Abstract
Streptococcus agalactiae is a poorly transformable bacterium and studies of molecular mechanisms are difficult due to the limitations of genetic tools. Employing the novel pGh9:ISS1 transposition vector we generated plasmid-based mutant libraries of S. agalactiae strains O90R and AC475 by random chromosomal integration. A screen for mutants with a nonhemolytic phenotype on sheep blood agar led to the identification of a genetic locus harboring several genes that are essential for the hemolytic function and pigment production of S. agalactiae. Nucleotide sequence analysis of nonhemolytic mutants revealed that four mutants had distinct insertion sites in a single genetic locus of 7 kb that was subsequently designated cyl. Eight different open reading frames were identified: cylX, cylD, cylG, acpC, cylZ, cylA, cylB, and cylE, coding for predicted proteins with molecular masses of 11, 33, 26, 11, 15, 35, 32, and 78 kDa, respectively. The deduced amino acid sequence of the protein encoded by cylA harbors a conserved ATP-binding cassette (ABC) motif, and the predicted proteins encoded by cylA and cylB have significant similarities to the nucleotide binding and transmembrane proteins of typical ABC transporter systems. Transcription analysis by reverse transcription-PCR suggests that cylX to cylE are part of an operon. The requirement of acpC and cylZABE for hemolysin production of S. agalactiae was confirmed either by targeted mutagenesis with the vector pGh5, complementation studies with pAT28, or analysis of insertion elements in naturally occurring nonhemolytic mutants.
Advances in understanding the function of bacterial genes depend largely on available genetic tools. Streptococcus agalactiae (group B streptococcus [GBS]) is an important human and animal pathogen (10, 20, 40), but very little is known about the genetic basis of its biological functions. The creation and screening of mutant libraries led to the identification and functional characterization of numerous genes in bacteria closely related to S. agalactiae, such as Streptococcus pneumoniae, Enterococcus faecalis, and Lactococcus lactis (4, 8, 27, 34). Characterization and identification of molecular mechanisms in S. agalactiae have been hampered by the limitations of available genetic tools. Tn916 mutagenesis has been applied successfully to identify the capsular genes (31), but it requires conjugation, and its usefulness has been limited by restricted target site specificity. Transposition appears to be nonrandom, occurring preferentially at a conserved consensus target sequence. The use of another transposon, Tn917, in GBS has been reported only recently (13). It appears to lead to efficient mutagenesis by random chromosomal integration and awaits further evaluation. Aside from these techniques no genetic method that has been used successfully for the creation of genomic mutant libraries in GBS has been published.
The hemolysin of S. agalactiae is expressed by the vast majority of all strains and was first described by Todd in 1934 (37). It appears to be a surface-associated molecule that can be extracted from the cells by a solution containing detergents and high-molecular-weight carrier molecules such as starch or albumin (23). Due to a rapid loss of hemolytic function in bacterial extracts, the biochemical nature of the molecule has not been elucidated yet. Attempts to identify and characterize the hemolysin by generating antibodies have not been successful (7). A strong correlation exists between the amount of hemolytic activity and the production of an orange-red pigment, suggesting a very close genetic linkage between these two properties (36).
The genetic basis for the hemolysin and pigment production of S. agalactiae remains unclear. A putative hemolysin gene has been identified by a screen for chromosomal GBS fragments that are able to confer a hemolytic phenotype to Escherichia coli (6). However, subsequent studies showed that a targeted mutation of the gene does not result in a loss of hemolytic function (26), indicating that the gene does not encode the major hemolysin of S. agalactiae. Studies of isogenic nonhemolytic and hyperhemolytic mutants revealed the presence of an 11-kDa protein that is overexpressed in hyperhemolytic mutants and absent in nonhemolytic mutants (26), but the corresponding gene has not been identified so far.
We report the successful application of the novel pGh9:ISS1 transposition vector to generate mutant libraries in different GBS strains. The mutant libraries were used to screen for genetic determinants of the hemolytic function of GBS, which led to the identification of a genetic locus that is essential for the production of hemolysin.
MATERIALS AND METHODS
Bacterial strains.
The E. coli and S. agalactiae strains used in this study are listed in Table 1. E. coli DH5 α served as a host for recombinant pGh5 and pAT28 plasmids; E. coli XL1-Blue MRF (Stratagene, Heidelberg, Germany) was used as a host for the phage Lambda ZAP Express.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5 α | endA1 hsdR17 supE44 ΔlacU169(φ80lacZDM15) recA1 gyrA96 thi-1 relA1 | Boehringer |
| XL1-Blue MRF | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| EC 101 | E. coli JM101 derivative with repA from pWV01 integrated into the chromosome | 21 |
| Streptococcus agalactiae | ||
| AC475 | Clinical wild-type isolate; Hly+ | Aachen collection |
| HLY26 | AC475 derivative acpC::pGh9:ISS1; Hly− | This study |
| HLY26ex | AC475 derivative acpC::ISS1; Hly− | This study |
| HLY27 | AC475 derivative cylE::pGh9:ISS1; Hly− | This study |
| HLY27ex | AC475 derivative cylE::ISS1; Hly− | This study |
| HLY33 | AC475 derivative cylE::pGh9:ISS1; Hly− | This study |
| HLY33ex | AC475 derivative cylE::ISS1; Hly− | This study |
| O90R (ATCC 12386) | R. Lancefield grouping strain; Hly+ | ATCCa |
| HLY22 | O90R derivative acpC::pGh9:ISS1; Hly− | This study |
| HLY22ex | O90R derivative acpC::ISS1; Hly− | This study |
| HLY226ex | O90R derivative acpC::ISS1 Δ cylZABE; Hly− | This study |
| CYLZ-K1 | O90R derivative cylZ::pGh5; Hly− | This study |
| CYLB-K2 | O90R derivative cylB::pGh5; Hly− | This study |
| BSP1969 | HLY26ex carrying plasmid pBS1964; Hly+ | This study |
| R268 | Clinical isolate; Hly− | Aachen collection |
| HLY6683 | Clinical isolate; Hly− | This study |
| AC 650 | Clinical isolate; Hly− | This study |
| HLY1000 | Clinical isolate; Hly− | This study |
| Plasmids | ||
| pGh9:ISS1 | Eryr ori Ts | 22 |
| pGh5 | Eryr ori pBR ori Ts | Appligene |
| pBS1890 | pGh5 derivative carrying an internal 247-bp fragment of cylZ | This study |
| pBS1913 | pGh5 derivative carrying an internal 428-bp fragment of cylB | This study |
| pAT28 | Specr ori pUC ori pAmβ1 | 38 |
| pBS1964 | pAT28 derivative carrying an 833-bp acpC-cylZ fragment | This study |
ATCC, American Type Culture Collection.
S. agalactiae isolates were cultured on Columbia agar (Oxoid, Basingstoke, England) supplemented with 3% sheep blood, in Todd-Hewitt broth (THB) (Oxoid) or in THB supplemented with 0.5% yeast (THY) at 37°C. Mutant strains harboring chromosomally integrated pGh5 and pGh9:ISS1 vectors were subcultured in medium containing erythromycin (5 mg/liter) at a temperature of ≥37°C. Mutant strains harboring pAT28 plasmids in the cytoplasm were grown in medium containing spectinomycin (60 mg/liter).
General DNA techniques.
Standard recombinant DNA techniques were employed for nucleic acid preparation and analysis. PCR was carried out with Taq polymerase according to the manufacturer’s protocol (Boehringer, Mannheim, Germany), with 35 cycles of amplification steps of 1 min at 94°C, 1 min at 50 to 56°C, and 1 to 3 min at 72°C depending on product size. Genomic streptococcal DNA was isolated as described previously (24). Plasmid DNA was isolated and purified using the Qiaprep Spin Miniprep kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Plasmids and PCR products were sequenced on an ABI 373 automated DNA sequencer using the ABI PRISM Dye terminator cycle sequencing kit (PE Applied Biosystems, Weiterstadt, Germany). GBS strains were transformed according to the protocol of Ricci et al. (30).
Phage techniques.
A Lambda ZAP Express library of strain AC475 was created as described by Podbielski et al. (28). Briefly, 200 μg of genomic DNA was digested with 0.2 U of Sau3A (Boehringer) for 30 min at 37°C. The resulting DNA fragments were separated according to size by a salt gradient technique (12). Fractions containing fragments 2 to 9 kb in length were ligated with BamHI-digested λ arms and packaged by using a Gigapack II packaging kit (Stratagene). Further processing and plaque lifting followed the manufacturer’s instructions. The library was screened by hybridization with PCR products at 65°C overnight. The PCR products were labeled by adding Dig-dUTP (Boehringer) to the PCR at a final concentration of 5 μM. Detection of positive plaques by CSPD (Boehringer) followed the manufacturer’s instructions.
Construction of mutants.
Mutant libraries of strain O90R and strain AC475 were constructed by chromosomal insertion of the novel pGh9:ISS1 vector (22). The vector system combines the insertion sequence ISS1 with the thermosensitive replicon pG+host, allowing the replication of plasmids in the streptococcal host at the permissive temperature of 30°C and chromosomal integration of the entire plasmid at temperatures of ≥37°C. S. agalactiae strains were transformed by electroporation with 1 μg of purified plasmid DNA, and plasmid-containing strains were selected on erythromycin (5 μg/ml)-containing blood agar plates at 30°C. To induce chromosomal integration of the plasmid, one of the isolates was grown overnight in THB supplemented with erythromycin (5 μg/ml) at 30°C. The saturated culture was diluted 1:100 in fresh THB medium without antibiotics and incubated at 30°C for 3 h. To reduce the plasmid copy number, the culture was transferred to a 38°C water bath and incubated for another 2 h. Samples were diluted, plated on erythromycin-containing blood agar plates, and grown overnight at ≥37°C.
To identify the chromosomal insertion site, total genomic DNA of the pGh9:ISS1 mutants was digested with EcoRI or HindIII, ligated, and transformed into E. coli EC101(8) (kindly provided by K. Leenhouts) (21). Erythromycin-resistant E. coli clones harboring pGh9:ISS1 plasmid with genomic streptococcal DNA located adjacent to the insertion site of the plasmid were selected on tryptic soy agar plates supplemented with erythromycin (150 mg/liter). Nucleotide sequencing of the inserted genomic DNA was performed with primers annealing to pGh9:ISS1 vector sequences (5′-CGA GGT CGA CGG TAT CG-3′, 5′-TAG ACT TAT CAG GAA ACT TTG C-3′).
The pGh9:ISS1 vector system facilitates the excision of chromosomally inserted plasmids by the induction of rolling-circle replication. This manipulation leads to stable mutants harboring a single ISS1 element at the initial insertion site. Stable mutants were obtained from the pGh9:ISS1 generated nonhemolytic strains.
The plasmid pGh5 was used for targeted genetic mutagenesis of cylZ and cylB. Two mutants of the O90R wild-type strain were created by plasmid insertion at nucleotides 3095 and 4786 of the cyl operon, respectively. Internal fragments of the cylZ and the cylB gene were amplified by PCR with primer 5′-GGC GGC GGA TCC GGT GGA TAG AGT GCA GAG-3′ paired with 5′-GGC GGC GAA TTC CAC ATC GGC TAA ATA ACC-3′ and primer 5′-CGC CGC GGA TCC TAA TTA TTG GCT CAT TGC AGG-3′ paired with 5′-CGC CGC GAA TTC CTA CCG AGC CAA TTG AGA G-3′, respectively; the newly introduced BamHI and EcoRI restriction sites are underlined. Resulting PCR products and the vector were digested with BamHI and EcoRI, ligated, and transformed into E. coli. Chromosomal integration into S. agalactiae O90R was performed as previously described (22). To confirm correct chromosomal insertion of the plasmid, PCR with primers annealing to vector sequences and genomic nucleotide sequence upstream or downstream of the duplication site followed by DNA sequencing of PCR products was employed to confirm chromosomal insertion for both mutants.
For complementation studies, recombinant pAT28 plasmids were transformed into the S. agalactiae strain HLY26ex. Recombinant plasmids harboring DNA fragments of nucleotides 2407 to 3231 of the cyl operon were constructed in E. coli by PCR of the genomic DNA of strain AC475 with primers 5′-GGC GGC GAA TTC AAG AGG TGG CTT GGC TCG-3′ and 5′-GGC GGC GGA TCC AAC ATC CTT CCT GAG GC-3′ and subsequent restriction digestion and ligation into the pAT28 plasmid. Purified plasmids were electroporated into S. agalactiae HLY26ex. Successful transfer of pAT28 plasmids into the cytoplasm of S. agalactiae was confirmed by plasmid preparation and subsequent hybridization with a probe to the inserted streptococcal DNA.
Amplification of the cyl operon in naturally occurring nonhemolytic mutants.
The nucleotide sequence of the cyl operon in nonhemolytic mutants was analyzed by generating a set of six different PCR products which comprise the entire DNA sequence of the operon. The following primers were used for amplification: PCR 1, 5′-TAA CTT GTG GGC TCT TGG-3′ and 5′-ACA TTG TTC ACA CCT ACT C-3′; PCR 2, 5′-ATG ATA TTT TAA TTA GAG TGT G-3′ and 5′-AAT CGT AAT ACC ATC TGA TGC-3′; PCR 3, 5′-GTA TTC TGA GTT TCT TAC GG-3′ and 5′-AAT CGT AAT ACC ATC TGA TGC-3′; PCR 4, 5′-CTG GTA TTG TTA GAG ATG GC-3′ and 5′-CAT GTG AAT AGT ATC ACT GG-3′; PCR 5, 5′-GAT GCA GGT TAT CTC AGA C-3′ and 5′-ATA TAA TTC ACT GTC TCT TGG-3′; and PCR 6, 5′-TCC CTT GGA AGA GGA GAC-3′ and 5′-GCA AGA TAA CCT TTC TCA CC-3′.
Determination of hemolytic activity in microtiter plate assays.
For the preparation of hemolysin extracts, bacteria were grown to mid-logarithmic phase in 50 ml of THY medium, pelleted by centrifugation, washed and resuspended in 500 μl of phosphate-buffered saline (PBS) containing 0.2% glucose, 1% dextran 500 (Serva, Heidelberg, Germany), and 3% Tween 80. After 30 min of incubation at 37°C in 5% CO2 the suspension was centrifuged and the supernatant containing the hemolysin was removed. For determination of hemolytic activity in microtiter plate assays 50 μl of hemolysin extracts was diluted up to a concentration of 1:1,024 in PBS. Fresh human erythrocytes from healthy volunteers were washed and adjusted to a 1% suspension in PBS, and then 15 μl was added to each well and incubation was done at 37°C for 2 h.
RNA preparation and analysis.
Total RNA was prepared from GBS strain AC475 grown to an OD of 0.6 to 0.8 in 10 ml of THB medium supplemented with 0.5% starch and 1% bovine serum. Cells were lysed mechanically by glass beads in a cell disrupter with the FastRNA kit (Bio 101, Vista, Calif.). Purification of the RNA followed the manufacturer’s instructions. Four reverse transcription (RT) reactions were carried out with 1 μg of RNA as the template and a solution containing 2 pmol of one of the indicated primers (A, 5′-GCT ACT ATT CCA ATC TAA ACA ACC-3′; B, 5′-CAT GTG AAT AGT ATC ACT GG-3′; C, 5′-AAC CGT ACA AAG CAC CTC-3′; D, 5′-AAT CGT AAT ACC ATC TGA TGC-3′) 0.1 M dithiothreitol (DTT), 10 mM deoxynucleoside triphosphate (dNTP) mix, and 200 U of SUPERSCRIPT II reverse transcriptase (Gibco BRL) in 50 mM Tris-HCl (pH 8.3), 75 mM KCl, and 3 mM MgCl2 at 42°C for 50 min in a 20-μl reaction volume. Five-microliter aliquots of the reaction mixtures were used as templates for a subsequent PCR with the following primers: A, 5′-TAT GGG ATG CTA TCG CAC-3′ and 5′-GCT ACT ATT CCA ATC TAA ACA ACC-3′; B, 5′-AAG AGG TGG CTT GGC TCG-3′ and 5′-CAT GTG AAT AGT ATC ACT GG-3′; C, 5′-TCA TAG AAA AGG ATG CGG AG-3′ and 5′-AAC CGT ACA AAG CAC CTC-3′; and D, 5′-ATG ATA TTT TAA TTA GAG TGT G-3′ and 5′-AAT CGT AAT ACC ATC TGA TGC-3′. The mapping of the transcription start site was performed with 5′-AAT CGT AAT ACC ATC TGA TGC-3′ as the reverse primer of the RT reaction; subsequent PCRs were carried out with the forward primers 5′-ATG ATA TTT TAA TTA GAG TGT G-3′, 5′-TAT GAA AGT GTT GAC AAA GAT G-3′, and 5′-GAG TGT TTA GAA GTT TTA GGC-3′ in combination with the reverse primer.
Nucleotide sequence accession number.
The nucleotide sequence of the coding regions for the S. agalactiae cyl genes has been submitted to the EMBL/GenBank/DDBJ nucleotide sequence data libraries and was assigned the accession no. AF093787.
RESULTS
Construction of S. agalactiae mutant libraries.
The novel pGh9:ISS1 vector was employed for the construction of mutant libraries from S. agalactiae strains O90R and AC475. To determine the transposition frequencies in S. agalactiae the number of Emr colonies at ≥37°C was measured. Transposition frequencies were 5 × 10−3 for strain O90R and 1.8 × 10−2 for strain AC475, indicating that the pGh9:ISS1 vector can be used for efficient mutagenesis in different GBS strains.
Identification of the cyl operon and analysis of ISS1 insertion sites.
To evaluate the usefulness of pGh9:ISS1 mutant libraries in S. agalactiae, we chose the loss of hemolysin production as a selection marker. Several thousand colonies of the pGh9:ISS1 mutant libraries from strain O90R and AC475 were screened for a loss of hemolytic activity on blood agar plates. Four mutants displayed a nonhemolytic phenotype and were selected for further characterization. Analysis of the DNA sequence at the insertion sites was performed as described by Maguin et al. (22) and revealed that two mutants (HLY22 and HLY26) harbored plasmid insertions in the same open reading frame; the mutants originated from different parent strains and the insertion sites were located 153 bp apart. Since these results indicated a close association between the hemolytic phenotype and the mutated genetic locus, further analysis of the nucleotide sequence adjacent to the insertion site was performed. It revealed that the plasmid integration sites of the two other mutants (HLY27 and HLY33) are also located in the same chromosomal region.
Chromosomal integration of the vector leads to a duplication of the ISS1 element and of an 8-bp target site sequence. To analyze the insertion sites of ISS1 in the nonhemolytic S. agalactiae mutant strains, a comparison of the target sites of the pGh9:ISS1 plasmid was performed. Each of the four different mutant strains contained a different target site (AATGATCA, GAAAATAG, ATAAGACA, and AATACGTT).
Nucleotide and protein sequence analysis.
Nucleotide sequence upstream and downstream of the initially identified chromosomal fragment was obtained by screening of a λ phage library. Based on the nucleotide sequence of this chromosomal fragment, PCR products were generated and used as hybridization probes. The nucleotide sequence of positive plaques was subsequently used to design primers and generate PCR products of the genomic DNA of strain AC475. DNA sequencing of the resulting PCR products revealed the presence of eight different open reading frames, which were designated cylX, cylD, cylG, acpC, cylZ, cylA, cylB, and cylE. A 21-bp inverted repeat (TGATATACTCCCCTTATAGTG....CACTATAAGGGGAGTATATCA) is located 552 to 482 bp upstream of the ATG start codon of cylX and presumably functions as a transcription terminator for genes upstream. All of the cyl genes are oriented in the same direction and have typical ribosome binding sites located 5 to 15 bp upstream of the ATG start codon. Deduced proteins were compared to those in the GenBank database. Percentages of identity and similarity of the amino acid sequence to similar protein sequences were calculated by gapped BLAST using the blastp 2.0 algorithm accessible at the NCBI website (1). The results of these analyses are summarized in Table 2. An analysis of the deduced proteins of cylA and cylB was performed with the PCgene program (Intelligenetics, Geneva, Switzerland). The 309-amino-acid CylA protein harbors a Walker A motif at amino acids 35 to 42 (GPNGAGKT) and a Walker B motif at amino acids 134 to 145 (LSGGQKRKVDIA) (39). Analysis of the deduced protein encoded by cylB for membrane associated regions was performed with the PCgene program using the method of Eisenberg et al. (9) and revealed the presence of six putative membrane-associated helices. These are amino acids 22 to 42 (SFLSVIILILVYQIFLGKIQL), 65 to 85 (WLIAGLTTIISMTSTLGAFGV), 114 to 134 (IFAVLFGIVMTMFSCIFAIGI), 154 to 174 (GIVSLGTVLSAAMILPILAFI), 185 to 205 (TIVGTFIGFISGVYLSIGSVG), and 267 to 287 (HFMLIYIIALILILLAIHFII) of the 292-amino-acid protein.
TABLE 2.
Comparison of deduced proteins of the cyl gene cluster with proteins in the GenBank databasea
| Gene | Length (aa) of deduced protein | Molecular mass (kDa) of deduced protein | Similar gene | Region of similarity (aa) | % Identity (% similarity) | Function |
|---|---|---|---|---|---|---|
| cylX | 101 | 11 | Unknown | |||
| cylD | 282 | 33 | B. subtilis fabD | 85–277 | 19 (46) | Malonyl-CoA-ACP transacylase |
| cylG | 240 | 26 | B. subtilis fabG | 2–238 | 45 (65) | 3-Ketoacyl-ACP reductase |
| acpC | 101 | 11 | S. coelicolor acp | 5–92 | 25 (51) | ACP |
| cylZ | 131 | 15 | E. coli fabZ | 4–126 | 39 (62) | Fatty acid elongation |
| cylA | 309 | 35 | Rhizobium sp. nodI | 3–224 | 35 (55) | ABC transporter (ATP-binding domain) |
| cylB | 292 | 32 | Rhizobium sp. nodJ | 150–236 | 22 (46) | ABC transporter (transmembrane domain) |
| cylE | 667 | 78 | Unknown |
Abbreviations: aa, amino acids; nt, nucleotides; CoA, coenzyme A.
Construction of cyl mutants.
To confirm the association between a nonhemolytic phenotype and the disruption of single genes in the cyl gene cluster, we generated additional mutants in two different ways. Targeted mutagenesis was performed by insertion duplication mutagenesis, and we obtained stable mutants by excision of the pGh9:ISS1 vector from the nonhemolytic mutant strains that were selected in the initial screen (Table 1). The plasmid pGh5 (2) was used for targeted genetic mutagenesis of cylZ and cylB. 247- and 428-bp internal fragments of cylZ and cylB, respectively, were amplified by PCR, subcloned into pGh5 in E. coli, and subsequently transformed and integrated into the genomic DNA of S. agalactiae O90R. The resulting mutants (CYLZ-K1 and CYLB-K2) are nonhemolytic and harbor the pGh5 plasmid integrated between the duplicated internal fragment of cylZ and cylB, respectively (Fig. 1).
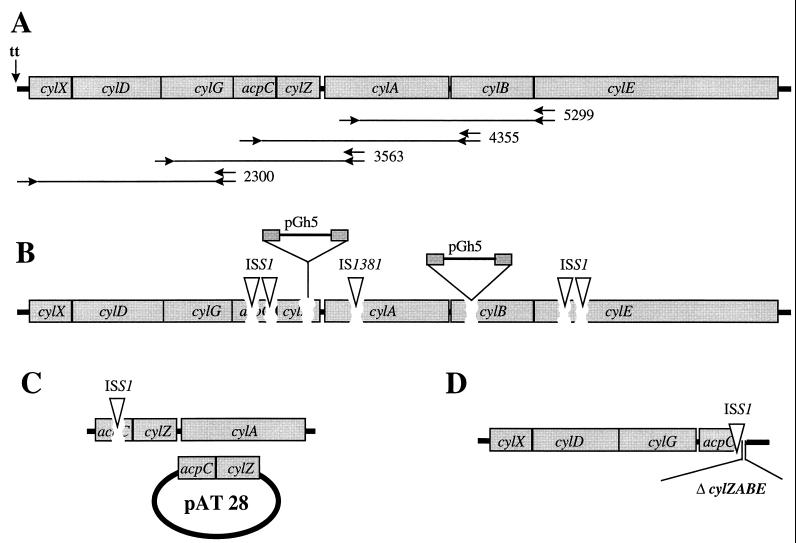
FIG. 1.
Graphic map of the cyl gene cluster. (A) The transcription terminator (tt) upstream of the putative transcription start site is indicated. Primer annealing sites for the RT-PCR experiments are represented by arrows, and the resulting PCR products are indicated. (B) DNA integration sites in nonhemolytic S. agalactiae mutants. Strains were either constructed as described in Materials and Methods or collected from routine clinical specimens. Mutants from the original libraries that were generated through pGh9:ISS1 integration and subsequent excision are indicated as ISS1; the IS1381-like insertion element in naturally occurring nonhemolytic mutants is shown as IS1381. For mutants that were generated through targeted pGh5 integration, the plasmid integration site is marked. (C) Strain BSP1969. The HLY26ex mutant (harboring a copy of the ISS1 element in acpC) was transformed with a copy of the acpC and cylZ gene integrated into the pAT28 plasmid. (D) Genomic structure of the HLY226ex deletion mutant.
Excision of the pGh9:ISS1 vector results in mutants with a single copy of the ISS1 element inserted at the initial integration site. These mutants are stable and do not require antibiotic pressure, and polar effects on downstream genes are reduced in comparison to the integration of the entire vector. Excision of the vector from the four pGh9:ISS1 strains with insertions in the cyl genes generated S. agalactiae mutants with a nonhemolytic phenotype. In one of these pGh9:ISS1 mutants (HLY22) the excision led to a deletion of genomic DNA at the integration site of the ISS1 element. The deletion comprises the last 28 nucleotides of the acpC gene and the entire region cylZ to cylE, demonstrating that these genes are not essential for regular growth and viability of S. agalactiae.
Hemolysin extracts.
The nonhemolytic phenotype of the mutant strains was confirmed by a microtiter plate assay employing hemolysin extracts. Extracts were obtained from the hemolytic wild-type strains (AC475 and O90R) and all of the nonhemolytic mutants listed in Table 1. Incubation with a 1% suspension of human erythrocytes confirmed that in contrast to the wild-type strains, no hemolytic activity can be extracted from the different mutant strains that we constructed or from the nonhemolytic clinical isolates (data not shown).
Transcription analysis.
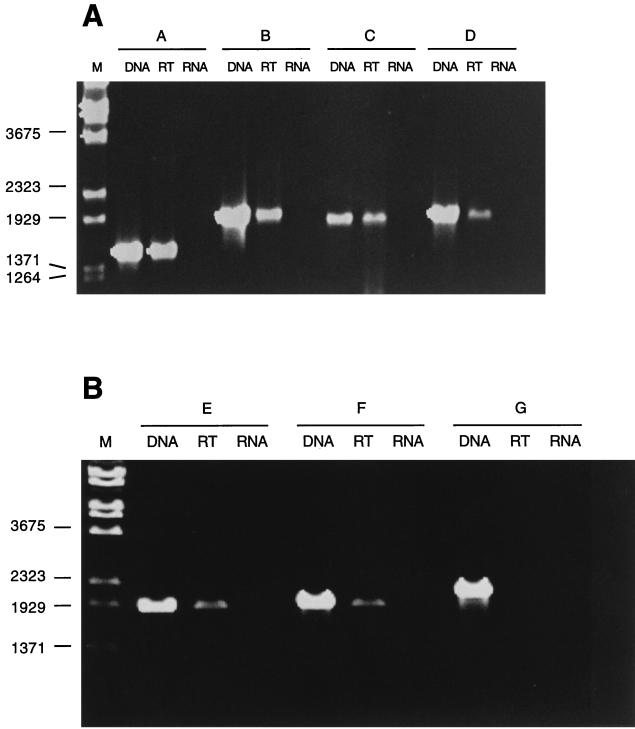
Transcription analysis of cyl genes was performed by RT-PCR. Based on the close association of open reading frames in this genetic locus, we hypothesized that several of the cyl genes are cotranscribed. RT was performed with primers annealing at nucleotides 5299 to 5276 (A) nucleotides 4355 to 4336 (B) nucleotides 3563 to 3546 (C), and nucleotides 2300 to 2280 (D) of the cyl locus (Fig. 1 and 2). The subsequent PCRs amplified PCR products between 1.5 and 2.0 kb from all of the RT reactions, indicating that genes cylX to cylE are located on a common transcript (Fig. 2). In an attempt to map the transcription start site RT-PCR was performed with primers annealing 45 to 25 bp upstream (G) and 152 to 173 (F) and 202 to 223 bp downstream (E) of the 21-bp inverted repeat, which presumably functions as a transcription terminator for genes upstream of the cyl gene cluster. PCR products could only be generated in reactions E and F in which the forward primers anneal downstream of the putative transcription terminator (Fig. 2). These results indicate that a major transcript for the cyl genes starts in a chromosomal region that is located between 482 and 331 bp upstream of the cylX start codon.
FIG. 2.
(A) Transcription analysis was performed by RT-PCR. Primers A to D, annealing at nucleotides 5299 to 5276, nucleotides 4355 to 4336, nucleotides 3563 to 3546, and nucleotides 2300 to 2280, respectively, were used for RT. PCR of chromosomal DNA was used as a positive control (lanes DNA). The PCR performed on mRNA subjected to an RT reaction is shown in lanes RT; PCR that was performed on mRNA without prior RT reaction (lanes RNA) served as a negative control. Lane M, molecular mass marker (in nucleotides). (B) RT-PCR analysis of the transcription start site. RT reactions were carried out with a primer annealing at nucleotides 2300 to 2280 of the cyl gene cluster. Subsequent PCRs were performed with forward primers annealing upstream (G) or downstream (E and F) of the putative transcription terminator at nucleotides 35 to 55, nucleotides 352 to 372, and nucleotides 301 to 321, respectively. Positive and negative controls are indicated.
Complementation studies.
The requirement of an acyl carrier protein (ACP) for the expression of a hemolytic phenotype has been shown for the E. coli hemolysin (19). In order to confirm that a functional acpC gene is also required for the hemolytic phenotype of S. agalactiae, we performed complementation studies in strain HLY26ex, which harbors an ISS1 copy integrated into the acpC gene. An 833-bp fragment of the cyl locus of strain AC475, comprising the open reading frames acpC and cylZ and 90 nucleotides of the chromosomal region upstream of the acpC startcodon, was amplified by PCR and subcloned into the plasmid pAT28 in E. coli (38) (Fig. 1). Upon transfer of the plasmid into S. agalactiae HLY26ex, the nonhemolytic strain partially regained a hemolytic phenotype. The blood agar underneath the colonies shows a beta-hemolytic zone the size of the removed colony (Fig. 3). These results support the hypothesis that acpC is required for hemolysin production and that the nonhemolytic phenotype of acpC knockout mutants is not caused by polar effects on the putative ATP-binding cassette (ABC) transporter genes, or cylE.
FIG. 3.
Hemolytic activity on blood agar of the AC 475 wild-type strain (A), the HLY26ex mutant (B), and the BSP1969 complementation mutant (C). The presence or absence of beta-hemolysis underneath removed colonies is indicated by arrows.
Analysis of naturally occurring nonhemolytic mutants.
Nine naturally occurring, nonhemolytic S. agalactiae strains were screened for the presence of the cyl genes by PCR. Six different PCRs spanning the 7-kb cyl operon were performed on each of the strains. Subsequent agarose gel electrophoreses demonstrated that in three of the strains the PCR amplifying the cylA gene generated a product which was about 1 kb bigger than the corresponding PCR product of the wild-type gene from strain AC475 (data not shown). DNA sequencing revealed the presence of a recently identified insertion sequence (IS1381 variant; GenBank accession no. AF064785) in these strains, integrated at codon 158 of the cylA gene, which codes for the nucleotide binding part of the putative ABC transporter. These strains were obtained from routine clinical specimens at Aachen University Hospital, and all three strains harbored the insertion element 473 bp downstream of the ATG start codon. The nonhemolytic strains were epidemiologically unrelated, and three out of nine harbored the insertion element at the same nucleotide position, indicating that the insertion of the IS1381 variant insertion element at codon 158 is a frequent event leading to the loss of hemolytic function.
Pigment production in nonhemolytic mutants.
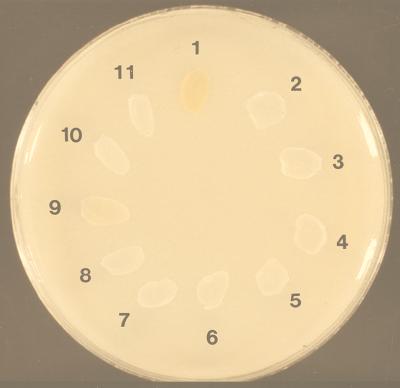
A close linkage between the hemolytic phenotype and the production of pigment has been reported by Tapsall (36). To investigate if the nonhemolytic mutants that we obtained were pigmented, the mutants were cultured on THB plates supplemented with 5% starch (Fig. 4) under anaerobic conditions. All mutants that displayed a nonhemolytic phenotype lost their ability to produce pigment. Our results suggest that the cyl locus is required for the hemolysin and pigment production of S. agalactiae.
FIG. 4.
Pigment production of hemolytic (Hly+) wild-type strains, nonhemolytic (Hly−) mutants, and nonhemolytic wild-type strains. Bacteria were grown anaerobically at 37°C on THB–5% starch under anaerobic conditions. Isolates: 1, AC 475 (Hly+); 2 to 5, clinical isolates (Hly−); CYLZ-K1 (Hly−); 7, CYLB-K2 (Hly−); 8, HLY22 (Hly−); 9, O90R wild type (Hly+); 10, HLY26ex (Hly−); 11, HLY26 (Hly−).
DISCUSSION
In the present study we employed the novel pGh9:ISS1 vector system to select nonhemolytic S. agalactiae mutants. Transposition frequencies of this plasmid in S. agalactiae strains and analysis of the duplicated target sequence demonstrate that efficient chromosomal integration can be achieved by pGh9:ISS1 mutagenesis. The vector has been shown to transpose randomly in L. lactis, Streptococcus thermophilus, and E. faecalis (22). Due to the limited number of analyzed mutants, a statistical analysis of the integration sites in the chromosomal DNA of S. agalactiae could not be performed. However, the presence of four distinct integration sites in a single genetic locus of 7 kb with different nucleotide sequences at each of the four target sites suggests that random integration of this plasmid also occurs in S. agalactiae. Using this novel system we could isolate a gene cluster that was subsequently designated cyl and is associated with the hemolytic phenotype of S. agalactiae.
The cyl locus harbors eight different open reading frames that were subsequently designated cylX, cylD, cylG, acpC, cylZ, cylA, cylB, and cylE. Screening of the initial S. agalactiae mutant library, analysis of nonhemolytic wild-type strains and the targeted mutagenesis of single genes in the acpC-cylE region led to the isolation of several different nonhemolytic mutants within the cyl gene cluster (Fig. 1). Attempts to generate targeted mutants of the genes cylX, cylD, and cylG by insertion duplication mutagenesis failed, which might indicate that either these mutations are lethal or that they could not be achieved for technical reasons, such as the size of the open reading frame.
The deduced amino acid sequence of the genes cylD, cylG, acpC, and cylZ demonstrate significant homologies to enzymes of prokaryotic fatty acid biosynthesis (Table 2). CylD and CylG resemble FabD and FabG of Bacillus subtilis (25) and could be essential for the constitutive fatty acid elongation reaction of S. agalactiae. In addition to the observed homologies to the fab genes of B. subtilis the genetic organization of the cyl genes in S. agalactiae (cylD, cylG, acpC) resembles the fab gene cluster of B. subtilis (fabD, fabG, acpP) (25).
AcpC has a 51% similarity with acp the ACP of S. coelicolor. Acylation is essential for the function of the E. coli hemolysin and other RTX toxins of gram-negative bacteria (for a recent review, see the work of Stanley et al. [35]), and the Nod factors of Rhizobium (33). Apart from their role in fatty acid synthesis, ACPs are required for the synthesis of the E. coli hemolysin (19), the polyketide synthesis of Streptomyces sp. (29), and the production of the cell host signaling Nod factors of Rhizobium species (32). In E. coli a single ACP appears to participate in fatty acid biosynthesis and hemolysin production. In contrast Rhizobium sp. and Streptomyces sp. have constitutive ACPs for fatty acid biosynthesis and specialized ACPs that allow the synthesis of unusual metabolites without interfering with the synthesis of essential cellular lipids. AcpC, a putative ACP, does not appear to be essential for regular fatty acid biosynthesis in S. agalactiae, since knockout mutants of this gene were isolated and show regular growth characteristics. Nizet et al. reported the increased production of an 11-kDa protein in hyperhemolytic S. agalactiae strains and the absence of this protein in nonhemolytic strains (26). Interestingly the deduced proteins of acpC and cylX both have a predicted molecular mass of 11 kDa.
The next open reading frame downstream of acpC is cylZ. The predicted protein encoded by cylZ exhibits a 62% similarity to FabZ, which participates in the elongation cycles of unsaturated fatty acids (17). The attempt to excise the pGh9:ISS1 vector from the initial HLY22 mutant led to the isolation of one nonhemolytic clone with a complete deletion of the cylZ gene (Fig. 1), a strong indication that cylZ cannot be essential for regular fatty acid synthesis in S. agalactiae. Our results suggest that the cylZ gene product has a specialized function that is required for the expression of a regular hemolytic phenotype.
The deduced amino acid sequences of cylA and cylB demonstrate significant homologies to prokaryotic and eukaryotic ABC transporters. ABC transporters are required for the export of hemolysin from E. coli (11), Bordetella pertussis (15) and E. faecalis (14). The typical ABC transporter consists of two transmembrane domains and two highly conserved ATP-binding domains; for a review see the work of Higgins (18). The ATP-binding domain of the transporter is characterized by the presence of two motifs, termed Walker A and B (39), and shows considerable protein sequence identities in prokaryotic and eukaryotic transporters. Both Walker motifs are present in the deduced protein of cylA which has significant similarity to the nucleotide binding region of ABC transporters of Rhizobium sp. (NodI) (5), B. subtilis (NatA) (3), and Streptomyces (DrrA) (16). The transmembrane domains of ABC transporters are highly hydrophobic, and the majority of transporters are predicted to have six membrane-spanning segments per domain. CylB codes for a hydrophobic protein with six potential membrane-associated segments. In addition considerable sequence similarities to NodJ and NatB, the transmembrane proteins of the Nod factor transporter of Rhizobium meliloti and the Na+ extruder of B. subtilis, were noted. Both genes, cylA and cylB, appear to be required for the hemolytic and pigmented phenotype of S. agalactiae wild-type strains, since several naturally occurring nonhemolytic S. agalactiae strains harbor an insertion sequence in the cylA gene and targeted pGh5 mutants of cylB exhibit a nonhemolytic and nonpigmented phenotype. We propose that CylA and CylB constitute a typical ABC transporter catalyzing an ATP-dependent export of hemolysin and pigment.
The function of the cylE gene that codes for a protein with a predicted molecular mass of 78 kDa remains unclear. Two independent ISS1 mutants of cylE are nonhemolytic; the predicted protein, however, exhibits no similarity to known hemolysins. Since attempts to raise antibodies against the S. agalactiae hemolysin have been unsuccessful (7) and hemolytic extracts of S. agalactiae do not reveal the presence of a large streptococcal protein it seems unlikely that the gene product of cylE represents the hemolysin.
Transcription analysis of the eight genes present in this chromosomal locus was performed by RT-PCR and suggests that all of the cyl genes are located on one major transcript. This does not however exclude the presence of additional transcription start sites within this gene cluster. The complementation studies showed that an acpC mutant could be partially complemented with a construct containing 90 nucleotides of the chromosomal region upstream of acpC, acpC, and cylZ. Complementation of the nonhemolytic phenotype by this plasmid suggests that at least a residual transcription of acpC can be initiated from the construct.
In conclusion we were able to show that the pGh9:ISS1 vector is a useful genetic tool for the study and analysis of S. agalactiae. Its application led to the identification of a genetic locus that is required for a hemolytic phenotype and involved in the production of pigment. Analysis of the disrupted open reading frames in this locus suggests that, in analogy to the hemolysin of E. coli or the Nod factor of Rhizobium sp., the hemolysin of S. agalactiae is transported via an ABC transporter and requires acylation.
ACKNOWLEDGMENT
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (Sp 511/2-1) to B.S.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biswas I, Gruss A, Ehrlich D, Maguin E. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J Bacteriol. 1993;175:3628–3635. doi: 10.1128/jb.175.11.3628-3635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng J, Guffanti A A, Krulwich T A. A two-gene ABC-type transport system that extrudes Na+ in Bacillus subtilis is induced by ethanol or protonophore. Mol Microbiol. 1997;23:1107–1120. doi: 10.1046/j.1365-2958.1997.2951656.x. [DOI] [PubMed] [Google Scholar]
- 4.Christie P, Dunny G. Identification of regions of the Streptococcus faecalis plasmid pCF-10 that encode antibiotic resistance and pheromone response functions. Plasmid. 1986;15:230–241. doi: 10.1016/0147-619x(86)90041-7. [DOI] [PubMed] [Google Scholar]
- 5.Cloutier J, Laberge S, Prevost D, Antoun H. Sequence and mutational analysis of the common nodBCIJ region of Rhizobium sp. (Oxytropis arctobia) strain N33, a nitrogen fixing microsymbiont of both arctic and temperate legumes. Mol Plant-Microbe Interact. 1996;9:523–531. doi: 10.1094/mpmi-9-0523. [DOI] [PubMed] [Google Scholar]
- 6.Conrads G, Podbielski A, Lütticken R. Molecular cloning and nucleotide sequence of the group B streptococcal hemolysin. Zentbl Bakteriol. 1991;275:179–194. doi: 10.1016/s0934-8840(11)80064-2. [DOI] [PubMed] [Google Scholar]
- 7.Dal M C, Monteil H. Hemolysin produced by group B Streptococcus agalactiae. FEMS Microbiol Lett. 1983;16:89–94. [Google Scholar]
- 8.Duwat P, Cochu A, Ehrlich S, Gruss A. Characterization of Lactococcus lactis UV-sensitive mutants obtained by ISS1 transposition. J Bacteriol. 1997;179:4473–4479. doi: 10.1128/jb.179.14.4473-4479.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisenberg D, Schwarz E, Komaromy M, Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- 10.Farley M M, Harvey R C, Stull T, Smith J D, Schuchat A, Wenger J D, Stephens D S. A population based assessment of invasive disease due to group B Streptococcus in nonpregnant adults. N Engl J Med. 1993;328:1807–1811. doi: 10.1056/NEJM199306243282503. [DOI] [PubMed] [Google Scholar]
- 11.Felmlee T, Pellet S, Welch R A. Nucleotide sequence of an Escherichia coli chromosomal hemolysin. J Bacteriol. 1985;163:5269–5273. doi: 10.1128/jb.163.1.94-105.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fink P. Using sodium chloride step gradients to fractionate DNA fragments. BioTechniques. 1991;10:447–449. [PubMed] [Google Scholar]
- 13.Framson P E, Nittayajarn A, Merry J, Youngman P, Rubens C E. New genetic techniques for group B streptococci: high efficiency transformation, maintenance of temperature-sensitive pWV01 plasmids, and mutagenesis with Tn917. Appl Environ Microbiol. 1997;63:3539–3547. doi: 10.1128/aem.63.9.3539-3547.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilmore M S, Segarra R A, Booth M C. An HlyB-type function is required for expression of the Enterococcus faecalis hemolysin/bacteriocin. Infect Immun. 1990;58:3914–3923. doi: 10.1128/iai.58.12.3914-3923.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glaser P, Sakamoto H, Bellalou J, Ullman A, Danchin A. Secretion of cyclolysin, the calmodulin-sensitive adenylate cyclase-haemolysin bifunctional protein of Bordetella pertussis. EMBO J. 1988;7:3997–4004. doi: 10.1002/j.1460-2075.1988.tb03288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guilfoile P G, Hutchinson C R. A bacterial analog of the mdr gene of mammalian tumor cells is present in Streptomyces peucetius, the producer of daunorubicin and doxorubicin. Proc Natl Acad Sci USA. 1991;88:8553–8557. doi: 10.1073/pnas.88.19.8553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heath R J, Rock C O. Roles of the FabA and FabZ β-hydroxylacyl-acyl carrier protein dehydratases in Escherichia coli fatty acid biosynthesis. J Biol Chem. 1996;271:27795–27801. doi: 10.1074/jbc.271.44.27795. [DOI] [PubMed] [Google Scholar]
- 18.Higgins C F. ABC transporters from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 19.Issartel J P, Koronakis V, Hughes C. Activation of Escherichia coli prohaemolysin to the mature toxin by acyl carrier protein-dependent fatty acylation. Nature. 1991;351:759–761. doi: 10.1038/351759a0. [DOI] [PubMed] [Google Scholar]
- 20.Keefe G P. Streptococcus agalactiae mastitis: a review. Can Vet J. 1997;38:429–437. [PMC free article] [PubMed] [Google Scholar]
- 21.Law J, Buist G, Haandrikman A, Kok J, Venema G, Leenhouts K. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J Bacteriol. 1995;177:7011–7018. doi: 10.1128/jb.177.24.7011-7018.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maguin E, Prevost H, Ehrlich S, Gruss A. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J Bacteriol. 1996;178:931–935. doi: 10.1128/jb.178.3.931-935.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchlewicz B A, Duncan J L. Properties of a hemolysin produced by group B streptococci. Infect Immun. 1980;30:805–813. doi: 10.1128/iai.30.3.805-813.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin N J, Kaplan E L, Gerber M A, Menegus M A, Randolph M, Bell K, Cleary P P. Comparison of epidemic and endemic group G streptococci by restriction enzyme analysis. J Clin Microbiol. 1990;28:1881–1886. doi: 10.1128/jcm.28.9.1881-1886.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morbidoni H R, de Mendoza D, Cronan J E J. Bacillus subtilis acyl carrier protein is encoded in a cluster of lipid biosynthesis genes. J Bacteriol. 1996;178:4794–4800. doi: 10.1128/jb.178.16.4794-4800.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nizet V, Gibson R, Rubens C. The role of GBS β-hemolysin expression in newborn lung injury. Adv Exp Med. 1997;418:627–630. doi: 10.1007/978-1-4899-1825-3_146. [DOI] [PubMed] [Google Scholar]
- 27.Pearce B J, Yin Y B, Masure H R. Genetic identification of exported proteins in Streptococcus pneumoniae. Mol Microbiol. 1993;9:1037–1050. doi: 10.1111/j.1365-2958.1993.tb01233.x. [DOI] [PubMed] [Google Scholar]
- 28.Podbielski A, Flosdorff A, Weber-Heynemann J. The group A streptococcal virR49 gene controls expression of four structural vir regulon genes. Infect Immun. 1995;63:9–20. doi: 10.1128/iai.63.1.9-20.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Revill W P, Bibb M J, Hopwood D A. Relationships between fatty acid and polyketide synthases from Streptomyces coelicolor A3(2): characterization of the fatty acid synthase acyl carrier protein. J Bacteriol. 1996;178:5660–5667. doi: 10.1128/jb.178.19.5660-5667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ricci M L, Manganelli R, Berneri C, Orefici G, Pozzi G. Electrotransformation of Streptococcus agalactiae with plasmid DNA. FEMS Microbiol Lett. 1994;119:47–52. doi: 10.1111/j.1574-6968.1994.tb06865.x. [DOI] [PubMed] [Google Scholar]
- 31.Rubens C E, Heggen L M, Haft R F, Wessels M R. Identification of cpsD, a gene essential for type III capsule expression in group B streptococci. Mol Microbiol. 1993;8:843–855. doi: 10.1111/j.1365-2958.1993.tb01631.x. [DOI] [PubMed] [Google Scholar]
- 32.Shearman C A, Rossen L, Johnston A W B, Downie J A. The Rhizobium leguminosarum nodulation gene nodF encodes a polypeptide similar to acyl carrier protein and is regulated by NodD plus a factor in pea root exudate. EMBO J. 1986;5:647–652. doi: 10.1002/j.1460-2075.1986.tb04262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spaink H P, et al. A novel highly unsaturated fatty acid moiety of lipo-oligosaccharide signals determines host specificity of Rhizobium. Nature. 1991;354:125–130. doi: 10.1038/354125a0. [DOI] [PubMed] [Google Scholar]
- 34.Spellerberg B, Cundell D R, Sandros J, Pearce B J, Idänpään-Heikkilä I, Rosenow C, Masure H R. Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol Microbiol. 1996;19:803–813. doi: 10.1046/j.1365-2958.1996.425954.x. [DOI] [PubMed] [Google Scholar]
- 35.Stanley P, Koronakis V, Hughes C. Acylation of Escherichia coli hemolysin: a unique protein lipidation mechanism underlying toxin function. Microbiol Mol Biol Rev. 1998;62:309–333. doi: 10.1128/mmbr.62.2.309-333.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tapsall J W. Pigment production by Lancefield group B streptococci (Streptococcus agalactiae) J Med Microbiol. 1986;21:75–81. doi: 10.1099/00222615-21-1-75. [DOI] [PubMed] [Google Scholar]
- 37.Todd E. A comparative serological study of streptolysins derived from human and animal infections with note on pneumococcal hemolysin, tetanolysin and streptococcal toxin. J Pathog Bacteriol. 1934;39:299–321. [Google Scholar]
- 38.Trieu-Cuot P, Carlier C, Poyart-Salmeron C, Courvalin P. A pair of mobilizable shuttle vectors conferring resistance to spectinomycin for molecular cloning in Escherichia coli and in Gram-positive bacteria. Nucleic Acids Res. 1990;18:4296. doi: 10.1093/nar/18.14.4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the α- and β-subunits of the ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weisman L E, Stoll B J, Cruess D F, Hall R T, Merenstein G B, Hemming V G, Fischer G W. Early-onset group B streptococcal sepsis: a current assessment. J Pediatr. 1992;121:428–433. doi: 10.1016/s0022-3476(05)81801-3. [DOI] [PubMed] [Google Scholar]