Abstract
A 5.8-kb fragment of the large conjugative plasmid pAW63 from Bacillus thuringiensis subsp. kurstaki HD73 containing all the information for autonomous replication was cloned and sequenced. By deletion analysis, the pAW63 replicon was reduced to a 4.1-kb fragment harboring four open reading frames (ORFs). Rep63A (513 amino acids [aa]), encoded by the largest ORF, displayed strong similarity (40% identity) to the replication proteins from plasmids pAMβ1, pIP501, and pSM19035, indicating that the pAW63 replicon belongs to the pAMβ1 family of gram-positive theta-replicating plasmids. This was confirmed by the facts that no single-stranded DNA replication intermediates could be detected and that replication was found to be dependent on host-gene-encoded DNA polymerase I. An 85-bp region downstream of Rep63A was also shown to have strong similarity to the origins of replication of pAMβ1 and pIP501, and it is suggested that this region contains the bona fide pAW63 ori. The protein encoded by the second large ORF, Rep63B (308 aa), was shown to display similarity to RepB (34% identity over 281 aa) and PrgP (32% identity over 310 aa), involved in copy control of the Enterococcus faecalis plasmids pAD1 and pCF10, respectively. No significant similarity to known proteins or DNA sequences could be detected for the two smallest ORFs. However, the location, size, hydrophilicity, and orientation of ORF6 (107 codons) were analogous to those features of the putative genes repC and prgO, which encode stability functions on plasmids pAD1 and pCF10, respectively. The cloned replicon of plasmid pAW63 was stably maintained in Bacillus subtilis and B. thuringiensis and displayed incompatibility with the native pAW63. Hybridization experiments using the cloned replicon as a probe showed that pAW63 has similarity to large plasmids from other B. thuringiensis subsp. kurstaki strains and to a strain of B. thuringiensis subsp. alesti.
The gram-positive bacterium Bacillus thuringiensis is of great industrial interest because of its production of insect toxins (δ-endotoxins) during sporulation. The toxin genes are often located on large self-transmissible or mobilizable plasmids and are active against a variety of insects (for a recent review, see reference 49). Besides the toxin gene-bearing plasmids, B. thuringiensis strains harbor a complex array of cryptic plasmids with sizes ranging from 2 to more than 600 kb (18, 29).
Several B. thuringiensis plasmids have been analyzed, and their mode of replication has been studied. The smaller plasmids (<15 kb), like most other small gram-positive plasmids, use rolling-circle replication (RCR), in which replication occurs through a single-stranded DNA (ssDNA) intermediate (for a review, see reference 37). Examples of these plasmids are pTX14-1 and pTX14-3 from B. thuringiensis subsp. israelensis (2, 8, 41), pGI2 and pGI3 from B. thuringiensis subsp. thuringiensis H1.1. (32, 42), and pHD2 from B. thuringiensis subsp. kurstaki HD1 (45). So far these plasmids remain cryptic since no function other than their replication machinery or mobilization activity has been associated with them. Plasmid pHT1030 (15 kb) from B. thuringiensis subsp. thuringiensis LM2 (39) and the four large plasmids from B. thuringiensis for which replication mechanisms have been analyzed (p43, p44, and p60 from B. thuringiensis subsp. kurstaki HD263 [4] and pHT73 from strain HD73 [28]) replicate via theta replication. Of these, only plasmid p43 has been shown to belong to the pAMβ1 family of gram-positive plasmids, which require host gene-encoded DNA polymerase I (PolI) (35). The replicon of pHT73 has high similarity to the origin of replication from plasmid p44 (ori44) (28); hence, it can be assumed that this plasmid also belongs to the non-pAMβ1 family of theta-replicating plasmids. Plasmids p44, p60, and pHT73 harbor genes for insect toxin production, while p43 remains cryptic. The capacity for transfer by conjugation has been established for p43 (4) and pHT73 (30), and p44 has been determined to be Tra+ (4), which presumably means that it is conjugative as well, while p60 is Tra− (4).
In this study the replication region of pAW63, a 70-kb broad-host-range conjugative plasmid from B. thuringiensis subsp. kurstaki HD73 (54), was cloned, sequenced, and analyzed. pAW63 is self-transmissible to several Bacillus species and able to mobilize nonconjugative plasmids in liquid medium very efficiently. Like several other B. thuringiensis plasmids, pAW63 also contains sequences with similarity to transposons and insertion sequences, but unlike the coresident plasmid pHT73, pAW63 is apparently devoid of insecticidal toxin genes (54). Based on DNA and protein homology, replication mechanism, and dependence on host gene-encoded PolI, the pAW63 replicon was classified as a new member of the pAMβ1 family of theta-replicating plasmids.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Bacterial strains and plasmids are listed in Tables 1 and 2. All cultures were grown in Luria-Bertani (LB) medium (48) containing antibiotics (Sigma), when appropriate, at the following concentrations: 6 μg of chloramphenicol per ml, 100 μg of ampicillin per ml, 4 μg of tetracycline per ml, 15 μg of nalidixic acid per ml, and 20 μg of rifampin per ml.
TABLE 1.
Strains and plasmidsa
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| Bacillus thuringiensis subsp. kurstaki | ||
| HD73 | 4D4 | BGSC |
| AW05 | HD73 cured of pAW63 | 54 |
| AW06 | HD73 cured of pHT73 | 54 |
| AW43 | HD73 cured of pAW63 and pHT73, Nalr | 54 |
| AW48 | AW43 containing pAW63::Tn5401 | 54 |
| AW120 | AW43 containing pAW105 | This study |
| AW121 | AW48 containing pAW105 | This study |
| B. thuringiensis subsp. israelensis | ||
| GBJ001 | Plasmid-cured derivative of 4Q7 (BGSC), Strr | 36 |
| AW57 | GBJ001 (pAW63::Tn5401) | This study |
| Bacillus subtilis | ||
| AND1014 | DN1885 | P. L. Jørgensen |
| AW61 | AND1014 containing pAW002 | This study |
| AW109 | AND1014 containing pAW105 | This study |
| 1A224 | PolI proficient | BGSC |
| 1A226 | polA5 mutant of 1A224 | BGSC |
| Escherichia coli | DH5α | BRL |
| Plasmids | ||
| pUC19 | E. coli vector, Ampr | 55 |
| pC194 | Natural Staphylococcus aureus plasmid, Camr | 34 |
| pAW001 | pUC19 containing the cat gene from pC194 | This study |
| pAW002 | pAW001 containing a 5.8-kb fragment from pAW63 | This study |
| pAW101 | pAW001 + 5.35-kb KpnI fragment from pAW002 | This study |
| pAW103 | pAW002 with the 2.6-kb HindIII fragment deleted | This study |
| pAW105 | pAW002 with the 1.7-kb HindIII-BglII fragment deleted | This study |
| pHV1610 | pC194::pUC19 | 7 |
| pHV1611 | pC194::pUC19, including sso of pUB110 | 7 |
BGSC, Bacillus Genetic Stock Center, Columbus, Ohio; P. L. Jørgensen, NOVO-Nordisk A/S, Bagsværd, Denmark; BRL, Bethesda Research Laboratories, Gaithersburg, Md.
TABLE 2.
Plasmid homology to the pAW63 replicon
| Strain | Sero-type(s) | Strain | Sourcea | Hybridization (size [kb] of plasmidb) |
|---|---|---|---|---|
| B. cereus | ATCC 10876 | Kolstø | − | |
| B. cereus | ATCC 10987 | Kolstø | − | |
| B. cereus | ATCC 11778 | Kolstø | − | |
| B. cereus | ATCC 14579 | Kolstø | − | |
| B. cereus | F837/76 | Kolstø | − | |
| B. thuringiensis subsp. | ||||
| thuringiensis | 1 | 4A4 | BGSC | − |
| thuringiensis | 1 | AH265 | Kolstø | − |
| finitimus | 2 | 4B2 | BGSC | − |
| alesti | 3a | 4C3 | BGSC | + (70) |
| kurstaki | 3a, 3b | HD1-NB168 | NOVO | + (75) |
| kurstaki | 3a, 3b | 4D1 | BGSC | + (75) |
| kurstaki | 3a, 3b | 4D4 | BGSC | + (70) |
| kurstaki | 3a, 3b | AW005 | 54 | − |
| kurstaki | 3a, 3b | AW006 | 54 | + (70) |
| kurstaki | 3a, 3b | KTo | 54 | − |
| kurstaki | 3a, 3b | 4D12 | BGSC | + (75) |
| kurstaki | 3a, 3b | 4D14 | BGSC | + (75) |
| kurstaki | 3a, 3b | 4D15 | BGSC | + (75) |
| kurstaki | 3a, 3b | 4D16 | BGSC | + (75) |
| kurstaki | 3a, 3b | 4D17 | BGSC | + (75) |
| kurstaki | 3a, 3b | 67R1 | DMU | + (75) |
| sotto | 4a, 4b | 4E4 | BGSC | − |
| canadensis | 4a, 4c | 4H2 | BGSC | − |
| galleriae | 5a, 5b | 4G5 | BGSC | − |
| entomocidus | 6 | 4I4 | BGSC | − |
| aizawai | 7 | 4J4 | BGSC | − |
| morrisoni | 8 | 4K1 | BGSC | − |
| tenebrionis | 8a, 8b | NB74 | NOVO | − |
| tolworthi | 9 | 4L3 | BGSC | − |
| darmstadiensis | 10 | 4M1 | BGSC | − |
| toumanoffi | 11 | 4N1 | BGSC | − |
| kysushensis | 11a, 11c | 4U1 | BGSC | − |
| thompsoni | 12 | 4O1 | BGSC | − |
| pakistani | 13 | 4P1 | BGSC | − |
| israelensis | 14 | 4Q2 | BGSC | − |
| dakota | 15 | 4R1 | BGSC | − |
| indiana | 16 | 4S2 | BGSC | − |
| tochigiensis | 19 | 4Y1 | BGSC | − |
| mexicanensis | 27 | 4AC1 | BGSC | − |
| amagiensis | 29 | 4AE1 | BGSC | − |
| toguchini | 31 | 4AD1 | BGSC | − |
| wuhanensis | Nonmotile | 4T1 | BGSC | − |
| roskildiensis | 45 | 39 | DMU | − |
Abbreviations are the same as in Table 1, footnote a. Additionally, Kolstø is A. B. Kolstø, Oslo, Norway, and DMU is the Danish National Environmental Research Institute.
Plasmid hybridizing to the pAW63 replicon.
DNA manipulations.
Restriction enzymes were purchased from New England Biolabs Inc. (Beverly, Mass.) or GIBCO-BRL, and T4 DNA ligase was purchased from GIBCO-BRL. These enzymes were used as specified by the suppliers. DNA fragments were isolated from agarose gels by using a Qiagen extraction kit. Plasmids from B. thuringiensis were extracted as described by Andrup et al. (1) or Jensen et al. (36). Plasmids from Escherichia coli and Bacillus subtilis were isolated as described by Sambrook et al. (48). Total DNA from B. subtilis was isolated by the method of Boe et al. (7). DNA was analyzed by horizontal gel electrophoresis (6 to 10 V/cm) in 0.5 to 1.0% agarose (SeaKem GTG) with 1× TBE buffer (48) for 1.5 to 2 h. After electrophoresis, the gel was stained in 1 μg of ethidium bromide per ml for 5 to 10 min and destained in water.
DNA was blotted from the agarose gel to Hybond N+ (pore size, 0.45 μm; Amersham International plc, Little Chalfont, Buckinghamshire, United Kingdom) according to the method of Southern (51). Probe labeling with fluorescein, DNA hybridization, and washing steps were performed with the Gene Images random prime labeling module and the Gene Images CDP-Star detection module from Amersham.
For detection of ssDNA, overnight cultures of B. subtilis were grown to mid-log phase and rifampin (20 μg/ml) was added for 1 h to half of the cultures before whole-cell DNA preparation. Two identical gels were run, and before Southern blotting, one of these was treated with HCl that denatures double-stranded DNA (dsDNA) to ssDNA.
DNA sequencing.
DNA was sequenced with a Thermo Sequenase Kit (Amersham) and fluorescein end-labeled oligonucleotide primers (DNA Synthesis Laboratory at the Biotechnology Centre of Oslo) with an A.L.F. automated sequencer (Pharmacia, Uppsala, Sweden).
DNA and translated protein sequences were analyzed with the Wisconsin Package, version 9.1, Genetics Computer Group, Madison, Wis., and the IBI Sequence Analysis Program (International Biotechnologies, Inc.).
Electroporation and transformation.
Electroporation of B. thuringiensis was performed as described previously (54) with a Gene Pulser apparatus (Bio-Rad Laboratories, Richmond, Calif.). Electroporation of E. coli was conducted as described in Current Protocols in Molecular Biology (3).
Competent B. subtilis cells were made in the following way: an overnight culture was diluted 100-fold into 25 ml of KM-1 medium (0.4% glucose, 0.02% Casamino Acids, 0.1% yeast extract, and 30 mM MnCl2 in KM stock [see below]). Sixty to 75 min after the end of exponential growth, the culture was diluted 10-fold into 180 ml of KM-2 medium (0.4% glucose, 0.02% Casamino Acids, 0.1% yeast extract, 0.001% saltmix [see below], 0.5 mM CaCl2, and 0.8 mM MgCl2 in KM stock) and grown for 45 to 60 min. The culture was centrifuged, and the pellet was resuspended in a solution containing 16 ml of the supernatant and 4 ml of glycerol (99 or 100%) and kept at −80°C. KM stock consisted of 10% 10× MM, 0.1% Na-citrate, and 2 mM MgSO4 in H2O; 10× MM is 20 g of (NH4)2SO4, 60 g of KH2PO4, and 140 g of K2HPO4 · 3H2O adjusted to 1,000 ml with H2O. Saltmix consisted of 10 mM CaCl2, 1 mM FeCl3, and 1 mM MnCl2 in H2O.
For transforming B. subtilis, 50 μl of competent cells stored at −80°C were thawed on ice and heated at 42°C and 50 μl of BTF (0.32% glucose, 40 mM MgCl2, 0.2 mM EGTA in 0.1% saltmix in KM stock) was added. Three microliters of DNA solution was mixed with competent cells and incubated at 37°C for 20 min. One volume of LB medium (50 μl) was added, and the cells were incubated for an additional 20 min before being plated on selective medium.
Stability and incompatibility test.
Overnight cultures of B. thuringiensis or B. subtilis grown with antibiotics were diluted 1,000-fold into fresh prewarmed LB medium (7 ml) without antibiotics and grown exponentially for about 40 generations. Appropriate dilutions of the cultures were plated onto LB medium, and after overnight incubation, 100 colonies were transferred with toothpicks onto LB medium to which the appropriate antibiotics had been added. Stability was estimated as the percentage of cells containing the plasmid (Cmr) after about 40 generations.
Nucleotide sequence accession number.
The DNA sequence reported in this paper has been deposited in the EMBL nucleotide sequence database under the accession no. AJ011655.
RESULTS
Cloning of a replicon from plasmid pAW63.
In order to facilitate the cloning of replicative regions of B. thuringiensis native plasmids, we constructed a vector that required the insertion of a gram-positive replicon to be transformable in B. subtilis. The 1,310-bp ClaI-MspI fragment from plasmid pC194 (34) containing the chloramphenicol acetyltransferase gene was cloned into the NarI site of the E. coli cloning vector pUC19 (55) to provide a selectable marker functional in gram-positive bacteria. The resulting plasmid, pAW001, was unable to transform B. subtilis to Cmr.
The replication region of pAW63 was cloned in the following way. Plasmid pAW63 tagged with Tn5401 (54) was transferred by conjugation (1) from B. thuringiensis subsp. kurstaki HD73 to a plasmid-cured derivative of B. thuringiensis subsp. israelensis, resulting in strain AW57. A plasmid preparation of strain AW57 was partially digested with HindIII and ligated to pAW001 linearized with HindIII. The ligation mixture was used to transform competent B. subtilis cells to chloramphenicol resistance. One transformant was obtained, and a plasmid preparation of this transformant was electroporated into E. coli for further analysis. The recombinant plasmid, designated pAW002, harbored a 5.8-kb insert consisting of two HindIII fragments of 2.6 and 3.2 kb. A restriction map of the 5.8-kb insert is shown in Fig. 1.
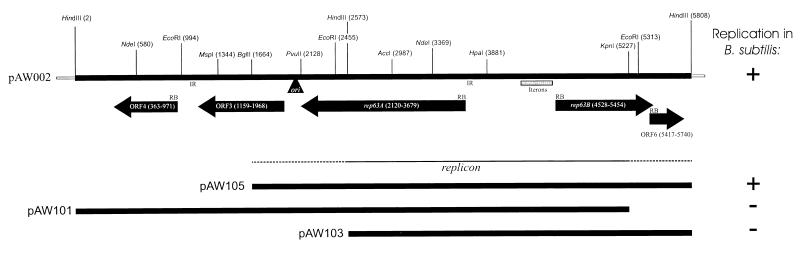
FIG. 1.
Map of the replication region of pAW63. Relevant restriction sites and ORFs larger than 100 codons are shown, and potential ribosome binding sites (RB) are marked. The locations of the origin of replication, two inverted repeats (IR), and a region containing multiple iterons are indicated. Three derivatives of the cloned fragment pAW002 and their replication capacities in B. subtilis are shown.
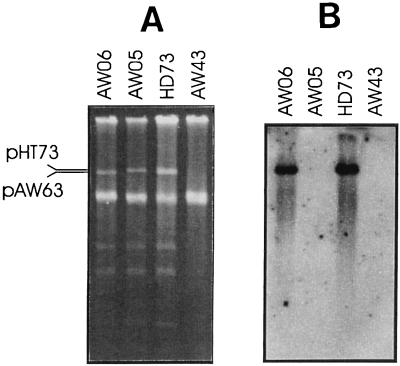
To verify that the cloned fragment originated from pAW63, pAW002 was used as a probe and hybridized to plasmid preparations from wild-type HD73, strain AW05 (HD73 cured of pAW63), strain AW06 (HD73 cured of pHT73), and strain AW43 (cured of both pAW63 and pHT73). As seen in Fig. 2, a hybridization signal was obtained only from strains containing pAW63, confirming that the isolated replicon originated from pAW63.
FIG. 2.
Origin of the cloned replicon. (A) Agarose gel electrophoresis of plasmids isolated from strains AW06 (pAW63), AW05 (pHT73), wild-type strain HD73 (pAW63 and pHT73), and AW43 (cured of all large plasmids). (B) Autoradiograph of a Southern blot of the gel shown in panel A in which the 3.3-kb HindIII fragment from pAW002 was used as a probe.
Deletion derivatives of pAW002 were constructed and tested for replication in B. subtilis. As shown in Fig. 1, a construct with the 1.7-kb HindIII-BglII fragment deleted (plasmid pAW105) was able to replicate in B. subtilis, whereas the two other derivatives shown in Fig. 1 (pAW101 and pAW103) were unable to replicate in B. subtilis. It can therefore be concluded that the replicon is located within a 4.1-kb BglII-HindIII fragment and that the minimal replicon is larger than 2.7 kb.
Sequence of the 5.8-kb fragment containing the pAW63 replicon.
The 5.8-kb fragment from pAW63, harboring the replication functions, was completely sequenced (EMBL accession no. AJ011655). The nucleotide sequence was determined to be 5,812 bp and the G+C content was 33.3%, well within the range characteristic of B. thuringiensis (20). Analysis of this sequence revealed the presence of seven open reading frames (ORFs) encoding putative proteins of more than 100 amino acids (aa). Except for ORF5 (nucleotides 3943 to 4317) and the truncated ORF7 (nucleotide 304 to beginning of cloned fragment), these ORFs had high coding probabilities according to Fickett’s TESTCODE program (27). For the five smaller ORFs, no significant similarity to available sequences could be observed in the current databases. In Fig. 1 are shown the five ORFs most likely to be genuine protein-coding regions. The two large ORFs, designated rep63A and rep63B, are essential for replication and are transcribed divergently. We suggest that the coding sequence of rep63A starts with the ATG codon located at position 3661. This results in a protein of 513 aa and a ribosome binding site with the sequence GAAAGGAGGT and a spacer region of 7 bp, as calculated according to the method of Andrup et al. (2). The second large ORF, rep63B, starts at position 4528 with a GTG codon and terminates at position 5454. A ribosome binding site with the sequence AAAAAGGAG has a spacer region of 10 bp.
Rep63A belongs to the pAMβ1 family of theta-replicating proteins.
Comparison of Rep63A, the 513-aa ORF of the pAW63 replicon, with sequences in databases revealed 40% identity to a group of closely related (>97% identical) replication proteins from gram-positive plasmids: the RepE protein of pAMβ1, a 26.5-kb conjugative plasmid from Enterococcus faecalis (52); the RepR protein of pIP501, a conjugative plasmid from Streptococcus agalactiae (12); and the RepS protein from pSM19035, a nonconjugative plasmid from Streptococcus pyogenes (12, 35). A weaker similarity (23% identity) was also found between Rep63A and the 510-aa replication protein from the self-transmissible 43-MDa (65-kb) plasmid p43 from B. thuringiensis subsp. kurstaki HD263 (4). The conservation among these replication proteins suggests that pAW63 belongs to the pAMβ1 family of gram-positive theta-replicating plasmids (17).
Origin of replication.
Replication of theta-replicating plasmids initiates at the origin (ori), a cis-functioning locus, probably via binding of the Rep protein. The replication origins of pAMβ1 and pIP501, which are almost identical, are located immediately downstream of their replication genes, repE and repR, respectively (10, 17). A region homologous to these sequences is also present downstream of repS from pSM19035 (19) and downstream of rep from p43 (4), indicating that these plasmids have the same organization. As shown in Fig. 3, the sequence just downstream of rep63A in pAW63 contains a region with similarity to the origins of pAMβ1 and p43. For the sake of clarity the ori’s from pSM19035 and pIP501, which are nearly identical to the ori from pAMβ1, are not included. Based on the similarity of this region in pAW63 to other origins of replication, the similar organizations of pAMβ1, pSM19035, p43, and pIP501 in the area of the rep gene, and the fact that this region was required for replication, we suggest that this region contains the origin of replication of pAW63.
FIG. 3.
Alignment of the origin of replication of plasmid pAMβ1 and the regions downstream of the rep genes from plasmids pAW63 and p43. The starting point of leading-strand synthesis of pAMβ1 is indicated as a triangle, and termination of its lagging-strand synthesis maps a further 16 bp downstream (14). The stop codons of the rep genes are underlined.
Rep63B, the second essential replication protein, is distantly related to plasmid copy control proteins from Enterococcus and Lactobacillus.
The second large ORF indispensable for replication, Rep63B (308 aa), showed similarity to the copy control proteins PrgP (32% identity over 310 aa) and RepB (34% identity over 281 aa) from plasmids pCF10 (31) and pAD1 (53), respectively. Plasmids pCF10 and pAD1 are conjugative pheromone-inducible plasmids from E. faecalis (for a review, see reference 25). We also found similar homology (34% identity over 295 aa) to the pTE15 replication-associated protein A (RepA) from Lactobacillus reuteri, reported to be involved in the control of plasmid copy number (EMBL accession no. AF036766). At a lower identity threshold (<22%), three other proteins could also be added to the cluster: two proteins of unknown functions flanking an erythromycin resistance gene in Clostridium perfringens (6) and the nearly identical delta protein of pSM19035 from S. pyogenes (19). However, the significance of these relationships remains dubious. Concerning the smallest ORF (ORF6) located in the minimal replication region, no significant homology to known DNA or protein sequences could be detected.
Other features of the sequence.
Repeated DNA motives, the so-called iterons, have been reported to be involved in the replication of theta-replicating plasmids and in regulating stability and/or copy control (for a review, see reference 24). Upstream of rep63B a sequence of 8 bp (consensus sequence, AAAGATAC) is repeated 10 times in the direct orientation and 6 times as inverted repeats (Fig. 4). The region also contains several shorter parts of the consensus sequence and a direct repeat of 23 bp. Three regions of AT-rich DNA (83 to 87%) were found (Fig. 4). However, unlike the organizations of replicons of several other theta-replicating plasmids, no DnaA boxes could be found in the replicon of pAW63. Furthermore, inverted repeats were located upstream the rep63A gene and between ORF3 and ORF4 (Fig. 1).
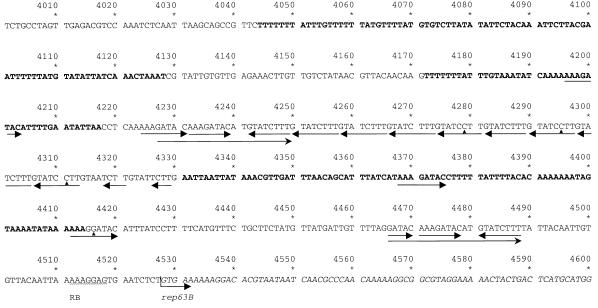
FIG. 4.
Region of pAW63 containing multiple iterons. (A) The repeated sequences are underlined, and arrows show the orientations. Mismatches from the consensus sequence are indicated with triangles. Sequences highlighted in boldface type represent regions of high A+T content (83 to 87%). The putative initiation codon and ribosome binding site (RB) of rep63B are also shown.
pAW63 replication mechanism.
To substantiate the possibility that pAW63 belongs to the family of theta-replicating plasmids, as indicated by its similarity to other rep genes and ori’s, we wanted to exclude the production of ssDNA molecules during replication. To test whether pAW002 makes ssDNA during replication, indicating that the plasmid uses RCR, Southern blot and hybridization experiments were performed as described in Materials and Methods. Plasmids pHV1610 (pC194 ligated to pUC19) and pHV1611 (containing the single-strand origin from pUB110) in B. subtilis strain SB202 were used as positive controls of ssDNA formation (7). Two B. subtilis strains containing either pAW002 (AW61) or pAW105 (AW109) were tested. With plasmid pAW002 as the probe, no ssDNA or high-molecular-weight molecules could be detected in the strain harboring pAW002 or in the strain carrying pAW105 (data not shown), indicating that pAW63 does not use RCR. Rifampin inhibits the RNA polymerase that is important for some RCR plasmids (including pUB110 and pC194) in converting ssDNA to dsDNA (7). The presence of rifampin had no influence on the formation of ssDNA in the strains containing the replicon from pAW63. Plasmid pHV1610 gave rise to ssDNA both with and without rifampin, whereas pHV1611 produced only detectable levels of ssDNA in the presence of rifampin. This result is consistent with the data presented by Boe et al. (7).
Unlike gram-positive theta-replicating plasmids belonging to the non-pAMβ1 family, the pAMβ1-like plasmids depend on PolI for replication. In order to test the dependence of pAW63 on PolI, plasmids pAW002 and pAW105 were tested for their ability to transform the polA5 B. subtilis strain 1A226, which lacks PolI (43, 44), and the isogenic PolI-proficient strain 1A224. RCR plasmids are independent of PolI, so plasmid pHV1610 was used as a positive control. As shown in Table 3, plasmid pHV1610 transformed both the polA5 mutant (1A226) and the PolI-proficient strain (1A224) efficiently to Cmr whereas pAW002 and pAW105 were inserted only in strain 1A224, indicating that the replicon from plasmid pAW63 required PolI for replication. Plasmid preparation confirmed that the transformed strains harbored the expected plasmids (data not shown).
TABLE 3.
Dependence on host gene-encoded PolI
| Plasmid | Transformation efficiency (CFU/ml)a of strain:
|
|
|---|---|---|
| 1A224 | 1A226 (PolI−) | |
| pAW002 | 20 | <3.3 |
| pAW105 | 80 | <3.3 |
| pHV1610 | 3 × 103 | 5 × 103 |
| None | <3.3 | <3.3 |
Data are the means of results of two independent experiments.
Stability and incompatibility of pAW002.
First, we examined the incompatibility between pAW105 and its native plasmid pAW63 in B. thuringiensis subsp. kurstaki (AW121). Plasmid pAW105 was lost at a high frequency (>90% after 10 generations) when there was no selection for the plasmid, verifying that pAW105 harbors replication functions incompatible with those of pAW63.
Large plasmids, replicating via theta replication, are generally more stable than plasmids replicating via RCR (26). To ascertain whether this was the case for pAW63, its stability in B. subtilis and B. thuringiensis was tested (Table 4). The two B. subtilis strains AW61 and AW109, containing pAW002 and pAW105, respectively, where found to retain the plasmid with a high frequency at 37°C (more than 92% of the cells harbored the plasmids after 40 generations) without selective pressure. Strain AW61 was also tested at 42°C, and it was found that pAW002 was very unstable at this temperature (about 1% retained the plasmid after 10 generations). In B. thuringiensis subsp. kurstaki HD73, with strain AW120 that had been cured for the parental plasmid pAW63, pAW105 was stably maintained at 30°C whereas its stability dropped significantly at 37°C.
TABLE 4.
Stability of the pAW63 replicon
| Strain | % of cells containing the plasmid after about 40 generationsa at:
|
|
|---|---|---|
| 30°C | 37°C | |
| AW61 (B. subtilis, pAW002) | ND | 92 |
| AW109 (B. subtilis, pAW105) | ND | 94 |
| AW120 (B. thuringiensis HD73, pAW63) | 87 | 56 |
Data presented are the averages of results from three independent experiments. ND, not determined.
Distribution of pAW63-related replicons among B. cereus and B. thuringiensis strains.
The presence of DNA sequences among B. thuringiensis and B. cereus strains with similarity to the replicon from pAW63 was assessed by hybridization experiments with the 3.2-kb HindIII fragment from pAW002 (Fig. 1) as the probe. Plasmid DNAs of 35 B. thuringiensis and 5 B. cereus strains were analyzed. As shown in Table 2, hybridization signals were detected in B. thuringiensis subsp. alesti and in eight strains of B. thuringiensis subsp. kurstaki in addition to B. thuringiensis subsp. kurstaki HD73, from which the replicon was isolated. B. thuringiensis subsp. kurstaki KTo did not show any hybridization signal, confirming that this strain does not harbor plasmid pAW63 (54). The hybridizing B. thuringiensis subsp. kurstaki plasmids were all of the same size, slightly larger than pAW63, whereas the hybridizing B. thuringiensis subsp. alesti plasmid was of a size similar to that of pAW63 (about 70 kb).
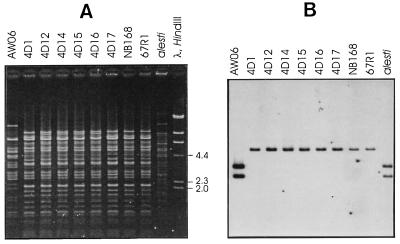
In order to test the relationship between the plasmids, total plasmid preparations of the hybridizing strains were digested with HindIII and probed with pAW105 containing the minimal replicon of pAW63. As can be seen from Fig. 5, pAW105 hybridized, as expected, to the two HindIII fragments (2.6 and 3.2 kb) from pAW63. A similar pattern was observed with plasmids from B. thuringiensis subsp. alesti, whereas only one fragment from the kurstaki plasmids (about 5.5 kb) gave a signal. The plasmids in the eight B. thuringiensis subsp. kurstaki strains displaying similarity to plasmid pAW105 have similar restriction patterns, and it is likely that they all contain copies of the same plasmids.
FIG. 5.
Strains harboring plasmids with similarity to the pAW63 replicon. (A) Agarose gel electrophoresis of a total plasmid preparation from B. thuringiensis subsp. kurstaki and B. thuringiensis subsp. alesti strains with regions displaying similarity to the 5.8-kb HindIII fragment from the pAW63 replicon, restricted with HindIII. (B) Autoradiograph of a Southern blot of the gel shown in panel A in which pAW105 was used as a probe. The strain numbers (Tables 1 and 2) are indicated above each lane.
DISCUSSION
The DNA sequence of the replication region of the large conjugative plasmid pAW63 from B. thuringiensis subsp. kurstaki HD73 has been cloned and determined. Deletion analysis minimized the region necessary for maintenance in B. subtilis to 4.1 kb. Within this region two large ORFs (rep63A and rep63B) and two smaller ones (ORF5 and ORF6) were detected. The replicon of plasmid pAW63 comprises genes with similarities to other plasmids from different plasmid families of various bacterial species.
The gene rep63A encodes a replication protein (Rep63A) with high similarity to replication proteins from the pAMβ1 family of theta-replicating plasmids. In addition, immediately downstream of rep63A a region with high similarity to the origins of replication of pAMβ1 and pIP501 was found, indicating that pAW63 belongs to the pAMβ1 family of gram-positive theta-replicating plasmids. Similar origins were found immediately downstream of the replication genes of the S. pyogenes plasmid pSM19035 and the B. thuringiensis plasmid p43, both of which belong to the pAMβ1 family. The relationship of pAW63 to the pAMβ1 family was further substantiated by the fact that pAW63 did not accumulate ssDNA during replication and was dependent on host gene-encoded PolI for stable inheritance, which is characteristic of this family (17).
Replication proteins from the pAMβ1 family are rate-limiting factors for plasmid replication (11), which implies that their expression must be strictly controlled. The regulation of the rep gene of pIP501 has been extensively studied and is negatively controlled by a long-lived antisense RNA (13) and the CopR protein, which represses transcription from the rep promoter (9, 14). No homology with the CopR protein was found in the sequenced region of pAW63, but the gene may reside on another part of the plasmid. In many cases, the origins of theta-replicating plasmids contain directly repeated sequences that are the binding sites for the plasmid-borne Rep protein genes. Among plasmids with the narrow host ranges of enterobacteria, iterons have been described for replicons from P1, F, and pSC101, among others (for a recent review, see reference 24). Iterons are also found in the replicons of theta-replicating broad-host-range plasmids such as RK2 or RP4, pCU1, and pSa and in non-theta-replicating plasmids (24, 38, 40, 50). For several large low-copy-number plasmids from gram-negative bacteria (R1, F, and P1) iterons have been found to be involved in a centromere-like function which, in conjunction with two plasmid-borne genes for proteins, comprises a partitioning system (15). The iterons have been shown to be required for maximum stability and incompatibility in the R1 system and to repress parA expression via autoregulation (15). A similar function may be performed by the iteron region in pAW63, but additional experiments are required to establish that.
The other large ORF in the replicon of pAW63 (Rep63B) showed distinct similarity to copy control proteins from the Enterococcus faecalis conjugative pheromone plasmids pAD1 and pCF10. These proteins, including Rep63B, have regions of homology to the ParA family of gram-negative partition proteins, including ATPase motifs suggested to be important for biological activity (22, 23, 46).
The small putative coding sequence ORF6, found in the replication region of pAW63, can be compared to repC from pAD1 and prgP from pCF10. The corresponding proteins have approximately the same size, they are all hydrophilic, and their genes overlap the 3′ ends of, and are transcribed in the same direction as, the copy control genes. The protein RepC from pAD1 has been implicated in regulating stable inheritance of pAD1 (53).
Plasmids of the pAMβ1 family have a broad host range, presumably because of their low requirement for host gene-encoded proteins (35) and their less specialized mechanism of conjugation (e.g., the requirement of pAMβ1 and pIP501 for solid surfaces to sustain efficient conjugation). Plasmid pAW63 is a conjugative plasmid whose host range comprises various Bacillus species (54). In contrast to this, plasmids pAD1 and pCF10 are restricted in their host range to E. faecalis and depend on the recipient gene-encoded pheromone to be transferred (for a review, see reference 21). An interesting observation is that host-produced pheromones may also play a role in plasmid replication of pAD1 and pCF10, contributing to the narrow host range of these plasmids (31). Both conjugation and replication of pAW63 are, apparently, independent of pheromones that may account for the broad host range of pAW63. Contrary to most plasmids from the pAMβ1 family, pAW63, pAD1, and pCF10 are capable of sustaining conjugation in liquid media, with a high frequency of plasmid transfer. It is therefore interesting that the DNA sequence of the pAW63 replicon displays similarity to plasmids from several different bacterial species containing fundamentally different conjugative and replication systems. Hybridization experiments demonstrated similarity between pAW63 and plasmids from various B. thuringiensis subsp. kurstaki strains and a plasmid from B. thuringiensis subsp. alesti. Contrary to what may be suspected, restriction digests gave similar hybridization patterns for pAW63 and the alesti plasmid, whereas the non-HD73 kurstaki plasmids were different, indicating that a replicon similar to the replicon of pAW63 is present on the alesti plasmid. Reddy et al. have previously demonstrated the presence of a self-transmissible plasmid, pXO15, in B. thuringiensis subsp. alesti YAL of about 50 MDa (47), which is about the same size as pAW63 and the plasmid present in B. thuringiensis subsp. alesti 4C3, used in this study.
In 1992, Baum and Gonzalez (5) reported a similar nonrandom distribution of plasmid replicon groups among B. thuringiensis subspecies and found, when probing with replicons from large kurstaki plasmids, almost exclusively hybridization to other kurstaki subspecies. They speculated that this nonrandom distribution might be due to a limited genetic exchange among subspecies of B. thuringiensis in infected insect larvae. Previous work (54) has shown that pAW63 can be transferred not only to kurstaki subspecies but also to B. thuringiensis subsp. israelensis, to B. cereus, and even to less related species such as Bacillus sphaericus and Bacillus licheniformis. Consequently, this limited distribution of pAW63-like replicons among B. thuringiensis and B. cereus is not due to an incapability of pAW63 to exist in these strains but rather to the fact that different subspecies of B. thuringiensis are quite separated in nature, having different host ranges of target insects and ecological niches.
Finally, a recent report by Hoover (33) on the replication origin of the capsule-bearing plasmid pXO2 from Bacillus anthracis indicated the presence of two ORFs of divergent orientations apparently showing the same similarities to other gram-positive rep genes as those described for pAW63. Moreover, the reported structural features of the pXO2 replicon are reminiscent of those observed in pAW63, including iterons, AT-rich regions, and inverted repeats. It will be particularly interesting to further investigate this apparent relationship between these two large plasmids originating from the two most distinctive members of the B. cereus group.
ACKNOWLEDGMENTS
We thank Esben Kjær Sørensen and Anne-Lise Rishovd for technical assistance.
The Danish Natural Science Research Council supported A.W., and grants from the Plasmid Foundation and The Beckett Foundation supported L.A. This work has also benefited from grants from the National Fund for Scientific Research (Brussels, Belgium), for which J.M. is a research associate, and grants from the Norwegian Research Council to A.-B.K.
REFERENCES
- 1.Andrup L, Damgaard J, Wassermann K. Mobilization of small plasmids in Bacillus thuringiensis subsp. israelensis is accompanied by specific aggregation. J Bacteriol. 1993;175:6530–6536. doi: 10.1128/jb.175.20.6530-6536.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrup L, Damgaard J, Wassermann K, Boe L, Madsen S M, Hansen F G. Complete nucleotide sequence of the Bacillus thuringiensis subsp. israelensis plasmid pTX14-3 and its correlation with biological properties. Plasmid. 1994;31:72–88. doi: 10.1006/plas.1994.1008. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. Brooklyn, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 4.Baum J A, Gilbert M P. Characterization and comparative sequence analysis of replication origins from three large Bacillus thuringiensis plasmids. J Bacteriol. 1991;173:5280–5289. doi: 10.1128/jb.173.17.5280-5289.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baum J A, Gonzalez J M. Mode of replication, size and distribution of naturally occurring plasmids in Bacillus thuringiensis. FEMS Microbiol Lett. 1992;96:143–148. doi: 10.1016/0378-1097(92)90394-4. [DOI] [PubMed] [Google Scholar]
- 6.Berryman D I, Rood J I. The closely related ermB-ermAM genes from Clostridium perfringens, Enterococcus faecalis (pAMβ1), and Streptococcus agalactiae (pIP501) are flanked by variants of a directly repeated sequence. Antimicrob Agents Chemother. 1995;39:1830–1834. doi: 10.1128/aac.39.8.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boe L, Gros M F, te Riele H, Ehrlich S D, Gruss A. Replication origins of single-stranded-DNA plasmid pUB110. J Bacteriol. 1989;171:3366–3372. doi: 10.1128/jb.171.6.3366-3372.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boe L, Nielsen T T, Madsen S M, Andrup L, Bolander G. Cloning and characterization of two plasmids from Bacillus thuringiensis in Bacillus subtilis. Plasmid. 1991;25:190–197. doi: 10.1016/0147-619x(91)90012-l. [DOI] [PubMed] [Google Scholar]
- 9.Brantl S. The copR gene product of plasmid pIP501 acts as a transcriptional repressor at the essential repR promoter. Mol Microbiol. 1994;14:473–483. doi: 10.1111/j.1365-2958.1994.tb02182.x. [DOI] [PubMed] [Google Scholar]
- 10.Brantl S, Behnke D. Characterization of the minimal origin required for replication of the streptococcal plasmid pIP501 in Bacillus subtilis. Mol Microbiol. 1992;6:3501–3510. doi: 10.1111/j.1365-2958.1992.tb01785.x. [DOI] [PubMed] [Google Scholar]
- 11.Brantl S, Behnke D. The amount of RepR protein determines the copy number of plasmid pIP501 in Bacillus subtilis. J Bacteriol. 1992;174:5475–5478. doi: 10.1128/jb.174.16.5475-5478.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brantl S, Behnke D, Alonso J C. Molecular analysis of the replication region of the conjugative Streptococcus agalactiae plasmid pIP501 in Bacillus subtilis. Comparison with plasmids pAMβ1 and pSM19035. Nucleic Acids Res. 1990;18:4783–4790. doi: 10.1093/nar/18.16.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brantl S, Birch-Hirschfeld E, Behnke D. RepR protein expression on plasmid pIP501 is controlled by an antisense RNA-mediated transcription attenuation mechanism. J Bacteriol. 1993;175:4052–4061. doi: 10.1128/jb.175.13.4052-4061.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brantl S, Nuez B, Behnke D. In vitro and in vivo analysis of transcription within the replication region of plasmid pIP501. Mol Gen Genet. 1992;234:105–112. doi: 10.1007/BF00272351. [DOI] [PubMed] [Google Scholar]
- 15.Breüner A, Jensen R B, Dam M, Pedersen S, Gerdes K. The centromere-like parC locus of plasmid R1. Mol Microbiol. 1996;20:581–592. doi: 10.1046/j.1365-2958.1996.5351063.x. [DOI] [PubMed] [Google Scholar]
- 16.Bruand C, Ehrlich S D, Janniere L. Unidirectional theta replication of the structurally stable Enterococcus faecalis plasmid pAMβ1. EMBO J. 1991;10:2171–2177. doi: 10.1002/j.1460-2075.1991.tb07752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bruand C, Lechatelier E, Ehrlich S D, Janniere L. A fourth class of theta-replicating plasmids. The pAMβ1 family from Gram-positive bacteria. Proc Natl Acad Sci USA. 1993;90:11668–11672. doi: 10.1073/pnas.90.24.11668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlson C R, Johansen T, Lecadet M M, Kolstø A B. Genomic organization of the entomopathogenic bacterium Bacillus thuringiensis subsp. berliner 1715. Microbiology. 1996;142:1625–1634. [Google Scholar]
- 19.Ceglowski P, Boitsov A, Chai S H, Alonso J C. Analysis of the stabilization system of pSM19035-derived plasmid pBT233 in Bacillus subtilis. Gene. 1993;136:1–12. doi: 10.1016/0378-1119(93)90441-5. [DOI] [PubMed] [Google Scholar]
- 20.Claus D, Berkeley R C W. Genus Bacillus. In: Sneath P A, Mair N S, Sharpe M E, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 2. Baltimore, Md: Williams & Wilkins; 1984. pp. 1105–1139. [Google Scholar]
- 21.Clewell D B. Bacterial sex pheromone-induced plasmid transfer. Cell. 1993;73:9–12. doi: 10.1016/0092-8674(93)90153-h. [DOI] [PubMed] [Google Scholar]
- 22.Davis M A, Martin K A, Austin S J. Biochemical activities of the ParA partition protein of the P1 plasmid. Mol Microbiol. 1992;6:1141–1147. doi: 10.1111/j.1365-2958.1992.tb01552.x. [DOI] [PubMed] [Google Scholar]
- 23.Davis M A, Radnedge L, Martin K A, Hayes F, Youngren B, Austin S J. The P1 ParA protein and its ATPase activity play a direct role in the segregation of plasmid copies to daughter cells. Mol Microbiol. 1996;21:1029–1036. doi: 10.1046/j.1365-2958.1996.721423.x. [DOI] [PubMed] [Google Scholar]
- 24.del Solar G, Giraldo R, Ruiz-Echevarria M J, Espinosa M, Diaz-Orejas R. Replication and control of circular bacterial plasmids. Microbiol Mol Biol Rev. 1998;62:434–464. doi: 10.1128/mmbr.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunny G M, Leonard B A B, Hedberg P J. Pheromone-inducible conjugation in Enterococcus faecalis: interbacterial and host-parasite chemical communication. J Bacteriol. 1995;177:871–876. doi: 10.1128/jb.177.4.871-876.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ehrlich S D, Bruand C, Sozhamannan S, Dabert P, Gros M F, Janniere L, Gruss A. Plasmid replication and structural stability in Bacillus subtilis. Res Microbiol. 1991;142:869–873. doi: 10.1016/0923-2508(91)90067-k. [DOI] [PubMed] [Google Scholar]
- 27.Fickett J W. Recognition of protein coding regions in DNA sequences. Nucleic Acids Res. 1982;10:5303–5318. doi: 10.1093/nar/10.17.5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gamel P H, Piot J C. Characterization and properties of a novel plasmid vector for Bacillus thuringiensis displaying compatibility with host plasmids. Gene. 1992;120:17–26. doi: 10.1016/0378-1119(92)90004-9. [DOI] [PubMed] [Google Scholar]
- 29.Gonzalez J M, Jr, Carlton B C. Patterns of plasmid in crystalliferous and acrystalliferous strains of Bacillus thuringiensis. Plasmid. 1980;3:92–98. doi: 10.1016/s0147-619x(80)90038-4. [DOI] [PubMed] [Google Scholar]
- 30.Gonzalez J M, Jr, Carlton B C. Plasmid transfer in Bacillus thuringiensis. In: Streips U N, Goodgal S H, Guild W R, Wilson G A, editors. Genetic exchange: a celebration and a new generation. New York, N.Y: Dekker; 1982. pp. 85–95. [Google Scholar]
- 31.Hedberg P J, Leonard B A B, Ruhfel R E, Dunny G M. Identification and characterization of the genes of Enterococcus faecalis plasmid pCF-10 involved in replication and in negative control of pheromone-inducible conjugation. Plasmid. 1996;35:46–57. doi: 10.1006/plas.1996.0005. [DOI] [PubMed] [Google Scholar]
- 32.Hoflack L, Seurinck J, Mahillon J. Nucleotide sequence and characterization of the cryptic Bacillus thuringiensis plasmid pGI3 reveal a new family of rolling circle replicons. J Bacteriol. 1997;179:5000–5008. doi: 10.1128/jb.179.16.5000-5008.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoover T A. Abstracts of the 3rd International Conference on Anthrax. DERA Chemical and Biological Defence Sector, Porton Down, United Kingdom, and the Society for Applied Microbiology, Plymouth, United Kingdom. 1998. Characterization of a region of Bacillus anthracis capsule-encoding plasmid pXO2 capable of autonomous replication; p. 52. [Google Scholar]
- 34.Horinouchi S, Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982;150:815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jannière L, Gruss A, Ehrlich D. Plasmids. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C: American Society for Microbiology; 1993. pp. 625–644. [Google Scholar]
- 36.Jensen G B, Wilcks A, Petersen S S, Damgaard J, Baum J A, Andrup L. The genetic basis of the aggregation system in Bacillus thuringiensis subsp. israelensis is located on the large conjugative plasmid pXO16. J Bacteriol. 1995;177:2914–2917. doi: 10.1128/jb.177.10.2914-2917.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khan S A. Rolling-circle replication of bacterial plasmids. Microbiol Mol Biol Rev. 1997;61:442–455. doi: 10.1128/mmbr.61.4.442-455.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lacks S A, Lopez P, Greenberg B, Espinosa M. Identification and analysis of genes for tetracycline resistance and replication functions in the broad-host-range plasmid pLS1. J Mol Biol. 1986;192:753–765. doi: 10.1016/0022-2836(86)90026-4. [DOI] [PubMed] [Google Scholar]
- 39.Lereclus D, Guo S, Sanchis V, Lecadet M M. Characterization of two Bacillus thuringiensis plasmids whose replication is thermosensitive in B. subtilis. FEMS Microbiol Lett. 1988;49:417–422. [Google Scholar]
- 40.Lin L S, Meyer R J. Directly repeated, 20-bp sequence of plasmid R1162 DNA is required for replication, expression of incompatibility, and copy-number control. Plasmid. 1986;15:35–47. doi: 10.1016/0147-619x(86)90012-0. [DOI] [PubMed] [Google Scholar]
- 41.Madsen S M, Andrup L, Boe L. Fine mapping and DNA sequence of replication functions of Bacillus thuringiensis plasmid pTX14-3. Plasmid. 1993;30:119–130. doi: 10.1006/plas.1993.1039. [DOI] [PubMed] [Google Scholar]
- 42.Mahillon J, Seurinck J. Complete nucleotide sequence of pGI2 a Bacillus thuringiensis plasmid containing Tn4430. Nucleic Acids Res. 1988;16:11827–11829. doi: 10.1093/nar/16.24.11827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mazza G, Galizzi A. The genetics of DNA replication, repair and recombination in Bacillus subtilis. Microbiologica. 1978;1:111–135. [Google Scholar]
- 44.Mazza P, Galizzi A. Revised genetics of DNA metabolism in Bacillus subtilis. Microbiologica. 1989;12:157–179. [PubMed] [Google Scholar]
- 45.McDowell D G, Mann N H. Characterization and sequence analysis of a small plasmid from Bacillus thuringiensis var. kurstaki HD1-DIPEL. Plasmid. 1991;25:113–120. doi: 10.1016/0147-619x(91)90022-o. [DOI] [PubMed] [Google Scholar]
- 46.Motallebi-Veshareh M, Rouch D A, Thomas C M. A family of ATPases involved in active partitioning of diverse bacterial plasmids. Mol Microbiol. 1990;4:1455–1463. doi: 10.1111/j.1365-2958.1990.tb02056.x. [DOI] [PubMed] [Google Scholar]
- 47.Reddy A, Battisti L, Thorne C B. Identification of self-transmissible plasmids in four Bacillus thuringiensis subspecies. J Bacteriol. 1987;169:5263–5270. doi: 10.1128/jb.169.11.5263-5270.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 49.Schnepf E, Crickmore N, van Rie J, Lereclus D, Baum J, Feitelson J, Zeigler D R, Dean D H. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol Mol Biol Rev. 1998;62:775–806. doi: 10.1128/mmbr.62.3.775-806.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scholz P, Haring V, Wittmann-Liebold B, Ashman K, Bagdasarian M, Scherzinger E. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene. 1989;75:271–288. doi: 10.1016/0378-1119(89)90273-4. [DOI] [PubMed] [Google Scholar]
- 51.Southern E. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 52.Swinfield T, Oultram J D, Thompson D E, Brehm J K, Minton N P. Physical characterisation of the replication region of the Streptococcus faecalis plasmid pAMβ1. Gene. 1990;87:79–90. [PubMed] [Google Scholar]
- 53.Weaver K E, Clewell D B, An F. Identification, characterization, and nucleotide sequence of a region of Enterococcus faecalis pheromone-responsive plasmid pAD1 capable of autonomous replication. J Bacteriol. 1993;175:1900–1909. doi: 10.1128/jb.175.7.1900-1909.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilcks A, Jayaswal N, Lereclus D, Andrup L. Characterization of plasmid pAW63; a second self-transmissible plasmid in Bacillus thuringiensis subspecies kurstaki HD-73. Microbiology. 1998;144:1263–1270. doi: 10.1099/00221287-144-5-1263. [DOI] [PubMed] [Google Scholar]
- 55.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]