properties of an aerobic, extremely thermophilic, alkaliphilic Bacillus strain, TA2.A1. This isolate can grow over a pH range from 7.7 to 10.5, with an optimum of pH 9.2 at 70°C. The growth of this bacterium and glutamate transport were completely dependent on sodium, indicating that this organism uses sodium rather than protons in bioenergetic processes.
MATERIALS AND METHODS
Chemicals.
l-[U-14C]glutamic acid was from Amersham International (Little Chalfont, Buckinghamshire, England). Amiloride-HCl, carbonyl cyanide m-chlorophenylhydrazone (CCCP), and monensin were obtained from Sigma Chemical Co. (St. Louis, Mo.). N,N-Dicyclohexylcarbodiimide (DCCD) and sodium vanadate were from BDH Chemicals Ltd. (Poole, England).
Growth and maintenance.
Bacillus strain TA2.A1 was isolated from a continuously enriched pool sample. The growth medium contained (per liter) 0.5 g of Na2SO4, 0.1 g of (NH4)2SO4, 0.1 g of MgSO4 · 7H2O, 0.2 g of K2HPO4, 9.0 g of NaHCO3, 5.0 ml of dictyglomous trace elements (23), 0.1 g of Trypticase (Oxoid), and 2 g of glutamate (BDH). The pH of the medium was adjusted to 10 (20°C), which equates to a pH of 9.2 at 70°C after autoclaving, using 2 N NaOH. Then 100-ml aliquots of the medium were dispensed into 500-ml flasks, which were stoppered with nonabsorbent cotton wool bungs and autoclaved at 15 lb/in2 for 15 min. Cells were cultured in a 65°C orbital shaking incubator and aerated by shaking at 100 rpm. In a typical experiment, flasks were inoculated (1% inoculum) from an overnight culture and cells were grown to mid-exponential phase (0.2 to 0.25 units of optical density at 450 nm).
Glutamate transport assays.
Cells were harvested by centrifugation (12,000 × g, 5 min, 4°C) during exponential growth (0.22 mg of protein/ml) and washed twice in Tris-HCl buffer (50 mM, pH 9.2; 70°C). The cell pellet was resuspended in the same buffer to achieve a concentration of 14 to 20 mg of protein per ml. Aliquots (200 μl) of cell suspension were placed into tubes in a shaking (70 rpm) water bath (Julabo; Labortechnik, GmbH) at 60°C, and transport was initiated by the addition of 100 nCi of l-[U-14C]glutamate (270 mCi/mmol). After 0 to 60 s, transport was terminated by the addition of ice-cold LiCl (2 ml, 100 mM) and rapid filtration (0.45-μm-pore-size cellulose-nitrate filter). Experiments were carried out where the transport rate was first order with respect to protein, and initial rates are reported here. The filters were washed once with 2.0 ml of LiCl, dried for 30 min at 105°C, and counted by liquid scintillation. Cells which were treated with monensin (10 μM) or incubated in the absence of sodium showed essentially no [14C]glutamate uptake, and this result indicated that there was little nonspecific binding of [14C]glutamate to cells.
Artificial membrane potentials.
To create an artificial ΔpNa and membrane potential (ΔΨ), washed cells (100 mM Tris-HCl containing 100 mM KCl, pH 9.2) from exponentially growing cultures were loaded with potassium by valinomycin treatment (10 μM, 0°C, 30 min) and diluted 50-fold (4 μl) into 200 μl of Tris-HCl buffer containing 100 mM NaCl (pH 9.0) plus 1 μM [14C]glutamate. Potassium-loaded cells were diluted into either 100 mM Tris-HCl buffer containing 100 mM NaCl and 100 mM KCl to create a ΔpNa in the absence of ΔΨ or 100 mM Tris-HCl buffer alone to create a ΔΨ. Controls (no driving force) were loaded with K+ or K+ and Na+ and diluted into K+ or K+ and Na+, respectively. Transport was initiated by a 50-fold dilution of concentrated cells (4 μl) into buffer (200 μl) containing the radioactive glutamate (see above).
Competition and metabolic inhibitor experiments.
Competitive substrates (amino acid) and metabolic inhibitors, tested as potential inhibitors of glutamate uptake, were added to the transport assay medium 5 min before [14C]glutamate. Unlabeled amino acids were added at a final concentration of 5 mM. Metabolic inhibitors were added at the final concentrations indicated in the text. All of the water-insoluble inhibitors were dissolved in 95% ethanol and compared with ethanol-treated controls. The results of competition experiments were expressed as the mean of three determinations, and the level of inhibition was expressed as the percent inhibition of the initial rate of uptake compared to controls (nominally 100%) in the absence of competitive substrate.
RESULTS
Growth of Bacillus strain TA2.A1.
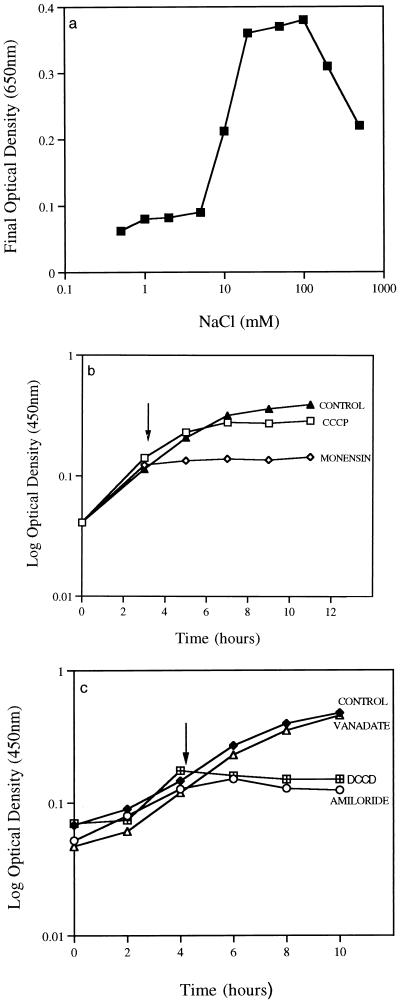
Bacillus strain TA2.A1 grew rapidly on minimal medium containing l-glutamate as the sole carbon and energy source. The optimal conditions for growth were pH 9.2 and 70°C, and the maximum specific growth rate was 0.35 h−1 (data not shown). Sodium was an absolute requirement for cell growth (Fig. 1a). Growth was barely evident below a concentration of 5 mM NaCl, and the final optical density increased as the sodium concentration in the growth medium was increased from 5 to 100 mM. Sodium concentrations greater than 100 mM were inhibitory to cell growth. These results suggested that the growth of Bacillus strain TA2.A1 may depend on sodium for energy generation and bioenergetic processes (i.e., transport, motility, etc.). To investigate this possibility in more detail, we tested the effect of specific metabolic inhibitors on Bacillus strain TA2.A1 to determine whether a Δp or ΔpNa was required for growth.
FIG. 1.
Effects of sodium ion concentration (a), monensin (0.1 μM) and CCCP (100 μM) (b), and vanadate (500 μM), DCCD (500 μM), and amiloride (500 μM) (c) on the growth of Bacillus strain TA2.A1 in minimal medium containing glutamate; 100 μl of a 100% ethanol solution was added to controls where inhibitor was dissolved in ethanol. Arrows indicate addition of inhibitor or ethanol.
The Δp, and therefore the energized state of the membrane, can be abolished by proton conductors or uncouplers such as CCCP (11). Monensin is a carboxylic ionophore that disrupts sodium or potassium gradients or both across bacterial membranes (21). When monensin was added to exponential-phase cells growing on glutamate, there was an immediate cessation of growth, even when as little as 0.1 μM was added (Fig. 1b). Growth was completely inhibited, and cells did not become resistant to monesin even after prolonged incubation. Cells challenged with CCCP (100 μM) in the exponential phase of growth were not initially inhibited, but after 2 h of further incubation there was a significant decline in the growth rate (Fig. 1b). This effect was more dramatic if higher concentrations of CCCP (500 μM) were added. Because CCCP is a weak acid and causes a collapse of the Δp by cycling between the protonated and unprotonated states, this process may be somewhat reduced at high pH and hence the small effect seen on growth. When amiloride (500 μM), an inhibitor of Na+/H+ antiporters (12), was added to exponentially growing cells, there was an immediate cessation of growth (Fig. 1c). Similarly, DCCD (25 μM), an inhibitor of the F1F0-ATPase, also caused growth to cease (Fig. 1c). Vanadate (500 μM), an inhibitor of V-type ATPases, had no effect on the growth of Bacillus strain TA2.A1 (Fig. 1c).
[14C]glutamate transport.
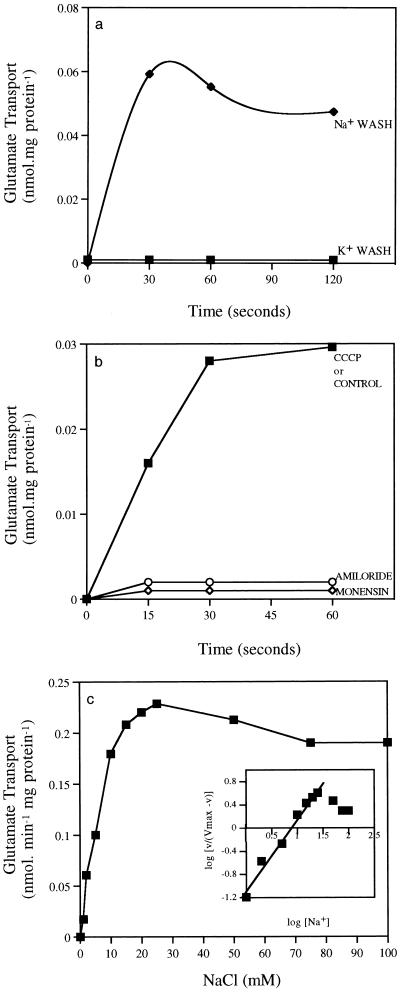
Cells grown in minimal medium containing glutamate and washed twice in Tris-HCl (100 mM, pH 9.0) containing 100 mM NaCl transported [14C]glutamate at an initial rate of 0.12 nmol · min−1 mg of protein (Fig. 2a). However, if cells were washed in Tris-HCl (100 mM, pH 9.0) containing 100 mM KCl, little if any uptake was observed (Fig. 2a). When sodium-washed cells were preincubated with either monensin (10 μM) or amiloride (500 μM), the uptake of [14C]glutamate was completely abolished (Fig. 2b). Preincubation with CCCP (10 μM) had no effect on [14C]glutamate transport. To better understand this requirement for extracellular sodium, we performed experiments in which the extracellular concentration of sodium was varied. Cells resuspended in Tris-HCl (pH 9.0) containing 100 mM KCl did not transport [14C]glutamate at a rate that could be differentiated from that of nonspecific [14C]glutamate binding to cells (Fig. 2c). When the extracellular concentration of NaCl was increased to greater than 5 mM, the rate of [14C]glutamate uptake increased and reached a maximum at 25 mM extracellular NaCl (Fig. 2c). Further increases in sodium concentration did not increase the rate of [14C]glutamate transport. The Km for sodium as calculated from an Eadie-Hofstee plot was 5.6 mM, and the slope of the Hill plot was approximately 1.0, suggesting that sodium binding was noncooperative (Fig. 2c, inset).
FIG. 2.
(a) [14C]glutamate transport by washed cells of Bacillus strain TA2.A1 with either 100 mM NaCl or 100 mM KCl (without sodium). The final concentration of [14C]glutamate was 1 μM. (b) Effects of monensin (10 μM), CCCP (100 μM), and amiloride (100 μM) on [14C]glutamate (1 μM) transport by washed cells of Bacillus strain TA2.A1. The assay buffer contained 100 mM NaCl. (c) Effect of extracellular sodium chloride concentration on [14C]glutamate (1 μM) uptake by washed cells of Bacillus strain TA2.A1. A Hill plot of the data is shown in the inset.
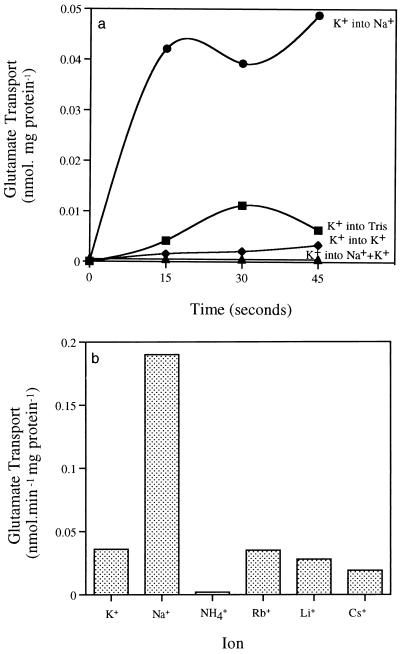
To demonstrate that sodium and therefore a ΔpNa was the driving force for [14C]glutamate transport, we used an artificially generated ΔΨ and ΔpNa to study [14C]glutamate transport. When K+-loaded cells were diluted into Tris-HCl containing 100 mM NaCl to create a ΔpNa and ΔΨ, the rate of [14C]glutamate uptake was 0.16 nmol · min−1 mg of protein (Fig. 3a). K+-loaded cells diluted in Tris-HCl containing both 100 mM NaCl and 100 mM KCl to create a ΔpNa in the absence of ΔΨ did not transport [14C]glutamate, suggesting that ΔΨ was required for glutamate transport. An artificially generated ΔΨ (K+-loaded cells diluted in Tris-HCl) in the absence of a ΔpNa was able to drive [14C]glutamate transport, but the level was fourfold lower, indicating that sodium was required in addition to a ΔΨ. No [14C]glutamate transport was observed when K+-loaded cells were diluted into Tris-HCl containing KCl (no driving force). Other ions were tested for the ability to act as a coupling ion for [14C]glutamate transport (Fig. 3b). Cells (K+ loaded) diluted into buffer containing 100 mM NaCl transported [14C]glutamate at a rate of 0.19 nmol · min−1 mg of protein (Fig. 3b). The ions Rb+, Li+, Cs+, and NH4+ were ineffective in driving [14C]glutamate transport (Fig. 3b).
FIG. 3.
(a) Transport of [14C]glutamate by valinomycin-treated and potassium (100 mM KCl)-loaded TA2.A1 cells. K+-loaded cells were diluted into 100 mM Tris-HCl buffer (pH 9.2) to create a ΔΨ, Tris-HCl buffer containing either 100 mM KCl (no driving force), 100 mM NaCl to create a ΔΨ and ΔpNa, or 100 mM NaCl and 100 mM KCl to create a ΔpNa in the absence of ΔΨ. (b) Cation specificity of the glutamate uptake system of Bacillus strain TA2.A1. Transport of [14C]glutamate (1 μM) was measured in valinomycin-treated, potassium-loaded cells, which were diluted into Tris-HCl (pH 9.2) containing 100 mM KCl, NaCl, NH4Cl, RbCl, LiCl, or CsCl.
[14C]glutamate transport versus glutamate concentration, temperature, and pH.
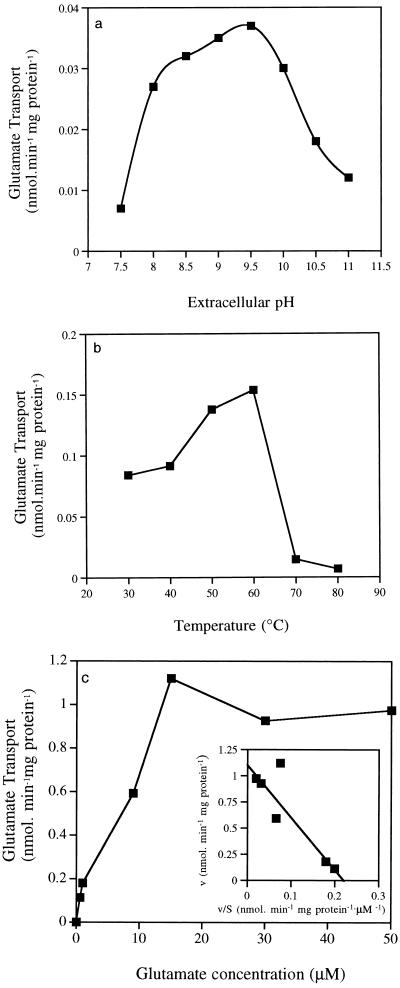
The rate of glutamate uptake was studied over a range of glutamate concentrations, pH values, and temperatures. The pH range for glutamate uptake correlated well with the pH profile for growth, with the greatest uptake at pH 9.5 and low levels of uptake at pH 7.5 and 11.0 (Fig. 4a). Maximum [14C]glutamate uptake was observed at 60°C, and the rate decreased rapidly above this temperature. Temperatures below 40°C decreased the rate by 50% (Fig. 4b). When the extracellular glutamate concentration was increased from 1 to 50 μM, the rate of glutamate transport increased rapidly and saturation kinetics were observed (Fig. 4c). The Eadie-Hofstee plot was linear; the Km for glutamate was 2.90 μM, and the Vmax was 0.7 nmol · min−1 mg of protein (Fig. 4c, inset).
FIG. 4.
Effects of extracellular pH (a), temperature (b), and glutamate concentration (c) on the rate of [14C]glutamate (1 μM) transport by washed cells of Bacillus strain TA2.A1. An Eadie-Hofstee plot of glutamate transport is shown in the inset in panel c.
Competitive amino acids of [14C]glutamate transport.
The specificity of the glutamate transport system was determined by measuring the uptake of [14C]glutamate in the presence of a 50-fold excess of other amino acids (Table 1). [14C]glutamate uptake was specifically inhibited by l-glutamate, d-proline, and the glutamate analogues dl-α-methylglutamate, l-cysteate, and d- and l-aspartate (Table 1). d-Glutamate had no effect on [14C]glutamate uptake, suggesting that the transporter was specific for the l isomer.
TABLE 1.
Effects of competing amino acids on the uptake of [14C]glutamate by Bacillus strain TA2.A1a
| Competing amino acid (5 mM) | Inhibition (% of control level) |
|---|---|
| d-Aspartate | 57 |
| l-Aspartate | 60 |
| l-Cysteate | 50 |
| l-Glutamate | 93 |
| α-Methylglutamate | 45 |
| d-Proline | 38 |
| l-Serine | 50 |
The following amino acids had no (<10%) effect on [14C]glutamate uptake: d-arginine, l-arginine, dl-asparagine, d-glutamate, l-glutamine, l-proline, and d-serine.
DISCUSSION
Bacteria that grow at extremes of pH and temperature are confronted by two bioenergetic problems. First, at high growth temperatures, the cytoplasmic membrane becomes more permeable to ions, including protons, and therefore the use of protons as coupling ions for solute transport and ATP generation has limitations (20, 27, 28). The use of sodium as a coupling ion for bioenergetic processes can be an important energetic advantage because phospholipid membranes are 6 to 10 orders of magnitude less permeable for sodium compared to protons (5, 28). The second bioenergetic problem is that of alkaline pH. The acidification of cytoplasmic pH at alkaline extracellular pH creates special bioenergetic problems. If the pH gradient is reversed (pHin < pHout), the electrical potential must increase to prevent an overall decline in the total Δp. This chemiosmotically adverse pH gradient is bypassed by the use of an electrochemical gradient of Na+ rather than of protons to energize solute uptake and motility (1, 16, 17, 22). In this study, we describe the growth of an obligate aerobic, alkaliphilic, thermophilic Bacillus isolate, strain TA2.A1, which has a pH and temperature optimum of 9.2 and 70°C, respectively. Microbial growth under these conditions has not been previously described. Growth of Bacillus strain TA2.A1 was completely dependent on sodium (>5 mM) and was inhibited by monensin and amiloride, inhibitors that collapse the ΔpNa and inhibit Na+/H+ antiporters, respectively (12, 21).
Because Bacillus strain TA2.A1 is an obligate aerobe and has a growth requirement for sodium, it was of further interest to determine what coupling ion this bacterium uses to drive bioenergetic processes such as transport. Bacillus strain TA2.A1 grew on glutamate as the sole carbon and energy source in minimal medium. To study the precise nature of the driving force for glutamate transport, glutamate uptake in response to an artificially imposed ion gradient was used. Glutamate transport could be driven by an artificially created ΔpNa but only in the presence of a ΔΨ. ΔΨ alone was a weak driving force for glutamate transport. When both gradients, i.e., ΔpNa and ΔΨ, were applied, the highest rate of glutamate transport was observed. These results demonstrate that glutamate uptake is driven by ΔΨ with sodium as a coupling ion. Because CCCP had no effect on glutamate transport, the transmembrane pH gradient (ZΔpH) does not seem to play an important role in glutamate uptake. The use of different monovalent cations to create an artificial membrane potential demonstrated that only sodium was able to couple effective glutamate uptake by Bacillus strain TA2.A1. Two glutamate transport mechanisms have been described for thermophilic bacteria. In Bacillus stearothermophilus, l-glutamate (or l-aspartate) transport proceeds via a sodium/proton symport mechanism with a 1:1:1 stoichiometry (3, 4, 8). Glutamate transport in other thermophilic bacteria has also been shown to depend on an electrochemical gradient of sodium. For example, Clostridium fervidus, a fermentative bacterium, has been shown to transport glutamate electrogenically (ΔΨ driven) in symport with two Na+ molecules and not by ZΔpH (26). Glutamate transport by the aerobic thermophilic bacterium Thermus thermophilus was also shown to be catalyzed by a sodium/glutamate symport mechanism (9).
Na+ is the predominant ion for solute transport in obligately alkaliphilic bacteria, and often in these bacteria the Na+-solute symporter has a low affinity for sodium (16). The Na+–H+–l-glutamate transport system in the thermophilic organism B. stearothermophilus has a very high affinity for sodium (Km < 5.5 μM) (8). In the present study, the effect of sodium ion concentration on glutamate transport followed Michaelis-Menten kinetics. The Hill plot suggested that the glutamate transporter had only one binding site for sodium. The Km for sodium was 5.6 mM, and glutamate transport was completely abolished by 0.1 μM monensin but not the protonophore CCCP. The results of this study indicate that Bacillus strain TA2.A1 transports glutamate electrogenically (ΔΨ driven) in symport with sodium. Like other sodium/glutamate symporters (5), the affinity for sodium is low (>5 mM).
Initial rates of l-glutamate uptake in B. stearothermophilus have been shown to be strongly dependent on the medium pH. Glutamate uptake was highest at low external pH (5.5 to 6.0) and declined with increasing pH (4, 8). Glutamate transport by Bacillus strain TA2.A1 was also affected by the extracellular pH, and the maximum rate was observed at an extracellular pH of 9.5. The Km for glutamate was 2.90 μM, and glutamate uptake by Bacillus strain TA2.A1 was specifically inhibited by the glutamate analogues dl-α-methylglutamate, l-cysteate, and d- and l-aspartate. The closely related amines l-glutamine and dl-asparagine did not inhibit glutamate transport. The glutamate uptake system of C. fervidus is not competitively inhibited by α-methylglutamate (26), suggesting that the glutamate transporter from Bacillus strain TA2.A1 is more like the glutamate transporter in Escherichia coli (6, 7) that is specifically inhibited by this analogue (24). The glutamate transporter in B. stearothermophilus is specific for acidic amino acids and is also most similar to that of E. coli (7, 24).
Despite the inverted pH gradient and the reduced Δp in alkaliphiles, ATP synthesis occurs via completely proton-coupled oxidative phosphorylation, but the mechanism for this remains unknown (14, 15). The Na+-ATPase from the thermophile C. fervidus functions as a Na+-extruding ATPase, stimulated to the same extent by both LiCl and NaCl (25). Speelmans et al. (25) found no evidence for an additional H+-pumping ATPase. Bacillus strain TA2.A1 grows aerobically and presumably generates the bulk of its Δp by respiration. CCCP had no effect on glutamate transport, but growth did slow in the presence of this protonophore, suggesting that the collapse of the Δp causes a decrease in ATP synthesis and hence anabolic reactions such as growth. DCCD, an inhibitor of the F1F0-ATPase (H type), inhibited growth, but vanadate, an inhibitor of V-type ATPases, had no effect on the growth of Bacillus strain TA2.A1. Further studies are needed to determine the precise mode of ATP generation in Bacillus strain TA2.A1.
ACKNOWLEDGMENTS
C. J. Peddie was supported by a University of Waikato Post-Graduate Fees Scholarship. G. M. Cook was supported by a travel grant from the School of Medical Sciences, University of Otago, Dunedin, New Zealand.
REFERENCES
- 1.Booth I R. Regulation of cytoplasmic pH. Microbiol Rev. 1985;49:359–378. doi: 10.1128/mr.49.4.359-378.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cook G M, Russell J B, Reichert A, Wiegel J. The intracellular pH of Clostridium paradoxum, an anaerobic, alkaliphilic, and thermophilic bacterium. Appl Environ Microbiol. 1996;62:4576–4579. doi: 10.1128/aem.62.12.4576-4579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Vrij W, Speelmans G, Heyne R I R, Konings W N. Energy transduction and amino acid transport in thermophilic aerobic and fermentative bacteria. FEMS Microbiol Rev. 1990;75:183–200. [Google Scholar]
- 4.de Vrij W, Bulthuis R A, van Iwaarden P R, Konings W N. Mechanism of l-glutamate transport in membrane vesicles from Bacillus stearothermophilus. J Bacteriol. 1989;171:1118–1125. doi: 10.1128/jb.171.2.1118-1125.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dimroth P. Sodium ion transport decarboxylases and other aspects of sodium cycling in bacteria. Microbiol Rev. 1987;51:320–340. doi: 10.1128/mr.51.3.320-340.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujimura T, Yamato I, Anraku Y. Mechanism of glutamate transport in Escherichia coli B. 1. Proton-dependent and sodium-ion dependent binding of glutamate to a glutamate carrier in the cytoplasmic membrane. Biochemistry. 1983;22:1954–1959. doi: 10.1021/bi00277a033. [DOI] [PubMed] [Google Scholar]
- 7.Fujimura T, Yamato I, Anraku Y. Mechanism of glutamate transport in Escherichia coli B. 2. Kinetics of glutamate transport driven by artificially imposed proton and sodium ion gradients across the cytoplasmic membrane. Biochemistry. 1983;22:1959–1965. doi: 10.1021/bi00277a034. [DOI] [PubMed] [Google Scholar]
- 8.Heyne R I R, de Vrij W, Crielaard W, Konings W N. Sodium ion-dependent amino acid transport in membrane vesicles of Bacillus stearothermophilus. J Bacteriol. 1991;173:791–800. doi: 10.1128/jb.173.2.791-800.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holtom G J, Sharp R J, Williams R A D. Sodium-stimulated transport of glutamate by Thermus thermophilus strain B. J Gen Microbiol. 1993;139:2245–2250. [Google Scholar]
- 10.Horikoshi K. Microorganisms in alkaline environments. Tokyo, Japan: Kodansha Ltd.; 1991. , and VCH Publishers Inc., Weinheim, Germany. [Google Scholar]
- 11.Kaback H R, Reeves J P, Short S A, Lombardi F J. Mechanisms of active transport in isolated bacterial membrane vesicles. XVIII. The mechanism of action of carbonylcyanide m-chlorophenylhydrazone. Arch Biochem Biophys. 1974;160:215–222. doi: 10.1016/s0003-9861(74)80028-7. [DOI] [PubMed] [Google Scholar]
- 12.Kleyman T R, Cragoe E J J. Amiloride and its analogs as tools in the study of ion transport. J Membr Biol. 1988;105:1–21. doi: 10.1007/BF01871102. [DOI] [PubMed] [Google Scholar]
- 13.Kroll R G. Alkalophiles. In: Edwards C, editor. Microbiology of extreme environments. New York, N.Y: McGraw-Hill; 1990. pp. 55–92. [Google Scholar]
- 14.Krulwich T A, Ito M, Gilmour R, Sturr M G, Guffanti A A, Hicks D B. Energetic problems of extremely alkaliphilic aerobes. Biochim Biophys Acta. 1996;1275:21–26. doi: 10.1016/0005-2728(96)00044-8. [DOI] [PubMed] [Google Scholar]
- 15.Krulwich T A, Guffanti A A. Proton-coupled bioenergetic processes in extremely alkaliphilic bacteria. J Bioenerg Biomembr. 1992;24:587–599. doi: 10.1007/BF00762351. [DOI] [PubMed] [Google Scholar]
- 16.Krulwich T A, Guffanti A A. The Na+ cycle of extreme alkalophiles: a secondary Na+/H+ antiporter and Na+/solute symporters. J Bioenerg Biomembr. 1989;21:663–678. doi: 10.1007/BF00762685. [DOI] [PubMed] [Google Scholar]
- 17.Krulwich T A. Bioenergetics of alkalophilic bacteria. J Membr Biol. 1986;89:113–125. doi: 10.1007/BF01869707. [DOI] [PubMed] [Google Scholar]
- 18.Li Y, Engle M, Weiss N, Mandelco L, Wiegel J. Clostridium thermoalcaliphilium sp. nov., an anaerobic and thermotolerant facultative alkaliphile. Int J Syst Bacteriol. 1994;44:111–118. doi: 10.1099/00207713-44-1-111. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Mandelco L, Wiegel J. Isolation and characterization of a moderately thermophilic anaerobic alkalophile, Clostridium paradoxum sp. nov. Int J Syst Bacteriol. 1993;43:450–460. [Google Scholar]
- 20.Nichols J W, Deamer D W. Net proton-hydroxyl permeability of large unilamellar liposomes measured by an acid-base titration technique. Proc Natl Acad Sci USA. 1980;77:2038–2042. doi: 10.1073/pnas.77.4.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pressman B C. Biological applications of ionophores. Annu Rev Biochem. 1976;45:501–530. doi: 10.1146/annurev.bi.45.070176.002441. [DOI] [PubMed] [Google Scholar]
- 22.Prowe S G, van de Vossenburg J L, Driessen A J, Antranikian G, Konings W N. Sodium-coupled energy transduction in the newly isolated thermoalkaliphilic strain LBS3. J Bacteriol. 1996;178:4099–4104. doi: 10.1128/jb.178.14.4099-4104.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saiki T, Kobayashi Y, Kawagoe K, Beppu T. Dictyoglomus thermophilum gen. nov., sp. nov., a chemoorganotrophic, anaerobic, thermophilic bacterium. Int J Syst Bacteriol. 1985;35:253–259. [Google Scholar]
- 24.Schellenberg G D, Furlong C E. Resolution of the multiplicity of the glutamate and aspartate transport systems of Escherichia coli. J Biol Chem. 1977;252:9055–9064. [PubMed] [Google Scholar]
- 25.Speelmans G, Poolman B, Abee T, Konings W N. The F- or V-type Na+-ATPase of the thermophilic bacterium Clostridium fervidus. J Bacteriol. 1994;176:5160–5162. doi: 10.1128/jb.176.16.5160-5162.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Speelmans G, Poolman B, Konings W N. Amino acid transport in the thermophilic anaerobe Clostridium fervidus is driven by an electrochemical sodium gradient. J Bacteriol. 1993;175:2060–2066. doi: 10.1128/jb.175.7.2060-2066.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tolner B, Poolman B, Konings W N. Adaptation of microorganisms and their transport systems to high temperatures. Comp Biochem Physiol A. 1997;118:423–428. doi: 10.1016/s0300-9629(97)00003-0. [DOI] [PubMed] [Google Scholar]
- 28.van de Vossenburg J L, Ubbink-Kok T, Elferink M G, Driessen A J, Konings W N. Ion permeability of the cytoplasmic membrane limits the maximum growth temperature of bacteria and archaea. Mol Microbiol. 1995;18:925–932. doi: 10.1111/j.1365-2958.1995.18050925.x. [DOI] [PubMed] [Google Scholar]