Abstract
Metabolomics studies in the context of ophthalmology have largely focused on identifying metabolite concentrations that characterize specific retinal diseases. Studies involving mass spectrometry (MS) and nuclear magnetic resonance (NMR) spectroscopy have shown that individuals suffering from retinal diseases exhibit metabolic profiles that markedly differ from those of control individuals, supporting the notion that metabolites may serve as easily identifiable biomarkers for specific conditions. An emerging branch of metabolomics resulting from biomarker studies, however, involves the study of retinal metabolic dysfunction as causes of degeneration. Recent publications have identified a number of metabolic processes—including but not limited to glucose and oxygen metabolism—that, when perturbed, play a role in the degeneration of photoreceptor cells. As a result, such studies have led to further research elucidating methods for prolonging photoreceptor survival in an effort to halt degeneration in its early stages. This review will explore the ways in which metabolomics has deepened our understanding of the causes of retinal degeneration and discuss how metabolomics can be used to prevent retinal degeneration from progressing to its later disease stages.
Keywords: Metabolomics, Retinal degeneration, Biomarkers, Photoreceptors, Age-related macular degeneration, Retinitis pigmentosa
Introduction
Over the past few decades, metabolomics has proven itself to be a highly versatile tool in medicine and translational research. The aim of metabolomics is to identify, quantify, trace, and interpret real-time metabolite concentrations in a given biological tissue sample [1]. Though the main goal of metabolomics is rather simple, the outcomes of such large-scale analyses offer researchers the information needed to optimize diagnosis, identify therapeutic targets, monitor disease stage-specific biomarkers, and develop biochemical solutions to cure, slow, or prevent the progression of complex diseases [2, 3]. Because metabolites are the products and intermediates of metabolism, any metabolite concentration that is abnormally high or low in relation to others reveals clues to dysfunctional or perturbed metabolic pathways that may underlie the disease in question. Knowledge of such pathways can then enable researchers to develop a metabolic approach towards rectifying these ailments. With the simple knowledge of metabolite identities and quantities residing within healthy versus diseased tissue, researchers can account for the diagnosis, underlying causes, and potential treatments of various diseases.
Ophthalmology is just one of many fields in which metabolomics has expanded the scope of possible treatment options for diseases. Using high-throughput screening technologies such as mass spectroscopy (MS) and nuclear magnetic resonance (NMR) spectrometry, researchers have identified key biomarkers and metabolic pathways that underpin the pathogenesis of major retinal degenerative diseases. One such disease is age-related macular degeneration (AMD), which is currently the leading cause of adult blindness in the developed world [4]. Anticipated to affect over 196 million people by 2020 [5], AMD is an incurable neurodegenerative disorder that mainly leads to loss of central vision and a significant loss of independence in activities of daily living over time. Given that AMD may result in a number of distinct molecular phenotypes in patients, investigators have conducted a myriad of biomarker studies to identify potential metabolic pathways that may be targeted for future therapeutic studies.
Retinitis pigmentosa (RP) is another category of major retinal diseases for which a growing number of metabolomics studies exists. Characterized by initial symptoms of night blindness followed by tunnel vision and photophobia, RP refers to a group of genetically heterogeneous diseases caused by the progressive loss of rod and cone photoreceptors. An estimated 1.5 million people worldwide bear the burdens of RP, preventing them from leading independent lives [6]. Studies that aim to develop non-gene-specific treatment options using metabolomics have especially been prevalent in the realm of RP research. Unlike gene therapy approaches, which are often gene-specific and only beneficial to those who exhibit the “correct” genetic mutation, metabolic approaches to ocular diseases may be applied to patients regardless of their mutation, as long as the underlying disease-causing pathways involved are identical. Though the process of reprogramming metabolism may necessitate gene-specific procedures, the resultant changes in cellular metabolism have the potential to repair the effects of any of the 60 genes known to be involved in RP pathogenesis. Consequently, this allows for a larger number of people to be treated by a single treatment intervention.
In this review, we will briefly discuss the utility and consequences of the most commonly employed technologies in metabolomics research. This will be followed by an overview of metabolomics studies that have identified key pathways involved in AMD and RP pathogenesis. Lastly, we will explore the ways in which researchers have reprogrammed the metabolome of photoreceptors in an effort to rescue photoreceptors from cell death.
Tools for identifying biomarkers in metabolism
One of the most common metabolomics studies conducted by researchers today is the discovery of metabolite biomarkers that correlate with specific disease phenotypes. Advancements in high-throughput screening technologies have enabled researchers to capture the metabolic profiles or “fingerprints” of individuals suffering from a particular disease. In ophthalmology, retinal degenerative diseases have been linked to a number of metabolic pathways essential for energy metabolism and cell survival [7–9]. To study the metabolic basis of these diseases, however, researchers must first be able to identify and quantify metabolites in tissues.
Mass spectrometry (MS)
Mass spectrometry (MS) is a frequently employed tool in the identification and quantification of metabolic biomarkers. Used to measure molecular masses by ionizing chemical samples and measuring its mass-to-charge ratio, MS is a highly selective, sensitive, and powerful technique that aids in the identification of metabolites [10]. Indeed, the emergence of metabolomics as a field of study was in large part due to the high analytical power of MS, which is capable of analyzing several hundred metabolites in a given sample simultaneously.
In general, MS strategies can be divided into two categories: direct MS, which involves the analysis of metabolites without chromatographic separations, and MS coupled to a chromatographic technique, which involves the separation of metabolites in addition to their analysis and quantification [11]. Although direct MS presents with a number of inherent challenges, such as its inability to differentiate between isomers and its reduced accuracy in metabolite quantitation due to ion suppression effects [11, 12], it is nevertheless highly practical for studies that require analysis of thousands of samples.
Nuclear magnetic resonance spectroscopy
Compared to MS, nuclear magnetic resonance (NMR) spectroscopy is the preferred choice of method for obtaining detailed structural information of metabolites. NMR relies on the magnetic resonance properties of atoms within molecules to determine their physical and chemical properties. The main advantages of NMR include data reproducibility and reliability in its structural analysis of compounds—metabolites that are difficult to identify and distinguish by MS, such as those with identical masses, can easily be resolved with NMR spectra [13]. However, an increasing number of metabolomics studies have gradually transitioned to MS analysis in part due to the lower sensitivity of NMR—an average of 60 metabolites are analyzed in NMR studies compared to several hundreds in studies using MS [14].
Stable isotope tracers
In metabolomics studies of the retina, both MS and NMR have been used to capture the metabolic profiles of diseased retinas. Of equal importance, however, is the ability to trace the products and intermediates of metabolic pathways that are altered by disease. Stable isotope tracers such as 13C, 2H, 15N, and 18O can be used to map key pathways in the retina such as the tricarboxylic acid (TCA) cycle, the pentose phosphate pathway (PPP), and glycolysis, among others [15]. Isotope labeling has been applied to studies of metabolic interactions between retinal glia and neurons [16], nutrient consumption in the retinal pigment epithelium (RPE) [17], and phototransduction and metabolic flux in mice retina [18]. Given the strong link between aberrant retinal metabolism and retinal degeneration [7–9], there is great potential for the application of stable isotope tracers to studies of retinal degeneration as well.
Metabolomics elucidates key pathways involved in retinal degenerative diseases
Biomarkers for age-related macular degeneration (AMD)
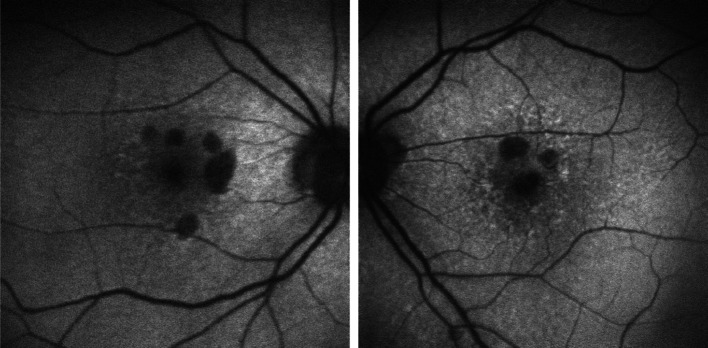
Studies that aim to establish disease biomarkers are of great clinical interest, as their results may help identify new therapeutic targets and diagnose specific conditions in clinical settings. Understandably, much of biomarker research in ophthalmology has been dedicated to AMD (Fig. 1): recent genome-wide association studies (GWAS) have pinpointed four distinct single-nucleotide polymorphisms (SNPs) present in the AMD-associated CFH, ARMS2, and HTRA1 genes [19], each of which may result in distinct metabolic fingerprints and thus requires different therapeutic treatments. The identification of AMD biomarkers is, therefore, a crucial stepping stone to future studies that pinpoint the most appropriate treatment option according to the patient’s specific disease phenotype.
Fig. 1.
Short-wave fundus autofluorescence images of right and left eye retina from consented patients seen at our clinic (IRB-AAAB6560) with age-related macular degeneration. Round hypoautofluorescent areas represent RPE death
A comprehensive overview of potential AMD biomarkers was done by Hoyng et al. in their 2017 review, which focused on systemic and ocular fluid compounds [20]. Unsurprisingly, the compounds with significantly differing levels between control and AMD subjects are those involved in key metabolic pathways known to underpin AMD. For example, several studies detected increased levels of oxidation products such as malondialdehyde (MDA) [21], a by-product of polyunsaturated fatty acid (PUFA) oxidation, and nitric oxide (NO) in patients with wet and dry forms of AMD [22]. Given that oxidative stress is one of the known causes underlying AMD [23], we would expect to observe abnormally high concentrations of oxidation products in retinal tissue. Other AMD biomarkers include vascular endothelial growth factor (VEGF) [24], which is involved in neovascularization, and low-density lipoprotein (LDL) [25], which suggests potential abnormalities in retinal lipid metabolism.
Metabolic processes involved in AMD and RP pathogenesis
An advantage of metabolomics in the context of therapeutic treatments lies in the fact that all tissues rely on the proper functioning of metabolic processes. There are as many as 140 genes that, when mutated, can cause retinal degeneration [26], but research suggests that only a handful of metabolic pathways underlie most of the effects of these mutated genes [27, 28]. For this reason, understanding the altered metabolome pathways responsible for retinal degeneration can serve as a gateway to non-gene-specific treatments that account for a large diversity of inherited retinal disorders.
Aerobic glycolysis
One of several metabolic pathways in the retina that are of interest to researchers is aerobic glycolysis, also known as the Warburg effect. Though commonly known to take place in rapidly dividing cancer cells [29], aerobic glycolysis was also found to be crucial for non-dividing cells like light-adapted photoreceptors in the retina, whose outer segments undergo high rates of anabolism due to daily photoreceptor turnover [30].
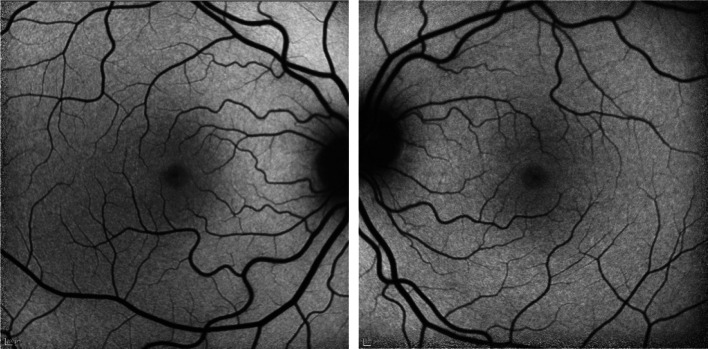
A recent study by Kanow et al. has highlighted the importance of maintaining a gradient of aerobic glycolytic activity between the retina and the RPE [31]. In a normal retina (Fig. 2), aerobic glycolysis is rapid and relatively robust in the outer retina, whereas the opposite can be said of the RPE. This seemingly paradoxical phenomenon can be explained by what the authors describe as a “metabolic ecosystem,” in which glucose is transported from the RPE to the outer retina for glycolysis, and the lactate produced by glycolysis in the outer retina is shuttled into the RPE, suppressing glycolytic activity in the RPE. When glycolytic activity is abnormally high in the RPE, less glucose is available for consumption in the retina, also leading to photoreceptor death [28].
Fig. 2.
Short-wave fundus autofluorescence images of normal retina in the right and left eye, respectively, from consented patients seen at our clinic (IRB-AAAB6560)
This ecosystem model is one example of a metabolic process that could underlie major retinal diseases that correlate with aging. As aging progresses in humans, glycolysis tends to decline in the outer retina and increase in the RPE, which may lead to a disruption in the retinal metabolic ecosystem and hence contribute to age-related vision loss [31]. Kanow et al. also suggest that this model could be used to explain the connection between mitochondrial DNA damage in RPE cells and AMD [32]—as the RPE becomes more dependent on glycolysis over time, less glucose remains for photoreceptor consumption in the retina, leading to AMD, and the RPE mitochondria become dysfunctional, leading to mitochondrial DNA damage.
Hypoxia-induced metabolic stress
Besides aging, another factor that could contribute to dysfunctional glucose metabolism is abnormal retinal oxygen levels [33]. Kurihara et al. demonstrated that a number of metabolic processes, including glucose and lipid metabolism, can be affected by hypoxic conditions in the RPE [34]. Hyperactivation of hypoxia-inducible factor alpha subunits (HIF-αs) via the alteration of VHL/HIF/VEGF pathways led to not only photoreceptor degeneration and structural changes in the RPE but also altered lipid oxidation and a reduction in oxidative phosphorylation in the RPE. These findings relate in particular to AMD, which is caused by low oxygen levels in the retina due to the thickening of Bruch’s membrane [35].
Oxidative stress
Cellular mechanisms that defend against oxidative stress have been of high interest in retinal research given the key role oxidative stress plays in the progression of retinal diseases [23]. Caused by a deficient antioxidant defense system against high levels of reactive oxygen species (ROS), oxidative stress accelerates the development of multiple diseases such as AMD, glaucoma, and diabetic retinopathy. The retina is especially vulnerable to oxidative stress due to its naturally high oxygen tension and high levels of polyunsaturated fatty acids, which increases retinal sensitivity to oxidative damage [36, 37].
A key metabolic pathway that may protect against oxidative stress is reductive carboxylation. In brief, reductive carboxylation is a process that occurs in select cells under hypoxic conditions; instead of citrate production via the TCA cycle, citrate is generated via the reductive carboxylation of α-ketoglutarate (αKG) to isocitrate by NADPH-dependent isocitrate dehydrogenases (IDH) [38]. Using 13C as an isotope tracer in human RPE cells, Du et al. discovered that the RPE possesses a high capacity for reductive carboxylation and, consequently, is protected against the loss of redox homeostasis [39]. They purported that the RPE may be further protected from oxidative stress by enhancing reductive carboxylation via an excess supplementation of αKG, a substrate of IDH2, and the restoration of NAD+, an important substrate of glycolysis shown to override the effects of ROS on reductive carboxylation. These findings reveal potential inroads into therapeutics that may rescue RPE cell death and hence prevent the progression of retinal diseases induced by oxidative stress.
Reprogramming the metabolome to halt retinal degeneration
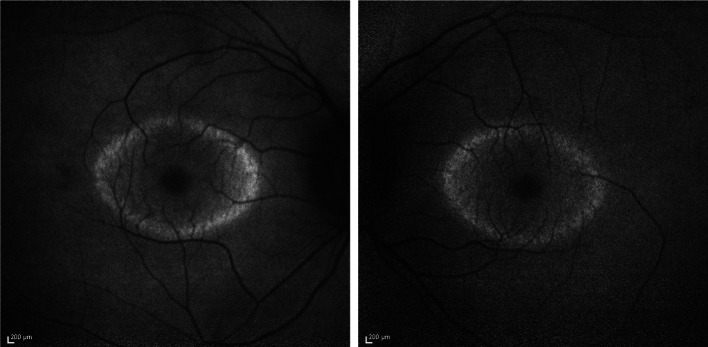
Recently, gene-editing tools such as clustered regularly interspaced short palindromic repeats (CRISPR) have gained enormous popularity in their potential to precisely repair genetic mutations and alter the course of disease in animal models and humans. While gene therapy confers endless clinical applications for many genetically inherited diseases, it is compromised by its impracticality when applied to highly diverse genetic disorders such as RP (Fig. 3), for which there are mutations in more than 60 known genes. For this reason, researchers have turned to metabolomics to not only uncover the underlying mechanisms behind retinal degeneration but also exploit metabolic processes to reprogram cells, delay cell death, and halt the progression of disease (Table 1). Comparatively less progress has been made towards developing therapeutics for AMD via metabolomics.
Fig. 3.
Short-wave fundus autofluorescence images of consented patients from our clinic (IRB-AAAB6560) with retinitis pigmentosa caused by mutations in the gene encoding PDE6B [48]. A hyperautofluorescent ring is usually observed in patients with retinitis pigmentosa, while the retinal area enclosed has preserved visual function. Peripheral to the high density ring, no visual function can be detected. Hence, this patient experiences “tunnel vision”
Table 1.
Reprogramming the metabolome in preclinical RP models to rescue retinal degeneration
| Targeted metabolic pathway | Treatment | Purpose | Result | References |
|---|---|---|---|---|
| Insulin/mTOR | Intraperitoneal injection of insulin in four RP mouse models | Prevent cone starvation | Prolonged cone survival | [27] |
| Insulin/mTOR | Ablation of mTOR inhibitor Tsc1in Pde6bH620Q/H620Q mice | Upregulation of mTOR to enhance rod anabolism | Prolonged rod and cone survival | [41] |
| Insulin/mTOR | Conditional deletion of Pten in rd1 mice | Activation of mTORC1 to improve glucose metabolism and increase NADPH levels in cones | Prolonged cone survival | [45] |
| Glycolysis | Ablation of glycolytic enzyme repressor Sirt6 in Pde6bH620Q/H620Q mice | Increase rod glycolytic activity | Rescue of retinal layers, cellular OS, and improved photoreceptor function | [28] |
| MEF2D-nuclear factor (erythroid-derived 2)-like 2 (NRF2) pathway | Treatment with carnosic acid in Mef2d+/− mice | Upregulation of endogenous NRF2 pathway in photoreceptors | Rescue of photoreceptors from light-induced oxidative stress | [46] |
| Reactive oxygen metabolism | Overexpression of SOD1 in β-actin promoter/sod1 transgenic mice | Protection against hyperoxia-induced oxidative retinal damage | Reduced oxidative stress levels in retina | [47] |
Typically, RP is a rod-cone dystrophy, in which rod degeneration occurs first prior to cone degeneration [6]. However, it remains unclear why cone degeneration almost always follows rod degeneration, especially given that diseases caused by mutations in cone-specific genes do not always result in rod cell death. To answer this question, Punzo et al. measured the gene expression changes of four RP mouse models with mutations in rod-specific genes and identified the cellular processes to which the changes in expression pattern corresponded [27]. Based on microarray analysis data obtained from the onset phase of cone cell death, almost 35% of the genes annotated were found to play roles in cellular metabolism, in particular the insulin/mechanistic target of rapamycin (mTOR) signaling pathway. A key regulator of metabolism, the insulin/mTOR pathway promotes anabolic processes such as protein synthesis and ribosome biogenesis under conditions of high cellular energy [40]. In addition, the authors observed that cone cell death was characterized by cone autophagy, which occurs during cellular starvation. These findings suggested that, in RP pathogenesis, nutrient deficiency may be a key factor fundamental to cone degeneration. To confirm the effect of mTOR-associated starvation on cone degeneration, the authors tested the effect of insulin level on cone survival using one of the mouse models. They discovered that, as hypothesized, daily systemic treatment of mice with insulin led to prolonged cone survival, in contrast to the effects of endogenous depletion of insulin, which led to cell death.
Building upon Punzo et al.’s results on the importance of the insulin/mTOR pathway in cone cell survival, Zhang et al. revealed another way to slow photoreceptor degeneration using Cre-lox to ablate the mTOR-negative regulator tuberous sclerosis complex 1 (Tsc1) in the rods of a Pde6bH620Q/H620Q preclinical RP mouse model [41, 42]. Among the genes involved in RP pathogenesis, mutations in the phosphodiesterase 6 (PDE6) gene disturb the balance between anabolic and catabolic processes in photoreceptors, resulting in the progressive shortening of photoreceptor outer segments (OS) and, ultimately, cell death [6, 34, 43]. To rectify this imbalance, the authors aimed to enhance rod anabolism in Pde6bH620Q/H620Q mice by upregulating mTOR via Tsc1 knockout. Interestingly, Tsc1 knockout led to not only phenotypic and functional rescue of rods in the early stages of RP but also that of cones; rods were preserved for up to 12 weeks, while cones were preserved for up to 20 weeks. In line with Punzo et al.’s hypothesis on cone starvation and degeneration, Zhang et al.’s data supported the notion that cone survival may be dependent upon rod-derived nutritional factors.
In a different study, Zhang et al. additionally focused on manipulating a different metabolic pathway, glycolysis, to achieve the same aforementioned goal of slowing the progression of photoreceptor degeneration throughout the course of RP [28]. Their study generated Sirt6−/−Pde6bH620Q/H620Q mice that lacked sirtuin 6 (Sirt6), a transcriptional repressor of glycolytic enzymes [44], which caused rods to enter a state of perpetual glycolysis. Given that glycolytic intermediates promote the survival of rods [30], the authors hypothesized that knockout of Sirt6 should increase glycolytic activity in rods, thereby protecting rods against OS degeneration and prolonging the life of rod cells. Indeed, ablation of Sirt6 preserved cellular OS, rescued the morphology of retinal layers, and improved photoreceptor function in Sirt6−/−Pde6bH620Q/H620Q mice.
Concluding remarks
Metabolomics studies of the retina have much to offer to the field of ophthalmology, given its potential to improve diagnosis of retinal diseases and reveal avenues for possible therapeutic targets. Recent advancements in MS and NMR technology have allowed the metabolomics field to develop into what it is today: a versatile discipline adaptable to both research and clinical settings. In this review, we explored the common techniques employed in the identification and quantification of metabolites, which allow researchers to establish disease biomarkers. In addition to being utilized as diagnostic tools, biomarkers are also invaluable in that they pinpoint specific signaling pathways and cellular processes that may underpin diseases of interest. As the mechanisms of disease pathogenesis become better understood, researchers can then develop strategies to exploit naturally occurring metabolic pathways and reprogram the metabolome with the goal of reversing retinal degeneration. The appeal of metabolomics lies in its capacity to account for a number of diseases because it is non-gene-specific; though gene therapy treatments, which usually account for single genes, may prove effective in treating disease, only a small fraction of patients would benefit from it for genetically heterogeneous disorders like RP. The metabolomics field should, therefore, be seen as a tool to complement current existing forms of gene therapy that may fall short in disorders with genetic heterogeneity.
Acknowledgements
Supported in part by Grants from the National Eye Institute, NIH; P30EY019007, R01EY018213, R01EY024698, R01EY026682, R21AG050437, National Cancer Institute Core (5P30CA013696), the Research to Prevent Blindness (RPB) Physician-Scientist Award, and unrestricted funds from RPB. S.H.T. is a member of the RD-CURE Consortium and is supported by the Tistou and Charlotte Kerstan Foundation, the Schneeweiss Stem Cell Fund, New York State (C029572), the Foundation Fighting Blindness New York Regional Research Center Grant (C-NY05-0705-0312), the Crowley Family Fund, and the Gebroe Family Foundation.
References
- 1.Young SP, Wallace GR. Metabolomic analysis of human disease and its application to the eye. J Ocular Biol Dis Inform. 2009;2(4):235–242. doi: 10.1007/s12177-009-9038-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gowda GA, Zhang S, Gu H, Asiago V, Shanaiah N, Raftery D. Metabolomics-based methods for early disease diagnostics. Expert Rev Mol Diagn. 2008;8(5):617–633. doi: 10.1586/14737159.8.5.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kosmides AK, Kamisoglu K, Calvano SE, Corbett SA, Androulakis IP. Metabolomic fingerprinting: challenges and opportunities. Crit Rev Biomed Eng. 2013;41(3):205–221. doi: 10.1615/CritRevBiomedEng.2013007736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim LS, Mitchell P, Seddon JM, Holz FG, Wong TY. Age-related macular degeneration. Lancet. 2012;379(9827):1728–1738. doi: 10.1016/S0140-6736(12)60282-7. [DOI] [PubMed] [Google Scholar]
- 5.Wong WL, Su X, Li X, Cheung CM, Klein R, Cheng CY, Wong TY. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Global Health. 2014;2(2):e106–e116. doi: 10.1016/S2214-109X(13)70145-1. [DOI] [PubMed] [Google Scholar]
- 6.Hamel C. Retinitis pigmentosa. Orphanet J Rare Dis. 2006;1:40. doi: 10.1186/1750-1172-1-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hartong DT, Dange M, McGee TL, Berson EL, Dryja TP, Colman RF. Insights from retinitis pigmentosa into the roles of isocitrate dehydrogenases in the Krebs cycle. Nat Genet. 2008;40(10):1230–1234. doi: 10.1038/ng.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maurer CM, Schonthaler HB, Mueller KP, Neuhauss SC. Distinct retinal deficits in a zebrafish pyruvate dehydrogenase-deficient mutant. J Neurosci. 2010;30(36):11962–11972. doi: 10.1523/JNEUROSCI.2848-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aherne A, Kennan A, Kenna PF, McNally N, Lloyd DG, Alberts IL, Kiang AS, Humphries MM, Ayuso C, Engel PC, Gu JJ, Mitchell BS, Farrar GJ, Humphries P. On the molecular pathology of neurodegeneration in IMPDH1-based retinitis pigmentosa. Hum Mol Genet. 2004;13(6):641–650. doi: 10.1093/hmg/ddh061. [DOI] [PubMed] [Google Scholar]
- 10.Dettmer K, Aronov PA, Hammock BD. Mass spectrometry-based metabolomics. Mass Spectrom Rev. 2007;26(1):51–78. doi: 10.1002/mas.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lei Z, Huhman DV, Sumner LW. Mass spectrometry strategies in metabolomics. J Biol Chem. 2011;286(29):25435–25442. doi: 10.1074/jbc.R111.238691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Annesley TM. Ion suppression in mass spectrometry. Clin Chem. 2003;49(7):1041–1044. doi: 10.1373/49.7.1041. [DOI] [PubMed] [Google Scholar]
- 13.Markley JL, Brüschweiler R, Edison AS, Eghbalnia HR, Powers R, Raftery D, Wishart DS. The future of NMR-based metabolomics. Curr Opin Biotechnol. 2017;43:34–40. doi: 10.1016/j.copbio.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scalbert A, Brennan L, Fiehn O, Hankemeier T, Kristal BS, van Ommen B, Pujos-Guillot E, Verheij E, Wishart D, Wopereis S. Mass-spectrometry-based metabolomics: limitations and recommendations for future progress with particular focus on nutrition research. Metabolomics. 2009;5(4):435. doi: 10.1007/s11306-009-0168-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du J, Linton JD, Hurley JB. Probing metabolism in the intact retina using stable isotope tracers. Methods Enzymol. 2015;561:149–170. doi: 10.1016/bs.mie.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindsay KJ, Du J, Sloat SR, Contreras L, Linton JD, Turner SJ, Sadilek M, Satrustegui J, Hurley JB. Pyruvate kinase and aspartate–glutamate carrier distributions reveal key metabolic links between neurons and glia in retina. Proc Natl Acad Sci USA. 2014;111(43):15579–15584. doi: 10.1073/pnas.1412441111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chao JR, Knight K, Engel AL, Jankowski C, Wang Y, Manson MA, Gu H, Djukovic D, Raftery D, Hurley JB, Du J. Human retinal pigment epithelial cells prefer proline as a nutrient and transport metabolic intermediates to the retinal side. J Biol Chem. 2017;292(31):12895–12905. doi: 10.1074/jbc.M117.788422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du J, Rountree A, Cleghorn WM, Contreras L, Lindsay KJ, Sadilek M, Gu H, Djukovic D, Raftery D, Satrustegui J, Kanow M, Chan L, Tsang SH, Sweet IR, Hurley JB. Phototransduction influences metabolic flux and nucleotide metabolism in mouse retina. J Biol Chem. 2016;291(9):4698–4710. doi: 10.1074/jbc.M115.698985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang J, Li Y, Chan L, Tsai YT, Wu WH, Nguyen HV, Hsu CW, Li X, Brown LM, Egli D, Sparrow JR, Tsang SH. Validation of genome-wide association study (GWAS)-identified disease risk alleles with patient-specific stem cell lines. Hum Mol Genet. 2014;23(13):3445–3455. doi: 10.1093/hmg/ddu053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kersten E, Paun CC, Schellevis RL, Hoyng CB, Delcourt C, Lengyel I, Peto T, Ueffing M, Klaver CCW, Dammeier S, den Hollander AI, de Jong EK. Systemic and ocular fluid compounds as potential biomarkers in age-related macular degeneration. Surv Ophthalmol. 2017 doi: 10.1016/j.survophthal.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 21.Ates O, Azizi S, Alp HH, Kiziltunc A, Beydemir S, Cinici E, Kocer I, Baykal O. Decreased serum paraoxonase 1 activity and increased serum homocysteine and malondialdehyde levels in age-related macular degeneration. Tohoku J Exp Med. 2009;217(1):17–22. doi: 10.1620/tjem.217.17. [DOI] [PubMed] [Google Scholar]
- 22.Evereklioglu C, Er H, Doganay S, Cekmen M, Turkoz Y, Otlu B, Ozerol E. Nitric oxide and lipid peroxidation are increased and associated with decreased antioxidant enzyme activities in patients with age-related macular degeneration. Doc Ophthalmol Adv Ophthalmol. 2003;106(2):129–136. doi: 10.1023/A:1022512402811. [DOI] [PubMed] [Google Scholar]
- 23.Masuda T, Shimazawa M, Hara H. Retinal diseases associated with oxidative stress and the effects of a free radical scavenger (Edaravone) Oxidative Med Cell Longev. 2017;2017:14. doi: 10.1155/2017/9208489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tong J-P, Chan W-M, Liu DTL, Lai TYY, Choy K-W, Pang C-P, Lam DSC. Aqueous humor levels of vascular endothelial growth factor and pigment epithelium-derived factor in polypoidal choroidal vasculopathy and choroidal neovascularization. Am J Ophthalmol. 2006;141(3):456–462. doi: 10.1016/j.ajo.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 25.Qin L, Mroczkowska SA, Ekart A, Patel SR, Gibson JM, Gherghel D. Patients with early age-related macular degeneration exhibit signs of macro- and micro-vascular disease and abnormal blood glutathione levels. Graefe’s Arch Clin Exp Ophthalmol. 2014;252(1):23–30. doi: 10.1007/s00417-013-2418-0. [DOI] [PubMed] [Google Scholar]
- 26.Bramall AN, Wright AF, Jacobson SG, McInnes RR. The genomic, biochemical, and cellular responses of the retina in inherited photoreceptor degenerations and prospects for the treatment of these disorders. Annu Rev Neurosci. 2010;33:441–472. doi: 10.1146/annurev-neuro-060909-153227. [DOI] [PubMed] [Google Scholar]
- 27.Punzo C, Kornacker K, Cepko CL. Stimulation of the insulin/mTOR pathway delays cone death in a mouse model of retinitis pigmentosa. Nat Neurosci. 2009;12(1):44–52. doi: 10.1038/nn.2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L, Du J, Justus S, Hsu CW, Bonet-Ponce L, Wu WH, Tsai YT, Wu WP, Jia Y, Duong JK, Mahajan VB, Lin CS, Wang S, Hurley JB, Tsang SH. Reprogramming metabolism by targeting sirtuin 6 attenuates retinal degeneration. J Clin Investig. 2016;126(12):4659–4673. doi: 10.1172/JCI86905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science (New York, NY) 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chinchore Y, Begaj T, Wu D, Drokhlyansky E, Cepko CL. Glycolytic reliance promotes anabolism in photoreceptors. eLife. 2017 doi: 10.7554/eLife.25946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanow MA, Giarmarco MM, Jankowski C, Tsantilas K, Engel AL, Du J, Linton JD, Farnsworth CC, Sloat SR, Lindsay KJ, Parker ED, Brockerhoff SE, Sadilek M, Chao JR, Hurley JB (2017) Biochemical adaptations of the retina and retinal pigment epithelium support a metabolic ecosystem in the vertebrate eye. bioRxiv:10.1101/143347 [DOI] [PMC free article] [PubMed]
- 32.Terluk MR, Kapphahn RJ, Soukup LM, Gong H, Gallardo C, Montezuma SR, Ferrington DA. Investigating mitochondria as a target for treating age-related macular degeneration. J Neurosci. 2015;35(18):7304–7311. doi: 10.1523/JNEUROSCI.0190-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu DY, Cringle SJ. Retinal degeneration and local oxygen metabolism. Exp Eye Res. 2005;80(6):745–751. doi: 10.1016/j.exer.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 34.Kurihara T, Westenskow PD, Gantner ML, Usui Y, Schultz A, Bravo S, Aguilar E, Wittgrove C, Friedlander M, Paris LP, Chew E, Siuzdak G, Friedlander M. Hypoxia-induced metabolic stress in retinal pigment epithelial cells is sufficient to induce photoreceptor degeneration. eLife. 2016 doi: 10.7554/eLife.14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feigl B. Age-related maculopathy—linking aetiology and pathophysiological changes to the ischaemia hypothesis. Prog Retinal Eye Res. 2009;28(1):63–86. doi: 10.1016/j.preteyeres.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 36.Whitehead AJ, Mares JA, Danis RP. Macular pigment: a review of current knowledge. Arch Ophthalmol (Chicago, Ill: 1960) 2006;124(7):1038–1045. doi: 10.1001/archopht.124.7.1038. [DOI] [PubMed] [Google Scholar]
- 37.De La Paz MA, Anderson RE. Lipid peroxidation in rod outer segments. Role of hydroxyl radical and lipid hydroperoxides. Invest Ophthalmol Vis Sci. 1992;33(7):2091–2096. [PubMed] [Google Scholar]
- 38.Wise DR, Ward PS, Shay JES, Cross JR, Gruber JJ, Sachdeva UM, Platt JM, DeMatteo RG, Simon MC, Thompson CB. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci. 2011;108(49):19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du J, Yanagida A, Knight K, Engel AL, Vo AH, Jankowski C, Sadilek M, Tran VTB, Manson MA, Ramakrishnan A, Hurley JB, Chao JR. Reductive carboxylation is a major metabolic pathway in the retinal pigment epithelium. Proc Natl Acad Sci. 2016;113(51):14710–14715. doi: 10.1073/pnas.1604572113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iadevaia V, Huo Y, Zhang Z, Foster LJ, Proud CG. Roles of the mammalian target of rapamycin, mTOR, in controlling ribosome biogenesis and protein synthesis. Biochem Soc Trans. 2012;40(1):168–172. doi: 10.1042/BST20110682. [DOI] [PubMed] [Google Scholar]
- 41.Zhang L, Justus S, Xu Y, Pluchenik T, Hsu C-W, Yang J, Duong JK, Lin C-S, Jia Y, Bassuk AG, Mahajan VB, Tsang SH. Reprogramming towards anabolism impedes degeneration in a preclinical model of retinitis pigmentosa. Hum Mol Genet. 2016;25(19):4244–4255. doi: 10.1093/hmg/ddw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsang SH, Chan L, Tsai YT, Wu WH, Hsu CW, Yang J, Tosi J, Wert KJ, Davis RJ, Mahajan VB. Silencing of tuberin enhances photoreceptor survival and function in a preclinical model of retinitis pigmentosa (an american ophthalmological society thesis) Trans Am Ophthalmol Soc. 2014;112:103–115. [PMC free article] [PubMed] [Google Scholar]
- 43.Punzo C, Xiong W, Cepko CL. Loss of daylight vision in retinal degeneration: are oxidative stress and metabolic dysregulation to blame? J Biol Chem. 2012;287(3):1642–1648. doi: 10.1074/jbc.R111.304428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez-Pastor B, Mostoslavsky R. Sirtuins, metabolism, and cancer. Front Pharmacol. 2012;3:22. doi: 10.3389/fphar.2012.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Venkatesh A, Ma S, Le YZ, Hall MN, Ruegg MA, Punzo C. Activated mTORC1 promotes long-term cone survival in retinitis pigmentosa mice. J Clin Investig. 2015;125(4):1446–1458. doi: 10.1172/JCI79766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagar S, Noveral SM, Trudler D, Lopez KM, McKercher SR, Han X, Yates JR, Piña-Crespo JC, Nakanishi N, Satoh T, S-i Okamoto, Lipton SA. MEF2D haploinsufficiency downregulates the NRF2 pathway and renders photoreceptors susceptible to light-induced oxidative stress. Proc Natl Acad Sci. 2017;114(20):E4048–E4056. doi: 10.1073/pnas.1613067114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dong A, Shen J, Krause M, Akiyama H, Hackett SF, Lai H, Campochiaro PA. Superoxide dismutase 1 protects retinal cells from oxidative damage. J Cell Physiol. 2006;208(3):516–526. doi: 10.1002/jcp.20683. [DOI] [PubMed] [Google Scholar]
- 48.Shen S, Sujirakul T, Tsang SH. Next-generation sequencing revealed a novel mutation in the gene encoding the beta subunit of rod phosphodiesterase. Ophthalmic Genet. 2014;35(3):142–150. doi: 10.3109/13816810.2014.915328. [DOI] [PMC free article] [PubMed] [Google Scholar]