Abstract
Peptidoglycan hydrolase, LytF (CwlE), was determined to be identical to YhdD (deduced cell wall binding protein) by zymography after insertional inactivation of the yhdD gene. YhdD exhibits high sequence similarity with CwlF (PapQ, LytE) and p60 of Listeria monocytogenes. The N-terminal region of YhdD has a signal sequence followed by five tandem repeated regions containing polyserine residues. The C-terminal region corresponds to the catalytic domain, because a truncated protein without the N-terminal region retained cell wall hydrolase activity. The histidine-tagged LytF protein produced in Escherichia coli cells hydrolyzed the linkage of d-γ-glutamyl-meso-diaminopimelic acid in murein peptides, indicating that it is a d,l-endopeptidase. Northern hybridization and primer extension analyses indicated that the lytF gene was transcribed by EςD RNA polymerase. Disruption of lytF led to slightly filamentous cells, and a lytF cwlF double mutant exhibited extraordinary microfiber formation, which is similar to the cell morphology of the cwlF sigD mutant.
Bacillus subtilis produces peptidoglycan hydrolases, some of which are autolysins (34, 38). Two vegetative autolysins, a major 50-kDa N-acetylmuramoyl-l-alanine amidase (amidase, CwlB [LytC]) and a 90-kDa endo-β-N-acetylglucosaminidase (glucosaminidase, CwlG [LytD]), have been studied at the molecular level (20, 24, 26, 31). Recently, two minor autolysins produced during vegetative growth were reported (32). CwlF is a 35-kDa protein, and its production is unaffected by the sigma D and flaD1 (sinR) mutations. The other one, CwlE (LytF), is a 50-kDa protein, and it is not produced by the sigD null mutant. CwlE (LytF) overlapped CwlB in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (32). Very recently, it was reported that CwlF is identical to PapQ and LytE (13, 28). The cells of the cwlF-deficient mutant were about twice as long as those of the wild-type strain, and the cwlF sigD double-mutant cells exhibited extraordinary microfiber formation (13). B. subtilis genome-sequencing analysis indicated the existence of many paralogs of cell wall hydrolases (17). One large group among the paralogs includes the cell wall-lytic enzyme, p60, of Listeria monocytogenes (4, 16), CwlF (PapQ, LytE), and YhdD.
In this study, we identified yhdD as a new peptidoglycan hydrolase gene, cwlE (lytF), expressed during the vegetative growth phase, characterized the gene expression, and determined the role of cell separation in B. subtilis. Moreover, we report that CwlE (LytF) is an endopeptidase which digests the linkage of d-γ-glutamyl-meso-diaminopimelic acid in muramic acid peptides.
(Preliminary data were presented at the International Conference on Bacilli, Japan [Osaka, Japan, 12 to 15 July 1998]. After the submission of this paper, Margot et al. [27] published the function of YhdD and designated the gene lytF.)
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains of B. subtilis and Escherichia coli and the plasmids used in this study are listed in Table 1. B. subtilis 168 was the parent strain throughout this study, and mutants having the 168 background were constructed. B. subtilis was grown on Luria-Bertani (LB) agar medium (35) at 37°C for about 10 h and was then incubated in Schaeffer medium (36) at 30°C unless otherwise noted. When necessary, chloramphenicol, tetracycline, and erythromycin were added to the medium to final concentrations of 3, 5, and 0.3 μg/ml, respectively. E. coli was grown in LB medium (35) at 37°C. When necessary, ampicillin and kanamycin were added to final concentrations of 50 or 100 μg/ml and 25 μg/ml, respectively.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relative genotype(s) | Source or referencea |
|---|---|---|
| B. subtilis | ||
| 168 | trpC2 | 42 |
| 168S | trpC2 strA smo-1 | 20 |
| 327SDC | purB his-1 smo-1 sigD::cat | M. H. Rashid |
| 327SD1 | purB his-1 smo-1 sigD::tet | M. H. Rashid |
| 168SDC | trpC2 sigD::cat | 327SDC→168 |
| 168SD1 | trpC2 sigD::tet | 327SD1→168 |
| FTD | trpC2 cwlF (papQ)::tet | pUCFTET→168 |
| FTDSDC | trpC2 cwlF (papQ)::tet sigD::cat | pUCFTET→168SDC |
| AN8 | purB his-1 smo-1 cwlB::cat | A. Kuroda |
| EN8 | trpC2 cwlB::cat | AN8→168 |
| ED | trpC2 yhdD (lytF, cwlE)::erm | pM2-HDD→168 |
| FED | trpC2 cwlF (papQ)::tet yhdD (lytF, cwlE)::erm | ED→168FTD |
| BED | trpC2 cwlB::cat yhdD (lytF, cwlE)::erm | AN8→ED |
| M. luteus ATCC 4698 | Sigma | |
| E. coli | ||
| JM109 | recA1 endA1 gyrA96 thi-1 hsdR17 relA1 supE44 Δ(lac-proAB)/F′ [traD36 proAB lacIqlacZ ΔM15] | Takara |
| C600 | supE44 hsdR17 thi-1 thr-1 leuB6 lacY1 tonA21 | Laboratory stock |
| M15 | Nals Strs Rifslac ara gal mtl F−recA+ uvr+ | Qiagen |
| Plasmids | ||
| pUC118/119 | lacZ bla | Takara |
| pMUTIN2 | lacZ lacI erm bla | 42 |
| pDG1515 | lacZ bla tet | BGSC |
| pQE-30 | bla cat | Qiagen |
| pREP4 | lacI kan | Qiagen |
| pM2-HDD | lacZ lacI erm bla ΔyhdD (lytF, cwlE) | This study |
| pU8cF2 | lacZ bla cwlF (papQ) | This study |
| pUCFTET | lacZ bla cwlF (papQ)::tet | This study |
| pUCEtCTD | lacZ bla yhdD (lytF, cwlE) | This study |
| pQECEtCTD | bla cat H-lytF | This study |
Arrows indicate construction by transformation. BGSC, Bacillus Genetic Stock Center, the Ohio State University.
Plasmid construction.
To construct a B. subtilis lytF (yhdD, cwlE) mutant, an internal fragment of the lytF gene was amplified by PCR with two primers, forward primer h-YHDD (5′-GCGCAAGCTTA30GCATCTGCGATTGTCGG47; the internal sequence of the yhdD (lytF, cwlE) region is italicized, the numbering is with respect to the first A of the translational start codon of lytF, and the HindIII site is underlined) and reverse primer b-YHDD (5′-GCGCGGATCCG275AACTTCCGCTCTTCATG258; the sequence complementary to the internal region of lytF is italicized, and the BamHI site is underlined), with B. subtilis 168 DNA as a template. The PCR fragment was digested with HindIII and BamHI. pMUTIN2 was also digested with HindIII and BamHI and was then ligated to the digested PCR fragment, followed by the transformation of E. coli JM109. The resultant plasmid, pM2-HDD, was used for the transformation of E. coli C600 to produce concatemeric DNAs (6). To construct a B. subtilis cwlF (papQ, lytE) mutant, an internal fragment of the cwlF gene was amplified by PCR with two primers, forward primer cFSDBF (5′-GCGCGGATCCT−26AGAGTTAACATTTGGGGAG−7; the upstream sequence of cwlF is italicized, the numbering is with respect to the first A of the translational start codon of cwlF, and the BamHI site is underlined) and reverse primer cFSDSR (5′-GCGCCCCGGGT1005TAGAATCTTTTCGCACCG987; the sequence complementary to the downstream sequence of cwlF is italicized, and the SmaI site is underlined), with B. subtilis 168 DNA as a template. The PCR fragment was digested with BamHI and SmaI and was then ligated to BamHI- and HincII-digested pUC118, followed by the transformation of E. coli JM109. The DNA of the resultant plasmid, pU8cF2, was digested with ClaI and SpeI, followed by ligation to the ClaI- and XbaI-digested pDG1515 DNA containing the tetracycline resistance gene. The resultant plasmid, pUCFTET, was used for the construction of cwlF mutants. To construct a lytF gene encoding a histidine-tagged protein (H-lytF), a region corresponding to the catalytic domain of LytF was amplified by PCR with forward primer BF-CWLE2 (5′-GCGCGGATCCA1105CGAGTGCGAAGATTAACAC1124; the BamHI site is underlined) and reverse primer KR-CWLE (GCGCGGTACCC1529ATCAACGTCTTTAGGCTCT1512; the KpnI site is underlined), with B. subtilis chromosomal DNA as a template. Then the amplified 445-bp fragment was digested with BamHI and KpnI, followed by ligation to the corresponding sites of pUC118. After the transformation of E. coli JM109, an ampicillin-resistant plasmid, pUCEtCTD, was extracted from the transformant. After reconfirmation of the sequence, the BamHI-KpnI fragment of pUCEtCTD was ligated into the corresponding site of a histidine-tag-encoding plasmid, pQE-30 (Qiagen), followed by the transformation of E. coli M15(pREP4). E. coli cells harboring the resultant plasmid, pQECEtCTD, were used for the production of H-LytF (134 amino acids, including a 12-histidine-tagged amino acid sequence; Mr, 14,616).
Mutant construction.
To construct isogenic strains, DNAs from B. subtilis 327SD1 (32) and 327SDC (33) were used for the transformation of B. subtilis 168; the resultant strains, 168SD1 and 168SDC, were selected with tetracycline and chloramphenicol, respectively. B. subtilis EN8 was also constructed through the transformation of B. subtilis 168 with B. subtilis AN8 DNA. For the construction of cwlF and cwlF sigD mutants, B. subtilis 168 and 168SDC were transformed with ScaI-digested pUCFTET DNA and transformants (FTD and FTDSDC) were selected with tetracycline. To obtain a lytF mutant, B. subtilis 168 was transformed with pM2-HDD DNA and a transformant (ED) was selected with erythromycin. To obtain a lytF cwlF mutant, B. subtilis FTD was transformed with B. subtilis ED DNA and a transformant (FED) was selected with erythromycin. To obtain a lytF cwlB mutant, B. subtilis ED was transformed with B. subtilis AN8 DNA and a transformant (BED) was selected with chloramphenicol. All of the mutants constructed in this study were confirmed to be properly constructed by PCR or Southern blot analysis.
Transformation of E. coli and B. subtilis.
E. coli transformation was performed as described by Sambrook et al. (35), and B. subtilis transformation was performed by the competent cell method (1).
Preparation of cell wall binding proteins.
To prepare cell wall binding proteins, B. subtilis 168, EN8, and BED cells were cultured in modified Spizizen medium (32) at 37°C to an optical density at 600 nm (OD600) of 1.5 to 1.8. Then cultures (40 ml each) were centrifuged at 8,000 × g for 5 min at 4°C, and the cells were resuspended in distilled water, followed by the addition of SDS-PAGE sample buffer as described previously (32). The cell suspensions were then boiled for 5 min at 100°C, and the cells were removed by centrifugation. The supernatants were used as SDS-extracted samples (extract S).
Preparation of cell walls.
B. subtilis 168S and Micrococcus luteus ATCC 4698 cell walls were prepared as described previously (18). For determination of the cleavage site of the enzyme, the partially purified B. subtilis cell walls were incubated in a 10% trichloroacetic acid solution at 4°C for 2 days. After a washing with deionized water, the cell walls were suspended in 0.1 M Tris-HCl (pH 7.5) containing α-amylase (0.1 mg/ml) and were then incubated at 37°C for 2 h. Then CaCl2 and trypsin were added to final concentrations of 10 mM and 0.1 mg/ml, respectively, followed by incubation at 37°C for 16 h. After the enzymatic reactions, SDS (final concentration, 1%) was added to the solution, followed by boiling for 15 min. After centrifugation, the purified cell wall peptidoglycan was washed with deionized water and 0.1 M EDTA and then with ultrapure water.
Zymography.
Zymography was performed essentially as described previously (9, 25, 32), using an SDS-polyacrylamide (12 or 10%) gel (23) containing 0.1% (wt/vol) B. subtilis and M. luteus cell walls.
Production of H-LytF in E. coli.
E. coli M15(pREP4, pQECEtCTD) was cultured in LB medium containing ampicillin, kanamycin, and 2% glucose at 37°C. When cell growth reached an OD600 of 0.7 to 0.9, isopropyl-β-d-thiogalactopyranoside (IPTG; final concentration, 2 mM) was added to the culture. After a 30-min incubation, the cells were harvested by centrifugation and resuspended in a 10 mM imidazole NPB solution (10 mM imidazole and 0.5 M NaCl in 20 mM sodium phosphate buffer [pH 7.4]). After ultrasonication, the suspension was centrifuged and the supernatant was filtered through a 0.45-μm-pore-size membrane filter (Nalgene), followed by loading onto a HiTrap chelating column (1 ml of resin; Pharmacia). Then the column was washed with 20 ml of the above-described buffer, and H-LytF was eluted with 10 ml each of 60, 100, 150, 200, 250, 300, and 500 mM imidazole NPB solutions. Imidazole in the enzyme solutions was removed with a HiTrap desalting kit (Pharmacia).
Effect of pH on enzyme activity.
For determination of the optimal pH of the cell wall hydrolase activity of H-LytF, the following buffers (20 mM) containing 100 mM KCl and B. subtilis cell wall (10 mg/ml) were used: citrate buffer for pHs 3.0, 4.0, 5.0, and 5.5; Good’s buffer for pHs 5.5, 6.0, 6.5, 7.5, 8.5, 9.5, and 10.5; and phosphate buffer for pHs 10.5, 11.5, and 12.5. Purified H-LytF was added to the buffers to a final concentration of 10 μg per ml, followed by incubation at 37°C, and the decrease in OD540 was measured with a Shimadzu UV-1200 spectrometer. One unit of enzyme was defined as the amount of enzyme necessary to decrease the OD540 by 0.001 in 1 min.
Determination of the cleavage site of cell wall peptidoglycan.
To determine the cleavage site of the H-LytF protein, B. subtilis cell wall peptidoglycan (3.3 mg) and the purified H-LytF protein (30 μg) were added to 10 ml of Good’s buffer (20 mM MES [morpholinoethanesulfonic acid], pH 6.5) containing 100 mM KCl. After enzymatic reaction at 37°C for 0, 10, 20, or 60 min, 1.5-ml samples were boiled at 100°C for 10 min. After centrifugation, the supernatants (released fractions) were filtered through a membrane filter (0.45 μm pore size). For detection of free amino groups in the released fractions, samples (500 μl each) were mixed with 60 μl of 10% K2B4O7 and 50 μl of 0.1 M 1-fluoro-2,4-dinitrobenzene and were then incubated for 45 min at 65°C in the dark. Then dinitrophenyl (DNP) derivatives were hydrolyzed in 4 M HCl for 12 h at 95 to 100°C. The hydrolyzed samples were dried under a vacuum and were then resuspended in 500 μl of a mixture (4:1, vol/vol) of buffer A (10% acetonitrile and 0.02 N acetic acid) and buffer B (90% acetonitrile and 0.02 N acetic acid). The hydrolyzed DNP compounds were analyzed by high-performance liquid chromatography (HPLC) on a reverse-phase column (Wakosil-II5 C18; 4.0 by 250 mm; Wako, Kyoto, Japan). The release of free reducing groups during the enzymatic reaction was assayed by the Thompson and Shockman (41) modification of the Park and Johnson method by using N-acetylglucosamine as the standard.
Northern blot and primer extension analyses.
B. subtilis 168 and 168SD1 (OD600 of 15 to 20) cells cultured in Schaeffer medium were harvested at various times. RNA was prepared as described previously (14). Agarose-formaldehyde gel electrophoresis was performed as described by Sambrook et al. (35), and the transfer of the RNAs onto a nylon membrane was performed as described previously (14). The DNA fragment used for preparing an RNA probe was amplified by PCR with PM-FK (5′-CGGGGTACCG−113TGTGGAATTGTGAGCG−97; the pMUTIN2 sequence is italicized, the numbering is with respect to the first G of the translational start codon of lacZ, and the KpnI site is underlined) and PM-T7 (5′-TAATACGACTCACTATATA−36GTGTATCAACAAGCTGG−53; the sequence complementary to the pMUTIN2 sequence is italicized, and the T7 promoter is underlined) as primers and with pM2-HDD DNA, containing the internal region of lytF, as a template. The amplified fragment was digested with HindIII, and then the fragments were purified by phenol-chloroform treatment, followed by precipitation with ethanol. The RNA probe was prepared with a digoxigenin RNA labeling kit (Boehringer Mannheim), and Northern (RNA) hybridization was performed according to the manufacturer’s instructions. Primer extension analysis was performed as described previously (14), using primer PEX-HDD (5′-GACCTTAATCGTTGCTGC; the 5′ and 3′ ends correspond to the complementary nucleotides at positions 93 and 76 with respect to the 5′ end of the lytF gene).
Microscopic observation and determination of cell density.
Cells were shake cultured at 120 strokes per min in test tubes (17-mm diameter) containing 5 ml of LB medium at 37°C. The cell morphology was observed by phase-contrast microscopy. The OD600 was measured after strong vortexing of samples. In the case of sigD cwlF and lytF cwlF mutants, a small amount of lysozyme was added to the samples just before vortexing.
RESULTS AND DISCUSSION
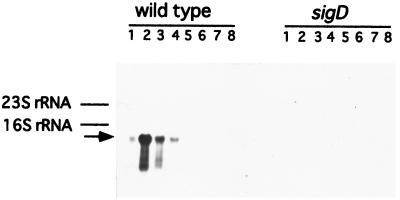
The B. subtilis genome project has revealed the existence of many cell wall hydrolase homologs. Since there was a possibility that LytF (CwlE) corresponds to one of the homologs, we selected three candidates, i.e., an approximately 50-kDa polypeptide (YrvJ [518 amino acids], YhdD [488 amino acids], and YvcE [473 amino acids]) (17, 37). Among these candidates, complete loss of RNA expression by the sigD mutation was observed only for the yhdD gene on the Northern blot analysis with the internal region of yhdD as a probe (Fig. 1).
FIG. 1.
Northern blot analysis of the yhdD region. Northern hybridization was performed with the yhdD-specific RNA probe as described in Materials and Methods. The lanes contained 10 μg of RNA from B. subtilis 168 (wild type) and 168SD1 (sigD) obtained at t−2 (lanes 1), t−1 (lanes 2), t−0.5 (lanes 3), t0 (lanes 4), t1.5 (lanes 5), t3 (lanes 6), t4.5 (lanes 7), or t6 (lanes 8). The arrow indicates a hybridizing RNA.
Identity of LytF (CwlE) to the yhdD gene product.
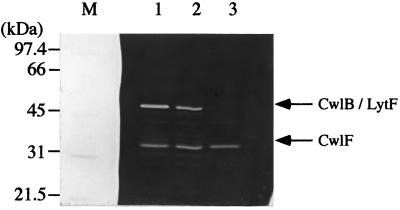
Zymography of cell wall extract (extract S) proteins from the 168 (wild type), EN8 (cwlB), and BED (cwlB lytF) strains was carried out, and the results are shown in Fig. 2. The 50-kDa protein, having cell wall hydrolase activity, was present in smaller amounts in the EN8 strain (Fig. 2, lane 2) and was completely lacking in the BED strain (lane 3). Since CwlB is a 50-kDa protein, the activity band at 50 kDa in lane 2 corresponds to the activity of LytF, and disruption of the yhdD gene led to the loss of LytF activity. These results indicate that YhdD is identical to LytF.
FIG. 2.
Zymography of proteins from B. subtilis 168, EN8 (cwlB), and BED (cwlB yhdD) cells. Electrophoresis was performed in an SDS–10% polyacrylamide gel containing 0.1% (wt/vol) B. subtilis cell wall as a substrate. Samples were prepared and subjected to zymography as described in Materials and Methods. Equal amounts of proteins (equivalent to 10 OD600 units of cells) were applied to the lanes. Lane M contained the protein standards (Bio-Rad), the molecular masses of which are shown on the left. Lanes 1 to 3, extracts S of the 168, EN8, and BED strains, respectively.
Amino acid sequence similarity of LytF (YhdD, CwlE) with other proteins.
The lytF (yhdD, cwlE) gene encodes a 488-amino-acid polypeptide with a molecular mass of 51,397 Da (17). LytF has three positively charged amino acids, K2, K3, and K4, in the N-terminal region, followed by a hydrophobic core (from L5 to G16) and a deduced signal peptidase cleavage site (A24EA↓A27; the arrow indicates the cleavage site). LytF also contains five tandem repeated regions with five polyserine regions and a C-terminal domain. The C-terminal domain, consisting of 118 amino acid residues, exhibits 67.0 and 45.2% identities over 115 amino acids with those of CwlF (13, 28) and the p60 protein (Iap) of L. monocytogenes (16, 43), respectively. The C-terminal region of LytF also exhibits high sequence similarity with the C-terminal regions of p60s from different Listeria species (4). E. coli NlpC and Haemophilus influenzae NlpC also exhibit high sequence similarities (35.8 and 33.9% identities over 123 and 115 amino acid residues, respectively) with the C-terminal domain of LytF (8, 15). Moreover, Bacillus sphaericus endopeptidase, EnpII, exhibits high sequence similarity (32.0% identity over 103 amino acids) with the C-terminal region of LytF (12). On the other hand, the repeated sequence in the N-terminal region of LytF exhibits similarity with the repeated sequences in the C-terminal regions of Lactococcus lactis muramidase AcmA (5), Streptococcus faecalis autolysin (2), and Enterococcus hirae muramidase-2 (7). These three cell wall hydrolases contain regions showing high sequence similarities in their N termini, which encompass the active-site regions (5). The amino acid sequence of LytF indicates that it is the second example of a novel type of peptidoglycan hydrolase (probably endopeptidase) in B. subtilis.
Alignment of the amino acid sequences of LytF paralogs in B. subtilis indicated that the amino acid sequence of paralog YojL is entirely the same as those of LytF and CwlF and that the sequences of the four paralogs (YddH, YvcE, YkfC, and YwtD) are similar to the C-terminal catalytic domain of LytF (27, 29).
Production of the histidine-tagged catalytic domain of LytF in E. coli.
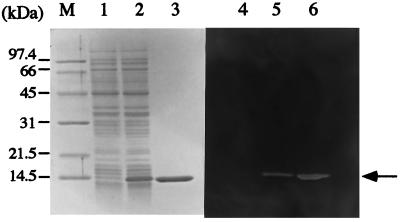
When we constructed a histidine-tagged fusion with CwlF, E. coli cells harboring a plasmid containing the gene were dramatically lysed, and thus it was difficult to produce a significant amount of the protein (29). Moreover, the purified CwlF easily aggregated during preservation, but the truncated form, which lacked the N-terminal cell wall binding domain, did not aggregate under such conditions (29). Therefore, we prepared the catalytic domain of LytF fused with the histidine-tagged sequence (H-LytF). A considerable amount of H-LytF was produced in E. coli cells after a 30-min induction with IPTG (Fig. 3). The protein was purified on a nickel column, and the purified protein showed a single band in SDS-PAGE, corresponding to the cell wall hydrolyzing band observed by zymography (Fig. 3). The size of 14.5 kDa corresponds with that calculated from the amino acid sequence (Mr, 14,616). The optimal pH of the enzyme activity specific for B. subtilis cell wall was 6.5, and the specific activity was 1,560 U/mg of protein. Although H-LytF is a histidine-tagged truncated enzyme, this specific activity was comparable with those of CwlA (2,500 U/mg of protein) (19) and CwlB (1,460 U/mg of protein) (20) but was much less than that of CwlG (26,000 U/mg of protein) (31). H-CwlF poorly digested M. luteus cell wall under conditions that were optimal for B. subtilis cell wall digestion. Among B. subtilis cell wall hydrolases, CwlA and CwlG (LytD) were able to digest M. luteus cell wall (21, 30), but CwlB (LytC) (30) did not digest M. luteus cell wall. Therefore, the poor activity of H-LytF for M. luteus cell wall is not a rare case, although the N-terminal region of LytF may affect the activity for M. luteus cell wall.
FIG. 3.
SDS–12% PAGE and zymography of proteins produced by E. coli M15 harboring a lacI plasmid, pREP4, and pQECEtCTD containing H-lytF. Lanes M and 1 to 3, SDS-PAGE; lanes 4 to 6, zymography. M, protein standards; lanes 1 and 4, cell lysate without induction; lanes 2 and 5, cell lysate with a 30-min induction; lanes 3 and 6, purified protein after HiTrap column chromatography. Proteins equivalent to 0.2 OD660 unit of cells were applied to lanes 1, 2, 4, and 5. The purified protein (2 μg) was applied to lanes 3 and 6.
Determination of the peptidoglycan cleavage site of H-LytF.
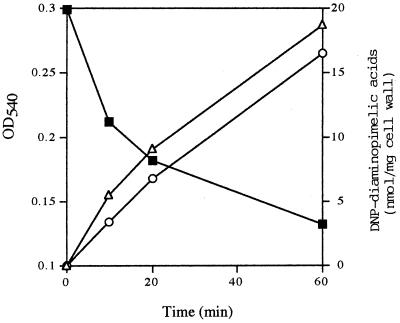
The purified peptidoglycan from B. subtilis cell wall was digested with H-LytF, but an increase in free reducing groups derived from peptidoglycan was not observed, thus indicating that the enzyme is neither an endo-N-acetylglucosaminidase nor an endo-N-acetylmuramidase. Moreover, the enzyme was not an N-acetylmuramoyl-l-alanine amidase (29), as determined by the method of Ghuysen et al. (10, 20). Since LytF exhibits high amino acid sequence similarity with B. sphaericus d-γ-glutamyl-meso-diaminopimelic acid endopeptidase II, free amino groups of the released compounds (supernatant fraction) derived from peptidoglycan after enzyme digestion were labeled with 1-fluoro-2,4-dinitrobenzene, followed by hydrolysis with 4 M HCl. The DNP-labeled and hydrolyzed compounds were separated by HPLC as described in Materials and Methods. After a 60-min digestion, the cell wall density was reduced by 57% and the amounts of mono-DNP-diaminopimelic acid and bis-DNP-diaminopimelic acid increased (Fig. 4). If the enzyme is assumed to be an endopeptidase which digests the d-alanine-meso-diaminopimelic acid cross-linkage, then only mono-DNP-diaminopimelic acid should be detected. However, both mono- and bis-DNP-diaminopimelic acid were formed, thus suggesting that the enzyme is a d-γ-glutamyl-meso-diaminopimelic acid endopeptidase.
FIG. 4.
Time course of the hydrolyzed compounds released from peptidoglycan with the H-LytF enzyme. The time course of the absorbance of peptidoglycan on H-LytF digestion is also shown. After 0-, 10-, 20-, and 60-min digestions of peptidoglycan, the reaction mixtures were centrifuged, and the amino groups of the released peptides and/or mucopeptides in the supernatants were labeled with 1-fluoro-2,4-dinitrobenzene. The labeled compounds were hydrolyzed with a high concentration of HCl, followed by separation by reverse-phase HPLC. Only the amounts of DNP derivatives of diaminopimelic acid were considerably increased during incubation. ■, cell wall turbidity; ▵, mono-dinitrophenyl diaminopimelic acid; ○, bis-dinitrophenyl diaminopimelic acid.
Determination of the size and the 5′ end of lytF RNA.
Northern blot analysis of RNAs from the wild type also showed that one transcription band hybridized to a probe containing the internal region of the lytF gene (Fig. 1). This transcript, estimated to be approximately 1.5 kb, was detected at t−2 to t0, but not after t1.5. Since yhdD comprises 1,464 bp, it was expressed as a monocistronic operon, and this result was supported by the existence of two deduced rho-independent terminators (ΔG = −15.3 and −10.8 kcal/mol) just upstream and downstream of yhdD (lytF, cwlE) (17).
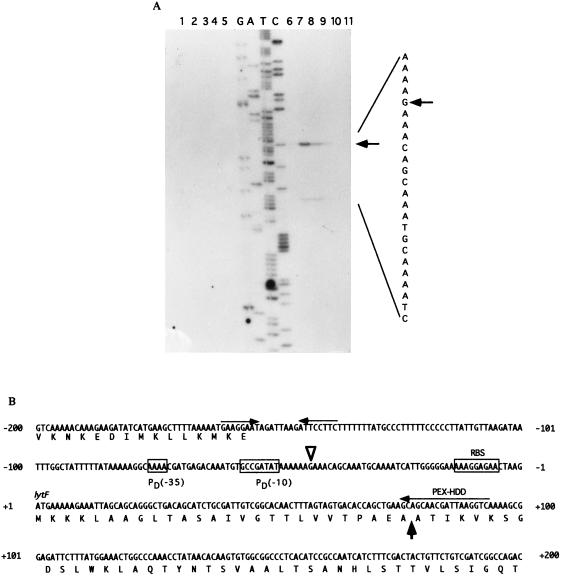
Primer extension analysis was performed with an oligonucleotide primer (PEX-HDD) that is complementary to the 5′ region of lytF (Fig. 5). A strong transcriptional signal starting at G−44 (the nucleotide being numbered with respect to the translational start point [+1] of lytF) was observed with RNA from the wild-type cells at t−1 (Fig. 5A, lane 7), t−0.5 (lane 8), and t0 (lane 9). A weak signal starting at A−29 was also observed at t−1 (lane 7) and t−0.5 (lane 8), but no signals were detected for the sigD-deficient mutant. When we used a different primer for the primer extension analysis, the weak signal was not found, thus suggesting that it is a misannealing one. From the similarities in the length and the timing of the appearance of the transcript, the strong primer extension product seemed to correspond to the 5′ end of the 1.5-kb RNA. The −35 region (AAAA) and the −10 region (GCCGATAT), with a spacing of 15 bp, were almost identical to those of the ςD consensus sequence (TAAA for the −35 region and GCCGATAT for the −10 region, with a spacing of 15 bp) (Fig. 5B) (11).
FIG. 5.
(A) Determination of the 5′ end of the lytF transcript, by primer extension analysis (10 μg), isolated from the wild-type strain 168SD1 (sigD) at t−2 (lane 1), t−1 (lane 2), t−0.5 (lane 3), t0 (lane 4), or t1.5 (lane 5) and from 168 (wild type) at t−2 (lane 6), t−1 (lane 7), t−0.5 (lane 8), t0 (lane 9), or t1.5 (lane 10). Arrows indicate the transcriptional start site and the position of the product. The dideoxy DNA-sequencing reaction mixture with the same primer (PEX-HDD) was electrophoresed in parallel (lanes G, A, T, and C). (B) Nucleotide sequence of the putative promoter region of the lytF gene. The nucleotides are numbered with respect to the translational start point (+1) of lytF. PD (−35) and PD (−10) represent the −35 and −10 regions of the ςD-like promoter. The open arrowhead indicates the transcriptional start site. PEX-HDD is the primer used for primer extension analysis. The rho-independent terminator (ΔG = −15.3 kcal/mol) is indicated by opposing arrows. The thick arrow indicates the deduced signal sequence cleavage site. RBS, ribosome binding site.
Cell morphology of the lytF and lytF cwlF disruptants.
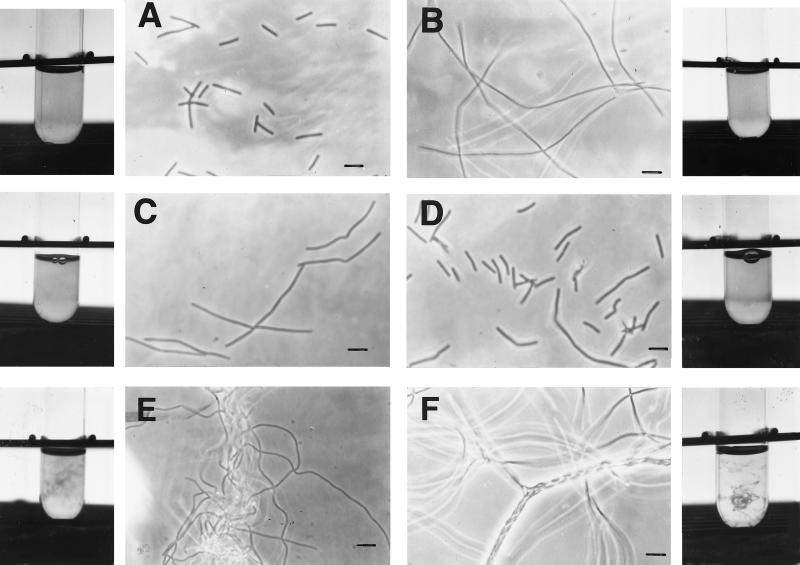
B. subtilis mutant cells which have deficiencies in the major autolysin gene (cwlB) and/or the glucosaminidase gene (cwlG) are rod shaped, while the sigD mutant forms filamentous cells, especially during exponential growth (20, 30, 32). Both autolysin genes are mainly transcribed by EςD RNA polymerase (22, 24, 26, 31). These results suggest that an unknown gene regulated by SigD is important for cell morphology. Moreover, we reported that cwlF mutant cells were only twice as long as wild-type ones but that cwlF sigD mutant cells showed extraordinarily dense microfiber formation and looked like cotton waste in a transparent culture (13). Since lytF is regulated by SigD, we compared the cell morphology among six strains, including the lytF, lytF cwlF, sigD, and cwlF sigD mutants. The lytF mutant cells were approximately 4.6 and 3.4 times longer than the wild-type and cwlF mutant cells, respectively (41.1 ± 25.7 μm for ED, 8.9 ± 3.8 μm for 168, and 12.1 ± 5.8 μm for FTD) (Fig. 6). The lytF cwlF mutant (FED) cells showed extraordinarily dense fiber formation and looked like cotton waste, like the cwlF sigD (FTDSDC) cells (Fig. 6). These results indicate that the effect of the sigD deficiency depends mainly on the effect of the lytF deficiency. However, the morphological difference between ED and 168SDC was still present, because the filamentation of the ED strain was not as great as that of 168SDC (Fig. 6). Therefore, other autolysins regulated by SigD may still have minor effects on cell separation. Although LytF (CwlE) and CwlF mainly play roles in cell separation, LytF and CwlF in combination with other cell wall hydrolases are still important for cell separation in B. subtilis. Cell wall hydrolases AcmA, p60, and Atl are involved in the cell separation of L. lactis, L. monocytogenes, and Staphylococcus aureus, respectively (5, 43, 44). Atl is a bifunctional protein which has an amidase domain and a glucosaminidase domain, and it undergoes proteolytic processing into two extracellular cell wall hydrolases (amidase and glucosaminidase). These enzymes synergistically act on cell separation (40). Blackman and colleagues and Smith and colleagues reported that the cwlB (lytC) sigD double mutant and the cwlB cwlG (lytD) sigD triple mutant formed typical long chains and that the cwlB cwlG mutant also formed long chains (3, 39). But our results, obtained under the conditions used in this study, indicated that the cwlB cwlG mutant does not form long chains (Fig. 6). The difference is probably due to the culture conditions, because those researchers used very gentle shaking (35 or 45 rpm) for the culture (3, 39).
FIG. 6.
Phase-contrast microscopy of B. subtilis 168 (wild type) (A), 168SDC (sigD) (B), ED (lytF) (C), FTD (cwlF) (D), FED (lytF cwlF) (E), and FTDSDC (cwlF sigD) (F) cells, as well as pictures of their corresponding test-tube cultures. The pictures of 168, 168SDC, ED, FTD, FED, and FTDSDC were taken at OD600s of 0.475, 0.465, 0.550, 0.400, 0.462, and 0.457, respectively. B. subtilis FED (lytF cwlF) and FTDSDC (cwlF sigD) cells exhibited a superfilamentous morphology, and their cells looked like cotton waste in the test-tube cultures. Bar, 10 μm.
Genome sequencing of the B. subtilis chromosome indicated seven p60 paralogs, including LytF (CwlE) and CwlF. Therefore, research on the combinations of cell wall peptidases will definitely reveal their cellular functions. Moreover, the use of combinations of other types of cell wall hydrolases (amidase, d,d-endopeptidase, glucosaminidase, muramidase, and lytic transglycosylase) will be very valuable for elucidating their functions.
ACKNOWLEDGMENTS
This research was supported by grant JSPS-RFTF96L00105 from the Japan Society for the Promotion of Science.
We thank Yasuhiro Yamada for valuable help and suggestions on experimental analyses.
REFERENCES
- 1.Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Béliveau C, Potvin C, Trudel J, Asselin A, Bellemare G. Cloning, sequencing, and expression in Escherichia coli of a Streptococcus faecalis autolysin. J Bacteriol. 1991;173:5619–5623. doi: 10.1128/jb.173.18.5619-5623.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackman S A, Smith T J, Foster S J. The role of autolysins during vegetative growth of Bacillus subtilis 168. Microbiology. 1998;144:73–82. doi: 10.1099/00221287-144-1-73. [DOI] [PubMed] [Google Scholar]
- 4.Bubert A, Kuhn M, Goebel W, Köhler S. Structural and functional properties of the p60 proteins from different Listeria species. J Bacteriol. 1992;174:8166–8171. doi: 10.1128/jb.174.24.8166-8171.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buist G, Kok J, Leenhouts K J, Dabrowska M, Venema G, Haandrikman A J. Molecular cloning and nucleotide sequence of the gene encoding the major peptidoglycan hydrolase of Lactococcus lactis, a muramidase needed for cell separation. J Bacteriol. 1995;177:1554–1563. doi: 10.1128/jb.177.6.1554-1563.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canosi U, Morelli G, Trautner T A. The relationship between molecular structure and transformation efficiency of some Streptococcus aureus plasmids isolated from Bacillus subtilis. Mol Gen Genet. 1978;166:259–267. doi: 10.1007/BF00267617. [DOI] [PubMed] [Google Scholar]
- 7.Chu C-P, Kariyama R, Daneo-Moore L, Shockman G D. Cloning and sequence analysis of the muramidase-2 gene from Enterococcus hirae. J Bacteriol. 1992;174:1619–1625. doi: 10.1128/jb.174.5.1619-1625.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fleischmann R D, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 9.Foster S J. Analysis of the autolysins of Bacillus subtilis 168 during vegetative growth and differentiation by using renaturing polyacrylamide gel electrophoresis. J Bacteriol. 1992;174:464–470. doi: 10.1128/jb.174.2.464-470.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghuysen J-M, Tipper D J, Strominger J L. Enzymes that degrade bacterial cell walls. Methods Enzymol. 1966;8:685–699. [Google Scholar]
- 11.Haldenwang W G. The sigma factors of Bacillus subtilis. Microbiol Rev. 1995;59:1–30. doi: 10.1128/mr.59.1.1-30.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hourdou M-L, Duez C, Joris B, Vacheron M-J, Guinand M, Michel G, Ghuysen J-M. Cloning and nucleotide sequence of the gene encoding the γ-d-glutamyl-l-diamino acid endopeptidase II of Bacillus sphaericus. FEMS Microbiol Lett. 1992;91:165–170. doi: 10.1111/j.1574-6968.1992.tb05203.x. [DOI] [PubMed] [Google Scholar]
- 13.Ishikawa S, Hara Y, Ohnishi R, Sekiguchi J. Regulation of a new cell wall hydrolase gene, cwlF, which affects cell separation in Bacillus subtilis. J Bacteriol. 1998;180:2549–2555. doi: 10.1128/jb.180.9.2549-2555.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishikawa S, Yamane K, Sekiguchi J. Regulation and characterization of a newly deduced cell wall hydrolase gene (cwlJ) which affects germination of Bacillus subtilis spores. J Bacteriol. 1998;180:1375–1380. doi: 10.1128/jb.180.6.1375-1380.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kadner, R. J. 1 November 1991, posting date. Sequences. [Online.] http://www.tokyo-center.genome.ad.jp. [5 April 1999, last date accessed.]
- 16.Köhler S, Leimeister-Wächter M, Chakraborty T, Lottspeich F, Goebel W. The gene coding for protein p60 of Listeria monocytogenes and its use as a specific probe for Listeria monocytogenes. Infect Immun. 1990;58:1943–1950. doi: 10.1128/iai.58.6.1943-1950.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunst F, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 18.Kuroda A, Asami Y, Sekiguchi J. Molecular cloning of a sporulation-specific cell wall hydrolase gene of Bacillus subtilis. J Bacteriol. 1993;175:6260–6268. doi: 10.1128/jb.175.19.6260-6268.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuroda A, Imazeki M, Sekiguchi J. Purification and characterization of a cell wall hydrolase encoded by the cwlA gene of Bacillus subtilis. FEMS Microbiol Lett. 1991;81:9–14. doi: 10.1016/0378-1097(91)90462-j. [DOI] [PubMed] [Google Scholar]
- 20.Kuroda A, Sekiguchi J. Molecular cloning and sequencing of a major Bacillus subtilis autolysin gene. J Bacteriol. 1991;173:7304–7312. doi: 10.1128/jb.173.22.7304-7312.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuroda A, Sekiguchi J. Characterization of the Bacillus subtilis CwbA protein which stimulates cell wall lytic amidases. FEMS Microbiol Lett. 1992;95:109–114. doi: 10.1016/0378-1097(92)90745-a. [DOI] [PubMed] [Google Scholar]
- 22.Kuroda A, Sekiguchi J. High-level transcription of the major Bacillus subtilis autolysin operon depends on expression of the sigma D gene and is affected by a sin (flaD) mutation. J Bacteriol. 1993;175:795–801. doi: 10.1128/jb.175.3.795-801.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Lazarevic V, Margot P, Soldo B, Karamata D. Sequencing and analysis of the Bacillus subtilis lytRABC divergon: a regulatory unit encompassing the structural genes of the N-acetylmuramoyl-l-alanine amidase and its modifier. J Gen Microbiol. 1992;138:1949–1961. doi: 10.1099/00221287-138-9-1949. [DOI] [PubMed] [Google Scholar]
- 25.Leclerc D, Asselin A. Detection of bacterial cell wall hydrolases after denaturating polyacrylamide gel electrophoresis. Can J Microbiol. 1989;35:749–753. doi: 10.1139/m89-125. [DOI] [PubMed] [Google Scholar]
- 26.Margot P, Mauël C, Karamata D. The gene of the N-acetylglucosaminidase, a Bacillus subtilis 168 cell wall hydrolase not involved in vegetative cell autolysis. Mol Microbiol. 1994;12:535–545. doi: 10.1111/j.1365-2958.1994.tb01040.x. [DOI] [PubMed] [Google Scholar]
- 27.Margot P, Pagni M, Karamata D. Bacillus subtilis 168 gene lytF encodes a γ-d-glutamate-meso-diaminopimelate muropeptidase expressed by the alternative vegetative sigma factor, ςD. Microbiology. 1999;145:57–65. doi: 10.1099/13500872-145-1-57. [DOI] [PubMed] [Google Scholar]
- 28.Margot P, Wahlen M, Gholamhuseinian A, Piggot P, Karamata D. The lytE gene of Bacillus subtilis 168 encodes a cell wall hydrolase. J Bacteriol. 1998;180:749–752. doi: 10.1128/jb.180.3.749-752.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohnishi, R., S. Ishikawa, and J. Sekiguchi. Unpublished results.
- 30.Rashid M H, Kuroda A, Sekiguchi J. Bacillus subtilis mutant deficient in the major autolytic amidase and glucosaminidase is impaired in motility. FEMS Microbiol Lett. 1993;112:135–140. doi: 10.1111/j.1574-6968.1993.tb06438.x. [DOI] [PubMed] [Google Scholar]
- 31.Rashid M H, Mori M, Sekiguchi J. Glucosaminidase of Bacillus subtilis: cloning, regulation, primary structure and biochemical characterization. Microbiology. 1995;141:2391–2404. doi: 10.1099/13500872-141-10-2391. [DOI] [PubMed] [Google Scholar]
- 32.Rashid M H, Sato N, Sekiguchi J. Analysis of the minor autolysins of Bacillus subtilis during vegetative growth by zymography. FEMS Microbiol Lett. 1995;132:131–137. [Google Scholar]
- 33.Rashid M H, Sekiguchi J. flaD (sinR) mutations affect SigD-dependent functions at multiple points in Bacillus subtilis. J Bacteriol. 1996;178:6640–6643. doi: 10.1128/jb.178.22.6640-6643.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rogers H J, Perkins H R, Ward J B. Microbial cell walls and membranes. London, England: Chapman and Hall; 1980. [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 36.Schaeffer P, Millet J, Aubert J P. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci USA. 1965;54:704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sekiguchi, J., S. Ishikawa, and R. Ohnishi. Unpublished data.
- 38.Shockman G D, Höltje J-V. Microbial peptidoglycan (murein) hydrolases. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier; 1994. pp. 131–166. [Google Scholar]
- 39.Smith T J, Blackman S A, Foster S J. Peptidoglycan hydrolases of Bacillus subtilis 168. Microb Drug Resist. 1996;2:113–118. doi: 10.1089/mdr.1996.2.113. [DOI] [PubMed] [Google Scholar]
- 40.Sugai M, Komatsuzawa H, Akiyama T, Hong Y-M, Oshida T, Miyake Y, Yamaguchi T, Suginaka H. Identification of endo-β-N-acetylglucosaminidase and N-acetylmuramyl-l-alanine amidase as cluster-dispersing enzymes in Staphylococcus aureus. J Bacteriol. 1995;177:1491–1496. doi: 10.1128/jb.177.6.1491-1496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson J S, Shockman G D. A modification of the Park and Johnson reducing sugar determination suitable for the assaying of insoluble materials: its application to bacterial cell walls. Anal Biochem. 1968;22:260–268. doi: 10.1016/0003-2697(68)90315-1. [DOI] [PubMed] [Google Scholar]
- 42.Vagner V, Dervyn E, Ehrlich S D. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology. 1998;144:3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 43.Wuenscher M D, Köhler S, Bubert A, Gerike U, Goebel W. The iap gene of Listeria monocytogenes is essential for cell viability, and its gene product, p60, has bacteriolytic activity. J Bacteriol. 1993;175:3491–3501. doi: 10.1128/jb.175.11.3491-3501.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamada S, Sugai M, Komatsuzawa H, Nakashima S, Oshida T, Matsumoto A, Suginaka H. An autolysin ring associated with cell separation of Staphylococcus aureus. J Bacteriol. 1996;178:1565–1571. doi: 10.1128/jb.178.6.1565-1571.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]