Abstract
Oncolytic adenoviruses (OAd) represent an attractive treatment option for cancer. Clinical efficacy of commonly utilized human adenovirus type 5 (Ad5)-based oncolytic viruses is limited by variable expression levels of the coxsackie- and adenovirus receptor (CAR) in tumor cells and high prevalence of neutralizing antibodies against human Ad5. However, previous studies have highlighted alternative human Ad types as promising candidates for oncolytic therapy. In this study, we generated novel OAds based on Ad1, -2, -5, and -6 derived from species C Ads. These OAds contain a 24-bp deletion in the early gene E1A for tumor selective replication and express the RNAi inhibitor P19. We examined these OAds for in vitro anticancer activity on various cancer cell lines derived from lung, colon, gynecologic, bone, and pancreatic carcinoma. In most surveyed cell lines, OAds based on Ad1, -2, and -6 demonstrated higher cell lysis capability compared with Ad5, suggesting enhanced oncolytic potential. Moreover, enhanced oncolytic activity was associated with P19 expression in a cell type–dependent manner. We further explored a A549 tumor xenograft mouse model to compare the novel OAds directly with Ad5 and H101, an oncolytic adenovirus used in clinical trials. These P19-containing OAds based on Ad1, -2, and -6 showed significantly decelerated tumor progression compared with H101, indicating better antitumor potency in vivo. Our studies provide a novel path for OAd development based on alternative Ad types with improved effectiveness by RNA interference suppression.
Introduction
Standard cancer therapy usually combines different treatment options including surgery, chemo-, and radiotherapy and increasingly targeted therapy. Viruses are a promising treatment modality as they have a working mechanism distinct from existing treatments, including targeted therapy. Oncolytic adenoviruses (OAd) are a promising novel treatment option for many human cancers. They target cancer cells by direct lysis and an indirect immunostimulatory effect (1). OAds have shown to be safe in numerous clinical trials and demonstrated modest antitumor activity in different cancers as single agents and an improved treatment efficacy in combination therapy (2). Despite some remarkable successes, concerns remain that need to be addressed.
First, viruses are immunogenic and previous infection with the same Ad type as the therapeutic virus triggers the generation of neutralizing antibodies, precluding systemic treatment with the same virus. Most OAds existing today are based on adenovirus type 5 (HAd5). Studies revealed high preexisting antibody prevalence for HAd5, ranging from 25% in a cohort of individuals from sub-Saharan Africa to 100% in children with cystic fibrosis (3, 4). In contrast, the seroprevalence for HAd6 was reported between 2% and 15%, the least among species C adenoviruses (4, 5). Generating oncolytic viruses based on less seroprevalent types should result in reduced preexisting immunity and therefore improve systemic therapy. Moreover, the availability of similar oncolytic viruses harboring different Ad type-specific surface molecules may allow repeated administration of these viruses in an immunocompetent patient.
Secondly, Ad types were shown to display varying antitumor effects in different tumor cells (6–8). Up to date, 103 human Ad types (Ad1 to Ad103) have been identified (http://hadvwg.gmu.edu/) and classified into seven species (A to G). Previous work demonstrated for instance that internalization efficiencies differ for the different Ad types in melanoma cells (8) and in breast cancer cells (9). Therefore, a library of several OAds based on multiple Ad types will allow screening for optimal antitumor effects for any cancer subtype.
As a third point, it needs to be mentioned that current oncolytic viruses, although having excellent safety and specificity, lack the ability in achieving long-term remission. Strategies intended at improving antitumor efficacy have focused on stimulating antitumor immunity, angiostasis, or direct tumor-killing effects (10). Inhibiting cellular antiviral mechanisms by RNAi suppression is a novel mechanism that was shown to be successful in inhibiting antiviral cellular response in insect cells (11, 12). RNAi suppression describes a mechanism that inhibits the innate antiviral response mediated by RNA molecules that lead to degradation of viral mRNA molecules. P19 is a potent RNAi inhibitor derived from a plant virus that was shown to successfully cleave miRNA, resulting in improved viral replication (13). P19-expression was shown to enhance virus replication for adenovirus and baculovirus (11, 14) and it was speculated that it may improve the oncolytic effect if used to arm oncolytic viruses.
Here we generated P19-expressing, selectively replicating (oncolytic) adenoviruses based on HAd1, -2, -5, and -6 by a novel high-throughput recombineering technique (15). We show that Ad types 1-, 2-, and 6-based viruses display higher cell lysis capacity in several tumor cell lines of different origins. Furthermore, the expression of P19 resulted in up to 10-fold higher oncolytic capacity in cell culture assays compared with control viruses lacking P19 in cell culture assays. Expression of P19 translated into significantly higher replication rates in lung cancer cells. Finally, treatment with P19 expressing oncolytic Ad based on types 1, 2, and 6 resulted in significantly reduced tumor growth and improved survival compared with existing oncolytic virus H101 (16) in a tumor xenograft lung cancer mouse model. Further studies will have to show whether using a combination or repeated application of viruses based on different Ad types will improve outcomes in an immunocompetent animal model compared with single-agent treatment.
Materials and Methods
Vector cloning procedures
Previously published cloned species C Human Ad (Ad1, Ad2, Ad5, Ad6; ref. 15) referring to the plasmids p15A-cm-Ad1, p15A-cm-Ad2, p15A-cm-Ad5, and p15A-cm-Ad6 were used to construct the OAds. The pCR2.1 BluntII-Topo based plasmid pFIPP was used to clone P19 containing viruses derived from HAd species C (AdC.P19). This plasmid was used as a donor of the FIPP cassette containing a 256-bp homology arm to the 3′ end of the hAd5 fiber gene, an encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES), p19, and a poly A signal were previously published (14). PFIPP was used as template for amplification of the FIPP cassette to introduce P19 into the fiber region of all species C HAd. All recombineering steps were performed as described before (17). The plasmid p15A-amp-ccdB (15) was used as a PCR template to amplify the ccdB-Amp cassette for counterselection for all recombineering steps. Prior to the PCR reaction, BseRI digestion was used to excise a segment of 43 bp from the vector backbone to prevent restoration of the plasmid.
All PCR steps for recombineering were performed using Phusion High-Fidelity DNA Polymerase (New England Biolabs) following the manufacturer's instructions. The PCR reaction was carried out with an initial denaturation step of 98°C for 30 seconds followed by 30 cycles of denaturation at 98°C for 10 seconds, annealing at 56°C for 30 seconds, and elongation at 72°C for 40 seconds. A final elongation step was conducted at 72°C for 5 minutes. Primers used to amplify the amp-ccdB cassette including the respective homology arms for recombineering are listed in Supplementary Table S1. The PCR product was then analyzed by agarose gel electrophoresis and column purified. Next, electro-competent cells for recombineering were prepared as described previously (17). The intermediate cloning vectors p15A-cm-amp-ccdB-Ad1, -2, -5, and -6 were generated via coelectroporation of the PCR fragments with the corresponding homology arms into the RecET expressing E. coli strain GBdir-gyrA462, which has the gyrAArg462Cys mutation rendering Gyrase A resistant to ccdB toxicity. After recovery, a suitable number of colonies was grown overnight in selective medium at 37°C and 900 rpm in 2mL microcentrifuge tubes. Then, plasmid DNA was isolated according to a standard protocol described elsewhere (http://www.biotec.tu-dresden.de/research/stewart/group-page/recombineering-guide.html) and subjected to PCR and restriction enzyme analysis. If confirmed by these methods, it was retransformed to remove remaining original plasmid and confirmed by sequencing.
For the second recombineering (counterselection) step to replace the amp-ccdB cassette with the P19 encoding sequence, pSC101-ccdA-gb was used instead of GBdir-gyrA462 as omission of Redα expression results in less intramolecular recombination (18). SphI digested and column-purified pFIPP was used as a PCR template to amplify the FIPP cassette with homology arms to the accepting intermediate plasmids of approximately 60 bp in length. E. coli containing the thermolabile pSC101-ccdA-gb that expresses the λ phage redβ and redγ genes under the control of the rhamnose-inducible PRhaB promoter were induced by adding rhamonose and made electrocompetent according to the standard protocol. The resulting plasmids p15A-cm-FIPP-AdC1, -2, and -6 were generated via coelectroporation of the PCR fragments with the corresponding homology arms into the electrocompetent E. coli containing the recombinase expressing pSC101-ccdA-gb plasmid. For generation of p15a-cm-FIPP-AdC5, the SphI-digested and column purified plasmid was used as donor plasmid of the FIPP cassette as it contains homology arms to HAd-5. After recovery at 37°C for 2hours, transformed cells were concentrated by centrifugation at 9,000 rpm for 30 seconds and then entirely transferred on chloramphenicol (Cm) containing Luria broth (LB) plates. Plates were incubated overnight at 37°C and clones subsequently picked, expanded, and DNA-purified for further analysis by PCR and restriction enzyme digest. Positive clones were then retransformed to purge remaining donor plasmid, midiprep was performed using the PureYield Plasmid Midiprep System (Promega), and clones were confirmed by sequencing. This resulted in the plasmids p15A-cm-FIPP-AdC1, -2, -5, and -6 referring to viruses Ad1.P19, Ad2.P19, Ad5.P19, and Ad6.P19 (Fig. 1).
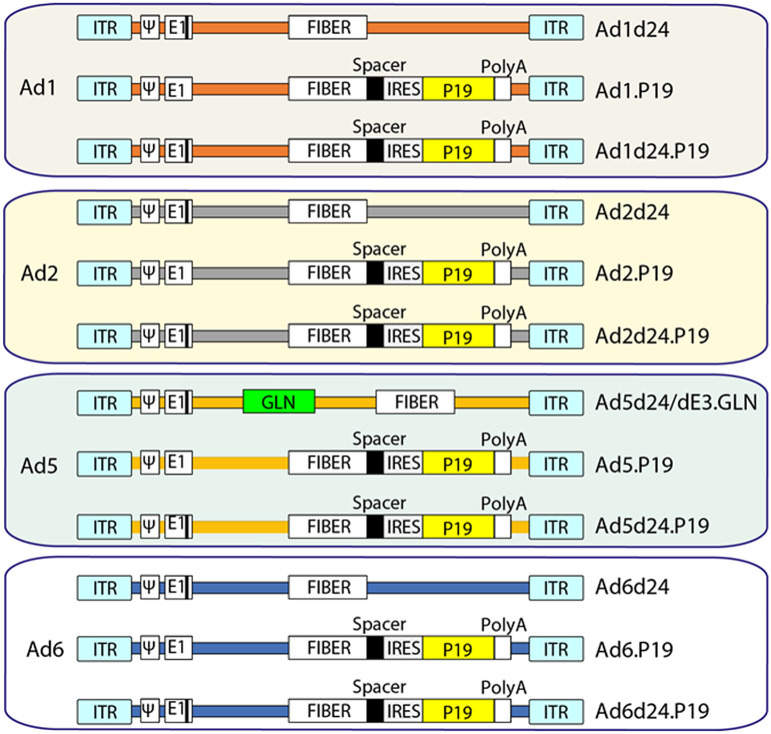
Figure 1.
Generation of eight OAds. Two groups of OAds were generated: 1, OAds containing the 24-bp deletion in the adenoviral early region E1A; 2, OAds with the 24-bp deletion as (1) and/or in addition express the RNAi inhibitor P19. The P19 is under the control of the major late promoter and is connected to the fiber via a spacer and an IRES and the SV40 polyadenylation signal (polyA). IRES: internal ribosome entry site, ITR: invert terminal repeat, 4: packaging signal.
The plasmids containing the Ad genomes carrying the 24-bp deletion of the pRB binding region in E1A (p15A-cm-AdCd24 and p15A-cm-AdCd24.P19) were generated as follows. In the first step, the 24 bp to be deleted were replaced with the amp-ccdB cassette by creating a PCR product using the primer pair ccdB-AdC-E1A-HR3′ and ccdB-AdC-E1A-HR5′. For the second step, viral DNA purified from Ad5d24.Luc was used as a template to create a PCR product of 555-bp length comprising the area of the AdC E1A gene including the deleted pRB binding region. Primer pair AdC_E1A5′ and AdC_E1A3′ was used for this purpose. Then, the amp-ccdB cassette was replaced with the PCR product in the second counterselection step. The same primer pair was used to screen for clones including the 24-bp deletion, resulting in a slightly but visibly larger fragment for plasmids with wild-type (WT) E1A. BssSI was used to confirm the 24-bp deletion as one of its cutting sites lies within the deleted sequence. This resulted in the final plasmids p15A-cm-Ad1d24, p15A-cm-Ad1d24.P19, p15A-cm-Ad2d24, p15A-cm-Ad2d24.P19, p15A-cm-Ad5d24.P19, p15A-cm-Ad6d24, and p15A-cm-Ad6d24.P19 referring to the viruses Ad1d24, Ad2d24, Ad5d24.P19, Ad6d24, Ad1d24.P19, Ad2d24.P19, and Ad6d24.P19.
For Ad5 carrying the 24-bp deletion without P19, the plasmid p15A-cm-Ad5/dE3.GLN was used containing a TurboGFP/NanoLuc luciferase/neomycin-kanamycin (GLN) cassette encoding eGFP, luciferase, and neomycin cassette in the E3 region based on a previously published plasmid (pAd5d24/dE3.GLN; ref. 15), resulting in the virus Ad5d24/dE3.GLN. All plasmids, E. coli strains, and viruses used in this study are outlined in detail in Supplementary Table S2. All plasmid maps used in this study were created with SnapGene Vector mapping software.
Plasmid DNA transfection and virus rescue
Plasmids containing the complete sequence of the adenoviral genome were linearized by restriction enzyme digest. Linearized virus genomes were then transfected into HEK293 cells, reconstituted, and purified. Approximately 50 μg of the p15A-based adenovirus genome containing plasmids were digested with either PacI (HAd-5 genome containing plasmids except for Ad5-dE3/GLN), I-SceI (Ad1, -2, and Ad5-dE3/GLN genome containing plasmids), or AsiSI & I-CeuI (HAd-6 genome containing plasmids) to release the complete adenovirus genome, or to linearize the p15A-based adenovirus genome containing plasmid. To purify and concentrate digested DNA, phenol/chloroform extraction followed by ethanol precipitation was performed for digested DNA. For virus rescue HEK293 cells were plated in 6-cm cell culture dishes and at 50% to 80% confluency transfected with 6 to 12 μg of digested viral DNA using a calcium-phosphate based standard method. After 24 to 48 hours, when the cells grew to up to more than 90% confluency, the medium was changed to 2% FBS-supplemented DMEM. Transfected cells were maintained for up to 2 weeks, until cytopathic effect (CPE) was observed. When CPE was visible, the cell lysate was collected and used to infect a new well of cells at a confluency of 90% to 95%. To release the virus from infected cells, the crude lysate was subjected to three freeze/thaw cycles in liquid nitrogen and in a 37°C water bath respectively. A small aliquot of lysate was collected for PCR analysis. As most viruses did not express a marker protein, PCR analysis of purified Viral DNA (vDNA) was performed using virus specific primer pairs. vDNA isolation was performed as outlined elsewhere (19). Serial infection circles to achieve 90% CPE were then performed for each virus. After a first passage to another 6-cm cell culture plate that resulted in 100% CPE after 3 to 7 days, 90% confluent cells in a 15-cm tissue culture dish were infected. From that stage, rescue was upscaled to 2 to 3 15-cm tissue culture dishes and finally to 10 or 20 15-cm tissue culture dishes. For the last amplification step, DMEM supplemented with 5% FBS was used. —Forty-eight to 72 hours postinfection, when 80% to 90% CPE was visible, cells were harvested and purified by a cesium chloride (CsCl) gradient-based ultracentrifugation method, followed by dialysis in a Slide-A-Lyzer dialysis cassette (Thermo Fisher Scientific), resulting in a final volume of approximately 1,5 mL virus preparation (19). Finally, sterile glycerol was added to a concentration of 10% and the virus preparations were aliquoted and stored at −80°C.
Titration and characterization of recombinant viruses
We determined the physical titer of final Ad preparations by optical density measurements. For the quantification of viral preparations, the amount of optical particle units (OPU) present in 1 mL of concentrated virus [also referred to as optical density (OD) titer] was determined as described by (19). Briefly, 10 μL of final vector preparation was diluted with 90 μL of lysis buffer [10 mmol/L Tris-HCl (pH 7.5), 10 mmol/L EDTA (pH 8.0), 0.5% SDS] and incubated at room temperature with shaking at 1,000 rpm for 20 minutes. Then, the absorbance of the sample at 260 nm was measured using an Eppendorf BioPhotometer plus (Eppendorf). Optical titer was then calculated using the following formula: OPU ml−1 = (absorbance at 260 nm) × (dilution factor) × (1.1 × 1012) × (36)/(size of Ad in kb).
To further characterize viral particles, we isolated Ad DNA from purified viral particles and performed restriction enzyme digests to confirm Ad viral genome integrity. To purify virus DNA, 100 to 150 μL of virus preparation was incubated with 200 to 250 μL of proteinase-K-SDS-TE solution [proteinase K (0.5 mg/mL), 10 mmol/L Tris-HCl (pH 7.5), 0.5% SDS, 10 mmol/L EDTA (pH 8.0)] at 56°C and shaking at 500 rpm for 2 hours. Then, lysate was purified by phenol/chlorophorm/isoamylalcohol (25:24:1; Roth) extraction followed by ethanol precipitation. After dissolution in 25 μL of water, 2 μL of dissolved DNA was used for overnight restriction digest with the enzymes BssSI and HindIII (New England BioLabs). The DNA fragments were then separated by agarose gel electrophoresis.
Cell culture procedures
Human embryonal kidney cells (HEK293) were obtained from the ATCC. Human lung carcinoma (A549) cells were supplied by Carsten Rudolph (Ludwig-Maximilians-University Munich, Munich, Germany) and certified by PCR-single-locus-technology for authentication (Eurofins Genomics Germany GmbH). MIA PaCa-2 and PANC-1 pancreas ductal adenocarcinoma cell lines were kindly provided by Antonio Marchini (German Cancer Research Center, Heidelberg, Germany). Caco-2 human epithelial colorectal adenocarcinoma cells were provided by Jan Postberg (Department of Pediatrics, Helios University Hospital Wuppertal, Germany) and human osteosarcoma cell lines Saos-2 and U-2 OS were provided by Claudia Hagedorn (Chair for Biochemistry and Molecular Medicine, Center for Biomedical Education and Research, Witten/Herdecke University, Witten, Germany). All cell lines were tested for presence of Mycoplasma using a PCR-based assay (Minerva Biolabs). All cell lines were cultured at 37°C in a humidified atmosphere with 5% CO2. HEK293, A549, PANC-1, MIA PaCa-2, and Huh7 hepatocellular carcinoma cells cell lines were cultured in DMEM medium, Saos-2, and U2OS cells in McCoy's 5A Modified Medium and Caco-2 cells in minimum essential medium (MEM), all supplemented with 10% FBS and 1% penicillin-streptomycin. Huh-7 cells were also supplemented with 1% nonessential amino acids (NEAA). Cell culture reagents were obtained from PAN Biotech and plasticware from Sarstedt.
Oncolysis assays
We used various cancer cell lines from different origin to perform oncolysis assays. A549, PANC-1, MIA-PaCa-2, Caco-2, Huh-7, Saos-2, and U-2 OS cells were seeded in 24-well tissue culture dishes and infected with Ad1d24, Ad2d24 Ad5d24/E3.GLN, or Ad6d24 and Ad1d24.P19, Ad2d24.P19, Ad5d24.P19, and Ad6d24.P19. For comparison of the first-generation OAd (selectively replicating WT Ad containing a 24-bp deletion in E1A), viral particle (vp) to cell ratios ranging from 10,000 to 0.1 were used, with 10 times serial dilutions at each step. For the P19-expressing, selectively replicating Ad, vp to cell ratios of 1,000, 100, 10, and 1 were used to infect the cells. Briefly, viruses were normalized to similar concentrations and diluted with elution buffer (50 mmol/L HEPES, 150 mmol/L NaCl, 10% glycerol). Plates were monitored daily for cytopathic effect, and the assay was terminated when total cytolysis was observed at a multiplicity of infection (MOI) of 0.1 with one of the viruses. Cells were fixed with phosphate buffered 10% formalin solution and stained with 0.5% crystal violet solution (0,5% crystal violet, 25% methanol, 75% H2O). Plates were then dried, and pictures taken with a digital camera.
Analysis of adenovirus replication in A549 cells
A549 cells were seeded in 24-well plates and infected at a confluency of 80% to 90% with the different OAd at 100 vp/cell. Infections were performed in triplicates. After 2 hours, the complete medium including the virus (500 μL) was harvested to analyze genome copy numbers at baseline and replaced with fresh DMEM, supplemented with 2% FBS. Medium including the released viral progeny was replaced completely every 24 hours, and starting at 48 hours, 250 μL of the growth medium was stored at −80°C for subsequent analysis. Thus, each sample contained the viral progeny released into the growth medium during the over the 24 hours before harvest. The experiment was terminated after 5 days when cells started to show significant cytopathic effect. If cytopathic effect was observed, the growth medium including detached cells was centrifuged at 3,500 g for 1 minute to pellet cells and only 250 μL of the supernatant were used for analysis. vDNA was then isolated from the growth medium as described before (19). Briefly, the growth medium was incubated with an equal amount of proteinase K -SDS-Solution [proteinase K, 0.5 mg/mL), 10 mmol/L Tris-HCl (pH 7.5), 0.5% SDS, 10 mmol/L EDTA (pH 8.0)] at 56°C, 1,000 rpm for 2 hours. Then, vDNA was precipitated by adding 30 μL of 3 mol/L sodium acetate (pH 5) and 750 μL of EtOH (>99.8%, stored at −20°C). After centrifugation for 8 minutes at full speed, DNA pellet was washed with 700 μL of 70% ethanol, dried and dissolved in 25 μL DNAse free H2O. For analysis of viral DNA copy numbers 2.5 μL of vDNA was used for analysis. We performed qRT-PCR analysis (Bio-Rad real time thermal cycler CFX96TM; Bio-Rad) to evaluate viral DNA copy numbers using consensus primers and dual-labeled fluorescent probes. The qRT-PCR was performed in a total volume of 10 μL using 5 μL SSo Advanced Universal Probes Supermix (Bio-Rad Laboratories), 300 nmol/L of each primer, 200 nmol/L of probe, and 2.5 μL vDNA. PCR was carried out with an initial denaturation step of 95°C for 3 minutes followed by 40 cycles of denaturation at 95°C for 15 seconds, annealing and elongation at 60°C for 30 seconds. To generate a standard curve DNA derived from the plasmid p15a-cm-hAd6 was used. According to the standard curve the copy numbers of vDNA from the different samples was interpolated.
Animal experiments
All experiments involving animals were conducted in accordance with the institutional guidelines of the University of Washington (Seattle, WA). The University of Washington is an Association for the Assessment and Accreditation of Laboratory Animal Care International (AALAC)–accredited research institution and all live animal work conducted at this university is in accordance with the Office of Laboratory Animal Welfare (OLAW) Public Health Assurance (PHS) policy, U.S. Department of Agriculture (USDA) Animal Welfare Act and Regulations, the Guide for the Care and Use of Laboratory Animals, and the University of Washington's Institutional Animal Care and Use Committee (IACUC) policies. The studies were approved by the University of Washington IACUC. Immunodeficient NOD.CB17-Prkdcscid/J (CB17) mice were obtained from the Jackson Laboratory. Mice were housed in specific-pathogen-free facilities. A549 xenograft tumors were established by subcutaneous injection of 2 × 106 cells. Ad (4 × 1010 vp/mouse) was injected intratumorally. Tumor volumes were calculated using the formula [largest diameter x (smallest diameter)2]. Animals were sacrificed, and the experiment terminated when tumors reached a volume of 1,000 mm3 or when tumors displayed ulceration.
Analytical and statistical methods
Statistical significance of in vivo data was calculated by the two-sided Student t test (Microsoft Excel). P values > 0.05 were considered not statistically significant.
Results
Generation and production of selectively replicating adenoviruses expressing p19
To examine, whether OAds based on other serotypes than type 5 have improved oncolytic potential, we generated selectively replicating adenoviruses based on Ad types 1, 2, and 6 and compared it with HAd type 5. Tumor selectivity was achieved by deleting a 24-bp segment of the pRB binding region in the early Ad gene E1A. As E1A in species C Ads are identical in analyzed Ad sequences, the deletion was achieved applying a high-throughput cloning strategy using identical primer pairs for cloning in all viruses. The resulting viruses Ad1d24, Ad2d24, and Ad6d24 had no further alterations from their WT genome sequence except for the 24-bp E1A deletion (Fig. 1; Supplementary Fig. S1). For Ad5carrying the 24-bp deletion, we used a previously published construct in which E3 was replaced by a GLN cassette encoding luciferase, eGFP, and neomycin (15) resulting in the virus Ad5d24/dE3.GLN. In a further step, the RNAi inhibitor P19 was included to the fiber region to increase the oncolytic effect by enhancing virus replication, as was previously shown in Ad5 (14).
The viruses lacking the pRB binding site showed comparable cytopathic effect during reconstruction compared with WT viruses and could be purified to similar titers. Virus genome integrity was then confirmed using vDNA purification, restriction enzyme digest, and agarose gel electrophoresis (Supplementary Fig. S2). Digested DNA patterns were predicted according to vDNA sequences published previously (15). Potential recombination of viral E1A with E1A from the producer cell line that has a 4kb-fragment containing early adenoviral genes stably integrated was excluded by restriction enzyme digest of vDNA (Supplementary Fig. S2), PCR spanning a small segment including the deleted 24-bp sequence (Supplementary Fig. S3), and DNA sequencing.
Tumor-selective OAds based on serotypes 1, 2, and 6 show enhanced oncolytic potential in different cancer cell lines compared with Ad5
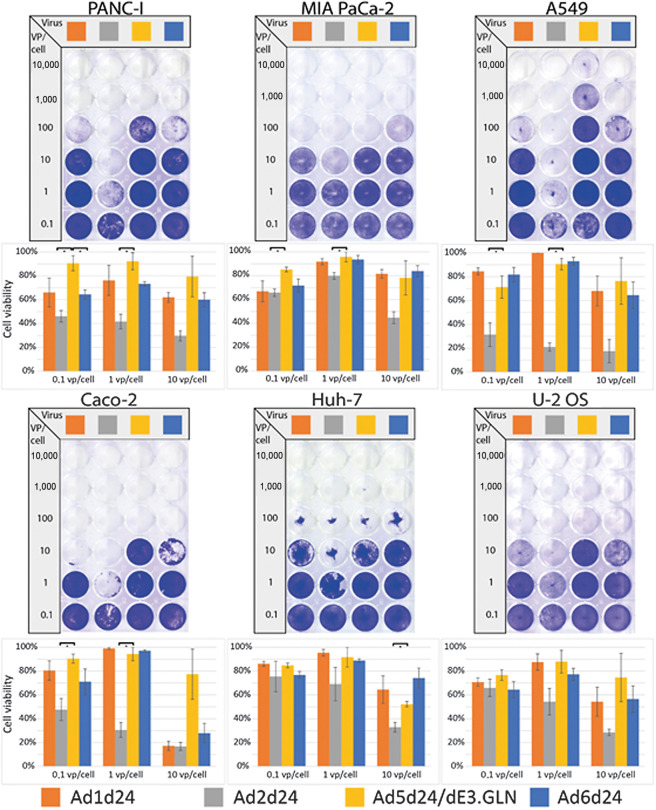
After titration, the oncolytic effect of the different viruses was determined by head-to-head comparison of the viruses in different tumor cell lines. All cell lines were tested for coxsackie- and adenovirus receptor (CAR) expression levels, the major entry receptor for species C adenoviruses (Supplementary Table S3). In most cell lines tested, Ad2 showed an increased oncolytic effect compared with Ad5 (Fig. 2). We also observed an increased oncolytic effect for Ad6 in Panc-I, but not in MIA PaCa-2 pancreatic cancer cells. Ad1 and Ad6 also showed a trend towards improved oncolysis in most cell lines, including PANC-I, A549, U-2 OS, and Caco-2 cells (both lung cancer), although not statistically significant. The most notable difference in oncolytic effect between Ad2 and Ad5 was observed in A549 lung cancer cells, where a 100 to 1,000 times lower dose of Ad2d24 resulted in similar cell lysis as Ad5d24/E3.GLN. These results underline the potential role of Ad2 and Ad6 as oncolytic viruses for many cancers. Moreover, a standardized cloning procedure resulted in tumor selective oncolytic viruses with the identical pRB binding defect but potentially different serologic and biologic features.
Figure 2.
Evaluation of selectively replicating oncolytic viruses based on group C Ad in vitro. Comparison of oncolytic effect of selectively replicating Ad based on Ad 1, 2, 5, and 6 carrying the 24-bp deletion in vitro. To perform an oncolysis assay, cancer cell lines representing different primary solid tumors were infected and stained after 5 to 7days. Representative slides are displayed. Oncolytic effect was quantified by measuring intensity and is displayed as cell viability 5 to 7 days after infection with different vp per cell ratios. Ad2-based oncolytic viruses showed significantly improved oncolytic effect in all cell lines except U2-OS. In PANC-I cells Ad 6 showed significantly enhanced oncolytic effect compared with Ad5. *, P < 0.05. Mean of three biological replicates per group is shown.
The RNAi suppressor P19 enhances oncolytic effect in vitro
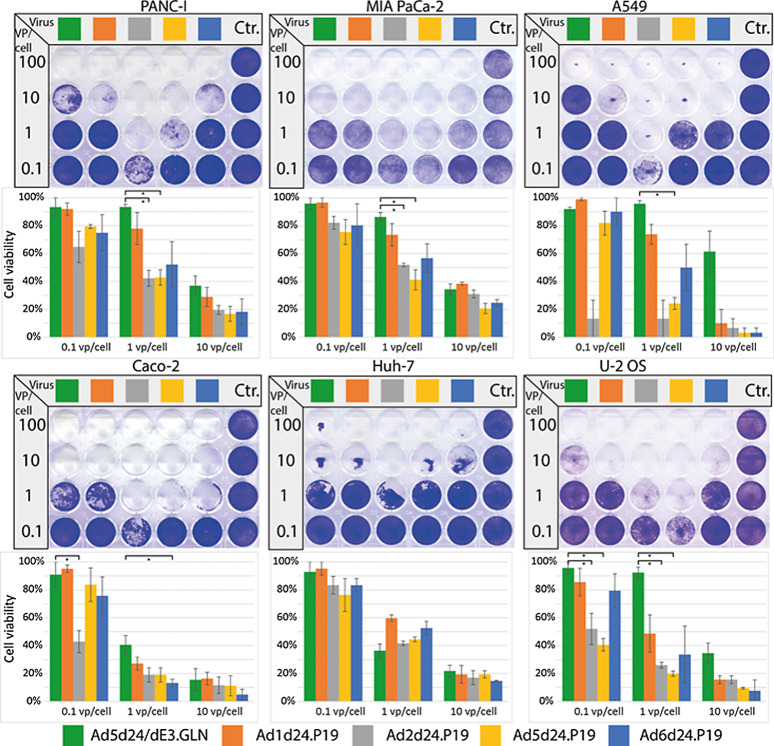
To further improve the oncolytic effects of the viruses based on previously unexploited group C Ad, we generated tumor-selective oncolytic Ad based on human serotypes 1, 2, 5, and 6 that express the RNAi suppressor protein P19, which is derived from the tomato bushy stunt virus. Previous experiments have shown that the expression of P19 resulted in significantly higher replication efficiency in otherwise unaltered Ad5 (14). We hypothesized that these findings can be transferred to other group C serotypes. Since high P19 expression is critical for its function (13), we coupled P19 to the major late promoter via an IRES, which has been shown to result in high transgene expression (20). The resulting viruses include a 24-bp deletion in E1A to confer tumor selectivity and a P19 expression cassette including a spacer, an IRES, and the P19 sequence, resulting in AdCd24.P19, with C indicating serotypes 1, 2, 5, and 6, respectively (Fig. 1). The viruses were reconstituted, and genomic integrity confirmed with PCR, sequencing and restriction enzyme digest of extracted vDNA (Supplementary Figs. S2 and S3). Viruses containing P19 could be produced to similar titers as their WT correspondent. The oncolytic effect of the viruses was then determined by head-to-head comparison of viruses from each group C serotype with each other and the control virus Ad5d24/dE3.GLN. For all viruses, the physical to infectious titer ratios as determined by qRT-PCR were similar. Viruses were compared for their oncolytic effect in tumor cell lines including lung (A549), pancreatic, and colon cancer cell lines. In most cell lines, P19 expressing group C viruses including Ad1, 2, 5, and 6 showed enhanced oncolytic effect, with Ad2d24.P19 showing an up to 100-times higher oncolytic effect in PANC-I and A549 cells compared with the control virus Ad5d24/dE3.GLN (Fig. 3). Results from these experiments confirmed the previous findings of the enhanced oncolytic effect of Ad2 compared with Ad5-based virus. Based on these findings, we selected A549 as the tumor model to investigate the oncolytic effect in further in vitro and in vivo studies.
Figure 3.
P19-expressing oncolytic viruses show improved oncolytic effect in vitro. The P19-containing viruses display increased oncolytic potential in comparison with control virus Ad5dFL. To perform an oncolysis assay, different cancer cell lines were infected and stained after 5 to 7days. Representative stainings are displayed. Oncolytic effect was quantified by measuring intensity and is displayed as cell viability 5to 7 days after infection with different vp per cell ratios. *, P > 0.05. Mean of three biological replicates per group is shown.
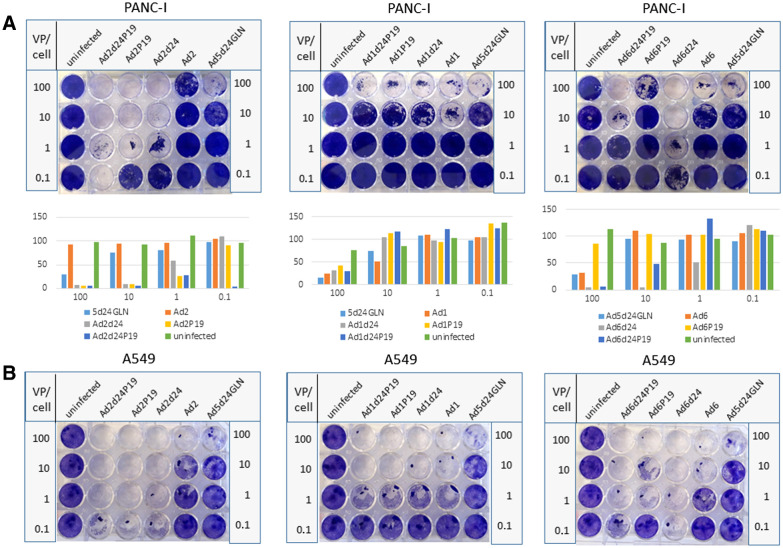
Next, we directly compared P19 and d24 viruses in PANC-1 (Fig. 4A) and A549 cells (Fig. 4B). We found that oncolytic activity was enhanced in A549 cells with Ad2 representing the most efficient candidate. In PANC-1 cells, the double mutant displayed improved oncolysis compared to the single mutants for Ad2 but not Ad1 or Ad6.
Figure 4.
The oncolytic effect in vitro is vector-type– and cell line–dependent. Comparison of oncolytic effect in vitro of selectively replicating Ad based on Ad 1, 2, and 6 carrying the 24-bp deletion and/or encoding P19. As controls, respective WT viruses (Ad1, Ad2, Ad6) and an Ad5-based vector carrying the 24-bp deletion were explored. Uninfected cells were used as negative control. To perform an oncolysis assay, PANC-I cell (A) and A549 cells (B) were infected at different viral particle numbers per cell (VP/cell) and stained after 4 to 7 days. Representative slides are displayed. Oncolytic effect was quantified by measuring intensity and is displayed as cell viability 4 to 7 days after infection. In PANC-I cells, modified Ad2 viruses showed most potent oncolytic activity and the Ad2 OAd containing P19 and 24-bp deletion revealed highest oncolytic potency. In A549 cells, oncolysis potency was cell line– and Ad-type–dependent. One biological replicate is shown.
Expression of P19 translates into enhanced release of OADs from A549 lung cancer cells
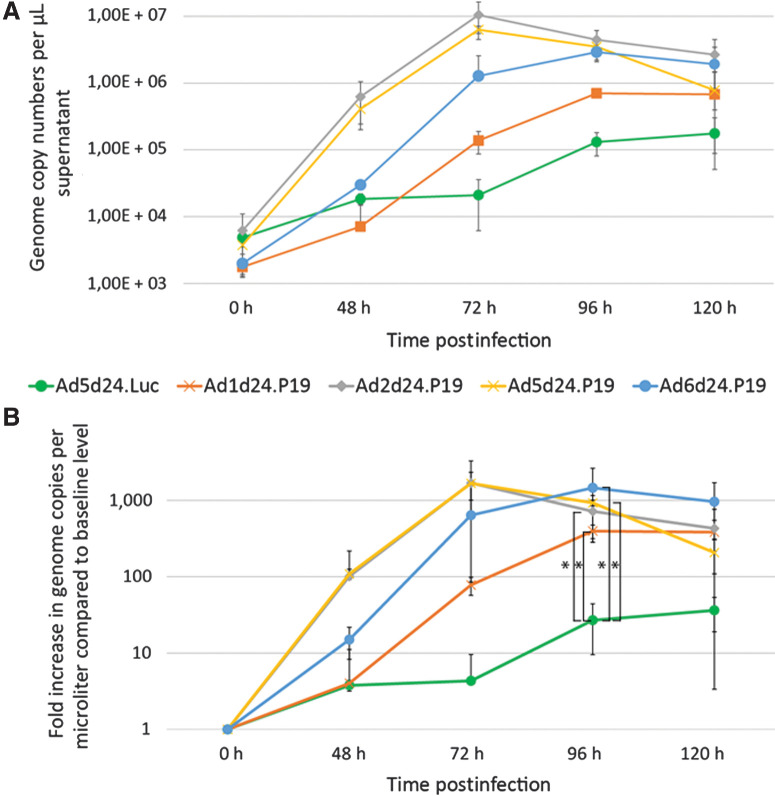
To determine the quantitative effect of the expression of P19 on the production and release of progeny in lung cancer cells, we investigated the number of viral copies released into the cell medium every 24 hours by P19-expressing OAd based on Ad1, 2, 5, and 6 and the control virus Ad5d24/dE3.GLN. We therefore isolated viral DNA from cell medium of lung cancer cells infected with similar titers of viruses at different time points after infection. Then, isolated DNA was subjected to qRT-PCR, using virus specific primers. Increased total numbers of virus genome copies were detected in cells treated with P19-expressing viruses compared with the control virus Ad5d24/dE3.G (Fig. 5). More than 30 times higher numbers of viral copies at 96 hours after infection were detected for Ad2d24.P19 [mean: 4,45E+06 copy numbers; 95% confidence interval (CI), 3,50E+05–8,55E+06; P = 0,0453] and more than 20 times higher copy numbers for Ad5d24.P19 (mean: 4,45E+06 copy numbers; 95% CI, 3,21E+05–6,64E+06; P = 0,0447) and Ad6d24.P19 (2,90E+06 copy numbers; 95% CI, 8,42E+05–4,96E+06; P = 0,0282) compared with Ad5d24/dE3.GLN (mean: 1,31E+05 copy numbers; 95% CI, 6,74E+03–2,55E+05; Fig. 5B) Decreasing numbers of viral copy numbers were observed at 120 hours postinfection, probably due to cell death because of virus infection. Interestingly, dynamics of viral progeny production for Ad2d24.P19 and Ad5d24.P19 were highly similar, whereas Ad1d24.P19 and Ad6d24.P19 multiplied at a similar rate, however with lower titers for Ad1d24.P19 compared with Ad6d24.P19 (Fig. 5A and B). While genome copy numbers released into the supernatant of the non–P19-expressing control virus remains stable for the time periods from 24 to 48 and 48 to 72 hours, virus particle release increased by a factor of at least 10-fold for the P19-expressing viruses in the same time window (Fig. 5B).
Figure 5.
P19 expressing viruses show enhanced replication rates in vitro. A, Virus particle production: Virus DNA copies released into the cell medium per 24 hours at different time points after infection of A549 cells with 10 vp/cell as determined by quantitative PCR of vDNA purified from the cell supernatant. Mean of three biological replicates with n = 3 technical replicates were used. B, Increase in virus genome copy numbers per microliter in relation to the baseline level. Significantly increased numbers of genome copies were detected 96 hours postinfection in cells treated with P19-expressing viruses compared with the control virus. Virus replication dynamics of Ad5dFIPP and Ad2dFIPP were similar. *, P < 0.05. h, hours.
P19-expressing Ad1-, 2-, and 6–based OAd suppress lung cancer growth in vivo
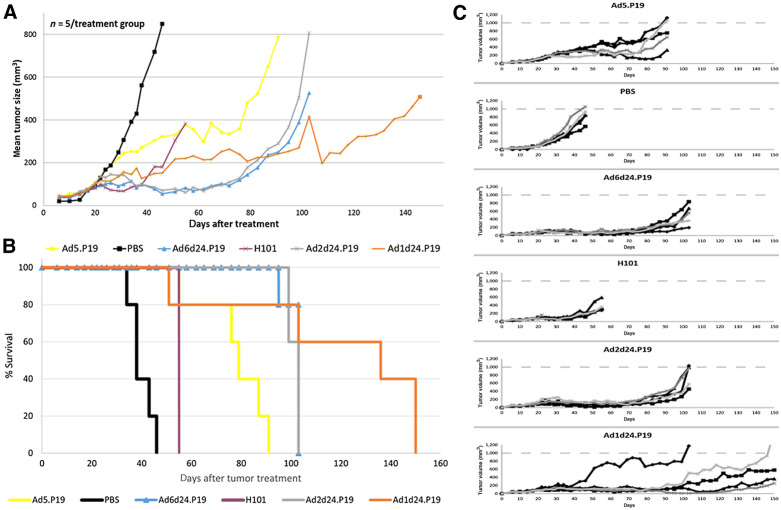
We next went on to study the potential of P19-expressing group C Ad to control tumor growth in vivo. We established subcutaneous tumors from the human lung cancer cell line A549 in CB17 mice. In a single experiment, mice (n = 5 per group) were treated with intratumoral injections of P19-expressing viruses Ad1d24.P19, Ad2d24.P19, Ad5.P19, Ad6d24.P19, H101 (16), and control (PBS) when tumors reached a volume of approximately 150 mm3 and tumor volume was determined for up to 160 days using caliper measurements. H101 contains partial deletion in the early gene E1 and E3 transcription units rendering this virus cancer cell-specific (16). In mice treated with P19-expressing Ad, tumor growth was delayed, and survival was significantly prolonged compared with mice that were injected with H101 (P < 0.005) or PBS (Fig. 6A). In 2 out of 5 mice that were treated with Ad1d24.P19, tumors almost had disappeared at day 100, but relapsed 1 week later, suggesting residual tumor burden. Mice treated with PBS rapidly grew tumors and had to be sacrificed by day 46.
Figure 6.
P19-expressing oncolytic Ad based on serotypes C 1, 2, 5, and 6 show higher potency in vivo compared with H101. Mice (n = 5 per treatment group) with preestablished A549 tumors (approximately 150 mm3) were intratumorally injected with 4 × 1010 vp of H101 or P19-expressing OAd based on serotypes C 1, 2, 5, and 6. A summary of the results of a single independent experiment is shown. A, Mean tumor volumes according to treatment group. The difference between the Ad2d24.P19, Ad6d24.P19 and the H101 group is significant at 55 days (P < 0.005). While all tumors eventually regrew, tumor regrowth was delayed in Ad1d24.P19. B, Kaplan–Meier survival plot using the day when tumors reached a volume of 500 mm3 or when mice had to be sacrificed due to tumor ulceration as endpoints. C, Individual animals and tumor growth kinetics in the different groups.
As demonstrated in Fig. 6B, treatment with Ad1d24.P19, Ad2d24.P19, and Ad6d24.P19 significantly prolonged survival in mice carrying A549 lung cancer cell subcutaneous tumors up to three times compared with control treatment with PBS. Median survival in mice treated with Ad1d24.P19 was significantly longer (132 days) compared with mice treated with H101 (51 days) and PBS (36 days; Fig. 6B). Median survival of mice treated with Ad2d24.P19, Ad5.P19, and Ad6d24.P19 was 99 days, 76 days, and 99 days, respectively. Tumor growth kinetics of each individual mouse are shown in Fig. 6C. Of note, mice treated with H101 developed moldering ulcers at the injection sites and 1 mouse treated with H101 was found dead despite low tumor burden. Therefore, treated mice could not be maintained for a longer time period. We have not performed toxicity studies and can thus not comment on toxicity of the used viruses. This study shows that tumor growth was significantly delayed in mice treated with a single injection of P19-carrying OAd and that antitumor effects were superior to the established OAd H101. Future studies will have to determine the safety profile of the novel viruses as well as compare single dose with multiple dose regimens. Moreover, we presume that studying different viruses expressing the same transgene in a rotating regimen in an immune competent animal model may be of interest.
Discussion
In this study, we generated tumor-selective OAd based on all group C Ad except HAd57 using a novel high throughput cloning strategy. With a strategy based on recombineering we simultaneously cloned several viruses at the same time, sharing primers and reagents (15). We successfully produced the modified viruses to high titers and confirmed the performed changes on sequence level including deletions and the addition of P19, an RNAi suppressor derived from a plant virus. We show differences in antitumor activity between serotypes 1, 2, 5, and 6 in different tumor cell lines (Fig. 1), a finding that potentially results in targeted tumor therapy for many human cancers, allowing the use of the optimal serotype for each tumor entity. Using cloned selectively replicating viruses based on several serotypes will allow using the most efficacious serotype for the targeted tumor. This study builds upon a previously cloned adenovirus library that contains many known human serotypes (15). Most existing oncolytic Ad are based on human Ad5, reasons for which are mostly historical. Most previous basic research on human Ad was performed in serotype 5 (and to a lesser extent in serotype 2), resulting in cloning platforms for this serotype, but not others. Studies comparing antitumor effect of WT Ad serotypes in various tumor cell lines have demonstrated lower seroprevalent type 6 to have comparable antitumor effect to type 5, thus highlighting its potential use for systemic application (7, 8, 21).
However, systematic comparison of selectively replicating OAd serotypes for antitumor effect have not yet been conducted. As we have demonstrated, high-throughput cloning of vectorized tumor-selective serotypes 1, 2, 5, and 6 enabled direct comparison of antitumor effect in vitro and in vivo. We hereby show that the previously unexploited human Ad types 1 and 6 may be superior platforms for anticancer virus development against many common human cancers, including pancreatic and lung cancer (Fig. 1). We thus confirmed earlier studies that suggested the development of Ad6 as an oncolytic vector (21). Future studies will build upon a library of tumor-selective viruses for vector selection in cancer cell lines, identifying the most active virus for each cancer subtype. Positively screened viruses may then be further developed by adding anticancer transgenes, such as GM-CSF.
One of the major limitations of oncolytic virotherapy is the recognition of vectors based on the same serotype by neutralizing antibodies, precluding repeat systemic administration (22). Therefore, previous efforts have concentrated on modification of immune-relevant surface molecules and particle shielding (23). Viruses that express identical transgenes but are based on distinct serotypes may allow repeated administration of the same active compound, potentiating their anticancer effect. Further studies are needed to investigate the value of repeated administration of different Ad types carrying identical antitumor transgenes in immunocompetent animal models.
Moreover, we took advantage of the RNAi suppressor protein P19 to improve the antitumor effect in OAd. Previous studies have shown that Ad5 replicates significantly faster and grows to higher titers in the presence of P19, suggesting a key role of RNAi in Ad life cycle (14). Further studies are warranted to explore whether improved activity is also derived from aspects of the cellular context, because at the time it cannot be ruled out that other cellular factors may be responsible for the observed improved activity. Additional studies indicate that baculovirus yields and transgene expression can be improved by the expression of P19. P19 was discovered in tombusvirus and was shown to suppress RNAi by appropriating siRNA directed against viral mRNA, thus suppressing an important innate antiviral immune mechanism (13). We hereby show that the expression of P19 in selectively replicating OAd resulted in significantly higher antitumor effect compared with non–P19-expressing Ad in vitro and to the established OAd H101 in vivo (Fig. 3 and Fig. 6). Expression of P19 translated into a more than 100-fold higher progeny production in A549 lung cancer cell lines compared with control virus Ad5d24/dE3.GLN (Fig. 4B). These results are consistent with earlier findings describing a 100-fold increased Ad5 genome replication rate in the presence of P19 (14). The molecular mechanisms responsible for increased virus replication and genome replication in P19-expressing Ad are unknown. Further studies will help elucidate whether similar mechanisms are functional in other virus species, as is suggested by recent findings showing that P19 improves baculovirus yield and transgene expression in sf9 (Spodoptera frugiperda, the fall armyworm, a commonly used cell line for recombinant protein production using baculovirus) insect cells. Thus, combination of P19 in oncolytic viruses with additional antitumor transgenes may result in improved replication and transgene expression, potentiating oncolytic effect.
Note that a significant difference in replication efficiencies is not necessarily expected for OAds with and without the d24 deletion. The deletion in the early region E1 leads to expression of a mutated version of E1A protein, which cannot form a complex with Rb and therefore this virus cannot drive cells into the S-phase required for virus replication. According to the literature A549 cells express WT RB (24) and A549 cells and PANC-I cells contain deletions in the CDKN2A/p16INK4A gene leading to impairment of Rb (25).
Finally, mice that were treated with P19-expressing OAd had a more than twice as long survival in an in vivo lung cancer model compared with mice treated with established selectively replicating oncolytic virus H101. Improved survival was observed for all P19-carrying OAd compared with H101. Note that experiments analyzing in vivo efficacy of the different oncolytic viruses in mice summarize a single experiment with n = 5 mice per group. This represents one limitation of our study and further experiments are mandatory including different tumor mouse models. Interestingly, the vectors used in this study did not express antitumor transgenes like many existing oncolytic Ad, including H101. Lack of immunostimulatory transgenes may result in an improved safety profile of P19-carrying viruses without additional transgene expression. On the other hand, if enhanced antitumor effect through immune stimulation is desirable, for instance in tumors exhibiting immunosuppressive features including pancreatic cancer, combination with P19 may provide additional benefit.
Supplementary Material
Acknowledgments
The authors would like to thank Dirk Nettelbeck for advice and for providing original material. Moreover, the authors would like to acknowledge excellent technical assistance of Annika Bremke and Fatima Arakrak.
This work was supported, in part, by DFG grant (grant no. EH 192/5-3 to (A. Ehrhardt) and received funding from the Ph.D. program of the Faculty of Health at Witten/Herdecke University (to J. Doerner).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Authors' Disclosures
M. Solanki reports no relationship. No disclosures were reported by the other authors.
Authors' Contributions
J. Doerner: Conceptualization, resources, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. E. Sallard: Formal analysis, investigation, methodology. W. Zhang: Resources, supervision, investigation, methodology. M. Solanki: Resources, methodology. J. Liu: Resources, methodology. E. Ehrke-Schulz: Resources, data curation, supervision. H. Zirngibl: Supervision, funding acquisition, writing–review and editing. A. Lieber: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, investigation, methodology, writing–review and editing. A. Ehrhardt: Conceptualization, resources, formal analysis, supervision, funding acquisition, visualization, writing–original draft, writing–review and editing.
References
- 1. Alemany R, Balague C, Curiel DT. Replicative adenoviruses for cancer therapy. Nat Biotechnol 2000;18:723–7. [DOI] [PubMed] [Google Scholar]
- 2. Chu RL, Post DE, Khuri FR, Van Meir EG. Use of replicating oncolytic adenoviruses in combination therapy for cancer. Clin Cancer Res 2004;10:5299–312. [DOI] [PubMed] [Google Scholar]
- 3. Abbink P, Lemckert AA, Ewald BA, Lynch DM, Denholtz M, Smits S, et al. Comparative seroprevalence and immunogenicity of six rare serotype recombinant adenovirus vaccine vectors from subgroups B and D. J Virol 2007;81:4654–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Piedra PA, Poveda GA, Ramsey B, McCoy K, Hiatt PW. Incidence and prevalence of neutralizing antibodies to the common adenoviruses in children with cystic fibrosis: implication for gene therapy with adenovirus vectors. Pediatrics 1998;101:1013–9. [DOI] [PubMed] [Google Scholar]
- 5. D'ambrosio E, Grosso ND, Chicca A, Midulla M. Neutralizing antibodies against 33 human adenoviruses in normal children in Rome. J Hyg (Lond) 1982;89:155–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Weaver EA, Hillestad ML, Khare R, Palmer D, Ng P, Barry MA. Characterization of species C human adenovirus serotype 6 (Ad6). Virology 2011;412:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen CY, Weaver EA, Khare R, May SM, Barry MA. Mining the adenovirus virome for oncolytics against multiple solid tumor types. Cancer Gene Ther 2011;18:744–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hoffmann D, Bayer W, Heim A, Potthoff A, Nettelbeck DM, Wildner O. Evaluation of twenty-one human adenovirus types and one infectivity-enhanced adenovirus for the treatment of malignant melanoma. J Invest Dermatol 2008;128:988–98. [DOI] [PubMed] [Google Scholar]
- 9. Mach N, Gao J, Schaffarczyk L, Janz S, Ehrke-Schulz E, Dittmar T, et al. Spectrum-wide exploration of human adenoviruses for breast cancer therapy. Cancers 2020;12:1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cattaneo R, Miest T, Shashkova EV, Barry MA. Reprogrammed viruses as cancer therapeutics: targeted, armed and shielded. Nat Rev Microbiol 2008;6:529–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu Y, Zhang L, Zhang Y, Liu D, Du E, Yang Z. Functional analysis of RNAi suppressor P19 on improving baculovirus yield and transgene expression in Sf9 cells. Biotechnol Lett 2015;37:2159–66. [DOI] [PubMed] [Google Scholar]
- 12. Hodgson JJ, Wenger LW, Clem RJ, Passarelli AL. Inhibition of dicer activity in lepidopteran and dipteran cells by baculovirus-mediated expression of Flock House virus B2. Sci Rep 2019;9:14494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Scholthof HB. The Tombusvirus-encoded P19: from irrelevance to elegance. Nat Rev Microbiol 2006;4:405–11. [DOI] [PubMed] [Google Scholar]
- 14. Rauschhuber C, Mueck-Haeusl M, Zhang W, Nettelbeck DM, Ehrhardt A. RNAi suppressor P19 can be broadly exploited for enhanced adenovirus replication and microRNA knockdown experiments. Sci Rep 2013;3:1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang W, Fu J, Liu J, Wang H, Schiwon M, Janz S, et al. An engineered virus library as a resource for the spectrum-wide exploration of virus and vector diversity. Cell Rep 2017;19:1698–709. [DOI] [PubMed] [Google Scholar]
- 16. Tuve S, Liu Y, Tragoolpua K, Jacobs JD, Yumul RC, Li ZY, et al. In situ adenovirus vaccination engages T effector cells against cancer. Vaccine 2009;27:4225–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang H, Bian X, Xia L, Ding X, Müller R, Zhang Y, et al. Improved seamless mutagenesis by recombineering using ccdB for counterselection. Nucleic Acids Res 2014;42:e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bird AW, Erler A, Fu J, Hériché J-K, Maresca M, Zhang Y, et al. High-efficiency counterselection recombineering for site-directed mutagenesis in bacterial artificial chromosomes. Nat Methods 2012;9:103–9. [DOI] [PubMed] [Google Scholar]
- 19. Jager L, Hausl MA, Rauschhuber C, Wolf NM, Kay MA, Ehrhardt A. A rapid protocol for construction and production of high-capacity adenoviral vectors. Nat Protoc 2009;4:547–64. [DOI] [PubMed] [Google Scholar]
- 20. Rivera AA, Wang M, Suzuki K, Uil TG, Krasnykh V, Curiel DT, et al. Mode of transgene expression after fusion to early or late viral genes of a conditionally replicating adenovirus via an optimized internal ribosome entry site in vitro and in vivo. Virology 2004;320:121–34. [DOI] [PubMed] [Google Scholar]
- 21. Shashkova EV, May SM, Barry MA. Characterization of human adenovirus serotypes 5, 6, 11, and 35 as anticancer agents. Virology 2009;394:311–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Atasheva S, Shayakhmetov DM. Adenovirus sensing by the immune system. Curr Opin Virol 2016;21:109–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jönsson F, Kreppel F. Barriers to systemic application of virus-based vectors in gene therapy: lessons from adenovirus type 5. Virus Genes 2017;53:692–9. [DOI] [PubMed] [Google Scholar]
- 24. Mack PC, Gandara DR, Bowen C, Edelman MJ, Paglieroni T, Schnier JB, et al. RB status as a determinant of response to UCN-01 in non-small cell lung carcinoma. Clin Cancer Res 1999;5:2596–604. [PubMed] [Google Scholar]
- 25. Gradiz R, Silva HC, Carvalho L, Botelho MF, Mota-Pinto A. MIA PaCa-2 and PANC-1 - pancreas ductal adenocarcinoma cell lines with neuroendocrine differentiation and somatostatin receptors. Sci Rep 2016;6:21648. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.