Abstract
The development of a method for genome editing based on CRISPR–Cas9 technology was awarded The Nobel Prize in Chemistry in 2020, less than a decade after the discovery of all principal molecular components of the system. For the first time in history a Nobel prize was awarded to two women, Emmanuelle Charpentier and Jennifer Doudna, who made key discoveries in the field of DNA manipulation with the CRISPR–Cas9 system, so-called “genetic scissors”. It is difficult to overestimate the importance of the technique as it enables one not only to manipulate genomes of model organisms in scientific experiments, and modify characteristics of important crops and animals, but also has the potential of introducing revolutionary changes in medicine, especially in treatment of genetic diseases. The original biological function of CRISPR–Cas9 system is the protection of prokaryotes from mobile genetic elements, in particular viruses. Currently, CRISPR–Cas9 and related technologies have been successfully used to cure life-threatening diseases, make coronavirus detection tests, and even to modify human embryo cells with the consequent birth of babies carrying the introduced modifications. This intervention with human germplasm cells resulted in wide disapproval in the scientific community due to ethical concerns, and calls for a moratorium on inheritable genomic manipulations. This review focuses on the history of the discovery of the CRISPR–Cas9 system with some aspects of its current applications, including ethical concerns about its use in humans.
Keywords: CRISPR–Cas9, genome editing, genetic scissors”, ethical considerations
A HISTORY OF THE DISCOVERY OF THE MAIN COMPONENTS OF THE CRISPR–Cas9 SYSTEM
CRISPR – clustered regularly interspaced short palindromic repeats – were first discovered in the sequences of DNA from Escherichia coli bacteria and described in 1987 by Ishino et al. [1] from Osaka University (Japan). At that time sequencing of these difficult-to-study DNA fragments took several months, but neither their origin nor their significance in the bacterial cell were understood by their discoverers. Although in the early work in this field, the biological function of the CRISPR system had not yet been elucidated, scientists had already proposed a way to use the information encoded in CRISPR loci in medical research, namely, for genotyping various strains of bacteria: initially on Mycobacterium tuberculosis [2], and later on Streptococcus pyogenes [3]. As it turned out, CRISPR loci had a high degree of polymorphism in different strains of the same species of pathogenic bacteria, which enabled the identification of bacterial strains in clinical conditions.
A significant breakthrough in understanding the biological function of CRISPR loci occurred with the discovery of Francisco Mojica of the University of Alicante (Spain), who came across similar structures in the archaeal genome of Haloferax mediterranei in 1995 [4]. Their presence in two evolutionarily remote domains of life suggested these elements’ great functional significance, and served as an impetus for further research. Mojica noticed the similarity of the elements he described in archaea with previously found DNA repeats in bacterial genomes, and was one of the first scientists to hypothesize that these unusual loci include fragments of foreign DNA and are, in fact, a part of the immune system of bacteria and archaea [5]. In the same year as Mojica, two other laboratories independently reached similar conclusions [6, 7], announcing the beginning of an era of active research into this extraordinary natural phenomenon. In line with the theory of the prokaryotic immune system, viral DNA fragments (“spacers” 17-84 bases long), separated by short palindromic repeats (23-50 bases [8]) and grouped into clusters in intergenic regions, represent a library of potentially dangerous genetic information (for an overview of the microbial antiviral arsenal, see reviews by Isaev et al. [9, 10]). Initially, it was assumed that such a system would work by the mechanism of RNA interference. However, in the publication of Marraffini and Sontheimer, it was experimentally demonstrated for the first time that the actual target of the immune system of prokaryotes was foreign DNA [11], and not mRNA, and, therefore, the use of such a system in the laboratory could represent a potential tool for genomic editing. Interestingly, later studies demonstrated that some of the described CRISPR systems do work with RNA molecules directly [12, 13] and, therefore, can be used to deactivate specific transcripts inside the cell in a selective way [14, 15].
The first experimental information about the mechanism of action of the CRISPR system was obtained in 2007 in the studies of two French food scientists, Rodolphe Barrangou and Philippe Horvath, who worked with yoghurt cultures of bacteria Streptococcus thermophilus for the Danish company Danisco [16]. Due to the company’s rich collection of bacterial strains collected since the 1980s, scientists have been able to trace the historical course of the bacterial acquisition of spacers at the CRISPR locus in response to viral attacks by bacteriophages. The addition of new spacers in this work caused acquired immunity to the corresponding new types of bacteriophages in S. thermophilus: observation which subsequently led to the authors obtaining one of the first patents in this area [17] and the start of bacterial cultures’ “vaccination” with the use of CRISPR-based technology by Danisco in 2005 [18].
Currently, CRISPR repeats have been found in most archaeal genomes and nearly half of the studied bacterial ones, but they have not been found in eukaryotic or viral DNA sequences. The existence of CRISPR repeats in mitochondria was suggested in one of the earliest publications on the subject (the same article described CRISPR in cyanobacteria for the first time) [19]. The authors used a set of previously published data on the sequencing of mitochondrial plasmids from Vicia faba L. beans [20], and their conclusions were further cited by Mojica et al. [21], but these observations were not confirmed in later studies [8].
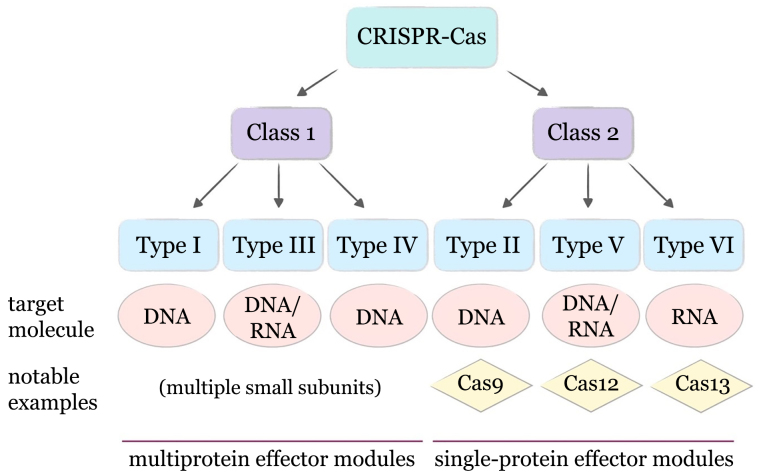
At the time of initial discoveries, a variety of different acronyms was used for CRISPR by individual scientific groups, which presently complicates the search for early articles on the topic. The current name for CRISPR first appeared in Jansen et al. [22] in 2002 and was suggested by Mojica in correspondence between the two collaborating scientific groups. The same publication was the first one to describe the presence of genes associated with CRISPR repeats (named by the authors cas1-4, CRISPR-associated genes). These genes were found in close proximity to the CRISPR loci of various prokaryotes, and two of them contained motifs characteristic of helicase and nuclease, which supported the authors’ hypothesis about the non-random association of the cas genes with the CRISPR locus, and their involvement in DNA metabolism. Also in 2002, the same neighborhood of genes was described by a team of scientists led by Eugene Koonin from the NCBI Institute (Bethesda, USA), but the association of these genes with CRISPR arrays was not discerned by them at the time [23]. From the moment of the first discovery of genes associated with the CRISPR system, to the present day, their truly extraordinary abundance and diversity have been found in prokaryotic cells, including representatives of the families of helicases, nucleases, polymerases, and others. Proteins associated with this system can be assigned to either the adaptive module (participating in the acquisition of immunity, main representatives – Cas1 and Cas2), or the effector module (directly involved in the destruction of mobile genetic elements through their recognition and cleavage), with some additional and regulatory proteins also found to be associated with the system [24]. At present, a way of classification is recognized in which all currently known CRISPR–Cas systems are divided into 2 classes and 6 types, which, in turn, are also divided into numerous subtypes: at the time of writing the review, Makarova et al. [25] described >30 subtypes (Fig. 1). The main difference between the classes is that the effector module of Class 1 systems is represented by a complex of several proteins, while in Class 2 it is a single multidomain protein (Cas9, Cas12, or Cas13) [26-28].
Fig. 1.
Conventional classification of known CRISPR–Cas systems.
Of all the known Cas proteins, the most studied ones are the proteins belonging to the system of directional cutting of foreign DNA (and, as it was found out later, in some cases, RNA), the so-called “genetic scissors”, among which is the nuclease Cas9. This protein was first described in connection with its association with CRISPR repeats in an article by Bolotin et al. [6], where it was originally named Cas5 (other alternative names are Csn1 and Csx12). In addition, the authors identified the presence of the HNH motif (His-Asn-His), which is also found in other nucleases. Another important observation made by Bolotin et al. was the discovery of a specific pattern in the nucleotide sequences on one side of the described spacers of the CRISPR arrays, but the understanding of the role for this phenomenon was only revealed in later studies. Currently, short motifs adjacent to protospacers but absent in the original spacers of the CRISPR locus are called PAMs (protospacer adjacent motifs) [29]. Protospacers are DNA fragments that are attacked by the immune system of prokaryotes, and are identical to the corresponding spacers at the CRISPR locus, except for the PAM motif. These motifs are important at the stage of recognition of potentially dangerous genetic information; their presence at the end of the sequence signals that the DNA fragment is foreign and needs to be destroyed, while the DNA sequences stored in the CRISPR locus as spacers and not containing PAM motifs are not attacked by the prokaryotic immune system.
A crucial player in the CRISPR–Cas9 system turned out to be a short RNA molecule, a processed product of transcription from the CRISPR locus that directs proteins of the prokaryotic immune system to foreign molecules with genetic information. A group of researchers led by John van der Oost (Wageningen University, the Netherlands), who described the existence of such RNA molecules, gave them the name crRNA (CRISPR-associated RNA). It was also noted that the initial result of transcription from the CRISPR locus is a pre-crRNA precursor molecule consisting of several spacers and repeats, which is later cleaved into individual fragments [30]. In the work of the group led by Virginijus Siksnys (Vilnius University, Lithuania), it was demonstrated that the length of the actual “guide” crRNA sequence of 20 base pairs, complementary to the target DNA, is necessary and sufficient for the nuclease activity of the CRISPR–Cas complex, even if the spacer in CRISPR locus is represented by a longer sequence of nucleotides [31]. This publication was one of two in vitro studies, carried out in parallel and independently in competing laboratories, that described, for the first time, how the Cas9 enzyme uses crRNA to attack foreign DNA.
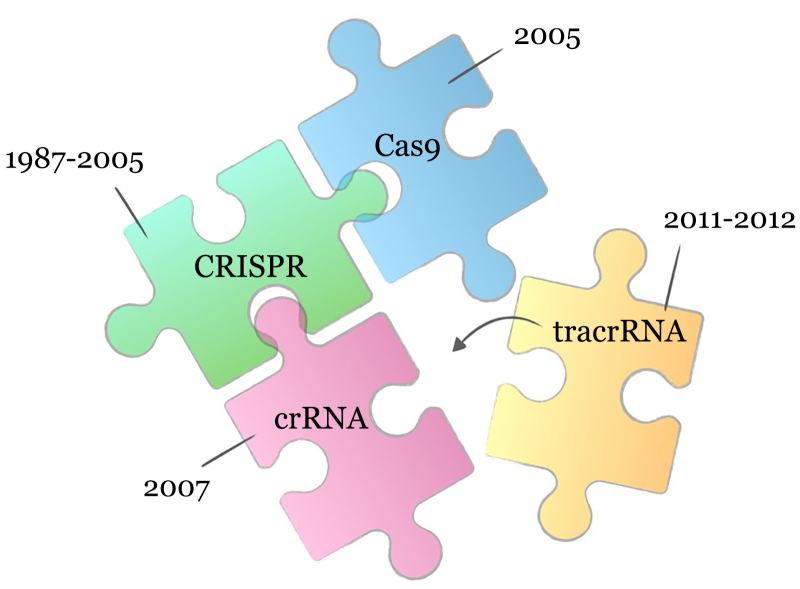
The final missing piece in the puzzle, without which it is impossible to assemble a working CRISPR–Cas9 system in vitro, turned out to be another short RNA molecule, discovered in connection with its participation in crRNA processing by Emmanuelle Charpentier’s group in 2011 [32]. This molecule, essential for nuclease activity, was named tracrRNA (trans-activating CRISPR RNA). In subsequent work, ultimately acknowledged by the Nobel Prize, the role of tracrRNA in the mechanism of target DNA cutting was shown. It was also proposed at the time that two RNA molecules, crRNA and tracrRNA, could be combined into one chimeric molecule (sgRNA – single guide RNA), which greatly facilitated the practical use of the CRISPR–Cas9 system in subsequent applications [33]. Figure 2 shows the timeline of the historical events in the discovery of the CRISPR–Cas9 system’s components: initially the CRISPR locus itself, then the proteins associated with it, including Cas9, and later, two RNA molecules necessary for the formation of the ribonucleoprotein complex and recognition of substrate DNA.
Fig. 2.
Historical timeline of discoveries of the components of the CRISPR–Cas9 system. 1987 – Short DNA repeats, later called CRISPR, were first noticed in bacterial genomes, and, in 1995, also found in archaea. 2005 – The role of CRISPR loci in the protection of prokaryotes from foreign genetic information was proposed, and the Cas9 protein was described for the first time (initial information on proteins associated with the CRISPR locus appeared in 2002). Two RNA molecules, crRNA and tracrRNA, were discovered as part of the complex in 2007 and 2011, respectively. The Nobel Prize-winning work, where all of the components were assembled in vitro and two RNA molecules combined into one strand for the ease of use of the system, was published in 2012.
USE OF THE CRISPR–Cas9 SYSTEM IN EUKARYOTIC CELLS
The discovery of the necessary and sufficient components of the CRISPR–Cas9 system started a race to be the first to apply the system to the genetic editing of human and animal cells. In January 2013, almost simultaneously, five research articles authored by different research teams appeared, all reporting that they had achieved the goal. Two publications from the same issue of the journal Science, offering probably the best approach to the problem had been produced by the laboratories of George Church (Harvard University, USA) and Feng Zhang (Broad Institute, USA). In these publications, it was shown that for successful DNA editing in human cells, it was necessary to carry out several steps: these include codon optimization and the addition of a nuclear localization signal to the cas9 gene, lengthening of the sgRNA molecule (to improve the efficiency of the system), as well as the possible addition of a DNA template for homologous recombination with which the cells can repair the DNA double break (the last step was described only by the group of G. Church) [34, 35]. Also in January 2013, similar publications came out from the laboratories of Jennifer Doudna (Berkeley College, USA) [36], Jin-Soo Kim (Seoul University, South Korea) [37] and J. Keith Joung (Harvard School of Medicine, USA) [38]. In the last article [38], the described work was carried out on zebrafish rather than human cells but, importantly, the use of the CRISPR–Cas9 system on germline cells was demonstrated for the first time.
FIRST CRYSTALLOGRAPHIC STUDIES
The most studied protein from the Cas group is the Cas9 nuclease; in the ~20 years since the discovery of the cas genes more than 20,000 articles in the PubMed system mention the name Cas9 in one context or another. Attempts to obtain detailed information about the structure of this protein resulted in the first two crystallographic studies being published almost simultaneously: in February 2014 two crystal structures of Cas9 appeared in the database PDBe (“Protein Data Bank in Europe”), and the accompanying articles were published in the journals Nature and Cell [39, 40]. The structure that came out of the laboratory of Jennifer Doudna was of an apo-protein (PDBe ID 4cmp, PDBe DOI: 10.2210/pdb4cmp/pdb), while the research group of Osamu Nureki (University of Tokyo, Japan) succeeded in crystallising the protein in a complex with a “guide”-RNA and “target”-DNA (PDBe ID 4oo8, PDBe DOI: 10.2210/pdb4oo8/pdb).
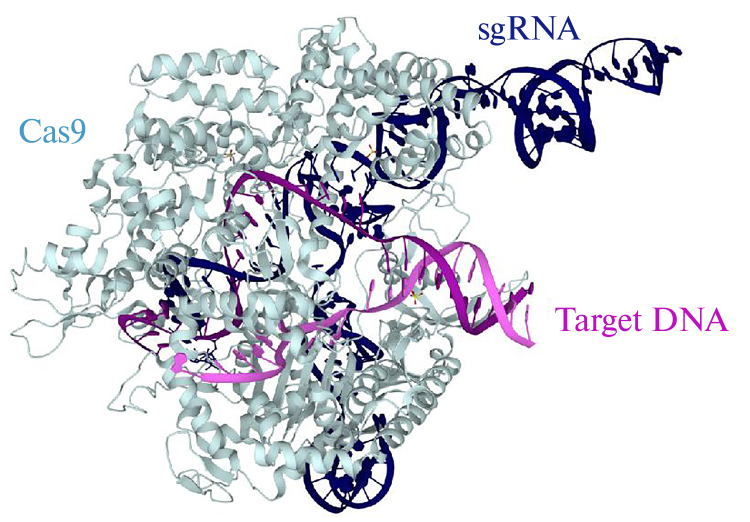
These, as well as many subsequent studies, used the Cas9 protein from S. pyogenes, SpCas9, which consists of 1368 amino acids and is a multidomain and multifunctional endonuclease. Crystal structures revealed that the Cas9 protein is spatially divided into 2 lobes: a target recognition lobe and a nuclease lobe, with the guide RNA and target DNA occupying the positively charged groove at their interface. The key structures of the nuclease lobe of SpCas9 are 2 domains: HNH and RuvC, each of them cleaves one of the target DNA strands. Figure 3 shows the general architecture of the SpCas9–sgRNA–DNA complex, where the complex secondary structure of the bound RNA molecule, and the unwound state of the double-stranded DNA molecule with the formation of a DNA–RNA heteroduplex can be seen (PDB ID 5F9R, PDB DOI: 10.2210/pdb5F9R/pdb, [41]). At the time of writing, hundreds of crystal structures of the Cas9 family proteins are available from the PDB, PDBe, and PDBj databases.
Fig. 3.
Three-dimensional organization of the Cas9 protein in the complex with “guide” RNA (sgRNA) and substrate (Target DNA), crystallographic data (PDB ID 5F9R, PDB DOI: 10.2210/pdb5F9R/pdb).
PATENT DISPUTE
The understandable motive of individual scientists, as well as organizations involved in the study of the CRISPR–Cas9 system, was the possible financial gain potentially obtainable from the use of this promising technology. One of the first patent applications was filed jointly by the University of California at Berkeley, representing Doudna, the University of Vienna (where one of the two lead authors from the key publication on CRISPR–Cas9 worked [33]), and Charpentier as an individual inventor in accordance with the rules of the University of Umeå (Sweden), where Charpentier worked at the time of publication of the article [18]. This patent application was filed in May 2012 [42], while in December 2012 Zhang and the Broad Institute also submitted a patent application [43] simultaneously with the acceptance of Zhang’s paper on human cells’ editing for publication in Science [35]. Initially, it was Zhang’s application that turned out to be successful and resulted in a patent in April 2014, while Doudna’s application was still pending at that time. Doudna’s team disagreed with the decision, after which a long dispute between the two parties followed, including appeals and court hearings which ultimately led to an ambiguous situation in CRISPR–Cas9 licensing. Due to the fact that by 2019 both competing parties had patents in this area, some of the biotech companies that used the CRISPR–Cas9 system on human cells received a license from the team of Doudna, while others – from Zhang. However, the U. S. Patent and Trademark Office Appeal Board in February 2022 again confirmed the priority of Zhang and the Broad Institute in the position of the patent holder for the use of CRISPR–Cas9 in human cells, which caused disappointment and frustration from the opposing side, and financial complications for companies licensed by the team of Doudna [44]. Doudna and Charpentier, however, won a similar dispute in Europe, and also hold major patents on the use of technology in the U.K., China, Japan, Australia, New Zealand, and Mexico [18].
GENE THERAPY AND ETHICAL ISSUES ASSOCIATED WITH IT
The haste with which competing laboratories sought to bring their research to the public’s attention, as well as the race to patent this technology, were indicators of the significance of this scientific breakthrough. Undoubtedly, one of the main driving forces that motivated many scientists to take part in research using this particular technology was the potential of modifying human cells, both somatic and germline. However, despite the apparent advantages of the CRISPR–Cas9 system, numerous ethical and technical difficulties stand in the way of researchers who dream of curing life-threatening diseases, especially if the genetic changes resulting from such manipulations can be inherited.
Gene therapy was administered for the first time in September 1990: a four-year-old girl suffering from adenosine deaminase (ADA) deficiency received an infusion of genetically engineered T-lymphocytes. Cells taken from the girl’s blood were modified using a viral vector – a deactivated virus that carries a healthy copy of the gene. As journalists who covered the story noted “rarely in modern medicine has an experiment been filled with so much hope”, and the doctor who performed this procedure, W. French Anderson, became known as the “father of gene therapy”. As time went on, however, the disturbing evidence of the adverse side effects of some attempts at gene therapy in both animals and humans began to accumulate. The tragic story of Jesse Gelsinger, an American teenager from Philadelphia who died from the effects of gene therapy in 1999, shocked the world and caused widespread skepticism and a significant delay in the development of the technology. In the case of Gelsinger, a large-scale autoimmune response of the body to a viral vector carrying the ornithine transcarbamylase gene led to a sharp increase in body temperature, renal and pulmonary failure, jaundice, impaired blood clotting, and subsequent death within only four days from the moment of gene therapy administration [45].
Extensive discussions of the safety and, importantly, the ethical issues arising from the possibility of potential gene therapy with CRISPR–Cas9 began soon after the first publications showing this system’s use in human cells. One of the first steps in initiating formal discussions was taken by Doudna, who organized a conference on scientific, medical, legal, and ethical issues related to the genomic modification, held in the Napa Valley in California in January 2015. A subsequent report of the results of the conference was published in March 2015 in the journal Science [46], which essentially carried recommendations to strongly discourage work on introducing heritable changes in human embryonic cells, at least for the duration of active discussions of the social, environmental and ethical consequences of such manipulations. Almost simultaneously with this report, a comment was also published in the journal Nature about the serious risks linked to creating heritable changes in human embryos [47]. The authors expressed concerns that premature work on embryonic cells could have a negative impact on the field of gene therapy in general, and could set back the work of researchers attempting to treat genetic and infectious diseases in somatic cells for years. The March 2015 report from the Napa conference and the commentary in Nature urging not to edit the human embryonic genome were released amidst growing agitation in the scientific community over leaked news that such experiments had actually already been carried out. A group of scientists from Sun Yat-sen University (Guangzhou, China), after unsuccessful attempts to get their manuscript accepted by the journals Nature and Science, in April 2015 finally published their article on the use of the CRISPR–Cas9 system on human embryonic cells [48]. The researchers emphasized that they used non-viable embryos obtained by the fusion of two sperm cells with one egg and, therefore, discarded by in vitro fertilization (IVF) laboratories. The main conclusion of the article was that the CRISPR–Cas9 technology at the time of the study was not yet ready for use on human embryonic cells due to the identified shortcomings in the system’s efficiency and specificity. A comment of the journal Protein & Cell (Beijing, China), that published this work, stated that the article (in addition to its scientific value) would promote an open exchange of information about current research in the area; and despite the ambiguity of the issue and conflicting opinions on the topic, the publication would stimulate the necessary discussions about genomic editing of germline cells. Interestingly, the manuscript had been sent to Protein & Cell together with the references obtained during previous attempts to publish the work, and was accepted by the editors for publication within two days from the date of submission. The subsequent debate in the scientific community was described as “epic” [49] and provoked interest in this complex issue from the wider public, as well as in governmental and regulatory organizations in various countries.
The notorious scandals caused by the conduct of medical experiments on humans in the past have led to the creation of general international guidelines on bioethics. The best-known documents in this area are the Nuremberg Code, developed after the trial of Nazi doctors in 1947, and the subsequent Declaration of Helsinki from 1964, which expanded the principles of the code and detailed the application of these principles to clinical research. Another important document, the Belmont Report, was issued by the National Commission for the Protection of Human Subjects of Biomedical and Behavioral Research in the United States in 1978. This commission was created in the wake of shocking revelations of an inhumane syphilis study from 1932 to 1972 in Tuskegee. For decades, hundreds of impoverished African-American men infected with syphilis have been studied for the progression of their disease. Although penicillin had become the standard treatment for syphilis by 1947, it was not offered to study participants, despite the obvious physical suffering of the patients and the continued spread of the infection in their families.
The Nuremberg Code, the Declaration of Helsinki and the Belmont Report are based on the basic ethical principles of biomedical research, such as respect for the individual, informed consent of the patient, understanding of the risks and benefits, voluntary participation, fairness in the conduct of experiments, maximum professionalism of the researchers, etc. These principles, and their application in medical practice, are relevant to the events of November 2018, when the Chinese scientist Jiankui He announced the birth of babies who, for the first time, had undergone gene modification using the CRISPR–Cas9 system. The injection of this system into the mother’s egg was made at the stage of the IVF procedure immediately after the fusion of the sperm, and therefore all the changes potentially introduced into the genome during this procedure would be heritable. The world scientific community was shocked at how premature such medical experiments were, and the high degree of risk taken by the researchers conducting the experiment. In particular, scientists were worried about the possibility of creating unplanned (“off-target”) mutations in the genome of future babies. At the time of the experiment He (also known under the shortened name JK – from Jiankui) was not a well-known figure in the CRISPR–Cas9 community, however, after the announcement of his experiments, he attracted world-wide attention. He studied physics at the University of Science and Technology (Hefei, China) and then moved to the United States, where he received his PhD under the supervision of Michael Deem, Professor of Physics, Astronomy and Bioengineering at Rice University (Houston, Texas), and later worked as a post-doc at Stanford University (California) in the laboratory of Professor Stephen Quake. In the group of Deem He used the methods of theoretical biophysics, mathematical modelling and computer simulations, publishing papers on, among other things, influenza virus strains and spacer sequences in CRISPR loci [50, 51], while in the laboratory of Quake, he learned the methods of molecular biology and became interested in the innovative technologies of Silicon Valley. Returning to China, He continued his collaboration with Deem, and also successfully implemented the innovative ideas in the field of DNA sequencing of his second supervisor, Quake, creating a successful company Direct Genomics based on the technology [18, 52]. In China, he became quite famous as a young scientist and successful entrepreneur who had returned from abroad under the Thousand Talents program. He received a position and a laboratory at the Southern University of Science and Technology (SUStech, Shenzhen), and participated in the creation of several start-up companies [53]. The next step in his career resulted in the biggest medical scandal of the last decade. In 2017 on WeChat social media platform, He announced that he was recruiting volunteers from among married couples who wanted to produce children genetically modified to be resistant to the human immunodeficiency virus (HIV). Among the conditions of recruitment was that in the couple who wished to participate in the experiment both people had a university degree, so that they had enough educational background to understand the basics of science and medicine. A second condition was for the man to be HIV-positive and for the woman – HIV-negative: a situation in which the risk of transmitting the virus to the baby would be minimal (provided that the sperm was “washed” during the IVF procedure), but made it likely that the couple’s motivation to participate in the experiment would be high [53]. He planned to modify the CCR5 gene, a known receptor on the cell surface, through binding to which the human immunodeficiency virus enters the cell. About 300 people responded to the advertisement, of these, 20 couples were selected for the next round of consultations, during which the participants learned about the procedure and the possible risks. From these consultations 11 couples agreed to participate in the studies, of which seven were ultimately selected by the researchers for the next stage – the IVF procedure with an additional step of genome editing. The motivation of individual participants was, apparently, not only the possibility of having children (the IVF procedure in China is prohibited if one of the parents has HIV infection), but also the desire to take part in an “historic” experiment designed to benefit future generations [53]. Ultimately, after several unsuccessful attempts, from a selected group of participants 2 pregnancies led to the birth of babies who had undergone a genomic modification procedure using the CRISPR–Cas9 system. Quite a lot is known about the first pregnancy, which resulted in the birth of two twin girls, Lulu and Nana (pseudonyms used in the press and scientific literature in order to protect their identity). Very little information is available on the second pregnancy, which resulted in the birth of another child. Since this event occured after the scandal caused by the birth of the first twins, many details of the second pregnancy remained a secret. A manuscript written by He, based on the results of the first pregnancy and named “Birth of twins after genome editing for HIV resistance” remains unpublished, but has been leaked to the scientific community [54, 55]. It has become known, for example, that in one of the embryos both copies of CCR5 were inactivated (Nana), while in the second, only one was modified (Lulu) [56]. Therefore, only Nana has a chance to be protected from HIV infection in the future, at least from the main variants of the virus that enter the cell through binding to the CCR5 receptor. In the case of Lulu, unfortunately, the treatment will provide no protection, since one copy of the CCR5 gene is enough to produce the corresponding receptor on the membrane. It is believed that two embryos were implanted in the uterus of a future mother in the hope that at least one of them will lead to the birth of a genetically modification baby. The twins were born premature (at 31 weeks) and spent the first weeks of their lives in neonatal incubators but were otherwise described as “healthy” [53]. Scientists who had gained access to the unpublished manuscript of He, also noted that several cells selected for sequencing early in embryonic development were in fact mosaics, an observation that led to increased criticism of He’s work. In the case of mosaicism, any information obtained during the sequencing of selected cells cannot be extrapolated to the entire embryo as a whole. Therefore, at the time of the key decision of whether to transfer the embryos into the womb, the researchers could not be sure that the CRISPR–Cas9 system did not produce any dramatic off-target mutations in the remaining cells of the embryos, even if the sequencing results showed the absence of such modifications in the selected cells. Many other aspects of the conduct of the study also received harsh criticism from the scientific and medical community [54], including the questionable circumstances of obtaining permission from the ethics committee of a hospital in Shenzhen, the level of qualification of He for clinical research (lack of medical education and adequate experience in the field), the choice of the gene that has undergone editing (social rather than medical reasons for patients seeking help), possible side effects from the lack of a valid copy of CCR5, etc. According to an American cardiologist and Professor of Medicine at the University of Pennsylvania Kiran Musunuru, the first babies of “the CRISPR generation”, unfortunately, were born not as a result “of a historic scientific achievement, but rather a historic ethical fiasco” [56]. A preceding PR-campaign conducted by He and his team resulted in fairly flattering initial news coverage of his work in the People’s Daily (the largest newspaper group in China). However, the following international scandal led to the placement of He under house arrest, and then to a 3-year prison sentence. He has already been released from prison, but little is known about his whereabouts and future plans [57].
A few months after the described scandal the Russian scientist Denis Rebrikov stirred up the international scientific community with a statement about his intention to become the second scientist in the world to create genetically modified babies. Rebrikov, a Professor at the Pirogov Russian National Research Medical University and Head of the Laboratory of Genomic Editing at the Center for Obstetrics, Gynecology and Perinatology, announced that his research facility was potentially ready to transfer modified embryos into the mother’s womb in June 2019 [58]. As in the experiments of He, he was planning to edit the CCR5 gene, and the preliminary work from his laboratory on non-viable embryos was published in the Bulletin of the Russian National Research Medical University [59]. The reaction of the scientific community to the statement was heated and primarily negative. In October 2019 the journals Nature and Science published news feeds reporting that at that time, Rebrikov had already switched to editing the GJB2 gene associated with inherited deafness, and was in the process of selecting couples who would agree to take part in the experiment [60, 61]. However, in numerous interviews with journalists Rebrikov emphasized that he would only conduct such experiments after obtaining all necessary permits from both regulatory and ethical authorities. This significantly distinguished his approach from He’s, who informed the scientific community about the birth of babies with a modified genome post factum. The Ministry of Health of the Russian Federation (following the recommendation of the World Health Organisation) later made a statement that the decision to grant permission for such a study would be premature and irresponsible, which prevented the further development of the situation at least until the situation in the regulatory sphere changes [62].
At the time of writing this review, the state of the legal framework that regulates the issue of genomic editing of human embryonic cells varies greatly in different countries. Thus, genomic modification of embryos for purposes other than reproductive is allowed in at least 11 countries, including China, the U.S., and the U.K. Nineteen countries, including Belarus, Canada, Sweden, and Switzerland, prohibit such experiments. Many other countries (Russia among them) take an intermediate or indeterminate position. The situation with the introduction of inherited genomic changes into embryos subsequently used for reproductive purposes is even more complicated [63].
MEDICAL APPLICATIONS WITH HUMAN SOMATIC CELLS
Despite increased attention to the introduction of heritable changes in germline cells, the less controversial and currently more common use of CRISPR–Cas9 for medical purposes is the modification of human somatic cells. As described above, in the first attempts at gene therapy (1990) an adeno-associated viral vector was used that delivered a healthy copy of the gene into cells (in the U.S. this technology was finally approved for clinical use only in 2017 [64]). The next step in the development of gene therapy was the introduction of genomic editing with the use of Homing Endonucleases (HEs), Zinc Fingers Nucleases (ZFNs), Transcription Activator-Like Effector Nucleases (TALENs), and later also CRISPR–Cas9 [65]. The first human clinical studies using CRISPR–Cas9 commenced in October 2016 in China [66]. The PD-1 gene was inactivated ex vivo in blood cells in the hope that such modified cells, would attack the non-small-cell lung cancer that the patient suffered from when returned to circulation. In the U.S., ex vivo therapy using CRISPR–Cas9 was first performed in July 2019 on a patient with sickle cell anemia (CRISPR Therapeutics, founded by Charpentier). The therapy significantly improved the patient’s condition for at least a few months after the procedure, however the cost of such treatment at the time of its implementation in the United States was estimated to be in the region of 0.5-1.5 million U.S. dollars. The high current cost of CRISPR–Cas9 therapy will probably act as an obstacle to its widescale use, even if clinical trials confirm the efficacy and safety of such treatment [18]. Currently, the most expensive drug on the market is Zolgensma, another gene therapy treatment used for spinal muscular atrophy ($2.125 million per dose). Zolgensma directly delivers a working copy of the defective gene into cells with the use of adeno-associated virus, a method different from genomic editing using nucleases [67].
The first example of an in vivo clinical study in which cells undergo in situ genomic editing with nucleases was performed using the ZFNs technology. Sangamo Therapeutics first performed this procedure in July 2017 on a patient suffering from Hunter syndrome (a rare genetic disease, form of mucopolysaccharidosis). The pioneers in using CRISPR–Cas9 for in vivo genomic editing were Editas Medicine (March 2020) [68]. A drug called EDIT-101 was injected locally into the retina of a patient suffering from a form of inherited blindness caused by a mutation in the CEP290 gene. Currently, various clinical studies are underway on the use of CRISPR–Cas9 for the treatment of diseases such as Alzheimer’s disease, various types of cancers, high cholesterol, angioedema, acute myeloid leukemia, and even androgenetic alopecia (baldness). Another promising application for CRISPR–Cas9 in the future could be the treatment of infectious diseases caused by such pathogens as, for example, HIV and human papillomavirus [65].
CONCLUSIONS
The discovery of CRISPR–Cas9 as an immune system in prokaryotes at the turn of the 20th-21st centuries – a finding at first glance only relevant to microbiology – has led to a revolution in the field of genomic manipulations. New opportunities have opened up in multiple areas of biomedicine, such as molecular diagnostics of infectious and non-infectious diseases (e.g., genotyping of bacterial strains, detection of viruses, and identification of genetic mutations in circulating extracellular DNA in patients with lung cancer [69]), as well as in the development of a potentially new method of immunization, DNA vaccines [18]. One of the more unusual examples of the application of the CRISPR–Cas9 system was the cultivation of brain-like organelles carrying different variants of the important NOVA1 gene characteristic of modern humans, Neanderthals, and Denisovans [70]. The development of CRISPR–Cas9 technology is a good example of how discoveries made in the course of basic research can change entire fields of science and technology, expanding the horizons of the possible. This ground-breaking technique is a worthy continuation of such exciting scientific events as the publication of the double-stranded structure of DNA by Watson and Crick in 1953, the birth of the first child by in vitro fertilization in 1978, and the cloning of Dolly the sheep in 1996. In the coming years the scientific community will watch with interest the development of legislation and ethical principles in the application of the CRISPR–Cas9 system in genome editing, as well as in what other areas of science this promising technology will find its application.
Acknowledgments
The author recalls with warmth and gratitude the years spent in the laboratory of Andrei Dmitrievich Vinogradov at the Department of Biochemistry of Moscow State University. The experiments conceived by Andrei Dmitrievich invariably brought interesting results, while his vast knowledge in various fields of science enabled staff and students to feel confident that any questions would be answered, and the time spent in the laboratory would bring well-deserved results. The publication of the results of the work carried out under the supervision of Andrei Dmitrievich gave the author the necessary start in scientific life and the opportunity to continue research in other laboratories and other fields of knowledge. A unique team of scientists, selected by Andrei Dmitrievich: Vera Georgievna Grivennikova, Tatiana Vadimovna Zharova, and Eleonora Vladimirovna Gavrikova, provided a family atmosphere of trust and support in the laboratory, for which the author is very grateful.
Abbreviations
- cas
CRISPR-associated genes
- CRISPR
clustered regularly interspaced short palindromic repeats
- crRNA
CRISPR-associated RNA
- PAM
protospacer adjacent motif
- sgRNA
single guide RNA
- SpCas9
Cas9 protein from Streptococcus pyogenes
- tracrRNA
trans-activating CRISPR RNA
Ethics declarations
The author declares no conflicts of interest. This article does not contain a description of the studies performed by the author with the participation of people or animals as objects.
References
- 1.Ishino Y., Shinagawa H., Makino K., Amemura M., Nakatura A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isoenzyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987;169:5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Groenen P. M., Bunschoten A. E., van Soolingen D., van Embden J. D. Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis; application for strain differentiation by a novel typing method. Mol. Microbiol. 1993;10:1057–1065. doi: 10.1111/j.1365-2958.1993.tb00976.x. [DOI] [PubMed] [Google Scholar]
- 3.Hoe N., Nakashima K., Grigsby D., Pan X., Dou S. J., Naidich S., et al. Rapid molecular genetic subtyping of serotype M1 group A Streptococcus strains. Emerg. Infect. Diseases. 1999;5:254–263. doi: 10.3201/eid0502.990210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mojica F. J. M., Juez G., Rodriguez‐Valera F. Transcription at different salinities of Haloferax mediterranei sequences adjacent to partially modified PstI sites. Mol. Microbiol. 1993;9:613–621. doi: 10.1111/j.1365-2958.1993.tb01721.x. [DOI] [PubMed] [Google Scholar]
- 5.Mojica F. J. M., Díez-Villaseñor C., García-Martínez J., Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 6.Bolotin A., Quinquis B., Sorokin A., Dusko Ehrlich S. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 7.Pourcel C., Salvignol G., Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 8.Popkov V. A., Zorova L. D., Korvigo I. O., Silachev D. N., Jankauskas S. S., et al. Do mitochondria have an immune system? Biochemistry (Moscow) 2016;81:1229–1236. doi: 10.1134/S0006297916100217. [DOI] [PubMed] [Google Scholar]
- 9.Isaev A. B., Musharova O. S., Severinov K. V. Microbial arsenal of antiviral defenses. Part I. Biochemistry (Moscow) 2021;86:319–337. doi: 10.1134/S0006297921030081. [DOI] [PubMed] [Google Scholar]
- 10.Isaev A. B., Musharova O. S., Severinov K. V. Microbial arsenal of antiviral defenses. Part II. Biochemistry (Moscow) 2021;86:449–470. doi: 10.1134/S0006297921040064. [DOI] [PubMed] [Google Scholar]
- 11.Marraffini L. A., Sontheimer E. J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shmakov S., Abudayyeh O. O., Makarova K. S., Wolf Y. I., Gootenberg J. S., et al. Discovery and functional characterization of diverse class 2 CRISPR–Cas systems. Mol. Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shmakov S., Smargon A., Scott D., Cox D., Pyzocha N. V., et al. Diversity and evolution of class 2 CRISPR–Cas systems. Nat. Rev. Microbiol. 2017;15:169–182. doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abudayyeh O. O., Gootenberg J. S., Konermann S., Joung J., Slaymaker I. M., et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abudayyeh O. O., Gootenberg J. S., Essletzbichler P., Han S., Joung J., et al. RNA targeting with CRISPR–Cas13. Nature. 2017;550:280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 17.Horvath, P., Barrangou, R., Fremaux, C., Boyaval, P., and Romero, D. (2007) Use of a Cas Gene in Combination with CRISPR Repeats for Modulating Resistance in a Cell. US Patent Application No. PCT/US2006/033167 (initially filed on 26.08.2005).
- 18.Isaacson, W. (2021) The Code Breaker: Jennifer Doudna, Gene Editing, and the Future of the Human Race, Simon & Schuster (New York, USA).
- 19.Masepohl B., Görlitz K., Böhme H. Long tandemly repeated repetitive (LTRR) sequences in the filamentous cyanobacterium Anabaena sp. PCC 7120. Biochim. Biophys. Acta Gene Struct. Expr. 1996;1307:26–30. doi: 10.1016/0167-4781(96)00040-1. [DOI] [PubMed] [Google Scholar]
- 20.Flamand M.-C., Goblet J.-P., Duc G. r., Briquet M., Boutry M. Sequence and transcription analysis of mitochondrial plasmids isolated from cytoplasmic male-sterile lines of Vicia faba. Plant Mol. Biol. 1992;19:913–923. doi: 10.1007/BF00040524. [DOI] [PubMed] [Google Scholar]
- 21.Mojica F. J. M., Díez-Villaseñor C., Soria E., Juez G. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol. Microbiol. 2000;36:244–246. doi: 10.1046/j.1365-2958.2000.01838.x. [DOI] [PubMed] [Google Scholar]
- 22.Jansen R., Embden J. D. A. v., Gaastra W., Schouls L. M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 23.Makarova K. S., Aravind L., Grishin N. V., Rogozin I. B., Koonin E. V. A DNA repair system specific for thermophilic Archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res. 2002;30:482–496. doi: 10.1093/nar/30.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shmakov S. A., Makarova K. S., Wolf Y. I., Severinov K. V., Koonin E. V. Systematic prediction of genes functionally linked to CRISPR–Cas systems by gene neighborhood analysis. Proc. Natl. Acad. Sci. USA. 2018;115:E5307–E5316. doi: 10.1073/pnas.1803440115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makarova K. S., Wolf Y. I., Iranzo J., Shmakov S. A., Alkhnbashi O. S., et al. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020;18:67–83. doi: 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makarova K. S., Wolf Y. I., Koonin E. V. The basic building blocks and evolution of CRISPR–Cas systems. Biochem. Soc. Trans. 2013;41:1392–1400. doi: 10.1042/BST20130038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koonin E. V., Makarova K. S., Zhang F. Diversity, classification and evolution of CRISPR–Cas systems. Curr. Opin. Microbiol. 2017;37:67–78. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koonin E. V., Makarova K. S. Origins and evolution of CRISPR–Cas systems. Philos. Trans. R. Soc. B Biol. Sci. 2019;374:20180087. doi: 10.1098/rstb.2018.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mojica F. J. M., Díez-Villaseñor C., García-Martínez J., Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 30.Brouns S. J. J., Jore M. M., Lundgren M., Westra E. R., Slijkhuis R. J., et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gasiunas G., Barrangou R., Horvath P., Siksnys V. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deltcheva E., Chylinski K., Sharma C. M., Gonzales K., Chao Y., et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mali P., Yang L., Esvelt K. M., Aach J., Guell M., et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cong L., Ran F. A., Cox D., Lin S., Barretto R., et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:1–9. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cho S. W., Kim S., Kim J. M., Kim J. S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 38.Hwang W. Y., Fu Y., Reyon D., Maeder M. L., Tsai S. Q., et al. Efficient genome editing in zebrafish using a CRISPR–Cas system. Nat. Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jinek M., Jiang F., Taylor D. W., Sternberg S. H., Kaya E., et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishimasu H., Ran F. A., Hsu P. D., Konermann S., Shehata S. I., et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang F., Taylor D. W., Chen J. S., Kornfeld J. E., Zhou K., et al. Structures of a CRISPR–Cas9 R-loop complex primed for DNA cleavage. Science. 2016;351:867–871. doi: 10.1126/science.aad8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jinek, M., Doudna, J., Charpentier, E., and Chylinski, K. (2013) Methods and Composition for RNA-Directed Target DNA Modification and for RNA-Directed Modulation of Transcription. US Patent Application No. 61/652,086 (initially filed on 25.05.2012).
- 43.Zhang, F. (2014) CRISPR–Cas9 Systems and Methods for Altering Expression of Gene Products. US Patent No. 8,697,359 (provisional application No. 61/736,527 filed on 12.12.2012).
- 44.Campbell, M. (2022) Broad Institute Wins CRISPR Patent Case, Technology Networks. Genomics Research, Published online on 2 March 2022.
- 45.Stolberg, S. G. (1999) The biotech death of Jesse Gelsinger, New York Times Magazine, 136-150. [PubMed]
- 46.Baltimore D., Berg P., Botchan M., Carroll D., Charo R. A., et al. A prudent path forward for genomic engineering and germline gene modification. Science. 2015;348:36–38. doi: 10.1126/science.aab1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lanphier E., Urnov F., Haecker S. E., Werner M., et al. Don’t edit the human germ line. Nature. 2015;519:410–411. doi: 10.1038/519410a. [DOI] [PubMed] [Google Scholar]
- 48.Liang P., Xu Y., Zhang X., Ding C., Huang R., et al. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell. 2015;6:363–372. doi: 10.1007/s13238-015-0153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cyranoski D., Reardon S. Embryo editing sparks epic debate. Nature. 2015;520:593–594. doi: 10.1038/520593a. [DOI] [PubMed] [Google Scholar]
- 50.He J., Deem M. W. Heterogeneous diversity of spacers within CRISPR (clustered regularly interspaced short palindromic repeats) Phys. Rev. Lett. 2010;105:128102. doi: 10.1103/PhysRevLett.105.128102. [DOI] [PubMed] [Google Scholar]
- 51.He J., Deem M. W. Low-dimensional clustering detects incipient dominant influenza strain clusters. PEDS. 2010;23:935–946. doi: 10.1093/protein/gzq078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cyranoski D. Direct genomics revives Helicos sequencing system for China’s hospitals. Nat. Biotechnol. 2016;34:122–123. doi: 10.1038/nbt0216-122b. [DOI] [PubMed] [Google Scholar]
- 53.Kirksey, E. (2021) The Mutant Project. Inside the Global Race to Genetically Modify Humans, Bristol University Press, U.K.
- 54.Regalado, A. (2019) China’s CRISPR babies: read exclusive excerpts from the unseen original research, MIT Technology Review. Biotechnology.
- 55.Regalado, A. (2019) Why the paper on the CRISPR babies stayed secret for so long, MIT Technology Review. Biotechnology.
- 56.Musunuru, K. (2019) The CRISPR Generation: The Story of the World’s First Gene-Edited Babies, BookBaby, N.J., U.S.A.
- 57.Regalado, A. (2022) The creator of the CRISPR babies has been released from a Chinese prison, MIT Technology Review. Biotechnology.
- 58.Cyranoski D. Russian biologist plans more CRISPR-edited babies. Nature. 2019;570:145–146. doi: 10.1038/d41586-019-01770-x. [DOI] [PubMed] [Google Scholar]
- 59.Kodyleva T. A., Kirillova A. O., Tyschik E. A., Makarov V. V., Khromov A. V., et al. The efficacy of CRISPR-Cas9-mediated induction of the CCR5delta32 mutation in the human embryo. Bull. RSMU. 2018;4:70–74. doi: 10.24075/brsmu.2018.052. [DOI] [Google Scholar]
- 60.Cohen, J. (2019) Embattled Russian scientist sharpens plans to create gene-edited babies, Science. News. Published online on 21 Oct 2019, 10.1126/science.aaz9337.
- 61.Cyranoski D. Russian “CRISPR-baby” scientist has started editing genes in human eggs with goal of altering deaf gene. Nature. 2019;574:465–466. doi: 10.1038/d41586-019-03018-0. [DOI] [PubMed] [Google Scholar]
- 62.Meyer M. The CRISPR babies controversy: responsibility and regulation in the spotlight. EMBO Rep. 2020;21:e50307. doi: 10.15252/embr.202050307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baylis F., Darnovsky M., Hasson K., Krahn T. M. Human germline and heritable genome editing: the global policy landscape. CRISPR J. 2020;3:365–377. doi: 10.1089/crispr.2020.0082. [DOI] [PubMed] [Google Scholar]
- 64.Smalley E. First AAV gene therapy poised for landmark approval. Nat. Biotechnol. 2017;35:998–999. doi: 10.1038/nbt1117-998. [DOI] [PubMed] [Google Scholar]
- 65.Porteus M. H. A new class of medicines through DNA editing. N. Engl. J. Med. 2019;380:947–959. doi: 10.1056/NEJMra1800729. [DOI] [PubMed] [Google Scholar]
- 66.Cyranoski D. CRISPR gene-editing tested in a person for the first time. Nature. 2016;539:479–479. doi: 10.1038/nature.2016.20988. [DOI] [PubMed] [Google Scholar]
- 67.Editorial Gene therapy’s next installment. Nat. Biotechnol. 2019;37:697. doi: 10.1038/s41587-019-0194-z. [DOI] [PubMed] [Google Scholar]
- 68.Ledford H. CRISPR treatment inserted directly into the body for first time. Nature. 2020;579:185. doi: 10.1038/d41586-020-00655-8. [DOI] [PubMed] [Google Scholar]
- 69.Chertow D. S. Next-generation diagnostics with CRISPR. Science. 2018;360:381–382. doi: 10.1126/science.aat4982. [DOI] [PubMed] [Google Scholar]
- 70.Remmel A. Neanderthal-like “mini-brains” created in lab with CRISPR. Nature. 2021;590:376–377. doi: 10.1038/d41586-021-00388-2. [DOI] [PubMed] [Google Scholar]