Abstract
Recent understanding of how the systemic environment shapes the brain throughout life has led to numerous intervention strategies to slow brain ageing 1–3. Cerebrospinal fluid (CSF) makes up the immediate environment of brain cells providing them with nourishing compounds 4,5. We discovered that infusing young CSF directly into aged brains improves memory function. Unbiased transcriptome analysis of the hippocampus identified oligodendrocytes to be most responsive to this rejuvenated CSF environment. We further revealed that young CSF boosts oligodendrocyte progenitor cell (OPC) proliferation and differentiation in the aged hippocampus and in primary OPC cultures. Using SLAMseq to metabolically label nascent mRNA, we identified serum response factor (SRF), a transcription factor that drives actin cytoskeleton rearrangement, as a mediator of OPC proliferation following young CSF exposure. With age, SRF expression decreases in hippocampal OPCs, and the pathway is induced by acute young CSF injection. We screened for potential SRF activators in CSF and found that fibroblast growth factor 17 (Fgf17) infusion is sufficient to induce OPC proliferation and long-term memory consolidation in aged mice while Fgf17 blockade impairs cognition in young mice. These findings demonstrate the rejuvenating power of young CSF and identify Fgf17 as a key target to restore oligodendrocyte function in the ageing brain.
Brain ageing underlies dementia and neurodegenerative diseases, imposing an immense societal burden. Systemic interventions in model organisms have shown great promise in reversing ageing related decline of various tissues, including the brain1. For example, heterochronic parabiosis and young plasma transfer rejuvenated the aged brain and restored memory function 2,3. Nevertheless, the brain is protected with barriers which may limit access of these factors, presumably impeding their rejuvenation potential. Cerebrospinal fluid (CSF), which is in close association with brain cells, carries signals that instruct neuronal progenitor proliferation and specification during development 6. However, CSF protein composition changes with human ageing 7, marked by an increase in inflammatory proteins 8 and decrease in growth factors such as BDNF 9. Whether these CSF changes contribute to age-related cognitive decline is unknown. Testing this functionally by performing CSF transfers in vivo has been difficult due to technical limitations in CSF collection and direct CSF infusion to the brain. We hypothesized that intracerebroventricular (i.c.v) administration of young CSF to aged mice will have rejuvenating effects on the brain (Fig. 1a).
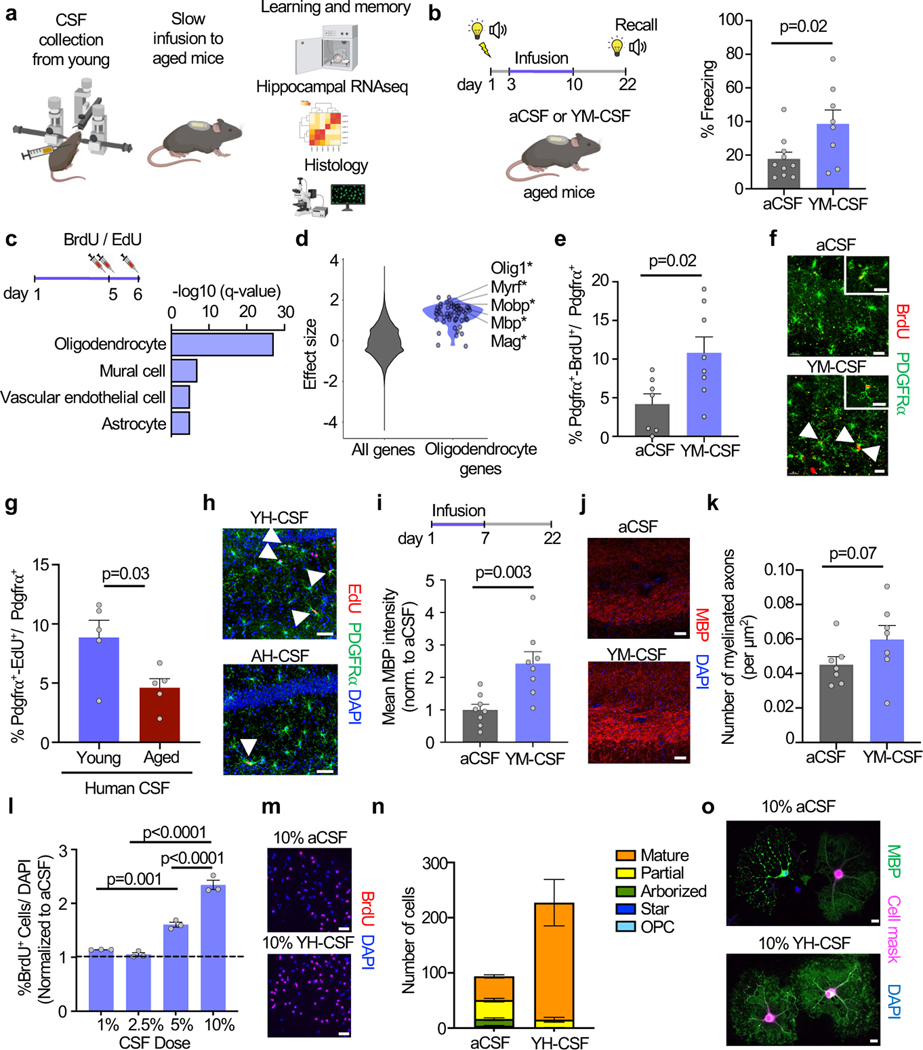
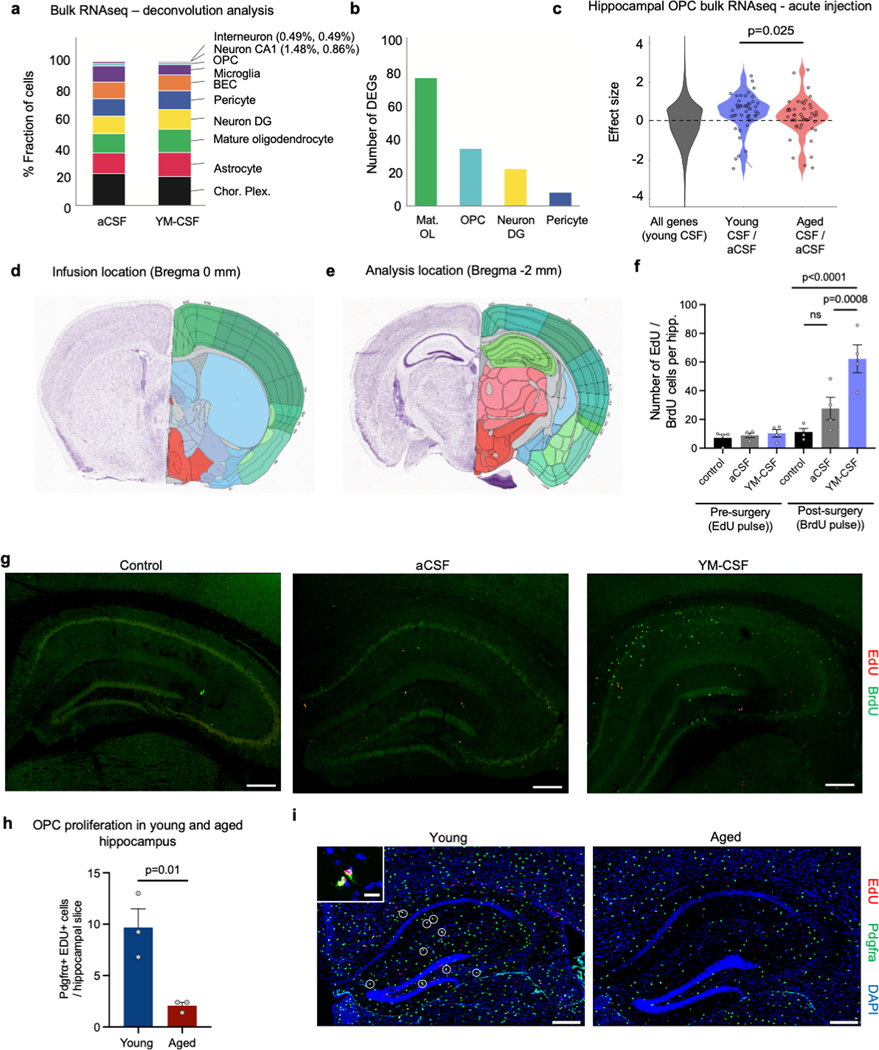
Figure 1. Young CSF improves memory consolidation and promotes OPC proliferation and differentiation.
a, Overview of the experimental paradigm.
b, Percentage of freezing of 20-month-old mice in the remote recall contextual fear conditioning test (aCSF n=10, YM-CSF n=8; two-sided t-test; mean ± s.e.m.).
c, Gene set enrichment analysis of hippocampal bulk RNAseq identifies oligodendrocytes genes as highly upregulated following 6 days of YM-CSF infusion.
d, Effect size of oligodendrocyte genes in YM-CSF vs. aCSF hippocampus compared to all genes in the dataset (asterisk indicates FDR<0.1) (aCSF n=8, YM-CSF n=7).
e, Quantification of OPC proliferating cells in the hippocampus following a week of aCSF or YM-CSF infusion to 20-month-old mice (aCSF n=7, YM-CSF n=8; two-sided t-test; mean ± s.e.m.).
f, Representative images of e. Arrowheads pointing at proliferating OPCs. Scale bar, 20μm. inserts 5μm.
g, As in e but for YH-CSF and AH-CSF (n=5; two-sided t-test; mean ± s.e.m.).
h, Representative images of g. Scale bar, 50μm.
i, Hippocampal MBP stain following the long-term paradigm of aCSF or YM-CSF infusion (n=8; two-sided t-test; mean ± s.e.m.).
j, Representative images of i. Scale bar, 50μm.
k, Quantification of number of myelinated axons per μm2 in the hippocampus of aged mice following the long-term paradigm of aCSF or YM-CSF infusion (n=7; one-sided t-test; mean ± s.e.m.).
l, Ratio of percentage of BrdU+/DAPI primary rat OPCs treated with indicated % YHCSF over matching aCSF as control (n=3; one-way ANOVA followed by Tukey’s post-hoc test; mean ± s.e.m.).
m, Representative images of l. Scale bar, 50μm.
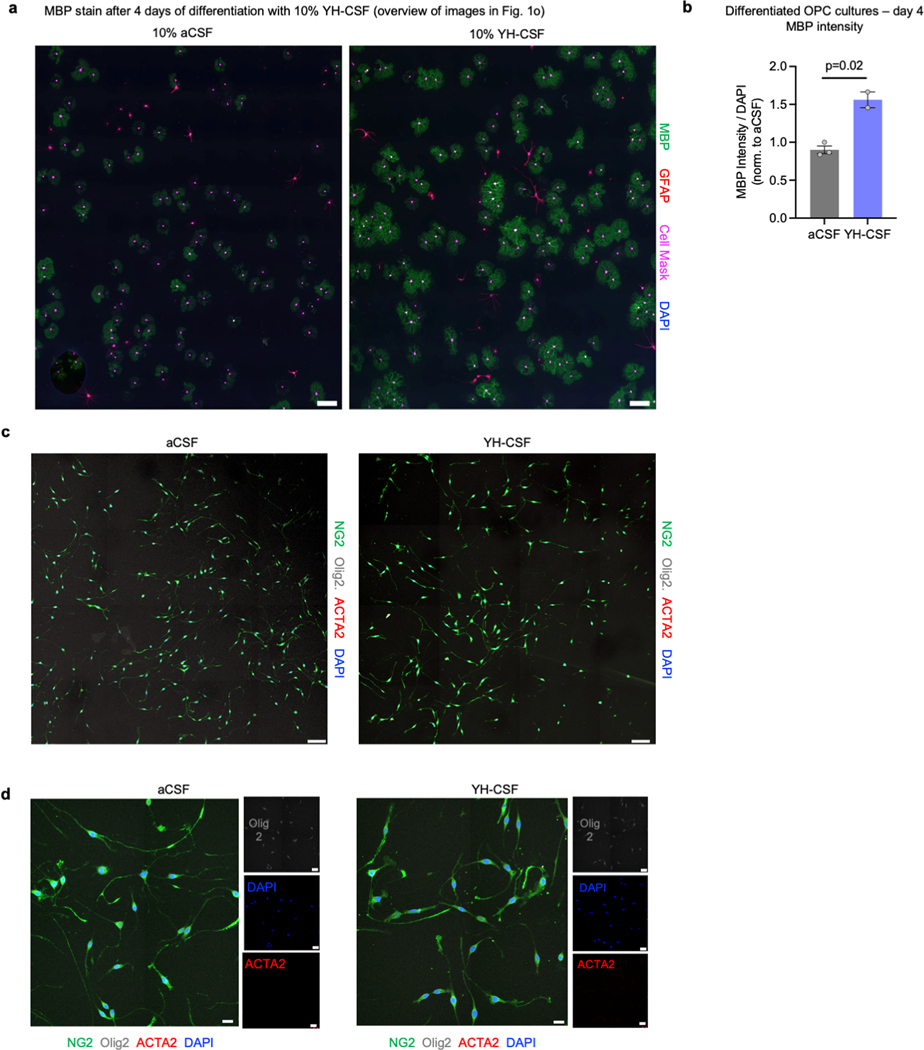
n, Stacked bar plot of average number of cells in each differentiation state at day 4 of differentiation with 10% aCSF or 10% YH-CSF (aCSF n=281 cells analyzed in 3 coverslips, YH-CSF n=454 cells analyzed in 2 coverslips).
o, Representative images of n. Scale bar, 20μm. See Supplementary videos S1-2.
Regulation of oligodendrogenesis by young CSF
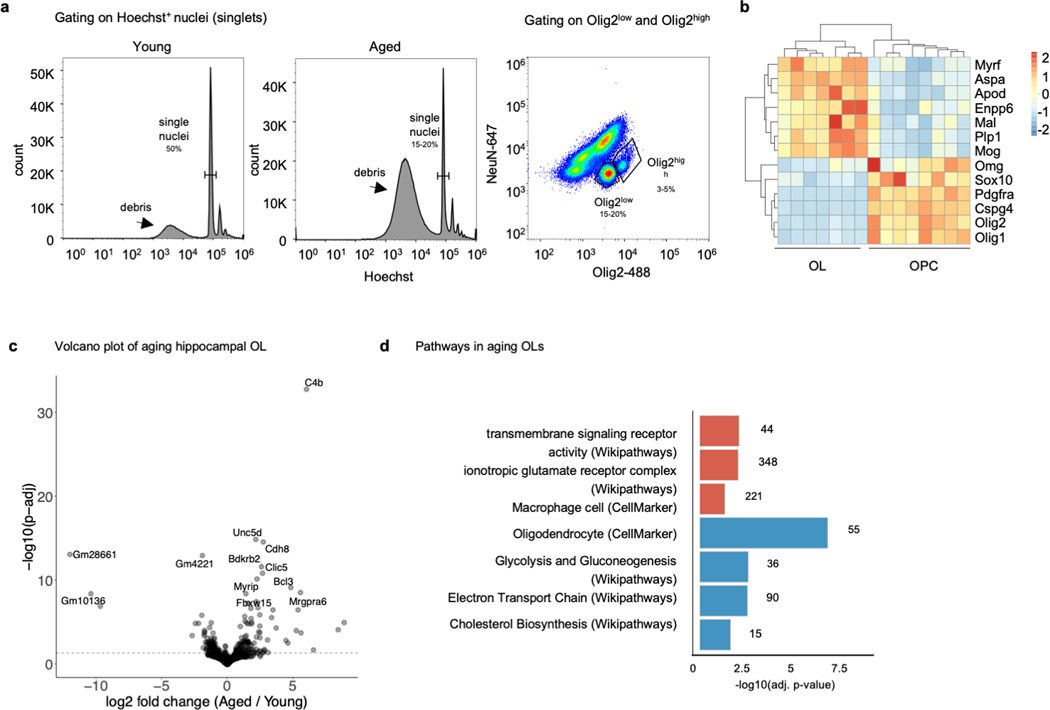
We first sought to test if young CSF infusion to aged mice could improve ageing-related impairments in hippocampal dependent learning and memory tasks 10. 20-month-old mice received three foot-shocks associated with a tone and a flashing light. Mice were then randomly split to two groups and infused with either artificial CSF (aCSF) or young mouse CSF (YM-CSF) for one week and remote memory recall was tested three weeks after memory acquisition. YM-CSF infusion resulted in higher average freezing rates following exposure to the tone and light, suggesting better preservation of the remote fear memory (Fig. 1b). We selected this paradigm because it allows to test interventions that affect remote memory formation and consolidation 11, a process implicated in ageing related cognitive decline 12. Notably, aged mice infused with aCSF had comparable freezing rates to age-matched naïve mice (aCSF 17.82±0.93, naïve 17.73±0.93; mean + SEM). Because the hippocampus is central to age-related cognitive decline and is in close apposition to CSF, we measured the effect of young CSF infusion on the hippocampal transcriptome by bulk RNA sequencing. Differential gene expression analysis between aCSF and YM-CSF identified 271 differentially expressed genes (DEGs) significantly altered (FDR<0.1) after young CSF treatment with 115 downregulated and 156 upregulated (Supplementary Table 1). Strikingly, oligodendrocyte genes were highly upregulated identifying this cell type as a major cellular substrate for CSF (Fig. 1c and Extended data Fig. 1a-b). Specifically, young CSF promoted upregulation of transcription factors driving oligodendrocyte differentiation and major myelin protein components (for example, Olig1, Myrf, Mag, Mbp and Mobp) (Fig. 1d). Consistent with these findings, even acute single infusions of young CSF, but neither aCSF nor aged mouse CSF (AM-CSF) resulted in an upregulation of these oligodendrocyte marker genes 16 hrs after injection (Extended data Fig. 1c and Supplementary Table 2).
Neuronal activity, induced by optogenetic tools or by learning tasks, was shown to promote OPC proliferation and differentiation, as well as to regulate myelin plasticity in mature oligodendrocytes 13. To determine whether OPC proliferation could be underlying the transcriptomic signature observed following young CSF infusion, we labeled dividing cells in the last 2 days of young CSF infusion with thymidine analogues (BrdU or EdU). Interestingly, while overall cell proliferation was very low in these aged brains, we discovered a surge in overall cell proliferation specifically in the hippocampus, relatively distant from the infusion site (Extended data Fig. 1d-i). Young CSF induced a 2.35-fold increase in the percentage of proliferating OPCs (Pdgfrα+BrdU+ / Pdgfrα+) in the CA1 region of the hippocampus but not in the cortex (Fig. 1e-f and Extended data Fig. 2a-f). In an additional cohort of mice, we infused human CSF pooled from healthy young (YH-CSF, mean age 24.6) or aged donors (AH-CSF, mean age 69). YH-CSF induced OPC proliferation at comparable levels to YM-CSF while AH-CSF induced only half as many cells (Fig. 1g-h). Notably, CSF infusions also triggered EdU incorporation in astrocytes and microglia (Extended data Fig. 2g-i). Next, we allowed these proliferating cells to mature for 3 weeks to assess effects of young CSF on hippocampal myelination by MBP staining and transmission electron microscopy (TEM). We found an increase in MBP intensity (Fig. 1i-j) and in the number of myelinated axons (Fig. 1k and Extended data Fig. 3) in the molecular layer of the hippocampus. This indicates that young CSF contains signals that promote cell growth for the oligodendrocyte lineage or substances that neutralize inhibitory factors.
To assess if young CSF can stimulate OPC proliferation and differentiation directly, we used an established primary rat OPC culture system 14. Cells were grown in full proliferation media supplemented with human CSF pools (three pools as replicates, each pool from three young adult healthy males (YH-CSF, mean age 24.6) or aCSF as control). We used human instead of rodent CSF because we can obtain much larger volumes necessary for these studies. Similar to the in vivo infusions, BrdU pulsing confirmed a dose-dependent increase in OPC proliferation (Fig. 1l-m). Excitingly, when OPCs were deprived of mitogens to promote cell differentiation over 4 days, YH-CSF not only induced a 2-fold increase in cell survival – a process in which a significant fraction of cells typically undergo apoptotic cell death 15 – but also promoted a prominent expansion of the more differentiated mature cell morphology 16 with an overall increase in MBP intensity per cell (Fig. 1n-o, Extended data Fig. 4a-b and Supplementary Video 1-2). These results are in line with previous work showing that human CSF from young healthy patients induced neuronal viability 17 while CSF from multiple sclerosis patients was toxic to neuronal 18 and OPC cultures 19.
SRF mediates young CSF effects on OPCs
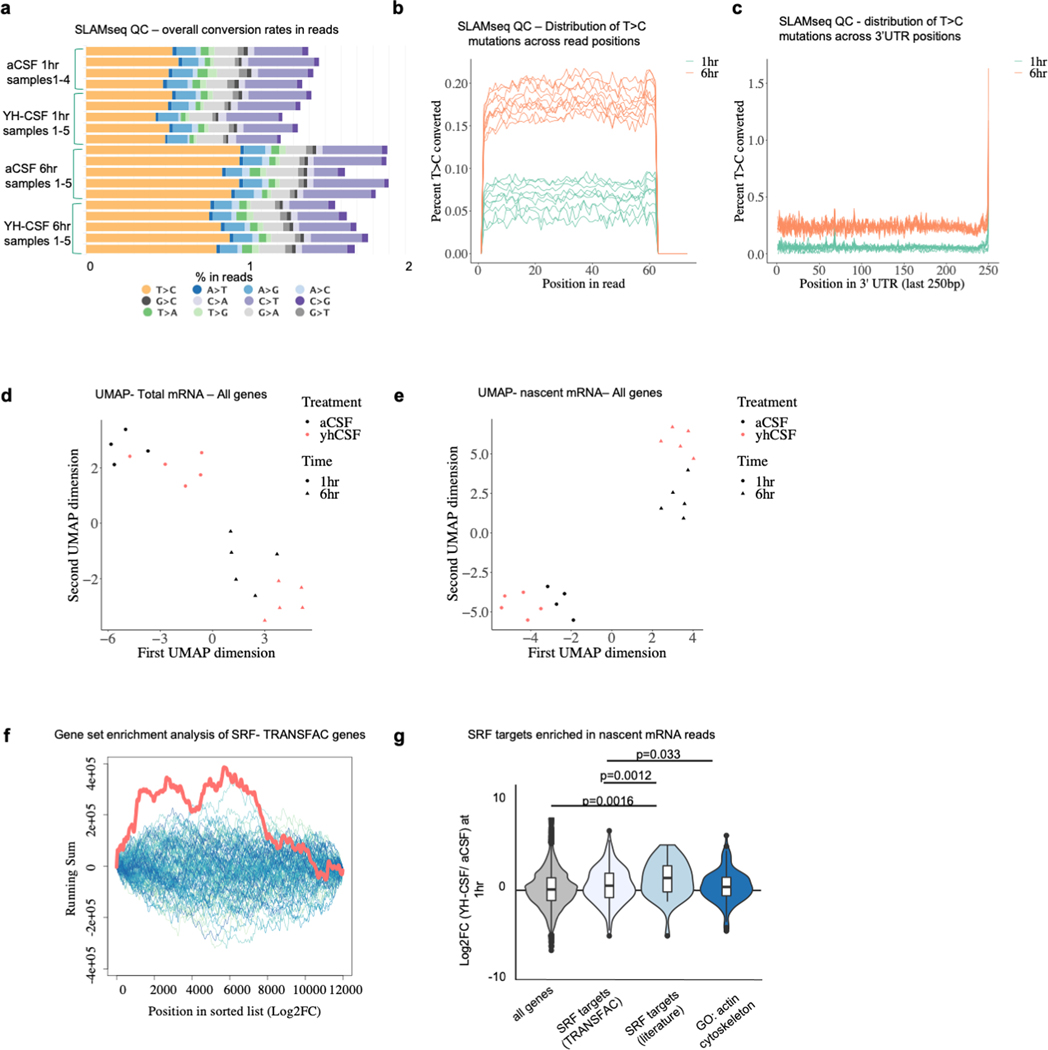
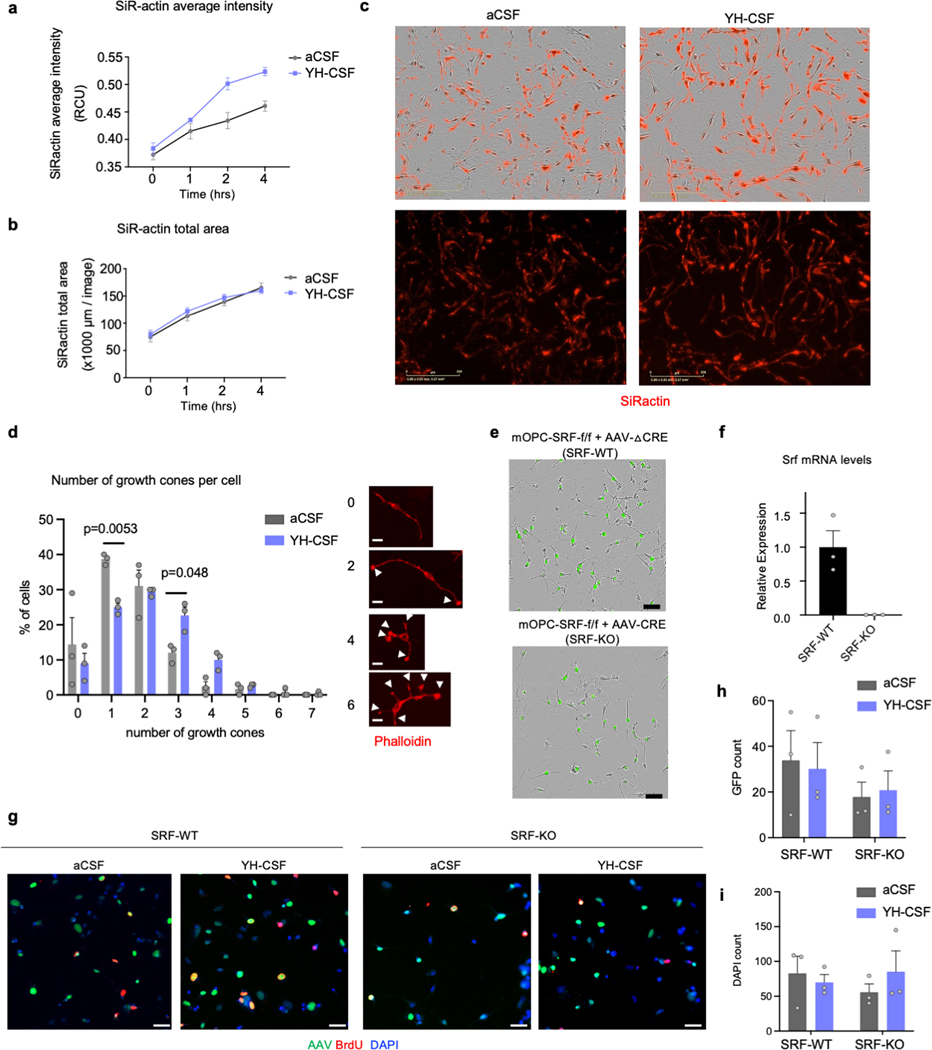
To gain a deeper mechanistic understanding of the cellular processes induced by young CSF in OPCs we metabolically labeled nascent mRNA with 4-thiouridine (s4U) using thiol(SH)-linked alkylation and sequenced RNA (SLAMseq) from cultured OPCs 1- or 6 hr after exposure to YH-CSF. The top gene induced after 1 hr was Serum Response Factor (SRF) (Fig. 2a, Extended data Fig. 5a-e and Supplementary Table 3), a transcription factor studied extensively in skeletal muscle 20, heart 21 and in neurons in the brain 22. SRF binds to the serum response element (SRE) promoter sequences to induce cell motility, proliferation, and differentiation through modulation of immediate early genes (such as Egr1) and the actin cytoskeleton 23. We also noted a dramatic downregulation of the negative regulator of Wnt signaling and pro-apoptotic factor Bcl7b as well as the DNA repair protein Rpa3, in line with an overall pro-survival response following YH-CSF exposure. Nascent SRF mRNA transcripts peaked at 1 hr and returned to baseline by 6 hr (Fig. 2b). Strikingly, many of the DEGs peaking at 6 hr are known SRF target genes (Fig. 2c, marked in red). Indeed, the most strongly increased genes were enriched as “target genes of SRF” based on gene set enrichment analysis (GSEA) and the TRANSFAC database (Extended data Fig. 5f) while the cells maintained OPC identity as determined by NG2 expression (Extended data Fig. 4c-d). The combined log2FC of the TRANSFAC SRF predicted targets (423 genes), validated SRF targets (74 genes, curated list from literature) and actin genes (212 genes) in the dataset indicated an overall activation of the SRF pathway and actin cytoskeleton transcripts (Extended data Fig. 5g and Supplementary Table 4). To test whether CSF-induced SRF expression regulates the OPC actin cytoskeleton, we exposed OPCs to YH-CSF in the presence of SiR-actin, a fluorescent probe used to label actin filaments for live imaging, or we fixed and stained OPCs with the actin filament dye phalloidin. Within hours of YH-CSF stimulation, SiR-actin intensity increased, without change in total area, indicating an increase in cellular actin filament levels (Extended data Fig. 6a-c). In fixed cells, OPCs exposed to YH-CSF for 6 hrs expressed twice as much phalloidin per cell as controls (Fig. 2d-e). The increase in phalloidin intensity was confirmed in hippocampal OPCs in aged mice infused with YM-CSF for 1 week (Fig. 2f-g).
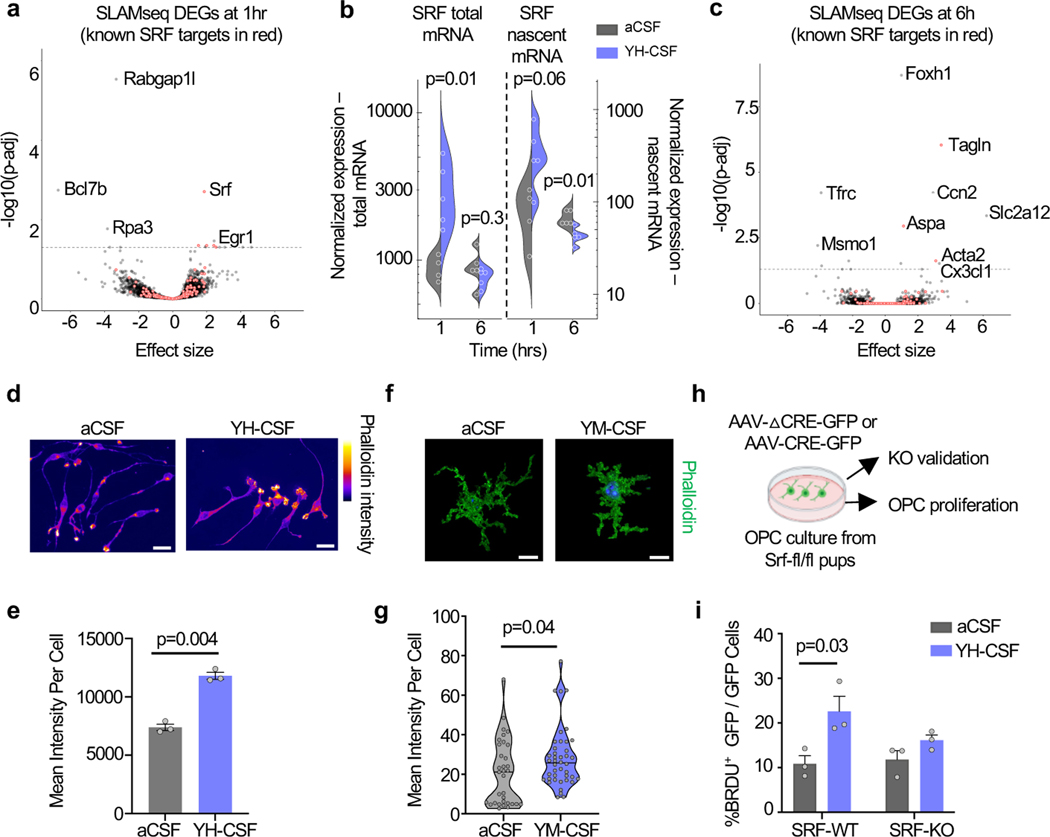
Figure 2. SRF is induced by young CSF and mediates CSF-induced OPC proliferation.
a, Volcano plot of DEGs at 1hr following YH-CSF addition (SRF targets marked in red) (YH-CSF 1hr n=4, rest n=5; p. adjusted value by Wald test in DESeq2; mean ± s.e.m).
b, Normalized expression levels of SRF in nascent mRNA counts and total counts (YH-CSF 1hr n=4, rest n=5; Two-way ANOVA Sidak’s post-hoc test (nascent and total reads separately)).
c, Volcano plot of DEGs at 6hr following YH-CSF addition (SRF targets marked in red). (n=5; p. adjusted value by Wald test in DESeq2; mean ± s.e.m).
d, Representative images of e. Scale bar, 20μm.
e, Mean phalloidin intensity in OPCs 6hr following YH-CSF exposure. (n=3 coverslips per condition; two-sided t-test; mean ± s.e.m.).
f, Representative images of g. Scale bar, 10μm.
g, Mean phalloidin intensity in hippocampal OPCs (Pdgfrα+ following aCSF or YM-CSF infusion for 6 days. (n=3 mice per group, total of 33–42 single cells measured per condition; two-sided t-test; mean ± s.e.m.).
h, Schematic of mouse OPC primary cultures from SRF-fl/fl pups infected with CRE-GFP and △CRE-GFP AAVs to induce recombination.
i, Percentage of proliferating cells (BRDU+/GFP+ cells) in SRF-WT and SRF-KO cells treated with 10% aCSF or YH-CSF (n=3; two-way ANOVA followed by Sidak’s post-hoc test; mean ± s.e.m.).
Because SRF is necessary for the formation of actin filaments in axonal growth cones in neurons 24 we tested if it has a similar role in OPCs and quantified the number of growth cones per OPC. We found YH-CSF induced significantly more growth cones per cell (Extended data Fig. 6d). To test if SRF is mediating the effect of young CSF we infected cultured OPCs from SRF-floxed mice with AAVs expressing Cre-GFP (to create SRF-KO OPCs) or a truncated Cre-GFP as a control (SRF-WT). We repeated the YH-CSF proliferation experiment and showed that CSF induced proliferation is dependent on SRF (Fig, 2h-i and Extended data Fig. 6e-i). These results shed light on the possible mechanisms underlying the effect of young CSF on OPCs and point towards SRF and actin cytoskeleton regulation, as potential mediators of young CSF effects in vivo.
Deletion of SRF signaling specifically in muscle cells causes accelerated ageing phenotypes in skeletal muscle of mice 25 and worms 26. We thus asked whether SRF signaling may be downregulated in OPCs in the ageing brain. Indeed, the fraction of Srf positive OPCs (Srf+ Pdgfrα+/Pdgfrα+) in the CA1 region of the hippocampus detected by in situ hybridization decreased dramatically with age (Fig. 3a-b). To expand this analysis to other SRF targets and cellular processes, we sorted hippocampal OPC and OL nuclei by expression of the oligodendrocyte transcription factor Olig2 (Olig2high for OPCs and Olig2low for OLs) from young (3m) and aged (25m) mice and performed bulk RNAseq (Extended data Fig. 7a-b and Supplementary Table 5). The top pathways downregulated in OPCs with ageing were related to oligodendrocyte cell markers, regulation of glial cell differentiation, cellular respiration and metabolism and protein folding. Conversely, immune-related pathways and microglia-specific genes were upregulated as previously reported for OPCs in ageing 27 and multiple sclerosis 28 (Fig. 3c-d and Extended data Fig. 7c-d). A focused analysis of SRF TRANSFAC genes in ageing OPCs indicated an overall downregulation with age (Fig. 3e, left box plot).
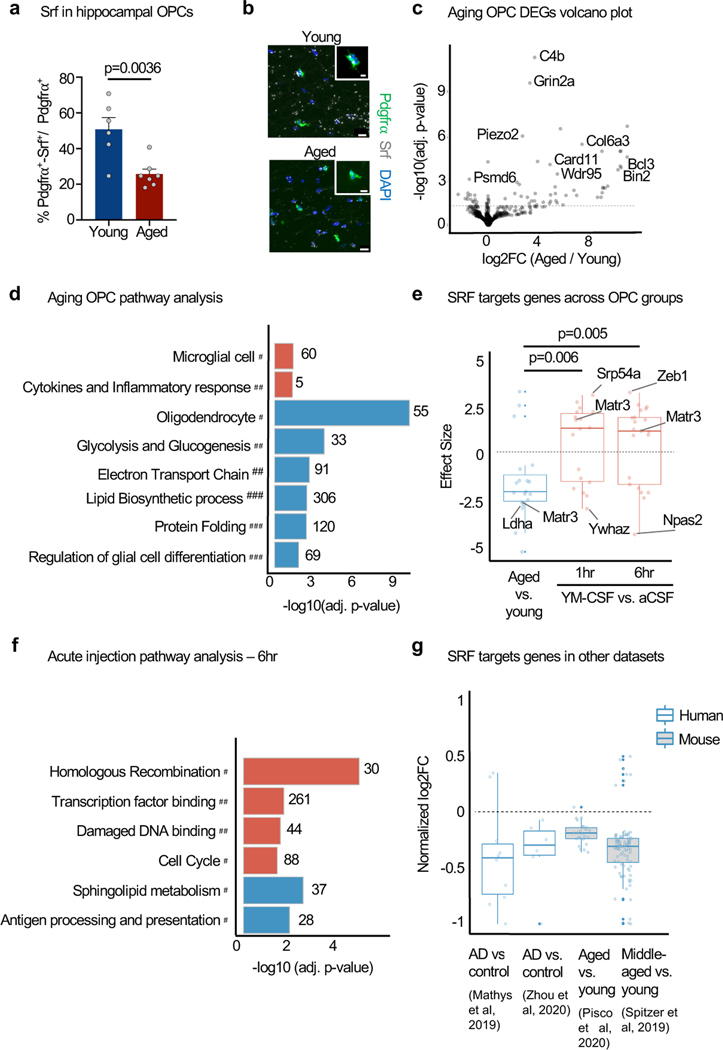
Figure 3. SRF signaling is downregulated in hippocampal OPCs with ageing and induced by acute young CSF injection.
a, Srf mRNA quantified in OPC (Pdgfrα+ nuclei) in the CA1 region of the hippocampus of young (3 months) and aged (22 months) mice. (young n=6, aged n=7; two-sided t-test; mean ± s.e.m.).
b, Representative images of a. Scale bar, 10μm and 5μm in insert.
c, Volcano plot of DEGs of aged vs. young hippocampal OPC nuclei. Dashed line represents p.adj=0.05 (n=4).
d, Pathways enriched (red) or depleted (blue) in hippocampal OPCs with age. Resource categories; # CellMarker; ## Wikipathways; ### GO BP (n=4, unweighted Kolmogorov-Smirnow test).
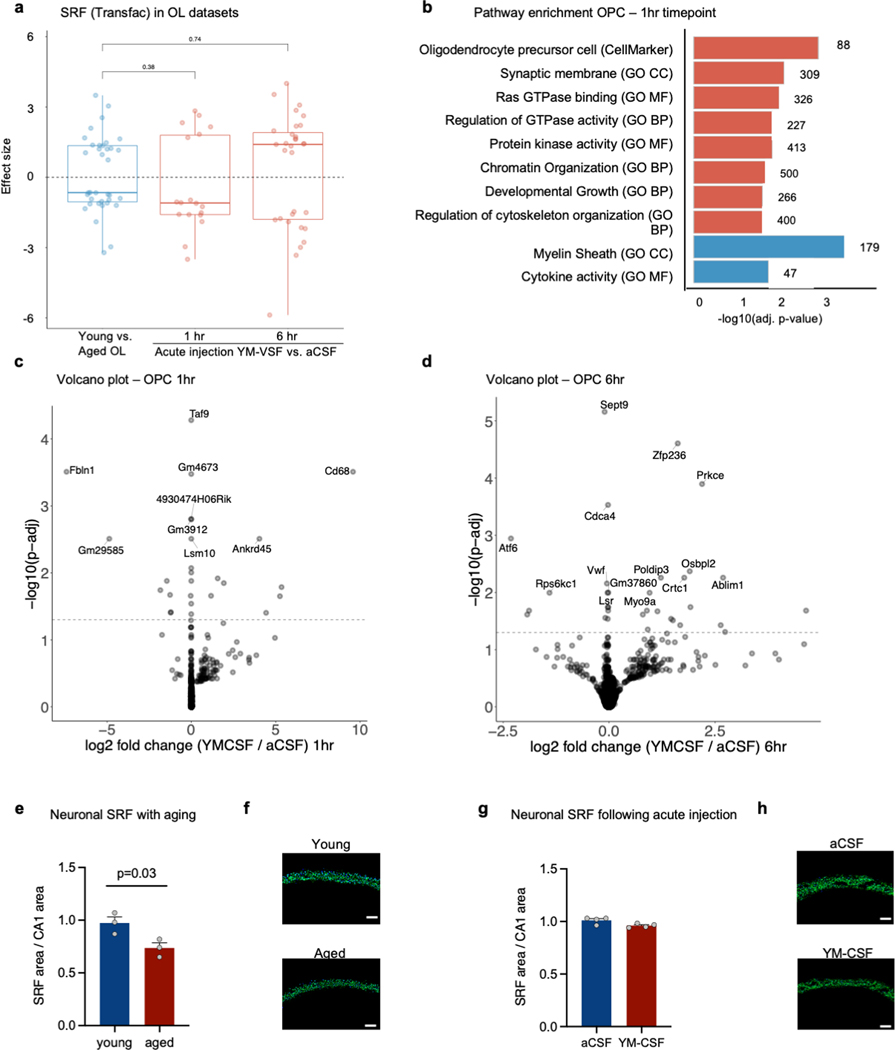
e, Box plot of effect size of Srf targets (TRANSFAC database) in hippocampal OPCs from aged vs. young, YM-CSF vs. aCSF at 1hr and 6hr timepoints (n=4; genes pre-filtered by p<0.05 cutoff; Wilcoxon rank sum test; box show the median and the 25–75th percentiles, and the whiskers indicate values up to 1.5-times the interquartile range).
f, Pathways enriched (red) or depleted (blue) in hippocampal OPCs following 6 hrs of aCSF or YM-CSF injection (n=4). Resource categories; # KEGG; ## GP MF (unweighted Kolmogorov-Smirnow test).
g, Meta-analysis of log2FC of SRF target genes (TRANSFAC) in human AD vs. control and mouse aged vs. young ageing datasets (genes pre-filtered by p<0.05 cutoff; box show the median and the 25–75th percentiles, and the whiskers indicate values up to 1.5-times the interquartile range).
We next aimed to test if young CSF induces SRF pathway activation in vivo in the aged brain. Due to the transient nature of SRF induction in the SLAMseq experiment, we designed an acute injection paradigm where CSF was injected into the lateral ventricle of 18-month-old mice and contralateral hippocampi were dissected 1 and 6 hrs post injection for OPC and OL nuclei RNAseq (Supplementary Table 6). We found again genes predicted to be targets of SRF (based on TRANSFAC) were upregulated in in OPCs at both timepoints (Fig. 3e and Extended data Fig. 8a). At the 6 hr timepoint, OPC genes were linked to SRF-related pathways such as “Regulation of GTPase activity”, “chromatin organization”, “transcription factor binding”, “cell cycle” and “regulation of cytoskeleton organization” (Fig. 3f and Extended data Fig. 8b-d). Notably, SRF target genes were also downregulated in other published human and mouse datasets of OPCs with age and Alzheimer’s disease (AD) (Fig. 3g). Notably, SRF mRNA levels in neurons decrease with ageing and are not changed following CSF acute injection (Extended data Fig. 8e-h). Altogether, these experiments indicate that SRF signaling in OPCs is downregulated with ageing and induced following in vivo CSF infusion.
Fgf17 mimics CSF effects on OPCs and memory
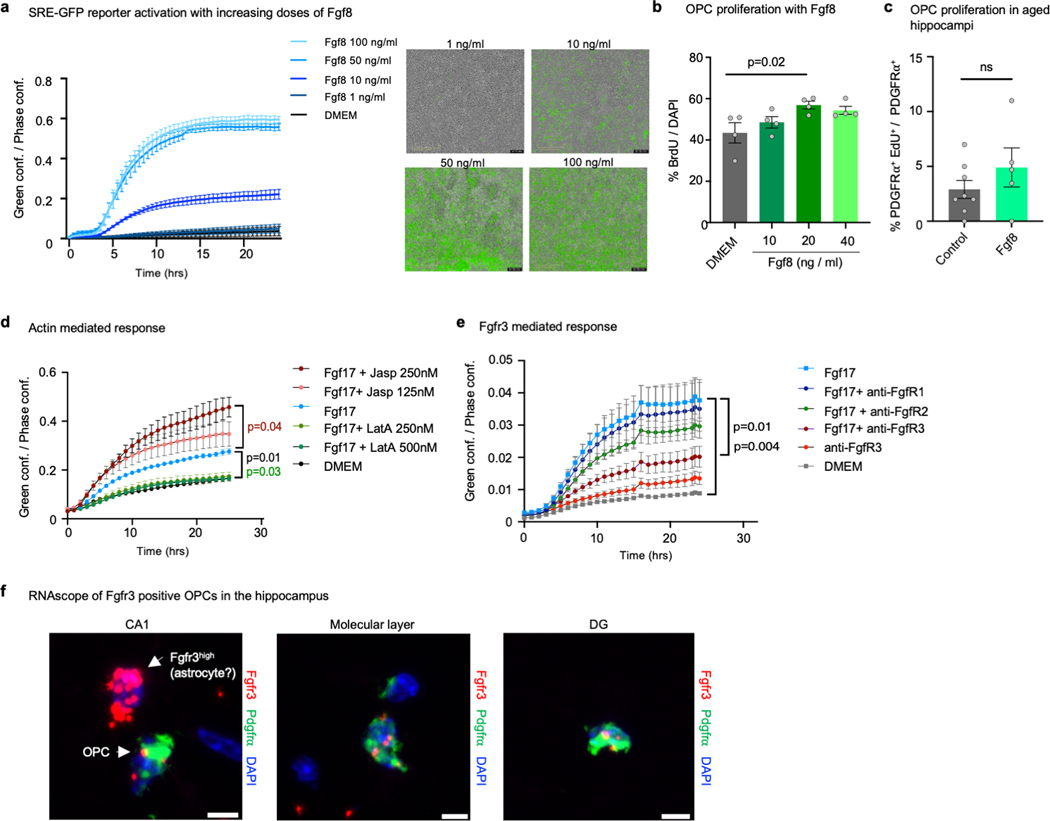
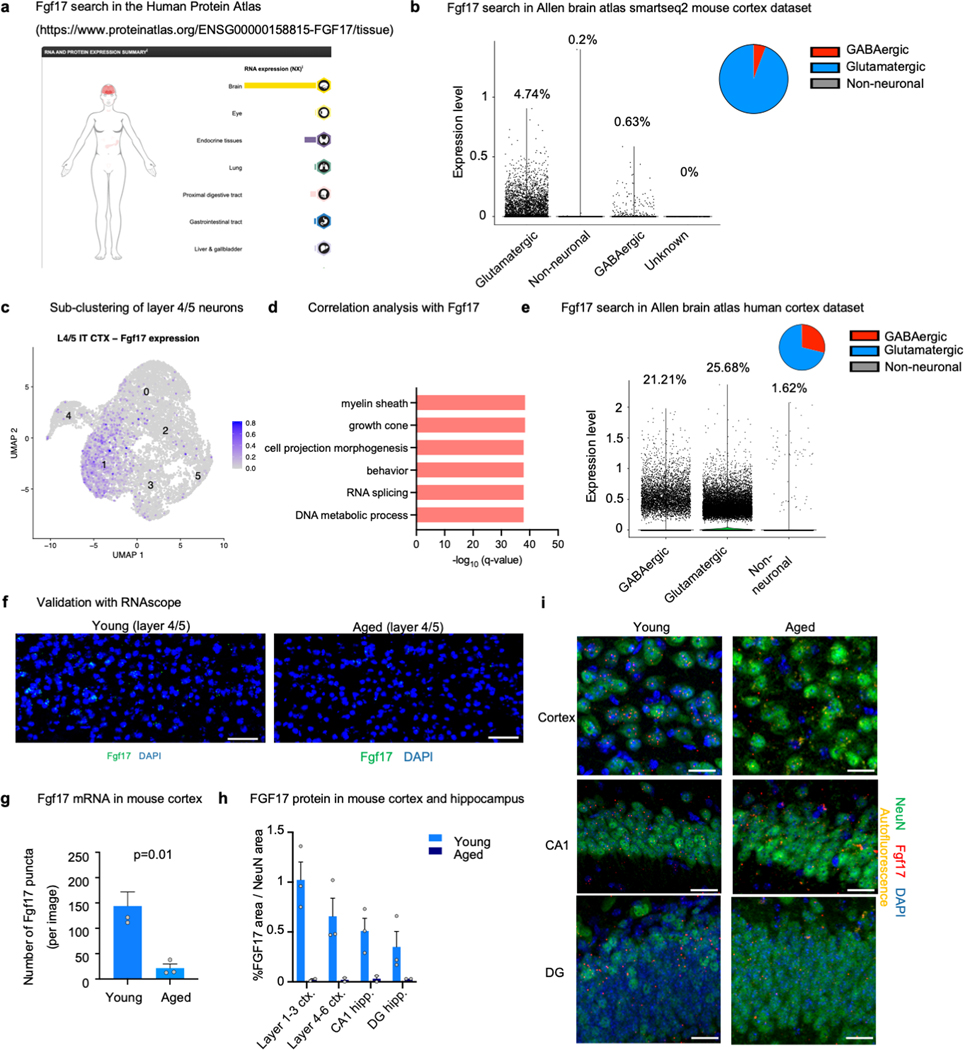
CSF contains hundreds of proteins that could potentially induce SRF signaling. We noticed that several SRF target genes are also known upstream inducers of SRF itself such as BDNF and IGF1 29,30. Based on this observation, we cross-referenced two published CSF proteomic datasets 9,31 with the predicted TRANSFAC SRF target list and generated a list of 35 potential SRF inducers (Supplementary Table 7). To test their activity, we transfected HEK293 cells with an SRE-GFP reporter and added proteins at different concentrations (Fig. 4a). Fibroblast growth factor 8 (Fgf8) and Fgf17 induced the strongest dose-dependent responses (Fig. 4b-d and Extended data Fig. 9a-b). We chose to proceed with Fgf17 because it is a brain enriched protein (Extended data Fig. 11a), and its levels decrease with age in human CSF (Fig. 4e), in human plasma in males and females 32 and in mouse neurons (Fig. 4f-g and extended data Fig. 10b-i). To gain better mechanistic understanding, we enhanced or inhibited actin polymerization using Jasplakinolide (Jasp) or Latrunculin A (LatA), respectively, and found that Fgf17 activates SRF signaling through actin modulation (Extended data Fig. 9d). This indicates, as previously described for other growth factors, that Fgf17 activates the SRF pathway through Rho GTPase activation and modulation of actin dynamics 33. We next performed the reporter experiment in the presence of blocking antibodies for Fgfr1, Fgfr2 or Fgfr3 and found that Fgf17 reporter activation was dependent on signaling through Fgfr3 (Extended data Fig. 9e). Interestingly, while Fgfr3 is very highly expressed by astrocytes we also detected a fraction of Fgfr3 positive hippocampal OPCs (Extended data Fig. 9f).
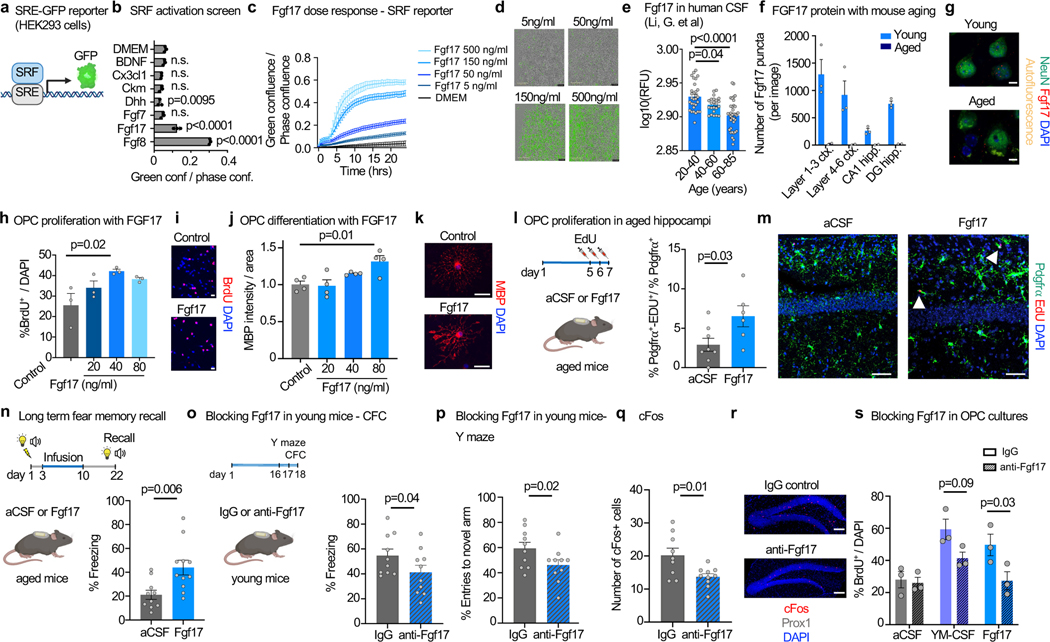
Figure 4. Fgf17 induces OPC proliferation and improves memory.
a, Diagram of SRE-GFP reporter in HEK293 cells.
b, SRE-GFP activation by CSF ligands (500 ng/ml; n=3; One-way ANOVA with Dunnett’s multiple comparisons test; mean ± s.e.m.).
c, Dose-dependent activation of SRE-GFP reporter by Fgf17 (n=3; one-way ANOVA with Tukey’s post-hoc test; mean ± s.e.m.).
d, Representative images of c. Scale bar, 400μm.
e, Meta-analysis of Fgf17 levels in healthy human CSF 9. (ages 20–40 n=30, ages 40–60 n=23, ages 60–85 n=36; One-way ANOVA with Dunnett’s post-hoc test; mean ± s.e.m.)
f, Number of Fgf17 protein puncta in the cortex and hippocampus of young (3 months) and aged (25 months) mice. (young n=3, aged n=2; mean ± s.e.m.)
g, Representative image of f. Scale bar, 5μm.
h, Percentage proliferating OPCs treated with Fgf17 under proliferation conditions (n=3; one-way ANOVA with Tukey’s post-hoc test; mean ± s.e.m.).
i, Representative images of h. Scale bar, 20μm.
j, MBP intensity per area in OPCs treated with Fgf17 under differentiation conditions (day 3) (n=4; one-way ANOVA with Tukey’s post-hoc test; mean ± s.e.m.).
k, Representative images of h. Scale bar, 20μm.
l, Quantification of OPC proliferating cells in the hippocampus of 20-month-old mice following a week of aCSF or Fgf17 infusion. (aCSF n=8, Fgf17 n=6; two-sided t-test; mean ± s.e.m.).
m, Representative images of l. Scale bar, 50μm.
n, Percentage of freezing of 20-month-old mice in the remote recall contextual fear conditioning test (aCSF n=10, Fgf17 n=11; two-sided t-test; mean ± s.e.m.).
o, Percentage of freezing of 3-month-old mice in the short-term contextual fear conditioning test (n=10; one-sided t-test; mean ± s.e.m.).
p, Percentage of entries to the novel arm of the forced alternation Y maze (n=10; one-sided t-test; mean ± s.e.m.).
q, Average number of active cFos+ cells in the DG (IgG n=9, anti-Fgf17 n=10; two-sided t-test; mean ± s.e.m.).
r, Representative image of q. Scale bar, 100μm.
s, Percentage proliferating OPCs treated with aCSF, YH-CSF or Fgf17 in combination with IgG or anti-Fgf17 antibodies (n=3; two-way ANOVA with Sidak’s post-hoc test; mean ± s.e.m.).
When added to primary rat OPCs, Fgf17 (40 ng/ml) induced OPC proliferation (Fig. 4h-i) and differentiation (Fig. 4j-k). Prior work with cultured OPC suggested that Fgf17 slightly promotes proliferation although it may inhibit OPC differentiation in some contexts 34. To determine their in vivo activity we infused recombinant Fgf8 and Fgf17 over 7 days into aged mice similar to administration of CSF (Fig 1). Fgf17 induces OPC proliferation in the aged hippocampus, but Fgf8 does not (Fig. 4l-m and extended data Fig. 9c). We next tested the effect of Fgf17 infusion on cognition and found that it improves long-term memory performance in the remote memory recall paradigm described in Figure 1 (Fig. 4n). These experiments demonstrate that Fgf17 is sufficient to mimic the effect of young CSF on OPCs in the hippocampus and on memory consolidation in aged mice.
Lastly, we infused an anti-Fgf17 blocking antibody ICV to young mice to test if Fgf17 is necessary for normal memory function. We found that mice infused with anti-Fgf17, but not with control antibody, showed impaired performance in two hippocampal dependent cognitive tests (Y maze and contextual fear conditioning, Fig. 4o-p) and impaired neuronal plasticity measured by lower c-fos levels in dentate gyrus granule cells following behavioral tests (Fig. 4q-r). In OPC cultures, the same concentration of anti-Fgf17 antibody inhibits young CSF- and Fgf17-induced OPC proliferation. This indicates that the boost in proliferation is in part mediated by Fgf17 (Fig. 4s). Taken together, these experiments provide a link between Fgf17 and cognitive function, in young and aged mice.
Discussion
In summary, we discovered that young CSF is sufficient to improve memory function in aged mice along with an increase in OPC proliferation and hippocampal myelination. Fgf17, which decreases with age in mouse neurons and in human CSF, is sufficient and necessary to improve cognition in aged mice and promotes OPC proliferation in vivo and in vitro, suggesting it constitutes a major component of the rejuvenating effects of young CSF. Moreover, young CSF induces expression of the transcription factor SRF, and its actin cytoskeleton target genes to promote OPC proliferation (Extended data Fig. 11). SRF is a versatile regulator of neuronal development and activity dependent plasticity 24,35,36. Here, we find that it is also widely expressed by OPCs, where it is downregulated with ageing. It is interesting to speculate that SRF orchestrates plasticity in various cell types and its loss with ageing promotes neurodegeneration. Expanding our examination of CSF proteins and SRF signaling in oligodendrocytes with ageing to other cell types could illuminate complex regulatory interactions within parenchymal cells and with the environment during development, ageing and ageing-related diseases.
The CSF proteome consists of proteins secreted by the choroid plexus or transferred through it from the blood plasma, as well as proteins secreted from parenchymal and immune cells. Heterochronic CSF experiments with cortical explants first demonstrated that developmental stage-specific CSF composition is critical for proper stem cell proliferation and brain development 4. During ageing, improper signaling cues derived from the ageing choroid plexus were shown to lead to neuronal stem cell quiescence 5. However, OPCs, which account for the largest population of stem cells in the aged brain, were mostly overlooked. Emerging studies in young rodents are revealing that oligodendrogenesis, the formation of myelinating oligodendrocytes from OPCs, is essential for consolidation of newly formed memories, implicating their active role in cognitive function 37–39. A recent study showed that hippocampal oligodendrogenesis is dramatically reduced with age and that increasing it was sufficient to improve performance in learning and memory tasks in aged mice 40 and in AD mouse models 41. This is in line with studies showing that aged OPCs in white matter regions, are slow to proliferate and to differentiate following demyelination in diseases such as multiple sclerosis, and that local or systemic environmental manipulations restored their myelination capacity 42,43. It is possible that these systemic therapeutic strategies affect OPCs through changes in the CSF composition. Furthermore, mature oligodendrocytes are critical for maintenance of axonal health and for regulation of neuronal function. We cannot exclude the possibility that CSF proteins directly affect mature oligodendrocyte function with beneficial outcomes on neuronal health and cognitive function. Myelin plasticity is emerging as an important mechanism in learning and memory 44 and the study of myelin dysfunction in ageing related cognitive decline is gaining interest.
As a proof of concept, we infused Fgf17 to the CSF of aged mice and found that it partially phenocopied the effects of young CSF on OPC proliferation and long-term memory recall. Conversely, blocking Fgf17 by infusing an inhibitory antibody, resulted in impaired function in hippocampal dependent memory tests. Fgf17 is critical for embryonic brain development 45 but not much is known about its function in the adult nervous system. We found that in the young adult mouse Fgf17 is abundantly expressed by cortical neurons and its expression drops dramatically with ageing. Fgfr signaling is critical for oligodendrocyte development 46 with complex and diverse functions in disease processes such as demyelination and remyelination in multiple sclerosis 34. Specifically, studies using Fgf3r-null mice show a delay in the terminal differentiation of pro-oligodendrocytes 47 and transient expression of Fgfr3 in subventricular zone progenitors drives oligodendrogenesis and promotes remyelination following a demyelinating injury 48. Interestingly, young mice lacking Fgf17 have smaller brains 49 and an array of social behavior abnormalities coinciding with lower c-fos expressing cells in the prefrontal cortex following a novel social interaction test 50. Further research is needed to demonstrate if SRF, which regulates c-fos expression, is involved in those circuits and potentially in neurodevelopmental and neuropsychiatric disorders. This strengthens the notion that studying the CSF, the media that has adapted to the unique needs of the brain, holds the potential to expand our understanding of its intricate mechanisms and cellular interactions.
Combined, our results suggest that targeting hippocampal myelination through factors present in young CSF might be a therapeutic strategy to prevent or rescue cognitive decline associated with ageing and neurodegenerative diseases.
Materials and Methods
Animals.
Aged C57BL/6 mice (18–22 months old) were obtained from the National Institute on Aging rodent colony. Young male C57BL/6 mice (2 months old) were obtained from Charles River Laboratories or Jackson laboratories. All experiments used male mice. All of the mice were housed at the Palo Alto VA animal facility under a 12 h–12 h light–dark cycle with dark hours between 18:30–06:30 and housed at 68–73 °F under 40–60% humidity. We used published data 2,3,51 to determine an optimal n for our studies on the effects of young CSF on behavior. For the bulk RNAseq of sorted oligodendrocyte nuclei we performed preliminary studies to ensure we will capture enough nuclei for downstream library prep and statistical analysis. Same aged mice were allocated into groups to achieve an equal average weight. All other criteria were not considered and as such, were randomized. All animal care and procedures complied with the Animal Welfare Act and were in accordance with institutional guidelines and approved by the V.A. Palo Alto Committee on Animal Research and the institutional administrative panel of laboratory animal care at Stanford University.
Young mouse CSF collection.
CSF was collected as previously described with several adaptations 52. Briefly, ten-week-old mice were anesthetized intraperitoneally with Ketamine (120mg/kg) and xylazine (8mg/kg), and then placed in a stereotactic instrument (KOPF) with the head secured at a 45-degree angle facing downwards. An incision was made above the neck and muscles were held separated with microretractors allowing exposure of the cisterna magna by blunt forceps without any bleeding. CSF was pulled out of the cisterna magna cavity with a 20μl pipettor connected through an aspirator tube assembly (Sigma, A5177) to a pooled glass capillary (Borosilicate glass tubes, ID 1.30 mm, OD 1.70 mm, Length 4.00 in, type 8250, King precision glass) held secure by Model 1769 90° Electrode Holder (KOPF). CSF was kept in a low-protein bind tube on ice and spun in a cold centrifuge for 10 min at 1500 rpm to exclude CSF immune cells. Supernatant was collected and kept in −80 until use. Pellet was resuspended in 6ul of milli-q ultra-pure water for blood contamination quality control using the Nanodrop UV-vis setting with a 415nm wavelength for detection of oxyhemoglobin 53. A cutoff of below than 0.02 AU was used for CSF infusions.
Human CSF.
CSF samples of nine young heathy individuals (ages 24–26) were obtained through a collaboration with Dr. Henrik Zetterberg, University of Gothenburg, Sweden. The study was approved by the regional ethics committee at the University of Gothenburg and informed consent was obtained from all participants 54,55. The samples were baseline (normal sleep) lumbar CSF samples, collected in the morning, from healthy volunteers who took part in a study on sleep restriction-induced changes of CSF composition. For in vitro experiments, three pools consisting of three individuals each were made for each experiment, two pools from six male samples and one of female samples were each used in 3–4 technical triplicates. Aged human CSF from healthy individuals (ages 65–76) was obtained from the Stanford Alzheimer’s Disease Research Center.
Osmotic pump intracerebroventricular infusion.
To minimize the volume of mouse CSF infused per mouse, young CSF or artificial CSF were loaded to a coiled polyethylene (PE-60) catheter prepared in house following the lynch coil technique 56. In brief, the total length of the coil needed was calculated by 4.56ul internal volume per cm tube. Usually, 20cm were wound around a syringe of the same outside diameter as the pump and secured with tape. The syringe was submerged in boiling water for 1 min and immediately immersed in ice cold water for 1 min. Coils were disassembled and left to dry overnight. 90 μl of pooled young CSF or artificial CSF (Tocris) were loaded to the coiled catheter connected to 100ul osmotic pumps (Alzet, 1007D) with a 7-day infusion at a rate of 0.5 μl/hr. Osmotic pumps were connected to a cannula (Brain infusion kit III, Alzet) and incubated overnight in a 37°C water bath. Cannula was inserted at +1 mm medio-lateral, 0 mm anterior–posterior, and −3mm dorso-ventral relative to bregma in order to target the right lateral ventricle. The pump was placed subcutaneously and mice received post-surgical buprenorphine and Baytril. In the human CSF infusion experiments, a pool of three young or three aged human CSF, pooled YM-CSF or aCSF was loaded to an osmotic pump (with no coil) and surgery was performed as described above. Recombinant carrier free human /mouse Fgf8b (423-F8/CF, R&D) and mouse Fgf17 (7400-FG-025/CF, R&D) were resuspended in aCSF (Tocris) to a concentration of 25 μg/ml and loaded to an osmotic pump (Alzet, 1007D) with a 7-day infusion at a rate of 0.5μl / hr. In Fgf17 blocking antibody experiments, polyclonal rabbit-anti Fgf17 (PA5–109722, Thermo) and a rabbit IgG isotype control (31235, Thermo) were diluted in aCSF, loaded on a 3kD MWCO Amicon Ultra centrifugal filters (UFC500396, ThermoFisher) for buffer exchange and spun 30min 14,000g at 4 deg. This step was repeated twice. Osmotic pumps (Alzet, 1004) were loaded with 0.68 mg/ml antibody to achieve a final CSF steady state concentration of 5 μg/ml. Surgery was performed as described above. In all experiments, mice were split to groups to achieve an equal average body weight in all groups.
Acute intracerebroventricular injection.
18-month-old mice were anesthetized with 2.5% isoflurane and then placed in a stereotactic instrument (KOPF). 3μl of a pool of young mouse CSF (as described above) or aCSF were injected to the right lateral ventricle using a digital pump (WPI syringe pump with Micro4t controller model UMP3T-1) at a pace of 1 μl/min. 1hr or 6hr following the injection, mice were perfused, and contralateral hippocampus was dissected and used for RNAseq to avoid gene signatures induced by local immune response to the surgery. In labeled Fgf17 experiments, 10μg of mouse Fgf17 (7400-FG-025/CF, R&D) was conjugated to Alexa Fluor 647 using the lightning-link conjugation kit (ab269823, ThermoFisher) following the manufacturer’s instructions and then loaded twice on a 3kD MWCO Amicon Ultra centrifugal filters (UFC500396, ThermoFisher) and spun 30min 14,000g at 4 deg for free dye cleanup and buffer exchange with aCSF. As control, a parallel labeling reaction (dye only control) was carried with a similar volume of aCSF as input. In CSF labeling experiment, 1ml of human CSF was conjugated to Alexa Fluor 647 (ThermoFisher) using NHS chemistry and then loaded on a 3kD MWCO Amicon Ultra centrifugal filters (UFC500396, ThermoFisher) and spun 30min 14,000g at 4 deg for free dye cleanup and buffer exchange with aCSF and for protein concentration. 0.5μg in 2μl of labeled Fgf17, dye-only control or YH-CSF were injected at a pace of 1 μl/min to young (2–3 months) mice and mice were taken down 2 or 3hr post-injection. Mice received post-surgical buprenorphine and Baytril.
Behavioral assays.
In the remote fear-conditioning recall test mice were trained to associate cage context or an audiovisual cue with an aversive stimulus (foot shock). On day 1, mice were placed in a cage and exposed to three periods of 30 s of paired cue light and 1,000-Hz tone followed by a 2s foot shock (0.6 mA), with a 60s interval. On day 2 and day 22, mice were subjected to two trials. In the first trial assessing contextual memory, mice were re-exposed to the same cage context, and freezing behavior was measured during minute 390s using a FreezeScan tracking system (Cleversys). In the second trial measuring cued memory, mice were placed in a novel context and exposed to the same cue light and tone from day 1 on min 2, 3 and 4 of the trial. Freezing behavior was averaged across min 3–5. No significant differences in contextual fear conditioning were observed between groups at day 22. All experiments were performed by a blinded researcher. In the experiment presented in Fig. 1b initial group numbers were n=11 for aCSF and n=9 for YM-CSF (due to limited amounts of mouse CSF) and one mouse from each group died throughout the experiment. In the experiment presented in Fig. 4n, initial group numbers were n=11 for aCSF (supposed to be n=12 but one pump was not implanted due to an air bubble) and n=12 for Fgf17 and one mouse from each group died throughout the experiment.
In the Fgf17 blocking antibody experiments, the forced alternation Y-maze and contextual fear conditioning was performed as previously described 57. Forced alteration Y-maze test consisted of a 5-min training trial followed by a 5-min retrieval trail, with a 1-h inter-trial interval. For the training trial, one arm of the Y maze was blocked off and mice were allowed to explore the two open arms. One hour later, the mouse was again placed in the Y maze with all three arms open and a black and white pattern placed at the end of the novel arm. Between mice and trials, the maze was wiped with ethanol to remove odor cues. For analysis, video was analyzed by a blinded observer and both number of arm entries and time spent in each arm were quantified. In the fear-conditioning paradigm mice were trained to associate cage context or an audiovisual cue with an aversive stimulus (foot shock). The test was administered over two days. On day 1, mice were placed in a cage and exposed to two periods of 30 s of paired cue light and 1,000-Hz tone followed by a 2-s foot shock (0.6 mA), with a 180-s interval. On day 2, mice were subjected to two trials. In the first trial assessing contextual memory, mice were re-exposed to the same cage context, and freezing behavior was measured during minute 1–3 using a FreezeScan tracking system (Cleversys). In the second trial measuring cued memory, mice were placed in a novel context and exposed to the same cue light and tone from day 1 after 2 min of exploration. Freezing behavior was measured for 1–3 min following the cue. No significant differences in cued fear conditioning were observed between groups. Experimental groups consisted of n=10 mice and no mice were excluded throughout the experiment.
In vivo BRDU and EDU pulses.
To assess baseline proliferation, mice received one EdU injection 15 mins before the stereotactic surgery (Invitrogen, E10415, 100 mg / kg intraperitoneally). To assess cell proliferation post infusion, mice were pulsed three times with BrdU to label proliferating cells; twice on day 5 of the infusion 10 hrs apart and once on day 6 two hours before perfusion (100 mg /kg intraperitoneally; B5002–5G, Sigma-Aldrich). In all subsequent cohorts, mice received post-infusion pulses of EDU instead of BRDU in the same paradigm described above, with no baseline labeling prior to surgery.
Tissue processing prior to immunostaining.
Mice were anesthetized with Avertin (2,2,2-tribromoethanol: T48402, Sigma-Aldrich; 2-methyl-2-butanol: 240486, Sigma-Aldrich) (0.018 ml (2.5%) per gram of body weight) and perfused with 20 ml cold PBS. Brains were collected and divided sagittally. One hemisphere was used to dissect the hippocampus for RNA sequencing, which was snap frozen and stored at −80 °C. The second hemisphere was fixed in phosphate-buffered 4% paraformaldehyde overnight at 4 °C before transfer to 30% sucrose in PBS at 4 °C until sectioning. Brains were frozen at −30 °C and cryosectioned coronally at 40 μm with a microtome (Leica, SM2010R). Brain sections were stored in cryoprotectant (40% PBS, 30% glycerol, 30% ethylene glycol) and kept at −20 °C until staining.
Immunostaining.
Brain slices immunofluorescent staining.
Brain sections were washed three times for 10 mins in TBST and then blocked in TBS++ (TBS + 3% donkey serum (130787, Jackson ImmunoResearch) + 0.25% Triton X-100 (T8787, Sigma-Aldrich)) for 1 hr, followed by primary antibody incubation overnight on a rocking platform at 4 ˚C. The following primary antibodies were used in this study; PDGF Receptor α (D1E1E) XP® Rabbit mAb (1:500, Cell Signaling, 3174S), Rabbit-anti-MBP (1:100, MAB386, Millipore), Rabbit anti-Fgf17 (1:500, PA5–109722, Thermo), goat anti-PROX1 (1:500, AF2727, R&D), rabbit anti-c-Fos (1:500, 9F6, Cell Signaling), Rabbit anti-GFAP (1:500, Z0334, Dako), Goat-anti-IBA1 (1:500, ab5076, abcam), Rabbit anti-NG2-Alexa488 (1:200, AB5230A4, Millipore), Rabbit-anti Olig2 (1:500, AB9610, Millipore), anti-Acta2-Cy3 (1:500, C6198, Sigma), mouse-anti NeuN (1:500, MAB377, Sigma). For secondary staining, brain sections were washed three times for 10 min in TBST, followed by incubation for 1.5 hrs in Alexa Fluor-conjugated secondary antibodies (1:500). For in vivo phalloidin staining phallidin-Alexa488 58 (A12379, Thermo) was added 1:50 to the secondary antibody mix. Brain sections were washed and mounted on Superfrost microscope slides (12–550-15, Fisher Scientific) with Vectashield Hardset Antifade Mounting Medium with DAPI (Vector labs, H-1500/NC9029229). For MBP stains an additional step of tissue de-lipidation was added before blocking: tissues were incubated in 100% EtOH for 10min in room temperature and then washed twice with PBS.
EdU staining.
Brain sections were washed three times for 10 min in PBS, then permeabilized for 20 min in 0.1% Triton X-100 (T8787, Sigma-Aldrich), washed again three times and then blocked in TBS++ for 1hr. EdU staining was performed following the Click-iT® Plus EdU Alexa Fluor® 555 Imaging Kit instructions (C10638, Life Technologies). Sections were washed and stained with primary and secondary antibodies as described above.
BrdU staining- brain slices and 384-well plates.
Following staining with other primary and secondary antibodies, sections were incubated in 2N HCl for 30 min at 37 °C and then washed three times for 10 min in TBST. Sections were blocked for 1.5 hr in TBS++ and then transferred to primary antibody with Rat anti-BRDU antibody (1:500, ab6326, Abcam) overnight at 4 °C. Secondary staining started with three washes for 10 min in TBST, followed by incubation with secondary antibody mix for 1.5 h. After three 10-min washes in TBST, sections were mounted as described above. For 384-well plates, nuclei were stained and with Hoechst 33342 (1:2000, H3570, Thermo) and immediately imaged on a Keyence microscope (BZ-X800). In cases where BRDU and EDU staining was performed on the same sections, the sequence was as follows: permeabilization, HCL antigen retrieval, EDU Click-it reaction, blocking, primary and secondary antibody stain as described above in detail.
In situ RNA hybridization (RNAScope)
RNAScope was performed on fresh frozen coronal brain sections (10μm thick) using the Multiplex Fluorescence v.2 kit (Advanced Cell Diagnostics) according to the manufacturer’s protocol with minor modifications. Tissue fixation with 4% PFA was extended to 60 min at RT, and Protease IV treatment was shortened to 20 min to better preserve the hippocampal tissue. Probes for mouse Pdgfrα, SRF, Fgfr3 and Fgf17 were commercially available from the manufacturer and secondary Opal 690 and 520 reagents (FP1497001KT and FP1487001KT, Akoya Biosciences) were diluted at 1:1500 in TSA buffer.
Image analysis (IMARIS)
Brain slices.
Confocal z stacks of four coronal brain sections spanning the dorsal hippocampus were captured on a Zeiss confocal LSM880 microscope for each brain sample using a 20x magnification. Max projection files (at least 4 hippocampal coronal slices per mouse, 400μm apart) were analyzed in IMARIS by generating masks of two main regions of interest: (1) cornu ammonis area 1 (CA1) - stratum oriens, stratum pyramidale and stratum lacunosum-moleculare (SLM) combined, and and (2) cortex. The percentage of newly proliferated OPCs was analyzed by dividing cell counts of BRDU+ or EDU+ cells by PDGFRα+ cells. For cell density, BRDU+ or PDGFRα+ cell counts were divided by the corresponding area of the mask per slice.
For RNAscope analysis, similar hippocampal tiled z-stacks were taken (at least 4 hippocampal coronal slices per mouse, 100μm apart). Percentage of SRF+ OPCs was calculated by dividing the number of SRF+ PDGFRα+ nuclei by PDGFRα+ nuclei in the CA1 region of the hippocampus. For MBP analysis, confocal images of the molecular layer were taken using the 20x magnification. MBP intensity was measured using batch analysis in ImageJ. For in vivo phalloidin analysis, 63x magnification z-stacks of individual OPCs were obtained by Pdgfrα staining. Using IMARIS, 3D surface rendering was reconstructed by Pdgfrα signal and phalloidin intensity was measured only inside the surface. All analyses were performed by a blinded observer.
In vitro cell culture.
Three random 20x images of each well were analyzed using IMARIS batch by setting similar surfaces to automatically count BRDU+ and Hoechst+ nuclei. For each image percentage of proliferating cells was calculated by dividing BRDU+ counts by total Hoechst+ counts. In differentiation experiments, cell morphology state was assessed manually as previously described 16. Cellular phalloidin intensity and MBP intensity was measured by manually delineating cell borders and measuring intensity and cell area, then all cell measurements averaged per coverslip. Representative images of phalloidin intensity were made with Fire LUT in IMAGEJ. All other quantifications we done using IMAGEJ and manual cell counts. All analyses were performed by a blinded observer.
Transmission electron microscopy
Prefusion and sectioning
Mice were perfused with cold 20ml of EM fixation buffer consisted of EM grade 2% Glutaraldehyde (50–262-08, EMS / Fisher) 4% PFA (50–980-486, EMS / Fisher) in 0.2M sodium cacodylate (50–980-279, EMS / Fisher) and kept in fixation buffer until sectioning. Brains were sectioned coronally to 100 μm sections using a Leica VT1200S vibratome and kept in EM fixative until TEM processing.
High pressure freezing with freeze substitution
100 μm thick vibratomed sections were stained by using an osmium-thiocarbohydrazide-osmium (OTO) method 59,60 in combination with microwave-assisted processing, followed by high pressure freezing and freeze substitution (HPF-FS), as previous described 61. Briefly, samples were OTO stained, incubated with 2% aqueous uranyl acetate overnight, then subjected to HPF, followed by super quick FS 62 with 4% osmium tetroxide, 0.1% uranyl acetate and 5% ddH2O in acetone; they were then thin-layer embedded and polymerized in hard epon resin. Resin-embedded samples were precision cut off the glass slide and glued with cyanoacrylate onto a blank resin block for sectioning or glued with silver paint onto a stub for focused ion beam imaging.
Transmission electron microscopy
90 nm ultrathin sections were cut using a Leica UC6 ultramicrotome (Leica Microsystems, Vienna, Austria) and collected onto formvar-coated 50 mesh copper grids or copper-rhodium slot grids. Due to native contrast from volume EM processing, no post-stain was necessary. Sections were imaged using a Tecnai 12 120kV TEM (FEI, Hillsboro, OR, USA), data was recorded using an UltraScan 1000 with Digital Micrograph 3 software (Gatan Inc., Pleasanton, CA, USA), and SerialEM was used to collect montages covering an area of 143 sq. μm.
SRF reporter assay
HEK293 cells (ATCC) were plated at 50K cells/well in a 96-well plate in full media (DMEM, 10% FCS and 1% P/S) and transfected on day 2 with Cignal SRE Reporter Assay Kit (GFP) (CCS-010G, Qiagen) using lipofectamine P3000 in experimental media (DMEM, 0.5% FCS, 1% non-essential amino acids (M7145, Sigma)), following the manufacturer’s instructions. On day 3 media was changed to 90μm fresh experimental media and supplemented with 10μm of 10x concentration of indicated concentrations of recombinant carrier free Recombinant Human KGF/FGF-7 (251-KG-010, R&D Systems), human /mouse Fgf8b (423-F8/CF, R R&D Systems), mouse Fgf17 (7400-FG-025/CF, R&D Systems), Recombinant Human/Murine/Rat BDNF (450–02, Peprotech), Recombinant Human CX3CL1/Fractalkine (365-FR-025, R&D Systems), Recombinant Human Dhh protein (ab78682, Abcam) and Ckm (9070-CK-050, R&D Systems). Plates were incubated in the IncuCyte (Essen BioScience) and imaged every hour for 24 hours. In actin inhibitors experiments, on day 3 media was changed to fresh experimental media and pre-incubated for 30 min at 37ºC with Jasplakinolide (Jasp, 125 or 250nM, J7473, Fisher Scientific) or Latrunculin A (LatA, 250 or 500nM, L12370, Thermo), before adding Fgf17 at a final concentration of 200 ng/ml. Plates were incubated in the IncuCyte and imaged every hour for 24 hours. In FgfR blocking antibody experiments, on day 3 media was changed to fresh experimental media and pre-incubated at 37ºC with anti-FgfR1 (NBP2–12308), anti FGFR2 (MAB684–100) and anti-FGFR3 (MAB7661–100, all from Novus Biologicals) final concentration of 50 μg/ml, before adding Fgf17 at a final concentration of 200 ng/ml. Plates were incubated in the IncuCyte and imaged every hour for 24 hours. HEK293 cell were not authenticated and not tested for mycoplasma contamination
OPC primary cultures.
Rat OPC cultures
OPCs were isolated from P7-P8 Sprague-Dawley (Charles Rivers) rat brains by immunopanning and grown in serum-free defined medium, as previously described 63. Cell culture for proliferation and differentiation experiments was done following the protocol with several modifications. To use the least possible CSF of young healthy human subjects, we minimized the culture conditions to 384 well plates. In addition, to account for inter-subject variability, we pooled CSF of 3 subjects with similar ages, and used three such pools in each experiment in triplicates. Following initial growth of 4 days in 10cm dishes, cells were trypsinized and split to 384-well (Falcon® 384 Well Optilux, 353962), PDL-covered (PDL; Sigma-Aldrich P6407; molecular weight 70–150 kDa) plates. For proliferation experiments, 2,500 cells were plated in a total volume of 50μl of full proliferation medium supplemented with 10 ng/ml PDGF (Peprotech 100–13A), 10 ng/ml CNTF Peprotech 450–02, 4.2 μg/ml forskolin (Sigma-Aldrich F6886) and 1ng/ml NT3 (Peprotech 450–03) with 10% of YH-CSF or aCSF. Actin filaments were visualized by live imaging by addition of 500nM of SiR-Actin (Cytoskeleton, CY-SC002) added with 10% YH-CSF. Wells were imaged every hr for the remaining 6hrs of the experiment. In BRDU experiments, 18hrs after plating in 384-well plates (with 10% CSF), 5ul of 200μM BRDU (20μM final concentration) was added for a pulse of 6 hrs followed by fixation with 4% PFA for 20 min. BRDU experiments were performed with indicated concentrations of YH-CSF or Fgf17 and in Fig. 4s in combination with Rabbit anti-Fgf17 (PA5–109722, Thermo) or IgG isotype control (31235, Thermo) to achieve a final concentration of 5μg/ml antibody, 40ng/ml Fgf17 and 10% YH-CSF in full proliferation media as indicated above.
In experiments that required phalloidin staining, 10,000 OPCs were plated on PDL-covered 12mm coverslips in a 24-well dish in 90% full proliferation medium (315μl) and let to adhere overnight, then supplemented with 10% CSF (35μl) for 6 hs. Coverslips were fixed with 4% PFA for 20 mins, washed and stained with 555-phalloidin in PBS (1:143, Invitrogen) for 15 min. Coverslips were washed and mounted with Vectashield Hardset Antifade Mounting Medium with DAPI (Vector labs, H-1500/NC9029229). For differentiation experiments, 10,000 OPCs were plated on PDL-covered 12mm coverslips in a 24-well dish in full proliferation medium overnight. Proliferation medium was completely changed to differentiation media (basal growth media supplemented with 40 ng/ml T3 (Sigma-Aldrich T6397)) with 10% CSF with a 50% media change (with 10% CSF) on day 2 of differentiation. At day 4 of differentiation coverslips were fixed with 4% PFA for 20 mins, washed with PBS, permeabilized with 0.1% Triton X-100 for 3 mins followed by wash and blocking in 3% BSA for 1 hr. Primary antibodies; Rabbit-anti-MBP (1:100, abcam, ab7349, knock-out validated 16) and mouse anti-GFAP (1:500, Chemicon, MAB360) were incubated overnight at 4 °C. Coverslips were washed, stained with Alexa Fluor-conjugated secondary antibodies (1:500) followed by a 15 min stain with Cell mask (1:1000, Invitrogen, C10046) mounted and set on a coverslip before imaging on a Keyence microscope (BZ-X800) or confocal laser-scanning microscope (Zeiss LSM880).
Mouse OPC cultures
Mouse OPCs were purified from brains of SRF floxed mice (generated by David Ginty and kindly provided by Eric Small) by immunopanning as described above for rat OPCs 63.
On day 3 of the culture, SRF-f/f OPCs were split and plated in 384-well plates for proliferation experiments. When the cells were in suspension in proliferation media before plating, 1*1010 viral genomes of AAV DJ-CMV eGFP-deleted cre (GVVC-AAV-62) or AAV DJ-CMV eGFP-cre (GVVC-AAV-63) (both generated by the Stanford Gene Vector and Virus Core). The following day, media was fully replaced and 48hrs after infection 10% aCSF or YH-CSF were added to proliferation media with BRDU (20μM final concentration) for 16 hrs. Cells were fixed and cell proliferation was assessed as indicated above in the BRDU experiment.
SLAMseq experiment and data analysis.
Optimal s4U concentration was assessed following SLAMseq 64 Explorer Kit – Cell Viability Titration Module (061, Lexogen) following manufacturer’s recommendations. In brief, OPCs were incubated with increasing doubling concentrations of s4U (1.95–2000μM) and viability was assessed with live-dead ratio as described above and with ATP incorporation following manufacturer’s recommendations (Promega,G7571). A final concentration of 62.5uM was found as the highest concentration without compromising on cell viability within 12 hrs (twice the duration of the intended experiment). SLAMseq experiment was conducted following SLAMseq Kinetics Kit - Anabolic Kinetics Module protocol (061, Lexogen). In brief, following initial growth of 4 days in 10cm dishes, 30,000 OPCs were plated in 315μl full proliferation medium on PDL-covered 24-well plates (one plate per timepoint) overnight. 35μl of pooled YH-CSF or aCSF spiked in with 625μM S4U (10x concentration) were added gently to minimize confounding induction of gene expression. One- or six-hours later the medium was removed, and cells were scraped with 1ml Trizol (Thremo, 15596018), transferred to foil-covered tubes and frozen until RNA extraction. RNA extraction was done following anabolic kit protocol. All cell culture and RNA extraction steps were done in the dark under red-light following manufacturer’s recommendations. Library preparation was done with QuantSeq 3’ mRNA-Seq Library Prep Kit for Illumina (FWD) (015, Lexogen) with the indicated modifications to adjust to low-RNA input. After normalization and pooling, libraries were sequenced on a Nextseq550 (Illumina) using single-end 75-bp reads. Libraries were sequenced to a mean depth of ~30 million reads per sample. Raw sequencing files were demultiplexed with bcl2fastq and resulting FASTQ files combined across lanes and per sample. Sequencing quality control was performed using FastQC v0.11.8 and summary reports were generated with MultiQC v1.7. FASTQ files were then analyzed using the SLAM-DUNK pipeline v0.3.4 and the related alleyoop toolchain. The pipeline comprises 1) read mapping, 2) alignment filtering, 3) SNP calling and correction, and 4) 3’-UTR sequence counting. Using NextGenMap reads were aligned against the genome of Rattus norvegicus (release 6.0), which was downloaded in FASTA format from Ensembl release v97. For mapping the following parameters were set; ‘−5 12, -a 4, -n 1, -ss’ leaving rest at default. Resulting Binary sequence Alignment/Map (BAM) format files were filtered to remove low-quality alignments using the parameters ‘-mq 2, -mi 0.95, -nm −1’ and rest at default. Next, SNPs in alignments, in particular the T to C (T>C) conversions, were called with parameters ‘-c 10, -f 0.8’ and others remaining at default. Statistical independence of distributions of true SNP-callings identified by VarScan2 in relation to the number of T>C reads was assessed with a Mann-Whitney-U test for each sample as initially described in the alleyoop snpeval toolchain module. For the reference set of 3’-UTRs, a genome feature file (GFF3) for the Rattus norvegicus genome (release 6.0) was downloaded from Ensembl release v97, filtered to retain only three_prime_utr features and converted to Browser Extensible Data (BED) format using bedops v2.4.36. The BED file was used in combination with parameters ‘-c 1, -q 27’ for the SLAMDUNK count command, leaving other parameters at default. Resulting 3’-UTR T>C and total read counts were added up on gene-level along all transcripts and alternative 3’-UTRs available per transcript. Entire down-stream analysis was performed using R v3.6.1, data.table v1.12.2, and common tidyverse packages (tidyr, dplyr, purrr, stringr, ggplot2). Differential gene expression analysis was carried out using DESeq2 v1.26.0 on the T>C count and total count matrices separately with standard settings with alpha level set to 0.05. In total, six DESeq2 experiments were performed, two with pooled timepoints (1hr + 6hr) per read-type (design variables Treatment and Time) and four with separate timepoint per read-type (design variable Treatment only). Effect size was calculated using the implementation of cohen’s d from the effsize v0.7.6 package. Gene set enrichment and over-representation analyses were performed using GeneTrail 3.0 65. Category source databases were adjusted independently, and P-values were corrected using the false discovery rate controlling procedure by Benjamini-Hochberg and considered significant if smaller than 0.05.
Oligodendrocyte nuclei isolation and sorting.
Nuclei isolation from frozen dissected hippocampi was done as previously described 66 using the Nuclei EZ Prep Kit (Sigma-Aldrich, St. Louis, USA). Following the final PBS wash, nuclei were pelleted and resuspended in 100 μl of antibody mix (1:100, Anti-NeuN antibody-Alexa Fluor® 647, EPR12763, and 1:100, Anti-Olig2 antibody-Alexa Fluor® 488) with 0.2U/μl RNAse Inhibitor (Takara, 23138) in FACS buffer (0.5% BSA in PBS) and incubated on ice with intermittent shaking for 30 min. Nuclei were washed with 1ml FACS buffer and pelleted by centrifugation at 500 r.c.f. for 5 min, resuspended in FACS buffer with 0.2U/μl RNAse Inhibitor and Hoechst 33342 (1:2000, H3570, Thermo) and sorted on a Sony Sorter (SH800) based on Houechst+ NeuN- Olig2+high (OPC) and Houechst+ NeuN- Olig2+low (OL) gating to 350μl of RTL buffer and stored in −80°C until RNA extraction. Data were analyzed using FlowJo software (TreeStar).
RT-qPCR.
Oligodendrocyte nuclei were isolated by FACS and RNA was extracted with the RNeasy Plus Micro kit (Qiagen, 74034). cDNA was generated with qScript™ cDNA SuperMix (QuantaBio, 95048). Samples were diluted and mixed with SYBR green master mix before loading as technical triplicates for qPCR on a LightCycler 480 (Roche). ΔΔC T values normalized to Gapdh were used to assess relative gene expression between samples. The following validated primer pairs for mouse SRF were used: 5′- GGC CGC GTG AAG ATC AAG AT-3′ (forward) and 5′- CAC ATG GCC TGT CTC ACT GG-3′ (reverse).
Bulk RNA sequencing.
Hippocampal RNAseq
Frozen dissected hippocampi were thawed on ice and homogenized in 350μl RLT buffer by 20 strokes using a manual homogenizer and total RNA was isolated with the RNeasy Plus Micro kit (Qiagen, 74034). RNA quantity and quality were assessed by Agilent 2100 Bioanalyzer (Agilent Technologies). All samples passed a quality-control threshold (RIN ≥ 9) to proceed to library preparation and RNA-seq on Hiseq 4000 (Illumina) using paired-end 100-bp reads. Libraries were sequenced to a depth of >20 million reads per sample. Raw sequencing files were demultiplexed with bcl2fastq, reads were aligned using STAR, and counts of technical replicates were summed up using DESeq2 before performing normalization and differential expression analysis with standard settings.
For deconvolution analysis of bulk RNAseq, we utilized the CIBERSORTx algorithm 67 to deconvolve the bulk RNA seq data. First, we used single nuc-seq data describing the brain of 3-month-old young mice 66 to construct a cell-type-specific signature matrix with CIBERSORTx. We sampled 100 cells equally across the following cell types: astrocytes, choroid plexus, endothelial cells (BEC), interneurons., microglia, neurons of the trisynaptic loop (neuron CA), neurons of the dentate gyrus (neuron DG), oligodendrocytes, oligodendrocyte precursors, and pericytes. Sampling was done over the annotated, quality-controlled data provided by the authors of 66 to ensure the efficiency of CIBERSORTx. Next, we run CIBERSORTx on this sampled and CPM normalized dataset with default parameters and inferred a ‘signature matrix’ that provides gene signatures for each noted cell type. Next by following 68, we deconvolved CPM normalized YM-CSF and aCSF samples separately in S-mode owing to the possibility of high technical variance. First, we conducted this step to infer cell type fractions per sample. Next, we estimated cellular expression levels in ‘group mode’ in order to identify cell type-specific gene expression profiles per condition group. Finally, we run the differential expression code provided in 68 between the cell type specific expression profiles estimated for YM-CSF and aCSF. P-values were corrected with Benjamini-Hochberg procedure (FDR=0.05), and log2 fold changes reflect changes in the estimated expression levels of each gene between YM-CSF and aCSF. Single-cell subsampling and single cell data normalization was conducted with Python Scanpy 1.6.0 69. All CIBERSORTx specific analyses were done with the web service of the CIBERSORTx team (https://cibersortx.stanford.edu/). Differential gene expression was conducted in R 4.0.5.
Sorted OPC RNAseq
RNA was extracted with the RNeasy Plus Micro kit (Qiagen, 74034). cDNA and library synthesis were done inhouse using the Smart-seq2 protocol as previously described 70 (detailed protocol at https://doi-org.laneproxy.stanford.edu/10.17504/protocols.io.2uvgew6) with several modifications. Due to the low input RNA content, 2μl of RNA extracted from sorted nuclei was reversed transcribed using 16 cycles for OL samples and 18 cycles for OPC samples. Following bead cleanup using 0.7x ratio with AMPure beads (A63881, Fisher), cDNA concentration was measured using the Qubit 1x dsDNA HS kit (Q33231) and normalized to 0.4 ng/μl as input for library prep. 0.4 μl of each normalized sample was mixed with 1.2 μl of Tn5 Tagmentation mix (0.64 μl TAPS-PEG buffer (PEG 8000, V3011, PROMEGA, and TAPS-NaOH pH8.5, BB-2375, Boston Bioproducts), 0.46 μl H2O and 0.1 μl Tn5 enzyme (20034198, Illumina)), then incubated at 55 °C for 10 min. The reaction was stopped by adding 0.4 μl 0.1% sodium dodecyle sulfate (Fisher Scientific, BP166–500). Indexing PCR reactions were performed by adding 0.4 μl of 5 μM i5 indexing primer (IDT), 0.4 μl of 5 μM i7 indexing primer (IDT), and 1.2 μl of KAPA HiFi Non-Hot Start Master Mix (Kapa Biosystems) using 12 amplification cycles. Libraries were purified using two purification rounds with a ratio of 0.8x and 0.7x AMPure beads. Library quantity and quality was assessed using a Bioanalyzer (Agilent) and Qubit. All steps were done manually using 8-strip PCR tubes and PCR reactions were carried out on a 96-plate thermal cycler (Biorad). Libraries were pooled and sequenced on a Nextseq550 sequencer (Illumina) using single end 63bp for Read 1 and 12bp for index 1 with a high output 75bp kit (20024906, Illumina).
Libraries were sequenced to a depth of at least >10 million reads per sample. Raw sequencing files were demultiplexed and known adapters were trimmed with bcl2fastq. Data analysis of raw sequencing data was performed using the nextflow-core RNA-seq pipeline v3.0. Briefly, the core workflow of the pipeline maps filtered reads against the species reference genome using STAR and computes transcript counts using RSEM. For nuclear RNA-seq data, a custom reference genome was created where exon sequences in GTF files were modified to include all introns per transcript and used for the mapping instead. For mouse and rat sequencing data the reference genome GRCm38 and Rnor 6.0 provided by Illumina igenomes were used, respectively. All gene annotations were based on the Ensembl database. Obtained raw gene transcript counts per sample were loaded into DESeq2, performing normalization for transcript length and sequencing depth, and differential expression analysis with standard settings. Effect sizes for each gene were computed based on normalized counts computed by DESeq2 using the function cohen.d of the R package effsize. Gene set enrichment analysis was performed using GeneTrail 3 using BH-FDR p-value adjustment with all remaining parameters kept at default.
Meta-analysis of Srf targets in single-cell data sets from literature.
For each data set gene counts, fold-changes and adjusted p-values calculated between either case and control or old and young age groups were acquired from the publicly available supplement tables of the corresponding publications 71–73. Next, the list of Srf targets known in Homo sapiens (by TRANSFAC dataset 74) were mapped to orthologous gene identifiers in Mus musculus using the Ensembl database at release 100. The process described in the following was performed independently for oligodendrocyte precursor and mature oligodendrocyte clusters. For each data set, the list of genes was intersected with the organism matched list of Srf targets. Next, genes that did not pass the significance threshold for the adjusted p-values at cut-off 0.05 were discarded from subsequent analysis. Finally, fold-changes were normalized for each data set to obtain a comparable scale and to mitigate project-dependent fluctuations of fold-changes due to varying sample counts.
Allen brain atlas analysis.
For secondary analysis of Fgf17 positive cells and corresponding expression levels, we downloaded raw gene expression counts for two data sets from the Allen Brain Atlas data portal (https://portal.brain-map.org/). The first comprised human M1 cortex samples profiled with 10× 3’ gene expression yielding ~77k single-nuclei and mouse hippocampus and cortex samples profiled with smart-seq2 yielding ~77k single-cell transcriptomes. Each was loaded and analyzed separately using the R programming language (v4.0.5) together with the packages Seurat (v4.0.6), SeuratWrappers (v0.3.0), bioconductor-mast (v1.16.0), monocle3 (v1.0.0), data.table (v1.14.2), lsa (v0.73.2), umap (0.2.7.0), and ggplot2 (v3.3.5). The official metadata and annotations provided were used to construct the Seurat objects and dissect the data. A minimal number of 250 expressed features per-cell and at least 100 cells detected per-feature were enforced to filter the raw matrices. We then followed the official Seurat analysis workflow; normalization (step 1), selecting variable features using VST normalization and the number of features set to 2000 (step 2), scaling and centering of informative features (step 3), running PCA with 50 PCs (step 4), computing the nearest neighbor graph using 20 dimensions (step 5), identifying clusters in the graph setting resolution to 0.8, number of starts and iterations to 10 and 15, respectively (step 6), and performing UMAP-based dimension reduction with 20 input dimensions, the number of neighbors set to 30, the minimum distance set to 0.3 and the spread set to 1 (step 7). Fgf17 positive cells were defined through a normalized expression count larger than zero. For cell-type cluster or region-specific analyses, the respective cells were extracted and the analysis steps 2–7 from above were repeated. To compute enriched markers for the different subclusters we used the FindAllMarkers function from Seurat together with the MAST package requiring an absolute log-fold change of at least 0.25 and a min percentage expressed of 10%. To find a global transcriptome signature for the Fgf17 positive cells, we correlated its expression to all other expressed genes in selected clusters using the cosine similarity. Lists of genes ranked by decreasing correlation were then used to perform GSEA using GeneTrail 3.0.
Statistics and reproducibility.
All non-RNAseq analysis was done using GraphPad Prism 8 and 9 with the indicated statistical tests. Gene set enrichment and over-representation analyses were performed using GeneTrail 3.0 75. Category source databases were adjusted independently, and P-values were corrected using the false discovery rate controlling procedure by Benjamini-Hochberg and considered significant if smaller than 0.05. Box plots show the median and the 25–75th percentiles, and the whiskers indicate values up to 1.5-times the interquartile range. In in vivo experiments with same age mice, mice were selected to groups to achieve similar average weight across groups. All other criteria were not considered and as such, were randomized. Data in figure 1b, 1e, 1i, 1k, 4l and Extended data fig. 1a-f are combined raw data from two independent cohorts of mice. Data in Fig. 1l-o, 2e, 2i, 4b-c, 4f, 4h, 4j and 4s and Extended data 2b, 7a, 7b, 7d, 7e, 7f, 7h, 7i, 9a, 9b, 9d, 9e, were successfully replicated in two independent experiments. Figure schematics were created with BioRender and Inkscape.
Extended Data
Extended data Fig. 1. Bulk RNAseq, infusion site details and overall overview of proliferating cells.
a, Relative proportions of cell types as predicted by deconvolution analysis of bulk RNAseq of aged mice infused with aCSF or YM-CSF (aCSF n=8, YM-CSF n=7).
b, Predicted number of DEGs per cell type by deconvolution analysis of bulk RNAseq of aged mice infused with aCSF or YM-CSF (aCSF n=8, YM-CSF n=7).
c, Effect size of the subset of oligodendrocyte genes in Fig. 1d 16hrs following acute injection of YM-CSF or aged mouse CSF (AM-CSF) calculated over aCSF as control (n=4; Wilcoxon rank sum test).
d, Location of infusion site. Image source: Allen Institute, Mouse brain atlas (coronal).
e, Location of analysis site. Image source: Allen Institute, Mouse brain atlas (coronal).
f, Hippocampal slice of 10-month-old mice given an EdU pulse prior to surgery showing low baseline proliferation, and three pulses of BrdU at day 5 and 6 of infusion showing an overall increase in proliferating cells following YM-CSF infusion (n=4 per group; repeated measures two-way ANOVA followed by Sidak’s post-hoc test; Means ± SEM).
g, Representative images of EdU (red) and BrdU (green) cells in mice with no surgery or infused with aCSF or YM-CSF. Scale bar, 500μm.
h, RNAscope of Pdgfrα+Edu+ cells in hippocampus of 2-month-old (young) and 19-month-old (aged) mice (n=3; two-sided t-test; mean ± s.e.m.).
i, Representative images of analysis in panel h. Arrows pointing to Pdgfrα+Edu+ cells. Scale bar, 100μm.
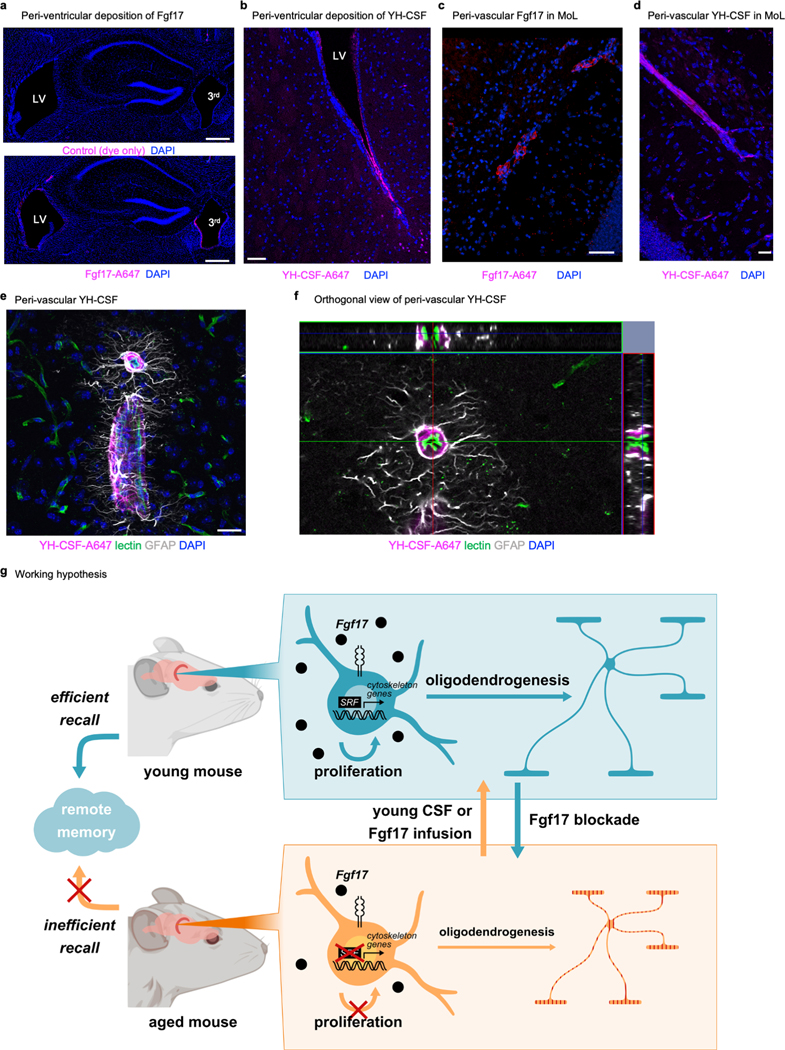
Extended data Fig. 2. Cortical Pdgfrα+EdU+ cells and identity of Pdgfrα- EDU+ cells.
a, Hippocampal density of Pdgfrα+ EdU+ cells per mm2 (aCSF n=7, YM-CSF n=8; two-sided t-test; mean ± s.e.m.).
b, Hippocampal density of Pdgfrα+ cells per mm2 (aCSF n=7, YM-CSF n=8; two-sided t-test; mean ± s.e.m.).
c, Location of region of interest in the cortex. Scale bar, 100μm.
d, Percentage of Pdgfrα+ EdU+ / Pdgfrα+ cells showing very low proliferation rates of OPCs in the cortex (n=6; two-sided t-test; mean ± s.e.m.).
e, Cortical density of Pdgfrα+ EdU+ cells per mm2 (n=6; two-sided t-test; mean ± s.e.m.).
f, Cortical density of Pdgfrα+ cells per mm2 (n=6; two-sided t-test; mean ± s.e.m.).
g, Percentage of Pdgfrα+ EdU+ / EdU+ in the hippocampus of aged mice infused with YM-CSF (n=3).
h, Example of IBA+ EdU+ cells in the hippocampus (n=3). Scale bar, 50μm. Insert, 10μm.
i, Example of GFAP+ EdU+ cells in the hippocampus (n=3). Scale bar, 50μm. Insert, 10μm.
Extended data Fig. 3. Young CSF increases number of myelinated axons in the molecular layer.
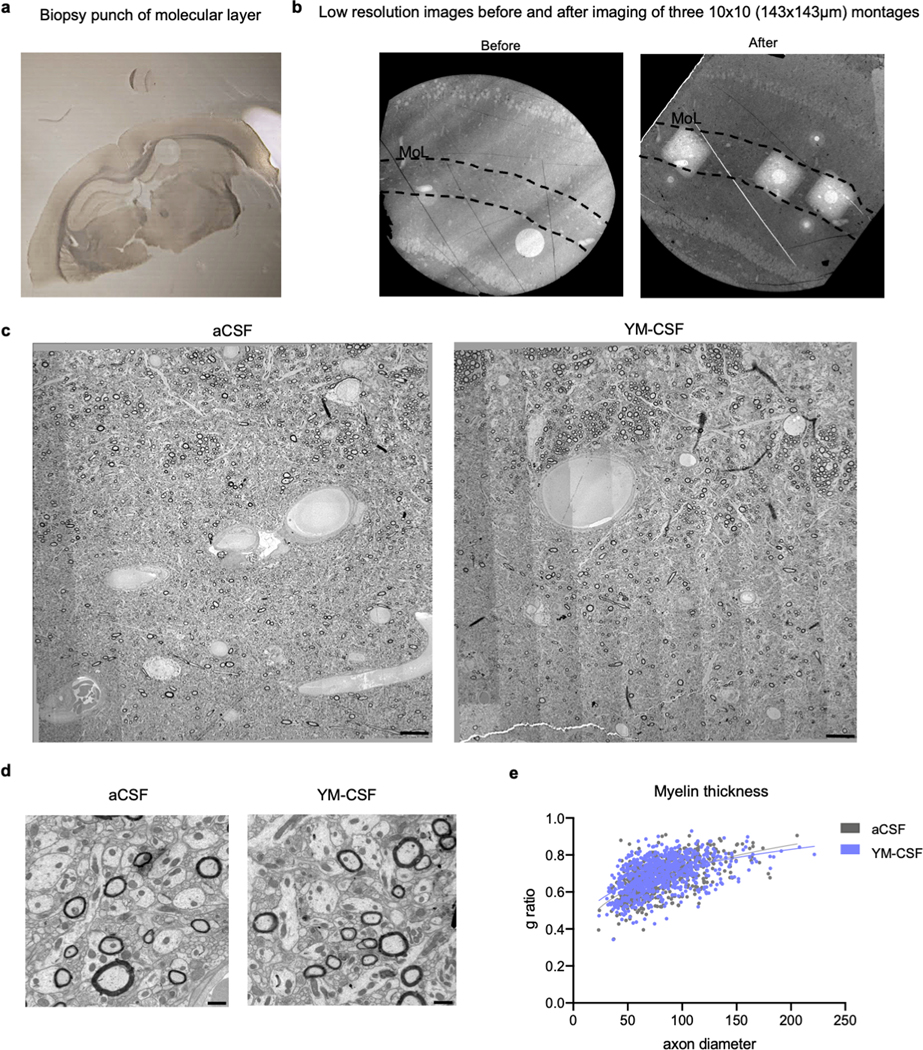
a, Representative overview of 1mm diameter biopsy punch in the hippocampus.
b, Representative overview of molecular layer (MoL, between dashed lines) before and after TEM imaging of three 10×10 montage squares (n=7).
c, Representative montage of MoL of aged mouse infused with aCSF and YM-CSF (n=7). Scale bar, 10 μm.
d, Representative higher resolution image of aged mouse infused with aCSF and YM-CSF (n=7). Scale bar, 1μm.
e, g-ratio analysis of myelinated axons in molecular layer. (n=3 mice per group, aCSF n=321 axons, YM-CSF n=291 axons).
Extended data Fig. 4. Young CSF boosts OPC differentiation in vitro and validation of OPC culture purity.
a, Related to images in Fig 1o. Overview of MBP stain of OLs at day 4 of differentiation supplemented with 10% aCSF or YH-CSF (aCSF n=3 coverslips, YH-CSF n=2 coverslips).
b, Quantification of MBP intensity of day 4 differentiated OLs. Scale bar, 200μm. (aCSF n=3 coverslips, YH-CSF n=2 coverslips; two-sided t-test; mean ± s.e.m).
c, Primary rat OPC cultures were supplemented with 10% aCSF or YH-CSF for 6 hrs and stained for NG2 (green), Olig2 (grey) and Acta2 (red). (n=3 coverslips; Scale bar, 100 μm)
d, Higher magnification of primary rat OPC cultures were supplemented with 10% aCSF or YH-CSF for 6 hrs and stained for NG2 (green), Olig2 (grey) and Acta2 (red). (n=3 coverslips; Scale bar, 20μm)
Extended data Fig. 5. SLAMseq QC and principal component analysis.
a, Overall conversion rates in all SLAMseq samples, showing an enrichment for T>C mutation rate (orange bar) which increases with longer incubation time (6hr).
b-c, Distribution of T>C mutations across b, read position and c, 3’UTR position indicating an equal distribution of s4U incorporation along the positive strand.
d-e, UMAP of aCSF and YH-CSF samples in both time points by all genes detected in the d, total and e, nascent mRNA counts. (young CSF 1hr n=4, all the rest n=5).
f, Gene set enrichment analysis (GSEA) of 6hr genes sorted by log2FC showing an enrichment for SRF target genes by TRANSFAC 74.
g, Overall log2FC enrichment indicating upregulation of SRF target genes (TRANSFAC and curated list) and actin cytoskeleton genes in YH-CSF treated OPCs over aCSF. (SRF TRANSFAC (423 genes), validated SRF targets from literature (74 genes) and actin genes (212 genes); Wilcoxon rank sum test; box show the median and the 25–75th percentiles, and the whiskers indicate values up to 1.5-times the interquartile range).
Extended data Fig. 6. YH-CSF induces actin cytoskeleton alterations in vitro.
a-b, Actin filament content measured by live imaging using SiR-actin (red) throughout 4hr of aCSF and YH-CSF exposure. Average SiR-actin a, intensity and b, area in rat OPC cultures exposed to aCSF or YH-CSF (n=6 wells per condition; Means ± SEM).
c, Representative images of experiment quantified in panel a and b. Scale bar 200μm.
d, OPC coverslips were treated with YH-CSF for 6 hrs and stained for phalloidin. Histogram of the percentage of OPC with the indicated number of growth cones per cell. YH-CSF treated cells show a shift towards more growth cones per cell (n=3 coverslips per condition, total of 200 cells analyzed per condition; two-way ANOVA followed by Sidak’s post-hoc test; Means ± SEM). Scale bar 20μm.
e, mouse OPC primary cultures from SRF-fl/fl pups infected with CRE-GFP and △CRE-GFP AAVs to induce recombination. Representative images of infected cells (green) 48 hours after infection. Scale bar, 100μm.
f, Normalized SRF mRNA levels as measured by RT-PCR (n=3 coverslips per condition; mean ± s.e.m.).
g, Representative image of data presented in figure 2h. Scale bar, 20μm.
h, Quantification of GFP+ cells per image in SRF-WT and SRF-KO cells treated with 10% aCSF or YH-CSF. (n=3; mean ± s.e.m.).
i, Quantification of number of DAPI cells per image in SRF-WT and SRF-KO cells treated with 10% aCSF or YH-CSF. (n=3; mean ± s.e.m.).
Data in panels a-i were replicated in two independent experiments.
Extended data Fig. 7. Bulk RNAseq of hippocampal OPC and OL nuclei from young and aged mice.
a, Gating strategy for sorting of hippocampal OPC and OL nuclei.
b, Heatmap of expression OPC and OL specific genes across young and aged OPC and OL samples (aged OL n=3, rest n=4).
c, Volcano plot showing OL genes up and downregulated with age (n=4; p. adjusted value by Wald test in DESeq2).
d, Pathways enriched (red) or depleted (blue) in hippocampal OLs with age (unweighted Kolmogorov-Smirnow test).
Extended data Fig. 8. Bulk RNAseq of hippocampal OPC and OL nuclei from aged mice following acute injection and Srf levels in neurons.
a, Box plot of effect size of Srf targets (TRANSFAC database) in hippocampal OLs from aged vs. young, YM-CSF vs. aCSF at 1hr and 6hr timepoints (n=4; genes pre-filtered by p<0.05 cutoff; Wilcoxon rank sum test, box show the median and the 25–75th percentiles, and the whiskers indicate values up to 1.5-times the interquartile range).
b, Pathways enriched (red) or depleted (blue) in hippocampal OPCs 1hr following injection of aCSF or YM-CSF (n=4; p. adjusted value by Wald test in DESeq2).
c, Volcano plot showing OPC genes up and down regulated 1hr following CSF injection (n=4; p. adjusted value by Wald test in DESeq2).
d, Volcano plot showing OPC genes up and down regulated 6hr following CSF injection (n=4; p. adjusted value by Wald test in DESeq2).
e, Neuronal Srf intensity in CA1 in young and aged mice. (n=3; two-sided t-test; mean ± s.e.m)
f, Representative image of panel e. Scale bar, 70μm.
g, Neuronal Srf intensity in CA1 in aged mice following YM-CSF infusion. (n=4; two-sided t-test; mean ± s.e.m)
h, Representative image of panel g. Scale bar, 70μm.
Extended data Fig. 9. Fgf8 induces OPC proliferation and Fgf17 induces SRF reporter activation mediated by actin dynamics and Fgfr3.
a, Dose-dependent activation of SRE-GFP reporter by increasing concentrations of Fgf8 and representative images of the experiment at 15.5 hrs. Scale bar, 400μm. (n=3; similar control as in Fig. 4c; one-way ANOVA followed by Sidak’s post-hoc test; mean ± s.e.m.).
b, Percentage of BRDU+/DAPI primary rat OPCs treated with 10, 20, 40 ng/ml Fgf8. (n=4; one-way ANOVA followed by Tukey’s post-hoc test; mean ± s.e.m.).
c, Quantification of OPC proliferating cells (Pdgfrα+EDU+ / Pdgfrα+ cells) in the CA1 region of the hippocampus of 20-month-old mice following a week of aCSF or Fgf8 infusion. (aCSF n=8 similar control as in Fig. 4l, Fgf8 n=4; two-sided t-test; mean ± s.e.m.).
d, SRE-GFP activation with 200 ng/ml Fgf17 following 30 min pre-treatment with Jasplakinolide (Jasp, 125 or 250nM) or Latrunculin A (LatA, 250 or 500nM). (n=3; Two-way ANOVA with Tukey’s multiple comparisons test; mean ± s.e.m.).
e, SRE-GFP activation with 200 ng/ml Fgf17 following 30 min pre-treatment with blocking antibodies for FgfR1, FgfR2, FgfR3 (all 50 μg/ml) or FgfR3 alone (n=3; One-way ANOVA with Sidak’s multiple comparisons test; mean ± s.e.m.).
f, Example of Pdgfr⍺+ Fgfr3+ cells in the hippocampus of young mice. (n=3). Scale bar, 5μm.
Extended data Fig. 10. Fgf17 is predominantly expressed in the brain by a subset of neurons and is downregulated with age.
a, Fgf17 is predominantly expressed in the brain based on the human protein atlas.
b, Fgf17 is expressed by cortical glutamatergic neurons in the young adult mouse (Allen brain atlas).
c, Sub-clustering of mouse cortical layer 4/5 neurons indicates expression by a subset of cortical neurons (Allen brain atlas).
d, Gene set enrichment analysis of genes mostly correlated with Fgf17 in layer 4/5 neurons (Allen brain atlas).
e, Fgf17 is expressed by cortical glutamatergic and GABAergic neurons in the human cortex (Allen brain atlas).
f, Representative image of analysis in panel g. Scale bar, 100μm.
g, Fgf17 mRNA expression in cortical neurons drops dramatically in aged mice. (n=3; two-way student t-test; mean ± s.e.m.).
h, Fgf17 protein expression in cortical and hippocampal neurons drops dramatically in aged mice. (n=3; mean ± s.e.m.).
i, Representative images of analysis in panel h and Fig. 4f. Scale bar, 20μm.
Extended data Fig. 11. Perfusion of labeled YH-CSF and mouse Fgf17 to the brain parenchyma and working model.
a, Deposition of labeled Fgf17 on ventricular walls 3 hours post ICV acute injection (n=3). Scale bar, 300μm.
b, Deposition of labeled YH-CSF on lateral ventricle walls 2 hours post ICV acute injection (n=3). Scale bar, 100μm.
c, Labeled Fgf17 in perivascular spaces in the molecular layer of the hippocampus (n=3). Scale bar, 50μm.
d, Labeled YH-CSF in perivascular spaces in the molecular layer of the hippocampus (n=3). Scale bar, 20μm.
e, YH-CSF in the perivascular space in between the vessel (green) and astrocyte endfeet (white; n=3). Scale bar, 20μm.
f, Orthogonal slice of YH-CSF (magenta) in perivascular space, in between the vessel (green) and astrocyte endfeet (white; n=3). Scale bar, 20μm.
g, Working model. OPC proliferation and differentiation (termed oligodendrogenesis) slow down with age 40–43. Re-exposure of the aged brain to young CSF or the brain-specific growth factor Fgf17 45, boost hippocampal oligodendorgenesis, concomitant with improvement in long term memory recall.
Supplementary Material
Acknowledgments:
We thank the members of the Wyss-Coray and Zuchero laboratories for feedback and support. Specifically, to Dr. H. Kantarci for advice on TEM tissue processing, imaging, and analysis. H. Zhang and K. Dickey for laboratory management. B. Carter for flow cytometry technical expertise. We thank Valentina Galata for graphical abstract design. Dr. Emmanuel Mignot and Dr. Josep Dalmau for providing human CSF samples. Thank you to Dr. Danielle Jorgens and the staff at the University of California Berkeley Electron Microscope Laboratory for electron microscopy sample preparation and data collection.
Funding:
This work was funded by the Department of Veterans Affairs (T.W.-C.), the National Institute on Aging (RF1-AG064897- 02 to T.W.-C., T32AG000266 to MH), the NOMIS Foundation (T.W.-C.), The Nan Fung Life Sciences Aging Research Fund (T.W.-C.), The Glenn Foundation for Aging Research (T.W.-C.), the Big Idea Brain Rejuvenation Project and Interdisciplinary Scholar fellowship from the Wu Tsai Neurosciences Institute (T.W.-C. and T.I.), the Zuckerman STEM leadership fellowship and Tel Aviv University President Award for women Postdoctoral scholars (T.I). The National MS Society Harry Weaver Neuroscience Scholar Award (J.B.Z.), The McKnight Scholar Award (J.B.Z.), The Myra Reinhard Family Foundation and National institute of Health (R01NS119823 to J.B.Z.) H.Z. is a Wallenberg Scholar supported by grants from the Swedish Research Council (#2018-02532), the European Research Council (#681712), Swedish State Support for Clinical Research (#ALFGBG-720931), the Alzheimer Drug Discovery Foundation (ADDF), USA (#201809-2016862), and the UK Dementia Research Institute at UCL;
Data and materials availability:
All data is available in the main text or the supplementary materials. Raw and processed sequencing data were deposited to NCBI’s SRA and GEO databases using the accession ID GSE198008.
Footnotes
Competing interests: T.W.-C. and T.I. are co-inventors on a patent application related to the work published in this paper (STDU2-39617.101, S21-153 - METHODS AND COMPOSITIONS FOR IMPROVED MEMORY IN THE AGING). HZ has served at scientific advisory boards and/or as a consultant for Abbvie, Alector, Annexon, Artery Therapeutics, AZTherapies, CogRx, Denali, Eisai, Nervgen, Novo Nordisk, Pinteon Therapeutics, Red Abbey Labs, Passage Bio, Roche, Samumed, Siemens Healthineers, Triplet Therapeutics, and Wave, has given lectures in symposia sponsored by Cellectricon, Fujirebio, Alzecure, Biogen, and Roche, and is a co-founder of Brain Biomarker Solutions in Gothenburg AB (BBS), which is a part of the GU Ventures Incubator Program (outside submitted work).
Code availability: All analyses have been carried out using freely available software packages. Custom code used to analyze the RNA-seq data and datasets generated and/or processed in the current study is available from the corresponding authors upon request.
Main text references
- 1.Pluvinage JV & Wyss-Coray T. Systemic factors as mediators of brain homeostasis, ageing and neurodegeneration. Nat Rev Neurosci 21, 93–102, doi: 10.1038/s41583-019-0255-9 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Castellano JM et al. Human umbilical cord plasma proteins revitalize hippocampal function in aged mice. Nature 544, 488–492, doi: 10.1038/nature22067 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Villeda SA et al. Young blood reverses age-related impairments in cognitive function and synaptic plasticity in mice. Nat Med 20, 659–663, doi: 10.1038/nm.3569 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lehtinen MK et al. The cerebrospinal fluid provides a proliferative niche for neural progenitor cells. Neuron 69, 893–905, doi: 10.1016/j.neuron.2011.01.023 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silva-Vargas V, Maldonado-Soto AR, Mizrak D, Codega P. & Doetsch F. Age-Dependent Niche Signals from the Choroid Plexus Regulate Adult Neural Stem Cells. Cell Stem Cell 19, 643–652, doi: 10.1016/j.stem.2016.06.013 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Fame RM & Lehtinen MK Emergence and Developmental Roles of the Cerebrospinal Fluid System. Dev Cell 52, 261–275, doi: 10.1016/j.devcel.2020.01.027 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Chen CP, Chen RL & Preston JE The influence of ageing in the cerebrospinal fluid concentrations of proteins that are derived from the choroid plexus, brain, and plasma. Exp Gerontol 47, 323–328, doi: 10.1016/j.exger.2012.01.008 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Baird GS et al. Age-dependent changes in the cerebrospinal fluid proteome by slow off-rate modified aptamer array. Am J Pathol 180, 446–456, doi: 10.1016/j.ajpath.2011.10.024 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li G. et al. Cerebrospinal fluid concentration of brain-derived neurotrophic factor and cognitive function in non-demented subjects. PLoS One 4, e5424, doi: 10.1371/journal.pone.0005424 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan S, Mayoral SR, Choi HS, Chan JR & Kheirbek MA Preservation of a remote fear memory requires new myelin formation. Nat Neurosci 23, 487–499, doi: 10.1038/s41593-019-0582-1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vetere G. et al. Chemogenetic Interrogation of a Brain-wide Fear Memory Network in Mice. Neuron 94, 363–374 e364, doi: 10.1016/j.neuron.2017.03.037 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Fogel SM et al. fMRI and sleep correlates of the age-related impairment in motor memory consolidation. Hum Brain Mapp 35, 3625–3645, doi: 10.1002/hbm.22426 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibson EM et al. Neuronal activity promotes oligodendrogenesis and adaptive myelination in the mammalian brain. Science 344, 1252304, doi: 10.1126/science.1252304 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dugas JC & Emery B. Purification of oligodendrocyte precursor cells from rat cortices by immunopanning. Cold Spring Harb Protoc 2013, 745–758, doi: 10.1101/pdb.prot070862 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Sun LO et al. Spatiotemporal Control of CNS Myelination by Oligodendrocyte Programmed Cell Death through the TFEB-PUMA Axis. Cell 175, 1811–1826 e1821, doi: 10.1016/j.cell.2018.10.044 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuchero JB et al. CNS myelin wrapping is driven by actin disassembly. Dev Cell 34, 152–167, doi: 10.1016/j.devcel.2015.06.011 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwarz N. et al. Human Cerebrospinal fluid promotes long-term neuronal viability and network function in human neocortical organotypic brain slice cultures. Sci Rep 7, 12249, doi: 10.1038/s41598-017-12527-9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wentling M. et al. A metabolic perspective on CSF-mediated neurodegeneration in multiple sclerosis. Brain 142, 2756–2774, doi: 10.1093/brain/awz201 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Mathur D. et al. Bioenergetic Failure in Rat Oligodendrocyte Progenitor Cells Treated with Cerebrospinal Fluid Derived from Multiple Sclerosis Patients. Front Cell Neurosci 11, 209, doi: 10.3389/fncel.2017.00209 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braun T. & Gautel M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat Rev Mol Cell Biol 12, 349–361, doi: 10.1038/nrm3118 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Guo Y. et al. Hierarchical and stage-specific regulation of murine cardiomyocyte maturation by serum response factor. Nat Commun 9, 3837, doi: 10.1038/s41467-018-06347-2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knoll B. & Nordheim A. Functional versatility of transcription factors in the nervous system: the SRF paradigm. Trends Neurosci 32, 432–442, doi: 10.1016/j.tins.2009.05.004 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Miralles F, Posern G, Zaromytidou AI & Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell 113, 329–342, doi: 10.1016/s0092-8674(03)00278-2 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Knoll B. et al. Serum response factor controls neuronal circuit assembly in the hippocampus. Nat Neurosci 9, 195–204, doi: 10.1038/nn1627 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Lahoute C. et al. Premature aging in skeletal muscle lacking serum response factor. PLoS One 3, e3910, doi: 10.1371/journal.pone.0003910 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mergoud Dit Lamarche A. et al. UNC-120/SRF independently controls muscle aging and lifespan in Caenorhabditis elegans. Aging Cell 17, doi: 10.1111/acel.12713 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ximerakis M. et al. Single-cell transcriptomic profiling of the aging mouse brain. Nat Neurosci 22, 1696–1708, doi: 10.1038/s41593-019-0491-3 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Falcao AM et al. Disease-specific oligodendrocyte lineage cells arise in multiple sclerosis. Nat Med 24, 1837–1844, doi: 10.1038/s41591-018-0236-y (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iacono G, Altafini C. & Torre V. Early phase of plasticity-related gene regulation and SRF dependent transcription in the hippocampus. PLoS One 8, e68078, doi: 10.1371/journal.pone.0068078 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuzniewska B. et al. Brain-derived neurotrophic factor induces matrix metalloproteinase 9 expression in neurons via the serum response factor/c-Fos pathway. Mol Cell Biol 33, 2149–2162, doi: 10.1128/MCB.00008-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasayama D. et al. Genome-wide quantitative trait loci mapping of the human cerebrospinal fluid proteome. Hum Mol Genet 26, 44–51, doi: 10.1093/hmg/ddw366 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Sathyan S. et al. Plasma proteomic profile of age, health span, and all-cause mortality in older adults. Aging Cell 19, e13250, doi: 10.1111/acel.13250 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Esnault C. et al. Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts. Genes Dev 28, 943–958, doi: 10.1101/gad.239327.114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fortin D, Rom E, Sun H, Yayon A. & Bansal R. Distinct fibroblast growth factor (FGF)/FGF receptor signaling pairs initiate diverse cellular responses in the oligodendrocyte lineage. J Neurosci 25, 7470–7479, doi: 10.1523/JNEUROSCI.2120-05.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramanan N. et al. SRF mediates activity-induced gene expression and synaptic plasticity but not neuronal viability. Nat Neurosci 8, 759–767, doi: 10.1038/nn1462 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Etkin A. et al. A role in learning for SRF: deletion in the adult forebrain disrupts LTD and the formation of an immediate memory of a novel context. Neuron 50, 127–143, doi: 10.1016/j.neuron.2006.03.013 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Pan S, Mayoral SR, Choi HS, Chan JR & Kheirbek MA Preservation of a remote fear memory requires new myelin formation. Nat Neurosci, doi: 10.1038/s41593-019-0582-1 (2020). [DOI] [PMC free article] [PubMed]
- 38.Xiao L. et al. Rapid production of new oligodendrocytes is required in the earliest stages of motor-skill learning. Nat Neurosci 19, 1210–1217, doi: 10.1038/nn.4351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steadman PE et al. Disruption of Oligodendrogenesis Impairs Memory Consolidation in Adult Mice. Neuron 105, 150–164 e156, doi: 10.1016/j.neuron.2019.10.013 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang F. et al. Myelin degeneration and diminished myelin renewal contribute to age-related deficits in memory. Nat Neurosci, doi: 10.1038/s41593-020-0588-8 (2020). [DOI] [PMC free article] [PubMed]
- 41.Chen JF et al. Enhancing myelin renewal reverses cognitive dysfunction in a murine model of Alzheimer’s disease. Neuron 109, 2292–2307 e2295, doi: 10.1016/j.neuron.2021.05.012 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Segel M. et al. Niche stiffness underlies the ageing of central nervous system progenitor cells. Nature 573, 130–134, doi: 10.1038/s41586-019-1484-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neumann B. et al. Metformin Restores CNS Remyelination Capacity by Rejuvenating Aged Stem Cells. Cell Stem Cell 25, 473–485 e478, doi: 10.1016/j.stem.2019.08.015 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonetto G, Belin D. & Karadottir RT Myelin: A gatekeeper of activity-dependent circuit plasticity? Science 374, eaba6905, doi: 10.1126/science.aba6905 (2021). [DOI] [PubMed] [Google Scholar]
- 45.Xu J, Liu Z. & Ornitz DM Temporal and spatial gradients of Fgf8 and Fgf17 regulate proliferation and differentiation of midline cerebellar structures. Development 127, 1833–1843 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Furusho M, Ishii A, Hebert JM & Bansal R. Developmental stage-specific role of Frs adapters as mediators of FGF receptor signaling in the oligodendrocyte lineage cells. Glia 68, 617–630, doi: 10.1002/glia.23743 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oh LY et al. Fibroblast growth factor receptor 3 signaling regulates the onset of oligodendrocyte terminal differentiation. J Neurosci 23, 883–894 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kang W, Nguyen KCQ & Hebert JM Transient Redirection of SVZ Stem Cells to Oligodendrogenesis by FGFR3 Activation Promotes Remyelination. Stem Cell Reports 12, 1223–1231, doi: 10.1016/j.stemcr.2019.05.006 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jen YH, Musacchio M. & Lander AD Glypican-1 controls brain size through regulation of fibroblast growth factor signaling in early neurogenesis. Neural Dev 4, 33, doi: 10.1186/1749-8104-4-33 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scearce-Levie K. et al. Abnormal social behaviors in mice lacking Fgf17. Genes Brain Behav 7, 344–354, doi: 10.1111/j.1601-183X.2007.00357.x (2008). [DOI] [PubMed] [Google Scholar]
Methods references
- 51.De Miguel Z. et al. Exercise plasma boosts memory and dampens brain inflammation via clusterin. Nature 600, 494–499, doi: 10.1038/s41586-021-04183-x (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu L. & Duff K. A technique for serial collection of cerebrospinal fluid from the cisterna magna in mouse. J Vis Exp, doi: 10.3791/960 (2008). [DOI] [PMC free article] [PubMed]
- 53.Smith A, Wu AH, Lynch KL, Ko N. & Grenache DG Multi-wavelength spectrophotometric analysis for detection of xanthochromia in cerebrospinal fluid and accuracy for the diagnosis of subarachnoid hemorrhage. Clin Chim Acta 424, 231–236, doi: 10.1016/j.cca.2013.06.017 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Olsson M, Arlig J, Hedner J, Blennow K. & Zetterberg H. Sleep deprivation and cerebrospinal fluid biomarkers for Alzheimer’s disease. Sleep 41, doi: 10.1093/sleep/zsy025 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Olsson M, Arlig J, Hedner J, Blennow K. & Zetterberg H. Sleep deprivation and plasma biomarkers for Alzheimer’s disease. Sleep Med 57, 92–93, doi: 10.1016/j.sleep.2018.12.029 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Lynch HJ, Rivest RW & Wurtman RJ Artificial induction of melatonin rhythms by programmed microinfusion. Neuroendocrinology 31, 106–111, doi: 10.1159/000123059 (1980). [DOI] [PubMed] [Google Scholar]
- 57.Pluvinage JV et al. CD22 blockade restores homeostatic microglial phagocytosis in ageing brains. Nature 568, 187–192, doi: 10.1038/s41586-019-1088-4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lukinavicius G. et al. Fluorogenic probes for live-cell imaging of the cytoskeleton. Nat Methods 11, 731–733, doi: 10.1038/nmeth.2972 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Friedman PL & Ellisman MH Enhanced visualization of peripheral nerve and sensory receptors in the scanning electron microscope using cryofracture and osmium-thiocarbohydrazide-osmium impregnation. J Neurocytol 10, 111–131, doi: 10.1007/BF01181748 (1981). [DOI] [PubMed] [Google Scholar]
- 60.Willingham MC & Rutherford AV The use of osmium-thiocarbohydrazide-osmium (OTO) and ferrocyanide-reduced osmium methods to enhance membrane contrast and preservation in cultured cells. J Histochem Cytochem 32, 455–460, doi: 10.1177/32.4.6323574 (1984). [DOI] [PubMed] [Google Scholar]
- 61.Ewald AJ et al. Mammary collective cell migration involves transient loss of epithelial features and individual cell migration within the epithelium. J Cell Sci 125, 2638–2654, doi: 10.1242/jcs.096875 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McDonald KL & Webb RI Freeze substitution in 3 hours or less. J Microsc 243, 227–233, doi: 10.1111/j.1365-2818.2011.03526.x (2011). [DOI] [PubMed] [Google Scholar]
- 63.Emery B. & Dugas JC Purification of oligodendrocyte lineage cells from mouse cortices by immunopanning. Cold Spring Harb Protoc 2013, 854–868, doi: 10.1101/pdb.prot073973 (2013). [DOI] [PubMed] [Google Scholar]
- 64.Muhar M. et al. SLAM-seq defines direct gene-regulatory functions of the BRD4-MYC axis. Science 360, 800–805, doi: 10.1126/science.aao2793 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stockel D. et al. Multi-omics enrichment analysis using the GeneTrail2 web service. Bioinformatics 32, 1502–1508, doi: 10.1093/bioinformatics/btv770 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Hahn O. et al. CoolMPS for robust sequencing of single-nuclear RNAs captured by droplet-based method. Nucleic Acids Res 49, e11, doi: 10.1093/nar/gkaa1127 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Newman AM et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol 37, 773–782, doi: 10.1038/s41587-019-0114-2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Steen CB, Liu CL, Alizadeh AA & Newman AM Profiling Cell Type Abundance and Expression in Bulk Tissues with CIBERSORTx. Methods Mol Biol 2117, 135–157, doi: 10.1007/978-1-0716-0301-7_7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wolf FA, Angerer P. & Theis FJ SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19, 15, doi: 10.1186/s13059-017-1382-0 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schaum N. et al. Ageing hallmarks exhibit organ-specific temporal signatures. Nature 583, 596–602, doi: 10.1038/s41586-020-2499-y (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Spitzer SO et al. Oligodendrocyte Progenitor Cells Become Regionally Diverse and Heterogeneous with Age. Neuron 101, 459–471 e455, doi: 10.1016/j.neuron.2018.12.020 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mathys H. et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337, doi: 10.1038/s41586-019-1195-2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou Y. et al. Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat Med 26, 131–142, doi: 10.1038/s41591-019-0695-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Matys V. et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res 34, D108–110, doi: 10.1093/nar/gkj143 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gerstner N. et al. GeneTrail 3: advanced high-throughput enrichment analysis. Nucleic Acids Res, doi: 10.1093/nar/gkaa306 (2020). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.