Abstract
Targeted radionuclide therapy (TRT) using probes labeled with Lutetium-177 (177Lu) represents a new and growing type of cancer therapy. We studied immunologic changes in response to TRT with 177Lu labeled anti-human CD20 camelid single domain antibodies (sdAb) in a B16-melanoma model transfected to express human CD20, the target antigen, and ovalbumin, a surrogate tumor antigen. High-dose TRT induced melanoma cell death, calreticulin exposure, and ATP-release in vitro. Melanoma-bearing mice received fractionated low and high-dose TRT via tumor targeting anti-human CD20 sdAbs, as opposed to control sdAbs. Tumor growth was delayed with both doses. Low- and high-dose TRT increased IL10 serum levels. Low-dose TRT also decreased CCL5 serum levels. At the tumor, high-dose TRT induced a type I IFN gene signature, while low-dose TRT induced a proinflammatory gene signature. Low- and high-dose TRT increased the percentage of PD-L1pos and PD-L2pos myeloid cells in tumors with a marked increase in alternatively activated macrophages after high-dose TRT. The percentage of tumor-infiltrating T cells was not changed, yet a modest increase in ovalbumin-specific CD8pos T-cells was observed after low-dose TRT. Contradictory, low and high-dose TRT decreased CD4pos Th1 cells in addition to double negative T cells. In conclusion, these data suggest that low and high-dose TRT induce distinct immunologic changes, which might serve as an anchoring point for combination therapy.
Introduction
Targeted radionuclide therapy (TRT) holds promise for treatment of cancer metastasis for which local radiotherapy is no longer suitable (1). In TRT, cytotoxic radiation is selectively delivered to cancer cells by tumor targeting agents coupled to α, β–, or Auger electron emitting radionuclides (1–3). Various targeting moieties have been tailored as TRT-agents, including radiolabeled mAbs, variants thereof, and peptides with recent clinical successes in various malignant diseases (4–7). However, use of radiolabeled mAbs has been hampered by poor tissue penetration and accumulation in bone marrow resulting in myelosuppression (8–10). So, mAb fragments have been evaluated, leading to development of single domain antibodies (sdAb), antigen binding fragments of light chain devoid mAbs of Camelidae. SdAbs have been shown to bind epitopes inaccessible to mAbs with high affinity and have been extensively studied for many cancer-related applications, including imaging (11–14). Imaging studies have shown high and specific accumulation of sdAbs in local and disseminated cancer deposits, a much desired trait for TRT agents (14, 15).
TRT with sdAbs labeled with β– emitting radionuclides (β–-TRT) is a promising field with various preclinical studies corroborating its therapeutic capacity in diverse cancer models (16–20). Recently, the first phase I clinical assessment with Iodine-131–labeled HER2-specific sdAbs in patients with breast cancer has been published (16–20). Despite ongoing clinical translation, immunologic changes in the periphery and tumor as a result of sdAb-mediated β–-TRT have not been studied. This warrants further investigation, as a recent study in the B16-melanoma model showed that low-dose β–-TRT with an Iodine-131–labeled melanin ligand or an Yttrium-90 (90Y)-labeled 2-(trimethylammonio)ethyl(18-(4-(2-(4,7,10-tris(carboxymethyl)-1,4,7,10-tetraazacyclo-dodecan-1-yl)acetamido)phenyl)octadecyl) phosphate resulted in increased immune-cell infiltration in the tumor with a marked increase in cytotoxic T cells (21). Moreover, several studies showed improved therapy outcome when β–-TRT was combined with blockade of inhibitory immune receptors (21–26). Whether immune activation occurs in response to β–-TRT performed with Lutetium-177 (177Lu)-labeled sdAbs has not yet been reported.
We used an immunocompetent B16-melanoma mouse model expressing human CD20 (huCD20), the target antigen, and ovalbumin, a surrogate tumor antigen, to assess the outcome and immune modifying properties of β–-TRT performed with a well-characterized anti-huCD20 sdAb labeled with 177Lu, later on referred to as 177Lu-9079 (16). While CD20 is expressed on a small (±2%) subpopulation of melanoma cells with stem cell like features, tumor initiating characteristics, and migratory capacity (27, 28), huCD20 was used in this study as a model surface antigen uniquely expressed on the transfected melanoma cells. We analyzed induction of cell death and hallmarks of immunogenic cell death (ICD) in vitro. We further studied tumor cell targeting in vivo and the ability of the therapy to delay tumor growth. Systemic effects upon 177Lu-9079 were studied through analysis of serum cytokine levels and tumor specificity of CD8pos splenocytes. The tumor environment was analyzed by gene expression profiling and flow cytometry. This analysis was performed in comparison with tumors treated with nontargeting 177Lu-sdAb R3B23 of similar size, as the tumor stage is a well-known confounding factor with regard to immune contexture.
Materials and Methods
Cell line
B16-melanoma cells, transfected to express huCD20 and ovalbumin, were provided by J. Tavernier (VIB-UGent; Ghent, Belgium). The antigen huCD20 serves as a target for TRT, while ovalbumin serves as an immunogenic model antigen that allows addressing activation of ovalbumin-specific T cells. Cells were cultured in DMEM (Sigma-Aldrich) containing 10% heat-inactivated FBS (Life Technologies), 2 mmol/L L-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin (Sigma-Aldrich). Cells were kept at 37°C in a humidified atmosphere with 5% CO2 and prior to use, tested for Mycoplasma contamination using the Venor GeM Classic kit (Minerva Biolabs). Mycoplasma-free cells were used at early passage (<20 passages upon thawing). To verify antigen expression, RNA was isolated using the Allprep DNA/RNA Mini kit (Qiagen). RNA was reverse transcribed to cDNA using the Verso cDNA Synthesis Kit (Thermo Fisher Scientific). Expression of gp100, Trp2, ADPGK, and ovalbumin was determined using the KAPA2G Fast HotStart PCR Kit (Sigma-Aldrich) on cDNA using primers for gp100 (FW: 5′-AGCACCTGGAACCACATCTA-3′; REV: 5′-GTTCCAGAGGGCTGTGTAGT-3′), Trp2 (FW: 5′-TTAGGTCCAGACGCCCC-3′; REV 5′-CTGTGCCACGTGACAAAGGC-3′), ADPGK (FW: 5′-CCAGAAGAGGATGTCACT-3′; REV 5′-CATCTTCTCTCACGA CCT-3′), and ovalbumin (FW: 5′-GAGCTCATCAATTCCTGGGT-3′; REV 5′-TACTCTCAAGCTGCTCAAGG-3′). A nonamplification control (NAC), consisting of B16-huCD20 RNA, and a nontemplate control (NTC), consisting of reaction mixture without B16-huCD20 cDNA, were included. Amplicons and a 1kb DNA-ladder (Thermo Fisher Scientific) were separated on a 2% agarose gel (Invitrogen) using a gel electrophoresis appliance (Labnet). Ethidium bromide (Apex) and an ultraviolet light analyzer (Vilber) were used for detection. Cell surface expression of huCD20 was assessed using antibodies that bind huCD20 (BV650, clone 2H7), acquired on the BD Celesta flow cytometer and data was analyzed using FlowJo X software (Tree Star Inc, RRID:SCR_008520). Besides assessment of B16-huCD20’s intra- and extracellular markers, no cell line authentication was performed.
SdAbs and 177Lu-radiolabeling
SdAb 9079 binds huCD20 and serves as a targeting moiety for TRT in the B16-huCD20 model and its sequence is available under patent application WO2017153345A1 (16). SdAb R3B23 binds the M-protein of 5T2MM cells and was used as a control (17). Both sdAbs were produced without a carboxyterminal tag. Purified sdAbs were reconstituted in 0.05M sodium carbonate buffer (pH 8.5) for conjugation with p-SCN-Bn-CHX-A”-DTPA (Macrocyclics). To that end, sdAbs were incubated with a 10-fold molar excess of chelator for 3 hours at room temperature. DTPA-conjugated sdAbs were purified via size exclusion chromatography (SEC) using Superdex peptide 10/300 column (GE Healthcare). Carrier-free 177LuCl3 (110–650 MBq; Isotope Technologies Garching) in 0.2M ammonium acetate (pH 5.0) was incubated with 50 to 150 μg DTPA-conjugated sdAbs for 30 minutes at 37°C. The resulting 177Lu-DTPA-sdAbs were purified via SEC using NAP5 columns. The eluate was passed over a 0.2-μm filter (Millipore). The radiochemical purity of 177Lu-DTPA-sdAbs was evaluated by instant thin layer chromatography using silica gel impregnated glass fiber sheets (Agilent Technologies) with 0.1M citrate buffer as eluent (pH 4.0). 177Lu-DTPA-sdAbs with a radiochemical purity of more than 95% and specific activity of 22.4 MBq/nmol were used for biodistribution purposes. 177Lu-DTPA-sdAbs with a radiochemical purity of more than 95% and specific activity of 186 MBq/nmol were used for the in vitro and in vivo therapy experiments. Henceforth, 177Lu-DTPA-sdAb9079 will be referred to as 177Lu-9079, 177Lu-DTPA-R3B23 as 177Lu-R3B23, and 177Lu-DTPA-sdAbs as 177Lu-sdAbs.
Detection of immunogenic cell death hallmarks
B16-huCD20 cells were plated at 105 cells in 1 mL culture medium per 24 wells and treated for 1 hour in 100 μL 37 to 370 MBq/mL 177Lu-9079 or 370 MBq/mL 177Lu-R3B23. Unbound 177Lu-sdAbs were removed, 1 mL fresh culture medium was added, and cells were incubated 48 hours at 37°C in a humidified atmosphere with 5% CO2. Cells treated for 48 hours with 2 μmol/L of ICD-inducing mitoxantrone (MTX) were used as a positive control. Live and dead cells were distinguished using SYTOX Blue (Thermo Fisher Scientific). Cell surface calreticulin was detected using an Alexa Fluor 488-labeled anti-calreticulin antibody (Abcam, clone EPR3924). Intracellular ATP levels were quantified using quinacrine dihydrochloride (Sigma-Aldrich). Cells were stained with the anti-calreticulin antibody or quinacrine for 30 minutes at 4°C, after which the cells were washed, stained with SYTOX Blue, and acquired on the BD Celesta flow cytometer to identify hallmarks of ICD (29).
Animal model
Female, 6- to 12-week old C57BL/6 mice (Charles River) were transplanted subcutaneously in the right thigh with 105 B16-huCD20 cells, while under sedation with isoflurane (Abbott); 5% for induction, 2.5% for maintenance with an oxygen flow of 1 L per minute. Animal weights and tumor volumes were recorded on alternate days to monitor the time to reach humane endpoints (loss of >20% of the initial body weight and tumor volume >1,500 mm³). The tumor length and width were measured with a digital caliper to calculate the volume. Experiments were performed in accordance with the European guidelines for animal experimentation and approved by the Ethical Committee for use of laboratory animals of the Vrije Universiteit Brussel (Brussels, Belgium; dossiers 15-272-4, 16-214-2).
Ex vivo biodistribution of 177Lu-sdAbs and dosimetry.
Mice bearing subcutaneous tumors of 250 ± 100 mm3 were injected intravenously with a single injection of 5 μg of 177Lu-9079 or 177Lu-R3B23 (3.08 ± 1.24 MBq for animals killed at 1 hour postinjection and 10.69 ± 0.210 MBq for animals killed 24, 48, and 72 hours postinjection) coinfused with 150 mg/kg Gelofusine, a plasma expander that reduces the kidney retention small radiotracers (18, 30). Different organs, tissues, and tumors were isolated, weighed, and their level of radioactivity was analyzed using a γ-counter (Cobra Inspector 5003, Canberra Packard), along with a standard of known radioactivity and corrected for decay. Results were expressed as percentage of injected activity per gram of tissue (%IA/g). To calculate tissue dosimetry, an S-factor of 2.330E-11 (Gy/Bq × s/g) was incorporated and performed using trapezoid fitting between timepoints and exponential-fitting on biodistribution data.
Therapy
On days 3, 9, and 14 of B16-huCD20 subcutaneous tumor growth, mice received an i.v. injection of 3.3 μg 177Lu-9079 coinfused with 150 mg/kg Gelofusin with a total cumulative radioactive dose of 50.7 ± 0.4 MBq (low-dose) or 141.3 ± 1.3 MBq (high-dose). Control mice received i.v. injections of 3.3 μg 177Lu-R3B23 coinfused with 150 mg/kg Gelofusin with a total cumulative radioactive dose of 143.2 ± 1.5 MBq. 3 days after inoculation, all mice displayed palpable subcutaneous tumors when therapy started. Gelofusin was employed to limit kidney toxicity by attenuating sdAb retention in kidneys.
Serum cytokine analysis
To analyze the immediate inflammatory response to TRT on serum cytokine levels, 50 to 100 μL blood was collected via retro-orbital bleeding 6 hours after the last treatment. Blood was transferred into a serum separation tube (Micro tube 1.1 mL Z-Gel, Sarstedt). The tubes were centrifuged at approximately 10,000 rcf for 5 minutes and serum was transferred into a clean Eppendorf tube. Cytokine content was determined using the Bio-Plex Pro Mouse Cytokine 23-plex Assay (Bio-Rad, RRID:AB_2857368) using a Luminex 200 System (Bio-Rad, RRID:SCR_018025).
Single cell suspensions
Tumors were cut in 5-mm pieces and immersed in 5-mL RPMI1640-medium (Sigma-Aldrich) with 5% fetal clone I serum (HyClone), 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mmol/L L-glutamine, 1 mmol/L sodium pyruvate, 1 mmol/L nonessential amino acids, and 50 μmol/L β-mercaptoethanol (Sigma-Aldrich). An enzymatic digestion with 2,000 U/mL DNase I (Sigma-Aldrich), 1 mg/mL Dispase II (Roche), and 300 U/mL collagenase I (Sigma-Aldrich) was performed for 1 hour at 37°C. The cell suspension was passed over a 70-μm cell strainer (Falcon) to obtain a single cell suspension. Red blood cell lysis was performed using 0.15M NH4Cl-0.017M Tris-solution.
Flow cytometry
Surface antigen staining was performed on 106 cells preincubated with anti-CD16/32 antibodies (Thermo Fisher Scientific) and stained with a live/dead marker (LIVE/DEAD Fixable Green Dead Cell Stain Kit, Thermo Fisher Scientific) for 30 minutes at 4°C to 8°C. Cells were washed and stained with antibodies that bind CD45.2 (APC-eF780, 104), CD3ε (BV605, 145-2C11), CD4 (AF700, RM4-5), CD8a (PerCP-Cy5.5, 53-6.7), CD11b (AF700, M1/70), CD11c (BV650, HL3), CD206 (PE, C068C2), CD274 (BV421, MIH5), CD279 (PE, J43), F4/80 (BB700, T45-2342), Ly6G (AF647, 1A8), and MHC-II (PE/Dazzle 594, M4/114.15.2). Antibodies were purchased from BD Biosciences except for antibodies that bind CD206 and MHC-II that were from Biolegend. Cells were acquired on the BD Celesta flow cytometer and data were analyzed using the FlowJo X software (Tree Star Inc, RRID:SCR_008520).
Restimulation of splenocytes
Spleens were transferred to a 40-μm cell strainer placed on top of a 50 mL tube containing 5 mL RPMI and mashed through the filter using a plunger from a disposable 3-mL syringe. Red blood cell lysis was performed, after which the cells were counted and plated at 250 thousand cells per well in a 96-well plate (Corning). Cells were stimulated with 100 pmol of the peptide OVA257–264, Trp2181–188, gp10025–33, or ADPGK299–307 dissolved in RPMI. Peptides were purchased from Anaspec. As a positive control, T-cells were stimulated with anti-CD3/CD28 antibody coated beads (Gibco) as instructed by the provider. As a negative control, T cells were cultured in RPMI. The 96-well plate was incubated 48 hours at 37°C in a humidified atmosphere with 5% CO2, after which the supernatant was collected for analysis. ELISA was performed to measure IFNγ in supernatant (mouse IFNγ ELISA, Invitrogen). The ELISA was performed as instructed by the manufacturer and analyzed using a spectrophotometer (GloMax, Promega) at λ = 450 nm.
Gene expression analysis
RNA isolation (RNeasy Plus Mini Kit, Qiagen), quality control on the Qubit4 (Qubit RNA HS Assay Kit, Thermo Fisher Scientific) and 2100 Bioanalyzer (RNA6000 Nano Kit, Agilent, RRID:SCR_019715), and analysis using the nCounter PanCancer Immune Profiling Panel on an nCounter Analysis System (Nanostring Technologies) was performed as described (31). Raw counts were normalized by the RuvSeq method adapted for Nanostring analysis (32). Normalized counts were subjected to gene set enrichment analysis (GSEA) using the canonical pathways of the BIOCARTA, KEGG, PID, REACTOME, and WikiPathways database from the Molecular Signatures Database (33, 34), only pathways exceeding a P value of 0.05 and FDR q value of 0.25 are displayed. More specifically, pathways of interest annotated with “*” and “**”, originate from the canonical pathways and the hallmarks databases respectively, whilst unmarked pathways originate from the Gene Ontology database. Scoring of tumor-infiltrating lymphocytes (TIL) in R was conducted as described (35, 36) using cell-type annotation, specified by the PanCancer Mouse Immune Profiling Panel. Cell scores were calculated as mean of the log2-normalized gene expression value of all genes related to a particular cell type. The total TIL-score was calculated as the mean of all cell scores, whose correlation with CD45 exceeds 0.6.
Statistical analysis
Kaplan–Meier curves that show the time to reach humane endpoints were statistically analyzed using log–rank (Mantel–Cox) and Gehan–Breslow–Wilcoxon tests. One-way ANOVA was performed for all other results and corrected for multiple comparisons via Tukey correction. All statistical analyses were conducted in Graphpad Prism 9.3.1. Sample sizes and the number of times experiments were repeated are indicated in the figure legends. The asterisks in the figures indicate the level of statistical significance: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Nonsignificant changes are not indicated.
Data availability statement
The data generated in this study are available within the article, its Supplementary data files, and Gene Expression Omnibus (GEO) online repository. Biodistribution files, multiplex data, fcs files from flowcytometric analysis are available upon request from the corresponding author. Raw and normalized nCounter Gene Expression Data obtained by analyzing the gene expression of treated B16-huCD20 melanoma samples using the nCounter Human PanCancer Immune Profiling panel, has been made publicly available via the GEO database (accession number: GSE199624).
Results
In vitro treatment of B16-huCD20 cells with 177Lu-9079 induces ICD
We showed that 177Lu-9079 at a dose of 370 MBq/mL, yet not 37 MBq/mL 177Lu-9079 or 370 MBq/mL 177Lu-R3B23, induced B16-huCD20 cell death (P < 0.0001; Fig. 1A; Supplementary Fig. S1), exposure of calreticulin on the cell surface on viable cells (P < 0.0001; Fig. 1B; Supplementary Fig. S1), and ATP-release from the cytosol by viable cells (P = 0.0482; Fig. 1C; Supplementary Fig. S1). These data suggest that 177Lu-9079 can induce ICD in a dose-dependent manner and warrant further in vivo evaluation.
Figure 1.
In vitro treatment of B16-huCD20 cells with 177Lu-9079 induces cell death that coincides with exposure of calreticulin and release of ATP. B16-huCD20 cells were treated for 1 hour with 370 MBq/mL 177Lu-R3B23 [CTRL (370 MBq/mL)] or 177Lu-9079 [TRT (dose)] or 48 hours of 2 μmol/L MTX (2 μmol/L), respectively. Flow cytometry was performed 48 hours after addition of 177Lu-sdAb to study the percentage of dead cells (A), cell surface calreticulin on viable cells (B), and ATP-release by viable cells (C). The bar graphs show individual datapoints and grouped mean ± SD (N = 1, n = 3).
177Lu-9079 specifically accumulates in B16-huCD20 tumors
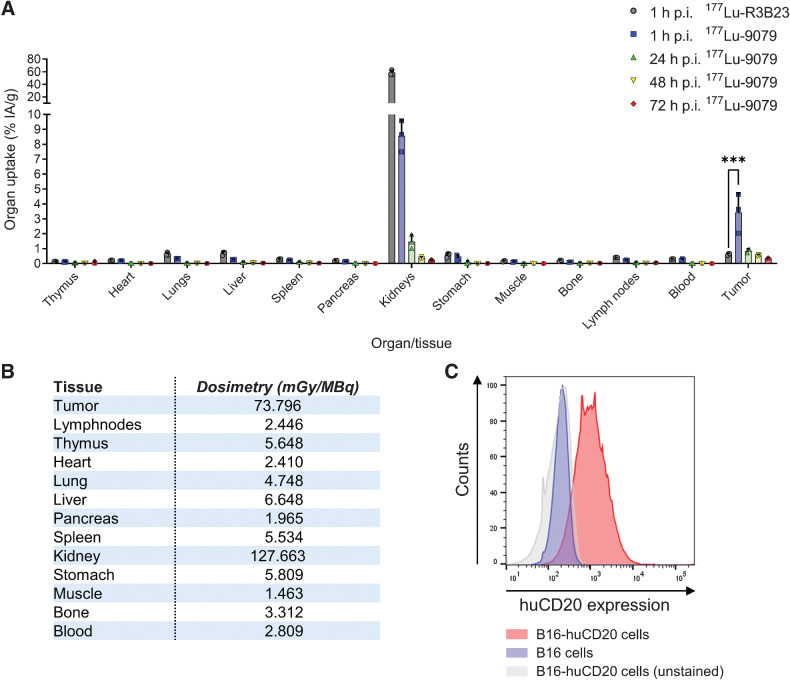
Ex vivo γ-counting showed a significant accumulation of 177Lu-9079 in tumor tissue, yet not 177Lu-R3B23, 1 hour postinjection (P < 0.0001). 177Lu-9079 was not retained over a long period of time as the percentage uptake declined from 3.440 ± 1.310%IA/g at 1 hour postinjection to 0.862 ± 0.127%IA/g at 24 hours. We observed little uptake and retention over time of 177Lu-9079 in nontumor tissues with the exception of the kidneys via which sdAbs are excreted from the body. Kidney accumulation of 177Lu-9079 was the highest 1 hour postinjection, decreasing from 8.576 ± 1.049%IA/g at 1 hour postinjection to 1.473 ± 0.464%IA/g at 24 hours postinjection (Fig. 2A). Dosimetry calculations showed increased doses delivered to B16-huCD20 tumors compared with spleen, bone marrow, and other tissues (excluding kidneys; Fig. 2B). Flowcytometric analysis confirmed surface expression of huCD20 on B16-huCD20 cells (Fig. 2C).
Figure 2.
Ex vivo biodistribution data show specific accumulation of 177Lu-9079 in B16-huCD20 tumors. A, Accumulation of 177Lu-9079 and 177Lu-R3B23 was quantified at 1, 24, 48, and 72 hours postinjection, and 1 hour postinjection, respectively (N = 1, n = 3 per time-point). The bar graphs show individual datapoints and grouped mean %IA/g ± SD. B, Tissue dosimetry upon 177Lu-TRT was performed using trapezoid-exponential fitting. C, Histograms show expression of huCD20 on B16-huCD20 cells. h, hours; p.i., postinjection.
Low- and high-dose 177Lu-9079 delays B16-huCD20 tumor growth
To evaluate the therapeutic potential of 177Lu-9079, we treated B16-huCD20 tumor-bearing mice by administering a cumulative dose of 50 or 140 MBq of 177Lu-9079 on days 3, 9, and 14 after inoculation, resulting in delivery of 4 or 10 Gy to B16-huCD20 tumors over the course of time, respectively (Fig. 3A). For comparison, mice received a cumulative dose of 140 MBq of 177Lu-R3B23 on days 3, 9, and 14 after tumor cell inoculation. Tumor growth was delayed at day 13 when mice were treated with 50 or 140 MBq of 177Lu-9079 compared with 140 MBq of 177Lu-R3B23 (P = 0.0065; P = 0.0169; Fig. 3B–D). However, an increased time to reach humane endpoints was only observed in mice treated with 140 MBq of 177Lu-9079 compared with mice treated with 140 MBq of 177Lu-R3B23 (P = 0.0090, Fig. 3E). No evidence of acute, systemic treatment related toxicity was observed, as indicated by the animals’ body weight (Fig. 3F).
Figure 3.
Therapy with low and high doses of 177Lu-9079 impacts on B16-huCD20 tumor growth. A, Schematic representation of the 177Lu-sdAbs treatment regimen detailing experimental conditions: 50 MBq of 177Lu-9079 [TRT (50 MBq)], 140 MBq of 177Lu-9079 [TRT (140 MBq)], and 140 MBq 177Lu-R3B23 [CTRL (140 MBq)]. Tumor kinetic after treatment are shown by group during the course of the experiment (B) and on day 13 (C), by individual mouse (D) and Kaplan–Meier curve showing the time at which mice reached humane endpoints by group (E). F, Weight of treated mice. The bar graphs show individual datapoints and grouped mean ± SD (N = 1, n = 6).
Systemic responses to low- and high-dose 177Lu-9079 differ
We analyzed 23 cytokines in serum collected 6 hours after the delivery of the last dose to reach 50 or 140 Mbq of 177Lu-9079, or 140 MBq of 177Lu-R3B23. IL10 was increased (P = 0.0270) and CCL5/RANTES decreased (P = 0.0133) upon therapy with 50 MBq 177Lu-9079 compared with 140 MBq 177Lu-R3B23 (Fig. 4A and B; Supplementary Fig. S2). No significant changes were observed for the cytokines IL-1α, IL6, IL9, IL12p40, IL12p70, IL13, TNFα, KC, eotaxin, G-CSF, GM-CSF, CCL3/MIP-1α, and CCL4/MIP-1β. The following cytokines were analyzed but not detected: IL1β, IL2, IL3, IL4, IL5, IL17, IFNγ, and CCL2/MCP-1. Therapy with 140 MBq of 177Lu-9079 resulted in serum cytokine levels that were similar to those detected after treatment with 140 MBq of 177Lu-R3B23 (Fig. 4A and B; Supplementary Fig. S2).
Figure 4.
Peripheral immune responses upon treatment with 177Lu-9079. Multiplex analysis of serum cytokine levels of IL10 (A) and CCL5 (B) was performed on serum isolated 6 hours after last administration of 50 MBq or 140 MBq of 177Lu-9079 [TRT (50 MBq) and TRT (140 MBq), respectively], or 140 MBq 177Lu-R3B23 [CTRL (140 MBq)]. The graphs represent the results as individual values with grouped mean ± SD (N = 1, n = 4). C, Agarose gel visualizing amplicons resulting from the PCR reaction with gp100 (90 bp), Trp2 (71 bp), ADPGK (400 bp), and ovalbumin (346 bp) specific primer sets. D, Analysis of IFNγ production by CD8pos splenocytes stimulated with antigen-derived peptides. E, Analysis of IFN-γ production by CD8pos splenocytes stimulated with anti-CD3/CD28 antibody-coated beads. D and E, The graphs represent the results as individual values with grouped mean ± SD (N = 1, n = 6).
The increase in IL10 and decrease in CCL5 after therapy with 50 MBq 177Lu-9079 prompted analysis of the antigen specificity of CD8pos splenocytes isolated when tumors reached 250 ± 100 mm3. These CD8pos splenocytes were stimulated with peptides, derived from antigens expressed by B16-huCD20 tumor cells, including ovalbumin, Trp2, gp100, and ADPGK (Fig. 4C). Production of IFNγ in response to the antigen-derived peptides, a measure of T-cell activation, was observed in some yet not all mice treated with 140 MBq 177Lu-9079 (Fig. 4D). IFNγ production was not observed in mice treated with 50 MBq 177Lu-9079 or 140 MBq 177Lu-R3B23 (Fig. 4D), despite T cells being able to produce IFNγ upon stimulation with anti-CD3/CD28 antibody–coated beads (Fig. 4E). These data suggest that low-dose 177Lu-9079 in the B16-huCD20 model does not induce tumor-specific T-cell responses, while high-dose has some T-cell activating capacity.
Low- and high-dose 177Lu-9079 induce a different immune signature
B16-huCD20 tumors from mice treated with 50 or 140 MBq of 177Lu-9079, or 140 MBq of 177Lu-R3B23 were isolated when they reached 250 ± 100 mm3. These were subjected to multiplex analysis to study the tumor immune signature. This analysis was performed on similar sized tumors, as the tumor stage can be considered a confounding factor.
Gene expression profiling followed by GSEA showed that tumors of mice treated with 140 MBq 177Lu-9079 displayed an immune signature characterized by upregulation of type I IFN, antigen processing and presentation, cell clearance and membrane invagination, and downregulation of TGFβ-signaling and Rho GTPase effectors compared with tumors of mice treated with 140 MBq 177Lu-R3B23 (Fig. 5A). In contrast, tumors treated with 50 MBq 177Lu-9079 were characterized by downregulated IL6, p73 and VEGF-signaling, and upregulated IL1-signaling, cytokine secretion, antigen processing and presentation, reactive oxygen species (ROS), and heat responses compared with tumors of mice treated with 140 MBq 177Lu-R3B23 (Fig. 5A). These differences in gene expression signature did not translate in major difference in total immune-cell infiltrate, yet translated in several nonstatistically significant differences in immune-cell distributions, determined using the nanostring nCounter scoring method (35, 36). The following immune cell types were decreased in tumors treated with 50 MBq 177Lu-9079 compared with tumors treated with 140 MBq 177Lu-9079 or 177Lu-R3B23: mast cells (P = 0.940; P = 0.918), exhausted CD8pos T-cells (P = 0.205; P = 0.190), cytotoxic cells (P = 0.684; P = 0.258), and linked herewith natural killer (NK) cells (P = 0.703; P = 0.0931; Fig. 5B; Supplementary Fig. S3). Notably, Th1 cells were significantly decreased in tumors treated with 50 MBq or 140 MBq 177Lu-9079 compared with tumors treated with 140 MBq 177Lu-R3B23 (P = 0.0347; P = 0.0390; Fig. 5B). Several immune cell types were increased, albeit not statistically significant, in tumors treated with 50 MBq or 140 MBq 177Lu-9079 compared with tumors treated with 140 MBq 177Lu-R3B23. These were CD8pos T-cells (P = 0.933; P = 0.583), dendritic cells (DC; P = 0.850; P = 0.901), neutrophils (P = 0.571; P = 0.743), and macrophages (P = 0.920; P = 0.768; Fig. 5B; Supplementary Fig. S3).
Figure 5.
NanoString RNA profiling of tumors treated with 50 or 140 MBq of 177Lu-9079 [TRT (50 MBq) and TRT (140 MBq), respectively], or 140 MBq 177Lu-R3B23 [CTRL (140 MBq)] shows a different gene expression profile. RNA was isolated from tumors with an average volume of 250 mm³. RNA samples with RNA Integrity values of 7.81 ± 0.35 were subjected to RNA profiling. A, Representation of the GSEA on gene expression changes in B16-huCD20 tumors treated with 50 MBq (right) or 140 MBq (left) 177Lu-9079 compared with 140 MBq 177Lu-R3B23. The color of the sphere indicates whether an increase (red) or decrease (blue) in the respective pathway has been observed. Immune and cell metabolism pathways are displayed. Pathways annotated with “*” and “**”, originate from the canonical pathways and hallmarks databases, while unmarked pathways originate from the Gene Ontology database. B, Immune cell scoring. The data are represented as 10 to 90 percentile Box and Whiskers (N = 1, n = 3). Tregs, regulatory T cells.
Multicolor flow cytometry analyses corroborated that tumors of an equal size treated with 50 or 140 MBq of 177Lu-9079, or 140 MBq of 177Lu-R3B23 showed similar immune cell levels (CD45pos, Fig. 6A; Supplementary Fig. S4). We observed a decrease in T cells (CD3pos, P = 0.0042; P = 0.0076) and increase in myeloid cells (CD11bpos, P = 0.0039; P = 0.0161) in tumors of mice treated with 50 and 140 MBq of 177Lu-9079 compared with tumors of mice treated with 177Lu-R3B23 (Fig. 6B). This decrease in T cells did not significantly influence the level of ovalbumin-specific CD8pos T cells (P = 0.0966; P = 0.5912; Fig. 6C). Within the T-cell population, a significantly lower percentage of CD3pos double negative (DN) T cells, lacking CD4 and CD8-expression, was observed in tumors of mice treated with 50 MBq 177Lu-9079 (P = 0.0035; Fig. 6D). Although not reaching significance, a similar observation was made with 140 MBq 177Lu-9079 (P = 0.0532; Fig. 6D). The fraction of CD4pos T cells was enhanced upon therapy with 50 MBq 177Lu-9079 (P = 0.0197; Fig. 6D). CD8pos T-cell levels were not significantly changed upon treatment with 50 and 140 MBq 177Lu-9079 (P = 0.1843; P = 0.1464; Fig. 6D). Analysis of the expression of CD279 (PD-1) on all T cells showed similarly low levels of PD-1 in all treatment conditions (Fig. 6E and F; Supplementary Fig. S5). For the myeloid cells, we observed an increase in myeloid cells and alternatively activated macrophages (M2 macrophages) in tumors treated with 140 MBq 177Lu-9079 (P = 0.0161 and P = 0.0505, respectively; Fig. 6G and H). Within the myeloid cells, we observed that CD274 (PD-L1) was significantly increased on conventional type 2 DCs (cDC2; P = 0.0286) and classically activated macrophages (M1 macrophages; P = 0.0286), while CD273 (PD-L2) was increased on non-cDC2 (P = 0.0082) in tumors treated with 140 MBq of 177Lu-9079 compared with 140 MBq 177Lu-R3B23 (Fig. 6I–L).
Figure 6.
Flow cytometry analysis of tumors treated with 177Lu-9079 versus 177Lu-R3B23 shows differences in immune composition. Multicolor flow cytometry analysis was performed on single-cell suspensions of tumors treated with 50 or 140 MBq of 177Lu-9079 [TRT (50 MBq) and TRT (140 MBq), respectively], or 140 MBq 177Lu-R3B23 [CTRL (140 MBq)] with an average volume of 250 mm³. A, Percentage of immune cells (CD45pos) within the tumor. B, Distribution of T cells (CD3εpos) versus myeloid cells [CD11bpos cells and nonconventional type 2 DCs (non-cDC2's)]. C, Percentage of CD8pos T cells able to recognize the surrogate antigen ovalbumin. D, Composition of the CD3εpos T-cell subpopulation, containing CD4pos, CD8pos, and CD4neg CD8neg DN T cells. E, Concatenated t-distributed Stochastic Neighbor Embedding (t-SNE) plot of all data acquired from the lymphoid lineage, overlayed with lymphoid cell populations (gating strategy, Supplementary Fig. S5A). F, Concatenated t-SNE heatmaps show the expression of PD-1 on T-cell subpopulations per treatment group. G, Percentage of myeloid cells amongst immune cells. H, Composition of the myeloid-cell population, containing CD11bpos Ly6Gpos neutrophils, Ly6Gneg CD11cpos MHC-IIhigh CD11bpos cDC2s, Ly6Gneg CD11cpos MHC-IIhigh CD11bneg (non-cDC2’s), Ly6Gneg CD11bpos F4-80pos MHC-IIhigh macrophages (M1 macrophages), Ly6Gneg CD11bpos F4-80pos MHC-IIlow macrophages (M2-macrophages), and the remaining cells are dubbed as miscellaneous myeloid cells (monocytes, mast cells, and eosinophils; gating strategy, Supplementary Fig. S5B). I, Concatenated t-SNE plot of all data acquired from the myeloid lineage, overlayed with myeloid-cell populations. J, Expression of PD-L1 and PD-L2 on myeloid subpopulations per treatment group. K, PD-L1 expression on myeloid-cell populations with significant upregulation upon TRT. L, PD-L2 expression on myeloid-cell populations with significant upregulation upon TRT. Results are presented as individual values with grouped mean ± SD (N = 1, n = 5–6).
Discussion
To date, 177Lu is a highly investigated therapeutic radionuclide in clinical practice, as evidenced by its inclusion in 70% of all ongoing TRT trials (37). 177Lu emits highly energetic β–-particles from its nucleus, which interact with cells and inflict damage. Cells in the vicinity of the β–-source and circumjacent nontargeted tissue suffer from its cross-firing effects (38). The β–-particles cause DNA-damage and induce ionization events, resulting in generation of ROS, which contributes to inducing cell damage (39, 40). It is further contended that immune activation upon TRT contributes to the therapy outcome (41–43). Compelling evidence for activation of CD8pos T cells that can recognize and kill tumor cells was recently delivered by Patel and colleagues, at a dose of 12 Gy (21). However, we showed that β–-TRT using 177Lu-sdAbs (4 to 10 Gy), although inducing ICD in vitro and tumor growth control in vivo, did not induce strong CD8pos T-cell responses, irrespective of the dose. Rather, we observed immune gene signatures that favor myeloid cells, that likely counter T-cell responses, both CD4 and CD8, due to high expression of PD-L1|2.
We evaluated accumulation of 177Lu-9079 in B16-huCD20 tumors and tissues that do not express huCD20 using ex vivo biodistribution analysis 1, 24, 48, and 72 hours after i.v. injection of 177Lu-sdAbs. 177Lu-9079 accumulates specifically in B16-huCD20 tumors when compared with 177Lu-R3B23, an ascertainment in line with previous work (16). Furthermore, we performed dosimetry and calculated that administration of 50 MBq of 177Lu-9079 corresponds to delivery of 4 Gy in tumor, with 140 MBq (10 Gy in tumor and 18 Gy in kidneys) being the highest radioactive dose that did not reveal kidney toxicity as demonstrated by a 99mTc-dimercaptosuccinic acid renal functionality assay in previous work (16). For therapeutic efficacy, a dose approximating 5 Gy was desired as literature described little therapeutic effect when β–-TRT is used as a monotherapy in that range, given solid tumors’ resilience upon radiation (21, 44). The observed delay in tumor growth with 177Lu-9079 is in line with already published preclinical and clinical studies that highlight the potential of 177Lu as a therapeutic radionuclide (4, 5, 16, 18, 45, 46). TRT in clinical practice consists of potent therapies employing peptides labeled with 177Lu such as Luthatera and 177Lu-PSMA-617, used to treat patients with somatostatin receptor positive neuroendocrine tumors and castration-resistant prostate cancer respectively (5, 47). These therapeutic agents are supplied in solution with an activity concentration in the same range as the one used in the in vitro TRT assay (around 370 MBq/mL). Hence, 177Lu-9079 described herein competes with the clinical state-of-the-art and remains a feasible application for clinical translation.
GSEA revealed a common disposition of both β–-TRT conditions to enhance antigen processing and presentation. Such functions are reserved to professional antigen-presenting cells (APC), e.g., macrophages and DCs, and corresponds with the higher myeloid cell infiltrate in tumor tissue (48). Intriguingly, the latter is no surprising observation as we demonstrated the ability of 177Lu-TRT in evoking ICD-hallmarks ATP-release and calreticulin, which in turn attract macrophages and DCs by binding to P2RY2 and CD91, respectively (49). Of note, subsequent RNA analyses showed distinct dose-dependent responses to β–-TRT, with 140 MBq eliciting an elevated IFNα signaling and a decreased TGFβ profile. Incidentally, recognition of cytosolic DNA fragments by cyclic GMP-AMP synthase (cGAS) incites type-I IFN production via the stimulator of IFN genes (STING) pathway (50). Furthermore, activation of cGAS-STING correlates with a repolarization of protumoral immune cells towards an antitumoral phenotype (51). STING activation upon β–-TRT has been described in a recent article, but no repolarization of M2 macrophages was observed in our model as the alternatively activated phenotype persisted upon β–-TRT (21). However, caution must be employed when comparing this recent work, as a single injection of 90Y-labeled NM600 was administered. Given the difference in isotope, treatment schedule, tumor model, and biodistribution, a number of factors come into play which can give rise to different outcomes. Complementary to IFNα-upregulation a decrease in TGFβ-signaling is a favorable outcome as well, given its tumor suppressive capacity (52). As for β–-TRT with 50 MBq, IL1, ROS, and overall cytokine secretion (except IL6) was upregulated, suggesting the involvement of inflammasomes over STING at a lower dose (53). An elevated ROS-signaling harbors either a pro- or an antitumoral effect. ROS is employed by antitumoral cells to dispose of malignant cells due to its toxic nature whilst a lower dose, unable to inflict cell death, promotes tumor growth (54). Altogether, the difference between the two treatment groups highlights a dose-dependent facet, which was also noted when assessing ICD hallmarks in the in vitro–TRT setting. Hence, surmising that the documented type-I IFN response could be linked to β–-TRT–induced ICD in the high-dose in vivo–TRT condition.
β–-TRT–mediated alteration of the tumor microenvironment (TME) is emphasized by an increase in immunosuppressive subsets of myeloid cells. More specifically, M2 macrophages are substantially increased in numbers. These cells are responsible for wound healing functions in physiologic conditions and constraining immune responses to prevent collateral tissue damage (55). In oncology, M2 macrophages are exploited to overexert immunosuppressive functions, subvert opposing antitumoral responses, mediate tumor progression, and metastasis (56). Further evidence supporting this statement is the elevated IL10-serum level upon β–-TRT, a recurring observation in literature (21). IL10 is a pleiotropic cytokine endowed with both pro- and antitumoral effects, shown to polarize macrophages to M2 phenotype and hampering Th1 functionality (57, 58). In this article, a significant decrease in Th1 cells was noted upon β–-TRT, which can be attributed to IL10 blunting the Th1 education (59–62).
Besides polarizing the TME in terms of immunosuppressive populations, β–-TRT alters the expression of immune checkpoints and serum cytokines as well. Assessment of immune checkpoint expression revealed an upregulation of PD-L1 and PD-L2 on various myeloid subsets, such as DCs and macrophages. DCs serve as a pivot in eliciting an antitumor immune response by presenting tumor antigens to T cells. Elevated PD-L1|2-expression interferes with this immune stimulating function, resulting in an impaired T-cell education (63–65). Similar observations have been described by Chen and colleagues, documenting the upregulation of PD-L1 on CD11bpos cells in the TME upon 177Lu-TRT directed against Integrin αvβ3 (26). CD11b is a marker for myeloid cells and represents not only cDC2 but macrophages, monocytes, and neutrophils alike. While we substantiate the results described by Chen and colleagues, we also provide a more detailed assessment of PD-L1 and PD-L2 expression. Notably, Chen and colleagues investigated tumors regardless of their volume, while we characterized tumors with a similar volume. Elevated PD-L1|2-expression on myeloid cells might signify a point of escape from an antitumor response, allowing tumors to persevere upon β–-TRT due to the inhibited T-cell education. These results might explain the success of combining blockade of PD-L1 with β–-TRT (26, 44).
The PD-L1 and PD-L2/PD-1-axis is potent in attenuating immune responses, hence disrupting anti-cancer T cells. Further aggravation of T-cell impairment is instilled due to the increased serum level of the immunosuppressive IL10 and decreased CCL5, the latter being required to attract T cells (62, 66). The B16-huCD20 tumor model expresses the immunogenic antigen ovalbumin of which the SIINFEKL peptide is presented in MHC-I molecules to the TCR of CD8pos T-cells. This antigen is an elegant tool to study tumor-specific responses, e.g., by restimulating CD8pos T cells from spleens with cells that present the SIINFEKL peptide. Ultimately, restimulation of splenocytes with antigen-derived peptides failed to elicit IFNγ-production, which was expected based on the serum cytokine profile and the observation that APCs residing in the tumor, therefore loaded with antigens such as ovalbumin, show high inhibitory PD-L1 and PD-L2-expression. Conclusively, these data hint towards β–-TRT‘s inability to activate T cells as a monotherapy in the experimental set-up described in this manuscript. Changes in immune-cell composition, favor myeloid cells that likely are unable to activate strong CD8pos T-cell responses, given the inhibitory role of PD-L1|2 on T-cell activation. Our observations, however, also pave the way for novel combination therapies in the field of TRT.
Such future perspectives for ameliorating the therapeutic index of β–-TRT, consists of combination strategies such as IL1 or IL10 blockade. Despite the complex nature of these ILs and the controversial effects as monotherapies, combination with other therapeutic instances mends these shortcomings (67, 68). However, caution should be employed since such therapies are often associated with immune-related adverse events (69). On a different track, macrophage repolarization could prove to be a useful itinerary towards novel β–-TRT immunotherapies as well. Such an approach was described in a recent study, in which shuttling SMAC-mimetics to CD206pos cells resulted in the repolarization of macrophages from M2 to M1 phenotype (70). Hence, β–-TRT might benefit from combining with these novel therapies, potentiating therapeutic effects.
Conclusion
To our knowledge, this is the first study addressing immune activation in a melanoma model upon β–-TRT with 177Lu-sdAbs. Our findings suggest that this therapy delays tumor growth and induces subtle changes in immune activity within the TME that form an anchoring point for combination therapies.
Supplementary Material
Acknowledgments
This study was supported by grants from the Belgian Foundation against Cancer (grant no. 2016-076 FAF/F/2016/798); Kom op tegen Kanker (Stand-up to Cancer): the Flemish Cancer Society and the Research Council of the Vrije Universiteit Brussel (Strategic Research Program 48 and 62). Furthermore, T. Ertveldt, R.M. Awad, Q. Lecocq, and L. De Beck are PhD fellows funded via the FWO-V (grant nos. 1S06622N, 1S05020N, 1S24220N, and 1142819N, respectively). Y. De Vlaeminck was supported by FWO-V during the execution of this work (grant no. 1S24817N). M. Keyaerts and M. D'Huyvetter are respectively senior clinical investigator (1801619N) and postdoctoral fellow (12H3619N) of FWO-V. The BD Celesta flow cytometer was funded via an FWO-Hercules grant (grant no. IOO1618N). The FLEX nCounter Analysis System was funded by the Research Council of the Vrije Universiteit Brussel (grant no. OZR2933). We also thank C. Peleman for her technical support with mouse handling and K. De Ridder for her expertise in RNA sequencing and big data analysis.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Authors’ Disclosures
T. Ertveldt reports grants from Research Foundation-Flanders (FWO), Belgian Foundation against Cancer; and grants from Stand-up to Cancer during the conduct of the study. L. De Beck reports grants from European Commission (ERA-NET Transcan-2 JTC 2015 - G0H7216N), FWO-V (predoctoral research grant - 1164918N); and grants from Vrije Universiteit Brussel (strategic research program - SRP48) during the conduct of the study. H. Ceuppens reports grants from Kom op Tegen Kanker; and grants from Stichting tegen Kanker during the conduct of the study. Y. De Vlaeminck reports grants from FWO during the conduct of the study. R.M. Awad reports grants from FWO during the conduct of the study. M. Keyaerts reports grants from Stichting tegen Kanker and FWO-Vlaanderen (FWO-V) during the conduct of the study; other support from Precirix and Abscint outside the submitted work; in addition, M. Keyaerts has a patent for Human PD-L1-binding immunoglobulins pending and issued and a patent for Radio-labelled antibody fragments for use in the prognosis, diagnosis of cancer as well as for the prediction of cancer therapy response issued and licensed to Abscint. N. Devoogdt reports grants, personal fees, and nonfinancial support from Precirix; and no-financial support from Abscint outside the submitted work; and to develop single domain antibodies to image and treat cancer; in addition, he is a founder, consultant, and holds stock of the companies Precirix and Abscint that respectively develop radiotherapeutics and radiodiagnostics. He is an inventor on patents related to these activities. None of these activities are directly related to the publication by Ertveldt and colleagues. M. D'Huyvetter reports grants from FWO-V during the conduct of the study; personal fees from Precirix SA outside the submitted work; in addition, M. D'Huyvetter has a patent for use of sdabs for diagnosis and therapy pending and a patent for use of sdabs for diagnosis and therapy issued. No disclosures were reported by the other authors.
Authors’ Contributions
T. Ertveldt: Data curation, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. L. De Beck: Data curation, software, formal analysis, validation, visualization, writing–review and editing. K. De Ridder: Data curation, software, formal analysis, validation, visualization, writing–review and editing. H. Locy: Data curation, formal analysis, writing–review and editing. W. de Mey: Methodology, writing–review and editing. C. Goyvaerts: Formal analysis, visualization, writing–review and editing. Q. Lecocq: Formal analysis, writing–review and editing. H. Ceuppens: Formal analysis, writing–review and editing. Y. De Vlaeminck: Formal analysis, writing–review and editing. R.M. Awad: Investigation, writing–review and editing. M. Keyaerts: Conceptualization, resources, funding acquisition, project administration, writing–review and editing. N. Devoogdt: Conceptualization, resources, funding acquisition, writing–review and editing. M. D'Huyvetter: Conceptualization, resources, funding acquisition, project administration, writing–review and editing. K. Breckpot: Conceptualization, resources, data curation, supervision, funding acquisition, project administration, writing–review and editing. A. Krasniqi: Conceptualization, resources, data curation, formal analysis, supervision, validation, visualization, methodology, writing–review and editing.
References
- 1. Malcolm J, Falzone N, Lee BQ, Vallis KA. Targeted radionuclide therapy: new advances for improvement of patient management and response. Cancers 2019;11:268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bavelaar BM, Lee BQ, Gill MR, Falzone N, Vallis KA. Subcellular targeting of theranostic radionuclides. Front Pharmacol 2018;9:996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Turner JH. An introduction to the clinical practice of theranostics in oncology. Br J Radiol 2018;91:20180440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gill MR, Falzone N, Du Y, Vallis KA. Targeted radionuclide therapy in combined-modality regimens. Lancet Oncol 2017;18:e414–23. [DOI] [PubMed] [Google Scholar]
- 5. Strosberg J, El-Haddad G, Wolin E, Hendifar A, Yao J, Chasen B, et al. Phase 3 trial of 177 Lu-Dotatate for midgut neuroendocrine tumors. N Engl J Med 2017;376:125–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sartor O, de Bono J, Chi KN, Fizazi K, Herrmann K, Rahbar K, et al. Lutetium-177–PSMA-617 for metastatic castration-resistant prostate cancer. N Engl J Med 2021;385:1091–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Emmanouilides C. Review of Y-ibritumomab tiuxetan as first-line consolidation radio-immunotherapy for B-cell follicular non-Hodgkin's lymphoma. Cancer Manag Res 2009;1:131–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Witzig TE, White CA, Gordon LI, Wiseman GA, Emmanouilides C, Murray JL, et al. Safety of Yttrium-90 ibritumomab tiuxetan radioimmunotherapy for relapsed low-grade, follicular, or transformed non-Hodgkin's lymphoma. J Clin Oncol 2003;21:1263–70. [DOI] [PubMed] [Google Scholar]
- 9. Scott AM, Wolchok JD, Old LJ. Antibody therapy of cancer. Nat Rev Cancer 2012;12:278–87. [DOI] [PubMed] [Google Scholar]
- 10. Marcucci F, Bellone M, Rumio C, Corti A. Approaches to improve tumor accumulation and interactions between monoclonal antibodies and immune cells. mAbs 2013;5:34–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xing Y, Chand G, Liu C, Cook GJR, O'Doherty J, Zhao L, et al. Early phase I study of a 99mTc-labeled anti-programmed death ligand-1 (PD-L1) single-domain antibody in SPECT/CT assessment of PD-L1 expression in non-small cell lung cancer. J Nucl Med 2019;60:1213–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lecocq Q, De Vlaeminck Y, Hanssens H, D'Huyvetter M, Raes G, Goyvaerts C, et al. Theranostics in immuno-oncology using Nanobody derivatives. Theranostics 2019;9:7772–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Debie P, Devoogdt N, Hernot S. Targeted Nanobody-based molecular tracers for nuclear imaging and image-guided surgery. Antibodies 2019;8:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Keyaerts M, Xavier C, Heemskerk J, Devoogdt N, Everaert H, Ackaert C, et al. Phase I study of 68Ga-HER2-Nanobody for PET/CT assessment of HER2 expression in breast carcinoma. J Nucl Med 2016;57:27–33. [DOI] [PubMed] [Google Scholar]
- 15. Vaneycken I, Govaert J, Vincke C, Caveliers V, Lahoutte T, De Baetselier P, et al. In vitro analysis and in vivo tumor targeting of a humanized, grafted Nanobody in mice using pinhole SPECT/micro-CT. J Nucl Med 2010;51:1099–106. [DOI] [PubMed] [Google Scholar]
- 16. Krasniqi A, D'Huyvetter M, Xavier C, Van der Jeught K, Muyldermans S, Van Der Heyden J, et al. Theranostic radiolabeled anti-CD20 sdAb for targeted radionuclide therapy of non-Hodgkin lymphoma. Mol Cancer Ther 2017;16:2828–39. [DOI] [PubMed] [Google Scholar]
- 17. Lemaire M, D'Huyvetter M, Lahoutte T, Van Valckenborgh E, Menu E, De Bruyne E, et al. Imaging and radioimmunotherapy of multiple myeloma with anti-idiotypic Nanobodies. Leukemia 2014;28:444–7. [DOI] [PubMed] [Google Scholar]
- 18. D'Huyvetter M, Vincke C, Xavier C, Aerts A, Impens N, Baatout S, et al. Targeted radionuclide therapy with A 177 Lu-labeled anti-HER2 Nanobody. Theranostics 2014;4:708–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Puttemans J, Dekempeneer Y, Eersels JL, Hanssens H, Debie P, Keyaerts M, et al. Preclinical targeted α- and β −-radionuclide therapy in her2-positive brain metastasis using camelid single-domain antibodies. Cancers 2020;12:1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. D'Huyvetter M, Vos J, Caveliers V, Vaneycken I, Heemskerk J, Duhoux F, et al. Phase I trial of 131 I-GMIB-anti-HER2-VHH1, a new promising candidate for HER2-targeted radionuclide therapy in breast cancer patients. J Nucl Med 2021;62:1097–105. [DOI] [PubMed] [Google Scholar]
- 21. Patel RB, Hernandez R, Carlson P, Grudzinski J, Bates AM, Jagodinsky JC, et al. Low-dose targeted radionuclide therapy renders immunologically cold tumors responsive to immune checkpoint blockade. Sci Transl Med 2021;13:eabb3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ghodadra A, Bhatt S, Camacho JC, Kim HS. Abscopal effects and Yttrium-90 radioembolization. Cardiovasc Intervent Radiol 2016;39:1076–80. [DOI] [PubMed] [Google Scholar]
- 23. Kwee SA, Lim J, Coel MN. Soft tissue response on 18F-fluorocholine PET/CT in metastatic castrate-resistant prostate cancer treated with 223Ra-Dichloride. Clin Nucl Med 2017;42:868–71. [DOI] [PubMed] [Google Scholar]
- 24. Wehrenberg-Klee E, Goyal L, Dugan M, Zhu AX, Ganguli S. Y-90 radioembolization combined with a PD-1 inhibitor for advanced hepatocellular carcinoma. Cardiovasc Intervent Radiol 2018;41:1799–802. [DOI] [PubMed] [Google Scholar]
- 25. Rouanet J, Benboubker V, Akil H, Hennino A, Auzeloux P, Besse S, et al. Immune checkpoint inhibitors reverse tolerogenic mechanisms induced by melanoma targeted radionuclide therapy. Cancer Immunol Immunother 2020;69:2075–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen H, Zhao L, Fu K, Lin Q, Wen X, Jacobson O, et al. Integrin αvβ3-targeted radionuclide therapy combined with immune checkpoint blockade immunotherapy synergistically enhances anti-tumor efficacy. Theranostics 2019;9:7948–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pinc A, Somasundaram R, Wagner C, Hörmann M, Karanikas G, Jalili A, et al. Targeting CD20 in melanoma patients at high risk of disease recurrence. Mol Ther 2012;20:1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lang D, Mascarenhas JB, Shea CR. Melanocytes, melanocyte stem cells, and melanoma stem cells. Clin Dermatol 2013;31:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kepp O, Senovilla L, Vitale I, Vacchelli E, Adjemian S, Agostinis P, et al. Consensus guidelines for the detection of immunogenic cell death. OncoImmunology 2014;3:e955691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. van Eerd JEM, Vegt E, Wetzels JFM, Russel FGM, Masereeuw R, Corstens FHM, et al. Gelatin-based plasma expander effectively reduces renal uptake of 111In-octreotide in mice and rats. J Nucl Med 2006;47:528–33. [PubMed] [Google Scholar]
- 31. Locy H, Correa RJM, Autaers D, Schiettecatte A, Jonckheere J, Waelput W, et al. Overcoming the challenges of high quality RNA extraction from core needle biopsy. Biomolecules 2021;11:621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bhattacharya A, Hamilton AM, Furberg H, Pietzak E, Purdue MP, Troester MA, et al. An approach for normalization and quality control for NanoString RNA expression data. Briefings Bioinf 2021;22:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011;27:1739–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Danaher P, Warren S, Dennis L, D'Amico L, White A, Disis ML, et al. Gene expression markers of tumor infiltrating leukocytes. J Immunother Cancer 2017;5:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Locy H, Verhulst S, Cools W, Waelput W, Brock S, Cras L, et al. Assessing tumor-infiltrating lymphocytes in breast cancer: a proposal for combining immunohistochemistry and gene expression analysis to refine scoring. Front Immunol 2022;13:794175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. National Institutes of Health. Clinical TRNT-trials | Oncology - List Results - ClinicalTrials.gov [Internet]. 2021[cited 2021 May 14]. Available from: https://clinicaltrials.gov/ct2/results?recrs=ab&cond=Oncology&term=targeted+radionuclide+therapy&cntry=&state=&city=&dist=.
- 38. Aghevlian S, Boyle AJ, Reilly RM. Radioimmunotherapy of cancer with high linear energy transfer (LET) radiation delivered by radionuclides emitting α-particles or Auger electrons. Adv Drug Deliv Rev 2017;109:102–18. [DOI] [PubMed] [Google Scholar]
- 39. Quintiliani M. The oxygen effect in radiation inactivation of DNA and enzymes. Int J Radiat Biol Relat Stud Phys Chem Med 2009;50:573–94. [DOI] [PubMed] [Google Scholar]
- 40. Rockwell S, Dobrucki IT, Kim EY, Marrison ST, Vu VT. Hypoxia and radiation therapy: past history, ongoing research, and future promise. Curr Mol Med 2009;9:442–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lumniczky K, Sáfrány G. The impact of radiation therapy on the antitumor immunity: local effects and systemic consequences. Cancer Lett 2015;356:114–25. [DOI] [PubMed] [Google Scholar]
- 42. Brady D, O'Sullivan JM, Prise KM. What is the role of the bystander response in radionuclide therapies? Front Oncol 2013;3:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marín A, Martín M, Liñán O, Alvarenga F, López M, Fernández L, et al. Bystander effects and radiotherapy. Rep Pract Oncol Radiother 2015;20:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Choi J, Beaino W, Fecek RJ, Fabian KPL, Laymon CM, Kurland BF, et al. Combined VLA-4–targeted radionuclide therapy and immunotherapy in a mouse model of melanoma. J Nucl Med 2018;59:1843–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tagawa ST, Milowsky MI, Morris M, Vallabhajosula S, Christos P, Akhtar NH, et al. Phase II study of lutetium-177-labeled anti-prostate-specific membrane antigen monoclonal antibody J591 for metastatic castration-resistant prostate cancer. Clin Cancer Res 2013;19:5182–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Calais J, Gafita A, Eiber MR, Armstrong WR, Gartmann J, Thin P, et al. Prospective phase 2 trial of PSMA-targeted molecular RadiothErapy with 177 Lu-PSMA-617 for metastatic Castration-reSISTant Prostate Cancer (RESIST-PC): efficacy results of the UCLA cohort. J Nucl Med 2021;62:1440–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tatkovic A, McBean R, Wong D. Lu177-PSMA therapy for men with advanced prostate cancer: 18 months survival analysis in a single Australian tertiary institution. J Med Imaging Radiat Oncol 2021;65:740–7. [DOI] [PubMed] [Google Scholar]
- 48. Germic N, Frangez Z, Yousefi S, Simon H-U. Regulation of the innate immune system by autophagy: monocytes, macrophages, dendritic cells and antigen presentation. Cell Death Differ 2019;26:715–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fucikova J, Kepp O, Kasikova L, Petroni G, Yamazaki T, Liu P, et al. Detection of immunogenic cell death and its relevance for cancer therapy. Cell Death Dis 2020;11:1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Röhl I, et al. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 2013;498:380–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Downey CM, Aghaei M, Schwendener RA, Jirik FR. DMXAA causes tumor site-specific vascular disruption in murine non-small cell lung cancer, and like the endogenous non-canonical cyclic dinucleotide STING agonist, 2′3′-cGAMP, induces M2 macrophage repolarization. PLoS One 2014;9:e99988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Katz LH, Li Y, Chen J-S, Muñoz NM, Majumdar A, Chen J, et al. Targeting TGF-β signaling in cancer. Expert Opin Ther Targets 2013;17:743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chan AH, Schroder K. Inflammasome signaling and regulation of interleukin-1 family cytokines. J Exp Med 2020;217:e20190314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Weinberg F, Ramnath N, Nagrath D. Reactive oxygen species in the tumor microenvironment: an overview. Cancers 2019;11:1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kim SY, Nair DMG. Macrophages in wound healing: activation and plasticity. Immunol Cell Biol 2019;97:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Awad RM, De Vlaeminck Y, Maebe J, Goyvaerts C, Breckpot K. Turn back the TIMe: targeting tumor infiltrating myeloid cells to revert cancer progression. Front Immunol 2018;9:1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mannino MH, Zhu Z, Xiao H, Bai Q, Wakefield MR, Fang Y. The paradoxical role of IL-10 in immunity and cancer. Cancer Lett 2015;367:103–7. [DOI] [PubMed] [Google Scholar]
- 58. Lopes RL, Borges TJ, Zanin RF, Bonorino C. IL-10 is required for polarization of macrophages to M2-like phenotype by mycobacterial DnaK (heat shock protein 70). Cytokine 2016;85:123–9. [DOI] [PubMed] [Google Scholar]
- 59. Mills CD, Kincaid K, Alt JM, Heilman MJ, Hill AM. M-1/M-2 macrophages and the Th1/Th2 paradigm. J Immunol 2000;164:6166–73. [DOI] [PubMed] [Google Scholar]
- 60. Jenkins S, Allen J. Similarity and diversity in macrophage activation by nematodes, trematodes, and cestodes. J Biomed Biotechnol 2010;2010:262609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Saraiva M, O'Garra A. The regulation of IL-10 production by immune cells. Nat Rev Immunol 2010;10:170–81. [DOI] [PubMed] [Google Scholar]
- 62. Couper KN, Blount DG, Riley EM. IL-10: the master regulator of immunity to infection. J Immunol 2008;180:5771–7. [DOI] [PubMed] [Google Scholar]
- 63. Peng Q, Qiu X, Zhang Z, Zhang S, Zhang Y, Liang Y, et al. PD-L1 on dendritic cells attenuates T cell activation and regulates response to immune checkpoint blockade. Nat Commun 2020;11:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Oh SA, Wu D-C, Cheung J, Navarro A, Xiong H, Cubas R, et al. PD-L1 expression by dendritic cells is a key regulator of T-cell immunity in cancer. Nat Cancer 2020;1:681–91. [DOI] [PubMed] [Google Scholar]
- 65. Latchman Y, Wood CR, Chernova T, Chaudhary D, Borde M, Chernova I, et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat Immunol 2001;2:261–8. [DOI] [PubMed] [Google Scholar]
- 66. Huffman AP, Lin JH, Kim SI, Byrne KT, Vonderheide RH. CCL5 mediates CD40-driven CD4+ T cell tumor infiltration and immunity. JCI Insight 2020;5:e137263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Llopiz D, Aranda F, Díaz-Valdés N, Ruiz M, Infante S, Belsúe V, et al. Vaccine-induced but not tumor-derived Interleukin-10 dictates the efficacy of Interleukin-10 blockade in therapeutic vaccination. Oncoimmunology 2015;5:e1075113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kaplanov I, Carmi Y, Kornetsky R, Shemesh A, Shurin GV, Shurin MR, et al. Blocking IL-1β reverses the immunosuppression in mouse breast cancer and synergizes with anti–PD-1 for tumor abrogation. Proc Natl Acad Sci U S A 2019;116:1361–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rider P, Carmi Y, Cohen I. Biologics for targeting inflammatory cytokines, clinical uses, and limitations. Int J Cell Biol 2016;2016:9259646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. De Vlaeminck Y, Lecocq Q, Giron P, Heirman C, Geeraerts X, Bolli E, et al. Single-domain antibody fusion proteins can target and shuttle functional proteins into macrophage mannose receptor expressing macrophages. J Control Release 2019;299:107–20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available within the article, its Supplementary data files, and Gene Expression Omnibus (GEO) online repository. Biodistribution files, multiplex data, fcs files from flowcytometric analysis are available upon request from the corresponding author. Raw and normalized nCounter Gene Expression Data obtained by analyzing the gene expression of treated B16-huCD20 melanoma samples using the nCounter Human PanCancer Immune Profiling panel, has been made publicly available via the GEO database (accession number: GSE199624).



![Figure 1. In vitro treatment of B16-huCD20 cells with 177Lu-9079 induces cell death that coincides with exposure of calreticulin and release of ATP. B16-huCD20 cells were treated for 1 hour with 370 MBq/mL 177Lu-R3B23 [CTRL (370 MBq/mL)] or 177Lu-9079 [TRT (dose)] or 48 hours of 2 μmol/L MTX (2 μmol/L), respectively. Flow cytometry was performed 48 hours after addition of 177Lu-sdAb to study the percentage of dead cells (A), cell surface calreticulin on viable cells (B), and ATP-release by viable cells (C). The bar graphs show individual datapoints and grouped mean ± SD (N = 1, n = 3).](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/4128/9377759/a54415c705ae/1136fig1.jpg)
![Figure 3: Therapy with low and high doses of 177Lu-9079 impacts on B16-huCD20 tumor growth. A, Schematic representation of the 177Lu-sdAbs treatment regimen detailing experimental conditions: 50 MBq of 177Lu-9079 [TRT (50 MBq)], 140 MBq of 177Lu-9079 [TRT (140 MBq)], and 140 MBq 177Lu-R3B23 [CTRL (140 MBq)]. Tumor kinetic after treatment are shown by group during the course of the experiment (B) and on day 13 (C), by individual mouse (D) and Kaplan–Meier curve showing the time at which mice reached humane endpoints by group (E). F, Weight of treated mice. The bar graphs show individual datapoints and grouped mean ± SD (N = 1, n = 6).](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/4128/9377759/6e8025259f49/1136fig3.jpg)
![Figure 4: Peripheral immune responses upon treatment with 177Lu-9079. Multiplex analysis of serum cytokine levels of IL10 (A) and CCL5 (B) was performed on serum isolated 6 hours after last administration of 50 MBq or 140 MBq of 177Lu-9079 [TRT (50 MBq) and TRT (140 MBq), respectively], or 140 MBq 177Lu-R3B23 [CTRL (140 MBq)]. The graphs represent the results as individual values with grouped mean ± SD (N = 1, n = 4). C, Agarose gel visualizing amplicons resulting from the PCR reaction with gp100 (90 bp), Trp2 (71 bp), ADPGK (400 bp), and ovalbumin (346 bp) specific primer sets. D, Analysis of IFNγ production by CD8pos splenocytes stimulated with antigen-derived peptides. E, Analysis of IFN-γ production by CD8pos splenocytes stimulated with anti-CD3/CD28 antibody-coated beads. D and E, The graphs represent the results as individual values with grouped mean ± SD (N = 1, n = 6).](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/4128/9377759/e9452a464a89/1136fig4.jpg)
![Figure 5. NanoString RNA profiling of tumors treated with 50 or 140 MBq of 177Lu-9079 [TRT (50 MBq) and TRT (140 MBq), respectively], or 140 MBq 177Lu-R3B23 [CTRL (140 MBq)] shows a different gene expression profile. RNA was isolated from tumors with an average volume of 250 mm³. RNA samples with RNA Integrity values of 7.81 ± 0.35 were subjected to RNA profiling. A, Representation of the GSEA on gene expression changes in B16-huCD20 tumors treated with 50 MBq (right) or 140 MBq (left) 177Lu-9079 compared with 140 MBq 177Lu-R3B23. The color of the sphere indicates whether an increase (red) or decrease (blue) in the respective pathway has been observed. Immune and cell metabolism pathways are displayed. Pathways annotated with “*” and “**”, originate from the canonical pathways and hallmarks databases, while unmarked pathways originate from the Gene Ontology database. B, Immune cell scoring. The data are represented as 10 to 90 percentile Box and Whiskers (N = 1, n = 3). Tregs, regulatory T cells.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/4128/9377759/ad342e68d4bd/1136fig5.jpg)
![Figure 6. Flow cytometry analysis of tumors treated with 177Lu-9079 versus 177Lu-R3B23 shows differences in immune composition. Multicolor flow cytometry analysis was performed on single-cell suspensions of tumors treated with 50 or 140MBq of 177Lu-9079 [TRT (50MBq) and TRT (140MBq), respectively], or 140MBq 177Lu-R3B23 [CTRL (140MBq)] with an average volume of 250 mm³. A, Percentage of immune cells (CD45pos) within the tumor. B, Distribution of T cells (CD3εpos) versus myeloid cells [CD11bpos cells and nonconventional type 2 DCs (non-cDC2’)]. C, Percentage of CD8pos T cells able to recognize the surrogate antigen ovalbumin. D, Composition of the CD3εpos T-cell subpopulation, containing CD4pos, CD8pos, and CD4neg CD8neg DN T cells. E, Concatenated t-distributed Stochastic Neighbor Embedding (t-SNE) plot of all data acquired from the lymphoid lineage, overlayed with lymphoid cell populations (gating strategy, Supplementary Fig. S5A). F, Concatenated t-SNE heatmaps show the expression of PD-1 on T-cell subpopulations per treatment group. G, Percentage of myeloid cells amongst immune cells. H, Composition of the myeloid-cell population, containing CD11bpos Ly6Gpos neutrophils, Ly6Gneg CD11cpos MHC-IIhigh CD11bpos cDC2s, Ly6Gneg CD11cpos MHC-IIhigh CD11bneg (non-cDC2’s), Ly6Gneg CD11bpos F4-80pos MHC-IIhigh macrophages (M1 macrophages), Ly6Gneg CD11bpos F4-80pos MHC-IIlow macrophages (M2-macrophages), and the remaining cells are dubbed as miscellaneous myeloid cells (monocytes, mast cells, and eosinophils; gating strategy, Supplementary Fig. S5B). I, Concatenated t-SNE plot of all data acquired from the myeloid lineage, overlayed with myeloid-cell populations. J, Expression of PD-L1 and PD-L2 on myeloid subpopulations per treatment group. K, PD-L1 expression on myeloid-cell populations with significant upregulation upon TRT. L, PD-L2 expression on myeloid-cell populations with significant upregulation upon TRT. Results are presented as individual values with grouped mean ± SD (N = 1, n = 5–6).](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/4128/9377759/89554b988555/1136fig6.jpg)